Abstract
This study aimed to explore the molecular mechanism by which mesenchymal stem cells (MSCs) mediate lung cancer progression. Extracellular vesicles (EVs) were isolated from transfected or untransfected MSCs, and were co-cultured with lung cancer cells with/without microRNA-130b-3p (miR-130b-3p) inhibitor, mimic, overexpression plasmids of FOXO3/NFE2L2, or shRNAs. CCK-8 assay, colony formation, transwell assay, and flow cytometry were carried out to determine the biological functioning of lung cancer cells. Furthermore, FOXO3, Keap1, NFE2L2, and TXNRD1 expression was determined by qRT-PCR and western blot analysis. A tumor xenograft mouse model was used to determine role of EVs-miR-130b-3p and its target FOXO3 in lung cancer progression in vivo. miR-130b-3p was highly expressed in lung cancer tissues and MSC-derived EVs. Moreover, the MSC-derived EVs transferred miR-130b-3p to lung cancer cells to promote cell proliferation, migration, and invasion while repress cell apoptosis. miR-130b-3p directly targeted FOXO3, and FOXO3 elevated Keap1 expression to downregulate NFE2L2, thus inhibiting TXNRD1. FOXO3 overexpression or silencing of NFE2L2 or TXNRD1 diminished lung cancer cell proliferation, invasion, and migration but enhanced apoptosis. EV-delivered miR-130b-3p or FOXO3 silencing promoted lung cancer progression in vivo. In summary, MSC-derived EVs with upregulated miR-130b-3p suppressed FOXO3 to block the NFE2L2/TXNRD1 pathway, thus playing an oncogenic role in lung cancer progression.
Keywords: lung cancer, extracellular vesicles, mesenchymal stem cells, microRNA-130b-3p, FOXO3, Keap1, NFE2L2, TXNRD1
Graphical Abstract
The study mainly addresses the oncogenic role of miR-130b-3p contained in mesenchymal stem cell-derived extracellular vesicles in the malignant progression of lung cancer, which was associated with the FOXO3/NFE2L2/TXNRD1 axis, thus highlighting a potential therapeutic action against lung cancer.
Introduction
Lung cancer, which is classified as small cell lung cancer (SCLC) and non-small cell lung cancer (NSCLC), is the one of the most lethal cancers.1,2 Until now, the outcome of lung cancer patients remained very poor despite extensive advances in cancer surgery, chemotherapy, and molecular targeting therapies. Metastasis events of lung cancer cells are still the main challenge in lung cancer therapy.3 Therefore, there is an urgent need to investigate the molecular mechanism of lung cancer progression.
Previous work has shown that mesenchymal stem cells (MSCs) are involved in lung cancer progression.4 MSCs are multipotent cells existing in various tissues, which can differentiate into mesenchymal cells, including osteoblasts, adipocytes, and chondrocytes.5 A previous study demonstrated that MSCs could be recruited to the tumor microenvironment,6 which has come to be viewed as a possible route for drug delivery in cancer therapy.7 It is also widely known that MSCs can secrete extracellular vesicles (EVs), which are a storable and cell-free alternative to experimental therapy, possessing comparable therapeutic potential to their parent cells.8 Intriguingly, MSC-derived EVs have been reported to exert therapeutic effects in lung cancer.9 EVs play a pivotal role in cellular communication by transferring their contents, especially microRNAs (miRNAs), to target cells and the microenvironment.10 For example, Dong et al.11 observed that miR-410 expression was strongly enriched in MSC-derived EVs, and that MSC-derived EVs contributed to lung cancer development. They suggested that this effect may be mediated by transferring miR-410 via MSC-derived EVs to lung cancer cells, with the result of inhibited phosphatase and tensin homolog (PTEN) protein expression.
The diverse families of miRNAs participate in many biological processes.12 Tian et al.13 reported that miR-130b-3p, a potential therapy target and biomarker for lung cancer, was upregulated in lung cancer and promoted its progression in vitro and in vivo. Forkhead box O3 (FOXO3) is an anti-oncogene that is known to be downregulated in various cancer cell lines and clinical biopsy specimens.14,15 More important, FOXO3 has been reported to suppress lung cancer tumorigenesis by inhibiting STAT3 activation.16 Nuclear factor erythroid-2-related factor-2 (NFE2L2) has been widely recognized as an oncogene, which promotes anti-oxidative pathways and enhances cancer cell survival.17 NFE2L2 emerged as the key factor for P53-induced protein with a death domain (PIDD)-mediated lung cancer chemoresistance.18 Furthermore, Guan et al.19 revealed that FOXO3 downregulated NFE2L2 to suppress cholangiocarcinoma tumorigenesis and chemoresistance via Kelch-like ECH-associated protein 1 (Keap1). Interestingly, thioredoxin reductase 1 (TXNRD1) has been revealed to be a target gene of NFE2L2,20 thus implicating redox balance in cancer pathways. A previous study manifested that downregulation of TXNRD1 elevated the sensitivity of lung cancer cells to MK2206 treatment in vitro and in vivo.21
In this context, we hypothesized that MSC-derived EVs may be correlated with lung cancer progression by transferring miR-130b-3p to lung cancer cells to orchestrate oncogenic effects via the FOXO3/NFE2L2/TXNRD1 axis. To test this hypothesis, we conducted tissue, cell, and animal experiments using EVs isolated from MSCs.
Results
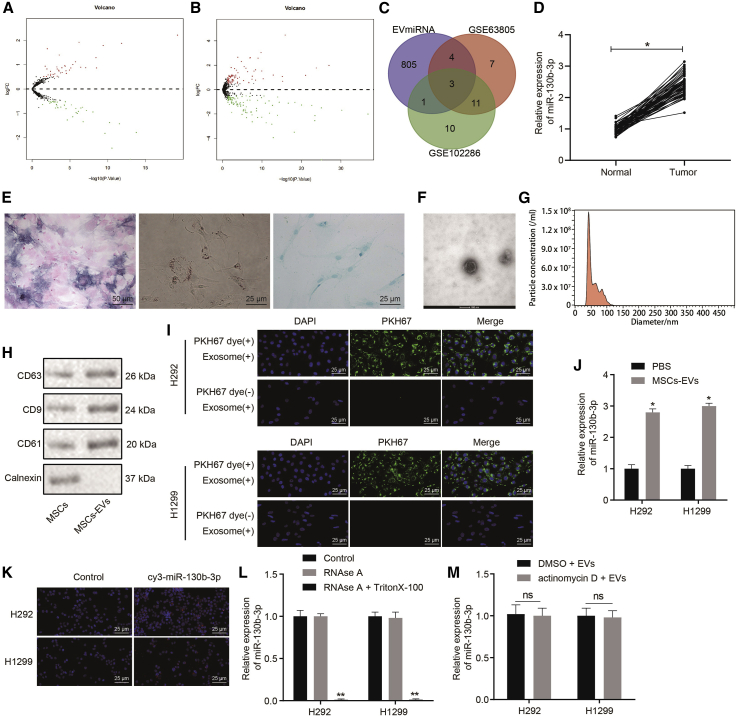
MSC-Derived EVs Contained Abundant miR-130b-3p and Delivered It to Lung Cancer Cells
A previous report revealed that MSC-derived EVs promoted lung cancer proliferation.4 To investigate the molecular mechanisms of lung cancer progression mediated by EVs, we accessed human lung cancer-related miRNA datasets GEO: GSE63805 and GSE102286 from the Gene Expression Omnibus (GEO) and then screened out the differentially expressed miRNAs with log2-fold changes (|logFC|) > 0.5 and p < 0.5 as criteria. Based on this analysis, 37 significantly upregulated miRNAs and 38 significantly downregulated miRNAs were found in GEO: GSE63805 (Figure 1A), whereas 75 significantly upregulated miRNAs and 76 obviously downregulated miRNAs were found in GEO: GSE102286 (Figure 1B). miR-31, miR-142-5p, and miR-130b-3p were identified by Venn analysis of overlapping miRNAs overexpressed in MSC-derived EVs according to the EVmiRNA database among the top 25 upregulated miRNAs in GEO: GSE63805 and GSE102286 (Figure 1C). Previous studies demonstrated that miR-130b-3p was upregulated in lung cancer and served as an oncomiRNA.13,22,23 To validate the elevated miR-130b-3p expression in lung cancer, we collected biopsy specimens from lung cancer patients and determined the miR-130b-3p expression by quantitative reverse transcription-polymerase chain reaction (qRT-PCR). The results showed that miR-130b-3p expression was higher in tumor tissues than in adjacent normal tissues (Figure 1D). Thus, we hypothesized that MSC-derived EVs promote lung cancer progression by transferring miR-130b-3p. To test this claim, we isolated human umbilical cord MSCs (hUCMSCs), which were identified by flow cytometry analysis of their expression of MSC surface markers, including high levels of CD105 (97.53%), CD73 (99.86%), and CD90 (99.64%), but low levels of CD45 (0.28%), CD34 (0.31%), CD14 (0.14%), CD19 (0.33%), and histocompatibility leukocyte antigen-DR isotype (HLA-DR) (0.26%) in the isolated cells. These isolated hUCMSCs had osteogenic, adipogenic, and chondrogenic differentiation capacities (Figure 1E). The data demonstrated the successful extraction of hUCMSCs. Based on this, we isolated EVs from the hUCMSCs and evaluated their characteristics as described below. The isolated EVs were cup shaped or spherical under transmission electron microscopy (TEM) observation (Figure 1F). Nanoparticle tracking analysis (NTA) showed that the diameters of the EVs ranged mainly from 40 to 120 nm (Figure 1G). Meanwhile, the surface markers (CD63, CD81, and CD9) of EVs were found to be much higher in the MSC-EVs than in MSCs (Figure 1H). After this successful isolation of EVs from hUCMSCs, H292 and H1299 were co-cultured with PKH67-labeled EVs for 24 h and then examined for the presence of green fluorescent protein (GFP), which was present in the cytoplasm of both cell lines under fluorescence microscopy (Figure 1I). Furthermore, miR-130b-3p was strikingly upregulated in H292 and H1299 cells incubated with MSC-derived EVs (Figure 1J). To confirm that the increase of miR-130b-3p in H292 and H1299 cells was directly transferred by EVs rather than arising endogenously, MSCs were transfected with Cy3-labeled miR-130b-3p mimic, and then EVs were extracted from these transfected MSCs and co-cultured with H292 and H1299 cells. The transference of Cy3-labeled miR-130b-3p mimic was observed under a fluorescence microscope. The results manifested that red fluorescence was found only in the H292 and H1299 cells incubated with EVs containing Cy3-labeled miR-130b-3p mimic (Figure 1K). Furthermore, MSCs were cultured with medium supplemented with RNase alone or RNase plus Triton X-100, and then RNAs were isolated to verify that miR-130b-3p was delivered to H292 and H1299 cells via EVs. As documented in Figure 1L, miR-130b-3p expression remained unchanged in cells after treatment with RNase alone, while miR-130b-3p was undetectable in cells treated with RNase plus Triton X-100. To exclude further the possibility that the upregulation of miR-130b-3p was due to endogenous induction, H292 and H1299 cells were treated with actinomycin D and MSC-derived EVs. The actinomycin D treatment did not change miR-130b-3p expression in either cell line, which indicated that miR-130b-3p expression was not induced endogenously (Figure 1M). Collectively, miR-130b-3p was upregulated in lung cancer. MSC-derived EVs contained abundant miR-130b-3p and successfully transferred it to lung cancer cells.
