Abstract
Long noncoding RNAs have been recently demonstrated to have an important role in fundamental biological processes, and their deregulated expression has been found in several human neoplasias. Our group has recently reported a drastic overexpression of the long noncoding RNA (lncRNA) RPSAP52 (ribosomal protein SA pseudogene 52) in pituitary adenomas. We have shown that this lncRNA increased cell proliferation by upregulating the expression of the chromatinic proteins HMGA1 and HMGA2, functioning as a competing endogenous RNA (ceRNA) through competitively binding to microRNA-15a (miR-15a), miR-15b, and miR-16. The aim of this work was to identify further mechanisms by which RPSAP52 overexpression could contribute to the development of pituitary adenomas. We investigated the involvement of RPSAP52 in the modulation of the expression of cell cycle-related genes, such as p21Waf1/CIP, whose deregulation plays a critical role in pituitary cell transformation. We report that RPSAP52, interacting with the RNA binding protein HuR (human antigen R), favors the delocalization of miR-15a, miR-15b, and miR-16 on the cyclin-dependent kinase inhibitor p21Waf1/CIP1 that, accordingly, results in downregulation in pituitary adenomas. A RNA immunoprecipitation sequencing (RIPseq) analysis performed on cells overexpressing RPSAP52 identified 40 messenger RNAs (mRNAs) enriched in Argonaute 2 (AGO2) immunoprecipitated samples. Among them, we focused on GAS8 (growth arrest-specific protein 8) gene. Consistently, GAS8 expression was downregulated in all the analyzed pituitary adenomas with respect to normal pituitary and in RPSAP52-overepressing cells, supporting the role of RPSAP52 in addressing genes involved in growth inhibition and cell cycle arrest to miRNA-induced degradation. This study unveils another RPSAP52-mediated molecular mechanism in pituitary tumorigenesis.
Key words: Pituitary adenomas, HMGA, p21Waf1/CIP, GAS8
INTRODUCTION
Great interest was recently raised from long noncoding RNAs (lncRNAs), RNA molecules with a length variable between 200 and 100,000 nucleotides. They are classified in five different groups (sense, antisense, bidirectional, intronic, and intergenic) in relation to their position on the DNA1,2, and unlike microRNAs (miRNAs), which exhibit highly conserved sequences across different species, lncRNAs are generally poorly conserved and they are preferentially expressed in a tissue-specific manner.
They play an essential role in the regulation of gene expression at the transcriptional or posttranscriptional level by modulating the function of transcription factors, directing chromatin reorganization and modification, or inhibiting the translation of messenger RNA3. Moreover, they can also regulate messenger RNA (mRNA) stability by interacting with miRNAs and RNA binding proteins. Accumulating evidence has shown that lncRNAs participate in a variety of neoplasias modulating the cell epigenome, leading then to cell transformation. Indeed, extensive genome-wide cancer mutation analyses have revealed that mutations within the noncoding genome influence the occurrence and development of a variety of human cancers, making these molecules attractive targets for cancer treatment4.
Our research group has recently analyzed the lncRNA expression profile of a panel of gonadotroph pituitary adenomas (PAs) in comparison with normal pituitaries, reporting the drastic expression of RPSAP52 (ribosomal protein SA pseudogene 52), a novel lncRNA overexpressed in pituitary tumors. Interestingly, RPSAP52 is the antisense of the HMGA2 gene, a member of the high mobility group A (HMGA) family that also includes HMGA1, previously demonstrated to have a causal role in pituitary tumorigenesis5–8.
We reported that RPSAP52 expression promotes cell proliferation by enhancing the G1/S transition of the cell cycle and increasing HMGA2 and HMGA1 protein levels in a competing endogenous RNA (ceRNA)-dependent way, acting as miRNA sponge for miR-15a, miR-15b, and miR-16. Consistently, an increased expression of HMGA1 and HMGA2, correlating with that of RPSAP52, was found in the large majority of PAs9.
Our study aimed at the definition of another RPSAP52-mediated molecular mechanism in pituitary tumorigenesis by modulating the expression of other cell cycle-related genes, whose deregulation plays a critical role in pituitary cell transformation.
Here we report that RPSAP52 induces the displacement of the HMGA-targeting miRNAs from HMGA proteins to the cyclin-dependent kinase inhibitor p21Waf1/CIP. We demonstrate that this mechanism was mediated by the interaction with HuR (human antigen R), a member of the ELAV (embryonic lethal abnormal vision) family of the RNA binding proteins (RBPs). Finally, RNA immunoprecipitation (RIP) expression profiling of HT-29 cells transfected with RPSAP52 and immunoprecipitated with AGO2 (Argonaute 2) protein identified the growth arrest-specific protein GAS8 as an mRNA modulated by RPSAP52 in the context of RNA-induced silencing complex (RISC). Consistently, GAS8 was found significantly downregulated in gonadotroph PAs and in HT-29 cells overexpressing RPSAP52.
Therefore, these results demonstrate that RPSAP52 overexpression contributes to pituitary tumorigenesis by different mechanisms.
MATERIALS AND METHODS
Tissue Samples, Cell Lines, Vectors, and Transfections
HT-29 and HCT-116 cells were cultured in McCoy’s, containing 10% fetal bovine serum, 1% glutamine, and 1% penicillin/streptomycin (Life Technologies, Monza, Italy) and were incubated at 37°C in a 5% CO2 atmosphere. The authenticity of the cell lines was confirmed by Cell IDTM System (Promega, Madison, WI, USA) in November 2013. Details on gonadotroph pituitary and normal pituitary tissue samples have been previously reported9.
