Abstract
CD8+ T cells are crucial to establish antitumor immunity, and their high infiltration associates with favorable prognoses. However, several CD8+ T cell subpopulations in the tumor microenvironment may play different roles in prognosis, progression, and immunotherapy. Here, we analyzed prior published single-cell RNA-sequencing (scRNA-seq) melanoma data to explore the heterogeneity of CD8+ T cell subpopulations and identified 7 major subpopulations. We found that high infiltration of exhausted CD8+ T cell subpopulation 2 would contribute to unfavorable prognoses. In contrast, a large proportion of naive/memory cells and cytotoxic CD8+ T cell subpopulation 3 would lead to favorable prognoses. Notably, the proportion of the cytotoxic CD8+ T cell subpopulation 3 would decrease in later-stage melanoma samples, while that of the exhausted CD8+ T cell subpopulation 2 would increase. We also found that high abnormal activities of metabolic pathways existed in exhausted CD8+ T cell subpopulation 1. Significantly, immunosuppressive checkpoints PD-1 and CTLA-4 signaling pathways were upregulated in exhausted CD8+ T cell subpopulations. In addition, a dynamic transcript landscape of immune checkpoints among different subpopulations was also depicted in this study. Moreover, we identified three overexpressed genes (PMEL, TYRP1, and EDNRB) that were significantly correlated to poor prognoses and only expressed in exhausted CD8+ T cell subpopulation 2. Importantly, they showed the highest expression in melanoma samples compared to other tumors. In general, we characterized the CD8+ T cell subpopulations in melanoma and identified that not only genes of immunosuppressive checkpoints but also PMEL, TYRP1, and EDNRB could serve as potential targets for melanoma therapy.
Keywords: single-cell RNA-sequencing, melanoma, CD8+ T cell subpopulations, immune checkpoints, immunotherapy, bioinformatics
Graphical Abstract
CD8+ T cells are crucial for antitumor immunity, yet different cell subpopulations have heterogeneous functions. Here, Deng et al. characterized the CD8+ T cell subpopulations in melanoma and identified that not only genes of immunosuppressive checkpoints but also PMEL, TYRP1, and EDNRB could serve as potential targets for melanoma therapy.
Introduction
Within the complex and heterogeneous tumor microenvironment (TME), CD8+ T cells play an essential role in eliminating tumors.1,2 The kill-tumor functions of CD8+ T cells are based on cell differentiation and infiltration.3 Following the infiltration, naive CD8+ T cells will initiate the differentiation program into effector, cytotoxic, and memory CD8+ T cells to achieve anti-tumor function.4,5 Several research studies revealed that cytotoxic CD8+ T cells in the TME could enhance anti-tumor outcomes in cervical cancer, breast cancer, and glioblastoma.6, 7, 8 However, a state of CD8+ T cell dysfunction or exhaustion is frequently observed in tumors.9,10 Chronic antigens will trigger the exhaustion program, and CD8+ T cells will reduce the functional effects during the program, which are likely involved in preventing excessive immune response.11 Therefore, to elucidate the differentiation program of CD8+ T cells in the TME can help offer a bird’s-eye view about the dynamic anti-tumor immune response of CD8+ T cells.
Checkpoint immunotherapy in tumors was established to overcome T cell exhaustion due to chronic antigen stimulation and immunosuppression in the TME.12 It is well known that PD-1, CTLA-4, LAG-3, and TIM-3/HAVCR2, co-inhibitory or checkpoint molecules, are expressed in tumor-infiltrating lymphocytes in the TME.13, 14, 15 To be more specific, PD-L1 and PD-L2 are the ligands of PD-1, and the interactions between them will promote tumor progression and dampen anti-tumor activities of T cells in the periphery.16,17 Similarly, ligand interactions with CTLA-4 are also involved in the inhibition of T cell activities, despite ambiguity about the exact mechanisms.18 LAG-3, the third clinical inhibitory receptor pathway, has the capabilities to dampen the excessive autoimmunology responses; unfortunately, it can also lead to a condition of T cell exhaustion or dysfunction.19 It is also recognized that Tim-3/HAVCR2, an essential immune checkpoint protein, is expressed on T cells and serves as an inhibitory receptor.20 In addition, TIGIT will also contribute to inhibitory activities in anti-tumor responses.21 Hence, understanding inhibitory checkpoint regulation during the differentiation program of CD8+ T cells may provide optimal treatment windows for immunotherapies and identify novel molecules that could serve as targets to overcome CD8+ T cell exhaustion or dysfunction.
Previous studies widely employed population-level mRNA profiling to reveal the transcriptome of tissues. However, conventional bulk RNA sequencing (RNA-seq) only can represent the average expression levels of all cells. It is not feasible to elucidate the heterogeneities among different subpopulations based on the transcriptome, let alone explore the subpopulations’ differentiation programs at a single-cell level. Nevertheless, as a technical development, single-cell RNA-sequencing (scRNA-seq) could provide an insight into the heterogeneities of different subpopulations and reveal the differentiation trajectory using algorithms in various tissues. Here, we unveiled the heterogeneities of different CD8+ T cell subpopulations using the sufficient high-quality scRNA-seq data of melanoma obtained from two previous studies.22,23 Our results revealed relatively large heterogeneities in CD8+ T cell subpopulations. We mainly found that different CD8+ T cell subsets would contribute to heterogeneous prognoses and pathway activities. We also identified the altered gene expression of cytotoxicity and immune checkpoints during CD8+ T cell differentiation. This study also found three novel potential therapeutic targets for melanoma (PMEL, TYRP1, and EDNRB).
Results
Identify CD8+ T cell subpopulations in melanoma
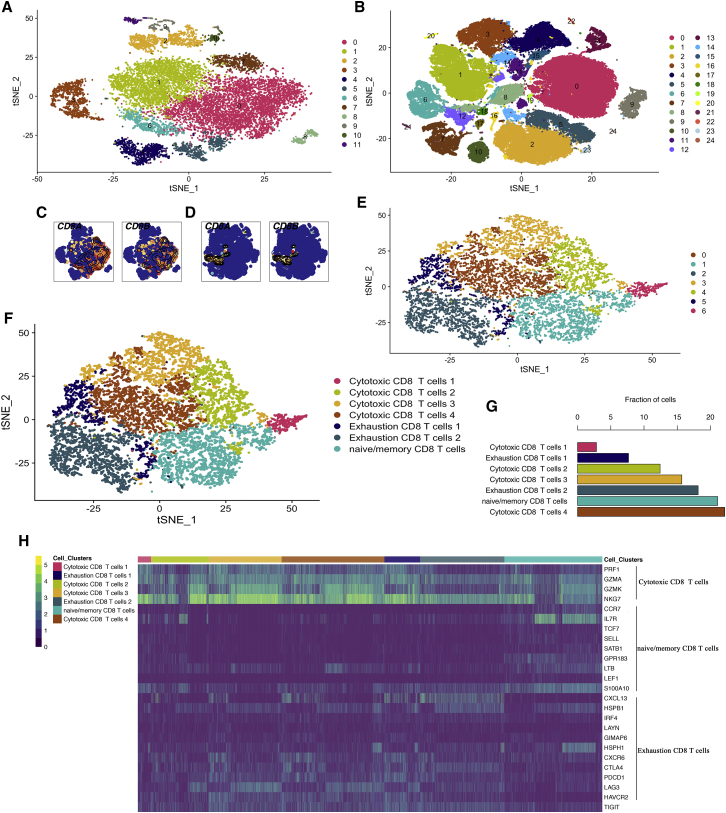
59 melanoma samples from smart-seq and 10× Genomics platforms were included to isolate CD8+ T cells in our research. We first separately found clusters for smart-seq and 10× Genomics data using t-distributed stochastic neighbor embedding (t-SNE) based on all gene expression levels (Figures 1A and 1B). The results of t-SNE showed that 12 clusters were identified in smart-seq data (Figure 1A), while 10× Genomics data included 25 clusters (Figure 1B) in melanoma tissues. Then, CD8+ T cell signatures were visualized to identify the varying degree of expression of specific cell clusters (Figures 1C and 1D). For smart-seq data, CD8A and CD8B were mainly highly expressed in cluster 0 (Figure 1C). However, 10× Genomics data had a specificity to several cell clusters (cluster 6, cluster 8, cluster 12, cluster 16, and cluster 17) (Figure 1D). Subsequently, these CD8+ T cell clusters were isolated from the two platforms by Seurat’s built-in algorithm and were integrated using the Harmony algorithm. Finally, 10,861 CD8+ T cells were isolated to perform subsequent steps of analyses. We observed that these CD8+ T cells highly expressed both CD8+ T cell markers and T cell markers (Figure S1A), and they had almost no expressions of the CD4+ T cell markers (Figure S1A) and melanocyte markers (Figure S1B). Thus, we identified that CD4+ T cells and melanocytes were excluded in our analyses (Figure S1).