Figure 1.
MSC-Derived EVs Transfers miR-130b-3p to Lung Cancer Cells and miR-130b-3p Is Upregulated in Lung Cancer
(A) miRNA expression in lung cancer datasets GSE63805. (B) miRNA expression in lung cancer datasets GSE102286. Black represents miRNAs without significant difference, green represents downregulated miRNAs, and red represents upregulated miRNAs in lung cancer. (C) The intersection of upregulated miRNAs in lung cancer-related datasets and miRNA expression in MSC-EVs through the EVmiRNA database. The red and green circles represent the top 25 upregulated miRNAs in the lung cancer datasets GSE63805 and GSE102286. The center represents the overlapped miRNAs in 3 sets of data. (D) miR-130b-3p expression in lung cancer and adjacent normal tissues (n = 50) determined by qRT-PCR. ∗p < 0.05 compared to adjacent normal tissues. (E) Analysis of MSC osteogenic differentiation assessed by alkaline phosphatase(ALP) staining (200×), adipogenic differentiation detected by oil red O staining (400×), and chondrogenic differentiation detected by Alcian blue staining (400×). (F) EV morphology observed under TEM (representative bar, 100 nm). (G) EV diameters distribution determined by NTA. (H) EV markers determined by western blot analysis. (I) The uptake of EVs by H292 and H1299 cells observed by the fluorescence microscope (400×). (J) miR-130b-3p expression in H292 and H1299 cells after co-culture with MSC-EVs determined by qRT-PCR. ∗p < 0.05 compared to PBS treatment. (K) The transference of cy3-labeled miR-130b-3p mimic into H292 and H1299 cells observed by the fluorescence microscope (400×). (L) miR-130b-3p expression after the MSC condition medium was treated with RNase or RNase + Triton X-100 determined by qRT-PCR. ∗p < 0.05 compared to control. (M) miR-130b-3p expression after H292 and H1299 cells were treated with actinomycin D or MSC-EVs determined by qRT-PCR. ∗p < 0.05 compared to treatment with DMSO + EVs. The data were shown as means ± standard deviations. The cancer tissues were compared with adjacent normal tissues using the paired t test, while the other 2 groups were compared by the unpaired t test. The cell experiment was repeated 3 times.
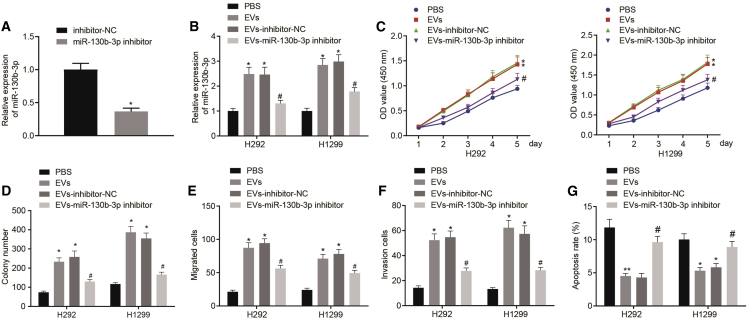
miR-130b-3p Delivered by MSC-Derived EVs Promoted Lung Cancer Cell Proliferation, Invasion, and Migration but Repressed Apoptosis
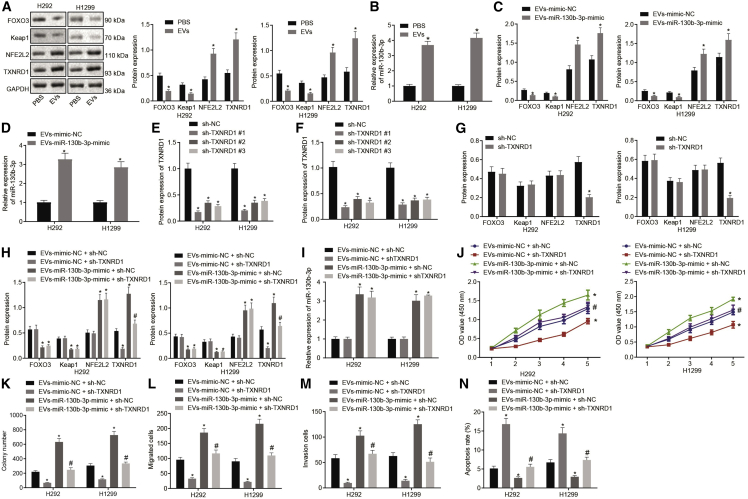
To investigate the role of MSC-EV-delivered miR-130b-3p in lung cancer progression, miR-130b-3p inhibitor or inhibitor-negative control (NC) was introduced to MSCs. Analysis revealed that the miR-130b-3p inhibitor significantly decreased miR-130b-3p expression in MSCs (Figure 2A). Based on this, EVs were extracted from untransfected MSCs (grouping as EVs) or MSCs transfected with miR-130b-3p inhibitor-transfected or inhibitor-NC-transfected MSCs (grouping as EVs-miR-130b-3p inhibitor or EVs-inhibitor-NC). Next, H292 and H1299 cells were treated with phosphate-buffered saline (PBS), EVs, EVs-miR-130b-3p inhibitor, or EVs-inhibitor-NC. After these incubations, qRT-PCR indicated that miR-130b-3p expression was much higher in H292 and H1299 cells incubated with EVs or EVs-inhibitor-NC compared to H292 and H1299 cells incubated with PBS. However, miR-130b-3p was found to be significantly downregulated in H292 and H1299 cells incubated with EVs-miR-130b-3p inhibitor (Figure 2B). Furthermore, cell viability was detected by Cell Counting Kit (CCK)-8 assay (Figure 2C), and colony formation was evaluated by the colony formation assay (Figure 2D). We observed from the results that incubation with EVs or the EVs-inhibitor-NC growth rate significantly increased H292 and H1299 cell viability and colony formation compared to PBS incubation, while H292 and H1299 cells incubated with EVs-miR-130b-3p inhibitor had the opposite effect as compared to H292 and H1299 cells incubated with EVs-inhibitor-NC. Next, a transwell assay revealed that treatment with EVs or EVs-inhibitor-NC elevated the migration and invasion of H292 and H1299 cells, while EVs-miR-130b-3p inhibitor reduced H292 and H1299 cell migration and invasion (Figures 2E and 2F). Consistently, EVs or EVs-inhibitor-NC treatment caused a reduction in H292 and H1299 cell apoptotic rates, whereas opposite effects were seen after treatment with the EVs-miR-130b-3p inhibitor (Figure 2G). In summary, in vitro biological function analysis revealed that MSC-EV-delivered miR-130b-3p augmented lung cancer cell proliferation, migration, and invasion and slowed apoptosis.
Figure 2.
miR-130b-3p Overexpression in MSC-Derived EVs Promotes Lung Cancer Cell Proliferation, Invasion, and Migration, but Reduces Cell Apoptosis
(A) miR-130b-3p expression in MSCs transfected with miR-130b-3p inhibitor determined by qRT-PCR. ∗p < 0.05 compared to MSCs treated with inhibitor-NC. (B) H292 and H1299 cells were treated with PBS, EVs, EVs-miR-130b-3p inhibitor, or EVs-inhibitor-NC. miR-130b-3p expression in H292 and H1299 cells determined by qRT-PCR. (C) H292 and H1299 cell viability determined by CCK-8 assay. (D) H292 and H1299 cell colony formation capacity determined by colony formation assay. (E) H292 and H1299 cell migration determined by transwell assay. (F) H292 and H1299 cell invasion determined by transwell assay. (G) H292 and H1299 cell apoptotic rate determined by flow cytometry. ∗p < 0.05 compared to H292 and H1299 cells treated with PBS. #p < 0.05 compared to H292 and H1299 cells treated with EVs-inhibitor-NC. The data were shown as means ± standard deviations. The 2 groups were compared by unpaired t test. Comparisons among multiple groups were analyzed by Tukey’s test-corrected one-way analysis of variance (ANOVA). Variables were analyzed at different time points using Bonferroni-corrected repeated-measures ANOVA. The cell experiment was repeated 3 times.
