Abstract
Blockade of the PD-1/PD-L1 immunologic checkpoint using monoclonal antibodies has provided breakthrough therapies against cancer in the recent years. Nevertheless, intrinsic disadvantages of therapeutic antibodies may limit their applications. Thus, blocking of the PD-1/PD-L1 interaction by small molecules may be a promising alternative for cancer immunotherapy. We used a docking-based virtual screening strategy to rapidly identify new small molecular inhibitors targeting PD-L1. We demonstrated that a small molecule compound (N-[2-(aminocarbonyl)phenyl][1,1′-biphenyl]-4-carboxamide [APBC]) could effectively interrupt the PD-1/PD-L1 interaction by directly binding to PD-L1, presenting the KD and IC50 values at low-micromolar level. Molecular docking study revealed that APBC may have function through a PD-L1 dimer-locking mechanism, occluding the PD-1 interaction surface of PD-L1. We further confirmed the ligand blocking activity and T cell-reinvigoration potency of APBC using cell-based assays. APBC could dose-dependently elevate cytokine secretions of the primary T-lymphocytes that are cocultured with cancer cells. Importantly, APBC displayed superior antitumor efficacy in hPD-L1 knock-in B16F10-bearing mouse model without the induction of observable liver toxicity. Analyses on the APBC-treated mice further revealed drastically elevated levels of infiltrating CD4+ and CD8+ T cells, and inflammatory cytokines production in tumor microenvironment. The APBC compound could serve as a privileged scaffold in the design of improved PD pathway modulators, thus providing us promising drug candidates for tumor immunotherapy.
Keywords: PD-1, PD-L1, Inhibitor, APBC, Cancer immunotherapy
Abbreviations: ALB, albumin; ALP, alkaline phosphatase; APBC, N-[2-(aminocarbonyl)phenyl][1,1′-biphenyl]-4-carboxamide; BMS, Bristol-Myers Squibb; DEGs, differentially expressed genes; FACS, fluorescence activated cell sorter; GOT, glutamic oxaloacetic transaminase; GPT, glutamic pyruvic transaminase; HTRF, homogenous time-resolved fluorescence; IHC, immunohistochemistry; mAbs, monoclonal antibody; RNA-seq, RNA-sequencing; SP, standard extra precision; SPR, surface plasmon resonance; TILs, tumor infiltrating lymphocytes
Introduction
Currently, immune checkpoint blockade is an advanced strategy, and has rapidly evolved into the most promising cancer immunotherapy with remarkable clinical benefit [1]. This approach is based on the idea that cancer cells can evade immune destruction through activation of inhibitory immune checkpoint proteins [2, 3]. Checkpoint inhibitors may stimulate immune responses by removing inhibition and subsequently restore T cell function to destruct cancer cells. Among the emerging checkpoint inhibitors, PD-1/PD-L1 blockade therapy has achieved an unprecedented, enduring response in clinical applications [4, 5].
PD-1 (also known as CD279) is a cell surface receptor mainly expressed on activated T cells, B cells, monocytes, natural killer T cells, and dendritic cells [6], [7], [8]. Its 2 known naturally occurring ligands are PD-L1 (B7-H1, CD274) and PD-L2 (B7-DC, CD273). PD-L1 is constitutively expressed by macrophages, B cells, activated T cells, and parenchymal cells, and is frequently upregulated in many types of cancers [9], [10], [11]. In cancer, when PD-1 comes into contact with its ligands, functional activities of T lymphocytes are inhibited, including proliferation, cytokine secretion, and cytolytic activity [12], [13], [14], thereby providing cancer cells the opportunity to avoid immune surveillance. To date, the remarkable clinical benefits and long-term remissions have been observed in 6 approved therapeutic antibodies targeting PD-1/PD-L1 including Nivolumab, Pembrolizumab, Avelumab, Atezolizumab, Cemiplimab, and Durvalumab for the treatment of a subset of patients with certain cancer types [15, 16]. And many more monoclonal antibody (mAbs) are expected to come shortly however, despite rapid advances of them, intrinsic disadvantages such as manufacturing costs, low tissues penetration, lacking oral bioavailability, immunogenicity, immune-related adverse events, and inaccessible to intracellular targets limit their applications [17, 18]. Thus, blocking of the PD-1/PD-L1 interaction by small molecules may be a promising alternative for tumor treatment.
Compared to mAbs, besides lower manufacturing costs and higher stability, small-molecule inhibitors can provide a better therapeutic index that not only allow for more flexible clinical dosing and oral administration based on the optimal pharmacodynamic parameter and also can maintain reasonable half-lives to avoid systemic immunogenicity. In addition, small-molecule inhibitors have a better tissue and tumor penetration that they can also stimulate intracellular pathways downstream of checkpoint proteins to overcome the drawbacks of mAbs. Therefore, small-molecule inhibitors provide an alternative treatment strategy either alone or complementary with therapeutic antibodies to address drug resistance and low clinical response [19], [20], [21]. However, the development of small-molecule drugs disrupting the PD-1/PD-L1 pathway is inherent challenging because of the relatively large and flat interface of the interaction without well-defined pockets [17, 22].
As of today, although a few patents and publications have disclosed series of small-molecule inhibitors targeting the PD-1/PD-L1 pathway [15, [23], [24], [25]], there are no FDA-approved small-molecule modulators for the PD-1/PD-L1. The most promising small-molecule antagonist is CA-170 against the PD-L1, PD-L2, and VISTA immune checkpoints, which is currently undergoing phase II clinical trials for head and neck/oral cavity cancer, lung cancer, MSI-H positive cancers and Hodgkin lymphoma in India (CTRI/2017/12/011026, ctri.nic.in). The structure of CA-170 has not been disclosed currently, and it is speculated to be a peptidomimetic derived from earlier peptide inhibitors from Aurigene [26]. However, Bogdan et al. recently reported the evidences that there is no direct binding between CA-170 and PD-L1 [27]. Recently, researchers from Bristol-Myers Squibb (BMS) company disclosed a series of small molecules with a tricyclic scaffold for the inhibition of the PD-1/PD-L1 interaction in patent applications [28, 29]. Moreover, cocrystal structures of hPD-L1 complexed with BMS small molecular offer several opportunities for the design of more potent inhibitors of PD-1/PD-L1 interaction [24, 30]. However, no further in vivo activity characterization including the efficacy and safety of these small molecules has been provided. More alternative compounds with novel scaffolds are eagerly needed for a future clinical application, considering that they may possess different pharmacodynamic and pharmacokinetic properties from the reported inhibitors.
Here, we presented the first report that N-[2-(aminocarbonyl)phenyl][1,1′-biphenyl]-4-carboxamide (APBC) could effectively block the PD-1/PD-L1 interaction by directly binding to PD-L1. Molecular docking studies indicated that APBC may have function through a PD-L1 dimer-locking mechanism, blocking the PD-1 interaction surface of PD-L1. The experiments outlined below systematically demonstrated that APBC could interfere with PD-1/PD-L1 biological function and, moreover, significantly inhibit the growth of tumors in mice, and in some cases complete regressions, by dramatically enhancing CD8+ effector T cells infiltration, activation, and cytokine production in tumor microenvironment. Therefore, APBC could be applied as a potential therapeutic candidate for immunotherapy of cancer.
Material and methods
Docking-based virtual screening
The whole virtual screening workflow was carried out with the Schrödinger 2015 software (Schrödinger, LLC, New York, 2015). The 3D structure of dimeric PD-L1 protein and BMS-202 complex (PDB ID: 5J89) was retrieved from Protein Data Bank, and the crystal structure was prepared by the Protein Preparation Wizard in Schrödinger. The docking grid file was generated to define the active binding site based on known ligand in the original complex. More than 200,000 compounds from the Specs database were preprocessed by the LigPrep module in Schrödinger. The ionized states, tautomers, and stereoisomers were predicted by using the Epik algorithm at pH = 7.0 implemented in Schrödinger. Docking was performed using the Glide-program with standard extra precision [31], and then the top compounds were redocked by Induced Fit Docking module of Schrödinger software. After docking calculation, small molecules were initially selected based on the top docking scores. Clustering analysis and binding mode analysis were further performed with the Canvas module of Schrödinger to evaluate the structural diversity and the rationality of the binding mode of candidate compounds. The cutoff of the Tanimoto coefficient used in the clustering analysis was set to 0.5.