Figure 1.
CD8+ T cell subpopulations in melanoma TME
(A and B) The t-SNE projection of all cells from smart-seq (A) and 10× Genomics platforms (B). (C and D) Single-cell transcript levels of CD8A and CD8B: left for smart-seq (C), right for 10× Genomics (D). (E and F) The t-SNE plots for CD8+ T cells; a total of 7 subpopulations were identified. (G) The bar graph showed the fraction of CD8+ T cell subpopulations. (H) Single-cell transcript levels of signature genes were presented by heatmap. Columns represented cells; rows represented genes. CD8+ T cell subpopulations were labeled by different colors.
These CD8+ T cells were then clustered into 7 subpopulations (Figure 1E). In order to annotate CD8+ T cells, we employed previously reported cell markers to distinguish them. The expression levels of signatures to annotate these subpopulations are shown in Figures 1H, S2, and S3. Ultimately, four cytotoxic subpopulations, two exhausted subpopulations, and one naive/memory subpopulation were identified (Figure 1F). The fraction of cells in each subpopulation revealed that cytotoxic subpopulation 4, naive/memory subpopulation, exhausted subpopulation 2, and cytotoxic subpopulation 3 were the most frequent cell subpopulations in melanoma TME (Figure 1G). We found that naive/memory signatures, including CCR7, IL7R, TCF7, and LEF1, were only expressed in naive/memory subpopulations (Figures S2 and S3). However, cytotoxic signatures, including PRF1, GZMA, GZMK, and NKG7, were widely expressed in all CD8+ T cell subpopulations (Figures S2 and S3). Among them, PRF1, GZMK, and NKG7 showed the highest specificity to cytotoxic CD8+ T cell subpopulations 1 and 3, while PRF1, GZMA, and GZMK had the lowest expression in exhausted subpopulation 2 (Figures S2 and S3). The results indicated that heterogeneous cytotoxicity of CD8+ T cells existed in these subpopulations. Notably, we also found that immune checkpoints or exhausted markers (CTLA4, PDCD1, LAG3, HAVCR2, and TIGIT) were widely expressed in nearly every subpopulation, while only in naive/memory and cytotoxic subpopulation 1 did these markers have relatively lower expression (Figures S2 and S3). CXCL13, a chemokine, was highly expressed in exhausted subpopulation 1 and exhausted subpopulation 2 (Figures S2 and S3), which combine with its receptor CXCR5 and can promote cancer cell proliferation, migration, and invasion.24 Similarly, compared with other CD8+ T cell subpopulations, CTLA4 had relatively higher expression in exhausted subpopulation 1 and exhausted subpopulation 2 (Figures S2 and S3). In addition, cytotoxic subpopulation 3 also showed varying degrees of high expression for inhibitory checkpoints (CTLA4, PDCD1, LAG3, HAVCR2, and TIGIT) (Figures S2 and S3). This may reflect that high cytotoxicity needed to be regulated by inhibitory checkpoints to prevent excessive immune responses. These results revealed that different expression levels of cytotoxic and immune checkpoint molecules in CD8+ T cell subpopulations might contribute to heterogeneous anti-tumor immune response in melanoma TME.
CD8+ T cell subpopulations were correlated with prognoses
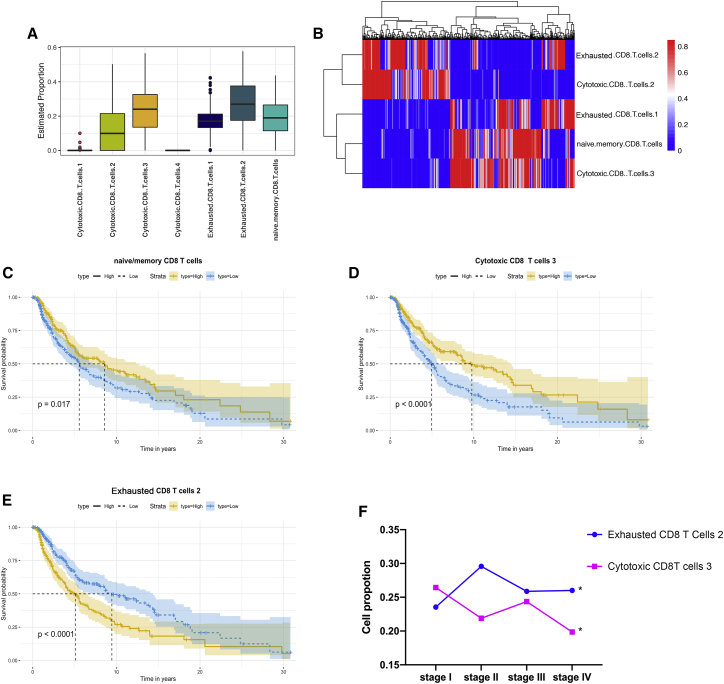
Given the heterogeneous cytotoxicity and checkpoints among different cell subpopulations, we hypothesized that these subpopulations would influence clinical outcomes. We derived signature genes from scRNA-seq data and utilized these matrixes to deconvolute 471 skin cutaneous melanoma (SKCM) and 1,809 normal bulk RNA-seq samples. In general, SKCM and normal samples contained naive/memory, cytotoxic subpopulation 2, cytotoxic subpopulation 3, exhausted subpopulation 1, and exhausted subpopulation 2 (Figures 2A and 2B; Figure S4B). However, cytotoxic subpopulations 1 and 4 were not found in SKCM and normal samples. Among the detected subpopulations, exhausted subpopulation 2 accounted for the largest proportions of CD8+ T cells (Figure 2A), while for normal samples, cytotoxic subpopulation 2 had the largest proportions followed by exhausted subpopulation 2 (Figure S4B).
Figure 2.
