Abstract
After being overlooked for decades, circular RNAs (circRNAs) have recently generated considerable interest. circRNAs play a role in a variety of normal and pathological biological processes, including hepatocarcinogenesis. Many circRNAs contribute to hepatocarcinogenesis through sponging of microRNAs (miRs) and disruption of cellular signaling pathways that play a part in control of cell proliferation, metastasis and apoptosis. In most cases, overexpressed circRNAs sequester miRs to cause de-repressed translation of mRNAs that encode oncogenic proteins. Conversely, low expression of circRNAs has also been described in hepatocellular carcinoma (HCC) and is associated with inhibited production of tumor suppressor proteins. Other functions of circRNAs that contribute to hepatocarcinogenesis include translation of truncated proteins and acting as adapters to regulate influence of transcription factors on target gene expression. circRNAs also affect hepatocyte transformation indirectly. For example, the molecules regulate immune surveillance of cancerous cells and influence the liver fibrosis that commonly precedes HCC. Marked over- or under-expression of circRNA expression in HCC, with correlating plasma concentrations, has diagnostic utility and assays of these RNAs are being developed as biomarkers of HCC. Although knowledge in the field has recently surged, the myriad of described effects suggests that not all may be vital to hepatocarcinogenesis. Nevertheless, investigation of the role of circRNAs is providing valuable insights that are likely to contribute to improved management of a serious and highly aggressive cancer.
Keywords: HCC, HBV, circRNA, ceRNA, microRNA, biomarkers, signaling pathways
Introduction
Hepatocellular carcinoma (HCC), which arises from malignant transformation of hepatocytes, accounts for approximately 75% of primary liver cancers (PLCs) (reviewed in1). Intrahepatic cholangiocarcinoma, the other major PLC, arises from cells of the bile ducts and accounts for 12–15% of hepatic malignancies. PLC is ranked as the seventh most common cancer in the world. Because of the aggressive nature of these hepatic malignancies, they are the fourth most common cause of cancer-related deaths. Equal annual HCC incidence and mortality resulting from the cancer further attests to the poor clinical prognosis of individuals with the disease. HCC typically develops in livers with chronic underlying disease and/or toxin exposure. Examples are persistent hepatitis B virus (HBV) infection, hepatitis C virus (HCV) infection, excessive alcohol consumption, smoking, aflatoxin B1 exposure and non-alcoholic fatty liver disease.2 Gender may also predispose to risk for HCC. Studies conducted throughout the world indicate that males have a higher risk than females. Although hormonal influences are likely to have an impact on HCC risk, gender-based differences in immune responses and epigenetic DNA modifications may play a part. Race and ethnicity have also been implicated in risk for HCC. However, such influences are difficult to separate from geographically linked risk factors.
Understandably, prevention of HCC has been aimed at limiting impact of risk factors. HBV remains the major risk factor, and it is estimated that there are 250 million chronic carriers of the virus who are at high risk for HCC.3–5 In parts of the world where HBV infection is endemic, such as sub-Saharan Africa, east and Southeast Asia and the western Pacific islands, prophylactic vaccination against HBV has been implemented to avert HCC. Particularly impressive cancer prevention achieved by vaccination against HBV has been reported in Taiwan.6 HCV infection is another important risk factor for HCC. Approximately 140 million people are infected with HCV and at risk for HCC but there is no effective vaccine against this virus.7,8 Both HBV and HCV cause chronic necro-inflammatory hepatic disease that contributes to malignant transformation. HBV is a DNA virus and integration of viral sequences into the host genome plays a part in malignant transformation. However, as an RNA virus with no DNA replication intermediate, HCV sequences are not found in the host genome. Exposure to aflatoxins formed by Aspergillus flavus, a fungus that grows on grain crops stored under humid conditions, is a well-established risk factor for HCC.9 Combination with exposure to HBV infection may further exacerbate HCC risk. Excessive alcohol consumption and tobacco use also place individuals at high risk.1,2 Fat deposition in the liver is now recognized as an important cause of HCC.2,10 Non-alcoholic fatty liver disease (NAFLD), associated with obesity, type II diabetes and metabolic syndrome, may progress to steatohepatitis, cirrhosis and HCC. So-called “westernization” of diets in many parts of the world together with sedentary lifestyles have compounded the issue. Increased prevalence of obesity and associated liver complications are contributing to increased incidence of HCC.
Diagnosis of HCC is based on clinical evaluation of patients with risk factors, symptoms, signs and presence of markers that indicate the cancer. In high risk groups, such as HBV chronic carriers, regular monitoring is advised to detect malignancy at an early stage.11 To monitor HBV carriers bi-annual measurement of alpha-fetoprotein and abdominal ultrasound are advised for early cancer detection. Management of the malignancy entails staging of the disease then selection of the appropriate treatment modality. Surgical resection or liver transplant may be considered if the cancer has not metastasized.12 Sorafenib, an inhibitor of several kinases and in particular Raf-1, has been used successfully to improve overall survival of patients with advanced HCC.13 Avoidance of risk factors, such as excessive alcohol consumption, smoking, aflatoxin, and curative treatment of HCV infection, are important to limit HCC development.
Liver cancer is a malignancy that is very difficult to treat. Cure is rare, and recurrence following resection or transplant of apparently localized tumors occurs commonly.12 Improved understanding of the basic molecular biology underlying HCC, diagnosis of the cancer, monitoring of disease and better treatment modalities are important to improve outcomes. Insights gained by research providing evidence of a role for circular RNAs (circRNAs) in the etiology of HCC, has generated considerable interest. This is evident from the flood of recent publications on the topic. Recently-described properties of circRNAs, which have potential for improved understanding and management of the cancer, are discussed in this review.
Biology of circRNAs
Regulatory non-coding RNAs (ncRNAs) have emerged as important participants in diverse normal and pathology-associated biological functions.14 These RNAs include micro RNAs (miRs), piwi-interacting RNAs (piRNAs), long ncRNAs (lncRNAs) and circRNAs. The covalently closed circular structure of circRNAs confers stability which is important to their biological properties. Without exposed 5ʹ and 3ʹ ends, circRNAs are resistant to ubiquitous exonucleases that rapidly degrade linear RNA sequences. Resulting extended half-lives augment functional efficacy of the circRNAs. Although circRNAs are classified as non-coding, protein translation by these sequences has been reported to occur rarely.15
Evidence for the existence of circRNAs was first reported in 1979.16 This was based on data generated using electron microscopy to study RNA extracted from HeLa cells. Significance of the observation was not immediately apparent, and it was thought that the circRNA could be an artefact. Understandably there was a subsequent hiatus in the field until 2013. At this time, the work of Memczak et al17 and Hansen et al18 reported on regulatory roles of circRNAs. In particular, they showed evidence for miR-binding capacity of the circular sequences. Several reasons have been put forward to explain the delayed emergence of evidence to support a role of circRNAs as regulators (reviewed in19). These include: 1) absence of a poly(A) sequence which excludes circRNAs from commonly used mRNA purification protocols, 2) assumption that the sequencing reads across the back-splicing junctions were artefact, and 3) inability of early RT-PCR procedures to distinguish between linear and circular RNAs. Since 2013, a large body of work supporting a role for circRNAs in normal processes and carcinogenesis has been published. The topic has been comprehensively covered in several recent reviews.14,19–23 The focus of this article is on recent progress, particularly reported during 2019 and 2020, that has been made through understanding mechanisms by which circRNAs contribute to hepatocarcinogenesis. Implications for diagnosis and therapy of the malignancy are also discussed.
Biogenesis of circRNAs
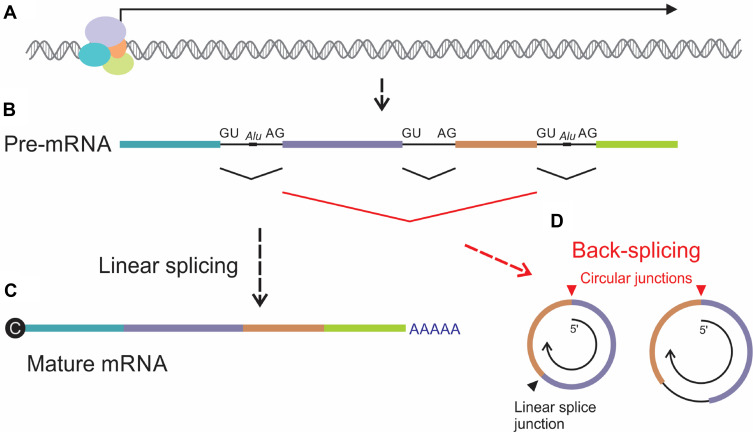
Although the essentials of circRNA biogenesis are established, incomplete understanding of the regulatory processes has been a challenge to the field. Cis and trans elements play a role in regulating circRNA formation24−26 but their mechanisms of action are yet to be fully characterized. Evidence indicates that disruption of circRNA formation is a common feature of many cancers, including HCC (reviewed in19). Back-splicing and processing of lariat structures generated during exon-skipping are the main mechanisms of circRNA genesis (Figure 1). In most cases, splicing to form circRNAs occurs at established exon boundaries or within sequences containing known splice signals.27 Covalent joining of downstream to upstream splice junctions, rather than the canonical upstream linkage to downstream junctions, underlies back-splicing. It appears that complementary Alu repeats at the 5ʹ and 3ʹ ends of the circularizing RNA facilitate the process.28 Although circRNAs typically comprise exons, intronic elements are also often detectable (Figure 1D). To add complexity, there is variability in the number of circRNAs produced by genes: some genes produce several different isoforms, while others produce unique circRNAs.27
Figure 1.
Schematic illustration of formation of circRNA by back-splicing. (A) Gene transcription generates (B) pre-mRNA sequences, which comprise exons (colored rectangles) and introns (intervening black lines). Alu repeats located within introns may play a role in circularization during back-splicing. (C) Canonical linear splicing of pre-mRNA joins upstream splice donors (GU) with downstream splice acceptors (AG) to link exons, remove introns and form mature mRNA with 5’ cap and 3’ poly(A). (D) Back-splicing, which involves circularization of RNA, entails coupling of downstream donor elements with upstream acceptors. Sequences at the resultant circular splice junctions (red arrowhead) are distinct from the sites of linear splicing (black arrowhead). circRNAs formed by back-splicing typically comprise combinations of exons but may also include introns. Arrow within the circRNAs represents 5’ to 3’ polarity of the pre-mRNA.
