Abstract
Pediatric gliomas (PGs) are the most common brain tumors in children and the leading cause of childhood cancer-related death. The understanding of the immune microenvironment is essential for developing effective antitumor immunotherapies. Transcriptomic data from 495 PGs were analyzed in this study, with 384 as a training cohort and 111 as a validation cohort. Macrophages were the most common immune infiltrates in the PG microenvironment, followed by T cells. PGs were classified into 3 immune subtypes (ISs) based on immunological profiling: “immune hot” (IS-I), “immune altered” (IS-II), and “immune cold” (IS-III). IS-I tumors, characterized by substantial immune infiltration and high immune checkpoint molecule (ICM) expression, had a favorable prognosis and were more likely to respond to anti-PD1 and anti-CTLA4 immunotherapies, whereas IS-III tumors, characterized by weak immune infiltration and low ICM expression, had a dismal prognosis and poor immunotherapy responsiveness. IS-II tumors represented a transitional stage. Immune classification was also correlated with somatic mutations, copy number alterations, and molecular pathways related to tumorigenesis, metabolism, and immune responses. Three predictive classifiers using eight representative genes were generated by machine learning methods for immune classification. This study established a reliable immunological profile-based classification system for PGs, providing implications for further immunotherapy strategies.
Keywords: pediatric glioma, immune classification, overall survival, immunotherapy responsiveness, tumor microenvironment
Graphical Abstract
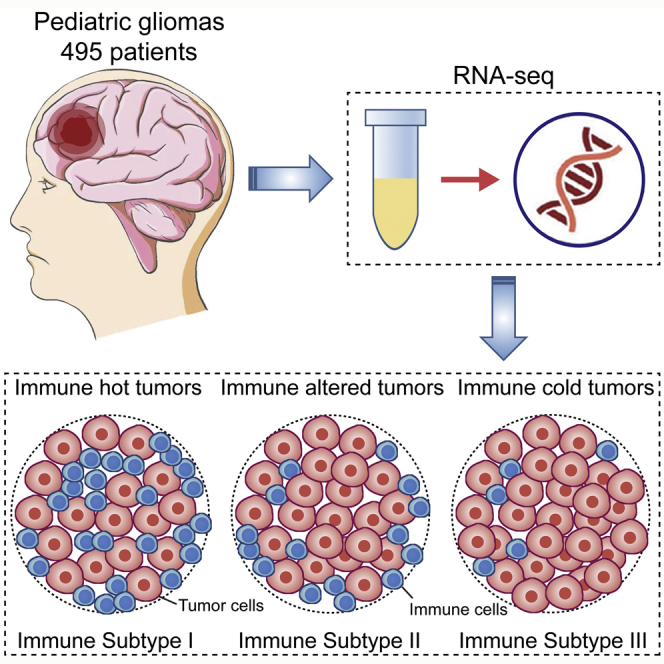
Pediatric gliomas (PGs) are the most common brain tumors in children and the leading cause of childhood cancer-related death. With the use of transcriptomic information, PGs can be classified into 3 immune subtypes: “immune hot” (IS-I), “immune altered” (IS-II), and “immune cold” (IS-III), based on immunological profiling, providing implications for further immunotherapy strategies.
Introduction
Brain tumors are the most common solid neoplasms in children and have overtaken leukemias as the leading cause of pediatric cancer-related death.1, 2, 3 Gliomas are among the most common forms of primary brain tumors arising in childhood.4 Unlike adult gliomas, the majority of pediatric gliomas (PGs) are classified as World Health Organization (WHO) grade I or II or low-grade gliomas (LGGs). Although some pediatric LGGs (pLGGs) are located deep and thus infrequently resectable, most of them are curable by surgery, with good clinical outcomes.1,5,6 The remaining PGs progress rapidly and are classified as WHO grade III or IV or high-grade gliomas (HGGs), with a dismal prognosis of a 5-year overall survival (OS) rate <20% despite the best available treatment modalities.7,8 Therefore, the main therapeutic strategies are to improve quality of life and prevent recurrence and late treatment-associated sequelae in pLGG patients and to prolong life expectancy in pediatric HGG (pHGG) patients.4 PGs are molecularly heterogeneous entities.5,8, 9, 10 Although recent studies on genomic alterations and molecular biology underlying PGs have expanded our understanding of tumor development and progression and offered great opportunities for large-scale therapeutic trials around the globe, improvements in patient survival are still unsatisfactory.4,6,7,10,11 Moreover, apart from therapeutic effects, adjuvant treatments involving radiotherapy and cytotoxic chemotherapy for pHGGs and unresectable/recurrent pLGGs also have considerable negative effects on children, including endocrine dysfunction, cognitive disorders, neuropsychologic diseases, and in some cases, secondary tumors.4,6,7 Therefore, there is an urgent need to explore new therapeutic approaches for PGs.
Recently, immunotherapy has demonstrated remarkable clinical efficacy in adult patients with refractory tumors, such as melanomas and renal cell carcinomas,12,13 with both satisfactory therapeutic effects and acceptable tolerability and thus, with potential to serve as a promising “fourth pillar” of treatment in addition to conventional surgery, radiotherapy, and chemotherapy in managing PGs.9,14, 15, 16 To date, several immunotherapy trials have been conducted for PGs, including trials on immune checkpoint blockade (ICB) therapy,14,17,18 chimeric antigen receptor T cell (CAR-T) therapy,15,19 and vaccination therapy.16,20,21 Although immunotherapy served as a powerful new approach in these PG studies, it worked only in a select subset of patients with unique features, e.g., vaccines for those carrying the human leukocyte antigen (HLA)-A2 allele20,21 and ICB for hypermutant tumors.14,17 More evidence is still needed from large trials to prove the population coverage and long-term efficacy of immunotherapy for PGs.9 A better understanding of the immune microenvironment of tumors is an essential first step for the rational development of effective cancer immunotherapy, and tumor classifications based on immunological profiling might serve as resources for invigorating targeted studies.22, 23, 24 In the field of gliomas, Wu et al.24 classified adult LGG patients into three groups based on the immune microenvironment, and the newly classified subgroups were correlated with distinct somatic alterations and patient prognosis. Recently, Plant et al.25 characterized the immune microenvironment of 22 pediatric brain tumors, including 13 PGs using flow cytometry and 59 PGs using immunohistochemistry. Despite this study, however, the understanding of the immune microenvironment and phenotype of PGs and their clinical and molecular relevance is still very limited.
In the current study, we classified PGs into 3 distinct subtypes based on the unsupervised consensus clustering of the immune-related transcriptome profiles of 384 tumor samples. We then proved the stability and reproducibility of this immune classification in an independent cohort of 111 PGs. Each subtype of PGs was demonstrated to be associated with distinct cellular and molecular features, including intratumoral microenvironment patterns, molecular pathways, genomic alterations, and immune checkpoint molecule (ICM) expression, as well as variations in patient survival and potential responsiveness to immunotherapy. Furthermore, candidate drugs and potential targeted mechanisms were predicted for PGs of each subtype. Finally, we constructed three formulas using 8 significant predictive genes to determine a PG classification for utility in clinical practice. The identification of immunological profile-based subtypes may facilitate the optimal selection of patients who are responsive to immunotherapeutic treatments and promote the further design of targeted immunotherapy.
Results
Three stable immune subtypes (ISs) of PGs were identified and correlated with patient OS, tumor histology, and WHO grade
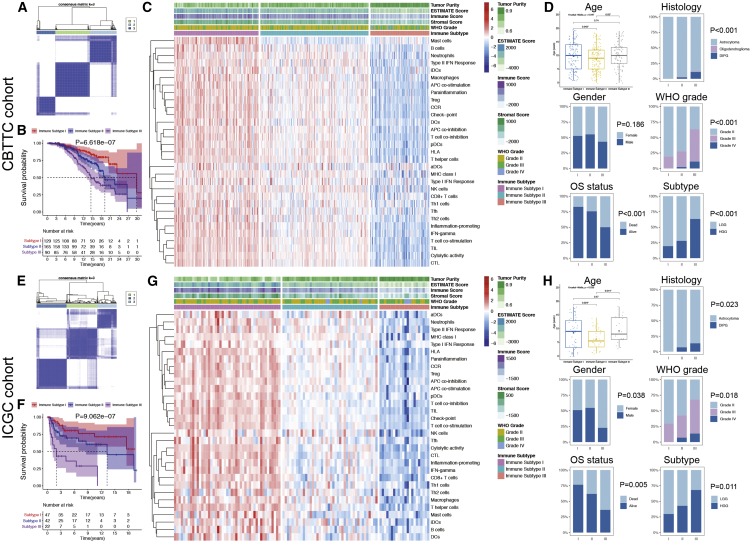
First, the single-sample gene set enrichment analysis (ssGSEA) enrichment levels of 31 immune metagene sets in each PG were quantified as immunoscores to represent the overall immune activity. The immunoscore is a robust and standardized scoring system used to quantify intratumoral immune activity and has been proven to be a reliable prognosis-predicting tool.26 Then, unsupervised consensus clustering of the immunoscores was performed, and the 384 PGs from the Children’s Brain Tumor Tissue Consortium (CBTTC) training cohort were divided into 3 clusters (Figure 1A). The cluster of gliomas with the highest immunoscores was defined as IS-I (n = 129, 33.6%), consisting of “immune-hot” tumors; the cluster with the lowest immunoscores was defined as IS-III (n = 90, 23.4%), consisting of “immune-cold” tumors; and the cluster with intermediate immunoscores was defined as IS-II (n = 165, 43.0%), consisting of “immune-altered” tumors (Figure 1C).