Cell composition of CD8+ T cell subpopulations in SKCM RNA-seq samples
(A and B) Boxplot (A) and heatmap (B) showed the proportions of CD8+ T cell subpopulations in SKCM. (C–E) Kruskal-Wallis curves for naive/memory CD8+ T cells (C), cytotoxic CD8+ T cell (D), and exhausted CD8+ T cell subpopulation 2 (E). (F) The average values of cell proportions in four pathology stages (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Next, we conducted the Kaplan-Meier survival analysis to characterize the prognostic roles of these subpopulations. As shown in Figures 2C and 2D, high proportions of naive/memory and cytotoxic subpopulation 3 were associated with favorable clinical outcomes. However, exhausted subpopulation 2 could contribute to an unfavorable prognosis (Figure 2E). In addition, cytotoxic subpopulation 2 and exhausted subpopulation 1 did not influence SKCM prognoses (Figure S4A). Importantly, there was more cytotoxic subpopulation 3 in SKCM than in normal samples (Figure S4C). Combined with the survival analysis results, we could have insight into the specific anti-melanoma functions of cytotoxic subpopulation 3. Higher proportions of exhausted subpopulation 1 were observed in SKCM, while cytotoxic subpopulation 2 had higher proportions in normal samples (Figure S4C). There was no difference in the proportions of naive/memory or exhausted subpopulation 2 between SKCM and normal samples (Figure S4C). Then, we explored dynamic changes of cytotoxic subpopulation 3 and exhausted subpopulation 2 in different pathology stages. Significantly, the results unveiled that the proportions of cytotoxic subpopulation 3 would decrease with tumor progression, although they increased in stage III (Figure 2F). For comparison, exhausted subpopulation 2 would elevate cell proportions in later-stage compared with stage I disease, especially in stage II, which contained the highest proportions (Figure 2F). These results highlighted that CD8+ T cell subpopulations correlated with melanoma prognoses and pathology progression. Also, these results revealed differential proportions of CD8+ T cell subpopulations between melanoma and normal samples.
Landscape of heterogeneous pathway activity
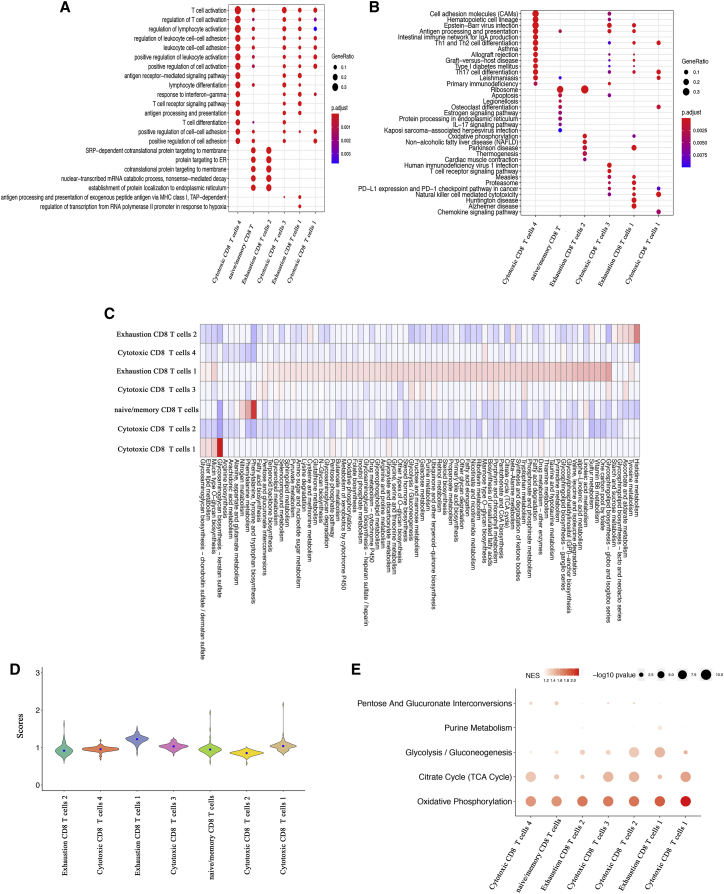
To explore if pathways existed heterogeneously in different CD8+ T cell subpopulations, we first performed Gene Ontology (GO) analysis, Kyoto encyclopedia of genes and genomes (KEGG) analysis, gene set enrichment analysis (GSEA), and pathway activity analyses. GO analysis revealed that exhausted subpopulation 2 had a distinct biology process, and few processes were enriched in the subpopulation (Figure 3A). Naive/memory and cytotoxic subpopulation 3 had similar biology process (Figure 3A). However, KEGG results (Figure 3B) identified considerable heterogeneity in each CD8+ T cell. Oxidative phosphorylation was significantly enriched in exhausted subpopulation 1 and exhausted subpopulation 2 (Figure 3B). Non-alcoholic fatty liver disease, thermogenesis, and cardiac muscle contraction were only enriched in exhausted subpopulation 2. Moreover, human immunodeficiency virus 1 infection and T cell receptor signaling pathway were only significantly enriched in cytotoxic subpopulation 3. The differences of these pathways in different cell subpopulations may provide us clues for heterogeneous outcomes. We then employed metabolic gene sets to score all cell subpopulations, and the deconvolution method was used to normalize scRNA-seq matrixes.25 Exhausted subpopulation 1 had the most substantial metabolic pathway activity, including oxidative phosphorylation, glycolysis/gluconeogenesis, and citrate cycle (TCA cycle) (Figure 3C). Histidine metabolism and tyrosine metabolism were significantly upregulated in exhausted subpopulation 2 (Figure 3C). Fatty acid biosynthesis induced the highest activity in cytotoxic subpopulation 3. Phenylalanine, tyrosine and tryptophan biosynthesis and glycosaminoglycan biosynthesis-keratan sulfate were significantly upregulated in naive/memory and cytotoxic subpopulation 1, respectively (Figure 3C). Further analysis of the distributions of metabolic pathway activities showed that exhausted subpopulation 1 had the highest activities (Figure 3D). The results of GSEA revealed that oxidative phosphorylation and glycolysis/gluconeogenesis showed highly upregulated activity in exhausted subpopulation 1, which was consistent with pathway activity analysis (Figure 3E). However, only oxidative phosphorylation was highly elevated in exhausted subpopulation 2 (Figure 3E). In addition, the upregulation of oxidative phosphorylation and the TCA cycle were observed in cytotoxic subpopulation 3 (Figure 3E). We also performed principal-component analysis (PCA) of metabolic genes to identify the variance of principal components (PCs) in different subpopulations (Figure S5A).
Figure 3.
Metabolic pathway activity of CD8+ T cell subpopulations
(A and B) GO analysis of biology process (A) and KEGG analysis (B). Node size represents numbers of genes. The color intensity is based on the adjusted p value. (C) Metabolic pathway activities in CD8+ T cell subpopulations. (D) Distributions of metabolic pathway activities in each subpopulation. (E) Top metabolic pathways enriched in all subpopulations. Node size represents −log10 p value. The color intensity is based on the scores of normalized enrichment scores (NES).
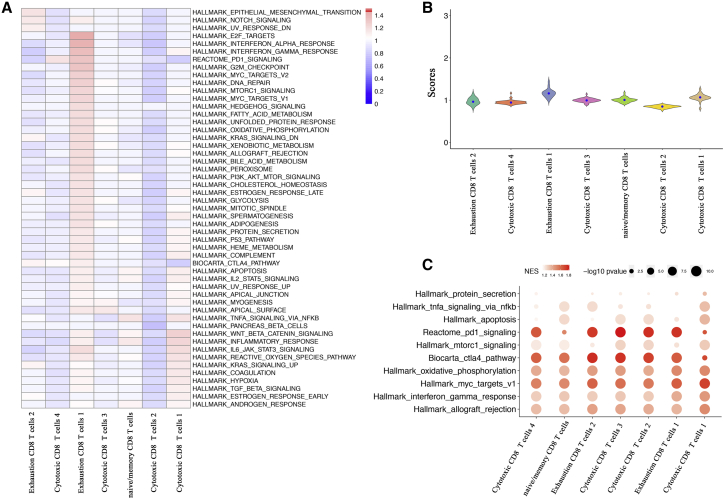
To further characterize the heterogeneity of different subpopulations, we also employed hallmark gene sets, PD-1, and CTLA-4 signaling pathways to perform pathway activity and GSEA analysis. Similar to the above-mentioned metabolic analysis, almost all hallmark pathways were generally upregulated in exhausted subpopulation 1 (Figures 4A and 4B). A more plausible explanation was that abnormal activity of metabolic and hallmark activity might appear in the exhausted subpopulation 1. Notably, both exhausted subpopulations had a high epithelial-mesenchymal transition (EMT) activity and Notch signaling (Figure 4A). We also found that immune checkpoint pathways, PD-1, and CTLA-4 signaling pathways were upregulated in each cell subpopulation (Figure 4C). Importantly, the highest activities of the immune checkpoint pathways were mainly enriched in cytotoxic subpopulation 2, cytotoxic subpopulation 3, exhausted subpopulation 1, and exhausted subpopulation 2 (Figure 4C). These results indicated that blockade immune checkpoints might be useful to promote anti-tumor immune responses of CD8+ T cells. The results may also explain the unfavorable prognostic effect of exhausted subpopulation 2. In addition, hallmark oxidative phosphorylation showed highly upregulated activity in exhausted subpopulation 1, while the ability of gamma interferon response was elevated across all subpopulations (Figure 4C). The variance of PCs based on hallmark gene sets is shown in Figure S5B.