Selective Detection of circRNAs
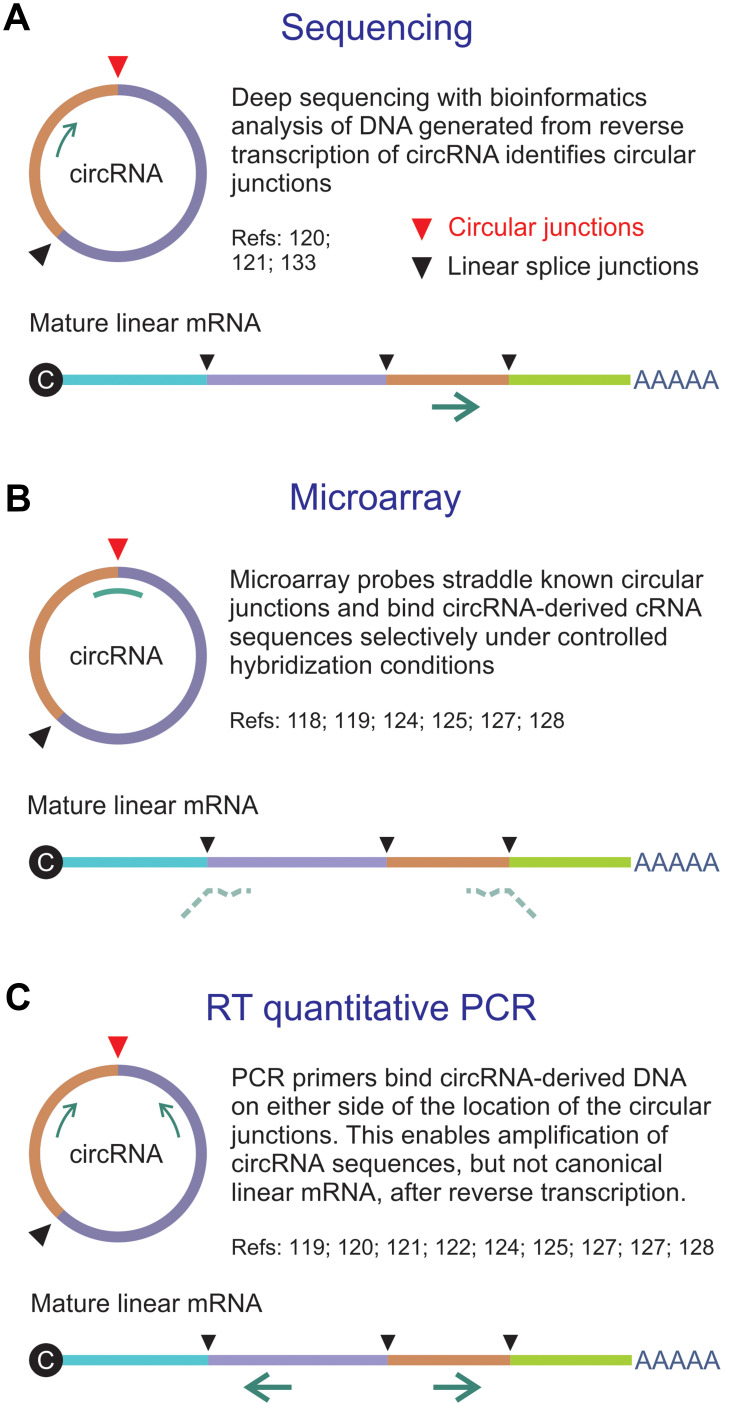
Reliable methods of discriminating circRNA from linear RNA are vital to explore functions of the regulatory RNA. Knowledge about the sequences of circRNAs, particularly at the circular junctions, has been useful to devise assays that each have valuable features (Figure 2). It was initially thought that next generation sequencing (NGS) of RNA isolates would provide the wherewithal for genome-wide circRNA identification (reviewed in27). However, several factors make reliable assay of circRNAs using NGS difficult. These include low abundance, formation of multiple circRNAs from single loci and limitations of algorithms used for bioinformatics analysis. circRNA-specific microarrays have been developed as an alternative genome-wide assay, and examples are the products offered by Arraystar (https://www.arraystar.com/). The method evidently has enhanced specificity and sensitivity when compared to NGS. Essentially the microarray assays are based on use of probes that straddle unique and defined circularization junctions. Typically, total RNA is extracted from cells, which is then treated with RNase R to eliminate linear RNA. The circRNA is amplified and transcribed to generate fluorescently labeled cRNA, which is then hybridized to the circRNA-specific probes. Availability of databases that provide detailed information about circRNA sequences is facilitating development of this type of assay.29,30 Validation and more detailed quantitation of circRNAs may be undertaken using standard PCR-based methods that include reverse transcription quantitative PCR (RT-qPCR) and droplet digital PCR. Primers designed for this type of assay typically span the putative splice junction of the circRNA. Other technologies such as nCounter© by NanoString© are likely to prove useful too (www.nanostring.com/)
Figure 2.
Methods for assay of circRNA. (A) Direct sequencing of a reverse transcribed circRNA typically employs a primer that is complementary to an exon. The sequencing reaction does not discriminate between linear RNA and circRNA, but bioinformatics analysis allows identification of circular junctions (red arrowhead). (B) Microarrays make use of probes that straddle unique and defined circularization junctions. Typically random primers are used to generate labeled cRNAs from circRNA templates. Probes span the circular junctions and hybridize to the cRNAs with higher Tms than the partly complementary sequences of linear mature RNA. (C) Reverse transcriptase (RT) quantitative PCR entails specific amplification of sequences derived from circRNAs by using primers that flank the circular junctions. The configuration of the amplifying primers is such that mature linear mRNA is not amplified.
Functions of circRNAs
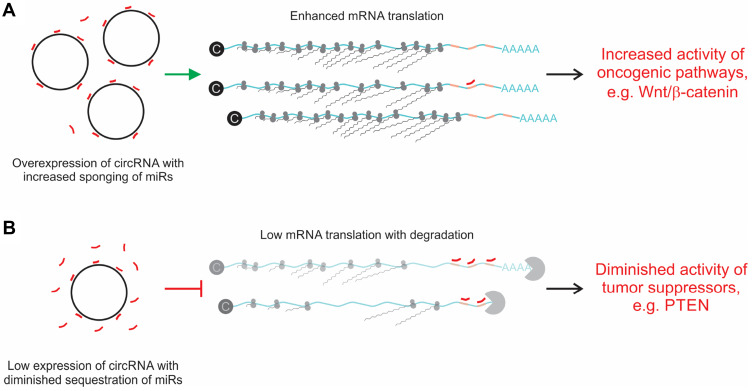
Many different functions of circRNAs have been described.21,22 These include actions as: 1) sponges of endogenous miRs, 2) templates for translation of short proteins, 3) signal transducers, 4) scaffolds for formation of protein complexes, 5) sponges of proteins, 6) nuclear regulators of splicing and 7) transcription. Action as competing endogenous RNAs (ceRNAs) that sequester miRs has been the most commonly described function, and was also the first reported effect of circRNAs on gene expression.17,18 It is well-established that miRs play important roles in cellular processes that lead to malignant transformation.31 These non-coding sequences partially hybridize to mRNA targets and inhibit translation through action of the RNA-Induced Silencing Complex (RISC). By sequestering miRs, circRNAs cause de-repression of target mRNA. If the target mRNA encodes an oncogenic protein, then circRNA sponging leads to overexpression of the potentially transforming protein (Figure 3A). This mechanism has recently been widely described in HCC and detailed information is provided below.
Figure 3.
Influence of sponging circRNA on mRNA translation. (A) Overexpression of circRNAs leads to increased sequestration of miRs (red lines). Resultant diminished free miR de-represses mRNA translation. This may lead to overexpression of genes that participate in pathways involving oncogenesis. An example is overexpression of the Wnt/β-catenin pathway that results from circ-DENND4C overexpression and sponging of miR-195-5p.67 (B) Lowered concentrations of circRNA diminish miR sponging. Increased concentrations of free miRs are available to bind to mRNA cognates, which in turn leads to inhibition of translation that is mediated by the RNA-Induced Silencing Complex (RISC). Hepatocarcinogenesis may be promoted if expression of a tumor suppressor gene is inhibited by this mechanism. An example is the effect of circMTO1 on miR-181b-5p, which leads to diminished expression of Phosphatase and Tensin homolog (PTEN) and promotes fibrosis in premalignant hepatic tissue.116
circRNAs Implicated in the Etiology of Hepatocellular Carcinoma
Recently published data provide compelling evidence that circRNAs play an important role in the etiology of HCC. Action as sponges of miRs that are responsible for controlling potentially oncogenic pathways in hepatocytes is the most frequently reported mechanism. Influence on other processes, such as regulation of immune responses to malignant cells, translation of truncated proteins and control of transcription, have also been shown to play a part. In addition to underlying the mechanisms of malignant transformation, circRNAs have emerged as a class of molecule that may be useful for diagnosing HCC and as targets for therapeutic intervention.
circRNAs as Sponges of microRNAs in HCC
Sponging of cellular miRs is by far the most common mechanism that implicates circRNAs in causing malignant transformation. An initial report on the role of circRNAs in disrupting miR function in HCC tissues was published in 2016.32 Expression of circRNA was analyzed in three patient-derived HCC tumor tissues and two tumor-adjacent samples. Three differentially expressed circRNAs were further analyzed using RT-qPCR in a larger panel of sixty matched samples. One of the circRNAs, hsa_circ_0005075, was consistently overexpressed in malignant cells and correlated with tumor size and other features of aggressiveness of the malignancy. To gain insight into the role of hsa_circ_0005075 in malignant transformation, a network of miRs that could be sequestered by hsa_circ_0005075 was constructed. miR-23b-5p, miR-93-3p, miR-581, miR-23a-5p were believed to be important because of a disruptive effect they had on cell adhesion and likely promotion of tumor cell invasion. Hsa_circ_0005075 was also thought to be a potential biomarker of HCC.
During 2019 and 2020, there have been approximately 70 reports that have supported increased or decreased sequestration of miRs as causative of hepatocarcinogenesis (Supplementary Table 1).32−105 Resulting pro-oncogenic effects include increased hepatocyte proliferation, invasion of surrounding tissues, metastasis and diminished apoptosis. Several different signaling pathways are disrupted by circRNA sponging of miRs. Effects of these circRNAs on endogenous miRs, and which have been reported during 2019 and 2020, are summarized in Table 1. Mechanisms were found to be highly varied and involved de-repression of tumor-promoting genes or inactivation of tumor suppressors. Feedback involving miR sponging by circRNAs to regulate expression of a parent oncogenic sequence, from which the circRNAs are derived, has also been described. One such investigation was carried out on hsa_circ_0003141, which is generated from the transcript of ubiquitin-associated protein 2 (UBAP2).106 Hsa_circ_0003141 was highly expressed in malignant liver tissue and sequestered miR-1827 to inhibit suppression of UBAP2. As a result, apoptosis was inhibited and tumor cell invasion with proliferation were enhanced in the malignant cells.