Molecular docking
To determine how APBC bound to PD-L1, the molecular docking calculation was carried out with the Schrödinger 2015 software. The crystal structure of PD-L1 with ligand complex (PDB ID: 5J89) was chosen as a docking model, and then be prepared with the Protein Preparation Wizard in Schrödinger. The structures of APBC were respectively constructed and minimization by Chemdraw and Chem3D software, and then preprocessed by LigPrep with OPLS-2005 force field. The docking grid box was generated to define the active binding site. APBC was unbiased docked into PD-L1 binding site using the Glide module with the standard precision score. The pose with the best score was chosen for further analysis.
Cell culture
Jurkat, NCI-H1975, B16F10, HEK293T, HeLa and BIC-1 were purchased from ATCC, MC38 cell line were obtained from Crisprbio Biotechnology company (Beijing, China), and Primary CD4+ T cells with the purity above 95% were purchased from LDEBIO company (Guangzhou, China). B16F10, MC38, HEK293T, HeLa, and BIC-1 were cultured in DMEM medium supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. NCI-H1975 and Jurkat cells were cultured in RPMI-1640 medium with 10% FBS and 1% penicillin/streptomycin. All cells were grown at 37°C in a humidified atmosphere containing 5% CO2. To generate CD4+ T cell blasts, cells were adjusted to 1 × 106 cells/mL in RPMI-1640 medium with 15% FBS and IL-2 (100 IU/mL; 202-IL-010, R&D), followed by the addition of human T-activator CD3/CD28 beads (beads to cells ratio was 1:1; 11131D, Gibco), and fresh medium was added every other day. After approximately 7 d, CD4+ T cells were harvested and frozen for later use.
Measuring binding affinity of APBC to PD-L1 by surface plasmon resonance (SPR)
APBC was purchased at TopScience Co., Ltd. (Shanghai, China). KD of APBC binding to hPD-L1 (Cat. 10084-H08H-200, SinoBiological) was evaluated by SPR (Biacore X100 plus, GE Healthcare). The PD-L1 protein was diluted to 50 μg/mL with 10 mM sodium acetate buffer (pH 4.5), then covalently immobilized on a CM5 sensor chip (Series S Sensor Chip, GE Healthcare) by the standard amine coupling reagent kit (GE Healthcare, PA), and the immobilization level reached approximately 6000 resonance units. Measurements were run in 1.05 × PBS buffer (pH 7.4, GE Healthcare) containing 5% DMSO at 25°C and a flow rate of 30 μL/min. Solvent correction was performed that 8 solutions with varying amounts of DMSO (4.5%–5.8%) were prepared. For binding experiments, APBC (10 mM in DMSO) was serially diluted with PBS buffer to give concentrations of 100, 50, 25, 12.5, 6.25, 3.125, and 0 μM in a final solution the same as the running buffer (1.05 × PBS buffer with 5% DMSO). A sample injection cycle consisted of a 90 s sample injection, 120 s of dissociation phase, and a 30 s buffer injection for checking sample carryover. And 0 concentration samples were used as blank control. The results were globally analyzed by a steady-state model using Biacore T200 Evaluation software, version 2.0.
PD-1/PD-L1 homogenous time-resolved fluorescence (HTRF) assay
The abilities of APBC to inhibit PD-1/PD-L1 interaction were determined by using the well-established HTRF assay. The assay kit of PD-1/PD-L1 interaction (64ICP01PEG) was purchased from Cisbio (Shanghai, China). Binding assays were performed according to the manufacturer's instructions (https://www.cisbio.cn/human-pd1-pd-l1-biochemical-binding-assay-40578). The HTRF unit was measured by a microplate reader (Tecan Spark, Tecan Trading AG) with the excitation wavelength located at 320 nm, and the emission wavelength of 665 and 620 nm, respectively. The inhibitory percentage of APBC was calculated following the instructions of the assay kit.
Flow cytometry measurements
Jurkat cells stably expressing human PD-1 (Jurkat-hPD-1) was established and used to determine the blocking activity of APBC by flow cytometry at the single cell level. The His-tagged PD-L1 protein (10084-H08H, Sino Biological) was diluted with PBS to give concentration of 10 ug/mL, then was formulated in 100 µL PBS with the tested compounds. Five tubes were prepared with different preincubated solutions (Tube 1: 100 µL PBS as the negative control; Tube 2: 50 µL PD-L1-His protein [10 ug/mL] +50 µL PBS as the positive control; Tube 3: 50 µL PD-L1-His protein +50 µL 80 µM inhibitor 1 compound [BMS-1 compound in WO 2015/034820 A1 patent [28], Selleck, S7911]; Tube 4 to 5: 50 µL PD-L1-His protein +50 µL 200 µM or 100 µM APBC compound). The mixtures were incubated for 30 min at 4°C in the dark. Meanwhile, cells were collected, washed with PBS, and resuspended in precooling PBS at concentration of 4 × 106 cells/mL. About 100 µL of cells was added to each above sample, mixed gently, and incubated on ice for additional 60 min. Subsequently, cells were collected, washed with PBS, and stained with Anti-His-APC (BioLegend) in buffer for 20 min on ice in the dark. Labeled cells were washed twice and measured by flow cytometry (Moflo XDP, Beckman) and the data were analyzed with FlowJo software (TreeStar, Ashland, OR). Blocking Efficiency (%) was calculated with the formula: 100% × (1-positive rate treatment/positive rate positive control).
T cell response assays
Human CD4+ T lymphocytes were expanded and activated as described above. T cell response assays were conducted by ELISA in coculture system. NCI-H1975 cells, PD-L1-negative wild type HEK293T, or HEK293T cells with hPD-L1 stable overexpression (HEK293T-hPD-L1) were plated in 96-well plates at a density of 10,000 cells per well and allowed to adhere for 24 h. Afterward, preactivated CD4+ T lymphocytes (2 × 104) were added to selected wells at a ratio of 2:1 effector to target (E:T), in the presence or absence of additives (Atezolizumab or APBC) in 200 μL complete RPMI-1640 medium, followed by coculturing at 37°C, 5% CO2 incubator for 48 h. Prestimulated T cells alone (2 × 104), in the presence or absence of additives in medium was also cultured at 37°C, 5% CO2 incubator for 48 h. Culture supernatants were harvested, and the levels of IFN-γ and TNF-α were detected by the Human IFN-γ ELISA kit (Cat. 88-7316-22, Invitrogen) and Human TNF-α ELISA kit (Cat. 88-7346-88, Invitrogen), respectively.
Cell viability assays
The viability of cells was determined by Cell Counting Kit 8 purchased from Beyotime (Shanghai, China). Briefly, 10,000 cells were seeded into transparent 96-well plates in 100 μL of media and cultured for 24 h or 48 h in the presence of increasing concentrations of the APBC compounds. Next, 10 μL Cell Counting Kit 8 reagent was added and the plate was incubated for another 2 h. The plate was measured by microplate reader (Enpire, PE).
In vivo effect of APBC
The animal protocol was approved by the Medical and Scientific Research Ethics Committee of Henan University (HUSOM2020-272) and conformed to guidelines for the Care and Use of Laboratory Animals of the Ministry of Science and Technology of the People′s Republic of China (2006-398). Five- to 6-wk-old female C57BL/6 mice (18–22 g) were purchased from Weitong Lihua Experimental Animal Technical Co., Ltd. (Beijing, China). The mice were housed at the animal care facility (School of Basic Medical Sciences, Henan University) in plastic cages under controlled temperature (23 ± 2°C), humidity (50% ± 10%) and with a 12 h light, 12 h dark cycle. The mice were allowed access to standard rodent diet and water ad libitum.
Here we constructed a hPD-L1-expressing mouse tumor cell model to study the therapeutic activity of APBC in mice. First, mPD-L1-deficient B16F10 cell lines were generated by CRISPR/Cas9 system with the guide sequences (5ʹ-CATAATCAGCTACGGTGGTG- 3ʹ). The deletion efficiency was identified by sequencing and flow cytometry. Lentiviral expression vector carrying human PD-L1 ORF was constructed and verified by sequencing. Then mPD-L1-deficient B16F10 were transduced with the human PD-L1 expressing lentivirus and selected by Puromycin to construct the hPD-L1 replacement B16F10 cell line (B16F10-hPD-L1). Successful replacing mPD-L1 by hPD-L1 was examined by fluorescence activated cell sorter (FACS) staining.