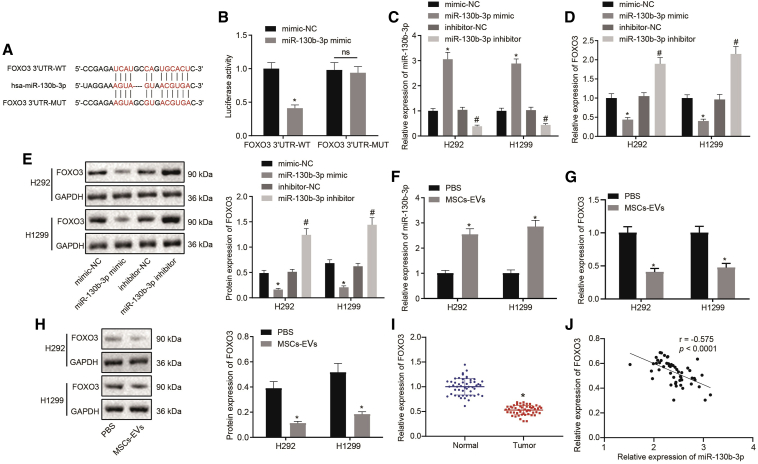
miR-130b-3p Directly Targeted FOXO3
To enable an in-depth study on the mechanism of miR-130b-3p in lung cancer progression, its downstream target genes were predicted by the StarBase database. Human lung cancer-related mRNA expression datasets GEO: GSE101929 and GSE118370 were retrieved from the GEO database. With |logFC| >0.5, p <0.01 (for GEO: GSE101929), and |logFC| >0.7, p <0.01 (for GEO: GSE118370) as screening criteria, differential analysis was conducted using the “limma” package of the R language. Finally, 2,256 significantly upregulated and 2,356 significantly downregulated mRNAs were obtained in lung cancer samples of GEO: GSE101929 (Figure S1A), whereas 683 significantly upregulated and 1,098 significantly downregulated mRNAs were obtained in lung cancer samples of GEO: GSE118370 (Figure S1B). Then, the top 1,600 significantly downregulated mRNAs in the GEO: GSE101929 and the top 1,000 significantly downregulated mRNAs in GEO: GSE118370 were intersected with the top 3,000 genes predicted by the StarBase database, and 118 genes were intersected in these 3 sets of data (Figures S1C and S1D). Through the STRING database (https://string-db.org/), the interaction between intersecting genes was evaluated and a protein-protein interaction (PPI) network was constructed (Figure S1E). The top 5 genes in the hub gene module were predicted from the PPI network using the maximal clique centrality (MCC) network topology algorithm in the cytoHubba application, and 5 reliable candidate target genes were finally identified: PIK3R1, FGF2, FOXO3, AKT3, and TNS1 (Figure S1F). Through the GEPIA database, it was found that the expression of FOXO3 in lung cancer samples was significantly decreased (Figure S1G). This investigation showed that FOXO3 was a direct downstream target gene of miR-130b-3p, and the database furthermore indicated binding sites at 3,838–3,862 between FOXO3 mRNA and miR-130b-3p (Figure 3A). A dual luciferase assay showed that treatment with miR-130b-3p mimic resulted in an obvious decline in luciferase activity in FOXO3 3′ untranslated region (3′ UTR)-wild-type (WT), but did not affect luciferase activity on the FOXO3 3′ UTR mutant type (MUT) sequence (Figure 3B). Next, H292 and H1299 cells were introduced with miR-130b-3p mimic and miR-130b-3p inhibitor. Through qRT-PCR and western blot analysis, we found that miR-130b-3p was appreciably upregulated (Figure 3C), but FOXO3 was remarkably downregulated (Figures 3D and 3E) in H292 and H1299 cells that were transfected with miR-130b-3p mimic; the opposite results were seen in H292 and H1299 cells transfected with miR-130b-3p inhibitor. Meanwhile, H292 and H1299 cells were co-cultured with PBS or EVs. Consistent with the above results, FOXO3 expression was found to be diminished, accompanied by an increase in miR-130b-3p expression in H292 and H1299 cells incubated with MSC-derived EVs compared to results in H292 and H1299 cells incubated with PBS (Figures 3F–3H). Furthermore, FOXO3 expression was analyzed in biopsy specimens from lung cancer patients, which revealed lower FOXO3 expression in lung cancer tissues than in adjacent normal tissue (Figure 3I). Pearson analysis documented an inverse correlation between FOXO3 and miR-130b-3p expression in lung cancer tissues (Figure 3J). In conclusion, FOXO3, which was downregulated in lung cancer tissues, was directly targeted by miR-130b-3p.
Figure 3.
FOXO3 Is Directly Targeted by miR-130b-3p in Lung Cancer Cells
(A) The specific binding sites of miR-130b-3p on FOXO3 predicted by online website. (B) The targeting relationship between miR-130b-3p and FOXO3 determined by dual luciferase assay. ∗p < 0.05 compared to mimic-NC treatment. (C) miR-130b-3p expression in H292 and H1299 cells transfected with miR-130b-3p mimic or inhibitor determined by qRT-PCR. (D) FOXO3 mRNA expression in H292 and H1299 cells transfected with miR-130b-3p mimic or inhibitor determined by qRT-PCR. (E) FOXO3 protein expression in H292 and H1299 cells transfected with miR-130b-3p mimic or inhibitor determined by western blot analysis. ∗p < 0.05 compared to H292 and H1299 cells transfected with mimic-NC. #p < 0.05 compared to H292 and H1299 cells transfected with inhibitor-NC. (F) miR-130b-3p expression in H292 and H1299 cells treated with PBS or MSC-EVs determined by qRT-PCR. (G) FOXO3 mRNA expression in H292 and H1299 cells treated with PBS or MSC-EVs determined by qRT-PCR. (H) FOXO3 protein expression in H292 and H1299 cells treated with PBS or MSC-EVs determined by western blot analysis. ∗p < 0.05 compared to cells treated with PBS. (I) FOXO3 mRNA expression in tumor and adjacent normal tissues (n = 50) determined by qRT-PCR. ∗p < 0.05 compared to adjacent normal tissues. (J) Correlation between miR-130b-3p and FOXO3 expression in lung cancer clinical samples determined by Pearson’s analysis (n = 50). The data were shown as means ± standard deviations. Cancer tissues were compared with adjacent normal tissues using the paired t test, while the other 2 groups were compared by the unpaired t test. Comparisons among multiple groups were analyzed by Tukey’s test-corrected one-way ANOVA. Pearson correlation analysis was used for the correlation between the observed indicators. The cell experiment was repeated 3 times.
miR-130b-3p in MSC-Derived EVs Inhibited FOXO3 to Promote Lung Cancer Proliferation, Invasion, and Migration but to Suppress Apoptosis
To confirm further whether EVs-miR-130b-3p promoted lung cancer progression by targeting FOXO3, we constructed FOXO3 overexpressing vector (OE-FOXO3) and first validated its overexpression efficiency. After H292 and H1299 cells were transfected with OE-FOXO3, FOXO3 expression was prominently enhanced (Figures 4A and 4B). Based on this result, H292 and H1299 cells were treated with EVs-miR-130b-3p mimic and/or OE-FOXO3. EVs-miR-130b-3p mimic treatment led to the upregulation of miR-130b-3p and the downregulation of FOXO3, whereas OE-FOXO3 treatment massively augmented FOXO3 expression in H292 and H1299 cells. However, in contrast to the treatment with EVs-miR-130b-3p mimic + OE-NC, FOXO3 expression was increased in H292 and H1299 cells treated with EVs-miR-130b-3p mimic + OE-FOXO3 (Figures 4C–4E). Through CCK-8 (Figure 4F) and colony formation (Figure 4G) assays, H292 and H1299 cell viability and colony formation were found to be elevated by treatment with the EVs-miR-130b-3p mimic, but diminished by OE-FOXO3; these effects were reversed by EVs-miR-130b-3p mimic + OE-FOXO3 treatment. In addition, the transwell assay illustrated that EVs-miR-130b-3p mimic treatment increased the migration and invasion of H292 and H1299 cells, but OE-FOXO3 treatment reduced these biological properties, which was negated by treatment with the EVs-miR-130b-3p mimic + OE-FOXO3 (Figures 4H and 4I). Consistent with these findings, H292 and H1299 cells treated with EVs-miR-130b-3p mimic had a decreased apoptotic rate, whereas the apoptosis rate was increased in FOXO3-overexpressed H292 and H1299 cells. Furthermore, reduced apoptotic rates caused by EVs-miR-130b-3p mimic treatment were also negated by FOXO3 overexpression (Figure 4J). Our data revealed the MSC-derived EVs containing miR-130b-3p overexpression promoted lung cancer progression by directly targeting FOXO3.
Figure 4.
miR-130b-3p Promotes Lung Cancer Cell Proliferation, Invasion, and Migration, but Decreases Apoptosis by Targeting FOXO3
(A) FOXO3 overexpression efficiency determined by qRT-qPCR after transfection. (B) FOXO3 overexpression efficiency determined by western blot after transfection. ∗p < 0.05 compared to H292 and H1299 cells treated with OE-NC. (C) H292 and H1299 cells were treated with EVs-mimic-NC + OE-NC, EVs-miR-130b-3p mimic + OE-NC, EVs-mimic NC + OE-FOXO3, or EVs-miR-130b-3p mimic + OE-FOXO3. miR-130b-3p expression in H292 and H1299 cells determined by qRT-PCR. (D) FOXO3 mRNA expression in H292 and H1299 cells determined by qRT-PCR. (E) FOXO3 protein expression in H292 and H1299 cells determined by western blot analysis. (F) H292 and H1299 cell viability determined by CCK-8 assay. (G) H292 and H1299 cell colony formation capacity determined by colony formation assay. (H) H292 and H1299 cell migration determined by transwell assay. (I) H292 and H1299 cell invasion determined by transwell assay. (J) H292 and H1299 cell apoptotic rate determined by flow cytometry. ∗p < 0.05 compared to H292 and H1299 cells treated with Evs-mimic-NC + OE-NC. #p < 0.05 compared to H292 and H1299 cells treated with Evs-miR-130b-3p mimic + OE-NC. The data were shown as means ± standard deviations. The 2 groups were compared by unpaired t test. Comparisons among multiple groups were analyzed by Tukey’s test-corrected one-way ANOVA. Variables were analyzed at different time points using Bonferroni-corrected repeated-measures ANOVA. The cell experiment was repeated 3 times.
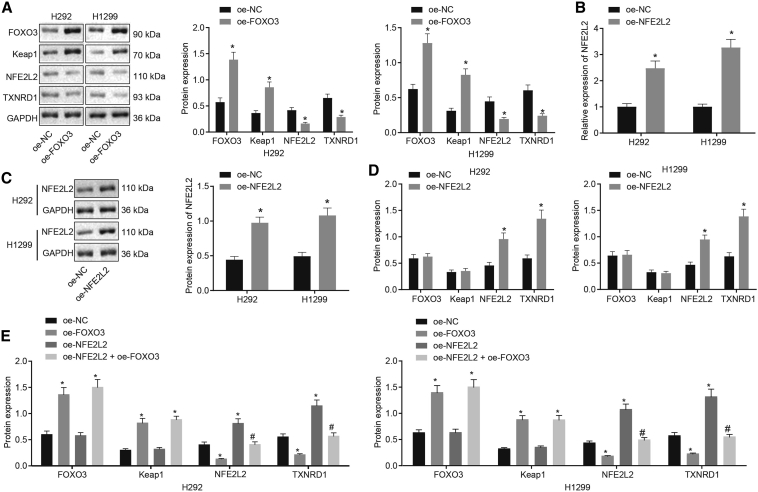
FOXO3 Inhibited TXNRD1 Expression by Reducing NFE2L2 Transcriptional Activity in Lung Cancer Cells
Previous research demonstrated that FOXO3 silencing downregulated Keap1 to activate the NFE2L2 pathway and that clinical cholangiocarcinoma biopsy specimens displayed FOXO3 and Keap1 downregulation and NFE2L2 hyperactivation.19 Meanwhile, activated NFE2L2 was reported to induce TXNRD1 expression.24 Another study revealed that TXNRD1 was overexpressed in lung cancer clinical samples.25 According to these findings, we assumed that FOXO3 may suppress TXNRD1 by orchestrating the Keap1-NFE2L2 pathway to inhibit lung cancer development. To test our hypothesis, FOXO3 was overexpressed in H292 and H1299 cells. Western blot analysis of these cells showed that ectopic expression of FOXO3 contributed to a reduction in NFE2L2 and TXNRD1 expression and the enhanced expression of FOXO3 and Keap1 in H292 and H1299 cells (Figure 5A). To confirm that TXNRD1 was downregulated by FOXO3 through NFE2L2, we constructed an NFE2L2 overexpressing vector (OE-NFE2L2) and introduced it into H292 and H1299 cells, which showed that the transfection of OE-NFE2L2 markedly elevated NFE2L2 and TXNRD1 expression without affecting FOXO3 and Keap1 expression (Figures 5B–5D). Based on this, we co-transfected H292 and H1299 cells with OE-FOXO3 and OE-NFE2L2. The results demonstrated that, compared with H292 and H1299 cells transfected with OE-FOXO3, NFE2L2 and TXNRD1 expression was increased in H292 and H1299 cells upon co-transfection with OE-FOXO3 and OE-NFE2L2 (Figure 5E). To sum up, FOXO3 suppressed TXNRD1 expression by downregulating NFE2L2 in lung cancer cells.