Figure 1.
Unsupervised consensus clustering of pediatric gliomas (PGs) based on 31 immune signatures and clinicopathological relevance
(A and E) Consensus matrix for k = 3, which was the optimal cluster number in the CBTTC training cohort (A) and ICGC validation cohort (E). (B and F) Kaplan-Meier survival analyses of patients with PGs of different immune subtypes (ISs) in the CBTTC training cohort (B) and ICGC validation cohort (F), indicating distinct clinical outcomes. (C and G) Heatmaps of the 31 immune signatures in the CBTTC (C) and ICGC (G) cohorts, with red indicating high immunoscores and blue indicating low immunoscores. Subtype I represented immune-hot tumors, subtype II represented immune-altered tumors, and subtype III represented immune-cold tumors. (D and H) Comparisons of clinicopathological variables among tumors of the three ISs in the CBTTC (D) and ICGC (H) cohorts.
Kaplan-Meier survival analysis indicated that PGs of different subtypes were correlated with distinct clinical outcomes (Figure 1B). Patients with IS-I tumors showed the best OS, whereas those with IS-III tumors showed the worst OS (p = 6.618 × 10−7). The median survival time of patients with IS-I, IS-II, and IS-III tumors was 29.8, 19.2, and 14.5 years, respectively.
In the International Cancer Genome Consortium (ICGC) validation cohort, unsupervised consensus clustering was performed, and the optimal number of clusters was also determined to be 3 (Figure 1E). All patients were classified as follows: IS-I, n = 47, 42.3%; IS-II, n = 42, 37.8%; and IS-III, n = 22, 19.8% (Figure 1G). Kaplan-Meier survival analysis also indicated that patients with IS-I tumors had the best OS, whereas those with IS-III tumors had the worst OS (p = 9.062 × 10−7; Figure 1F). The median survival time of patients with IS-I, IS-II, and IS-III tumors was >18, 13.3, and 1.99 years, respectively. These findings suggest that this classification based on intratumoral immune activity is robust and reliable in different patient populations and that greater immune activity in PG is correlated with more favorable clinical outcomes.
Subsequently, clinicopathological variables were compared among the three ISs (Table 1). Patients with IS-II tumors were the youngest at diagnosis, but there was no significant difference in age between patients with IS-I and IS-III tumors (Figures 1D and 1H). The rate of mortality (up to the time of this study), diffuse intrinsic pontine gliomas (DIPGs), and HGGs increased from IS-I to IS-III, as shown in both the training and validation cohorts (Figures 1D and 1H).
Table 1.
Demographics and clinicopathological characteristics of 495 patients with pediatric gliomas
| Variables | CBTTC cohort |
ICGC cohort |
||||||
|---|---|---|---|---|---|---|---|---|
| Total (n = 384) | IS-I (n = 129) | IS-II (n = 165) | IS-III (n = 90) | Total (n = 111) | IS-I (n = 47) | IS-II (n = 42) | IS-III (n = 22) | |
| Age at diagnosis | 9.3 ± 4.8 | 9.8 ± 5.1 | 8.7 ± 4.6 | 9.9 ± 4.7 | 8.2 ± 3.5 | 8.5 ± 5.7 | 6.2 ± 3.7 | 9.2 ± 4.6 |
| Gender | ||||||||
| Female | 186 | 61 | 74 | 51 | 59 | 23 | 19 | 17 |
| Male | 198 | 68 | 91 | 39 | 52 | 24 | 23 | 5 |
| OS status | ||||||||
| Dead | 107 | 22 | 40 | 45 | 41 | 11 | 16 | 14 |
| Alive | 277 | 107 | 125 | 45 | 70 | 36 | 26 | 8 |
| Histology | ||||||||
| Astrocytoma | 368 | 129 | 160 | 79 | 105 | 47 | 39 | 19 |
| Oligodendroglioma | 2 | 0 | 1 | 1 | 0 | 0 | 0 | 0 |
| DIPG | 14 | 0 | 4 | 10 | 6 | 0 | 3 | 3 |
| WHO grade | ||||||||
| Grade II | 256 | 104 | 119 | 33 | 64 | 33 | 24 | 7 |
| Grade III | 114 | 25 | 42 | 47 | 41 | 14 | 15 | 12 |
| Grade IV | 14 | 0 | 4 | 10 | 6 | 0 | 3 | 3 |
| Subtype | ||||||||
| LGG | 256 | 104 | 119 | 33 | 64 | 33 | 24 | 7 |
| HGG | 128 | 25 | 46 | 57 | 47 | 14 | 18 | 15 |
CBTTC, Children’s Brain Tumor Tissue Consortium; ICGC, International Cancer Genome Consortium; IS, immune subtype; OS, overall survival; DIPG, diffuse intrinsic pontine glioma; LGG, low-grade glioma; HGG, high-grade glioma.
PGs of different ISs had distinct microenvironments
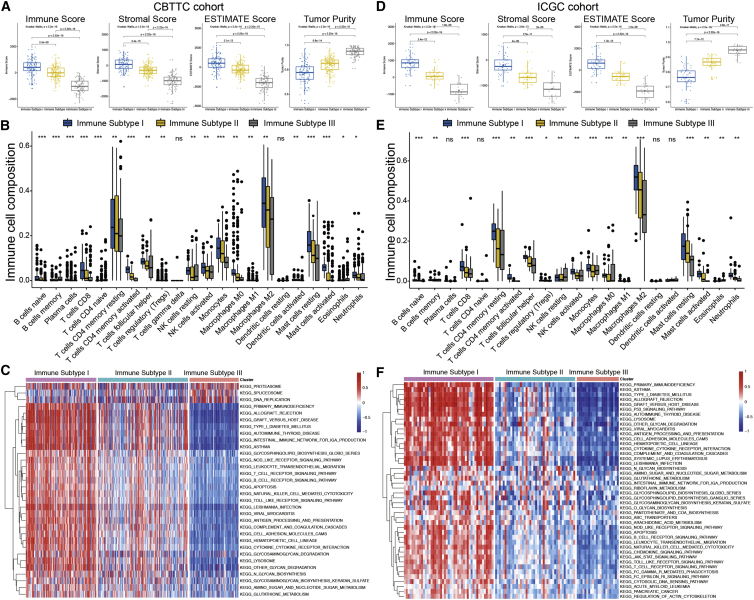
Estimation of Stromal and Immune Cells in Malignant Tumor Tissues using Expression Data (ESTIMATE) was performed to reveal the composite tumor microenvironment (TME) of PGs. As evaluated in both the CBTTC (Figure 2A) and ICGC (Figure 2D) cohorts, the immune and stromal scores were both significantly higher in IS-I tumors, indicating a high abundance of immune and stromal cells, and lower in IS-III tumors, indicating a small fraction of immune and stromal cells. In contrast, the ESTIMATE score decreased, whereas the tumor purity score increased from IS-I to IS-III (p < 2.2 × 10−16), indicating a high purity in IS-III tumors and a low purity in IS-I tumors. CIBERSORT was then applied to quantify in detail the abundance of 22 types of tumor-infiltrating immune cells (TIICs) in PGs. M2-type macrophages, CD4 memory resting T cells, and resting mast cells were among the most common cells in the PG microenvironment. The majority of TIICs were more abundant in IS-I tumors but less abundant in IS-III tumors, as validated in both the training (Figure 2B) and validation (Figure 2E) cohorts.
Figure 2.
Tumor microenvironment features and functional annotations of the three ISs
(A and D) Comparisons of tumor purity and the immune, stromal, and ESTIMATE scores among the three ISs in the CBTTC (A) and ICGC (D) cohorts. (B and E) Comparisons of 22 types of immune cells estimated by the CIBERSORT algorithm among the three ISs in the CBTTC (B) and ICGC (E) cohorts. (C and F) Heatmaps of the enrichment scores of critical molecular pathways of each IS by GSVA in the CBTTC (C) and ICGC (F) cohorts. Red represents high enrichment scores, and blue represents low enrichment scores.