The pRPSAP52 expression vector and the empty vector were purchased from OriGene (Rockville, MD, USA). The pRPSAP52-MUT-15/16 sequence was obtained from Eurofins Genomics (Ebersberg, Bayern, Germany)9. HT-29 cells were transfected with the above-mentioned expression vectors using FuGene transfection reagent (Promega) according to the manufacturer’s instructions. For silencing experiments, HCT-116 cells were transfected with RPSAP52 antisense LNA™ GapmeR and the relative negative control (Exiqon/Qiagen, Hilden, Germany) or with an anti-HuR siRNA or a scrambled oligonucleotide (Eurofins Genomics) using Lipofectamine RNAiMAX (Invitrogen, Carlsbad, CA, USA). Then 50 nmol/ml of pre-miRNA precursors or scrambled oligonucleotides (Thermo Fisher Scientific, Waltham, MA, USA) were transfected using siPORT neoFX Transfection Agent (Thermo Fisher Scientific).
The CDKN1A (cyclin-dependent kinase inhibitor 1A) 3′-untranslated region (3′-UTR) region was amplified by polymerase chain reaction (PCR) from human genomic DNA. The amplified DNA was cloned either in sense or in antisense orientation into the pmirGLO Dual-Luciferase Expression Vector (Promega) at the XbaI site immediately downstream from the stop codon of the luciferase gene.
RIP Assay
The RIP assay was performed according to the instructions of the Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Magnetic beads were incubated with anti-AGO2 or anti-HuR antibodies or immunoglobulin G (IgG) and cell lysate. The coprecipitated RNAs were detected by reverse transcription polymerase chain reaction (RT-PCR).
Reverse Transcription and Quantitative RT-PCR
RNAs were reverse transcribed to complementary DNA (cDNA) using a Reverse Transcription Kit (Qiagen). Real-time PCR analyses were performed using SYBR Green quantitative real-time PCR Master Mix (Bio-Rad, Hercules, CA, USA). Reverse transcription and qRT-PCR for mature miRNA were performed according to the manufacturer’s instructions of miScript system kits (Qiagen). To calculate the relative expression levels, we used the 2ΔΔCT method10.
The primer sequences used are shown in Table 1.
Table 1.
Primer Sequences Used
| RPSAP52 | Forward 5′-gcaaacacatcggagacaaa-3′ |
| Reverse 5′-tttctctttaagtcatgacaggaatc-3′ | |
| CDKN1A | Forward 5′-ccgaagtcagttccttgtgg-3′ |
| Reverse 5′-catgggttctgacggacat-3′ | |
| GAS8 | Forward 5′- tgaaggtggagctcaaggag-3′ |
| Reverse 5′-ggtgatctcctcggtgtgtt-3′ | |
| TYW1B | Forward 5′-aagagcaggacgaattgcat-3′ |
| Reverse 5′-ttcactggagctctcgaagg-3′ | |
| MAPK11 | Forward 5′-catgctcaactggatgcatta-3′ |
| Reverse 5′-cgcttcagctggtcaatgta-3′ | |
| AC006547.8 | Forward 5′-tggatggcatcagcacag-3′ |
| Reverse 5′-gggtgtgcgatggaaatg-3′ | |
| G6PD | Forward 5′-cagcggcaactaaactcaga-3′ |
| Reverse 5′-ttccctcaggatcccacac-3′ |
Protein Extraction, Western Blotting, and Antibodies
For protein extraction, cells were lysed in lysis buffer containing1% NP-40, 1 mM EDTA, 50 mM Tris-HCl (pH 7.5), and 150 mM NaCl, supplemented with complete protease inhibitor mixture (Roche, Mannheim, Germany). Protein concentration was determined by the Bradford assay (Bio-Rad) using bovine serum albumin as standard. Total proteins were separated on a 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (GE Healthcare, Chicago, IL, USA). Membranes were blocked with 5% nonfat dry milk.
After blotting, membranes were incubated with primary antibodies against p21 (DCS60; Cell Signaling Technology, Leiden, Netherlands) or β-actin (sc-1615; Santa Cruz Biotechnology, Dallas, TX, USA). The reaction was detected with a Western blotting detection system (Thermo Fisher Scientific).
Luciferase Assay
Cells were cotransfected with the CDKN1A 3′-UTR or with the empty luciferase vectors described above and the miRNA oligonucleotides. Firefly and Renilla luciferase activities were measured 48 h after transfection with the dual-luciferase reporter assay system (Promega). Firefly activity was normalized to Renilla activity as control of transfection efficiency9.
RIP Sequencing
Next-generation sequencing experiments, comprising samples quality control, were performed by Genomix4life S.R.L. (Baronissi, Salerno, Italy). Indexed libraries were prepared from 1 μg/ea purified RNA with TruSeq Stranded total RNA Sample Prep Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Libraries were quantified using the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and pooled such that each index-tagged sample was present in equimolar amounts, with final concentration of the pooled samples of 2 nM. The pooled samples were subject to cluster generation and sequencing using an Illumina HiSeq2500 System (Illumina) in a 2 × 100 paired-end format at a final concentration of 8 pmol. The raw sequence files generated (.fastq files) underwent quality control analysis using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
Statistical Analysis
Statistical analysis was performed using the GraphPad Prism (version 6) software. The Student’s t-test or analysis of variance (ANOVA) was used to determine the significance of quantitative experiments. Error bars represent the standard deviation (SD) or the standard error of mean (SEM). To evaluate the statistical correlation, the nonparametric Spearman correlation coefficient R was used. Values of p < 0.05 were considered significant.