Table 1.
Overview of Selected Recently Described circRNAs That are Implicated in Pathogenesis of HCC
| Mechanism of Action | circRNAa | miR Target | Observations |
|---|---|---|---|
| Pro-oncogenic | circRNA-103809 | miR-1270 | Competed with PLAGL2 for binding to miR-1270.34 |
| miR-620 | Downregulated in malignant liver cells resulting in increased miR-620.56 | ||
| miR-377-3p | Competing endogenous (ceRNA) of miR-377-3p. De-repression of the gene encoding FGFR1.105 | ||
| circRNA-ABCB10 | miR-670-3p | Sponging of miR-670-3p increased expression of HMG20A.40 | |
| miR-340-5p/miR-452-5p | Downregulated in HCC. Diminished sequestration of miR-340-5p and miR-452-5p and disrupted NRP1 and ABL2 expression.84 | ||
| circMYLK | miR-29a | Binding to miR-29a increased KMT5C gene expression.42 | |
| miR-362-3p | Decrease in miR-362-3p and associated increased Rab23 expression.58 | ||
| circ-0000517 | miR-326 | Sponging effect of miR-326 to increase activity of SMAD646 and IGF1R47 | |
| miR-1296-5p | Upregulation of TXDNC5 by targeting miR-1296-5p.87 | ||
| circPVT1 | miR-3666 | Sequestration of miR-3666 increased expression of SIRT7.57 | |
| miR-203 | ceRNA of miR-203. Stimulated expression of the HOXD3 gene.103 | ||
| circ-0008450 | miR-214-3p | Sponging of miR-214-3p increased expression of EZH2.61 | |
| miR-548p | Amount of circRNA correlated with clinicopathological features of poor prognosis. Effect thought to result from sponging of miR-548p.126 | ||
| circ-0015756 | miR-7 | Associated with diminished functional miR-7 and upregulation FAK.65 | |
| miR-610 | Inhibited miR-610 function. Increased cellular proliferation and invasion by a mechanism involving FGFR1.44 | ||
| circ-0001955 | miR-516a-5p | Binding of miR-516a-5p resulting in de-repression of TRAF6 and MAPK11.85 | |
| mi-R-145-5p | Binding of miR-145-5p augmented activity of the NRAS proto-oncogene.36 | ||
| Post-transcriptional modification | circ-KIAA1429 | - | The YTHDF3 protein is thought to interact with m(6)A modifications of the circRNA, leading to increased stability of Zeb1 mRNA.108 |
| Liver fibrosis | circMTO1 | miR-181b-5p | Reduced circMTO1 leads to disruption of PTEN expression.116 |
| Immunological mechanisms | circUHRF1 | miR-449-5p | Inhibited secretion of IFN-γ and TNF-α, increased expression of TIM-3.134 |
| circARSP91 | - | Upregulation of mRNA encoding ULBP1.136 | |
| circMET | miR-30-5p | Decreased CXCL10 in response to circMET-induced increased DPP4 activity.137 | |
| circ-0064428 | - | Low concentrations of circRNA in patients with increased TILs. High concentrations of circRNA in patients with low TILs.139 | |
| Translatable protein | circβ-catenin | n/a | Encodes 370 amino acid protein which inhibited phosphorylation and subsequent proteasomal degradation of β-catenin. Disruption of Wnt/β-catenin signaling.111 |
| Enhanced transcription | circRHOT1 | n/a | Acts as a scaffold. Recruits TIP60 to increase expression of NR2F6.106 |
Note: aNot all circRNAs with putative sponging mechanisms of action have been included in this table and the complete list is available in Supplementary Table 1.
A variety of experimental approaches was employed to characterize the actions of circRNAs.32–35,37–52,54–60,62,63,65–74,76–91,94 Many of the investigations used patient-derived tumor samples for analysis, which was often carried out in conjunction with information available in public databases such as from the Gene Expression Omnibus (GEO).107 Thereafter differential expression of circRNAs could be verified in HCC tissues or cell lines using standard procedures such as tailored RT-qPCR. Typically, RNA extracts were subjected to treatment with RNase R to remove linear transcripts. Effects of circRNAs on the proliferation of cells, invasion, migration and apoptosis were frequently investigated. Bioinformatics studies were employed to identify miR cognates on the circRNA that could function by sequestering and attenuating function of the miRs. Assessment of the functionality of circRNA and miR interaction was often assessed using well-established dual luciferase reporter assays. Bioinformatics analyses were also employed to identify mRNA targets of the miRs and thereby define regulatory pathway disruption. Functional significance has frequently been assessed by measuring concentrations of affected proteins. Several studies reversed effects of the circRNA by using RNAi-mediated inactivation. Cleavage of the circRNA rendered the sequences linear and therefore susceptible to degradation. In many studies, xenografting was used to assess effects of circRNAs on tumor growth in vivo. For example, malignant liver cells overexpressing circMYLK were grafted onto susceptible mice.58 Injection of circRNA-targeting siRNAs, with or without miR-362-3p agonist, allowed analysis of the role of the miR in tumor cell growth in vivo.
In addition to typical sponging mechanisms, post transcriptional modification of circRNAs may also play a role in influencing translation. This was recently demonstrated in a study on circ_KIAA1429 in HCC.108 circ_KIAA1429 was overexpressed in tumor tissue from patients. The effect was mediated by the YTH N6-Methyladenosine RNA Binding Protein 3 (YTHDF3), which was thought to interact with m(6)A modifications of the circRNA. The result was to increase stability of Zinc Finger E-Box Binding Homeobox 1 (Zeb1) mRNA, which encodes a transcription factor implicated in the loss of e-cadherin expression and epithelial mesenchymal transition (EMT).109
Oncogenic Signaling Pathways Affected by circRNAs
Pathways that have been reported to be disrupted by circRNAs’ effects on miRs are varied. These include signaling mechanisms that are well-established as promoters of tumor growth, such as MAPK,39,45,85 JUN,49,72 MYC,72 Wnt/β-catenin110,111 and JAK2 (Table 1).33,45 In addition, activation of other proteins such as Sirtuin 7 (SIRT7),57 prosaposin (PSAP)63 and hexokinase 2 (HK2)37 have been associated with circRNA-mediated oncogenesis of liver cells. Activation of the Met pathway by circPTGR1 was demonstrated in another study carried out on samples of HCC tissue.93 The mechanism involved sequestration of miR-449a, and intercellular transmission of exosomal circRNA to increase metastatic potential of cells. A study by Wang et al aimed to identify an HCC circRNA signature that was linked to activation of hub genes implicated in malignant transformation.95 A competitive endogenous RNA network was established using information from microarray analysis that was coupled to The Cancer Genome Atlas (TCGA) and the Genotype-Tissue Expression (GTEx) databases. Three hub genes were identified: Aurora Kinase A (AURKA), Kinesin-1 heavy chain encoding gene (KIF5B), and Ras homolog gene family member A (RHOA), which involved circHMGCS1 and circTMCO3. miR cognates of the circRNA and target genes were identified in processes that disrupt various hepatocarcinogenic signaling pathways.
The myriad of circRNAs with varied sponging effects that are associated with causing liver cancer is impressive. Although very interesting, the field has rapidly become complex and applicability to HCC diagnosis and treatment will require careful analysis. The next phase of research will no doubt be focused on detailed verification of physiological importance, correlating the findings with HCC of different etiologies and translating the findings to clinical application.
circRNAs as Therapeutic Targets
The large body of evidence that implicates circRNAs in causing HCC indicates that these sequences may also be targeted for therapy. Many of the studies analyzing the sponging mechanisms have shown that disabling the circRNAs reverses pro-oncogenic properties of overexpressed circRNAs. Delivery of mimics of the “sponged” miRs may be used to rescue effects of sequestration.42 Alternatively, inactivation by RNAi-based cleavage of circRNAs, antisense oligonucleotides and CRISPR/Cas13 could also be used to counter effects of circRNAs (reviewed in21). A recent study employed DNAzymes, which had circRNA-cleaving properties, to disable sequences implicated in HCC and inhibit proliferation and migration of HCC cells.112 circFN1, which is overexpressed in HCC cells that are resistant to sorafenib treatment, may be targeted to improve sensitivity to the multikinase inhibitor.96 Conversely, downregulation of hsa_circ_0006294 and hsa_circ_0035944 was consistently observed in sorafenib-resistant liver cancer-derived cells, and upregulation of these two circRNAs may improve sensitivity to the drug.113 Anticancer activity of another HCC-targeting agent, nitidine chloride, was also recently shown to be influenced by circRNA expression in cancerous tissues.114 Crosstalk between hsa_circ_0088364, hsa_circ_0090049 and eighteen hub genes that regulate cell signaling and cancer progression was demonstrated. Although these interesting findings may have application to the management of HCC, clinical translation of circRNA-disabling effects is currently some way from practical realization. Hurdles, such as efficient delivery of therapeutic sequences to target cells, need to be overcome before the technology can be used to treat HCC.
circRNAs That Promote Liver Fibrosis
Cirrhosis is a commonly observed phenomenon in livers of patients with HCC, and is considered to be a precursor of hepatocyte malignant transformation.115 The process is characterized by liver cell death, regeneration and accompanying accumulation of extracellular collagen. Hepatic stellate cells (HSCs) play an important role in regulating this process and miRs that affect HSC function may therefore influence the development of cirrhosis. To address this, Jin et al investigated the possible role of circMTO1, a circRNA derived from the mitochondrial tRNA translation optimization 1 (MTO1) gene, in the etiology of fibrosis and HCC.116 circMTO1 had previously been shown to act as a tumor suppressor of HCC.117 As in patients with HCC, circMTO1 was reduced in liver tissue from patients with hepatic fibrosis.116 Moreover, the circRNA was downregulated in HSCs and in fibrotic murine tissue, but activation of HSCs was reduced when circMTO1 was overexpressed. The mechanism underlying the process involved disruption of expression of Phosphatase and Tensin homolog (PTEN) (Figure 3B), which is targeted by miR-181b-5p and is involved in malignant transformation of several cancers. Naturally circMTO1 sequesters oncogenic miR-181b-5p. When concentrations of the circRNA were reduced, active miRs increased to diminish PTEN. The study emphasizes the importance of a role for circRNAs in the pre-cancerous state that may eventually lead to full-blown HCC.
circRNAs with Potential for Diagnosing Hepatocellular Carcinoma
HCC is an aggressive malignancy that is often irresectable. Rapid proliferation of the cancerous cells, invasion of surrounding tissue and spread to lymph nodes followed by metastasis to other organs contribute to a poor prognosis. Early diagnosis of HCC is thus crucial for more effective treatment, which may include surgical resection. Currently available and commonly used serum markers of HCC are alpha-fetoprotein and carbohydrate antigen 19–9 (CA19-9). Abdominal ultrasound, or other imaging techniques, is typically carried out in conjunction with serological markers to aid with tumor detection. Implication of circRNAs in the etiology of HCC has prompted investigating their utility as biomarkers for diagnosis of HCC. Stability conferred by the covalently closed circular structure is a useful property that is advantageous over HCC-specific linear RNAs. The field has been an area of vigorous investigation and many research articles on the topic have recently been published.118–128 Many circRNAs have shown potential utility for the diagnosis of HCC and the results from recent studies are summarized in Table 2.