Figure 4.
Hallmark and immune checkpoint pathway activity of CD8+ T cell subpopulations
(A) Hallmark and immune checkpoint pathway activities in CD8+ T cell subpopulations. (B) Distributions of hallmark and immune checkpoint pathway activities in different CD8+ T cell subpopulations. (C) Top hallmark and immune checkpoint pathways enriched in all subpopulations. Node size represents −log10 p value. The color intensity is based on the scores of NES.
Hypoxia, an environmental factor, has a significant impact on other pathways. Thus, we explored the relationships of hypoxia with metabolic and hallmark pathways. Oxidative phosphorylation, glycolysis/gluconeogenesis, and TCA cycle were positively correlated with hypoxia (Figure S5C). Significantly, we identified that apoptosis and p53 pathways showed the strongest positive correlation with hypoxia (Pearson’s R > 0.6, p < 0.001) (Figure S5D). However, hypoxia showed a weak yet positive correlation with PD-1 and CTLA-4 signaling pathways. Together, these results indicated that pathway activity existed heterogeneously in different CD8+ T cell subpopulations, which might be associated with differential prognoses of SKCM. Also, hypoxia interfered with certain pathway activity of CD8+ T cell subpopulations.
Pseudo-time trajectory revealed dynamic changes of immune checkpoints
We performed a pseudo-time trajectory to explore if the differentiation program of CD8+ T cells will contribute to heterogeneous prognoses. As shown in Figure 5A, the pseudo-time differentiation program was visualized by t-SNE. We found the following trajectories during CD8+ T cell differentiation: naive/memory CD8+ T cells tended to convert into exhausted subpopulation 2 (lineage 1) or cytotoxic subpopulation 1 (lineage 2) (Figure 5A). The results may explain why high proportions of exhausted subpopulation 2 would contribute to a poor prognosis, particularly because exhausted subpopulation 2, a terminal state of the subpopulations, showed low cytotoxicity and high immune checkpoint activity (Figures S2 and S3). In addition, exhausted subpopulation 1 with abnormal pathway activity (Figures 3C–3E and 4) may be an intermediate state to differentiate to the terminal exhausted subpopulation 2.
Figure 5.
The pseudo-time trajectory for cell subpopulations
(A) The t-SNE plot for cell differentiation trajectory. Naive/memory subpopulation was the initial subpopulation, which differentiates into cytotoxic subpopulation 1 and exhausted subpopulation 2. (B and C) Expression profiles of lineage 1 for cytotoxic genes (B) and immune checkpoints (C).
In the lineage 1 program, cytotoxic signatures (PRF1, GZMA, GZMK, and NKG7) would be upregulated during the process of naive/memory CD8+ T cells differentiating to cytotoxic subpopulation 3, and then these signatures tended to be downregulated until the least expression in exhausted subpopulation 2 (Figure 5B; Figure S6B). A similar tendency was observed in immune checkpoints (PDCD1, TIM-3/HAVCR2, and LAG3) and costimulatory molecules (CD27) (Figure 5C). However, exhausted-like signatures (CTLA4, TIGIT, and TNFRSF9) and a synergist checkpoint CD7 showed an upward tendency. Other immune checkpoints (CD244, TMIGD2, CD226, and KLRG1) did not show dynamic evolution over time in both lineages (Figures S6A and S6C). For the lineage 2 program, PRF1, GZMA, and NKG7 had an overall upward trend, while GZMK was on the decline (Figure S6B). Almost all inhibitory checkpoints were downregulated in lineage 2 process, apart from TIM-3/HAVCR2, TIGIT, and synergist checkpoint CD7 (Figure S6C). These results expounded the pseudo-time trajectory of CD8+ T cells in melanoma and the time-dependent changes of immune checkpoints.
Heterogeneous interacted pairs and transcription factors in CD8+ T cell subpopulations
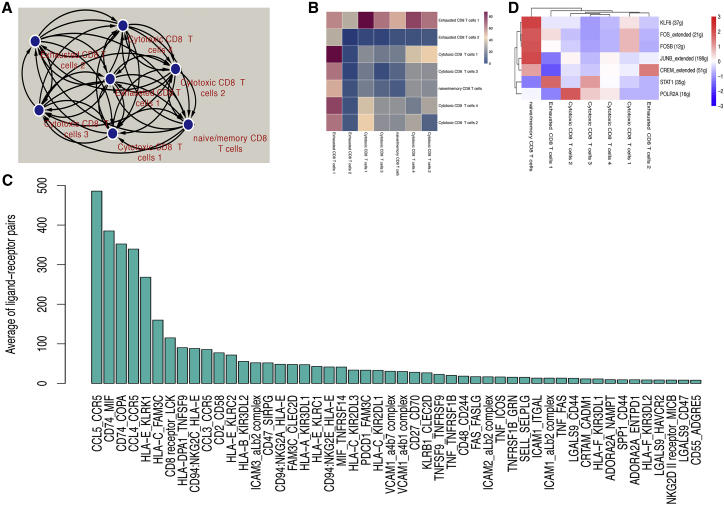
Given the heterogeneity for different CD8+ T cell subpopulations, we analyzed their communication network to identify key ligand-receptor pairs and cell subpopulations, which dominated the interaction. We found that exhausted CD8+ T cell subpopulation 1 played a dominant role in interacted pairs (Figures 6A and 6B). There were 83 significant ligand-receptor pairs among different CD8+ T cell subpopulations. Among them, the top 5 frequent ligand-receptor pairs were CCL5_CCR5, CD74_MIF, CD74_COPA, CCL4_CCR5, and HLA−E_KLRK1 (Figure 6C). The heatmap (Figure S7) showed the specific distribution of Top 50 frequent ligand-receptor pairs. These ligand-receptor pairs mainly interacted in exhausted CD8+ T cell subpopulation 1, suggesting its important crosstalk roles with other subpopulations (Figure S7). Therefore, exhausted CD8+ T cell subpopulation 1 may have a high degree of dysfunction.
Figure 6.
Cell-cell interactions and TFs within different subpopulations
(A) A network of cell-cell interactions. (B) A heatmap showed the counts of ligand-receptor pairs among different subpopulations. (C) Average counts of top 50 interactions across each subpopulation. (D) Expression levels of TFs.
A previously reported study has identified that transcription factors (TFs) can shape T cell phenotype and regulate gene expression.26 Thus, we employed SCENIC to explore underlying differential TFs within CD8+ T cell subpopulations. We found that several TFs expressed in specific subpopulations, such as high expression of KLF6, FOS, FOSB, JUNB, and CREM in the naive/memory subpopulation (Figure 6D). Also, STAT1 and CREM were observed in exhausted subpopulation 1 and exhausted subpopulation 2, respectively (Figure 6D). STAT1 and POLR2A (the gene that encodes RNA polymerase II complex) were both upregulated in cytotoxic subpopulation 3, and there was higher activity of POLR2A in cytotoxic subpopulation 2 (Figure 6D). TF analysis results may reveal the evidential facts that high activity of multiple TFs in the naive/memory subpopulation provided essential regulations for initial differentiation. Collectively, these analyses identified the dominant subpopulation (exhausted CD8+ T cell subpopulation 1), frequent ligand-receptor pairs, and differential regulation of TFs for CD8+ T cell subpopulations.