B16F10-hPD-L1 and MC38 tumor cells (5 × 106 and 3 × 106, respectively) were injected subcutaneously at sites on the right side of the abdomen. When the tumor volume reached approximately 50 to 70 mm3, mice were randomly divided into the experimental groups: (1) Vehicle (normal saline containing 8% ethanol and 8% glycerin), (2) Atezolizumab antibody (10 mg/kg), (3) APBC (10 mg/kg), (4) APBC (15 mg/kg). The APBC was administered via intraperitoneal injection every 2 d and the Atezolizumab antibody was administrated as a positive control via tail vein injection every 3 d. Tumor growth was measured every 2 d using a digital caliper, and the tumor size was calculated with the formula 1/2 × length × width2. Mice were sacrificed after 7 to 8 times treatment of the APBC (8 times for B16F10-hPD-L1 tumor and 7 times for MC38 tumor), the blood, tumor tissue, major organs (liver, spleen, and thymus) samples were collected for immune phenotype and function analyses.
Total growth inhibition (%) was calculated with the formula 100% × (Tvolcontrol –Tvoltreated)/(Tvolcontrol – Tvolpredose) [32].
Tvolcontrol –Tvoltreated: tumor volume of control mice after treatment– tumor volume of mice treated with APBC; Tvolcontrol – Tvolpredose: tumor volume of control mice after treatment– tumor volume of control mice before dosing.
RNA-seq and data analysis
On day 24 after B16F10-hPD-L1 tumor inoculation, tumors were harvested. Total RNA of APBC-treated group and vehicle group were isolated using TRIzol Reagent (Invitrogen), and quantified by NanoDrop (Thermo Fisher Scientific). Sequencing libraries were constructed using an MGI Easy mRNA Library Prep Kit (BGI, Wuhan, China) following the manufacturer's instructions. RNA-seq analysis was performed on BGIseq500 platform (BGI-Shenzhen, China). The sequencing data underwent quality trimming to remove sequencing adapter and low-quality bases using SOAPnuke (v1.5.2) [33], followed by aligned to the mouse genome (Mus musculus. GRCm38) using HISAT2 (v2.0.4) [34]. Bowtie2 (v2.2.5) [35] was performed to align the clean reads to the reference coding gene set, then expression level of gene was quantified by RSEM (v1.2.12) [36]. Furthermore, differentially expressed genes (DEGs) were screened by the DESeq2(v1.4.5) [37] with Q value ≤0.05. To take insight to the change of phenotype, comprehensive functional enrichment analysis of DEGs was performed by Metascape [38]. GSEA [39] was used to investigate the specific gene expression patterns and pathways. The significant levels of terms and pathways were corrected by Q value with a rigorous threshold (Q value ≤0.05). To estimate the immune infiltration, gene expression data were performed with CIBERSORTx analysis [40] in TIMER2.0 database (http://timer.comp-genomics.org/).
Flow cytometry of splenic lymphocytes
On day 24 after B16F10-hPD-L1tumor inoculation, murine spleens were collected. Each spleen was prepared by mechanical disruption and mashed through a 70-μm nylon filter and brought to a volume of 15 mL RPMI-1640 medium with 10% FBS. Single cell suspensions were centrifuged at 2000 rpm for 15 min, and suspended in 20 mL ACK lysis buffer (BioLegend) for 5 min, followed by centrifuged at 2000 rpm for 10 min. Then after washed with PBS to remove cell debris, the splenocytes were suspended. The concentration of the cells was adjusted to 1 × 106 cells in a final volume of 100 µL of flow cytometry staining buffer. Cells were then stained with CD3 (Cat.100203, BioLegend), CD4 (Cat. 100407, BioLegend), CD8 (Cat. 100707, BioLegend), and perforin (Cat. 154303, BioLegend) antibodies, and analyzed by a flow cytometer (CytoFLEX S, Beckman Coulter).
For intracellular staining of IFN-γ and TNF-α, isolated splenocytes were plated at a density of 2 × 106 per well into a 6-well plate. Next, Cell Activation Cocktail (1:500; Cat. 423301, BioLegend,) and Brefeldin A (1:1000; Cat. No. 420601, BioLegend) were added into the wells, and plates were incubated at 37°C for 3 to 4 h followed by one wash with PBS. Cells were stained with CD3, CD4, IFN-γ (Cat. 505809, BioLegend) and TNF-α (Cat. 506328, BioLegend) antibodies for flow cytometry analysis.
Flow cytometry of mouse tumor
B16F10-hPD-L1 tumor tissues from sacrificed mice were minced into small pieces and digested for 45 min in RPMI-1640 containing collagenase IV (200 U/mL; Invitrogen) and DNase I (40 U/mL; Sigma-Aldrich) with agitation at 37°C. Digested tissues were incubated with EDTA (0.5 M) for 5 min at 37°C to prevent DC/T-cell aggregates and then mashed through a 70-µm cell strainer. Cells were washed, resuspended in FACS buffer, and stained with CD3, CD4, CD8, perforin, and Granzyme B (Cat. 396413, BioLegend) antibodies for flow cytometry analysis.
Immunohistochemistry staining
For immunohistochemistry (IHC), sections (4 µm thickness) were cut from 4% paraformaldehyde-fixed paraffin-embedded spleen and B16F10-hPD-L1 tumor tissues. After deparaffinization and rehydration, the sections were incubated with primary antibodies against CD4 (Cat. ab183685, Abcam), CD8 (Cat. ab209775, Abcam), FoxP3 (Cat. ab215206, Abcam), overnight at 4°C. The slides were incubated with HRP-labeled secondary antibody for 30 min, followed by the DAB chromogen mixture. The slides were imaged on a Nikon fluorescence microscopy (NI-U, NIKON). CD4, CD8, FoxP3 stains were quantified by counting the number of positive particles in magnification.
Biochemical analysis
In a 1-wk toxicity study, B16F10-hPD-L1 tumor-bearing mice were randomized and dosed daily with vehicle control, 15 mg/kg and 50 mg/kg of the APBC, respectively. On days 7 post first dose, serum samples from tumor-bearing mice were analyzed for glutamic oxaloacetic transaminase (GOT), glutamic pyruvic transaminase (GPT), alkaline phosphatase (ALP), and albumin (ALB) activities using an automatic dry-type biochemical analyzer (DRI-CHEM NX500iVC, FUJI).
Stability assay in human blood plasma
Compound APBC was incubated at a concentration of 2 μM in human plasma (BioreclamationIVT, Cat. No. HUMANPLK2P2N, batch No. HMN8022) at 37°C. Samples were taken at each time point during the incubation (0, 10, 30, 60, and 120 min in duplicate). The reactions terminated with the addition of 400 μL of stop solution (200 ng/mL tolbutamide and 200 ng/mL Labetalol in 50% ACN/MeOH). The plates were centrifuged at 4000 rpm for 10 min, and the supernatants were submitted to LC-MS/MS analysis.
Statistical analysis
Results were presented as mean ± SD from independent triplicate experiments. All statistical analyses were performed using Prism 7.0 software (GraphPad). Figure legends showed the statistical analysis method performed. P values <0.05 were considered statistically significant.
Results
Computation-based APBC binding and interaction with PD-L1
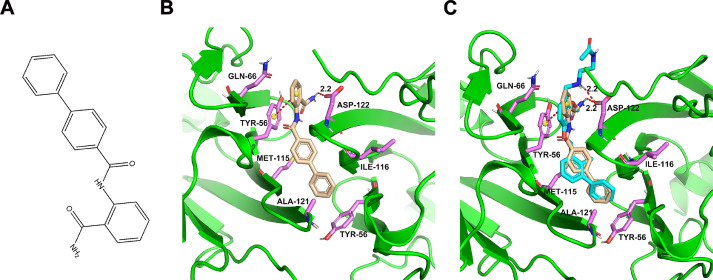
To discover potential small molecules targeting PD-L1, virtual screening based on molecular docking was performed on Specs database with more than 200,000 compounds. Eventually, the most promising compound APBC (SPECS No. AG-690/11449006, APBC) with snugly binding mode and good docking score was selected for further study (Fig. 1A). Molecular docking studies were performed to simulate the interactions between the APBC and the human PD-L1 extracellular domain protein (PDB ID: 5J89) using Schrödinger software package. As shown in Fig. 1B, APBC could bind into the hydrophobic site formed from the two PD-L1 monomers, especially from anchor residues Y56, M115, and A121. APBC formed one key hydrogen bond with the key residue D122 thus stabilizing the complex structures. In addition, the stability of complex was attributed to the π-π stacking interaction between aniline group of APBC and Y56 residue. The hydrophobic contacts formed between APBC and surrounded residues also contributed to the binding of small molecule and PD-L1. The result indicated that the APBC has similar binding mode with small-molecule inhibitors of BMS [24, 30] (Fig. 1C). A series of experimental in vitro and in vivo were conducted to evaluate the biological activity of APBC.