Table 2.
Recently Described circRNAs with Potential Application as Biomarkers of HCC
| circRNA | Mechanisms of Action and Evidence for Utility as Biomarkers |
|---|---|
| Circ_0016788 | Analysis was carried out on a panel of 278 tumor and 116 adjacent non-malignant resected tissues. Data revealed that Circ_0016788 was highly expressed in malignant cells and could reliably be used to distinguish cancerous from non-cancerous cells. Concentration of the circRNA correlated with tumor size, stage of disease and other markers of progression of the malignancy.118 |
| Several circRNAs | Meta-analysis was carried out on studies available in various databases, such as PubMed. Studies reporting on diagnosis, prognosis and clinicopathology were considered. Several upregulated and downregulated circRNAs were identified as potentially useful prognostic and diagnostic biomarkers of HCC.129–132 |
| hsa_circ_0078602 | Microarray analysis was carried out to identify circRNAs that were downregulated in malignant liver cells. Among those sequences identified as having potential for evaluating diagnosis and prognosis, diminished hsa_circ_0078602 was most consistently associated with poor clinical outcomes.119 |
| circ_0000798 | RNA sequencing analysis was carried out on extracts of peripheral blood mononuclear cells (PBMCs) from patients with HCC and healthy controls. Analysis showed that circ_0000798 concentrations could be used to discriminate patients with cancer from healthy controls.120 |
| Various circRNAs, as well as mRNAs and lncRNAs | Sequencing of RNA isolated from extracellular vesicles of 159 healthy individuals and 150 patients with five different cancers was undertaken. RNA species included mRNA, lncRNA and circRNA. Various RNAs were identified in the extracellular vesicles that had potential for diagnosis. Performance of the markers as classifiers of HCC was assessed. A support vector machine (SVM) algorithm showed that measurement of the vesicular RNAs could be used with 84% sensitivity and 94% specificity for diagnosis of HCC.133 |
| circSMARCA5 | circSMARCA5 was measured in tumor, tumor-adjacent and plasma of patients with and without liver cancer. Low concentrations of circSMARCA5 were found in malignant cells and correlated with indicators of tumor aggressiveness such as tumor, node, metastasis (TNM) and size of the primary malignancy. Moreover, low levels of circSMARCA5 were also detected in plasma and could be especially useful for diagnosis in patients where alpha-fetoprotein was below 200 ng/ml.121 |
| circ_0021093 | Increased expression of circ_0021093 was observed in 82 matched tumor and tumor-adjacent tissues, as well as in cultured HCC lines. The circRNA functioned as a sponge for miR-766-3p and caused de-repression of metastasis-associated protein 3. circ_0021093 was considered a useful therapeutic target and also diagnostic marker of HCC.64 |
| circAKT3 | Concentrations of circAKT3 were measured in circulating exosomes isolated from 124 patients with HCC and 100 healthy volunteers. Data revealed that a high concentration of the circRNA was associated with higher risk of mortality and recurrence of the malignancy following resection.122 |
| circ_0000267 | circ_0000267 was assayed in tumor samples from patients with HCC, and high concentration was found to be associated with severity of the disease and poorer prognosis. The effect was mediated by sequestration of miR-646.69 |
| Hsa_circ_0003998 | Hsa_circ_0003998 was measured in tissues and plasma of patients with HCC, chronic HBV infection and healthy subjects. Results showed that the circRNA was elevated in patients with the malignancy. The marker correlated with poor differentiation, vascular invasion, alpha-fetoprotein concentrations and could be used to predict prognosis.123 |
| circ_0005394 | Compared to non-cancerous samples, circ_0005394 was found to be elevated in malignant tissue. High concentration of the circRNA was associated with larger tumor size, metastasis and poorer prognosis. Mechanistically the effect was mediated by sponging of miR-507 and miR-515-5p to increase function of E2F Transcription Factor 3 (ETF3) and C-X-C motif chemokine ligand 6 (CXCL6).75 |
| circ_0009582, circ_0037120 and circ_0140117 | circ_0009582, circ_0037120 and circ_0140117 were found to be useful markers of HCC in tumor tissue of patients with newly diagnosed HCC. Assay in plasma verified utility of the markers. Sensitivity and specificity could be improved when used in combination with assay for alpha-fetoprotein, and the measurements could be used to distinguish HBV-infected patients with and without cancer.124 |
| hsa_circ_0000976, hsa_circ_0007750 and hsa_circ_0139897 | Microarray and RT-qPCR analysis was undertaken on RNA isolated from plasma to identify circRNAs that were upregulated in patients with HBV-related HCC. The data revealed that hsa_circ_0000976, hsa_circ_0007750 and hsa_circ_0139897 could be used to diagnose HCC, and the assays together were more reliable than measurement of alpha-fetoprotein assay.125 |
| circ_0008450 | circ_0008450 expression in HCC tissue was upregulated when compared to adjacent non-malignant tissue. The amount of circ_0008450 also correlated with clinicopathological features of poor prognosis. Silencing of the circRNA in cultured liver-derived cells reversed effects on cell viability, migration and invasion. The effect was thought to be a result of sponging of miR-548p.126 |
| circRNA_101237 | circRNA_101237 was upregulated in tumor tissue and plasma of patients with HCC, when compared to normal liver tissue and plasma from healthy controls. Overall survival of patients with high expression of circRNA_101237 was poor. Of interest was the observation that cisplatin induced expression of the circRNA.127 |
| hsa_circ_0027089 | Microarray analysis was carried out on plasma samples from 10 patients with HBV-related HCC and 5 patients with HBV-related cirrhosis. Of the panel of up- and down-regulated circRNAs, hsa_circ_0027089 was identified as a potentially useful diagnostic marker that distinguishes HCC from cirrhosis in patients with chronic HBV infection.128 |
RT-qPCR has been employed extensively to detect circRNAs that may be suitable for diagnosis and meta-analyses of these studies, using information in publicly available databases, have been carried out.129–132 Upregulated and downregulated circRNAs were identified, then measured in patients’ samples to correlate with features of tumor cell proliferation, invasion and spread. More recently screening of RNA extracted from tumor and normal tissue using microarray analysis119,124,125,127,128 has also been performed. These analyses were followed by verification using RT-qPCR. Many studies made use of tumor and adjacent normal tissue to compare levels of circRNAs. Again data showed compelling correlations between concentrations of specific circRNAs, tumor size, proliferation, features of apoptosis and metastasis.
Although promising, circRNA measurements in biopsy samples may not be ideal for routine diagnostic purposes. Obtaining liver tissue is an invasive procedure that has complications, which makes less invasive procedures preferable. To address this, investigators have measured circulating RNAs,122–125,127,128,133 which may be located in exosomes derived from malignant liver cells. Correlation between profiles observed in blood samples and malignant liver cells provided validation of circRNAs’ utility as biomarkers.123,124,127 In addition, relationships of circRNAs with mechanistic effects such as those resulting from sponging of miRs provided further corroboration.64,75,126 Assay of circRNAs in peripheral blood mononuclear cells has also been investigated as an indirect method of detecting HCC.120 Results from this study showed that circ_0000798 was a particularly useful indicator that also correlated with patients’ prognosis.
Examples of promising candidate circRNA biomarkers are Circ_0016788118 and a panel of three circulating circRNAs (hsa_circ_0000976, hsa_circ_0007750 and hsa_circ_0139897).125 These circRNAs reliably discriminated normal from malignant tissue and performed as well as conventional HCC markers such as alpha-fetoprotein and CA19-9. Utility of the candidate markers is not yet proven in a clinical setting, which will be important to establish.
Immunological Mechanisms by Which circRNAs Contribute to HCC
In addition to the direct effects of circRNAs on promoting hepatocyte malignancy, this class of molecule also influences normal immunological mechanisms that are responsible for tumor suppression. To achieve this, circRNAs may be secreted from hepatocytes via exosomes to exert effects on immune cells. Several recent studies have provided evidence in support of such mechanisms. To assess the role of circular ubiquitin-like with PHD and ring finger domain 1 RNA (circUHRF1) in suppressing anti-tumor immune surveillance, Zhang et al measured concentrations of the circRNA in tumor tissue, tumor-derived exosomes and cell lines.134 Elevated expression of circUHRF1 was observed in the malignant tissue, which was higher than that observed in non-malignant adjacent tissue. Moreover, the increased circUHRF1 concentration correlated with poor clinical prognosis. circUHRF1 was found in exosomes of patients’ plasma samples, and was associated with decreased infiltration of NK cells. circUHRF1 inhibited secretion of IFN-γ and TNF-α by NK cells through diminished action of miR-449-5p. Consequent increased expression of T cell immunoglobulin and mucin domain 3 (TIM-3), which plays a role in suppressing anti-tumor immunity,135 mediated the effect.134 The study suggests that circUHRF1 could serve as a target for therapy of HCC: inhibiting action of the circRNA or use of anti-TIM3 agents may promote NK and CD8+ T cell immunity to tumor cells. Another investigation into regulating NK cell function in HCC identified a role for circRNA of AR-suppressed PABPC1 91 bp (CircARSP91).136 By increasing susceptibility of HCC cells to NK cytotoxicity, CircARSP91 acted as a tumor suppressor. The mechanism involved upregulation of mRNA encoding UL16 binding protein 1 (ULBP1), which may activate NK receptors involved in eliminating malignant liver cells.
Another report on a function of circRNA in regulating immunity to malignant liver cells implicated HCC-produced circMET.137 This sequence is derived from chr.7q21-7q31, which is frequently amplified in HCC,138 and was overexpressed in malignant liver cells and associated with poor patient prognosis.137 Increased circMET promoted malignant transformation by inducing EMT and immunosuppression in the tumor microenvironment. The authors demonstrated that the mechanism of circMET action was through inhibitory sponging action on miR-30-5p, which in turn led to increased activity of the Snail transcription factor. Snail activated expression of dipeptidyl peptidase 4 (DPP4) through interaction with enhancer elements of DPP4. The substrate of DPP4 was identified as the C-X-C motif chemokine ligand 10 (CXCL10), which normally increases CD8+ lymphocyte infiltration of tumors in response to IFN-γ. Decreased CXCL10 was observed in response to circMET-induced increased DPP4 activity. Importantly this effect could be reversed in a xenograft model of HCC when sitagliptin, an inhibitor of DPP4, was given with a programmed death 1 (PD1) blocker.