PMEL, TYRP1, and EDNRB were novel therapeutic targets
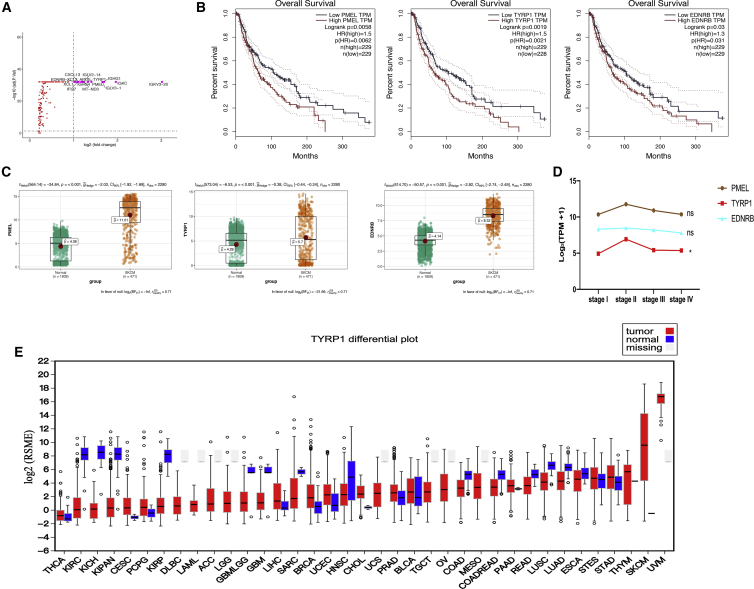
Given the poor prognosis in SKCM caused by high proportions of exhausted subpopulation 2, we analyzed the highest expression genes in this subpopulation to screen out the potential therapy targets (Figure 7A). Then, Kaplan-Meier survival analysis was conducted to explore the prognostic values of screened genes (Figure 7B). Finally, we screened out PMEL, TYRP1, and EDNRB, which may serve as novel targets, and high activity of them was significantly associated with a poor prognosis in SKCM (Figure 7B). Notably, PMEL, TYRP1, and EDNRB only highly expressed in exhausted subpopulation 2 (Figures S8A and S9A), suggesting their important role in prognoses. In addition, the three genes also showed high activity in bulk RNA-seq samples of SKCM (Figure 7C). We also explored if PMEL, TYRP1, and EDNRB would dynamically express in different pathology stages. The results showed that only TYRP1 might be involved in SKCM progression. Importantly, it would show higher activity in later stages than stage I; the highest expression was observed in stage II (Figure 7D).
Figure 7.
The novel therapeutic targets were screened out and validated
(A) Top expression genes within exhausted subpopulation 2. Gene symbols were labeled for adjusted p value < 0.05 and log2 fold change > 1. (B) Kruskal-Wallis curves were plotted to validate the survival values of PMEL, TYRP1, and EDNRB. (C) Gene expression of the novel targets within SKCM and normal RNA-seq samples. The ordinate showed values of log2(TPM + 1). (D) Average log2(TPM + 1) values among four pathology stages (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001). (E) The expression values of TYRP1 across 37 tumors and normal samples (download from FIREBROWSE database).
Next, transcriptional levels of the novel targets were investigated across various tumors by the FIREBROWSE database. Conspicuously, all novel targets had the highest variation in SKCM and uveal melanoma (UVM), revealing a tight connection between the novel targets and melanoma (Figure 7E; Figures S8B and S8C). Given the specific high expressions of the novel targets in melanoma samples and exhausted subpopulation 2, we explored if different scRNA-seq data of the T cells and CD8+ T cells in the tumor microenvironment of melanoma also had the property. Thus, we obtained the single-cell data of T cells,27 which were well clustered, to verify the accuracy of our results. In line with the above results, we observed that PMEL and TYRP1 were highly expressed in one CD8+ T cell cluster and were not highly expressed in a certain CD4+ T cell cluster (Figures S9B–S9D). However, we did not observe high EDNRB expression in the certain CD8+ T cell cluster. This may be due to low cell abundance—only 263 CD8+ T cells in cluster 3 (Figure S9B)—while there were 1,971 CD8+ T cells in exhausted subpopulation 2 (Figure 1F). Taken together, we identified that PMEL, TYRP1, and EDNRB, which were tightly associated with melanoma, would contribute to poor clinical outcomes and may serve as novel therapeutic targets.
Discussion
Several recent studies have analyzed tumor infiltration in melanoma and other tumors.22,23,27, 28, 29 These studies have provided some clues about the transcriptional roles in the regulation of T cells. However, the heterogeneity of CD8+ T cell subpopulations has not been fully described. Therefore, we collected scRNA-seq data of melanoma to analyze specific roles of CD8+ T cell subpopulations in prognoses and explore the source of heterogeneity. Exhausted CD8+ T cells expressed exhausted signatures and immune checkpoints along with producing CXCL13, suggesting an underlying feature of interaction with intertumoral cell subpopulations.30 Cytotoxic CD8+ T cells also elevated the activity of immune checkpoints to balance cytotoxicity in TME. Understanding the dynamic activity of immune checkpoints during CD8+ T cell differentiation can help pave the way for immune checkpoint blockade.
In this study, we annotated seven CD8+ T cell subpopulations in the melanoma TME and found that naive/memory and cytotoxic subpopulation 3 were associated with a favorable clinical outcome. However, exhausted subpopulation 2, a terminal state of CD8+ T cell subgroups, led to a poor prognosis. Differential activities of cytotoxicity and immune checkpoint molecules were only one side to explain the heterogeneous prognoses (Figures 5B and 5C; Figures S2, S3, and S6). On the other hand, differential pathways may account for heterogeneous regulations among all subpopulations (Figure S5; Figures 3 and 4,). Abnormal pathway activity in exhausted subpopulation 1 reflected a high dysfunction state during the process of subpopulation conversion to terminal-like CD8+ T cells (Figures 3 and 4). In addition, many other poorly investigated metabolic pathways, such as histidine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, and glycosaminoglycan biosynthesis-keratan sulfate showed high activities in specific cell subpopulations, which may provide novel insights into these pathways (Figure 3C). Remarkably, high activities of EMT and Notch signaling were observed in exhausted subpopulations (Figure 4A). EMT had an inhibitory effect for intratumoral CD8+ T cells, related to tumor progression and poor prognosis in non-small cell lung cancer.31, 32, 33, 34 Furthermore, EMT could elevate the expression of CTLA4.35 A recent study based on a murine model of colorectal cancer found that Notch signaling in the TME would contribute to a poor prognosis, highly penetrant metastasis, and potential targets for therapy.36 Abnormal pathway activity of EMT and Notch signaling in exhausted CD8+ T cells may enhance dysfunction of these cells; thus, high exhausted subpopulation 2 infiltration would be unfavorable for melanoma prognoses (Figures 2E and 2F). Importantly, since pathways and molecules of immune checkpoints showed a dynamic change during CD8+ T cell differentiation (Figures 4C and 5C), the optimal treatment time of immune checkpoint blockade may be obtained in our study (the optimal time was when there were the most subpopulations of exhausted CD8+ T cells with the high expression of immune checkpoints).