Figure 5.
FOXO3 Suppresses NFE2L2 and TXNRD1 Expression in Lung Cancer Cells
(A) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in H292 and H1299 cells after FOXO3 overexpression determined by western blot analysis. ∗p < 0.05 compared to H292 and H1299 cells treated with OE-NC. (B) NFE2L2 overexpression efficiency determined by qRT-PCR. (C) NFE2L2 overexpression efficiency determined by western blot analysis. (D) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in H292 and H1299 cells after NFE2L2 overexpression determined by western blot analysis. ∗p < 0.05 compared to H292 and H1299 cells treated with OE-NC. (E) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in H292 and H1299 cells after NFE2L2 and FOXO3 overexpression determined by western blot analysis. ∗p < 0.05 compared to H292 and H1299 cells treated with OE-NC. #p < 0.05 compared to H292 and H1299 cells treated with OE-FOXO3. The data were shown as means ± standard deviations. The 2 groups were compared by unpaired t test. Comparisons among multiple groups were analyzed by Tukey’s test-corrected one-way ANOVA. The cell experiment was repeated 3 times.
EVs-miR-130b-3p Activated the NFE2L2/TXNRD1 Pathway to Promote Lung Cancer Cell Proliferation, Invasion, and Migration but to Inhibit Apoptosis by Suppressing FOXO3
The above results demonstrated that EVs-miR-130b-3p downregulated FOXO3, while FOXO3 could suppress TXNRD1 expression by the Keap1-NFE2L2 pathway. Therefore, we supposed that EVs-miR-130b-3p facilitated TXNRD1 expression to promote lung cancer progression via the FOXO3-Keap1-NFE2L2 axis. We next proceeded to treat H292 and H1299 cells with EVs or PBS, which showed that EV treatment noticeably augmented miR-130b-3p, NFE2L2, and TXNRD1 expression and diminished FOXO3 and Keap1 expression in H292 and H1299 cells (Figures 6A and 6B). Meanwhile, EVs were extracted from MSCs transfected with miR-130b-3p mimic or mimic-NC and co-culture with H292 and H1299 cells. qRT-PCR and/or western blot analysis depicted that EVs-miR-130b-3p mimic greatly elevated miR-130b-3p, NFE2L2, and TXNRD1 expression but decreased FOXO3 and Keap1 expression (Figures 6C and 6D). Taken together, EVs-miR-130b-3p triggered an upregulation of TXNRD1 by suppressing FOXO3. To verify that EVs-miR-130b-3p promoted lung cancer progression through TXNRD1, 2 short hairpin RNAs (shRNAs) were constructed. qRT-PCR and western blot analysis were performed to evaluate the silencing efficiency of the shRNAs, which showed that shTXNRD1, especially shTXNRD1#1, discernibly reduced TXNRD1 expression in H292 and H1299 cells (Figures 6E and 6F). Therefore, we conducted the subsequent experiments with shTXNRD1#1. Moreover, western blot analysis manifested that shTXNRD1 treatment deceased TXNRD1 expression in H292 and H1299 cells, but did not affect FOXO3, NFE2L2, and Keap1 expression (Figure 6G). Subsequently, H292 and H1299 cells were treated with EVs-miR-130b-3p mimic and/or shTXNRD1. qRT-PCR and western blot analysis documented that miR-130b-3p, NFE2L2, and TXNRD1 were upregulated but FOXO3 and Keap1 were downregulated in H292 and H1299 cells via treatment with EVs-miR-130b-3p mimic in the presence of sh-NC. TXNRD1 expression in H292 and H1299 cells, however, was reduced by treatment with shTXNRD1 in the presence of EVs-mimic-NC. Furthermore, in comparison to H292 and H1299 cells treated with EVs-miR-130b-3p mimic + sh-NC, reduced TXNRD1 expression was observed in H292 and H1299 cells treated with EVs-miR-130b-3p mimic + shTXNRD1 (Figures 6H and 6I). In addition, EVs-miR-130b-3p mimic + sh-NC treatment substantially augmented the viability (Figure 6J), colony formation (Figure 6K), migration (Figure 6L), and invasion (Figure 6M) of H292 and H1299 cells but reduced their apoptosis (Figure 6N). Opposite results were seen following treatment with EVs-mimic NC + shTXNRD1. In the presence of EVs-miR-130b-3p mimic, cell viability (Figure 6J), colony formation (Figure 6K), migration (Figure 6L), and invasion (Figure 6M) also were diminished in H292 and H1299 cells, but cell apoptosis (Figure 6N) was accelerated by silencing TXNRD1. These results suggested that EVs-miR-130b-3p facilitated TXNRD1 expression to promote lung cancer progression by FOXO3/NFE2L2 axis.
Figure 6.
EVs-miR-130b-3p Upregulated NFE2L2 and TXNRD1 to Promote Lung Cancer Cell Proliferation, Invasion, and Migration, but to Inhibit Apoptosis by Suppressing FOXO3
(A) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in H292 and H1299 cells treated with EVs determined by western blot analysis. (B) miR-130b-3p expression in H292 and H1299 cells treated with EVs determined by qRT-PCR. ∗p < 0.05 compared to H292 and H1299 cells treated with PBS. (C) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in H292 and H1299 cells treated with EVs-miR-130b-3p mimic determined by western blot analysis. (D) miR-130b-3p expression in H292 and H1299 cells treated with EVs-miR-130b-3p mimic determined by qRT-PCR. ∗p < 0.05 compared to H292 and H1299 cells treated with EVs-mimic-NC. (E) TXNRD1 silencing efficiency determined by qRT-PCR. (F) TXNRD1 silencing efficiency determined by western blot analysis. (G) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in H292 and H1299 cells after TXNRD1 silencing determined by western blot analysis. ∗p < 0.05 compared to H292 and H1299 cells treated with short hairpin (sh)-NC. H292 and H1299 cells were treated with EVs-mimic-NC + sh-NC, EVs-miR-130b-3p mimic + sh-NC, EVs-mimic NC + shTXNRD1, or EVs-miR-130b-3p mimic + shTXNRD1. (H) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in H292 and H1299 cells determined by western blot analysis. (I) miR-130b-3p expression in H292 and H1299 cells determined by qRT-PCR. (J) H292 and H1299 cell viability determined by CCK-8 assay. (K) H292 and H1299 cell colony formation capacity determined by colony formation assay. (L) H292 and H1299 cell migration determined by transwell assay. (M) H292 and H1299 cell invasion determined by transwell assay. (N) H292 and H1299 cell apoptotic rate determined by flow cytometry, ∗p < 0.05 compared to H292 and H1299 cells treated with EVs-mimic-NC + sh-NC. #p < 0.05 compared to H292 and H1299 cells treated with EVs-miR-130b-3p mimic + sh-NC. The data were shown as means ± standard deviations. The 2 groups were compared by unpaired t test. Comparisons among multiple groups were analyzed by Tukey’s test-corrected one-way ANOVA. Variables were analyzed at different time points using Bonferroni-corrected repeated-measures ANOVA. The cell experiment was repeated 3 times.
EVs-miR-130b-3p Reduced FOXO3 Expression to Promote Lung Cancer Cell Tumorigenesis In Vivo
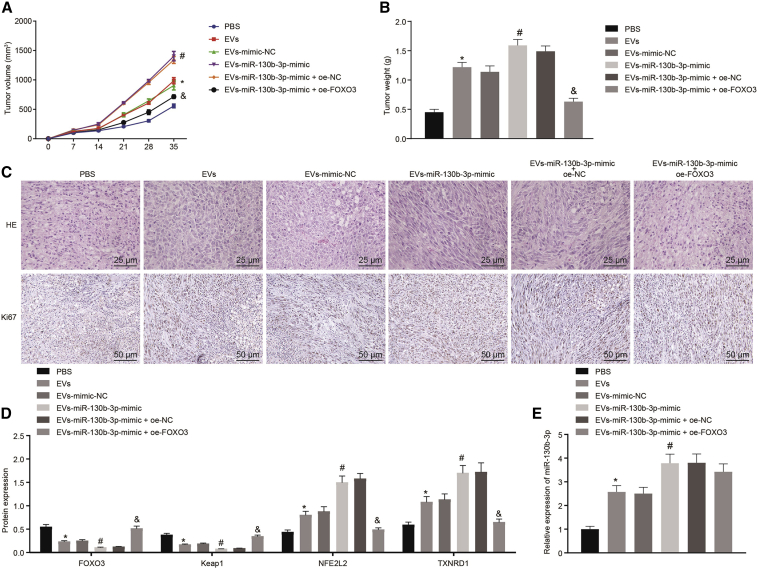
To investigate the role of EVs-miR-130b-3p in lung cancer progression in vivo, we established a xenograft tumor mouse model by subcutaneous injection of H1299 lung cancer cells pretreated with EVs and/or lentivirus encoding OE-FOXO3. Compared to PBS treatment, EV treatment, especially treatment with EVs-miR-130b-3p mimic, promoted H1299 cell growth in vivo. Notably, the effect of EVs-miR-130b-3p mimic on H1299 cell growth was annulled by overexpressing FOXO3 (Figure 7A). After cell injection for 35 days, mice were euthanized, and tumors were harvested for further analysis. Tumor weight was increased by EV treatment, especially by EVs-miR-130b-3p mimic treatment. Ectopic expression of FOXO3 abolished the effect of EVs-miR-130b-3p mimic on tumor weight (Figure 7B). Consistent with this, analysis of the xenograft tumor by hematoxylin and eosin (H&E) staining and immunohistochemistry showed that the number of Ki-67+ cells was significantly augmented after EV treatment, especially after EVs-miR-130b-3p mimic treatment, thus suggesting that cell proliferation was accelerated. an increase of Ki-67+ cells caused by EVs-miR-130b-3p mimic was neutralized by FOXO3 overexpression (Figure 7C). Furthermore, miR-130b-3p, FOXO3, Keap1, NFE2L2, and TXNRD1 expression was determined in tumor tissues, which revealed that EV treatment, especially EVs-miR-130b-3p mimic treatment, significantly reduced FOXO3 and Keap1 expression and enhanced miR-130b-3p, NFE2L2, and TXNRD1 expression. Meanwhile, the upregulation of FOXO3 gave rise to an elevation in FOXO3 and Keap1 expression but reduced NFE2L2 and TXNRD1 expression in tumor tissues in the presence of EVs-miR-130b-3p mimic (Figures 7D and 7E). EV treatment promoted lung cancer cell tumorigenesis in vivo and miR-130b-3p could enhance the effect of EVs on lung cancer tumorigenesis. Furthermore, our results suggested that EVs-miR-130b-3p promoted lung cancer progression in vivo by targeting FOXO3.