Fig. 1.
The result of molecular docking. (A) The chemical structure of APBC. (B) Docking analysis of APBC in the hydrophobic cleft formed by the dimeric hPD-L1. Salmon sticks represent the key residues. Sand yellow sticks represent the APBC small molecule. (C) Binding overlap of APBC (sand yellow stick) and BMS-202 (cyan stick) in the binding site. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Biochemical activity of APBC in blocking PD-1/PD-L1 interaction
The binding affinities of APBC to hPD-L1 were assessed with the SPR-based assay. Recombinant human PD-L1 ectodomain protein (residues 18–239) was immobilized on a CM5 sensor chip by a direct amine coupling method. Firstly, we determined the affinity of well-known inhibitor 1 (BMS-1 compound in WO 2015/034820 A1 patent [28]) to hPD-L1, setting as a positive reference to confirm that the immobilized hPD-L1 was functional. The results showed that the KD value of inhibitor 1 for the binding to hPD-L1 was 3.16 ± 0.16 μM (Supplementary Fig. 1A). As illustrated in Fig. 2A, the binding of APBC to hPD-L1 had a KD value of 49.50 ± 4.21 μM, indicating that this small molecule compound represented a new active ligand of hPD-L1 with detectable affinities.
Fig. 2.
Biochemical activity of APBC in blocking PD-1/PD-L1 interaction. (A) The binding affinity of compound APBC with hPD-L1 was determined by SPR method. (B) Concentration response curves of APBC examined in the HTRF PD-1/PD-L1 protein–protein interaction assay.
We next validated whether the APBC was effective in disrupting the human PD-1/PD-L1 interaction. The first was via the well-established HTRF assay that the interference of APBC as well as inhibitor 1 on the binding of PD-L1 to PD-1 was investigated. The result showed that the half-maximal inhibitory concentration (IC50) value for inhibitor 1 was 135.50 nM (Supplementary Fig. 1B). As revealed in Fig. 2B, the IC50 of the APBC was 27.82 μM with 65.1% blocking efficiency against the hPD-1/hPD-L1 at the concentration of 100 μM, and approximately 52.4% blocking efficiency at 50 μM.
Blockade of the PD-1/PD-L1 interaction of APBC at the cellular level
To evaluate whether APBC could block PD-1/PD-L1 complex in a physiological environment, we performed flow cytometry experiments (Fig. 3). For this, Jurkat cells were engineered to stably overexpress hPD-1-EGFP. Recombinant human His-tagged PD-L1 protein was incubated with these cells, with or without the APBC at different concentrations (25 or 50 μM). After the cell components were washed, APC labeled anti-His antibody was added to stain the remaining PD-L1 bound to PD-1 on the Jurkat cell surfaces. As revealed in Fig. 3A, no staining was observed in the negative group in which the mixture had no His-tagged PD-L1. While the positive control that the Jurkat-hPD-1 cells was incubated only with His-tagged PD-L1, displayed an average fluorescence intensity signal 2 orders of magnitude stronger than the negative control, suggesting that the staining was specific. When was added at 20 μM, inhibitor 1 showed an 85.6% blocking efficiency against hPD-L1/hPD-1 complex. And for APBC, when was added at 25 μM, it showed a weaker blocking efficiency (approximately 10.2%) against hPD-L1 binding to the Jurkat-PD-1 cells. Whereas when the concentration was increased to 50 μM, significant inhibition of the interaction by APBC (approximately 37.3%) was observed (Fig. 3B). Moreover, during the flow cytometric experiments, cell toxicity was not observed on the Jurkat cells at the concentration of APBC up to 50 μM, confirming that the APBC could serve as a potential lead compound for novel therapeutic agents.
Fig. 3.
Functional activity of APBC in cell-based bioassays. (A) Representative flow cytometry analysis for investigating the ability of APBC blocking the interaction of hPD-L1 and hPD-1 overexpressed Jurkat cells. (B) Flow cytometry data are shown in mean ± SEM from 3 separate experiments, ***P< 0.001, unpaired t test.
APBC activated the function of CD4+ T cells in vitro
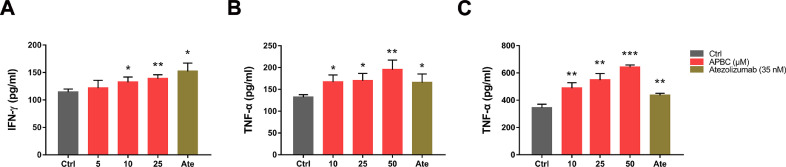
Cytokines secretion is an important indicator for T-cell activation evaluation. To investigate whether APBC can restore the repressed function of T cells, we examined the secretion of IFN-γ and TNF-α in CD4+ T cells by T cell-tumor coculture assay. Specifically, activated CD4+ T cells were cocultured with NCI-H1975 cells constitutively expressing human PD-L1, followed by the addition of APBC and Atezolizumab at the indicated concentrations. IFN-γ and TNF-α levels were analyzed 48 h after stimulation. As indicated in Fig. 4A and B, the cytokines IFN-γ and TNF-α levels showed a dose dependent increase in response to APBC treatment, which were verified by another cell coculture assay. We cocultured activated CD4+ T cells with well-established HEK 293T cells stably expressing human PD-L1, with or without addition of APBC and Atezolizumab. Similarly, at APBC concentration of 10 μM, TNF-α secretion elevated significantly by comparison, and this also occurs in a dose dependent manner. The results presented in Fig. 4C indicated that upon treatment with 10 µM APBC, the TNF-α level was increased by 41.8%, and in the presence of 50 μM APBC, TNF-α production was increased by 86.6%. To confirm whether our observations were dependent on blocking the PD-1/PD-L1 interaction, we performed the coculture experiments using PD-L1-negative HEK293T cells or prestimulated T cells alone treated with APBC compound. We found that APBC did not significantly affect IFN-γ and TNF-α production of the primary T-lymphocytes during culture alone (Supplementary Fig. 2A and C) or coculture with PD-L1-negative HEK293T cells (Supplementary Fig. 2B and D). These results indicated that APBC was capable of restoring the PD-L1-mediated suppression of T-cell function, thus the in vivo efficacy of APBC warrants further exploration.
Fig. 4.
Effects of APBC on T-cell function and cytokine release. (A) and (B) Effects of APBC on IFN-γ and TNF-α secretion from CD4+ T cells cocultured with NCI-H1975 cells. (C) Effects of APBC on TNF-α secretion from CD4+ T cells cocultured with hPD-L1 overexpressed HEK293T cells. Data are shown as mean ± SEM, *P< 0.05, **P < 0.01, ***P< 0.001, unpaired t test.
Antitumor efficacy of the APBC in tumor-bearing C57BL/6 mice
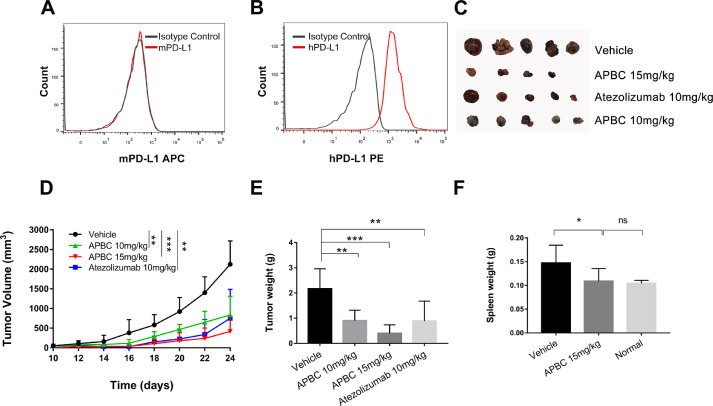
The amino acid sequences of murine and human PD-L1 proteins are 77% identical [41]. In order to more objectively examine the therapeutic efficacy of the APBC in vivo, we constructed a hPD-L1-expressing mouse tumor model which was engineered by replacing mouse PD-L1 with human PD-L1 in a transplantable mouse B16F10 tumor cell line. Firstly, we used CRISPR-Cas9 technology to knockout mPD-L1 and then overexpressed hPD-L1 in these mPD-L1 deletion cells (Fig. 5A and B). C57BL/6 mice were inoculated with B16F10-hPD-L1 cells, once the tumor volume reached approximately 50 mm3, the animals were dosed every 2 d with APBC. As Fig. 5C to E showed, APBC significantly inhibited the tumor growth compared with the vehicle control group. Upon treatment with 10 mg/kg of APBC, tumor growth inhibition reached 62.1%, which was slightly lower than the efficacy induced by Atezolizumab antibody (66.1% at 10 mg/kg). When animals were treated with 15 mg/kg of APBC, a dramatic efficacy of 82.2% tumor growth inhibition relative to the vehicle control was observed, and 4 of 10 treated animals were almost tumor free. Notably, a large number of tumor-bearing mice from vehicle group developed splenomegaly at last, resulting in a significant increase in spleen weight, while mice from the APBC treatment group had normal spleens (P< 0.05; Fig. 5F). In addition, to further verify whether the PD-1/PD-L1 blocking activity of the APBC was effective against tumors of a variety of histologic types, we examined the antitumor activity of APBC on another mice MC38 tumor model. As shown in Supplementary Fig. 3A and B, after 7 times of treatment, the growth of colon tumors was also inhibited significantly following APBC administration at the concentration of 10 mg/kg. Meanwhile, during the treatment period, APBC had no significant effect on the body weight of the mice (Supplementary Fig. 3C).