RESULTS
RPSAP52 Overexpression Enhances the Binding of miR-15a, miR-15b, and miR-16 to CDKN1A mRNA
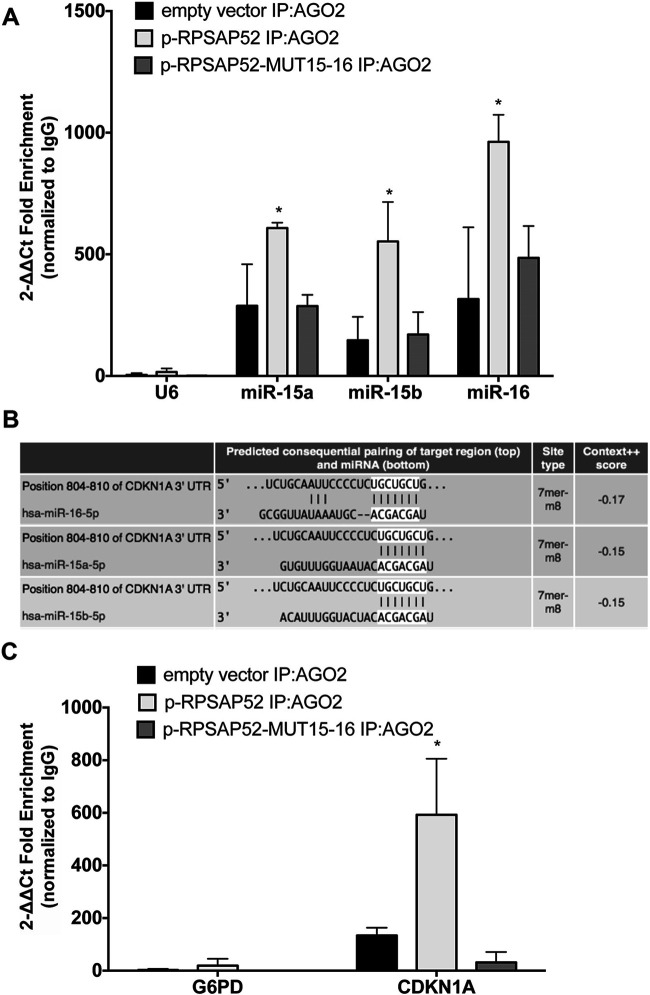
We have recently demonstrated that RPSAP52 binds miR-15a, miR-15b, and miR-16 and increases HMGA protein levels, releasing them from miRNA inhibition9. In order to determine whether the presence of RPSAP52 could induce changes in the association of miRNAs and their target genes, we performed an anti-AGO2 RIP analysis in the presence of RPSAP52. Indeed, AGO2 protein is considered the core component of the miRNA-induced silencing complex (RISC), and it is well known that miRNAs bind their targets on RISC in an AGO2-dependent manner, facilitating their interaction with target genes11. For this purpose, an empty vector, a vector containing the full sequence of lncRNA RPSAP52 (p-RPSAP52), and a vector containing RPSAP52 with mutations in the miR-15a, miR-15b, and miR-16 binding sites were transfected in HT-29 cells. As shown in Figure 1A, we found that miR-15a, miR-15b, and miR-16 levels were significantly enriched in the anti-AGO2 RNA immunoprecipitated from the p-RPSAP52-transfected cells, compared with empty vector and the corresponding mutated vector (p-RPSAP52-MUT-15-16).
Figure 1.
RPSAP52 (ribosomal protein SA pseudogene 52) enhances the binding of miR-15a, miR-15b, and miR-16 to CDKN1A messenger RNAs (mRNAs). (A) RNA immunoprecipitation sequencing (RIPseq) expression profile of anti-AGO2 RIP analysis in HT-29 cells transfected with p-RPSAP52, p-RPSAP52 vector containing mutated binding sites for miR-15a, miR-15b, and miR-16, or empty vector. Normal immunoglobulin Gs (IgGs) and U6 RNA enrichment were used as negative control. Error bars indicate the mean of three independent experiments performed in triplicate. *p < 0.05 compared with HT-29–empty vector-transfected cells immunoprecipitated with an anti-AGO2 antibody. (B) Representation of human CDKN1A 3′-untranslated region (3′-UTR) and the relative position of the predicted microRNA (miRNA) binding sites (http://www.targetscan.org). (C) RIP analysis, assessed by immunoprecipitation of AGO2 in HT-29 cells transfected with p-RPSAP52, p-RPSAP52 vector containing mutated binding sites for miR-15a, miR-15b, and miR-16, or empty vector. IgG and G6PD immunoprecipitation were used as a negative control. *p < 0.05 compared with HT-29–empty vector-transfected cells immunoprecipitated with anti-AGO2 antibody.
Subsequently, we searched for other targets in addition to the previously demonstrated HMGA proteins of these miRNAs by using the bioinformatic tools MiRwalk, miRanda, and TargetScan. Among the candidate miR-15a, miR-15b, and miR-16-target genes, we focused on the CDKN1A gene, which encodes the p21Waf1/CIP protein, a potent cyclin-dependent kinase inhibitor (Fig. 1B). A significant number of genetic or epigenetic alterations of regulators of the G1/S transition of the cell cycle have been described in pituitary tumors. These alterations induce a cell cycle hyperactivity that leads to increased proliferation and genomic instability in pituitary tumors12.
Interestingly, a CDKN1A mRNA enrichment in AGO2 pull-down was observed after transfection with RPSAP52 in comparison with empty and mutated vectors (Fig. 1C).