Figure 7.
EVs-miR-130b-3p Enhanced Lung Cancer Cell Tumorigenesis by Suppressing FOXO3 In Vivo
Nude mice were injected with H1299 cells pretreated with PBS, EVs, EVs-mimic-NC, EVs-miR-130b-3p mimic, EVs-miR-130b-3p mimic + OE-NC, or EVs-miR-130b-3p mimic + OE-FOXO3 for 12 h. (A) Tumor volume. (B) Tumor weight. (C) Tumor tissue morphology determined by H&E staining (400×) and Ki-67 expression determined by immunohistochemistry (200×). (D) FOXO3, Keap1, NFE2L2, and TXNRD1 expression in tumor tissues determined by western blot analysis. (E) miR-130b-3p expression determined by qRT-PCR. ∗p < 0.05 compared to mice injected with H1299 cell pretreated with PBS. #p < 0.05 compared to mice injected with H1299 cell pretreated with EVs-mimic-NC. &p < 0.05 compared to mice injected with H1299 cell pretreated with EVs-miR-130b-3p mimic + OE-NC. The data were shown as means ± standard deviations. Comparisons among multiple groups were analyzed by Tukey’s test-corrected one-way ANOVA. Variables were analyzed at different time points using Bonferroni-corrected repeated-measures ANOVA.
Discussion
MSCs were initially reported to be recruited to the site of tissue injury, where they could participate in injured tissue regeneration, which implied that MSCs had potential therapeutic applications,26,27 and MSCs are recruited to the cancer microenvironment.6 Although it has been demonstrated that MSC-EVs contributed to cancer progression,28 the molecular mechanism of this oncogenic action remained elusive. In our study, gain- and loss-of-function analyses showed that MSC-derived EVs-miR-130b-3p promoted lung cancer progression by activating the NFE2L2/TXNRD1 pathway through FOXO3 downregulation.
Our initial findings in this study make plain that miR-130b-3p was overexpressed in lung cancer tissues and cells and in MSC-derived EVs, and that MSC-EV treatment promoted lung cancer progression. Consistent with this result, data obtained by Mitra et al.29 showed a high expression of miR-130b-3p in lung cancer, which suggested miR-130b-3p to be an oncogenic miRNA. Moreover, consistent with our present results, a recent report demonstrated that human Wharton’s jelly MSCs (hWJMSCs) can increase the growth of the lung adenocarcinoma-derived cancer stem cell lines AC-229 and AC-223.30 More important, MSC-derived EVs could transfer an oncogenic miRNA, namely miR-410, to promote lung cancer progression.11 This observation partially supports our present results that MSC-derived EVs transferred miR-130b-3p into lung cancer cells to augment their proliferation, migration, and invasion while reducing cell apoptosis. Concurring with our new findings, Lv et al.31 revealed that miR-130b-3p upregulation promoted cell proliferation, invasion, and migration but diminished cell apoptosis in bladder cancer by targeting PTEN. Furthermore, another research group32 also uncovered that miR-130b-3p overexpression transferred by M2 macrophage-derived EVs accelerated the proliferation, migration, and invasion of gastric cancer cells.
In the subsequent experiment, the prediction that FOXO3 was a direct downstream target of miR-130b-3p was validated by dual luciferase assay. It is widely recognized that miRNAs assume a pivotal role in the posttranscriptional silencing of target genes.33 For instance, it was documented that miR-130b-3p was involved in tumor angiogenesis and progression of hepatocellular carcinoma by targeting HOXA5.34 Another study elaborated that miR-590-3p augmented ovarian cancer cell proliferation, invasion, and spheroid formation by targeting FOXO3.35 These findings indirectly support the targeting relationship between miR-130b-3p and FOXO3 that we see in lung cancer. Furthermore, through gain- and loss-of-function analyses, we demonstrated that MSC-derived miR-130b-3p promoted lung cancer cell proliferation, migration, and invasion and suppressed apoptosis by inhibiting FOXO3 expression. FOXO3 is a widely known tumor suppressive gene in various cancers.15 For example, FOXO3 has been reported to suppress breast cancer metastasisin vitro and in vivo by inhibiting miRNAs synthesis.36 Meanwhile, accumulating evidence has revealed that FOXO3 is involved in the regulation of lung cancer progression, such as proliferation, chemoresistance, migration, and invasion events.37, 38, 39
The final key finding in our study was that FOXO3 could also downregulate NFE2L2 by activating Keap1 in lung cancer cells to decrease TXNRD1 expression, thus inhibiting lung cancer progression. NFE2L2 is regarded as a critical transcriptional factor for anti-oxidation processes, which has been reported to be a therapy target and biomarker for diverse cancers.40 Previous data revealed NFE2L2 as a key regulator for lung cancer.41 Xu et al.42 reported that PAQR4 promoted chemoresistance in small cell lung cancer by preventing NFE2L2 from degradation by proteasomes. Zhou et al.43 demonstrated that NFE2L2 played a crucial role in lung cancer cell metastasis. Keap1, an E3 and tumor suppressor, has been reported to facilitate NFE2L2 degradation through ubiquitination.44 More important, recent research uncovered that FOXO3 could inhibit NFE2L2 transcriptional activity by promoting Keap1 transcription, by which means FOXO3 suppressed cancer cell chemoresistance.19 Intriguingly, another research study identified that TXNRD1 expression was induced by NFE2L2 upregulation and that FOXO3 disrupted the NFE2L2/TXNRD1 pathway to sensitize colorectal cancer cells to 5-fluorouracil.20 Building on this result, we found that MSC-derived EVs upregulated NFE2L2 and TXNRD1 by blocking the FOXO3-Keap1 axis via transferring miR-130b-3p to lung cancer cells.
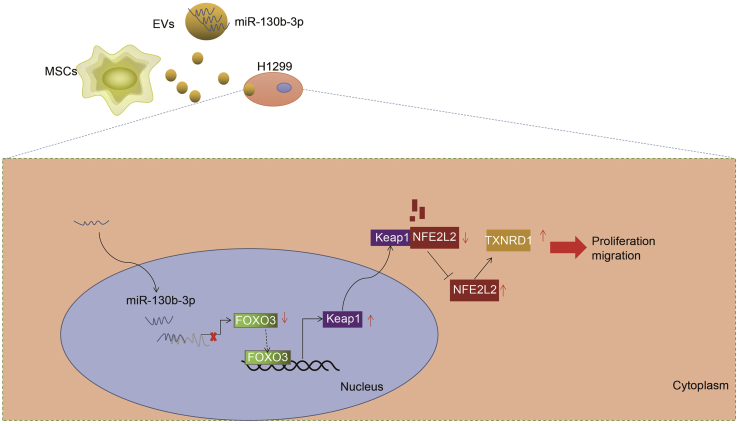
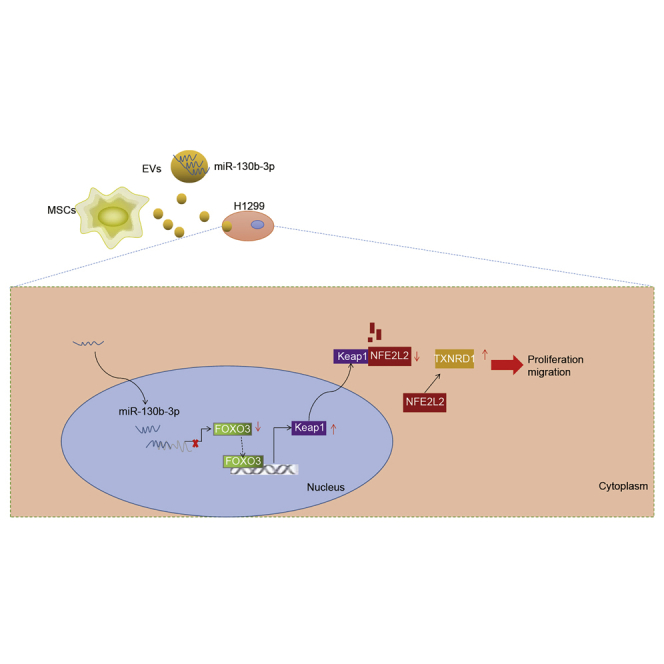
In summary, our study found that miR-130b-3p was enriched in MSC-derived EVs and could be transferred into lung cancer cells by MSC-derived EVs, by which means MSCs promoted lung cancer progression in vitro and in vivo. Notably, we found that miR-130b-3p overexpression in MSC-derived EVs directly targeted and suppressed FOXO3 to reduced Keap1 expression, thus upregulating NFE2L2 and its downstream target TXNRD1 to promote lung cancer progression (Figure 8). Our study revealed miR-130b-3p to be a potential therapy target and biomarker for lung cancer. Although we had demonstrated the molecular mechanism of MSC-derived EVs-mediated lung cancer progression, key questions remain unanswered about how the MSCs were recruited to accurate lung cancer. Further studies should focus on this topic, with the ultimate aim of establishing a new molecular therapy target for lung cancer.
Figure 8.