Fig. 5.
Antitumor activity of the APBC in B16F10-hPD-L1 tumor-bearing C57BL/6 mice models. (A) Flow cytometry analysis of mPD-L1 expression on B16F10-hPD-L1 cells. (B) Flow cytometry analysis of hPD-L1 expression on B16F10-hPD-L1 cells. (C) Representative images of subcutaneous tumors harvested at day 24. (D) Tumor volume changes measured over time in the vehicle (n = 10), APBC 10 mg/kg (n = 5), APBC 15 mg/kg (n = 10), Atezolizumab 10 mg/kg (n = 6) groups. (E) Weight of tumors at day 24 after tumor inoculation. (F) Spleen weight at day 24 after tumor inoculation. All quantitative data are represented as means ± SEM, *P< 0.05, **P< 0.01, ***P< 0.001, unpaired t test.
To further evaluate the toxicity and side effects of APBC, the serum biochemical indices of B16F10-hPD-L1-bearing mice, including GOT, GPT, ALP, and ALB, were analyzed. We found that serum activities of GOT and GPT both showed a significantly positive correlation with tumor size (Pearson r = 0.837, P< 0.01; and r = 0.810, P< 0.01, respectively), while ALP level showed a significantly negative correlation with tumor size (Pearson r = −0.902, P< 0.001). Compared to control group, there were no significant changes in the serum levels of ALP and ALB after APBC administration. Notably, there was a tendency to considerable decrease serum levels of GOT and GPT after APBC administration at concentrations up to 50 mg/kg (Supplementary Fig. 4), indicating that APBC treatment not only had no observable toxicity in murine models but also could reduce liver injury induced by tumor progression. We also evaluated the unspecific toxicity of APBC with the use of cell viability assays. For this, 4 kinds of cell lines were exposed to the increasing concentration of the APBC for 24 h and 48 h, respectively. As shown in Supplementary Fig. 5, significantly low toxicity of compound APBC was confirmed, as the inhibition ratios for the 4 cell lines (B16F10-hPD-L1, HeLa, BIC-1, and Primary CD4+ T cells) were less than 25.6% at the 200 μM concentration. According to these results, we proposed that the antitumor efficacy of APBC might depend on the reinvigoration immune–mediated tumor cell killing, rather than by directly killing tumor cells [42, 43].
APBC alters gene signatures in murine tumor tissue
For an unbiased assessment of the effect of APBC treatment on transcriptional signatures of tumor tissue, we also performed RNA-sequencing (RNA-seq) for isolated B16F10-hPD-L1 tumor tissue taken from APBC (15 mg/kg) and vehicle-treated mice (Fig. 6).
Fig. 6.
APBC alters gene signatures in murine tumor tissue. (A) A heatmap showing the top enrichment clusters of up- and downregulated genes in the RNA-seq data of isolated tumor tissue taken from APBC (15 mg/kg, n = 3) treated tumor-bearing mice using Metascape analysis. Gray color indicates a lack of significance. (B) Network of functional and pathway enrichment analyses were performed for differentially expressed genes (DEGs) using the Metascape tool. The sizes of the nodes reflect the enrichment significance of the terms, terms with up-/downregulated genes are shown in blue/red, respectively. (C) GSEA enrichment plot of KEGG pathway for genes in each group. (D) Tumor infiltration proportion of immune cells in each sample using CIBERSORT estimation. CD8+ TILs from APBC therapy groups showed significant enrichment compared to vehicle-treated ones. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
When the threshold was adjusted to Q value <0.05, 818 DEGs were identified, of these 342 were upregulated while 476 were downregulated in APBC-treated group compared to the vehicle group, the DEGs were listed in Supplementary Table 1. To examine biological processes and pathways which may be altered after APBC-treated, comprehensive functional enrichment analysis based on the DEGsDEGs was conducted by Metascape database. Results presented that the genes overexpressed in APBC-treated group were mainly associated with mRNA metabolic process and microtubule-based process. In comparison, several biological pathways known to be favorable for tumor growth such as regulation of epithelial to mesenchymal transition, autophagy, regulation of cell cycle, regulation of GTPase activity, regulation of cellular component organization, were found to be downregulated in the APBC-treated group (Fig. 6A and B). Considering the gene expression differences were minimal when compared the APBC treatment group and control group, GSEA [39] may be a more powerful tool to extract biological insight from the RNA-seq data. Through this analysis, several immune pathways associated with adaptive immune activation responses such as antigen processing and presentation, T-cell receptor signaling and TNF signaling pathways, Th1 and Th2 cell differentiation were markedly upregulated, while pathways related to Glycolysis or MAPK signaling pathway were markedly downregulated in tumor-bearing mice treated with the APBC (Fig. 6C and Supplementary Table 2). To further analyze the influence of APBC treatment on the tumor-associated immune system, we utilized a recently developed transcriptome deconvolution method called CIBERSORTx analysis [40]. Detailed analysis showed that a significant higher infiltration of CD8+ T cells (P< 0.05) in the tumor from APBC-treatment animals compared to vehicle-treated ones (Fig. 6D). Taken together, these results suggested that APBC treatment played an essential role in enhancing effector T-cell function relevant for tumor rejection.
APBC suppressed tumor growth via a T-cell-dependent mechanism
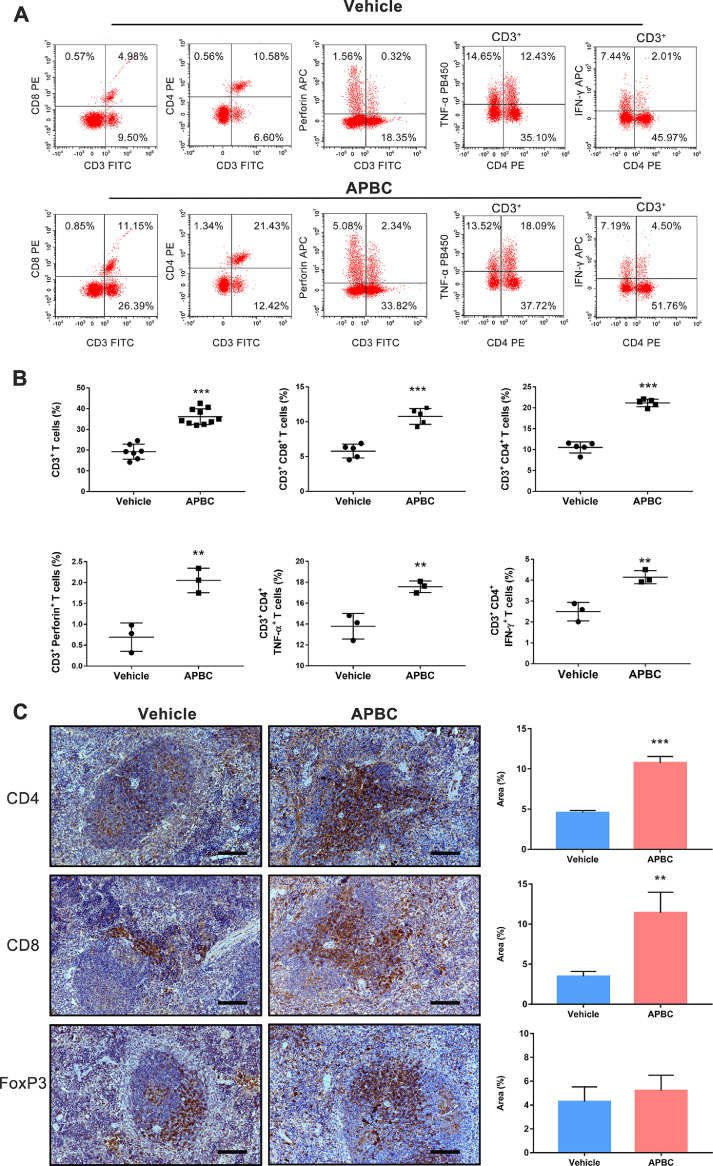
Analyses of responders to the anti-PD-L1 antibody, Atezolizumab, showed increased cytokines production such as IFN-γ and TNF-α in the post-treatment tumors [44]. We therefore analyzed whether similar phenomena could be observed in the APBC-treated tumor-bearing mice. At the end of the treatment, spleens were harvested from several B16F10-hPD-L1-bearing mice and detailed phenotypic biomarkers of the immune cells was examined by flow cytometry. As illustrated in Fig. 7A and B, significantly increased the percentages of CD3+, CD3+ CD4+, and CD3+ CD8+ T lymphocytes in the spleen (P < 0.001), as compared to vehicle-treated control group, were observed from the APBC-treated mice. These findings were related with the alterations in the levels of cytokine production. Thus, compared to the vehicle group, the perforin production from CD3+ cells was significantly increased in the APBC-treated animals (P< 0.01). Notably, the APBC treatment resulted in extremely significant increase in the percentage of IFN-γ and TNF-α producing CD4+ T cells within the spleens of mice when compared with vehicle control (Fig. 7A and B). Consistent with these flow cytometry results, the IHC analyses also showed a considerably higher density of CD4+ and CD8+ T lymphocytes in the spleen from APBC-treatment animals compared to vehicle-treated ones, however, there were no significant changes in the expression of FoxP3 which is a biomarker of CD4+CD25+ regulatory T cells (Fig. 7C), suggesting systemically enhanced host immune responses after APBC treatment.