ISs were correlated with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways
Gene set variation analysis (GSVA) was performed to explore the pathways and molecular mechanisms related to the immune classification of PGs. The enrichment scores of KEGG pathways of each IS were assessed. In the training cohort, 32 differentially enriched KEGG pathways were identified, including 29 pathways positively correlated with IS-I tumors and three positively correlated with IS-III tumors (Figure 2C). In the validation cohort, 44 differentially enriched pathways were identified, which were all positively correlated with IS-I tumors (Figure 2F). Finally, 28 KEGG pathways were determined as the most differentially enriched molecular pathways shared by both datasets. The GSVA results of PGs revealed positive correlations between elevated immune activity and molecular pathways related to tumorigenesis (e.g., apoptosis), metabolism (e.g., N-glycan biosynthesis), and immune responses (e.g., T/B cell receptor signaling pathway).
PGs of different ISs had distinct types and loads of somatic mutations and copy number alterations (CNAs)
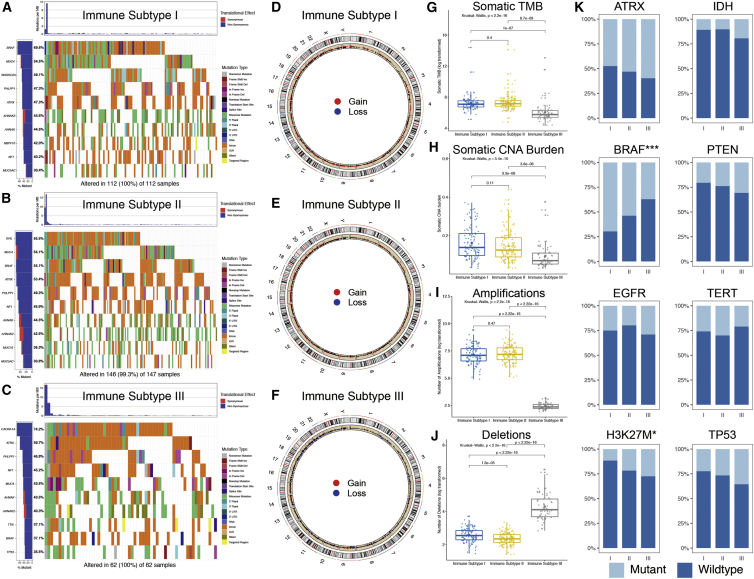
As shown in Figures 3A−3C, each IS had its specific top mutated gene, e.g., BRAF (69.6%) in IS-I tumors, SVIL (55.5%) in IS-II tumors, and CACNA1A (74.2%) in IS-III tumors. Moreover, MUC4, ATRX, PHLPP1, NF1, AHNAK, and AHNAK2 were among the top 10 mutated genes of PGs detected among all subtypes. IS-III tumors showed a lower somatic tumor mutation burden (TMB) than IS-I (p = 1.0 × 10−7) and IS-II (p = 8.7 × 10−9) tumors, whereas no significant difference was found between IS-I and IS-II tumors (p = 0.40; Figure 3G). In addition, regarding the representative molecular biomarkers for gliomas, the mutation frequency of BRAF showed a declining trend (p < 0.001), whereas that of H3K27M showed an increasing trend (p = 0.024) from IS-I to IS-III. No significant difference was found regarding the mutation frequencies of ATRX, EGFR, IDH, PTEN, TERT, or TP53 among the different subtypes (Figure 3K).
Figure 3.
Somatic variations of the three ISs
(A−C) Waterfall plots showing the top 10 mutated genes of each IS. (D−F) Circos plots of each IS revealing the gains and losses of chromosomes, with red dots representing gains, blue dots representing losses, and black dots representing no significant CNA. (G−J) Comparisons of the TMB, CNA burden, and number of amplifications and deletions among three ISs. (K) Mutation status of the eight representative molecular biomarkers among the ISs.
In terms of CNAs, as shown in Figures 3D−3F, IS-I and IS-II tumors tended to bear more gains and fewer losses, whereas IS-III tumors tended to bear more losses and fewer gains. IS-III tumors showed the lowest CNA burdens, fewest amplifications, and most deletions compared with IS-I and IS-II tumors (Figures 3H−3J).
Different ISs had varied immune checkpoint expression levels and immunotherapy responsiveness
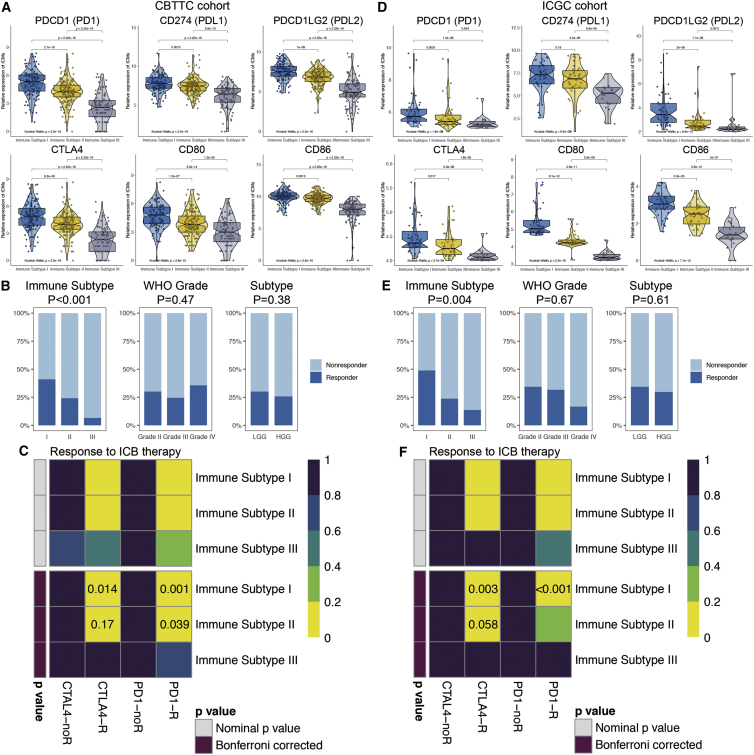
The expression levels of six ICMs, including PDCD1 (PD1), CD274 (PDL1), PDCD1LG2 (PDL2), CTLA4, CD80, and CD86, showed a descending trend among tumors from IS-I to IS-III as estimated in both the CBTTC (Figure 4A) and ICGC (Figure 4D) cohorts. Tumor Immune Dysfunction and Exclusion (TIDE) was used to predict the likelihood of a PG responding to immunotherapy. As shown in the training set (Figure 4B), the proportion of a response to immunotherapy was highest among IS-I tumors (41.1%), followed by IS-II tumors (24.2%) and IS-III tumors (6.7%) (p < 0.001). Similarly, IS-I tumors in the ICGC validation set (48.9%) were also more likely to respond to immunotherapy than IS-II (23.8%) and IS-III tumors (13.6%) (p = 0.004; Figure 4E). However, no significant differences were observed in the response rate among tumors of different WHO grades or between LGGs and HGGs (Figures 4B and 4E).
Figure 4.
Expression of immune checkpoint molecules (ICMs) and predictions of immunotherapy responsiveness of PGs
(A and D) Expression levels of six ICMs among ISs in both the CBTTC (A) and ICGC (D) cohorts. (B and E) Comparisons of the proportion of responders to immunotherapy among different subgroups, including the ISs and WHO grades in both the training (B) and validation (E) sets. (C and F) Subclass mapping analysis for predicting the likelihood of a response to ICB therapy for the ISs in the CBTTC (C) and ICGC (F) cohorts. R, response to ICB (Bonferroni corrected p value < 0.05); noR, no response to ICB (Bonferroni corrected p value > 0.05).
Subclass mapping analysis was applied to predict the responsiveness to ICB therapy of PGs among different ISs. As shown in Figures 4C and 4F, IS-I tumors were the most sensitive to anti-PD1 and anti-CTLA4 therapy (p < 0.05). IS-I tumors were likely to respond to PD1 inhibitors in the CBTTC cohort (p = 0.039), whereas this was not verified in the ICGC cohort (p = 0.21). Additionally, IS-III tumors, immune-cold tumors, tended to be resistant to ICB therapy, as shown in both cohorts (p > 0.05).