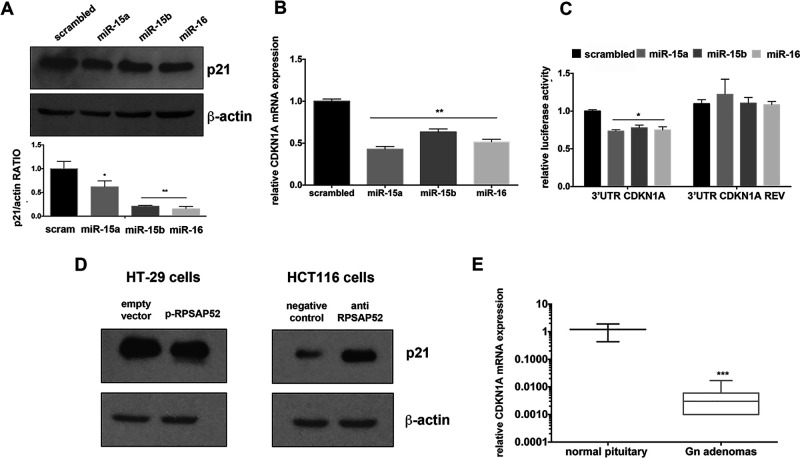
Then, to validate CDKN1A as target of miR-15a, miR-15b, and miR-16, we transfected them into HT-29 cells and evaluated the p21Waf1/CIP protein levels: a significant decrease in this protein levels compared with the scrambled-transfected cells was observed (Fig. 2A). Interestingly, a significant downregulation in CDKN1A mRNA levels was also observed in the same miRNA-transfected cells, suggesting that these miRNAs are also able to induce CDKN1A mRNA degradation (Fig. 2B). Finally, to demonstrate the direct interaction between the analyzed miRNAs and the CDKN1A mRNA, we performed a luciferase assay introducing 3′-UTR of CDKN1A downstream of the luciferase open reading frame, either in sense (3′-UTR CDKN1A) or in antisense (3′-UTR CDKN1A REV) orientation, and these reporter vectors were transfected in HT-29 cells together with miRNA oligonucleotide precursors or a scrambled oligonucleotide. As shown in Figure 2C, the luciferase activity was significantly diminished after transfection of each analyzed miRNA compared with the scrambled oligonucleotide, whereas the luciferase activity of the antisense construct did not vary, indicating that these miRNAs interfere with CDKN1A mRNA through direct interaction with its 3′-UTR.
Figure 2.
CDKN1A is target of miR-15a, miR-15b, and miR-16. (A) Western blot analysis of p21Waf1/CIP protein levels in HT-29 cells transfected with p-RPSAP52 or relative empty vector and HCT-116 cells transfected with anti-RPSAP52 or negative control oligonucleotide. β-Actin expression was analyzed as loading control. Densitometric analysis performed using ImageJ software and normalized to β-actin was also reported. The error bars represent the mean value ± standard error (SD). *p < 0.05 and **p < 0.01 compared to scrambled oligonucleotide-transfected cells. (B) qRT-PCR analysis of CDKN1A mRNA in HT-29 cells transfected with the indicated miRNAs. Relative CDKN1a mRNA levels between miRNA treated or scrambled oligonucleotide treated, normalized with G6PD. The error bars represent the mean value ± SE from three independent experiments performed in triplicate. **p < 0.01 compared to scrambled oligonucleotide-transfected cells. (C) Luciferase assay using a Luc-3′-UTR CDKN1A or a Luc-3′-UTR CDKN1A REV expression vector along with the indicated miRNA oligonucleotides or with a scrambled oligonucleotide. The relative activity of firefly luciferase expression was standardized to a transfection control using Renilla luciferase. The scale bars represent the mean ± SE of three independent experiments performed in triplicate. *p < 0.05 compared to scrambled-transfected cells. (D) Western blot analysis of p21Waf1/CIP protein levels in HT-29 cells transfected with p-RPSAP52 or relative empty vector and HCT-116 cells transfected with anti-RPSAP52 or negative control oligonucleotide. β-Actin expression was analyzed as loading control. (E) Box plot of RNA expression of CDKN1Ain gonadotroph pituitary adenomas (PAs) versus five normal pituitaries. The boxes define the interquartile range, and the thick line is the median. ***p < 0.001 compared with CDKN1A expression in normal pituitaries.
Consistent with the ability of RPSAP52 to readdress miR-15a, miR-15b, and miR-16 on CDKN1A mRNA, we found reduced p21Waf1/CIP protein levels after RPSAP52 overexpression, whereas the opposite effect was observed after silencing of RPSAP52 (Fig. 2D). Moreover, a downregulation of CDKN1A mRNA expression in gonadotroph PAs compared with normal pituitary gland was also observed (Fig. 2E).
These results support the hypothesis that RPSAP52 could release HMGA2 or HMGA1 from miRNA-inhibitory action also favoring their binding to other targets, such as CDKN1A.
RPSAP52 Expression Induces a Decrease in p21Waf1/CIP Levels by Interacting With the RBP HuR
It has been reported that lncRNAs can interact with RNA binding proteins to antagonistically or synergistically regulate gene transcription or protein stability13. HuR, also known as ELAV-like protein 1, is a ubiquitously expressed member of the ELAV/Hu family of RNA-binding proteins. In the nucleus, HuR regulates RNA splicing and nuclear export14. In the cytoplasm, HuR binds elements rich in adenylate and uridylate (AU-rich elements; ARE) in the 3′-UTR regions of target transcripts and protects them against degradation15. In this way, it regulates mRNA stability and translational efficiency of several genes. Interestingly, it was reported that HuR binds to the p21Waf1/CIP mRNA and promotes its stability, thus modulating its expression16,17.