Furthermore, cell-cell interaction analysis revealed top ligand-receptor pairs among all subpopulations. CCL5/CCR5 axis was the most frequent interactive ligand-receptor pairs in our study (Figure 6C), which promotes vascular endothelial growth factor-mediated tumor angiogenesis in the osteosarcoma TME.37 Inhibition of CD74/MIF will elevate the level of reactive oxygen species to induce cancer cell death in colon carcinomatosis.38 The enhancement of the CCL4/CCR5 axis will promote glioblastoma invasion.39 Importantly, these ligand-receptor pairs were mainly distributed across cytotoxic subpopulations 1, 2, 4, and exhausted subpopulation 1 (Figure S7). TFs such as STAT1 with the highest activity in exhausted subpopulation 1 (Figure 6D) can induce breast tumor proliferation and promote immunosuppressive TME.40 Therefore, these interacted pairs and TFs may also be potential therapeutic targets for melanoma.
It has been demonstrated that elevated activity of TYRP1 in metastatic melanoma will result in an unfavorable clinical outcome, which can also sequester miR-16 to induce a change of transcriptional program that drives melanoma expansion.41 Moreover, preventing miR-16 sequestration on TYRP1 would reduce tumor growth in vivo.41 It was found that PMEL was highly expressed in melanoma tissues, and target PMEL can induce apoptosis of melanoma cells in vitro.42 EDNRB, a marker of melanoma progression, was overexpressed in metastatic melanoma.43, 44, 45 Also, blockading EDNRB was effective to reduce the proliferation of melanoma in vitro and in vivo.46 These genes are highly expressed in melanoma tissues, but the transcript levels in CD8+ T cells remain unclear. Here, we identified that these genes were also overexpressed in a certain CD8+ T cell subpopulation (Figures S8A and S9). Thus, these genes may also influence the functions of CD8+ T cells, for example reducing tumor-killing ability. This may provide new potential therapy targets of melanoma, in which the tumor-killing ability of CD8+ T cells is enhanced by blocking these genes and relative proteins.
In conclusion, we characterized the heterogeneity among different CD8+ T cell subpopulations based on melanoma scRNA-seq data. Comprehensive analyses enabled us to understand the heterogenous functions of different CD8+ T cell subpopulations in the melanoma TME. Furthermore, we provided a dynamic gene expression landscape for immune checkpoints to optimize the treatment time window for immunotherapy. Most importantly, we identified that PMEL, TYRP1, and EDNRB could serve as potential therapeutic targets for melanoma. However, further experiments, such as knocking out and overexpressing of these genes in CD8+ T cells, are still needed to validate the hypotheses inferred from our work.
Materials and methods
scRNA-seq and RNA-seq data
The scRNA-seq data were downloaded from the GEO database (Smart-seq data, GEO: GSE120575 and 10× Genomics data, GEO: GSE139829). We also downloaded scRNA-seq data of T cells (Smart-seq data, GEO: GSE72056) to validate the single-cell transcript levels of the novel targets. T cells were classified by Tirosh et al.27 in those data. The RNA-seq data of 471 skin cutaneous melanomas and 1,809 normal samples were downloaded from TCGA and GTEx databases.
Cell subpopulation identification
We employed 48 smart-seq and 11 10× Genomics melanoma samples to perform CD8+ T cell analyses. Seurat’s (version 3.1.5) standard process was conducted. In brief, we removed low-quality cells according to the standard, which is cells with fewer than 200 unique molecular identifiers (UMIs) or mitochondrial gene expression exceeding 60%. Gene expression was then normalized using the default parameter of the Seurat package. FindVariableFeatures and ScaleData functions were used in the default parameter. Subsequently, PCA (dims = 20) and FindClusters (resolution = 0.4) were performed to identify cell clusters. Then, t-SNE was used to visualize the single cells. These parameters were also used in the following analyses of single-cell datasets. We classified CD8+ T cell subpopulations using straightforward analysis, in which single cells expressed high CD8A and CD8B (CD8+ T cell markers). Also, markers of T cells (CD3D, CD3E, and CD3G), CD4+ T cells (CD4), and melanocytes (DCT, BNC2, CRABP1, PTGDS, FILIP1L, DKK3, and AHNAK2) were used to validate if there were mixed CD4+ T cells and melanocytes in CD8+ T cells. These markers validated that CD8+ T cells (with high expressions in CD8+ T cell markers and T cell markers) were accurately classified in our study. We isolated 3,525 and 7,336 CD8+ T cells from Smart-seq and 10× Genomics, respectively. The Harmony algorithm (version 1.0) can accurately integrate single-cell data from different technology platforms and batches.47 Thus, we integrated a total of 10,861 CD8+ T cells from smart-seq and 10× Genomics platforms using the Harmony algorithm. Then, following the Seurat standard process, 7 CD8+ T cell subpopulations were identified. These subpopulations were annotated with data generated from the FindAllMarkers function in the Seurat package. Previously reported marker genes were used to annotate the following CD8+ T cell subpopulations: naive/memory CD8+ T cells (CCR7, IL7R, TCF7, SELL, SATB1, GPR183, LTB, LEF1, and S100A10), cytotoxic CD8+ T cells (PRF1, GZMA, GZMK, and NKG7), and exhausted CD8+ T cells (CXCL13, HSPB1, IRF4, LAYN, GIMAP6, HSPH1, CXCR6, CTLA4, PDCD1, LAG3, HAVCR2, and TIGIT).
Cell components of CD8+ T cell subpopulations in bulk RNA-seq
The CIBERSORT algorithm (https://cibersort.stanford.edu/) was developed to calculate the proportions of different cell types in bulk RNA-seq data based on the signature matrix. In order to detect cell components of CD8+ T cell subpopulations in the bulk RNA-seq, the expression matrix was normalized using transcripts per million (TPM). Then, log2(TPM + 1) values were calculated to scale the bulk RNA-seq matrix. For the signature matrix of each CD8+ T cell subpopulation, we employed Seurat FindMarkers function to obtain significantly overexpressed genes (adjusted p value < 0.05, log2 fold change > 0.25). Finally, cell proportions were identified using default parameters of CIBERSORT.
To explore the relationship between CD8+ T cell subpopulations and melanoma patients’ prognoses, we divided TCGA tumor samples into two subgroups: high-proportion CD8+ T cell subpopulations 50% and low-proportion CD8+ T cell subpopulations 50%. Subsequently, Kaplan-Meier curves were generated to identify the influence of CD8+ T cell subpopulations on prognoses using survival (version 3.2) and survminer (version 0.4.7) package in R. Also, we divided tumor samples into four pathology stages according to the information of TCGA. Then dynamic changes of CD8+ T cell subpopulations were identified in different pathology stages. Also, differential proportions of infiltrating cell subpopulations between normal and tumor samples were analyzed using ggstatsplot (version 0.5.0) package.
Pathway activity analysis
After characterizing prognostic effectors of CD8+ T cell subpopulations, we performed GO analysis, KEGG analysis, and GSEA to elucidate the mechanisms of different clinical outcomes at single-cell levels. GO and KEGG analyses were done by ClusterProfiler (version 3.14.3) package, while the GSEA analysis was conducted using the javaGSEA (https://www.gsea-msigdb.org/gsea/downloads.jsp). We also scored pathway activities of subpopulations using a recent algorithm.48 For the analyses of GSEA and pathway activities, metabolic and hallmark gene sets were downloaded from the GSEA database. Pearson’s correlation analysis between hypoxia and other pathways was conducted to identify the influence of hypoxia.