Candidate drugs and modes of action (MoAs) for PGs of each IS
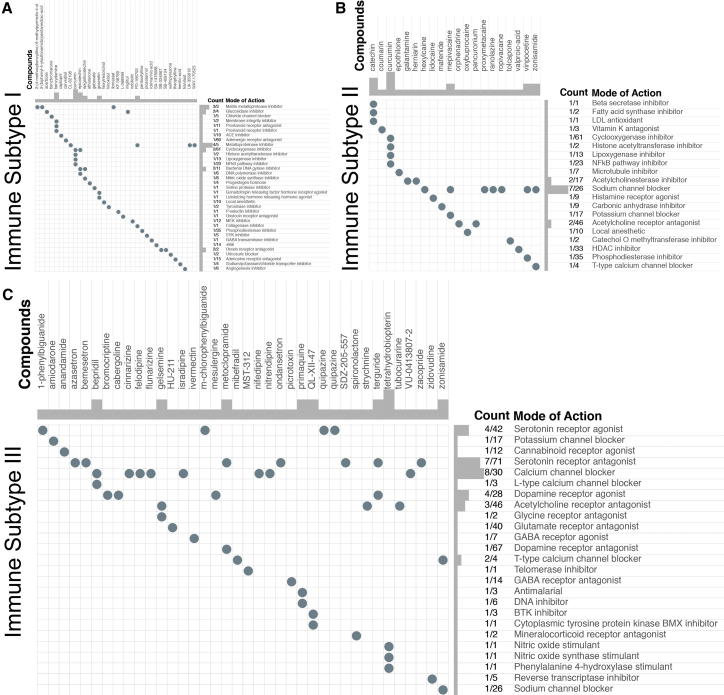
Following fold-change (FC) analysis and the Kruskal-Wallis test, 245, 154, and 327 genes were determined as representative genes for IS-I, -II, and -III tumors, respectively (Table S2). Then, Connectivity Map analysis was applied to explore the potential compounds targeting the molecular pathways and representative genes of each IS. MoA analysis revealed 35 molecular pathways targeted by 34 compounds in IS-I (Figure 5A), 20 pathways targeted by 20 compounds in IS-II (Figure 5B), and 25 pathways targeted by 38 compounds in IS-III (Figure 5C). Regarding the most critical MoAs for each subtype, there were 4 compounds sharing the same MoA as metalloproteinase inhibitors in IS-I, 7 compounds sharing the same MoA as sodium channel blockers in IS-II, and 8 compounds sharing the same MoA as calcium channel blockers in IS-III. Notably, there were no intersections in drugs and corresponding MoAs among the three subtypes of PGs.
Figure 5.
Exploration of candidate drugs and molecular pathways for each IS by Connectivity Map analysis
(A–C) MoA analysis revealed 35 molecular pathways targeted by 34 compounds in IS-I (A), 20 pathways targeted by 20 compounds in IS-II (B), and 25 pathways targeted by 38 compounds in IS-III (C).
Formulas and representative genes for identifying ISs of PGs
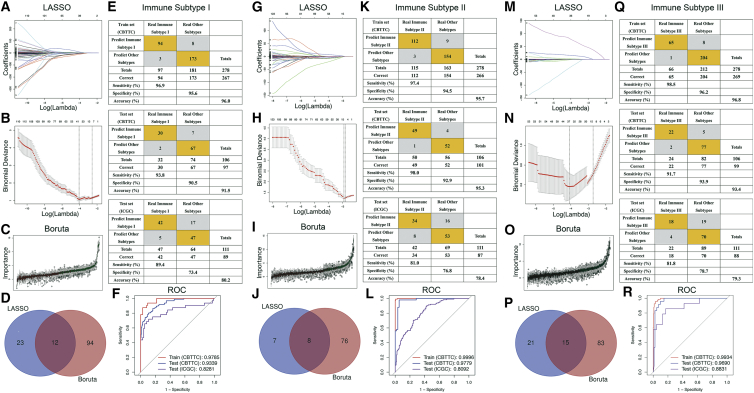
The most representative genes of each IS were used to identify IS-related features using machine learning methods. A total of 35 genes and 106 genes were identified by least absolute shrinkage and selection operator (LASSO), binomial deviance, and Boruta analyses, respectively (Figures 6A−6C), and the 12 genes shared by both methods were determined as the specific features of IS-I tumors (Figure 6D). Similarly, 8 genes (Figure 6J) and 15 genes (Figure 6P) were determined to be specific features of IS-II and IS-III tumors. Following univariate and multivariate analyses, 8 significant genes were selected to construct formulas for calculating predictor scores of ISs (Table 2).
Figure 6.
Construction and evaluation of three IS predictors based on the most representative genes of each subtype
(A−F) IS-I predictors. (G−L) IS-II predictors. (M−R) IS-III predictors. LASSO regression analysis (A, G, and M): coefficient values at varying levels of penalty. Each curve represents a gene. (B, H, and N) Ten-fold cross-validation was used to calculate the best lambda, contributing to the minimum mean cross-validated error (cvm). Red dots represent partial likelihood deviance; solid vertical lines indicate the corresponding 95% confidence interval (95% CI); the left dotted vertical line is the value of lambda that gives the minimum cvm; and the right dotted vertical line is the largest value such that error is within 1 standard error of the minimum. (C, I, and O) Importance plot of the genes. Green boxes represent important features (retained), and red boxes represent unimportant features (declined). (D, J, and P) Venn diagram identifying the most critical IS-specific variables that were shared by the LASSO and Boruta methods. (E, K, and Q) Confusion matrices of binary results of the IS predictors for the training set (CBTTC), CBTTC test set, and ICGC test set. (F, L, and R) ROC curves of the IS predictors for distinguishing each subtype and other subtypes in the training set and two test sets.
Table 2.
Univariate and multivariate logistic regression analyses for identifying risk factors and IS predictors for pediatric gliomas
| Variables | Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|---|
| Β | OR (95% CI) | p value | β | OR (95% CI) | p value | ||
| IS-I | |||||||
| Intercept | 1.34 | 3.81 (1.91−8.37) | 3.73e−04a | ||||
| AC060834.2 | 0.73 | 2.09 (1.01−12.10) | 0.24 | ||||
| AC113418.1 | −1.46 | 0.29 (0.10−2.37) | 0.28 | ||||
| AL353747.4 | −2.99 | 0.14 (0.08−5.39) | 0.17 | ||||
| CCL18 | 1.84 | 1.53 (1.32−1.73) | 0.041a | 4.87 | 1.34 (1.13−1.55) | 1.57e−03a | |
| FOXR2 | −1.46 | 0.46 (0.11−3.78) | 0.10 | ||||
| GTF3AP6 | −3.26 | 0.79 (0.24−1.35) | 0.39 | ||||
| IGHEP1 | 6.86 | 9.55 (1.48−1,842) | 0.88 | ||||
| IGKV2-29 | 2.84 | 17.10 (1.46−605) | 0.16 | ||||
| IGKV3-15 | 0.87 | 1.09 (0.19−15.6) | 0.95 | ||||
| IGLV2-11 | 0.29 | 1.34 (0.54−3.69) | 0.55 | ||||
| IGKV3D-20 | 0.38 | 1.46 (0.68−5.96) | 0.46 | ||||
| SIGLEC6 | 1.49 | 3.16 (2.38−3.93) | 3.48e−06a | 11.19 | 7.31 (4.90−9.52) | 1.05e−09a | |
| IS-II | |||||||
| Intercept | −1.59 | 0.20 (0.10−0.37) | 1.49e−06a | ||||
| POU4F1 | −0.65 | 0.53 (0.03−1.01) | 0.15 | ||||
| IGFN1 | −0.93 | 0.41 (0.32−0.49) | 0.033a | −1.27 | 0.28 (0.09−0.58) | 5.30e−03a | |
| IGHV3-7 | −0.70 | 0.79 (0.43−1.16) | 0.38 | ||||
| IGKV3-15 | −0.20 | 0.98 (0.67−2.19) | 0.98 | ||||
| SIGLEC6 | −7.01 | 0.57 (0.41−0.76) | 2.49e−04a | −7.23 | 0.72 (0.62−0.88) | 8.37e−06a | |
| WDR72 | −6.37 | 0.90 (0.55−1.27) | 0.12 | ||||
| AL020995.1 | 2.76 | 1.58 (0.63−7.59) | 0.11 | ||||
| GTF3AP6 | −2.61 | 0.84 (0.65−1.10) | 0.86 | ||||
| IS-III | |||||||
| Intercept | −1.17 | 0.38 (0.11−0.62) | 1.64e−10a | ||||
| POU4F1 | 1.06 | 2.88 (0.33−3.07) | 0.37 | ||||
| AL354863.1 | 1.37 | 3.02 (0.12−34.81) | 0.72 | ||||
| GLOD5 | −4.34 | 0.12 (0.01−3.96) | 0.35 | ||||
| DNAJC5G | −0.43 | 0.64 (0.09−20.07) | 0.69 | ||||
| CD3D | −5.34 | 0.48 (0.06−14.08) | 0.23 | ||||
| GZMH | −5.04 | 0.65 (0.53−0.77) | 0.012a | −3.07 | 0.46 (0.11−0.68) | 0.036a | |
| CD2 | −5.93 | 0.26 (0.21−0.42) | 0.023a | −6.08 | 0.23 (0.15−0.31) | 0.021a | |
| HLA-DQA1 | −4.31 | 0.44 (0.30−0.79) | 0.027a | −4.26 | 0.14 (0.08−0.21) | 4.42e−03a | |
| KRT34 | 7.23 | 1.38 (0.33−2.61) | 0.64 | ||||
| NEUROD6 | 3.95 | 5.19 (0.02−43.34) | 0.50 | ||||
| GTF3AP6 | 1.04 | 1.95 (0.77−3.45) | 0.99 | ||||
| FCER1A | −4.82 | 0.81 (0.73−0.91) | 0.003a | −3.88 | 0.21 (0.09−0.36) | 0.010a | |
| C7 | −0.20 | 0.82 (0.36−1.13) | 0.73 | ||||
| CLEC12A | −0.25 | 0.77 (0.01−16.93) | 0.89 | ||||
| MAGEA11 | 2.13 | 1.40 (0.60−7.45) | 0.86 | ||||
β, regression coefficient; OR, odds ratio; 95% CI, 95% confidence interval.
p < 0.05.