Fig. 7.
Antitumor activity of the APBC is associated with enhanced systemic immunity. (A) Flow cytometry analysis of splenic lymphocytes for T lymphocyte markers and cytokine production. Splenic lymphocytes were purified from APBC (15 mg/kg) and vehicle-treated tumor-bearing mice, and analyzed by flow cytometry for lymphocyte markers CD3, CD4, CD8, intracellular perforin, IFN-γ, TNF-α production. (B) All quantitative data of flow cytometry analysis are represented as means ± SEM, *P< 0.05; **P< 0.01, ***P< 0.001, unpaired t test. (C) Immunohistochemical analysis of spleen for T lymphocyte markers after APBC (15 mg/kg)/vehicle therapy. Representative images of specimen stained with anti-CD4, CD8, and FoxP3 antibody. Scale bars = 100 μm, × 200. Immunohistochemical quantification (n = 5) were used Image J. Data were shown as mean ± SEM, *P < 0.05, **P < 0.01, ***P< 0.001 unpaired t test.
We next examined whether the APBC-treated tumors had higher levels of tumor infiltrating lymphocytes (TILs) compared to vehicle-treated ones. Flow cytometry and IHC results demonstrated significantly increased CD4+ and CD8+ T-cell population in the APBC-treated tumors (Fig. 8), which was consistent with the results of bioinformatics analysis. A better penetration of CD8+ T cells into the tumor tissue was indeed observed in APBC-treated mice (Fig. 8C), which might be due to the better tumor penetration of the small compound, with potential benefits to promote the proliferation or infiltration of CD8+ T cells. Much as we observed in the spleens, APBC treatment did not significantly change FoxP3 expression levels within tumors. As previously reported that perforin and GzmB can be used to identify cytotoxic cells [45, 46], we thus examined the percentage of perforin+ T and GzmB+ T cells. Analysis of TILs by flow cytometry demonstrated that in APBC-treated tumors both the perforin and GzmB expression were markedly elevated in CD3+ T cells compared to controls (Fig. 8A and B). Overall, these findings highlighted the therapeutic potential of APBC, which could improve systemic and antitumor immune responses via elevated effector T cell function, enhanced infiltration and inflammatory cytokine production in the periphery and the tumor microenvironment.
Fig. 8.
APBC treatment induces increased lymphocyte infiltration into tumor and enhanced antitumor immunity. (A) Flow cytometry analysis of tumor-infiltrating lymphocytes (TILs) and cytokine production. TILs were isolated from tumors in APBC (15 mg/kg) and vehicle-treated tumor-bearing mice, and were analyzed by flow cytometry for lymphocyte markers CD3, CD4, CD8, intracellular Granzyme B and perforin production. (B) All quantitative data of flow cytometry analysis are represented as means ± SEM, *P< 0.05; **P< 0.01, *** P< 0.001, unpaired t test. (C) B16F10-hPD-L1-bearing C57BL/6 mice after APBC therapy (15 mg/kg) present an enriched lymphocyte infiltration measured by immunohistochemical staining. Representative images of tumor specimen stained with anti-CD8, CD4, and FoxP3 antibody. Scale bars = 100 μm, × 200. Immunohistochemical quantification (n = 5) were used Image J. Data were shown as mean ± SEM, *P < 0.05, **P < 0.01, unpaired t test.
Discussion
Agents directed against the PD-1/PD-L1 axis have shown promising clinical benefits in several indications, and small-molecules inhibitors that disrupt PD-L1-mediated tumor immune escape are considerably desirable [47, 48]. However, the development of effective small molecular agents for clinical use is extremely challenging, potential new chemical scaffolds are still needed for future drug design. Docking-based virtual screening approach on the basis of atomic-resolution biological structures is an efficient strategy for discovering drug candidates from small-molecule databases [49]. Here, we selected crystal structure of PD-L1 with ligand complex (PDB code: 5J89) as the basis of virtual screening to rational identify small molecular inhibitors against the PD-1/PD-L1 pathway. We demonstrated that a small molecule compound APBC can interrupt the PD-1/PD-L1 interaction by directly binding to PD-L1, leading to a potent antitumor activity via reactivation of the T cells.
Concerning the immune function has emerged as an essential part of therapy in cancer [50]. In the current study, with the combination of in vitro and in vivo platforms, we comprehensively evaluated the PD-1/PD-L1 blocking and immune-related activity of APBC at the molecular, cellular and in vivo levels. In our study, the APBC showed hPD-L1 binding and hPD-1/hPD-L1-inhibiting activity, presenting the KD and IC50 values at low-micromolar level (Fig. 2). And we also confirmed the ligand blocking and T cell-reinvigoration potency of APBC using cell-based blocking assays and cytokine release assays. Human gene expressing mouse tumor model is becoming routinely used for the preclinical investigation of therapeutic efficiency [51]. Here, we established the human PD-L1 knock-in B16F10-bearing mouse model to evaluate the ability of APBC to reinvigorate T cell–mediated tumor cell killing. And as we expected, the results demonstrated the APBC's superior ability to drastically reduce tumor size in mice (Fig. 5). Meanwhile, we also observed antitumor effect of APBC at the concentration of 10 mg/kg in another murine colon carcinoma MC38 tumor model (Supplementary Fig. 3). However, based on our SPR experiment, APBC exhibited a high KD value (>100 μM, data not shown), representing the weak binding affinity of APBC for murine PD-L1 protein. By the comparison of the 2 different tumor models, we found that APBC has a better antitumor therapeutic effect on the B16F10-hPD-L1 tumor model. We thus speculated that APBC may be a multifunctional molecule simultaneously targeting PD-L1 and other targets to collaboratively implicate in the treatment of tumor. We predicted other possible targets of APBC through molecular docking, and found that hPD-L1 presented the best docking score. Therefore, structural modifications of this parent compound for enhancement the PD-L1 binding affinity would be required and it is necessary to experimentally confirm our hypothesis using various target binding assays in future.