To identify circRNAs that may be involved with regulating infiltration of tumors by lymphocytes, patient samples were stained for CD8+ cells and categorized as having high or low numbers of tumor infiltrating lymphocytes (TILs).139 Expression profiles of circRNAs in plasma were then assessed using microarray analysis, and correlated with TIL numbers. Hsa_circ_0064428 concentrations were low in patients with increased TILs and conversely the circRNA was high in patients with low TILs. Large numbers of TILs were associated with a better prognosis, and hsa_circ_0064428 was thus proposed as a marker of HCC prognosis. An indirect role of circRNAs in regulating tumor development and spread by modulating immune surveillance is significant. As with the direct roles implicated through sequestration of miRs, immune modulation by circRNAs may have diagnostic and therapeutic utility.
Translatable circRNA Associated with Hepatocarcinogenesis
To date, one circRNA has been shown to translate a protein that is implicated in the etiology of HCC.111 Liang et al identified circβ-catenin, which is derived from the β-catenin gene, as playing such a role. The RNase R-resistant circRNA was initially isolated from cultured liver-derived cancer cells and was associated with oncogenic features, which could be reversed by silencing the circRNA. Circβ-catenin was also highly expressed in malignant liver tissue. Bioinformatics analysis revealed a putative internal ribosomal entry site (IRES) that may initiate translation from the circRNA. Indeed a 370 amino acid protein of approximately 41 kDa was encoded by circβ-catenin and was shown to be central to the oncogenic process. Sequences at the 5ʹ end of the open reading frames, including the translation initiation codons, were common to both full-length and 41 kDa variants. Premature termination of translation was caused by a stop codon that was generated during the circularization reaction. The circRNA-derived peptide inhibited proteasomal degradation of the full-length β-catenin protein. The mechanism involved substrate competition for glycogen synthase kinase 3β (GSK3β)-induced β-catenin phosphorylation. Diminished β-catenin phosphorylation led to its increased stability and nuclear translocation. Resultant disruption of Wnt/β-catenin signaling augmented the hepatocarcinogenic process.
Translation of proteins encoded by circRNAs are involved with causing glioblastomas, but interestingly the mechanism is different to what has been reported for HCC.140–143 The four glioblastoma-associated circRNAs have tumor suppressor functions that involve arrest of cell division and inhibition of the tyrosine kinase/phosphatidylinositol-3-kinase/protein kinase B (RTK/PI3K/AKT) signaling pathway have been reported. The observations on the role of circβ-catenin in causing HCC are therefore particularly interesting.111 hsa_circ_0079929, has recently been shown to be downregulated in HCC tissue,144 and also has an effect on AKT signaling in HCC but it is not yet clear whether the mechanism involves translation of the circRNA.
circRNA-Mediated Regulation of Transcription of Genes Associated with Hepatocarcinogenesis
In a study carried out on malignant tissues of patients with HCC, Wang et al showed that enhanced transcription of nuclear receptor subfamily 2, group F, member 6 (NR2F6), which is a gene implicated in hepatocarcinogenesis, was effected by circRHOT1.106 Bioinformatics was initially used to identify differentially expressed circRNAs in HCC. Thereafter, standard methods of molecular biology, which included RT-PCR, Northern blotting and RNA-FISH, verified that circRHOT1 was overexpressed in the malignant tissue. Knockout of the circRNA reversed features of malignancy in liver-derived cancer cells. circRHOT1 acted as a scaffold and recruited TIP60, a chromatin remodeler with transcription factor function, to the NR2F6 transcription initiation site and increased expression of NR2F6. circRHOT1 was associated with a poor prognosis in patients with HCC, indicating that the circRNA and upregulated NR2F6 protein could be pharmacological targets and useful biomarkers of HCC.
Discussion and Conclusions
Research on the role of circRNAs in hepatocarcinogenesis has generated considerable interest and recent progress has occurred at an impressive pace. Many studies have been published which implicate circRNAs in a variety of mechanisms underlying transformation of hepatocytes. Most work has demonstrated that derangement of circRNA production leads to altered sequestration of miRs with disruption of mRNA translation and altered functioning of pathways implicated in hepatocarcinogenesis (Table 1 and Supplementary Table 1). Both oncogenic and tumor-suppressor sponging functions of circRNAs have been described. Most reports demonstrate tumor promotion resulting from increased circRNA concentrations and sequestration of miRs that inhibit oncogenic pathways (Figure 3). In addition, by sponging of miRs, circRNAs may promote premalignant conditions such as hepatic fibrosis and disruption of immunological mechanisms responsible for eliminating malignant cells. The importance of circRNAs in hepatocarcinogenesis has led to assessment of their utility for HCC diagnosis. Several circRNAs appear in the blood plasma, probably located within exosomes, and direct measurement in the blood may in future be used conveniently to diagnose HCC. Many functional studies have made use of RNAi-based methods to inactivate circRNAs, and reverse oncogenic effects of the ceRNA. Functional efficacy indicates that targeting circRNAs has therapeutic potential and is a topic that will no doubt receive attention in the near future.
Although exciting, the field is faced with challenges. The lack of information about regulation of circRNA biogenesis has hampered an understanding of how processing of these sequences underlies hepatocarcinogenesis. Although there is overwhelming evidence that circRNAs play an important role in various normal and pathological biological processes, it is likely that in some cases circRNA formation is a by-product of imperfect splicing.145 Sequestration of miRs by circRNAs that function as ceRNAs is widely reported and particularly important for hepatocarcinogenesis (Table 1 and Supplementary Table 1). The large numbers of circRNAs that have been reported to have this property is impressive. However, absence of detailed information about molecular quantities of the circRNAs and abundance of conserved miR cognate sequences in the ceRNAs suggests that further verification will be important. Previous work that used RNA-Seq to analyze circRNA populations in unselected RNA from normal mammalian cells showed that circRNAs typically comprise a small proportion of isoforms derived from primary transcripts.145 Moreover, circRNA sequences were generally not more conserved than their neighboring linear exons. Although conservation of sequences located in circRNAs is considered supportive of the notion that they have important biological function, this interpretation may also be flawed. Efficiency of canonical splicing varies at splice acceptor and donor sites and may be influenced by surrounding RNA sequences. Inefficient splicing, which may be conserved at particular sites across species, is likely to favor formation of circRNA by-products. CDR1-AS (also known as CiRS-s) may be a notable exception.18 This circRNA has more than 60 conserved miR-7 recognition sites, which is compelling evidence for a role of distinct circRNA as a sponge. Low abundance of ceRNAs may nevertheless exert an effect on translation by participating in enzymatic catalytic processes.
Methods to assay circRNAs have improved considerably, but error and bias that influence accuracy are yet to be completely eliminated.27 Theoretically, RNA-seq provides the means to identify circRNAs accurately within populations of different RNAs that constitute complete transcriptomes. However, algorithms used to analyze the NGS data often apply different problem solving rules (heuristics) to minimize false positives and false negatives. This can lead to incorrect discoveries and variations in data interpretation. Other variables that may confound interpretation of data include introduction of sequence artefacts during library preparation, sequence-specific influences on splicing, biases resulting from rarity of circRNAs and difficulties with classifying splice junctions as true positives. Comprehensive evaluation of such problems and their relationship to computational approaches used to identify and assay circRNAs are provided in the review by Szabo and Salzman.27
circRNAs previously did not receive attention because methodology used to prepare RNA libraries excluded circRNA sequences by carrying out procedures such as poly(A) selection. Enriching circRNA populations by treating extracts with RNase R to eliminate linear RNAs is now widely used in studies employing microarray assays. Supplementing the technology to enable simultaneous assay of linear mRNAs may be valuable to standardize data, correlate quantitative information about sponging circRNAs, their miR cognates and the miR-targeted mRNAs. Direct assay of the fractions of circularized transcripts that are derived from parental genes will assist with interpretations about regulatory functions.
Evidence in support of circRNAs playing an important role in HCC has been overwhelming. The multitude of studies with compelling evidence in support of the class of sequences in HCC has been impressive. Not only are circRNAs implicated in the etiology of HCC, but they also have potential as markers of the malignancy, are possible targets of therapeutic intervention and therapeutic agents themselves. The field is at an interesting stage of its development and clarifying research in the immediate future will be important. Reconciling disparate observations, standardization of methodology and devising methods to exclude possible artefacts will be important.