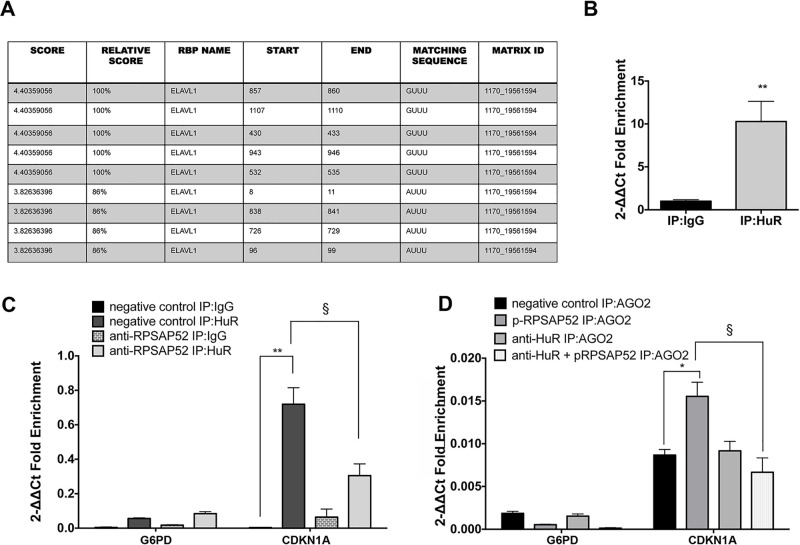
Therefore, we investigated whether the decreased p21Waf1/CIP expression was also dependent on the interaction of RPSAP52 with HuR. We first searched for the ELAV1 motif in the RPSAP52 sequence by the “RBPDB binding protein binding motifs database” (rbpdb.ccbr.utoronto.ca). As shown in Figure 3A, we identified nine RPSAP52–HuR potential interaction sites. To validate this interaction also in vitro, we performed a RIP analysis by using anti-HuR antibody. As shown in Figure 3B, RPSAP52 was highly enriched (ninefold) in the HuR-immunoprecipitated samples in comparison with the control IgG samples, indicating that HuR selectively associates with RPSAP52.
Figure 3.
RPSAP52 promotes the binding of miR-15a, miR-15b, and miR-16 to CDKN1A mRNAs by interacting with HuR RNA binding protein. (A) ELAV1/HuR binding motifs in RPSAP52 sequence by using the “RBPDB binding protein binding motifs database” (rbpdb.ccbr.utoronto.ca). (B) qRT-PCR analysis of RPSAP52 RNA levels in HT-29 cells immunoprecipitated with HuR antibody. IgG was used as a negative control of immunoprecipitation. **p < 0.01compared with IgG:IP samples. (C) qRT-PCR analysis of CDKN1A RNA levels in HCT-116 cells transfected with anti-RPSAP52 oligonucleotide or the relative negative control and immunoprecipitated with HuR antibody. IgG and G6PD enrichment were used as negative controls of immunoprecipitation. **p < 0.01 compared with HCT-116-negative control samples immunoprecipitated with IgG; §p < 0.05 compared with HCT-116 negative control HuR:IP. (D) qRT-PCR analysis of CDKN1A RNA levels in HT-29 cells transfected with scrambled, p-RPSAP52 vector, anti-HuR siRNA, or anti-HuR and p-RPSAP52 vector, and immunoprecipitated with anti-AGO2 antibody. IgG and G6PD enrichment were used as negative controls of immunoprecipitation. *p < 0.05 compared with HT-29 negative control:IP AGO2; §p < 0.05 compared with HT-29 p-RPSAP52:IP AGO2.
Then, to explore how the interaction between HuR and RPSAP52 affected p21Waf1/CIP levels, we performed a RIP assay with HuR antibody in the presence or in the absence of RPSAP52. As shown in Figure 3C, in the presence of RPSAP52, a CDKN1A mRNA enrichment was found in HuR immunoprecipitated samples compared with the IgG samples. Conversely, the silencing of RPSAP52 leads to less accumulation of CDKN1A in the HuR IP samples (Fig. 3C).
In order to strengthen these findings, we performed a RIP assay with AGO2 antibody in HT-29 cells after HuR silencing, in the presence or in the absence of RPSAP52. In the presence of RPSAP52, a CDKN1A enrichment in AGO2 pull-down was observed, as already reported in Figure 1C. When we silenced HuR, we found a reduction in CDKN1A enrichment (both in the presence and in the absence of RPSAP52), demonstrating that RPSAP52 needs to interact with HuR to address p21Waf1/CIP to RISC-mediated degradation.
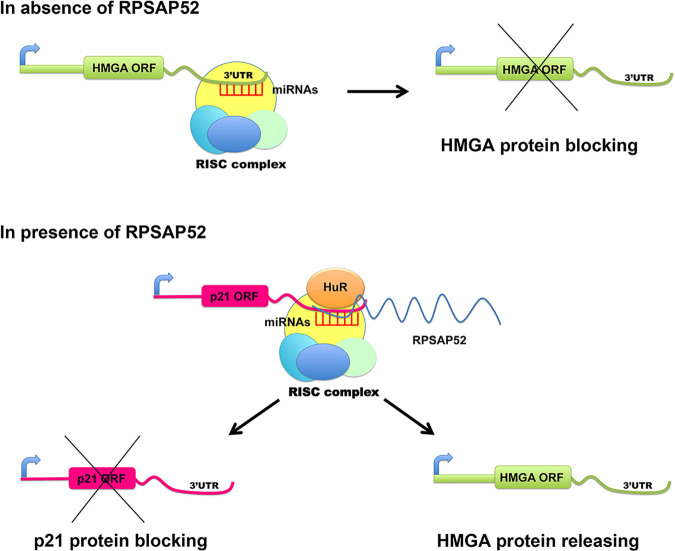
Based on these results, we hypothesize that RPSAP52 binds HuR, preventing the stabilization of CDKN1A mRNA induced by HuR alone. Therefore, RPSAP52 would readdress miRNAs from HMGAs to CDKN1A by interacting with HuR, with consequent reduction in p21Waf1/CIP protein levels (Fig. 4) and increase in HMGA proteins.
Figure 4.