Based upon the RNA-seq and predicted results, APBC induced a proinflammatory microenvironment characterized by increased antigen processing and presentation, TNF signaling pathway, cytokine-cytokine receptor interaction, Th1 and Th2 cell differentiation, IL-17 signaling pathway, Toll-like receptor signaling pathway, as well as CD8+ T-cell infiltration in tumor tissue (Fig. 6 and Supplementary Table 2). Several clinical evidences to date suggest that checkpoint inhibitors largely depend on restoring preexisting antitumor T-cell responses, and exhibit excellent therapeutic effect in inflamed tumors as characterized by positive PD-L1 in tumor, high CD8+ T-cell proliferation and infiltration, or the presence of an increased IFN-γ cytolytic T-cell signature [32, 44, 52, 53]. IFN-γ and GzmB, as biomarkers of T-cell activation, are believed to be secreted by CD8+ cytotoxic T cells or CD4+ Th1 cells [54, 55]. And as perforin can be used to identify activated cytotoxic cells [45, 46, 56], it is also worthy of evaluation. In our study, splenocytes harvested from mice treated with APBC showed remarkable increases in the CD4+ and CD8+ T cell subpopulations, and in the inflammatory cytokine production such as IFN-γ, TNF-α, and perforin, relative to those taken from mice treated with vehicle control (Fig. 7), which confirmed the potency of APBC to promote an on-going immune response. In addition, significantly greater immune cell infiltration, and drastically increased GzmB and perforin release in tumor microenvironment were also observed after APBC administration compared to the control (Fig. 8). These inflammatory cytokines might orchestrate together to recruit more T cells into the tumor microenvironment to form the positive feedback loop against tumor growth. These results supported the idea of the reinvigoration of T cell responses by PD-1/PD-L1 blockade [57, 58]. Small-molecule inhibitors against PD-1/PD-L1 interaction are attractive because their better tissue penetration may result in broader and stronger antitumor efficacy [59, 60]. And this may explain why APBC showed a weaker blocking efficiency against hPD-L1 at the molecular or cellular levels compared to the well-known monoclonal antibody, while a comparable antitumor efficacy to the antibody in vivo levels.
Previous trials have reported that enhanced response rates in patients appear to occur with riskily increased adverse events. As the outcomes of existing immune checkpoint agents are not universally optimistic and may be accompanied by toxicities [61], developing novel suitable chemical immunomodulators and combinatory therapies is a major priority [62]. Pharmacodynamic as well as pharmacokinetic properties of the small molecular inhibitors should be explored more comprehensively to identify a suitable candidate compound with the best balance of high efficacy and low toxicity. We examined the APBC stability in human blood plasma and found that it possessed relatively good blood plasma stability (Supplementary Table 3). It is noteworthy to mention that, in a 1-wk toxicity study, APBC was well tolerated when administrated daily up to 50 mg/kg (Supplementary Fig. 4). There were no drug-related deaths, and no drug-related toxic effects observed during the study. All these results above indicated that compound APBC could serve as a privileged scaffold for further development of potent biologically active PD pathway modulators. The pharmacokinetic parameters and the entirely accurate action mechanism of APBC deserve further exploration.
In summary, APBC exhibited specific binding affinity to human PD-L1, and significant inhibitory potency against PD-1/PD-L1 interaction, and thereby restoring PD-L1-mediated suppression of T-cell activation. Furthermore, APBC also displayed superior antitumor efficacy in human PD-L1 knock-in B16F10-bearing mouse model and its tumor-inhibitory activity coincided with the changes in TIL signatures after treatment. Thus, APBC with a new scaffold may represent a promising lead compound for immunotherapeutic drug discovery. This study also provides molecular insights into the screen of novel selective PD-1/PD-L1 inhibitors.
Competing interests
The authors declare that they have no conflicts of interest.
Authors’ contributions
In this study, FW performed the experiments, analyzed the data, and wrote the manuscript. HZ and YW performed in the molecular docking. SW, YZ, and JH took part in the in vitro studies. WY contributed to the in vivo experiments. YH, ZB, and SJ assisted with data analysis. HL and XY conceived the idea, supervised the experiments, and revised the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 21775060), and Shaanxi University of Chinese Medicine (No. 2020XG01). The funding bodies were not involved in the study design, data collection, analysis and interpretation of data, or in writing of this manuscript.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.01.001.
Contributor Information
Huanxiang Liu, Email: hxliu@lzu.edu.cn.
Xiaojun Yao, Email: xjyao@lzu.edu.cn.
Appendix. Supplementary materials
References
- 1.Gong J, Chehrazi-Raffle A, Reddi S, Salgia R. Development of PD-1 and PD-L1 inhibitors as a form of cancer immunotherapy: a comprehensive review of registration trials and future considerations. J ImmunoTher Cancer. 2018;6:8. doi: 10.1186/s40425-018-0316-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang J, Hu L. Immunomodulators targeting the PD-1/PD-L1 protein-protein interaction: from antibodies to small molecules. Med Res Rev. 2019;39:265–301. doi: 10.1002/med.21530. [DOI] [PubMed] [Google Scholar]
- 6.Cheng X, Veverka V, Radhakrishnan A, Waters LC, Muskett FW, Morgan SH, Huo J, Yu C, Evans EJ, Leslie AJ. Structure and interactions of the human programmed cell death 1 receptor. J Biol Chem. 2013;288:11771–11785. doi: 10.1074/jbc.M112.448126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riella LV, Paterson AM, Sharpe AH, Chandraker A. Role of the PD-1 pathway in the immune response. Am J Transplant. 2012;12:2575–2587. doi: 10.1111/j.1600-6143.2012.04224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wherry EJ. T cell exhaustion. Nat Immunol. 2011;12:492–499. doi: 10.1038/ni.2035. [DOI] [PubMed] [Google Scholar]
- 9.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL. Colocalization of inflammatory response with B7-H1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003689. 127ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Francisco LM, Salinas VH, Brown KE, Vanguri VK, Freeman GJ, Kuchroo VK, Sharpe AH. PD-L1 regulates the development, maintenance, and function of induced regulatory T cells. J Exp Med. 2009;206:3015–3029. doi: 10.1084/jem.20090847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie F, Xu M, Lu J, Mao L, Wang S. The role of exosomal PD-L1 in tumor progression and immunotherapy. Mol Cancer. 2019;18:146. doi: 10.1186/s12943-019-1074-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, Eppolito C, Qian F, Lele S, Shrikant P. Tumor-infiltrating NY-ESO-1–specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci. 2010;107:7875–7880. doi: 10.1073/pnas.1003345107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muenst S, Soysal SD, Gao F, Obermann EC, Oertli D, Gillanders WE. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2013;139:667–676. doi: 10.1007/s10549-013-2581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma P, Allison JP. The future of immune checkpoint therapy. Science. 2015;348:56–61. doi: 10.1126/science.aaa8172. [DOI] [PubMed] [Google Scholar]
- 15.Jiao P, Geng Q, Jin P, Su G, Teng H, Dong J, Yan B. Small molecules as PD-1/PD-L1 pathway modulators for cancer immunotherapy. Curr Pharm Des. 2018;24:4911–4920. doi: 10.2174/1381612824666181112114958. [DOI] [PubMed] [Google Scholar]
- 16.Alsaab HO, Sau S, Alzhrani R, Tatiparti K, Bhise K, Kashaw SK, Iyer AK. PD-1 and PD-L1 checkpoint signaling inhibition for cancer immunotherapy: mechanism, combinations, and clinical outcome. Front Pharmacol. 2017;8:1–15. doi: 10.3389/fphar.2017.00561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sasikumar PG, Ramachandra M. Small-molecule antagonists of the immune checkpoint pathways: concept to clinic. Future Med Chem. 2017;9:1305–1308. doi: 10.4155/fmc-2017-0107. [DOI] [PubMed] [Google Scholar]
- 18.Zhan MM, Hu XQ, Liu XX, Ruan BF, Xu J, Liao C. From monoclonal antibodies to small molecules: the development of inhibitors targeting the PD-1/PD-L1 pathway. Drug Discov Today. 2016;21:1027–1036. doi: 10.1016/j.drudis.2016.04.011. [DOI] [PubMed] [Google Scholar]
- 19.Kerr WG, Chisholm JD. The next generation of immunotherapy for cancer: small molecules could make big waves. J Immunol. 2019;202:11–19. doi: 10.4049/jimmunol.1800991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang T, Wu X, Guo C, Zhang K, Xu J, Li Z, Jiang S. Development of inhibitors of the programmed cell death-1/programmed cell death-ligand 1 signaling pathway. J Med Chem. 2019;62:1715–1730. doi: 10.1021/acs.jmedchem.8b00990. [DOI] [PubMed] [Google Scholar]
- 21.Chen S, Song Z, Zhang A. Small-molecule immuno-oncology therapy: advances, challenges and new directions. Curr Top Med Chem. 2019;19:180–185. doi: 10.2174/1568026619666190308131805. [DOI] [PubMed] [Google Scholar]
- 22.Zak KM, Grudnik P, Guzik K, Zieba BJ, Musielak B, Dömling A, Dubin G, Holak TA. Structural basis for small molecule targeting of the programmed death ligand 1 (PD-L1) Oncotarget. 2016;7:30323–30335. doi: 10.18632/oncotarget.8730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qin M, Cao Q, Zheng S, Tian Y, Zhang H, Xie J, Xie H, Liu Y, Zhao Y, Gong P. Discovery of [1,2,4]Triazolo[4,3-a]pyridines as Potent Inhibitors Targeting the Programmed Cell Death-1/Programmed Cell Death-Ligand 1 Interaction. J Med Chem. 2019;62:4703–4715. doi: 10.1021/acs.jmedchem.9b00312. [DOI] [PubMed] [Google Scholar]
- 24.Skalniak L, Zak KM, Guzik K, Magiera K, Musielak B, Pachota M, Szelazek B, Kocik J, Grudnik P, Tomala M. Small-molecule inhibitors of PD-1/PD-L1 immune checkpoint alleviate the PD-L1-induced exhaustion of T-cells. Oncotarget. 2017;8:72167–72181. doi: 10.18632/oncotarget.20050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shaabani S, Huizinga HPS, Butera R, Kouchi A, Guzik K, Magiera-Mularz K, Holak TA, Dömling A. A patent review on PD-1/PD-L1 antagonists: small molecules, peptides, and macrocycles (2015-2018) Expert Opin Ther Pat. 2018;28:665–678. doi: 10.1080/13543776.2018.1512706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin X, Lu X, Luo G, Xiang H. Progress in PD-1/PD-L1 pathway inhibitors: from biomacromolecules to small molecules. Eur J Med Chem. 2020;186 doi: 10.1016/j.ejmech.2019.111876. [DOI] [PubMed] [Google Scholar]
- 27.Musielak B, Kocik J, Skalniak L, Magiera-Mularz K, Sala D, Czub M, Stec M, Siedlar M, Holak TA, Plewka J. CA-170—a potent small-molecule PD-L1 inhibitor or not? Molecules. 2019;24:2804. doi: 10.3390/molecules24152804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chupak LS, Zheng X. Compounds Useful as Immunomodulators, WO2015034820A1. March 12, 2015.