The formulas for each subgroup were as follows: IS-I predictor score = 1.34 + 4.87 × (expression of CCL18) + 11.19 × (expression of SIGLEC6); IS-II predictor score = −1.59 − 1.27 × (expression of IGFN1) − 7.23 × (expression of SIGLEC6); and IS-III predictor score = −1.17 − 3.07 × (expression of GZMH) − 6.08 × (expression of CD2) − 4.26 × (expression of HLA-DQA1) − 3.88 × (expression of FCER1A). In the training set (CBTTC), the IS-I predictor formula yielded an area under the curve (AUC) of 0.979 for distinguishing IS-I tumors from others (Figures 6E and 6F), and patients with an IS-I predictor score of >45.1 (optimal cutoff value) were considered to have IS-I tumors. In addition, the IS-I predictor formula also had an AUC of 0.934 and 0.828 in the CBTTC test set and the ICGC test set, respectively (Figures 6E and 6F). Similarly, the IS-II predictor formula yielded an AUC of 0.9996 in the training set (CBTTC), and patients with an IS-II predictor score of >−40.1 were considered to have IS-II tumors (Figures 6K and 6L); the IS-III predictor formula demonstrated an AUC of 0.993, and patients with an IS-III predictor score of >−3901.6 were considered to have IS-III tumors (Figures 6Q and 6R). Moreover, both the IS-II and IS-III predictor formulas indicated excellent performance in discriminating tumor subtypes as evaluated in the test sets.
Discussion
Recently, several preliminary studies and ongoing trials of immunotherapies have demonstrated good tolerance and clinical benefits for PGs, especially for progressive pHGGs and recurrent pLGGs resistant to conventional treatments and associated with uniquely poor outcomes.14, 15, 16 Further, prospective phase 2 or 3 trials to prove the overall population coverage and long-term efficacy of immunotherapy are promising but challenging.9 Characterization of the intratumoral immune microenvironment serves as an initial and critical step for the design of successful antitumor immunotherapies.22,27 Herein, we characterized the immunological profiles of PGs and classified the tumors into three ISs accordingly. Each subtype was associated with distinct TME patterns, molecular pathways, somatic alterations, patient outcomes, and immunotherapy responsiveness. These results were reliable and validated in another PG cohort. This study adds to our understanding of the immune microenvironment of PGs and provides implications for further research on optimal immunotherapy strategies.
This classification system demonstrated perfect discrimination in predicting OS. IS-I tumors conferred the most favorable prognosis, whereas IS-III tumors had the worst clinical outcomes. Different from the conclusion drawn by Plant et al.25 from a small cohort that the immunophenotype of pediatric brain tumors had no correlation with patient survival, most studies have revealed that the density of TIICs is positively associated with prognosis in other types of cancer.28, 29, 30 Our result is in agreement with the majority of findings in the literature, indicating that immune-hot PGs predicted a favorable prognosis, whereas immune-cold PGs predicted a dismal prognosis.
Regarding the composition of the intratumoral immune microenvironment, our results revealed that the major immune infiltrates in PGs were macrophages and T cells, consistent with the TIIC patterns reported in adult gliomas.24,31 Furthermore, the fractions of different immune cells showed similar trends but distinct densities among PG subtypes, with IS-I immune-hot PGs having the highest fractions of almost all types of TIICs, followed by IS-II tumors, and IS-III PGs having the lowest TIIC fractions.
Nontumor cells, including stromal cells and immune cells, in the TME dilute the purity of tumors, and an immune-hot status is always correlated with low tumor purity.32 Recently, Zhang et al.33 characterized the purity of adult gliomas and demonstrated that low-purity gliomas were correlated with malignant progression and poor prognosis. However, our results in the context of PGs are in contrast with this conclusion, based on tumors in adults. As shown in our study, immune-hot PGs showed low tumor purity, high immune activity, and favorable clinical outcomes, whereas immune-cold PGs revealed high tumor purity, low immune activity, and dismal prognosis. One explanation might be that although low-purity adult gliomas had intensive immune infiltration (immune hot), they were “immune suppressive” with considerable proportions of regulatory T cells and tumorigenic M2-type macrophages and high levels of immunosuppressive ICMs,24,31,34,35 whereas low-purity PGs were both quantitatively immune hot and functionally immune active, as validated in this study.
The increasing proportions of DIPGs and pHGGs in the IS-III subtype paralleled the decreasing OS and intratumoral immune activity. DIPGs are the most devastating pediatric brain tumors, with a median OS of only 9−11 months.36 As shown in this study, all DIPGs were classified into IS-II and IS-III subtypes, with more classified into the latter, showing lower immune infiltration and fewer available ICMs to be targeted in DIPGs, in agreement with the conclusion in another study reached using flow cytometry and transcriptome analysis.37 The study by Lin et al.38 also conveyed a similar conclusion that DIPG contained few T cells and that intratumoral macrophages were relatively noninflammatory, indicating low antitumor immune activity. In terms of the WHO grade, one study with a small sample revealed that the immune classification was not correlated with the grade of pediatric brain tumors.25 However, our results show that pHGGs were more likely than pLGGs to be classified as IS-II or IS-III tumors, exhibiting low immune activity. This finding suggests a disappointing result for pHGGs and DIPGs, i.e., the relatively low immune infiltration and ICM expression might increase the difficulty of identifying successful alternative treatment targets. Thus, we propose that the histology-specific immune microenvironment needs to be taken into serious consideration when designing immunotherapies for PGs.
Mutation heterogeneity and high CNA loads are correlated with neoantigen abundance, strong immune infiltration, and favorable immunotherapy responses, as observed in several types of tumors.22,23,39, 40, 41, 42, 43 In contrast to adult gliomas, characterized by IDH1/2 mutation, EGFR amplification, TERT promoter mutation, etc., PGs have unique genomic alteration patterns.4,5,7 In pLGGs, genetic hits mostly occur in mitogen-activated protein kinase (MAPK) pathways, including BRAF fusion, neurotrophic receptor tyrosine kinase (NTRK)-family fusion, BRAFV600E mutation, FGFR1 mutation, and NF1 mutation,5,44 whereas in pHGGs, common genomic alterations include mutations in K27M, G34R/V, TP53, MYC, ATRX, FGFR1, and ACVR1.4,8 In this study, BRAF, SVIL, and CACNA1A were the most mutated genes in the IS-I, IS-II, and IS-III subtypes, respectively, and MUC4, ATRX, PHLPP1, NF1, AHNAK, and AHNAK2 were other top mutated genes among all subtypes of PGs. Moreover, CNA and TMB were found to be positively correlated with the immune activity of PGs. The immune-cold PGs were shown to have lower burdens of mutations and CNAs, whereas immune-hot and immune-altered tumors had higher genomic alteration burdens, implying as a whole that PGs with intense mutation and CNA burdens were more likely to have robust immune infiltration, abundant ICM expression, a favorable prognosis, and strong antitumor reactions to neoantigens. In addition, the substantial enrichment of alterations in MAPK pathways might also contribute to a better prognosis for patients with immune-hot PGs.45 Our results differ from those based on adult gliomas showing that the mutation burden was correlated with low immune infiltration24,46 but are consistent with those of a study on PGs revealing that hypermutated tumors had greater lymphocyte infiltration.10 Detailed analysis and recognition of IS-specific neoplastic antigens caused by gene mutations and CNAs are suggested to promote the design of CAR-T cell and vaccination therapies for PGs.
In comparison to the confusing two-sided immune microenvironment patterns of adult gliomas that were immune hot but immune suppressive,24,34,47 PGs of the same subtype showed identical immune patterns as both immune hot and immune active or both immune cold and immune suppressive. As shown in our study, immune-hot PGs expressed the highest levels of ICMs and were more likely to respond to anti-PD1 and anti-CTLA4 therapies, whereas IS-III immune-cold tumors exhibited the lowest levels of ICMs and were unlikely to respond to ICM-targeted treatments. This phenomenon increases the difficulty of ongoing immunotherapy research, especially the selection of immune targets for refractory PGs since more pHGGs and DIPGs, which are in urgent need of promising immunotherapies,8,10,36,38 were classified as IS-III.
The expression of ICMs serves as a potential tool in predicting responsiveness to ICB therapy.48 pHGGs have lower expression levels of PD1 than atypical teratoid/rhabdoid tumors and ependymomas.49 Responsiveness to immunotherapies was predicted in this study among different subtypes of PGs using TIDE.50 There existed a significant descending trend in the ICM expression levels and response rates from immune-hot to immune-cold tumors (pHGG proportion from low to high), indicating low immune infiltration and ICM expression correlated negatively with immunotherapy responsiveness. Different ISs of tumors respond differently to ICB therapy.23,24 According to our prediction, IS-I PGs could respond to both anti-PD1 and anti-CTLA4 therapies, IS-II PGs might respond to anti-PD1 therapy but not anti-CTLA4 therapies, whereas IS-III PGs were unlikely to have a favorable response to immunotherapy. The WHO grading system has been used to predict prognosis for gliomas.51 However, this system might not be proper for an immunotherapy response prediction for PGs. The intratumoral immune microenvironment and phenotype, but not grade, need to be underlined when predicting the response to immunologic treatments in PGs.