Acknowledgments
The authors gratefully acknowledge support from the National Research Foundation (Unique Grant Numbers 118022 and 120383), Poliomyelitis Research Foundation (PRF) and South African Medical Research Council.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978-2007. Int J Cancer. 2016;139(7):1534–1545. doi: 10.1002/ijc.30211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petrick JL, McGlynn KA. The changing epidemiology of primary liver cancer. Curr Epidemiol Rep. 2019;6(2):104–111. doi: 10.1007/s40471-019-00188-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cha C, DeMatteo RP. Molecular mechanisms in hepatocellular carcinoma development. Best Pract Res Clin Gastroenterol. 2005;19(1):25–37. doi: 10.1016/j.bpg.2004.11.005 [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Protzer U, Siddiqui A. Revisiting Hepatitis B virus: challenges of curative therapies. J Virol. 2019;93:20. doi: 10.1128/JVI.01032-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Revill PA, Chisari FV, Block JM, et al. A global scientific strategy to cure hepatitis B. Lancet Gastroenterol Hepatol. 2019;4(7):545–558. doi: 10.1016/S2468-1253(19)30119-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang MH, You SL, Chen CJ, et al. Long-term Effects of Hepatitis B immunization of infants in preventing liver cancer. Gastroenterology. 2016;151(3):472–480. doi: 10.1053/j.gastro.2016.05.048 [DOI] [PubMed] [Google Scholar]
- 7.de Oliveria Andrade LJ, D’Oliveira A, Melo RC, De Souza EC, Costa Silva CA, Parana R. Association between hepatitis C and hepatocellular carcinoma. J Glob Infect Dis. 2009;1(1):33–37. doi: 10.4103/0974-777X.52979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ringelhan M, McKeating JA, Protzer U. Viral hepatitis and liver cancer. Philos Trans R Soc Lond B Biol Sci. 2017;372:1732. doi: 10.1098/rstb.2016.0274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu HC, Santella R. The role of aflatoxins in hepatocellular carcinoma. Hepat Mon. 2012;12(10HCC):e7238. doi: 10.5812/hepatmon.7238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baffy G, Brunt EM, Caldwell SH. Hepatocellular carcinoma in non-alcoholic fatty liver disease: an emerging menace. J Hepatol. 2012;56(6):1384–1391. doi: 10.1016/j.jhep.2011.10.027 [DOI] [PubMed] [Google Scholar]
- 11.Marrero JA, Kulik LM, Sirlin CB, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the american association for the study of liver diseases. Hepatology. 2018;68(2):723–750. doi: 10.1002/hep.29913 [DOI] [PubMed] [Google Scholar]
- 12.Liu CY, Chen KF, Chen PJ. Treatment of liver cancer. Cold Spring Harb Perspect Med. 2015;5(9):a021535. doi: 10.1101/cshperspect.a021535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llovet JM, Ricci S, Mazzaferro V, et al. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359(4):378–390. doi: 10.1056/NEJMoa0708857 [DOI] [PubMed] [Google Scholar]
- 14.Zhang P, Wu W, Chen Q, Chen M. Non-Coding RNAs and their integrated networks. J Integr Bioinform. 2019;16:3. doi: 10.1515/jib-2019-0027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, Wang Z. Efficient backsplicing produces translatable circular mRNAs. RNA. 2015;21(2):172–179. doi: 10.1261/rna.048272.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsu MT, Coca-Prados M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature. 1979;280(5720):339–340. doi: 10.1038/280339a0 [DOI] [PubMed] [Google Scholar]
- 17.Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi: 10.1038/nature11928 [DOI] [PubMed] [Google Scholar]
- 18.Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi: 10.1038/nature11993 [DOI] [PubMed] [Google Scholar]
- 19.Kristensen LS, Hansen TB, Veno MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Shen Y, Li Z, et al. The biogenesis and biological functions of circular RNAs and their molecular diagnostic values in cancers. J Clin Lab Anal. 2020;34(1):e23049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holdt LM, Kohlmaier A, Teupser D. Circular RNAs as therapeutic agents and targets. Front Physiol. 2018;9:1262. doi: 10.3389/fphys.2018.01262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holdt LM, Kohlmaier A, Teupser D. Molecular roles and function of circular RNAs in eukaryotic cells. Cell Mol Life Sci. 2018;75(6):1071–1098. doi: 10.1007/s00018-017-2688-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi: 10.1038/s41576-019-0158-7 [DOI] [PubMed] [Google Scholar]
- 24.Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10(1):103–111. doi: 10.1016/j.celrep.2014.12.002 [DOI] [PubMed] [Google Scholar]
- 25.Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134. doi: 10.1016/j.cell.2015.02.014 [DOI] [PubMed] [Google Scholar]
- 26.Errichelli L, Dini Modigliani S, Laneve P, et al. FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun. 2017;8:14741. doi: 10.1038/ncomms14741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo L, Salzman J. Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet. 2016;17(11):679–692. doi: 10.1038/nrg.2016.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. doi: 10.1016/j.cell.2014.09.001 [DOI] [PubMed] [Google Scholar]
- 29.Liu Q, Cai Y, Xiong H, Deng Y, Dai X. CCRDB: a cancer circRNAs-related database and its application in hepatocellular carcinoma-related circRNAs. Database (Oxford). 2019;2019:2019. doi: 10.1093/database/baz063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rophina M, Sharma D, Poojary M, Scaria V. Circad: a comprehensive manually curated resource of circular RNA associated with diseases. Database (Oxford). 2020;2020:2020. doi: 10.1093/database/baaa019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. miRNA deregulation in cancer cells and the tumor microenvironment. Cancer Discov. 2016;6(3):235–246. doi: 10.1158/2159-8290.CD-15-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shang X, Li G, Liu H, et al. Comprehensive Circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine (Baltimore). 2016;95(22):e3811. doi: 10.1097/MD.0000000000003811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao S, Wang G, Wang J, Li C, Zhang L. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol Cell Biol. 2019;97(2):218–228. doi: 10.1111/imcb.12213 [DOI] [PubMed] [Google Scholar]
- 34.Cao Y, Tao Q, Kao X, Zhu X. Hsa-circRNA-103809 promotes hepatocellular carcinoma development via MicroRNA-1270/PLAG1 like zinc Finger 2 axis. Dig Dis Sci. 2020. doi: 10.1007/s10620-020-06416-x [DOI] [PubMed] [Google Scholar]
- 35.Chen Z, Zuo X, Pu L, et al. circLARP4 induces cellular senescence through regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular carcinoma. Cancer Sci. 2019;110(2):568–581. doi: 10.1111/cas.13901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding B, Fan W, Lou W. hsa_circ_0001955 enhances in vitro proliferation, migration, and invasion of HCC cells through miR-145-5p/NRAS axis. Molecular Therapy - Nucleic Acids. 2020;22:445–455. doi: 10.1016/j.omtn.2020.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding Z, Guo L, Deng Z, Li P. Circ-PRMT5 enhances the proliferation, migration and glycolysis of hepatoma cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. 2020;19(3):269–279. doi: 10.1016/j.aohep.2020.01.002 [DOI] [PubMed] [Google Scholar]
- 38.Fang Q, Liu H, Zhou A, Zhou H, Zhang Z. Circ_0046599 promotes the development of hepatocellular carcinoma by regulating the miR-1258/RPN2 network. Cancer Manag Res. 2020;12:6849–6860. doi: 10.2147/CMAR.S253510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu X, Zhang J, He X, et al. Circular RNA MAN2B2 promotes cell proliferation of hepatocellular carcinoma cells via the miRNA-217/MAPK1 axis. J Cancer. 2020;11(11):3318–3326. doi: 10.7150/jca.36500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fu Y, Cai L, Lei X, Wang D. Circular RNA ABCB10 promotes hepatocellular carcinoma progression by increasing HMG20A expression by sponging miR-670-3p. Cancer Cell Int. 2019;19:338. doi: 10.1186/s12935-019-1055-z [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Gao J, Dai C, Yu X, Yin XB, Zhou F. Circ-TCF4.85 silencing inhibits cancer progression through microRNA-486-5p-targeted inhibition of ABCF2 in hepatocellular carcinoma. Mol Oncol. 2020;14(2):447–461. doi: 10.1002/1878-0261.12603 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Gao J, Li E, Liu W, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression through the miR29a/KMT5C signaling pathway. Onco Targets Ther. 2020;13:8615–8627. doi: 10.2147/OTT.S258715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo J, Duan H, Li Y, Yang L, Yuan L. A novel circular RNA circ-ZNF652 promotes hepatocellular carcinoma metastasis through inducing snail-mediated epithelial-mesenchymal transition by sponging miR-203/miR-502-5p. Biochem Biophys Res Commun. 2019;513(4):812–819. doi: 10.1016/j.bbrc.2019.03.214 [DOI] [PubMed] [Google Scholar]
- 44.Guo W, Zhao L, Wei G, Liu P, Zhang Y, Fu L. Circ_0015756 aggravates hepatocellular carcinoma development by regulating FGFR1 via sponging miR-610. Cancer Manag Res. 2020;12:7383–7394. doi: 10.2147/CMAR.S262231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo X, Jin W, Chang C, et al. Large-scale quantitative genomics analyzes the circRNA expression profile and identifies the key circRNA in regulating cell proliferation during the proliferation phase of rat LR. Artif Cells Nanomed Biotechnol. 2019;47(1):2957–2966. doi: 10.1080/21691401.2019.1640710 [DOI] [PubMed] [Google Scholar]
- 46.He S, Guo Z, Kang Q, Wang X, Han X. Circular RNA hsa_circ_0000517 modulates hepatocellular carcinoma advancement via the miR-326/SMAD6 axis. Cancer Cell Int. 2020;20:360. doi: 10.1186/s12935-020-01447-w [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 47.He S, Yang J, Jiang S, Li Y, Han X. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020;20:404. doi: 10.1186/s12935-020-01496-1 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 48.Jia B, Yin X, Wang Y, et al. CircRNA-PTN sponges miR-326 to promote proliferation in hepatocellular carcinoma. Onco Targets Ther. 2020;13:4893–4903. doi: 10.2147/OTT.S251300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jia C, Yao Z, Lin Z, et al. circNFATC3 sponges miR-548I acts as a ceRNA to protect NFATC3 itself and suppressed hepatocellular carcinoma progression. J Cell Physiol. 2020;236(2):1252–1269. doi: 10.1002/jcp.29931 [DOI] [PubMed] [Google Scholar]
- 50.Jiang QL, Feng SJ, Yang ZY, Xu Q, Wang SZ. CircHECTD1 up-regulates mucin 1 expression to accelerate hepatocellular carcinoma development by targeting microRNA-485-5p via a competing endogenous RNA mechanism. Chin Med J (Engl). 2020;133(15):1774–1785. doi: 10.1097/CM9.0000000000000917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jin J, Liu H, Jin M, Li W, Xu H, Wei F. Silencing of hsa_circ_0101145 reverses the epithelial-mesenchymal transition in hepatocellular carcinoma via regulation of the miR-548c-3p/LAMC2 axis. Aging (Albany NY). 2020;12(12):11623–11635. doi: 10.18632/aging.103324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li D, Zhang J, Li J. Role of miRNA sponges in hepatocellular carcinoma. Clin Chim Acta. 2020;500:10–19. doi: 10.1016/j.cca.2019.09.013 [DOI] [PubMed] [Google Scholar]