- 29.LS Chupak, M Ding, SW Martin, X Zheng, P Hewawasam, TP Connolly, N Xu, KS Yeung, J Zhu, DR Langley, (2017). Compounds useful as immunomodulators, WO2015160641. October 22, 2015.
- 30.Guzik K, Zak KM, Grudnik P, Magiera K, Musielak B, Törner R, Skalniak L, Dömling A, Dubin G, Holak TA. Small-molecule inhibitors of the programmed cell death-1/programmed death-ligand 1 (PD-1/PD-L1) interaction via transiently induced protein states and dimerization of PD-L1. J Med Chem. 2017;60:5857–5867. doi: 10.1021/acs.jmedchem.7b00293. [DOI] [PubMed] [Google Scholar]
- 31.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 32.Zhang S, Zhang M, Wu W, Yuan Z, Tsun A, Wu M, Chen B, Li J, Miao X, Miao X. Preclinical characterization of Sintilimab, a fully human anti-PD-1 therapeutic monoclonal antibody for cancer. Antibody Therap. 2018;1:65–73. [Google Scholar]
- 33.Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- 34.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Newman AM, Steen CB, Liu CL, Gentles AJ, Chaudhuri AA, Scherer F, Khodadoust MS, Esfahani MS, Luca BA, Steiner D. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin DY-w, Tanaka Y, Iwasaki M, Gittis AG, Su HP, Mikami B, Okazaki T, Honjo T, Minato N, Garboczi DN. The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors. Proc Natl Acad Sci. 2008;105:3011–3016. doi: 10.1073/pnas.0712278105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deng L, Liang H, Burnette B, Beckett M, Darga T, Weichselbaum RR, Fu YX. Irradiation and anti–PD-L1 treatment synergistically promote antitumor immunity in mice. J Clin Investig. 2014;124:687–695. doi: 10.1172/JCI67313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Couzin-Frankel J. Cancer immunotherapy. Science. 2013;342:1432–1433. doi: 10.1126/science.342.6165.1432. [DOI] [PubMed] [Google Scholar]
- 44.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Griffiths GM, Mueller C. Expression of perforin and granzymes in vivo: potential diagnostic markers for activated cytotoxic cells. Immunol Today. 1991;12:415–419. doi: 10.1016/0167-5699(91)90145-J. [DOI] [PubMed] [Google Scholar]
- 46.Trapani JA, Smyth MJ. Functional significance of the perforin/granzyme cell death pathway. Nat Rev Immunol. 2002;2:735–747. doi: 10.1038/nri911. [DOI] [PubMed] [Google Scholar]
- 47.Konstantinidou M, Zarganes-Tzitzikas T, Magiera-Mularz K, Holak TA, Dömling A. Immune checkpoint PD-1/PD-L1: is there life beyond antibodies? Angew Chem Int Ed. 2018;57:4840–4848. doi: 10.1002/anie.201710407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu S, Yang J, Xu B, Magiera-Mularz K, Skalniak L, Musielak B, Kholodovych V, Holak TA, Hu L. Design, synthesis, evaluation, and structural studies of C2-symmetric small molecule inhibitors of programmed cell death-1/programmed death-ligand 1 protein–protein interaction. J Med Chem. 2019;62:7250–7263. doi: 10.1021/acs.jmedchem.9b00795. [DOI] [PubMed] [Google Scholar]
- 49.Shoichet BK. Virtual screening of chemical libraries. Nature. 2004;432:862–865. doi: 10.1038/nature03197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burr ML, Sparbier CE, Chan Y-C, Williamson JC, Woods K, Beavis PA, Lam EYN, Henderson MA, Bell CC, Stolzenburg S. et al CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature. 2017;549:101. doi: 10.1038/nature23643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang A, Peng D, Guo H, Ben Y, Zuo X, Wu F, Yang X, Teng F, Li Z, Qian X. A human programmed death-ligand 1-expressing mouse tumor model for evaluating the therapeutic efficacy of anti-human PD-L1 antibodies. Sci Rep. 2017;7:42687. doi: 10.1038/srep42687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372:2018–2028. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 53.Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61:1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li C, Zhang N, Zhou J, Ding C, Jin Y, Cui X, Pu K, Zhu Y. Peptide blocking of PD-1/PD-L1 interaction for cancer immunotherapy. Cancer Immunol Res. 2017;6:178–188. doi: 10.1158/2326-6066.CIR-17-0035. [DOI] [PubMed] [Google Scholar]
- 55.Schroder K, Hertzog PJ, Ravasi T, Hume DA. Interferon-γ: an overview of signals, mechanisms and functions. J Leukoc Biol. 2004;75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 56.Podack ER, Hengartner H, Lichtenheld MG. A central role of perforin in cytolysis? Annu Rev Immunol. 1991;9:129–157. doi: 10.1146/annurev.iy.09.040191.001021. [DOI] [PubMed] [Google Scholar]
- 57.Velu V, Titanji K, Zhu B, Husain S, Pladevega A, Lai L, Vanderford TH, Chennareddi L, Silvestri G, Freeman GJ. Enhancing SIV-specific immunity in vivo by PD-1 blockade. Nature. 2009;458:206–210. doi: 10.1038/nature07662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hams E, McCarron MJ, Amu S, Yagita H, Azuma M, Chen L, Fallon PG. Blockade of B7-H1 (programmed death ligand 1) enhances humoral immunity by positively regulating the generation of T follicular helper cells. J Immunol. 2011;186:5648–5655. doi: 10.4049/jimmunol.1003161. [DOI] [PubMed] [Google Scholar]
- 59.Dhanak D, Edwards JP, Nguyen A, Tummino PJ. Small-molecule targets in immuno-oncology. Cell Chem Biol. 2017;24:1148–1160. doi: 10.1016/j.chembiol.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 60.Cheng B, Yuan W-E, Su J, Liu Y, Chen J. Recent advances in small molecule based cancer immunotherapy. Eur J Med Chem. 2018;157:582–598. doi: 10.1016/j.ejmech.2018.08.028. [DOI] [PubMed] [Google Scholar]
- 61.Boutros C, Tarhini A, Routier E, Lambotte O, Ladurie FL, Carbonnel F, Izzeddine H, Marabelle A, Champiat S, Berdelou A. Safety profiles of anti-CTLA-4 and anti-PD-1 antibodies alone and in combination. Nat Rev Clin Oncol. 2016;13:473–486. doi: 10.1038/nrclinonc.2016.58. [DOI] [PubMed] [Google Scholar]
- 62.Mahoney KM, Rennert PD, Freeman GJ. Combination cancer immunotherapy and new immunomodulatory targets. Nat Rev Drug Discov. 2015;14:561–584. doi: 10.1038/nrd4591. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.