Pseudo-time trajectory, cell-cell interaction, and single-cell regulatory network analysis
We performed trajectory analysis using the slingshot (version 1.4.0) package for all CD8+ T cell subpopulations, setting the naive/memory subpopulation as the start subpopulation. Default parameters of the slingshot package were used during the process of analysis. After a pseudo-time trajectory, we analyzed the expression levels of cytotoxicity and checkpoint molecules during the process that naive/memory subpopulations differentiated to the exhausted subpopulation. To investigate the cell-cell interaction, CellphoneDB (version 2.0), a Python-based package, was used to identify the interacted pairs with default parameters. In addition, the gene regulatory network of the cell subpopulations was calculated using SCIENIC (version 1.1.2.2) package.
Novel targets analysis
In order to screen out novel targets for therapy, the overexpressed genes (adjusted p value < 0.05, log2 fold change > 1) in exhausted CD8+ T cell subpopulation 2 were analyzed by Kaplan-Meier survival analysis with GEPIA database (http://gepia.cancer-pku.cn/detail.php?gene=CUL3). Then, the expression levels of screened targets were validated in tumor and normal samples using ggstatsplot package. The data of TCGA and GTEx were normalized using TPM. log2(TPM + 1) values were calculated to scale the bulk RNA-seq matrix. The data were then mixed, and batch effects were corrected by the removeBatchEffect function in limma package (version 3.42.2). FIREBROWSE database (http://firebrowse.org/) was used to identify the expression values of screened targets in various tumor samples. Subsequently, we isolated 2,043 T cells from the data to validate the novel targets. These T cells were classified by Tirosh et al.,27 and they sorted these cells using fluorescence-activated cell sorting (FACS), which can exclude other cells. We also visualized these T cells using t-SNE, and scatterplots were performed to show the single-cell expression of the novel markers, T cell markers, CD8+ T cell markers, and melanocyte markers.
Statistical analysis
Differentially expressed genes among different subpopulations, GO, KEGG, and GSEA analyses were statistically analyzed using default methods in respective packages or software. A random permutation test generated statistical significance of pathway activities. A Student’s t test was used to detect differential cell frequency between normal and tumor samples; the statistical analysis was also used to test the expression levels in tumor and normal samples. A log-rank test was used to test the significance of Kaplan-Meier curves. Kruskal-Wallis test was used to detect dynamic changes in cell proportions and gene expression levels within different pathology stages.
Acknowledgments
This work was supported by Major Infectious Diseases Such as AIDS and Viral Hepatitis Prevention and Control Technology Major Projects (2018ZX10712001-011).
Author contributions
R.L. and J.C. designed and supervised the research activity planning. W.D. drafted the manuscript and finished the bioinformatics analysis. Y.M., Z.S., Y.L., P.L., C.H., X.L., Y.Z., K.Z., and J.S. collected data and revised the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.12.003.
Contributor Information
Jian Chen, Email: chenj8@sysu.edu.cn.
Ruoyu Li, Email: mycolab@126.com.
Supplemental information
References
- 1.Schreiber R.D., Old L.J., Smyth M.J. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 2.Chang C.-H., Qiu J., O’Sullivan D., Buck M.D., Noguchi T., Curtis J.D., Chen Q., Gindin M., Gubin M.M., van der Windt G.J. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nolz J.C. Molecular mechanisms of CD8(+) T cell trafficking and localization. Cell. Mol. Life Sci. 2015;72:2461–2473. doi: 10.1007/s00018-015-1835-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harty J.T., Badovinac V.P. Shaping and reshaping CD8+ T-cell memory. Nat. Rev. Immunol. 2008;8:107–119. doi: 10.1038/nri2251. [DOI] [PubMed] [Google Scholar]
- 5.Butler N.S., Nolz J.C., Harty J.T. Immunologic considerations for generating memory CD8 T cells through vaccination. Cell. Microbiol. 2011;13:925–933. doi: 10.1111/j.1462-5822.2011.01594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piersma S.J., Jordanova E.S., van Poelgeest M.I., Kwappenberg K.M., van der Hulst J.M., Drijfhout J.W., Melief C.J., Kenter G.G., Fleuren G.J., Offringa R., van der Burg S.H. High number of intraepithelial CD8+ tumor-infiltrating lymphocytes is associated with the absence of lymph node metastases in patients with large early-stage cervical cancer. Cancer Res. 2007;67:354–361. doi: 10.1158/0008-5472.CAN-06-3388. [DOI] [PubMed] [Google Scholar]
- 7.Kim P.S., Ahmed R. Features of responding T cells in cancer and chronic infection. Curr. Opin. Immunol. 2010;22:223–230. doi: 10.1016/j.coi.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kmiecik J., Poli A., Brons N.H., Waha A., Eide G.E., Enger P.Ø., Zimmer J., Chekenya M. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J. Neuroimmunol. 2013;264:71–83. doi: 10.1016/j.jneuroim.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 9.Baitsch L., Baumgaertner P., Devêvre E., Raghav S.K., Legat A., Barba L., Wieckowski S., Bouzourene H., Deplancke B., Romero P. Exhaustion of tumor-specific CD8+ T cells in metastases from melanoma patients. J. Clin. Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jadhav R.R., Im S.J., Hu B., Hashimoto M., Li P., Lin J.-X., Leonard W.J., Greenleaf W.J., Ahmed R., Goronzy J.J. Epigenetic signature of PD-1+ TCF1+ CD8 T cells that act as resource cells during chronic viral infection and respond to PD-1 blockade. Proc. Natl. Acad. Sci. USA. 2019;116:14113–14118. doi: 10.1073/pnas.1903520116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LaFleur M.W., Nguyen T.H., Coxe M.A., Miller B.C., Yates K.B., Gillis J.E., Sen D.R., Gaudiano E.F., Al Abosy R., Freeman G.J. PTPN2 regulates the generation of exhausted CD8+ T cell subpopulations and restrains tumor immunity. Nat. Immunol. 2019;20:1335–1347. doi: 10.1038/s41590-019-0480-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speiser D.E., Ho P.-C., Verdeil G. Regulatory circuits of T cell function in cancer. Nat. Rev. Immunol. 2016;16:599–611. doi: 10.1038/nri.2016.80. [DOI] [PubMed] [Google Scholar]
- 13.Fourcade J., Sun Z., Benallaoua M., Guillaume P., Luescher I.F., Sander C., Kirkwood J.M., Kuchroo V., Zarour H.M. Upregulation of Tim-3 and PD-1 expression is associated with tumor antigen-specific CD8+ T cell dysfunction in melanoma patients. J. Exp. Med. 2010;207:2175–2186. doi: 10.1084/jem.20100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matsuzaki J., Gnjatic S., Mhawech-Fauceglia P., Beck A., Miller A., Tsuji T., Eppolito C., Qian F., Lele S., Shrikant P. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc. Natl. Acad. Sci. USA. 2010;107:7875–7880. doi: 10.1073/pnas.1003345107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walunas T.L., Lenschow D.J., Bakker C.Y., Linsley P.S., Freeman G.J., Green J.M., Thompson C.B., Bluestone J.A. CTLA-4 can function as a negative regulator of T cell activation. Immunity. 1994;1:405–413. doi: 10.1016/1074-7613(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 16.Keir M.E., Liang S.C., Guleria I., Latchman Y.E., Qipo A., Albacker L.A., Koulmanda M., Freeman G.J., Sayegh M.H., Sharpe A.H. Tissue expression of PD-L1 mediates peripheral T cell tolerance. J. Exp. Med. 2006;203:883–895. doi: 10.1084/jem.20051776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wei S.C., Duffy C.R., Allison J.P. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 18.Rowshanravan B., Halliday N., Sansom D.M. CTLA-4: a moving target in immunotherapy. Blood. 2018;131:58–67. doi: 10.1182/blood-2017-06-741033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solinas C., Migliori E., De Silva P., Willard-Gallo K. LAG3: The biological processes that motivate targeting this immune checkpoint molecule in human cancer. Cancers (Basel) 2019;11:1213. doi: 10.3390/cancers11081213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan W., Liu X., Ma H., Zhang H., Song X., Gao L., Liang X., Ma C. Tim-3 fosters HCC development by enhancing TGF-β-mediated alternative activation of macrophages. Gut. 2015;64:1593–1604. doi: 10.1136/gutjnl-2014-307671. [DOI] [PubMed] [Google Scholar]