Schematic representation of the RPSAP52 mechanisms. In the absence of RPSAP52, miRNAs bind the 3′-UTR of the HMGA1 and HMGA2 gene, reducing the synthesis of the relative proteins. RPSAP52, overexpressed, is able to bind miRNAs and recruit HuR, readdressing miRNAs from the high mobility group A (HMGA) to CDKN1A, with reduction of p21Waf1/CIP protein levels and release of HMGA proteins.
RIPseq Analysis of AGO2-Binding RNAs Regulated by RPSAP52
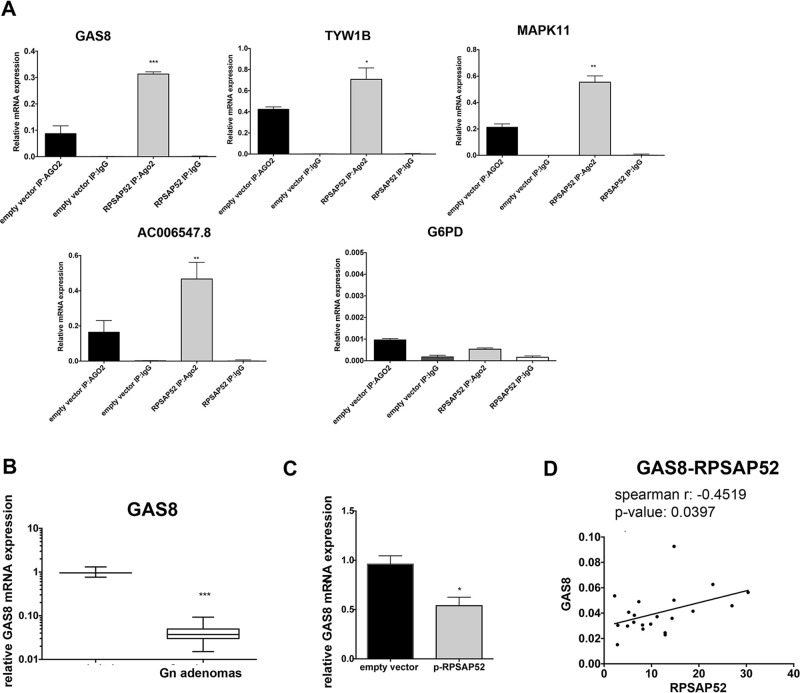
In order to extend our search for RNA target enrichment mediated by RPSAP52 expression, we performed RIP-coupled high-throughput sequencing (RIPseq) in HT-29 cells overexpressing or not RPSAP52. This analysis allowed to identify RNAs linked to AGO2 and regulated by RPSAP52. Indeed, we identified 40 RNAs enriched in AGO2 immunoprecipitated samples in the presence of RPSAP52, compared with the control-untransfected cells (Table 2). Next, we validated RNA–protein interactions choosing four differentially enriched RNAs (GAS8, TYW1B, MAPK11, and AC006547.8) by qRT-PCR. The increased expression of such RNAs was confirmed in AGO:IP-RPSAP52-overexpressing cells compared with the AGO:IP-empty vector cells (Fig. 5A). The G6PD expression was used as no-target RNA control. Among the validated RNAs, we focused our attention on the GAS8 gene, also known as growth arrest-specific protein 11 (GAS11), which encodes a putative tumor suppressor gene involved in cell cycle control and deleted in breast and prostate cancers18,19. Consistently, GAS8 was significantly downregulated in all the analyzed gonadotroph PAs (Fig. 5B), and RPSAP52 overexpression reduces the GAS8 levels in HT-29 cells (Fig. 5C).
Table 2.
List of RNAs Differentially Associated With AGO2 in HT-29–RPSAP52 Cells Versus HT-29–Empty Vector Cells
| Seqname | Gene Name | HT-29–Empty Vector IP:Ago2 | HT-29–RPSAP52 IP:Ago2 | Fold Enrichment | Prob. |
|---|---|---|---|---|---|
| 1. ENST00000493929 | RPSAP52 | 17.69999006 | 29,1467.0085 | 16,467.07 | 1.000 |
| 2. ENST00000264005 | LCAT | 4.164703544 | 25.93473228 | 6.23 | 0.914 |
| 3. ENST00000387365 | MT-TI | 59.34702551 | 14.4081846 | 4.12 | 0.961 |
| 4. ENST00000524964 | RP11-712L6.5 | 38.52350779 | 9.6054564 | 4.01 | 0.931 |
| 5. ENST00000330651 | MAPK11 | 9.370582975 | 35.54018868 | 3.79 | 0.917 |
| 6. ENST00000007414 | OSBPL7 | 12.49411063 | 43.2245538 | 3.46 | 0.926 |
| 7. ENST00000620995 | TYW1B | 15.61763829 | 53.79055584 | 3.44 | 0.942 |
| 8. ENST00000387392 | MT-TA | 66.63525671 | 22.09254972 | 3.02 | 0.939 |
| 9. ENST00000624592 | AC004623.3 | 27.07057304 | 78.76474248 | 2.91 | 0.942 |
| 10. ENST00000300714 | ENTHD2 | 19.78234184 | 54.75110148 | 2.77 | 0.919 |
| 11. ENST00000268699 | GAS8 | 63.51172905 | 175.7798521 | 2.77 | 0.944 |
| 12. ENST00000288022 | 19.78234184 | 54.75110148 | 2.77 | 0.919 | |
| 13. ENST00000640327 | PSMD5-AS1 | 34.35880424 | 92.21238144 | 2.68 | 0.935 |
| 14. ENST00000261507 | MSMO1 | 3092.292382 | 1,157.457496 | 2.67 | 0.941 |
| 15. ENST00000321612 | GLDC | 51.01761842 | 19.2109128 | 2.66 | 0.907 |
| 16. ENST00000506894 | GPM6A | 73.92348791 | 27.85582356 | 2.65 | 0.928 |
| 17. ENST00000387382 | MT-TW | 92.66465386 | 37.46127996 | 2.47 | 0.926 |
| 18. ENST00000412713 | AC006547.8 | 29.15292481 | 72.040923 | 2.47 | 0.918 |
| 19. ENST00000290418 | CCDC142 | 29.15292481 | 71.08037736 | 2.44 | 0.916 |
| 20. ENST00000381007 | SGTB | 58.30584962 | 24.013641 | 2.43 | 0.904 |
| 21. ENST00000300057 | MESP1 | 69.75878437 | 28.8163692 | 2.42 | 0.915 |
| 22. ENST00000381418 | HR | 46.85291488 | 111.4232942 | 2.38 | 0.925 |
| 23. ENST00000357179 | TOP3B | 36.44115601 | 86.4491076 | 2.37 | 0.919 |
| 24. ENST00000302609 | ZNF25 | 92.66465386 | 39.38237124 | 2.35 | 0.920 |