Molecular Mechanism Diagram
MSC-delivered EVs delivered miR-130b-3p into lung cancer cells to inhibit the expression of FOXO3. FOXO3 could promote the expression of Keap1. After inhibiting the expression of FOXO3, the inactivation of Keap1 resulted in the upregulation of NFE2L2, and promoted the expression of TXNRD1, thus promoting cell proliferation and migration but inhibiting cell apoptosis in lung cancer.
Materials and Methods
Ethical Approval
The experiments involving humans were approved by the Ethics Committee of Shenzhen Hospital, Southern Medical University, and Nanfang Hospital, Southern Medical University, and performed in strict accordance with the Declaration of Helsinki. All of the participants signed informed consent documentation before enrollment. The experimental procedures involving animals were ratified by the Animal Ethics Committee of Shenzhen Hospital, Southern Medical University, and performed in strict accordance with the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health. Extensive efforts were made to ensure the included animals experienced minimal discomfort.
Bioinformatics Analysis
Human lung cancer-related miRNA datasets (GEO: GSE63805 and GSE102286) and mRNA datasets (GEO: GSE101929 and GSE118370) were searched using the GEO database (https://www.ncbi.nlm.nih.gov/geo/). The differential analysis was conducted with the criteria of |logFC| >0.5 and p <0.5 (for GEO: GSE63805 and GSE102286) or |logFC| >0.5, p <0.01 (for GEO: GSE101929) and |logFC| >0.5, p <0.01 (for GEO: GSE118370) using the “limma” package in R language. miRNAs expressed in hMSC-derived EVs were acquired from the EVmiRNA database (http://bioinfo.life.hust.edu.cn/EVmiRNA). To determine the research objectives, we performed the Venn analysis on the significantly upregulated miRNAs screened from the lung cancer-related miRNA datasets and the miRNAs in the MSC-derived EVs.
Tumor Biopsy Specimens
Lung cancer and adjacent normal specimens were collected in Shenzhen Hospital, Southern Medical University, from 2017 to 2018 (n = 50). The collected tissues were confirmed by pathological diagnosis.
Isolation and identification of hUCMSCs
Fresh umbilical cords were obtained from Shenzhen Hospital, Southern Medical University, and Nanfang Hospital, Southern Medical University, and rapidly processed. The cords were rinsed twice with PBS supplemented with penicillin and streptomycin (GIBCO, Carlsbad, CA, USA), and the cord vessels were removed. The washed cords were subsequently cut into small fragments that were individually attached to the substrate of culture plates, followed by the addition of stem cell culture medium (Cyagen, Guangzhou, China) and incubation at 37°C with 5% CO2. The medium was replaced every 3 days after the initial culture, and well-developed colonies of fibroblast-like cells appeared ∼10 days later. The colonies were then trypsinized (GIBCO) and passaged using new plastic plates for further expansion. The following experiments were conducted on hUCMSCs at passages 3–7.
The osteogenic, adipogenic, and chondrogenic differentiations were identified by alizarin red staining, oil red O staining, and Alcian blue staining, respectively. To accomplish the differentiation, MSCs at passages 3–7 were cultured in OriCell hUCMSC osteogenic, adipogenic, or chondrogenic differentiation media (all from Cyagen).
After the 3rd passage, the hUCMSCs were trypsinized and washed twice with PBS. They were then stained with human antibodies to CD105, CD73, CD90, CD45, CD34, CD14, CD19, or HLA-DR. Identical concentrations of fluorescein isothiocyanate (FITC)- or phycoerythrine (PE)-conjugated mouse immunoglobulin G (IgG) isotype antibodies were used as NCs (all from BD Pharmingen, San Jose, CA, USA). The results were acquired on a FACSVerse instrument (BD Biosciences) using a FlowJo software (Tree Star, Ashland, OR, USA).
Isolation of EVs
MSCs were grown in 10-cm dishes with serum-free medium for 72 h. After the cells reached 90%–100% confluence, the medium was harvested and centrifuged at 3,000 × g and 4°C for 10 min to remove cells in the medium. Subsequently, the medium was centrifuged at 10,000 × g and 4°C for 30 min to remove large extracellular vesicles. Supernatant was obtained and filtered through 0.22-μm filters (Millipore, Burlington, MA, USA) to remove microvesicles and contaminating apoptotic bodies. The supernatant was then centrifuged at 100,000 × g and 4°C using Optima L-80XP Ultracentrifuge (Beckman Coulter, Brea, CA, USA) for 2 h. The supernatant was carefully removed, and the pellets were resuspended in 70 mL ice-cold PBS and then centrifuged at 100,000 × g and 4°C for 2 h. The PBS was carefully removed, and EVs were resuspended in 200 μL ice-cold PBS.
Identification of EVs
The dynamic light scattering (DLS) of the Nanosizer instrument (Malvern Instruments, Malvern, UK) was used to measure the size distribution of MSC-derived MSCs. EVs were diluted in 1 mL PBS and thoroughly mixed. Then, the diluted EVs were injected into the NanoSight NS300 instrument. Particles were automatically tracked, and their size calculated based on Brownian motion and the diffusion coefficient. EV morphology was observed using a Hitachi H-7650 TEM (Hitachi, Tokyo, Japan). A 10-μL volume EV pellet was placed on formvar carbon-coated 200-mesh copper electron microscopy grids, incubated for 5 min at room temperature, and then subjected to standard 1% uranyl acetate staining for 1 min at room temperature. The grid was washed 3 times with PBS and semi-dried at room temperature before observation under the TEM. The surface markers of EVs were determined by western blot analysis to identify the characteristics of EVs using primary rabbit antibodies (Abcam, Cambridge, UK) against CD63 (1:2,000, ab216130), CD9 (1:2,000, ab92726), CD81 (1:10,000, ab109201), and calnexin (1:100,000, ab92573).
Cell Culture
Human lung cancer cell lines (H292 and H1299, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China) were cultured with Dulbecco’s modified Eagle’s medium (DMEM; GIBCO) encompassing 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 μg/mL streptomycin (GIBCO). Cells were maintained at 37°C in a saturated humidity atmosphere containing 95% air and 5% CO2.
EV Uptake by Lung Cancer Cells
Purified EVs were labeled by PKH67 (Sigma, St. Louis, MO, USA). Lung cancer cells were planted into 8-well chamber slides (Thermo Scientific, Waltham, MA, USA) at 8,000 cells per well and then incubated with 5 μL PKH67 for 24 h to allow lung cancer cells to take up the EVs. After incubation, cells were rinsed by PBS twice and fixed by 4% paraformaldehyde for 15 min. The nucleus was stained by 4′,6-diamidino-2-phenylindole (DAPI, 0.5 mg/mL, Invitrogen, Carlsbad, California, USA). Photographs were taken using a Zeiss LSM 780 confocal microscope (Zeiss, Jena, Germany).
Cell Transfection
The MSCs H292 and H1299 at 60% confluence were transfected fusing Lipofectamine 2000 (Invitrogen), according to the manufacturer’s protocols. The synthetic miR-130b-3p mimic, miR-130b-3p inhibitor, mimic-NC, and inhibitor-NC were purchased from RiboBio (Guangzhou, China). FOXO3 and NFE2L2 overexpression plasmid and their control vector plasmid and shTXNRD1 and its control were purchased from Genecopoeia (Rockville, MD, USA).
MSCs or lung cancer cells were transfected with miR-130b-3p mimic, mimic-NC, miR-130b-3p inhibitor, or inhibitor-NC. In addition, lung cancer cells were co-cultured with PBS, EVs, EVs-inhibitor-NC, or EVs-miR-130b-3p inhibitor. Lung cancer cells were also co-cultured with EVs-mimic-NC + OE-NC, EVs-mimic-NC + OE-FOXO3, EVs-miR-130b-3p mimic + OE-NC, or EVs-miR-130b-3p mimic + OE-FOXO3. Meanwhile, lung cancer cells were transfected with OE-NC, OE-FOXO3, OE-NFE2L2, or OE-FOXO3 + OE-NFE2L2. Lung cancer cells were treated with EVs-mimic-NC + sh-NC, EVs-mimic-NC + shTXNRD1, EVs-miR-130b-3p mimic + sh-NC, EVs-miR-130b-3p mimic + shTXNRD1, PBS, EVs, EVs-mimic-NC, EVs-miR-130b-3p mimic, EVs-miR-130b-3p mimic + OE-NC, or EVs-miR-130b-3p mimic + OE-FOXO3.
CCK-8 Assay
Cell viability was measured using CCK-8 (Dojindo Laboratories). In brief, lung cancer cells were seeded in 96-well plates at the density of 5 × 103 cells per well. After cells were treated for 1, 2, 3, 4, and 5 days, 10 μL CCK-8 solution and 100 μL fresh medium were added into each well, followed by a 2-h incubation at 37°C. The optical density (OD) value was detected at 450 nm using a microplate reader (Bio-Rad 680, Bio-Rad, Hercules, CA, USA). The viability of the cells was expressed by subtracting the OD value of the blank well from the OD value of the experimental well.
Colony Formation Assay
For colony formation assay, transfected cells were plated into 6-well plates (1 × 103 cells per well) and maintained in DMEM containing 10% FBS at 37°C for 2 weeks. Cells were washed twice with PBS and fixed with 100% absolute methanol for 20 min. The colony cells were stained with 0.5% crystal violet for 10 min. The number of colonies with >50 cells was counted under a microscope (Leica, Wetzlar, Germany). Three recordings were performed for each sample.
Transwell Assay
Migration and invasion were determined by transwell assay. Cells (1.0 × 106 cells per milliliter) in serum-free medium were added to the top chamber (coated with Matrigel for invasion, but not coated with Matrigel for migration) of 24-well transwell plates (8 mm well size; Corning Star, Cambridge, MA, USA). Then, 600 μL complete medium with 10% FBS was supplemented into the bottom chamber. The assembled chamber was incubated at 37°C with 5% CO2 for 24 h, fixed with 10% formalin, and stained with crystal violet for visualization.