- 53.Li J, Qin X, Wu R, Wan L, Zhang L, Liu R. Circular RNA circFBXO11 modulates hepatocellular carcinoma progress and oxaliplatin resistance through miR-605/FOXO3/ABCB1 axis. J Cell Mol Med. 2020;24(9):5152–5161. doi: 10.1111/jcmm.15162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Q, Pan X, Zhu D, Deng Z, Jiang R, Wang X. Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 2019;70(4):1298–1316. doi: 10.1002/hep.30671 [DOI] [PubMed] [Google Scholar]
- 55.Li S, Weng J, Song F, et al. Circular RNA circZNF566 promotes hepatocellular carcinoma progression by sponging miR-4738-3p and regulating TDO2 expression. Cell Death Dis. 2020;11(6):452. doi: 10.1038/s41419-020-2616-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li X, Shen M. Circular RNA hsa_circ_103809 suppresses hepatocellular carcinoma proliferation and invasion by sponging miR-620. Eur Rev Med Pharmacol Sci. 2019;23(2):555–566. doi: 10.26355/eurrev_201902_16868 [DOI] [PubMed] [Google Scholar]
- 57.Li Y, Shi H, Yuan J, Qiao L, Dong L, Wang Y. Downregulation of circular RNA circPVT1 restricts cell growth of hepatocellular carcinoma through downregulation of Sirtuin 7 via microRNA-3666. Clin Exp Pharmacol Physiol. 2020;47(7):1291–1300. doi: 10.1111/1440-1681.13273 [DOI] [PubMed] [Google Scholar]
- 58.Li Z, Hu Y, Zeng Q, et al. Circular RNA MYLK promotes hepatocellular carcinoma progression by increasing Rab23 expression by sponging miR-362-3p. Cancer Cell Int. 2019;19:211. doi: 10.1186/s12935-019-0926-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Z, Liu Y, Yan J, et al. Circular RNA hsa_circ_0056836 functions an oncogenic gene in hepatocellular carcinoma through modulating miR-766-3p/FOSL2 axis. Aging (Albany NY). 2020;12(3):2485–2497. doi: 10.18632/aging.102756 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 60.Liao X, Zhan W, Tian B, Luo Y, Gu F, Li R. Circular RNA ZNF609 promoted hepatocellular carcinoma progression by upregulating PAP2C expression via sponging miR-342-3p. Onco Targets Ther. 2020;13:7773–7783. doi: 10.2147/OTT.S253936 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Lin T, Dai Y, Guo X, et al. Silencing of hsa_circ_0008450 represses hepatocellular carcinoma progression through regulation of microRNA-214-3p/EZH2 axis. Cancer Manag Res. 2019;11:9133–9143. doi: 10.2147/CMAR.S222716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Y, Huang G, Jin H, Jian Z. Circular RNA Gprc5a Promotes HCC progression by activating YAP1/TEAD1 signalling pathway by sponging miR-1283. Onco Targets Ther. 2020;13:4509–4521. doi: 10.2147/OTT.S240261 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 63.Liu C, Zhong X, Li J, Xu F. Circular RNA circVAPA promotes cell proliferation in hepatocellular carcinoma. Hum Gene Ther Clin Dev. 2019;30(4):152–159. doi: 10.1089/humc.2019.079 [DOI] [PubMed] [Google Scholar]
- 64.Liu L, Qi X, Gui Y, Huo H, Yang X, Yang L. Overexpression of circ_0021093 circular RNA forecasts an unfavorable prognosis and facilitates cell progression by targeting the miR-766-3p/MTA3 pathway in hepatocellular carcinoma. Gene. 2019;714:143992. doi: 10.1016/j.gene.2019.143992 [DOI] [PubMed] [Google Scholar]
- 65.Liu L, Yang X, Li NF, Lin L, Luo H. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle. 2019;18(21):2939–2953. doi: 10.1080/15384101.2019.1664223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu W, Yin C, Liu Y. Circular RNA circ_0091579 promotes hepatocellular carcinoma proliferation, migration, invasion, and glycolysis through miR-490-5p/CASC3 axis. Cancer Biother Radiopharm. 2020. doi: 10.1089/cbr.2019.3472 [DOI] [PubMed] [Google Scholar]
- 67.Liu X, Yang L, Jiang D, Lu W, Zhang Y. Circ-DENND4C up-regulates TCF4 expression to modulate hepatocellular carcinoma cell proliferation and apoptosis via activating Wnt/beta-catenin signal pathway. Cancer Cell Int. 2020;20:295. doi: 10.1186/s12935-020-01346-0 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 68.Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900. doi: 10.1038/s41419-019-2089-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pan H, Tang L, Jiang H, et al. Enhanced expression of circ_0000267 in hepatocellular carcinoma indicates poor prognosis and facilitates cell progression by sponging miR-646. J Cell Biochem. 2019;120(7):11350–11357. doi: 10.1002/jcb.28411 [DOI] [PubMed] [Google Scholar]
- 70.Pu J, Wang J, Li W, et al. hsa_circ_0000092 promotes hepatocellular carcinoma progression through up-regulating HN1 expression by binding to microRNA-338-3p. J Cell Mol Med. 2020. doi: 10.1111/jcmm.15010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qiu L, Huang Y, Li Z, et al. Circular RNA profiling identifies circADAMTS13 as a miR-484 sponge which suppresses cell proliferation in hepatocellular carcinoma. Mol Oncol. 2019;13(2):441–455. doi: 10.1002/1878-0261.12424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Qiu L, Wang T, Ge Q, et al. Circular RNA signature in hepatocellular carcinoma. J Cancer. 2019;10(15):3361–3372. doi: 10.7150/jca.31243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song LN, Qiao GL, Yu J, et al. Hsa_circ_0003998 promotes epithelial to mesenchymal transition of hepatocellular carcinoma by sponging miR-143-3p and PCBP1. J Exp Clin Cancer Res. 2020;39(1):114. doi: 10.1186/s13046-020-01576-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Su Y, Lv X, Yin W, et al. CircRNA Cdr1as functions as a competitive endogenous RNA to promote hepatocellular carcinoma progression. Aging (Albany NY). 2019;11(19):8182–8203. doi: 10.18632/aging.102312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun C, Li G, Liu M. A novel circular RNA, circ_0005394, predicts unfavorable prognosis and contributes to hepatocellular carcinoma progression by regulating miR-507/E2F3 and miR-515-5p/CXCL6 signaling pathways. Onco Targets Ther. 2020;13:6171–6180. doi: 10.2147/OTT.S256238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sun P, Fan X, Hu X, Fu X, Wei Q, Zang Y. circPCNX and pecanex promote hepatocellular carcinoma cell viability by inhibiting miR-506. Cancer Manag Res. 2019;11:10957–10967. doi: 10.2147/CMAR.S232940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sun Q, Yu R, Wang C, Yao J, Zhang L. Correction to: circular RNA circ-CSPP1 regulates CCNE2 to facilitate hepatocellular carcinoma cell growth via sponging miR-577. Cancer Cell Int. 2020;20:293. doi: 10.1186/s12935-020-01339-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sun Q, Yu R, Wang C, Yao J, Zhang L. Circular RNA circ-CSPP1 regulates CCNE2 to facilitate hepatocellular carcinoma cell growth via sponging miR-577. Cancer Cell Int. 2020;20:202. doi: 10.1186/s12935-020-01287-8 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 79.Tian F, Yu C, Wu M, Wu X, Wan L, Zhu X. MicroRNA-191 promotes hepatocellular carcinoma cell proliferation by has_circ_0000204/miR-191/KLF6 axis. Cell Prolif. 2019;52(5):e12635. doi: 10.1111/cpr.12635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang W, Li Y, Li X, et al. Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed Pharmacother. 2020;121:109517. doi: 10.1016/j.biopha.2019.109517 [DOI] [PubMed] [Google Scholar]
- 81.Wei X, Zheng W, Tian P, et al. Oncogenic hsa_circ_0091581 promotes the malignancy of HCC cell through blocking miR-526b from degrading c-MYC mRNA. Cell Cycle. 2020;19(7):817–824. doi: 10.1080/15384101.2020.1731945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie B, Zhao Z, Liu Q, Wang X, Ma Z, Li H. CircRNA has_circ_0078710 acts as the sponge of microRNA-31 involved in hepatocellular carcinoma progression. Gene. 2019;683:253–261. doi: 10.1016/j.gene.2018.10.043 [DOI] [PubMed] [Google Scholar]
- 83.Yang J, Li Y, Yu Z, et al. Circular RNA Circ100084 functions as sponge of miR23a5p to regulate IGF2 expression in hepatocellular carcinoma. Mol Med Rep. 2020;21(6):2395–2404. doi: 10.3892/mmr.2020.11069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang W, Ju HY, Tian XF. Circular RNA-ABCB10 suppresses hepatocellular carcinoma progression through upregulating NRP1/ABL2 via sponging miR-340-5p/miR-452-5p. Eur Rev Med Pharmacol Sci. 2020;24(5):2347–2357. doi: 10.26355/eurrev_202003_20501 [DOI] [PubMed] [Google Scholar]
- 85.Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi: 10.1038/s41419-019-2176-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yu X, Sheng P, Sun J, et al. The circular RNA circMAST1 promotes hepatocellular carcinoma cell proliferation and migration by sponging miR-1299 and regulating CTNND1 expression. Cell Death Dis. 2020;11(5):340. doi: 10.1038/s41419-020-2532-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zang H, Li Y, Zhang X, Huang G. Circ_0000517 contributes to hepatocellular carcinoma progression by upregulating TXNDC5 via Sponging miR-1296-5p. Cancer Manag Res. 2020;12:3457–3468. doi: 10.2147/CMAR.S244024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang PF, Wei CY, Huang XY, et al. Circular RNA circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress hepatocellular carcinoma progression. Mol Cancer. 2019;18(1):105. doi: 10.1186/s12943-019-1031-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang S, Liu Y, Liu Z, et al. CircRNA_0000502 promotes hepatocellular carcinoma metastasis and inhibits apoptosis through targeting microRNA-124. J BUON. 2019;24(6):2402–2410. [PubMed] [Google Scholar]
- 90.Zhang W, Zhu L, Yang G, et al. Hsa_circ_0026134 expression promoted TRIM25- and IGF2BP3-mediated hepatocellular carcinoma cell proliferation and invasion via sponging miR-127-5p. Biosci Rep. 2020;40:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhao M, Dong G, Meng Q, Lin S, Li X. Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to regulate the growth and aggressiveness of hepatocellular carcinoma cells. J Cell Biochem. 2020;121(11):4440–4449. doi: 10.1002/jcb.29672 [DOI] [PubMed] [Google Scholar]
- 92.Zou H, Xu X, Luo L, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle. 2019;18(19):2398–2413. doi: 10.1080/15384101.2019.1618120 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 93.Wang G, Liu W, Zou Y, et al. Three isoforms of exosomal circPTGR1 promote hepatocellular carcinoma metastasis via the miR449a-MET pathway. EBioMedicine. 2019;40:432–445. doi: 10.1016/j.ebiom.2018.12.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/beta-catenin pathway. Arch Biochem Biophys. 2019;661:196–202. doi: 10.1016/j.abb.2018.11.020 [DOI] [PubMed] [Google Scholar]