| 25. ENST00000306851 | B4GALT6 | 166.5881418 | 71.08037736 | 2.34 | 0.927 |
| 26. ENST00000247001 | SUGP1 | 43.72938722 | 101.8178378 | 2.33 | 0.921 |
| 27. ENST00000539298 | DHX37 | 90.58230209 | 208.4384039 | 2.30 | 0.924 |
| 28. ENST00000556936 | CCDC175 | 434.1703445 | 189.2274911 | 2.29 | 0.925 |
| 29. ENST00000247930 | ZNF777 | 61.42937728 | 140.2396634 | 2.28 | 0.921 |
| 30. ENST00000565624 | ZNF469 | 46.85291488 | 106.620566 | 2.28 | 0.917 |
| 31. ENST00000223129 | RPA3 | 135.3528652 | 60.51437532 | 2.24 | 0.917 |
| 32. ENST00000335999 | NCKAP5L | 60.38820139 | 133.515844 | 2.21 | 0.914 |
| 33. ENST00000256925 | CABLES1 | 83.29407089 | 182.5036716 | 2.19 | 0.914 |
| 34. ENST00000366758 | JMJD4 | 49.97644253 | 108.5416573 | 2.17 | 0.906 |
| 35. ENSPTRT00000076404 | MT-ND3 | 541.4114608 | 250.702412 | 2.16 | 0.912 |
| 36. ENST00000340368.8 | INSIG1 | 9,270.63009 | 4,306.126104 | 2.15 | 0.912 |
| 37. ENST00000231790 | MLH1 | 46.85291488 | 100.8572922 | 2.15 | 0.904 |
| 38. ENST00000483786 | SLC4A2 | 241.5528056 | 519.6551912 | 2.15 | 0.912 |
| 39. ENST00000361851 | MT-ATP8 | 319.640997 | 148.8845742 | 2.15 | 0.910 |
| 40. ENST00000315264 | KIF26A | 91.62347798 | 195.9513106 | 2.14 | 0.909 |
Figure 5.
RIPseq analysis of AGO2-binding RNAs regulated by RPSAP52. (A) qRT-PCR analysis of GAS8, TYW1B, MAPK11, AC006547.8, and G6PD expression in AGO:IP-RPSAP52-overexpressed cells compared with the AGO:IP-empty vector cells. Each bar represents the mean value ± SE from three independent experiments performed in triplicate. *p < 0.05, **p < 0.01, ***p < 0.001. (B) Box plot of GAS8 RNA expression gonadotroph PAs versus five normal pituitary samples. The boxes define the interquartile range, and the thick line is the median. (C) qRT-PCR analysis of GAS8 mRNA in HT-29 cells transfected with p-RPSAP52 or empty vector, normalized with G6PD. The error bars represent the mean value ± SE from three independent experiments performed in triplicate. *p < 0.05 compared to empty vector-transfected cells. (D) Spearman rank correlation graph shows the negative relationship between RPSAP52 and GAS8 on gonadotroph adenomas of (B) (Spearman r: −0.4519, p = 0.0397).
Therefore, these results support a role of RPSAP52 overexpression in pituitary tumorigenesis readdressing genes involved in growth inhibition and cell cycle arrest to miRNA degradation.
DISCUSSION
Recent studies have revealed that lncRNAs, a novel class of regulators of gene expression, are dysregulated in several neoplasias20. Therefore, lncRNAs can be considered new emerging interactors in the comprehension of the molecular features of cancer.
By the analysis of the lncRNA expression profile of gonadotroph PAs, we have very recently demonstrated a drastic overexpression of the RPSAP52 lncRNA. Interestingly, this lncRNA, located on chromosome 12, represents the antisense of the HMGA2 gene, whose overexpression is a feature of malignant neoplasias and has a driver role in pituitary tumorigenesis8,21,22. Indeed, HMGA2 is constantly overexpressed in PAs by different mechanisms such as genomic amplification, downregulation of miRNAs able to target it, and upregulation of the HMGA1 pseudogenes, HMGA1P6 and HMGA1P76,23,24. Consistently, transgenic mice overexpressing the HMGA2 gene develop PAs with an increased penetrance (85% vs 40%) and earlier latency period (6 vs. ≈18 months) in transgenic females with respect to the males with a strong similarity with the human pathology25.
RPSAP52, whose expression is positively correlated with that of HMGA1 and HMGA2 in PAs, enhances cell growth by upregulating the HMGA protein levels with a ceRNA-dependent mechanism releasing HMGA mRNAs from miRNAs (miR-15a, miR-15b, and miR-16) able to target these genes9.