The Connectivity Map, a pharmacogenome-based tool, was used to explore the potential drugs targeting immunophenotype-associated molecular pathways and genes and to understand the potential MoAs.52 The process of exploring candidate drugs and corresponding mechanisms could provide novel therapeutic options in addition to immunotherapy for PGs. Notably, no intersections in drugs and MoAs were found among the subtypes, suggesting that PGs of different ISs might have different favorable chemotherapy regimens.
To standardize the process of discriminating the ISs of PGs, predictive formulas were constructed and validated using the 8 most representative genes. With sequencing data on these genes, physicians could easily classify a resected PG into one subtype using the formulas and thus obtain related information on its TME pattern, intratumoral immune activity, predicted OS, and immunotherapy responsiveness.
In conclusion, the current study classified PGs from an immune perspective and proposed three subtypes: immune-hot (IS-I), immune-altered (IS-II), and immune-cold (IS-III) tumors. IS-I tumors had substantial immune infiltration, high ICM expression, a favorable prognosis, and proper responsiveness to immunotherapy; this subtype comprised more pLGGs and no DIPGs. IS-III tumors exhibited weak immune infiltration, low ICM expression, a dismal outcome, and poor responsiveness to immunotherapy; this subtype consisted of large fractions of pHGGs and DIPGs. IS-II tumors represented a transitional stage. Moreover, the immune classification was correlated with somatic mutations, CNAs, and several molecular pathways related to tumorigenesis, metabolism, and immune response. With reliable predictive formulas, the IS of PGs could be identified and thus assist in the further design of successful immunotherapies and the stratification of patients responsive to targeted therapies.
Materials and methods
Patient population and genomic data acquisition
Level 3 RNA sequencing data of 561 PGs and the corresponding clinical and follow-up information were downloaded from the CBTTC (https://cbttc.org/) and the ICGC (https://dcc.icgc.org/). Thirteen patients from the CBTTC and 53 from the ICGC were excluded due to lack of prognostic data or patient age at diagnosis >19 years. Finally, 384 patients with intact clinical data from the CBTTC dataset were enrolled and set as the training cohort, and 111 patients from the ICGC dataset were set as the validation cohort. In addition, somatic mutation and CNA data of 321 PGs from the CBTTC dataset were also included.
Informed consent was obtained from all participants by CBTTC and ICGC member institutions. This study was approved by the Institutional Ethics Committee of Peking Union Medical College Hospital in accordance with the ethical standards of the Institutional Ethics Committee and with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Immunoscores and unsupervised consensus clustering of PGs
Figure S1 presents the overall workflow of the present study. We obtained 31 immune metagene sets that represented the immune activity of the PGs based on the types, functions, and molecular pathways of the TIICs (Table S1).23 By applying the ssGSEA algorithm, the enrichment levels of the 31 immune signatures of each tumor were quantified as the immunoscores based on the expression profiles of the samples.53 Then, unsupervised consensus clustering was applied to explore a novel immune classification for PGs based on the immunoscores using the ConsensusClusterPlus package in R. The clustering procedure with 1,000 iterations was performed by sampling 80% of the data in each iteration. The optimal number of clusters was determined by the relative change in the area under the cumulative distribution function (CDF) curve, the proportion of ambiguous clustering algorithm, and the consensus heatmap.54 Hence, ISs with different immunoscores could demonstrate distinct immune characteristics. Kaplan-Meier survival analysis was performed to evaluate the OS of patients with tumors of different ISs.
TME patterns of PGs
PG TME patterns were estimated using the ESTIMATE and CIBERSORT algorithms. ESTIMATE was used for predicting tumor purity and the abundance of intratumoral stromal cells and immune cells based on the gene-expression profile.32 The algorithm can generate four scores: the immune score (positively reflecting the abundance of TIICs), the stromal score (positively reflecting the abundance of stromal cells), the ESTIMATE score (negatively reflecting tumor purity), and the tumor purity score (positively reflecting tumor purity). Then, the CIBERSORT algorithm was applied to quantify in detail the composition of 22 types of TIICs based on the gene-expression profile.55
Functional annotations of PG ISs
GSVA, an unsupervised gene set enrichment method, was applied to estimate the enrichment scores of KEGG pathways of each PG based on the gene-expression data using the GSVA package in R.39,53 Then, differential analysis of the enrichment scores of KEGG pathways among different ISs was performed using the limma package. FC analysis and the Kruskal-Wallis test were performed to determine the most differentially enriched KEGG pathways of each IS. KEGG pathways with |log2FC| >0.2, Benjamini-Hochberg false discovery rate (FDR) <0.05, and Kruskal-Wallis test p <0.05 were considered the most differentially enriched molecular pathways.56
Somatic mutation and CNA analysis
Since genomic alterations have been investigated to be correlated with tumor immunity and immune infiltration,22,57 we performed somatic mutation analysis and CNA analysis here to explore the distinct genomic variations of three ISs of PGs. The mutation type and frequency of the top mutated genes in each subtype were visualized by waterfall plots using the GenVisR package in R.58 The TMB was defined as the total number of nonsynonymous mutations in the coding region per megabase.59 Comparisons of the TMB among ISs were performed by the Kruskal-Wallis test.
Regarding the CNA data, loci with segment-mean values greater than 0.2 were defined as gains (amplifications), and those with values less than −0.2 were defined as losses (deletions).60 To better exhibit the gains and losses of chromosomes, CNA summary plots of each IS were visualized by circos plots using the RCircos package in R.61 The CNA burden was defined as the total number of genes with copy number changes.59 The CNA burden and number of gains and losses of chromosomes among ISs were compared by the Kruskal-Wallis test.
Prediction of immunotherapy responsiveness
The TIDE (http://tide.dfci.harvard.edu/) algorithm was developed based on the integration of the expression signatures of T cell dysfunction and T cell exclusion to model tumor immune evasion.50 The signatures of T cell dysfunction can be determined from large tumor cohorts by testing how the expression of each gene in tumors interacts with the cytotoxic T lymphocyte infiltration level to influence patient survival and response to immunotherapy. The clinical response to ICB can be predicted by the TIDE algorithm using the tumor transcriptomic profile. In addition, the unsupervised subclass mapping (https://cloud.genepattern.org/gp/pages/login.jsf) method was further applied to predict the responsiveness to ICB of different ISs in PG patients.62
Identification of the most representative genes of each subtype
FC analysis and the Kruskal-Wallis test were performed to determine the most representative genes in tumors of each IS. The differentially expressed genes (DEGs) among ISs were screened using the limma package in R. FDR < 0.05 and |log2FC| >3 were considered the cutoff criteria for determining DEGs, and subsequently, the DEGs with Kruskal-Wallis test p <0.05 were considered the most representative genes of each IS.
Connectivity Map analysis
The Connectivity Map database (https://clue.io/) was applied to explore potential compounds targeting the molecular pathways and genes associated with the IS of PGs.52 The Connectivity Map database not only predicts drugs based on gene-expression signatures but also reveals the MoA of compounds and related molecular pathways. The most representative genes of each IS were enrolled in querying the Connectivity Map database.
Construction and evaluation of predictors for IS identification by machine learning methods
In this section, the CBTTC dataset was randomly classified as the training set (n = 278) and the test set (n = 106) at a ratio of 7:3, and the ICGC dataset was selected as another test set (n = 111). LASSO regression and the Random Forest and Boruta analyses were applied to screen the most critical group-relevant features by calculating the importance score for each variable using the glmnet, rms, Random Forest, and Boruta packages in R.63,64 The expression levels of the most representative genes of each IS were selected as the input variables (independent variables), and the status of each IS (e.g., one subtype as 1 and other subtypes as 0) was selected as the outcome (binary dependent variables). Then, the genes intersecting between LASSO and Boruta analysis were considered the most critical IS-relevant variables for further analysis, which were visualized by a Venn diagram. First, univariate logistic regression analysis was performed on the most critical variables. According to the results, those genes with p <0.05 were further analyzed by multivariate logistic analysis to identify the risk factors and construct the predictors for IS identification.65 Then, the discriminative performance of the IS predictors was evaluated by receiver operating characteristic (ROC) curve analysis, and the optimal cutoff values, AUCs, sensitivity, specificity, and accuracy were determined. Subsequently, the performance of the predictors was also validated by the CBTTC and ICGC test sets in similar ways.
Acknowledgments
RNA sequencing data of all PGs were downloaded at the CBTTC database (https://cbttc.org/) and the ICGC database (https://daco.icgc.org/). Accession numbers are available on above websites. This study was supported by the Graduate Innovation Fund of the Chinese Academy of Medical Sciences and Peking Union Medical College (2019-1002-73).