- 95.Wang F, Xu X, Zhang N, Chen Z. Identification and integrated analysis of hepatocellular carcinoma-related circular RNA signature. Ann Transl Med. 2020;8(6):294. doi: 10.21037/atm.2020.03.06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang C, Dong Z, Hong H, et al. circFN1 mediates sorafenib resistance of hepatocellular carcinoma cells by sponging miR-1205 and regulating E2F1 expression. Molecular Therapy - Nucleic Acids. 2020;22:421–433. doi: 10.1016/j.omtn.2020.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wei Y, Chen X, Liang C, et al. A noncoding regulatory RNAs network driven by Circ-CDYL acts specifically in the early stages hepatocellular carcinoma. Hepatology. 2020;71(1):130–147. doi: 10.1002/hep.30795 [DOI] [PubMed] [Google Scholar]
- 98.Wu J, Liu S, Xiang Y, Qu X, Xie Y, Zhang X. Bioinformatic analysis of circular RNA-Associated ceRNA network associated with hepatocellular carcinoma. Biomed Res Int. 2019;2019:8308694. doi: 10.1155/2019/8308694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Xu Q, Zhou L, Yang G, et al. CircIL4R facilitates the tumorigenesis and inhibits ferroptosis in hepatocellular carcinoma by regulating the miR-541-3p/GPX4 axis. Cell Biol Int. 2020;44(11):2344–2356. doi: 10.1002/cbin.11444 [DOI] [PubMed] [Google Scholar]
- 100.Yang G, Wang X, Liu B, et al. circ-BIRC6, a circular RNA, promotes hepatocellular carcinoma progression by targeting the miR-3918/Bcl2 axis. Cell Cycle. 2019;18(9):976–989. doi: 10.1080/15384101.2019.1601477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond). 2019;133(13):1487–1503. doi: 10.1042/CS20190394 [DOI] [PubMed] [Google Scholar]
- 102.Zhang T, Zhang L, Han D, Tursun K, Lu X. Circular RNA hsa_Circ_101141 as a competing endogenous RNA facilitates tumorigenesis of hepatocellular carcinoma by regulating miR-1297/ROCK1 pathway. Cell Transplant. 2020;29:963689720948016. doi: 10.1177/0963689720948016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhu Y, Liu Y, Xiao B, et al. The circular RNA PVT1/miR-203/HOXD3 pathway promotes the progression of human hepatocellular carcinoma. Biol Open. 2019;8:9. doi: 10.1242/bio.043687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang Y, Gao R, Li J, et al. Circular RNA hsa_circ_0003141 promotes tumorigenesis of hepatocellular carcinoma via a miR-1827/UBAP2 axis. Aging (Albany NY). 2020;12(10):9793–9806. doi: 10.18632/aging.103244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhan W, Liao X, Chen Z, et al. Circular RNA hsa_circRNA_103809 promoted hepatocellular carcinoma development by regulating miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 2020;235(2):1733–1745. doi: 10.1002/jcp.29092 [DOI] [PubMed] [Google Scholar]
- 106.Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18(1):119. doi: 10.1186/s12943-019-1046-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Clough E, Barrett T. The gene expression omnibus database. Methods Mol Biol. 2016;1418:93–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang M, Yang Y, Yang J, Yang J, Han S. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m(6)A-YTHDF3-Zeb1. Life Sci. 2020;257:118082. doi: 10.1016/j.lfs.2020.118082 [DOI] [PubMed] [Google Scholar]
- 109.Eger A, Aigner K, Sonderegger S, et al. DeltaEF1 is a transcriptional repressor of E-cadherin and regulates epithelial plasticity in breast cancer cells. Oncogene. 2005;24(14):2375–2385. doi: 10.1038/sj.onc.1208429 [DOI] [PubMed] [Google Scholar]
- 110.Zhu YJ, Zheng B, Luo GJ, et al. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9(12):3526–3540. doi: 10.7150/thno.32796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Liang WC, Wong CW, Liang PP, et al. Translation of the circular RNA circbeta-catenin promotes liver cancer cell growth through activation of the Wnt pathway. Genome Biol. 2019;20(1):84. doi: 10.1186/s13059-019-1685-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ding J, Zhou W, Li X, Sun M, Ding J, Tandem ZQ. DNAzyme for double digestion: a new tool for circRNA suppression. Biol Chem. 2019;400(2):247–253. doi: 10.1515/hsz-2018-0232 [DOI] [PubMed] [Google Scholar]
- 113.Wu MY, Tang YP, Liu JJ, Liang R, Luo XL. Global transcriptomic study of circRNAs expression profile in sorafenib resistant hepatocellular carcinoma cells. J Cancer. 2020;11(10):2993–3001. doi: 10.7150/jca.39854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xiong DD, Feng ZB, Lai ZF, et al. High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma. Cell Death Dis. 2019;10(9):658. doi: 10.1038/s41419-019-1890-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O’Rourke JM, Sagar VM, Shah T, Shetty S. Carcinogenesis on the background of liver fibrosis: implications for the management of hepatocellular cancer. World J Gastroenterol. 2018;24(39):4436–4447. doi: 10.3748/wjg.v24.i39.4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jin H, Li C, Dong P, Huang J, Yu J, Zheng J. Circular RNA cMTO1 Promotes PTEN expression through sponging miR-181b-5p in liver fibrosis. Front Cell Dev Biol. 2020;8:714. doi: 10.3389/fcell.2020.00714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Han D, Li J, Wang H, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66(4):1151–1164. doi: 10.1002/hep.29270 [DOI] [PubMed] [Google Scholar]
- 118.Cheng F, Wang L, Circular ZJ. RNA 0016788 displays as a biomarker for tumor progression and poor prognosis in surgical hepatocellular carcinoma patients. J Clin Lab Anal. 2020;e23300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kou P, Zhang C, Lin J, Wang H. Circular RNA hsa_circ_0078602 may have potential as a prognostic biomarker for patients with hepatocellular carcinoma. Oncol Lett. 2019;17(2):2091–2098. doi: 10.3892/ol.2018.9863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lei B, Zhou J, Xuan X, et al. Circular RNA expression profiles of peripheral blood mononuclear cells in hepatocellular carcinoma patients by sequence analysis. Cancer Med. 2019;8(4):1423–1433. doi: 10.1002/cam4.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li Z, Zhou Y, Yang G, et al. Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta. 2019;492:37–44. doi: 10.1016/j.cca.2019.02.001 [DOI] [PubMed] [Google Scholar]
- 122.Luo Y, Liu F, Gui R. High expression of circulating exosomal circAKT3 is associated with higher recurrence in HCC patients undergoing surgical treatment. Surg Oncol. 2020;33:276–281. doi: 10.1016/j.suronc.2020.04.021 [DOI] [PubMed] [Google Scholar]
- 123.Qiao GL, Chen L, Jiang WH, et al. Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther. 2019;12:5849–5860. doi: 10.2147/OTT.S210363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wu C, Deng L, Zhuo H, et al. Circulating circRNA predicting the occurrence of hepatocellular carcinoma in patients with HBV infection. J Cell Mol Med. 2020;24(17):10216–10222. doi: 10.1111/jcmm.15635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu J, Ding WB, Wang MC, et al. Plasma circular RNA panel to diagnose hepatitis B virus-related hepatocellular carcinoma: a large-scale, multicenter study. Int J Cancer. 2020;146(6):1754–1763. doi: 10.1002/ijc.32647 [DOI] [PubMed] [Google Scholar]
- 126.Zhang J, Chang Y, Xu L, Qin L. Elevated expression of circular RNA circ_0008450 predicts dismal prognosis in hepatocellular carcinoma and regulates cell proliferation, apoptosis, and invasion via sponging miR-548p. J Cell Biochem. 2019;120(6):9487–9494. doi: 10.1002/jcb.28224 [DOI] [PubMed] [Google Scholar]
- 127.Zhou S, Wei J, Wang Y, Liu X. Cisplatin resistance-associated circRNA_101237 serves as a prognostic biomarker in hepatocellular carcinoma. Exp Ther Med. 2020;19(4):2733–2740. doi: 10.3892/etm.2020.8526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhu K, Zhan H, Peng Y, et al. Plasma hsa_circ_0027089 is a diagnostic biomarker for hepatitis B virus-related hepatocellular carcinoma. Carcinogenesis. 2020;41(3):296–302. doi: 10.1093/carcin/bgz154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hao Q, Han Y, Xia W, Wang Q, Qian H. Systematic review and meta-analysis of the utility of circular RNAs as biomarkers of hepatocellular carcinoma. Can J Gastroenterol Hepatol. 2019;2019:1684039. doi: 10.1155/2019/1684039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Huang X, Zhang W, Shao Z. Prognostic and diagnostic significance of circRNAs expression in hepatocellular carcinoma patients: A meta-analysis. Cancer Med. 2019;8(3):1148–1156. doi: 10.1002/cam4.1939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Yu G, Yang L, Zhou J, Zhang L, Xia L. Abnormally expressed circular RNAs are promising biomarkers for diagnosis of hepatocellular carcinoma: a meta-analysis. Clin Lab. 2019;65:11. doi: 10.7754/Clin.Lab.2019.190354 [DOI] [PubMed] [Google Scholar]
- 132.Wang Y, Li Z, Xu S, Guo J. Novel potential tumor biomarkers: circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J Clin Lab Anal. 2020;e23359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li Y, Zhao J, Yu S, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem. 2019;65(6):798–808. doi: 10.1373/clinchem.2018.301291 [DOI] [PubMed] [Google Scholar]
- 134.Zhang PF, Gao C, Huang XY, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19(1):110. doi: 10.1186/s12943-020-01222-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ju Y, Hou N, Meng J, et al. T cell immunoglobulin- and mucin-domain-containing molecule-3 (Tim-3) mediates natural killer cell suppression in chronic hepatitis B. J Hepatol. 2010;52(3):322–329. doi: 10.1016/j.jhep.2009.12.005 [DOI] [PubMed] [Google Scholar]
- 136.Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. 2019;17(1):388–397. doi: 10.3892/ol.2018.9606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Huang XY, Zhang PF, Wei CY, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 2020;19(1):92. doi: 10.1186/s12943-020-01213-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sy SM, Wong N, Lai PB, To KF, Johnson PJ. Regional over-representations on chromosomes 1q, 3q and 7q in the progression of hepatitis B virus-related hepatocellular carcinoma. Mod Pathol. 2005;18(5):686–692. doi: 10.1038/modpathol.3800345 [DOI] [PubMed] [Google Scholar]
- 139.Weng Q, Chen M, Li M, et al. Global microarray profiling identified hsa_circ_0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2019;56(1):32–38. doi: 10.1136/jmedgenet-2018-105440 [DOI] [PubMed] [Google Scholar]
- 140.Xia X, Li X, Li F, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18(1):131. doi: 10.1186/s12943-019-1056-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang Y, Gao X, Zhang M, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:3. doi: 10.1093/jnci/djx166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang M, Huang N, Yang X, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37(13):1805–1814. doi: 10.1038/s41388-017-0019-9 [DOI] [PubMed] [Google Scholar]
- 143.Zhang M, Zhao K, Xu X, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9(1):4475. doi: 10.1038/s41467-018-06862-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zheng H, Chen T, Li C, et al. A circular RNA hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma. Cancer Manag Res. 2019;11:443–454. doi: 10.2147/CMAR.S189338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15(7):409. doi: 10.1186/s13059-014-0409-z [DOI] [PMC free article] [PubMed] [Google Scholar]