Flow Cytometry
Apoptotic rates were determined by the BD Pharmingen FITC Annexin V Apoptosis Detection Kit I (556547, BD Biosciences). The cells were cultured in the 6-well plates and transfected or treated with EVs when the confluence reached 60%–70%. After the cells were treated for the indicated times, the old medium was collected into a new centrifuge tube. The adherent cells were rinsed by PBS once, followed by trypsinization. Then, cells were suspended in the collected old medium, transferred into a centrifuge tube, and centrifuged at 1,000 × g for 5 min. After the supernatant was discarded, the cells were resuspended in PBS and counted. Samples of 50,000–100,000 resuspended cells were centrifuged at 200 × g for 5 min and resuspended in 195 μL annexin V-FITC binding buffer. Then, 5 μL annexin V-FITC was added into the cells and mixed well, followed by a 10-min incubation at room temperature (20°C–25°C) in the dark. Cells were subjected to a 5-min centrifugation at 200 × g, followed by the removal of supernatant. The cells were resuspended by 190 μL annexin V-FITC binding buffer, then supplemented with 10 μL propidium iodide (PI) in the dark, gently mixed, and stored in an ice bath in the dark. Cells were detected by a flow cytometer (BD Biosciences) with annexin V-FITC as GFP and PI as red fluorescence protein. Results were analyzed by ModFit 3.0 software (Verify Software House, Topsham, ME, USA).
Dual Luciferase Assay
Human FOXO3 3′ UTR sequence or the mutant sequence of FOXO3 3′ UTR containing the predicted binding sites of miR-130b-3p was inserted into the pGL3 promoter vector (Genscript, Nanjing, China). The 293T cells (American Type Culture Collection, Manassas, VA, USA) were seeded in 24-well plates (5 × 105 cells per well) on the day before transfection. Subsequently, the cells were co-transfected with luciferase reporter vectors (0.12 μg) and miR-130b-3p mimic or mimic-NC using Lipofectamine 3000 Reagent (Invitrogen). The luciferase activity was analyzed 48 h after transfection.
qRT-PCR
Total RNA was extracted by using TRIzol (Invitrogen). cDNA from 1 μg mRNA was generated using the Revert Aid first-strand cDNA synthesis kit (Fermentas, Life Sciences, Canada). Then, mRNA expression was assessed by qRT-PCR using SYBR Premix ExTaq II in an ABI PRISM 7900HT System (Takara Biotechnology, Tokyo, Japan) normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). For miRNA expression, miRNA in EVs was isolated using the SeraMir Evssome RNA Purification Kit (System Biosciences, Mountain View, CA, USA). cDNA for miRNA detection was generated using the TaqMan miRNA assay kit (Applied Biosystems, Foster City, CA, USA). qRT-PCR was conducted using FastStart Universal SYBR Green Master Mix (Roche, Indianapolis, IN, USA) with the miRNA-specific forward primer (Sangon Biotech, Shanghai, China) and universal reverse primer provided by the TaqMan miRNA assay kit. The miR-130b-3p level was normalized to Cel-miR39-3p small nuclear RNA (snRNA). The results were calculated by using the 2−ΔΔCT method. The primers are listed in Table 1.
Table 1.
Primer Sequences Used for qRT-PCR
| Genes | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| miR-130b-3p | GGCAGTGCAATGATGAAAGG | GTGCAGGGTCCGAGGT |
| Cel-miR-39-3p | UCACCGGGUGUAAAUCAGCUUG | AACGCTTCACGAATTTGCGT |
| FOXO3 | CGGACAAACGGCTCACTCT | GGACCCGCATGAATCGACTAT |
| NFE2L2 | TACTCCCAGGTTGCCCACA | CATCTACAAACGGGAATGTCTGC |
| TXNRD1 | GGCAACACAGCTCACAAGAA | CGCTGTTTTCACAGAGGTCA |
| GAPDH | GGAGCGAGATCCCTCCAAAAT | GGCTGTTGTCATACTTCTCATGG |
Western Blot Analysis
The protein was separated using freshly prepared sodium dodecyl sulfate-polyacrylamide gel electrophoresis and electrotransferred onto polyvinylidene fluoride membranes, which were blocked by Tris-buffered saline with 5% milk and 0.1% Tween-20 for 1 h. The membranes were probed overnight at 4°C with primary antibodies from abclonal (Woburn, MA, USA) against Keap1 (A1820, 1:2,000, rabbit), NFE2L2 (A0674, 1:2,000, rabbit), TXNRD1 (A2100, 1:2,000, rabbit), and GAPDH (AC033, 1:5,000, mouse), and primary antibodies from Cell Signaling Technologies (Beverly, MA, USA) against FOXO3 (#2497, 1:1,000, rabbit). Then, the membranes were re-probed with horseradish peroxidase-conjugated anti-rabbit IgG (AS014, 1:10,000, abclonal) or anti-mouse IgG (AS003, 1:10,000, abclonal) secondary antibody at 37°C for 1 h. The protein bands were visualized with enhanced chemiluminescence detection reagents (Thermo Fisher Scientific), and the images were captured under the ChemiDoc XRS Plus luminescent image analyzer (Bio-Rad). The gray value of the target protein bands was quantified using Image-Pro plus 6.0 software, with GAPDH used for normalization.
Xenograft Tumor Mouse Model
Four-week-old male nude mice (BALB/CJNJU-FOXN1NU/NJU) were obtained from SJA Laboratory Animal Company (Changsha, Hunan, China). H1299 cells were pretreated with EVs or lentivirus encoding OE-NC or OE-FOXO3 for 12 h and then suspended into concentrations of 1 × 107 cells per milliliter. The 0.2-mL suspended cells were slowly inoculated into the right dorsal subcutaneous tissue of each nude mouse with a 1-mL syringe. After injection, mice were allowed to feed ad libitum. After daily injection for 7 days, the injected site was observed. The bidimensional tumor measurement (the product of the longest diameter and its longest perpendicular diameter for each tumor) was recorded every 7 days using Vernier calipers. The long and short diameters of each tumor mass were recorded as “A” and “B,” respectively, and the tumor volume was calculated as V = AB2/2. Mice were euthanized after 5 weeks, and the tumors were dissected and weighed.
H&E Staining and Immunohistochemistry
The tumor tissues were fixed by formalin, embedded by paraffin, and then sliced. The sections were stained with an H&E-staining kit (Beyotime, Shanghai, China). For immunohistochemistry, Ki-67 (A2094, 1:100, rabbit, abclonal) expression in sections was determined by staining with the SABA-POD kit (Boster, Wuhan, China) and visualized by a diaminobenzidine kit (Boster). Samples were counterstained with hematoxylin, dehydrated, and sealed. The sections were observed using a Leica microscope and analyzed by an experienced pathologist. The tumor cells were counted in three randomly selected views.
Statistical Analysis
All of the data were processed using SPSS 22.0 statistical software (IBM, Armonk, NY, USA). The data were shown as means ± standard deviations from at least 3 independent experiments. Cancer tissues were compared with adjacent normal tissues using a paired t test, while the other 2 groups were compared by unpaired t test. Comparisons among multiple groups were analyzed by Tukey’s test-corrected one-way analysis of variance (ANOVA). Variables were analyzed at different time points using Bonferroni-corrected repeated-measures ANOVA. The Pearson correlation analysis was used for the correlation between the observed indicators. p <0.05 was considered a level of statistical significance.
Author Contributions
Q.G., C.Z. and J.T. wrote the paper; J.Y., Y.M., and J.Z. conducted and designed the experiments; T.S. and D.L. analyzed the data; Q.G., J.K., and Z.S. collected and provided the sample for this study; K.C. revised the figures and table; all of the authors have read and approved the final submitted manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We acknowledge and appreciate our colleagues for their valuable efforts and comments on this paper. This work was supported by Shenzhen Hospital of Southern Medical University, Research Promotion Funds for the Key Discipline Construction Program (No. ZDXKKYTS009).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.09.005.
Contributor Information
Kaican Cai, Email: doc_caikc@163.com.
Jianhua Zhang, Email: zhangjianhua_smu@yeah.net.
Supplemental Information
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer Statistics, 2017. CA Cancer J. Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Filaire E., Dupuis C., Galvaing G., Aubreton S., Laurent H., Richard R., Filaire M. Lung cancer: what are the links with oxidative stress, physical activity and nutrition. Lung Cancer. 2013;82:383–389. doi: 10.1016/j.lungcan.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Y., Li S., Li J., Wang D., Li Q. Effect of microRNA-135a on Cell Proliferation, Migration, Invasion, Apoptosis and Tumor Angiogenesis Through the IGF-1/PI3K/Akt Signaling Pathway in Non-Small Cell Lung Cancer. Cell. Physiol. Biochem. 2017;42:1431–1446. doi: 10.1159/000479207. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L.N., Kong C.F., Zhao D., Cong X.L., Wang S.S., Ma L., Huang Y.H. Fusion with mesenchymal stem cells differentially affects tumorigenic and metastatic abilities of lung cancer cells. J. Cell. Physiol. 2019;234:3570–3582. doi: 10.1002/jcp.27011. [DOI] [PubMed] [Google Scholar]
- 5.Uccelli A., Moretta L., Pistoia V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 6.Loebinger M.R., Eddaoudi A., Davies D., Janes S.M. Mesenchymal stem cell delivery of TRAIL can eliminate metastatic cancer. Cancer Res. 2009;69:4134–4142. doi: 10.1158/0008-5472.CAN-08-4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kidd S., Spaeth E., Dembinski J.L., Dietrich M., Watson K., Klopp A., Battula V.L., Weil M., Andreeff M., Marini F.C. Direct evidence of mesenchymal stem cell tropism for tumor and wounding microenvironments using in vivo bioluminescent imaging. Stem Cells. 2009;27:2614–2623. doi: 10.1002/stem.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casado S., Lobo M.D.V.T., Paíno C.L. Dynamics of plasma membrane surface related to the release of extracellular vesicles by mesenchymal stem cells in culture. Sci. Rep. 2017;7:6767. doi: 10.1038/s41598-017-07265-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalimuthu S., Gangadaran P., Li X.J., Oh J.M., Lee H.W., Jeong S.Y., Lee S.W., Lee J., Ahn B.C. In vivo therapeutic potential of mesenchymal stem cell-derived extracellular vesicles with optical imaging reporter in tumor mice model. Sci. Rep. 2016;6:30418. doi: 10.1038/srep30418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohno S., Drummen G.P., Kuroda M. Focus on Extracellular Vesicles: Development of Extracellular Vesicle-Based Therapeutic Systems. Int. J. Mol. Sci. 2016;17:172. doi: 10.3390/ijms17020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong L., Pu Y., Zhang L., Qi Q., Xu L., Li W., Wei C., Wang X., Zhou S., Zhu J. Human umbilical cord mesenchymal stem cell-derived extracellular vesicles promote lung adenocarcinoma growth by transferring miR-410. Cell Death Dis. 2018;9:218. doi: 10.1038/s41419-018-0323-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rubens D., Sterns R.H., Segal A.J. Postpartum renal vein thrombosis. Urol. Radiol. 1985;7:80–84. doi: 10.1007/BF02926860. [DOI] [PubMed] [Google Scholar]