- 21.Harjunpää H., Guillerey C. TIGIT as an emerging immune checkpoint. Clin. Exp. Immunol. 2020;200:108–119. doi: 10.1111/cei.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sade-Feldman M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013.e20. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durante M.A., Rodriguez D.A., Kurtenbach S., Kuznetsov J.N., Sanchez M.I., Decatur C.L., Snyder H., Feun L.G., Livingstone A.S., Harbour J.W. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020;11:496. doi: 10.1038/s41467-019-14256-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kazanietz M.G., Durando M., Cooke M. CXCL13 and its receptor CXCR5 in cancer: inflammation, immune response, and beyond. Front. Endocrinol. (Lausanne) 2019;10:471. doi: 10.3389/fendo.2019.00471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lun A.T., Bach K., Marioni J.C. Pooling across cells to normalize single-cell RNA sequencing data with many zero counts. Genome Biol. 2016;17:75. doi: 10.1186/s13059-016-0947-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rothenberg E.V. The chromatin landscape and transcription factors in T cell programming. Trends Immunol. 2014;35:195–204. doi: 10.1016/j.it.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tirosh I., Izar B., Prakadan S.M., Wadsworth M.H., 2nd, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Azizi E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308.e36. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guo X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 30.Thommen D.S., Koelzer V.H., Herzig P., Roller A., Trefny M., Dimeloe S., Kiialainen A., Hanhart J., Schill C., Hess C. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 2018;24:994–1004. doi: 10.1038/s41591-018-0057-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsoukalas N., Aravantinou-Fatorou E., Tolia M., Giaginis C., Galanopoulos M., Kiakou M., Kostakis I.D., Dana E., Vamvakaris I., Korogiannos A. Epithelial–mesenchymal transition in non small-cell lung cancer. Anticancer Res. 2017;37:1773–1778. doi: 10.21873/anticanres.11510. [DOI] [PubMed] [Google Scholar]
- 32.Mahmood M.Q., Ward C., Muller H.K., Sohal S.S., Walters E.H. Epithelial mesenchymal transition (EMT) and non-small cell lung cancer (NSCLC): a mutual association with airway disease. Med. Oncol. 2017;34:45. doi: 10.1007/s12032-017-0900-y. [DOI] [PubMed] [Google Scholar]
- 33.Lou Y., Diao L., Cuentas E.R.P., Denning W.L., Chen L., Fan Y.H., Byers L.A., Wang J., Papadimitrakopoulou V.A., Behrens C. Epithelial–mesenchymal transition is associated with a distinct tumor microenvironment including elevation of inflammatory signals and multiple immune checkpoints in lung adenocarcinoma. Clin. Cancer Res. 2016;22:3630–3642. doi: 10.1158/1078-0432.CCR-15-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen L., Gibbons D.L., Goswami S., Cortez M.A., Ahn Y.-H., Byers L.A., Zhang X., Yi X., Dwyer D., Lin W. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat. Commun. 2014;5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chae Y.K., Chang S., Ko T., Anker J., Agte S., Iams W., Choi W.M., Lee K., Cruz M. Epithelial-mesenchymal transition (EMT) signature is inversely associated with T-cell infiltration in non-small cell lung cancer (NSCLC) Sci. Rep. 2018;8:2918. doi: 10.1038/s41598-018-21061-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackstadt R., van Hooff S.R., Leach J.D., Cortes-Lavaud X., Lohuis J.O., Ridgway R.A., Wouters V.M., Roper J., Kendall T.J., Roxburgh C.S. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36:319–336.e7. doi: 10.1016/j.ccell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S.-W., Liu S.-C., Sun H.-L., Huang T.-Y., Chan C.-H., Yang C.-Y., Yeh H.-I., Huang Y.-L., Chou W.-Y., Lin Y.-M., Tang C.H. CCL5/CCR5 axis induces vascular endothelial growth factor-mediated tumor angiogenesis in human osteosarcoma microenvironment. Carcinogenesis. 2015;36:104–114. doi: 10.1093/carcin/bgu218. [DOI] [PubMed] [Google Scholar]
- 38.Bozzi F., Mogavero A., Varinelli L., Belfiore A., Manenti G., Caccia C., Volpi C.C., Beznoussenko G.V., Milione M., Leoni V. MIF/CD74 axis is a target for novel therapies in colon carcinomatosis. J. Exp. Clin. Cancer Res. 2017;36:16. doi: 10.1186/s13046-016-0475-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Liu T., Yang N., Xu S., Li X., Wang D. Hypoxia and macrophages promote glioblastoma invasion by the CCL4-CCR5 axis. Oncol. Rep. 2016;36:3522–3528. doi: 10.3892/or.2016.5171. [DOI] [PubMed] [Google Scholar]
- 40.Hix L.M., Karavitis J., Khan M.W., Shi Y.H., Khazaie K., Zhang M. Tumor STAT1 transcription factor activity enhances breast tumor growth and immune suppression mediated by myeloid-derived suppressor cells. J. Biol. Chem. 2013;288:11676–11688. doi: 10.1074/jbc.M112.441402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gilot D., Migault M., Bachelot L., Journé F., Rogiers A., Donnou-Fournet E., Mogha A., Mouchet N., Pinel-Marie M.-L., Mari B. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat. Cell Biol. 2017;19:1348–1357. doi: 10.1038/ncb3623. [DOI] [PubMed] [Google Scholar]
- 42.Wang J.-J., Li Z.-F., Li X.-J., Han Z., Zhang L., Liu Z.-J. Effects of microRNA-136 on melanoma cell proliferation, apoptosis, and epithelial-mesenchymal transition by targetting PMEL through the Wnt signaling pathway. Biosci. Rep. 2017;37 doi: 10.1042/BSR20170743. BSR20170743. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Loftus S.K., Chen Y., Gooden G., Ryan J.F., Birznieks G., Hilliard M., Baxevanis A.D., Bittner M., Meltzer P., Trent J., Pavan W. Informatic selection of a neural crest-melanocyte cDNA set for microarray analysis. Proc. Natl. Acad. Sci. USA. 1999;96:9277–9280. doi: 10.1073/pnas.96.16.9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bagnato A., Rosanò L., Spinella F., Di Castro V., Tecce R., Natali P.G. Endothelin B receptor blockade inhibits dynamics of cell interactions and communications in melanoma cell progression. Cancer Res. 2004;64:1436–1443. doi: 10.1158/0008-5472.can-03-2344. [DOI] [PubMed] [Google Scholar]
- 45.Demunter A., De Wolf-Peeters C., Degreef H., Stas M., van den Oord J.J. Expression of the endothelin-B receptor in pigment cell lesions of the skin. Evidence for its role as tumor progression marker in malignant melanoma. Virchows Arch. 2001;438:485–491. doi: 10.1007/s004280000362. [DOI] [PubMed] [Google Scholar]
- 46.Lahav R., Heffner G., Patterson P.H. An endothelin receptor B antagonist inhibits growth and induces cell death in human melanoma cells in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 1999;96:11496–11500. doi: 10.1073/pnas.96.20.11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Korsunsky I., Millard N., Fan J., Slowikowski K., Zhang F., Wei K., Baglaenko Y., Brenner M., Loh P.R., Raychaudhuri S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiao Z., Dai Z., Locasale J.W. Metabolic landscape of the tumor microenvironment at single cell resolution. Nat. Commun. 2019;10:3763. doi: 10.1038/s41467-019-11738-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.