Author contributions
Z.W., X.G., B.X., and W.M. conceived and designed the study. Z.W., X.G., L.G., Y.W., and Y.G. performed most of the analyses. Z.W. and X.G. drafted the manuscript. B.X. and W.M. critically revised the study. All authors have confirmed the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2020.12.012.
Contributor Information
Bing Xing, Email: xingbingemail@aliyun.com.
Wenbin Ma, Email: mawb2001@hotmail.com.
Supplemental information
References
- 1.Ostrom Q.T., de Blank P.M., Kruchko C., Petersen C.M., Liao P., Finlay J.L., Stearns D.S., Wolff J.E., Wolinsky Y., Letterio J.J., Barnholtz-Sloan J.S. Alex’s lemonade stand foundation infant and childhood primary brain and central nervous system tumors diagnosed in the United States in 2007-2011. Neuro-oncol. 2015;16(Suppl 10):x1–x36. doi: 10.1093/neuonc/nou327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Curtin S.C., Miniño A.M., Anderson R.N. Declines in cancer death rates among children and adolescents in the United States, 1999–2014. NCHS Data Brief. 2016;257:1–8. [PubMed] [Google Scholar]
- 3.Pollack I.F., Agnihotri S., Broniscer A. Childhood brain tumors: current management, biological insights, and future directions. J. Neurosurg. Pediatr. 2019;23:261–273. doi: 10.3171/2018.10.PEDS18377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sturm D., Pfister S.M., Jones D.T.W. Pediatric gliomas: current concepts on diagnosis, biology, and clinical management. J. Clin. Oncol. 2017;35:2370–2377. doi: 10.1200/JCO.2017.73.0242. [DOI] [PubMed] [Google Scholar]
- 5.Packer R.J., Pfister S., Bouffet E., Avery R., Bandopadhayay P., Bornhorst M., Bowers D.C., Ellison D., Fangusaro J., Foreman N. Pediatric low-grade gliomas: implications of the biologic era. Neuro-oncol. 2017;19:750–761. doi: 10.1093/neuonc/now209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Blank P., Bandopadhayay P., Haas-Kogan D., Fouladi M., Fangusaro J. Management of pediatric low-grade glioma. Curr. Opin. Pediatr. 2019;31:21–27. doi: 10.1097/MOP.0000000000000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braunstein S., Raleigh D., Bindra R., Mueller S., Haas-Kogan D. Pediatric high-grade glioma: current molecular landscape and therapeutic approaches. J. Neurooncol. 2017;134:541–549. doi: 10.1007/s11060-017-2393-0. [DOI] [PubMed] [Google Scholar]
- 8.Jones C., Karajannis M.A., Jones D.T.W., Kieran M.W., Monje M., Baker S.J., Becher O.J., Cho Y.J., Gupta N., Hawkins C. Pediatric high-grade glioma: biologically and clinically in need of new thinking. Neuro-oncol. 2017;19:153–161. doi: 10.1093/neuonc/now101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang S.S., Bandopadhayay P., Jenkins M.R. Towards immunotherapy for pediatric brain tumors. Trends Immunol. 2019;40:748–761. doi: 10.1016/j.it.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Mackay A., Burford A., Molinari V., Jones D.T.W., Izquierdo E., Brouwer-Visser J., Giangaspero F., Haberler C., Pietsch T., Jacques T.S. Molecular, pathological, radiological, and immune profiling of non-brainstem pediatric high-grade glioma from the HERBY phase II randomized trial. Cancer Cell. 2018;33:829–842.e5. doi: 10.1016/j.ccell.2018.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartzentruber J., Korshunov A., Liu X.Y., Jones D.T., Pfaff E., Jacob K., Sturm D., Fontebasso A.M., Quang D.A., Tönjes M. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 12.Wolchok J.D., Chiarion-Sileni V., Gonzalez R., Rutkowski P., Grob J.J., Cowey C.L., Lao C.D., Wagstaff J., Schadendorf D., Ferrucci P.F. Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med. 2017;377:1345–1356. doi: 10.1056/NEJMoa1709684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motzer R.J., Tannir N.M., McDermott D.F., Arén Frontera O., Melichar B., Choueiri T.K., Plimack E.R., Barthélémy P., Porta C., George S., CheckMate 214 Investigators Nivolumab plus ipilimumab versus sunitinib in advanced renal-cell carcinoma. N. Engl. J. Med. 2018;378:1277–1290. doi: 10.1056/NEJMoa1712126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cacciotti C., Choi J., Alexandrescu S., Zimmerman M.A., Cooney T.M., Chordas C., Clymer J., Chi S., Yeo K.K. Immune checkpoint inhibition for pediatric patients with recurrent/refractory CNS tumors: a single institution experience. J. Neurooncol. 2020;149:113–122. doi: 10.1007/s11060-020-03578-6. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed N., Brawley V., Hegde M., Bielamowicz K., Kalra M., Landi D., Robertson C., Gray T.L., Diouf O., Wakefield A. HER2-specific chimeric antigen receptor-modified virus-specific T cells for progressive glioblastoma: a phase 1 dose-escalation trial. JAMA Oncol. 2017;3:1094–1101. doi: 10.1001/jamaoncol.2017.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pollack I.F., Jakacki R.I., Butterfield L.H., Hamilton R.L., Panigrahy A., Normolle D.P., Connelly A.K., Dibridge S., Mason G., Whiteside T.L., Okada H. Immune responses and outcome after vaccination with glioma-associated antigen peptides and poly-ICLC in a pilot study for pediatric recurrent low-grade gliomas. Neuro-oncol. 2016;18:1157–1168. doi: 10.1093/neuonc/now026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bouffet E., Larouche V., Campbell B.B., Merico D., de Borja R., Aronson M., Durno C., Krueger J., Cabric V., Ramaswamy V. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J. Clin. Oncol. 2016;34:2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 18.Gorsi H.S., Malicki D.M., Barsan V., Tumblin M., Yeh-Nayre L., Milburn M., Elster J.D., Crawford J.R. Nivolumab in the treatment of recurrent or refractory pediatric brain tumors: a single institutional experience. J. Pediatr. Hematol. Oncol. 2019;41:e235–e241. doi: 10.1097/MPH.0000000000001339. [DOI] [PubMed] [Google Scholar]
- 19.Mount C.W., Majzner R.G., Sundaresh S., Arnold E.P., Kadapakkam M., Haile S., Labanieh L., Hulleman E., Woo P.J., Rietberg S.P. Potent antitumor efficacy of anti-GD2 CAR T cells in H3-K27M+ diffuse midline gliomas. Nat. Med. 2018;24:572–579. doi: 10.1038/s41591-018-0006-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollack I.F., Jakacki R.I., Butterfield L.H., Hamilton R.L., Panigrahy A., Potter D.M., Connelly A.K., Dibridge S.A., Whiteside T.L., Okada H. Antigen-specific immune responses and clinical outcome after vaccination with glioma-associated antigen peptides and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in children with newly diagnosed malignant brainstem and nonbrainstem gliomas. J. Clin. Oncol. 2014;32:2050–2058. doi: 10.1200/JCO.2013.54.0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollack I.F., Jakacki R.I., Butterfield L.H., Hamilton R.L., Panigrahy A., Normolle D.P., Connelly A.K., Dibridge S., Mason G., Whiteside T.L., Okada H. Antigen-specific immunoreactivity and clinical outcome following vaccination with glioma-associated antigen peptides in children with recurrent high-grade gliomas: results of a pilot study. J. Neurooncol. 2016;130:517–527. doi: 10.1007/s11060-016-2245-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., Cancer Genome Atlas Research Network The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He Y., Jiang Z., Chen C., Wang X. Classification of triple-negative breast cancers based on Immunogenomic profiling. J. Exp. Clin. Cancer Res. 2018;37:327. doi: 10.1186/s13046-018-1002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu F., Wang Z.L., Wang K.Y., Li G.Z., Chai R.C., Liu Y.Q., Jiang H.Y., Zhai Y., Feng Y.M., Zhao Z., Zhang W. Classification of diffuse lower-grade glioma based on immunological profiling. Mol. Oncol. 2020;14:2081–2095. doi: 10.1002/1878-0261.12707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Plant A.S., Koyama S., Sinai C., Solomon I.H., Griffin G.K., Ligon K.L., Bandopadhayay P., Betensky R., Emerson R., Dranoff G. Immunophenotyping of pediatric brain tumors: correlating immune infiltrate with histology, mutational load, and survival and assessing clonal T cell response. J. Neurooncol. 2018;137:269–278. doi: 10.1007/s11060-017-2737-9. [DOI] [PubMed] [Google Scholar]
- 26.Galon J., Bruni D. Approaches to treat immune hot, altered and cold tumours with combination immunotherapies. Nat. Rev. Drug Discov. 2019;18:197–218. doi: 10.1038/s41573-018-0007-y. [DOI] [PubMed] [Google Scholar]