- 13.Tian J., Hu L., Li X., Geng J., Dai M., Bai X. MicroRNA-130b promotes lung cancer progression via PPARγ/VEGF-A/BCL-2-mediated suppression of apoptosis. J. Exp. Clin. Cancer Res. 2016;35:105. doi: 10.1186/s13046-016-0382-3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Yan R., Jiang Y., Lai B., Lin Y., Wen J. The positive feedback loop FOXO3/CASC11/miR-498 promotes the tumorigenesis of non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2019;519:518–524. doi: 10.1016/j.bbrc.2019.08.136. [DOI] [PubMed] [Google Scholar]
- 15.He X., Zou K. MiRNA-96-5p contributed to the proliferation of gastric cancer cells by targeting FOXO3. J. Biochem. 2020;167:101–108. doi: 10.1093/jb/mvz080. [DOI] [PubMed] [Google Scholar]
- 16.Lyu X., Zeng L., Zhang H., Ke Y., Liu X., Zhao N., Yuan J., Chen G., Yang S. Hydroxychloroquine suppresses lung tumorigenesis via inducing FoxO3a nuclear translocation through STAT3 inactivation. Life Sci. 2020;246:117366. doi: 10.1016/j.lfs.2020.117366. [DOI] [PubMed] [Google Scholar]
- 17.Smolková K., Mikó E., Kovács T., Leguina-Ruzzi A., Sipos A., Bai P. Nuclear Factor Erythroid 2-Related Factor 2 in Regulating Cancer Metabolism. Antioxid. Redox Signal. 2020 doi: 10.1089/ars.2020.8024. Published online March 18, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji L., Zhang R., Chen J., Xue Q., Moghal N., Tsao M.S. PIDD interaction with KEAP1 as a new mutation-independent mechanism to promote NRF2 stabilization and chemoresistance in NSCLC. Sci. Rep. 2019;9:12437. doi: 10.1038/s41598-019-48763-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guan L., Zhang L., Gong Z., Hou X., Xu Y., Feng X., Wang H., You H. FoxO3 inactivation promotes human cholangiocarcinoma tumorigenesis and chemoresistance through Keap1-Nrf2 signaling. Hepatology. 2016;63:1914–1927. doi: 10.1002/hep.28496. [DOI] [PubMed] [Google Scholar]
- 20.Liu C., Zhao Y., Wang J., Yang Y., Zhang Y., Qu X., Peng S., Yao Z., Zhao S., He B. FoxO3 reverses 5-fluorouracil resistance in human colorectal cancer cells by inhibiting the Nrf2/TR1 signaling pathway. Cancer Lett. 2020;470:29–42. doi: 10.1016/j.canlet.2019.11.042. [DOI] [PubMed] [Google Scholar]
- 21.Dai B., Yoo S.Y., Bartholomeusz G., Graham R.A., Majidi M., Yan S., Meng J., Ji L., Coombes K., Minna J.D. KEAP1-dependent synthetic lethality induced by AKT and TXNRD1 inhibitors in lung cancer. Cancer Res. 2013;73:5532–5543. doi: 10.1158/0008-5472.CAN-13-0712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu Y.L., Hung J.Y., Lee Y.L., Chen F.W., Chang K.F., Chang W.A., Tsai Y.M., Chong I.W., Kuo P.L. Identification of novel gene expression signature in lung adenocarcinoma by using next-generation sequencing data and bioinformatics analysis. Oncotarget. 2017;8:104831–104854. doi: 10.18632/oncotarget.21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirono T., Jingushi K., Nagata T., Sato M., Minami K., Aoki M., Takeda A.H., Umehara T., Egawa H., Nakatsuji Y. MicroRNA-130b functions as an oncomiRNA in non-small cell lung cancer by targeting tissue inhibitor of metalloproteinase-2. Sci. Rep. 2019;9:6956. doi: 10.1038/s41598-019-43355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen Z.H., Saito Y., Yoshida Y., Sekine A., Noguchi N., Niki E. 4-Hydroxynonenal induces adaptive response and enhances PC12 cell tolerance primarily through induction of thioredoxin reductase 1 via activation of Nrf2. J. Biol. Chem. 2005;280:41921–41927. doi: 10.1074/jbc.M508556200. [DOI] [PubMed] [Google Scholar]
- 25.Poerschke R.L., Moos P.J. Thioredoxin reductase 1 knockdown enhances selenazolidine cytotoxicity in human lung cancer cells via mitochondrial dysfunction. Biochem. Pharmacol. 2011;81:211–221. doi: 10.1016/j.bcp.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karp J.M., Leng Teo G.S. Mesenchymal stem cell homing: the devil is in the details. Cell Stem Cell. 2009;4:206–216. doi: 10.1016/j.stem.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 27.Ortiz L.A., Gambelli F., McBride C., Gaupp D., Baddoo M., Kaminski N., Phinney D.G. Mesenchymal stem cell engraftment in lung is enhanced in response to bleomycin exposure and ameliorates its fibrotic effects. Proc. Natl. Acad. Sci. USA. 2003;100:8407–8411. doi: 10.1073/pnas.1432929100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shojaei S., Hashemi S.M., Ghanbarian H., Salehi M., Mohammadi-Yeganeh S. Effect of mesenchymal stem cells-derived exosomes on tumor microenvironment: tumor progression versus tumor suppression. J. Cell. Physiol. 2019;234:3394–3409. doi: 10.1002/jcp.27326. [DOI] [PubMed] [Google Scholar]
- 29.Mitra R., Edmonds M.D., Sun J., Zhao M., Yu H., Eischen C.M., Zhao Z. Reproducible combinatorial regulatory networks elucidate novel oncogenic microRNAs in non-small cell lung cancer. RNA. 2014;20:1356–1368. doi: 10.1261/rna.042754.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vulcano F., Milazzo L., Ciccarelli C., Eramo A., Sette G., Mauro A., Macioce G., Martinelli A., La Torre R., Casalbore P. Wharton’s jelly mesenchymal stromal cells have contrasting effects on proliferation and phenotype of cancer stem cells from different subtypes of lung cancer. Exp. Cell Res. 2016;345:190–198. doi: 10.1016/j.yexcr.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 31.Lv M., Zhong Z., Chi H., Huang M., Jiang R., Chen J. Genome-Wide Screen of miRNAs and Targeting mRNAs Reveals the Negatively Regulatory Effect of miR-130b-3p on PTEN by PI3K and Integrin β1 Signaling Pathways in Bladder Carcinoma. Int. J. Mol. Sci. 2016;18:78. doi: 10.3390/ijms18010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y., Meng W., Yue P., Li X. M2 macrophage-derived extracellular vesicles promote gastric cancer progression via a microRNA-130b-3p/MLL3/GRHL2 signaling cascade. J. Exp. Clin. Cancer Res. 2020;39:134. doi: 10.1186/s13046-020-01626-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Lu T.X., Rothenberg M.E. MicroRNA. J. Allergy Clin. Immunol. 2018;141:1202–1207. doi: 10.1016/j.jaci.2017.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liao Y., Wang C., Yang Z., Liu W., Yuan Y., Li K., Zhang Y., Wang Y., Shi Y., Qiu Y. Dysregulated Sp1/miR-130b-3p/HOXA5 axis contributes to tumor angiogenesis and progression of hepatocellular carcinoma. Theranostics. 2020;10:5209–5224. doi: 10.7150/thno.43640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salem M., Shan Y., Bernaudo S., Peng C. miR-590-3p Targets Cyclin G2 and FOXO3 to Promote Ovarian Cancer Cell Proliferation, Invasion, and Spheroid Formation. Int. J. Mol. Sci. 2019;20:1810. doi: 10.3390/ijms20081810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L., Cai M., Gong Z., Zhang B., Li Y., Guan L., Hou X., Li Q., Liu G., Xue Z. Geminin facilitates FoxO3 deacetylation to promote breast cancer cell metastasis. J. Clin. Invest. 2017;127:2159–2175. doi: 10.1172/JCI90077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen G., Yu L., Dong H., Liu Z., Sun Y. MiR-182 enhances radioresistance in non-small cell lung cancer cells by regulating FOXO3. Clin. Exp. Pharmacol. Physiol. 2019;46:137–143. doi: 10.1111/1440-1681.13041. [DOI] [PubMed] [Google Scholar]
- 38.Li X., Yang B., Ren H., Xiao T., Zhang L., Li L., Li M., Wang X., Zhou H., Zhang W. Hsa_circ_0002483 inhibited the progression and enhanced the Taxol sensitivity of non-small cell lung cancer by targeting miR-182-5p. Cell Death Dis. 2019;10:953. doi: 10.1038/s41419-019-2180-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X., Zeng Q., Li Z., Yang X., Xia W., Chen Z. Adjudin synergizes with paclitaxel and inhibits cell growth and metastasis by regulating the sirtuin 3-Forkhead box O3a axis in human small-cell lung cancer. Thorac. Cancer. 2019;10:642–658. doi: 10.1111/1759-7714.12976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bi Z., Zhang Q., Fu Y., Wadgaonkar P., Zhang W., Almutairy B., Xu L., Rice M., Qiu Y., Thakur C., Chen F. Nrf2 and HIF1α converge to arsenic-induced metabolic reprogramming and the formation of the cancer stem-like cells. Theranostics. 2020;10:4134–4149. doi: 10.7150/thno.42903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Backman H., Lindberg A., Hedman L., Stridsman C., Jansson S.A., Sandström T., Lundbäck B., Rönmark E. FEV1 decline in relation to blood eosinophils and neutrophils in a population-based asthma cohort. World Allergy Organ. J. 2020;13:100110. doi: 10.1016/j.waojou.2020.100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu P., Jiang L., Yang Y., Wu M., Liu B., Shi Y. PAQR4 promotes chemoresistance in non-small cell lung cancer through inhibiting Nrf2 protein degradation. Theranostics. 2020;10:3737–3778. doi: 10.7150/thno.43142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou J., Zhang S., Xu Y., Ye W., Li Z., Chen Z., He Z. Cullin 3 overexpression inhibits lung cancer metastasis and is associated with survival of lung adenocarcinoma. Clin. Exp. Metastasis. 2020;37:115–124. doi: 10.1007/s10585-019-09988-9. [DOI] [PubMed] [Google Scholar]
- 44.He X., Ma Q. NRF2 cysteine residues are critical for oxidant/electrophile-sensing, Kelch-like ECH-associated protein-1-dependent ubiquitination-proteasomal degradation, and transcription activation. Mol. Pharmacol. 2009;76:1265–1278. doi: 10.1124/mol.109.058453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.