- 27.Bindea G., Mlecnik B., Angell H.K., Galon J. The immune landscape of human tumors: Implications for cancer immunotherapy. OncoImmunology. 2014;3:e27456. doi: 10.4161/onci.27456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z., Liu Z., Li M., Chen C., Wang X. Immunogenomics analysis reveals that TP53 mutations inhibit tumor immunity in gastric cancer. Transl. Oncol. 2018;11:1171–1187. doi: 10.1016/j.tranon.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ali H.R., Provenzano E., Dawson S.J., Blows F.M., Liu B., Shah M., Earl H.M., Poole C.J., Hiller L., Dunn J.A. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann. Oncol. 2014;25:1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 30.Garnelo M., Tan A., Her Z., Yeong J., Lim C.J., Chen J., Lim K.H., Weber A., Chow P., Chung A. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut. 2017;66:342–351. doi: 10.1136/gutjnl-2015-310814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Z., Hambardzumyan D. Immune microenvironment in glioblastoma subtypes. Front. Immunol. 2018;9:1004. doi: 10.3389/fimmu.2018.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang C., Cheng W., Ren X., Wang Z., Liu X., Li G., Han S., Jiang T., Wu A. Tumor purity as an underlying key factor in glioma. Clin. Cancer Res. 2017;23:6279–6291. doi: 10.1158/1078-0432.CCR-16-2598. [DOI] [PubMed] [Google Scholar]
- 34.Yang I., Han S.J., Sughrue M.E., Tihan T., Parsa A.T. Immune cell infiltrate differences in pilocytic astrocytoma and glioblastoma: evidence of distinct immunological microenvironments that reflect tumor biology. J. Neurosurg. 2011;115:505–511. doi: 10.3171/2011.4.JNS101172. [DOI] [PubMed] [Google Scholar]
- 35.Bieńkowski M., Preusser M. Prognostic role of tumour-infiltrating inflammatory cells in brain tumours: literature review. Curr. Opin. Neurol. 2015;28:647–658. doi: 10.1097/WCO.0000000000000251. [DOI] [PubMed] [Google Scholar]
- 36.Vitanza N.A., Monje M. Diffuse intrinsic pontine glioma: from diagnosis to next-generation clinical trials. Curr. Treat. Options Neurol. 2019;21:37. doi: 10.1007/s11940-019-0577-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lieberman N.A.P., DeGolier K., Kovar H.M., Davis A., Hoglund V., Stevens J., Winter C., Deutsch G., Furlan S.N., Vitanza N.A. Characterization of the immune microenvironment of diffuse intrinsic pontine glioma: implications for development of immunotherapy. Neuro-oncol. 2019;21:83–94. doi: 10.1093/neuonc/noy145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin G.L., Nagaraja S., Filbin M.G., Suvà M.L., Vogel H., Monje M. Non-inflammatory tumor microenvironment of diffuse intrinsic pontine glioma. Acta Neuropathol. Commun. 2018;6:51. doi: 10.1186/s40478-018-0553-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kanehisa M., Furumichi M., Tanabe M., Sato Y., Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45(D1):D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenberg J.E., Hoffman-Censits J., Powles T., van der Heijden M.S., Balar A.V., Necchi A., Dawson N., O’Donnell P.H., Balmanoukian A., Loriot Y. Atezolizumab in patients with locally advanced and metastatic urothelial carcinoma who have progressed following treatment with platinum-based chemotherapy: a single-arm, multicentre, phase 2 trial. Lancet. 2016;387:1909–1920. doi: 10.1016/S0140-6736(16)00561-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Champiat S., Ferté C., Lebel-Binay S., Eggermont A., Soria J.C. Exomics and immunogenics: Bridging mutational load and immune checkpoints efficacy. OncoImmunology. 2014;3:e27817. doi: 10.4161/onci.27817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder A., Makarov V., Merghoub T., Yuan J., Zaretsky J.M., Desrichard A., Walsh L.A., Postow M.A., Wong P., Ho T.S. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014;371:2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown S.D., Warren R.L., Gibb E.A., Martin S.D., Spinelli J.J., Nelson B.H., Holt R.A. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24:743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reitman Z.J., Paolella B.R., Bergthold G., Pelton K., Becker S., Jones R., Sinai C.E., Malkin H., Huang Y., Grimmet L. Mitogenic and progenitor gene programmes in single pilocytic astrocytoma cells. Nat. Commun. 2019;10:3731. doi: 10.1038/s41467-019-11493-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao J., Chen A.X., Gartrell R.D., Silverman A.M., Aparicio L., Chu T., Bordbar D., Shan D., Samanamud J., Mahajan A. Immune and genomic correlates of response to anti-PD-1 immunotherapy in glioblastoma. Nat. Med. 2019;25:462–469. doi: 10.1038/s41591-019-0349-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amankulor N.M., Kim Y., Arora S., Kargl J., Szulzewsky F., Hanke M., Margineantu D.H., Rao A., Bolouri H., Delrow J. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017;31:774–786. doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinton L., Masetto E., Vettore M., Solito S., Magri S., D’Andolfi M., Del Bianco P., Lollo G., Benoit J.P., Okada H. The immune suppressive microenvironment of human gliomas depends on the accumulation of bone marrow-derived macrophages in the center of the lesion. J. Immunother. Cancer. 2019;7:58. doi: 10.1186/s40425-019-0536-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibney G.T., Weiner L.M., Atkins M.B. Predictive biomarkers for checkpoint inhibitor-based immunotherapy. Lancet Oncol. 2016;17:e542–e551. doi: 10.1016/S1470-2045(16)30406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hwang K., Koh E.J., Choi E.J., Kang T.H., Han J.H., Choe G., Park S.H., Yearley J.H., Annamalai L., Blumenschein W. PD-1/PD-L1 and immune-related gene expression pattern in pediatric malignant brain tumors: clinical correlation with survival data in Korean population. J. Neurooncol. 2018;139:281–291. doi: 10.1007/s11060-018-2886-5. [DOI] [PubMed] [Google Scholar]
- 50.Jiang P., Gu S., Pan D., Fu J., Sahu A., Hu X., Li Z., Traugh N., Bu X., Li B. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat. Med. 2018;24:1550–1558. doi: 10.1038/s41591-018-0136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Louis D.N., Perry A., Reifenberger G., von Deimling A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 52.Subramanian A., Narayan R., Corsello S.M., Peck D.D., Natoli T.E., Lu X., Gould J., Davis J.F., Tubelli A.A., Asiedu J.K. A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell. 2017;171:1437–1452.e17. doi: 10.1016/j.cell.2017.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fang M., Zhou T., Yin J., Wang Y., Tao D. Data subset selection with imperfect multiple labels. IEEE Trans. Neural Netw. Learn. Syst. 2019;30:2212–2221. doi: 10.1109/TNNLS.2018.2875470. [DOI] [PubMed] [Google Scholar]
- 55.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y., Hoang C.D., Diehn M., Alizadeh A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang C., Huang X., Liu Z., Qin W., Wang C. Metabolism-associated molecular classification of hepatocellular carcinoma. Mol. Oncol. 2020;14:896–913. doi: 10.1002/1878-0261.12639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rooney M.S., Shukla S.A., Wu C.J., Getz G., Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Skidmore Z.L., Wagner A.H., Lesurf R., Campbell K.M., Kunisaki J., Griffith O.L., Griffith M. GenVisR: genomic visualizations in R. Bioinformatics. 2016;32:3012–3014. doi: 10.1093/bioinformatics/btw325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Budczies J., Seidel A., Christopoulos P., Endris V., Kloor M., Győrffy B., Seliger B., Schirmacher P., Stenzinger A., Denkert C. Integrated analysis of the immunological and genetic status in and across cancer types: impact of mutational signatures beyond tumor mutational burden. OncoImmunology. 2018;7:e1526613. doi: 10.1080/2162402X.2018.1526613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mermel C.H., Schumacher S.E., Hill B., Meyerson M.L., Beroukhim R., Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H., Meltzer P., Davis S. RCircos: an R package for Circos 2D track plots. BMC Bioinformatics. 2013;14:244. doi: 10.1186/1471-2105-14-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoshida Y., Brunet J.P., Tamayo P., Golub T.R., Mesirov J.P. Subclass mapping: identifying common subtypes in independent disease data sets. PLoS ONE. 2007;2:e1195. doi: 10.1371/journal.pone.0001195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sauerbrei W., Royston P., Binder H. Selection of important variables and determination of functional form for continuous predictors in multivariable model building. Stat. Med. 2007;26:5512–5528. doi: 10.1002/sim.3148. [DOI] [PubMed] [Google Scholar]
- 64.Kursa M.B., Rudnicki W.R. Feature selection with the Boruta package. J. Stat. Softw. 2010;36:1–13. [Google Scholar]
- 65.Xu R.H., Wei W., Krawczyk M., Wang W., Luo H., Flagg K., Yi S., Shi W., Quan Q., Li K. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat. Mater. 2017;16:1155–1161. doi: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.