However, these studies do not exclude that RPSAP52 might contribute to pituitary cell growth and tumorigenesis by further mechanisms in addition to the ceRNA-mediated upregulation of the HMGA proteins, since the miRNAs able to bind RPSAP52 can target several genes involved in cell proliferation.
Moreover, lncRNAs, frequently present at nuclear and cytoplasmic levels, as already reported for RPSAP52, are involved in the regulation of nuclear processes such as transcription and RNA processing and regulate mRNA translation or stability and protein localization by acting at the cytoplasmic level26–28.
Therefore, we performed an anti-AGO2 RIP analysis on cells transfected with RPSAP52 in order to search for mRNAs enriched on this complex in the presence of RPSAP52, focusing on candidate targets for miRNAs with seed sequences in the RPSAP52 sequence. Interestingly, we found in RPSAP52-transfected cells an enrichment in the CDKN1A mRNA, coding for a negative regulator of cell cycle, whose deregulation is a feature of PAs. Consistently, we confirmed that CDKN1A is a target of miR-15a, miR-15b, and miR-16, and is significantly downregulated in gonadotroph PAs.
Since it has been reported that HuR, a ubiquitous protein member of the Hu/ELAV family, binds CDKN1A mRNA and enhances its stability, we performed a RIP analysis by using anti-HuR antibody demonstrating that HuR associated with RPSAP52 and the interaction between HuR and RPSAP52 prevented the stabilization of CDKN1A mRNA induced by HuR alone.
Indeed, RPSAP52, interacting with HuR, shifted the localization of miR-15a, miR-15b, and miR-16 from HMGAs to CDKN1A, probably modifying the local RNA conformation and masking miRNA interaction sites on HMGAs while exposing them on CDKN1A. This ultimately leads to increased HMGA and decreased p21Waf1/CIP protein levels, respectively, and enhances the G1/S progression of the cell cycle.
Finally, the RIPseq analysis, performed on HT-29 cells immunoprecipitated with an anti-AGO2 antibody in the presence of RPSAP52, revealed that RPSAP52 induces, in the context of RISC, an increased binding and, consequently, a negative regulation of GAS8, whose protein product is involved in tumor suppression18,19. Consistently, we found that GAS8 is also downregulated in PAs and inversely correlated with RPSAP52, supporting the idea that RPSAP52 contributes to HMGA1 and HMGA2 overexpression in pituitary tumors by a mechanism of action that is not limited to the miRNA sponge activity.
It is noteworthy that this analysis revealed the enrichment of 40 genes in the RISC complex after RPSAP52 overexpression. Among them, we found that the MAPK11 gene was enriched in RPSAP52-transfected samples. MAPK11 (also known as p38) is a member of the mitogen-activated protein kinase (MAPK) cascade involved in several processes including cell survival, apoptosis, and immune function. MAPK11 activity is dysregulated in breast cancer, where it is responsible for breast cancer-induced osteolytic bone destruction29, and in advanced prostate cancer30.
We also found the TYW1B gene, coding for Wybutosine, a hypermodified guanosine found in phenylalanine tRNA that stabilizes codon–anticodon interactions during ribosome decoding and promotes the maintenance of the reading frame.
Other deregulated genes found derived from mitochondrial genome, such as MT-TI, MT-TA, MT-TW, mitochondrially encoded tRNA genes, or PDF, coding for peptide deformylase, a mitochondrial enzyme that removes the formyl group of N-terminally formylated peptides. PDF is overexpressed in breast, colon, lung, and hematopoietic cancers31,32. Its silencing in cancer cells blocks mitochondrial translation and leads to apoptotic cell death. These findings are noteworthy given the increasing amount of evidence that reports the implications of mitochondrial DNA (mtDNA) alterations in cancer and the possibility that lncRNAs may impact directly or indirectly on mitochondrial biology33–35. Further studies are required to assess the role of these genes in pituitary tumorigenesis.
Therefore, the results reported here confirm that a complex network involving lncRNAs, RNA binding proteins, and miRNAs acts at the transcriptional and posttranscriptional levels to enhance the G1/S transition in PAs, playing a critical role in the development of these neoplasias.
ACKNOWLEDGMENTS
This work was supported by grants from the PNR-CNR Aging Program 2012–2014, CNR Flagship Projects (Epigenomics-EPIGEN), Associazione Italiana per la Ricerca sul Cancro (AIRC IG 11477). D.D. and A.F. conceived the study and designed the experiments. D.D. performed the experiments. D.D. and C.A. analyzed the data. D.D. and A.F. wrote the manuscript. All authors contributed to the editing of the manuscript. All authors read and approved the final manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, Cabili MN, Jaenisch R, Mikkelsen TS, Jacks T, Hacohen N, Bernstein BE, Kellis M, Regev A, Rinn JL, Lander ES. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 2009;458:223–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gutschner T, Diederichs S. The hallmarks of cancer: A long non-coding RNA point of view. RNA Biol. 2012;9:703–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Silva A, Bullock M, Calin G. The clinical relevance of long non-coding RNAs in cancer. Cancers (Basel) 2015;7:2169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Meseure D, Drak Alsibai K, Nicolas A, Bieche I, Morillon A. Long noncoding RNAs as new architects in cancer epigenetics, prognostic biomarkers, and potential therapeutic targets. Biomed Res Int. 2015;2015:320214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. D’Angelo D, Borbone E, Palmieri D, Uboldi S, Esposito F, Frapolli R, Pacelli R, D’Incalci M, Fusco A. The impairment of the high mobility group A (HMGA) protein function contributes to the anticancer activity of trabectedin. Eur J Cancer 2013;49:1142–51. [DOI] [PubMed] [Google Scholar]
- 6. D’Angelo D, Esposito F, Fusco A. Epigenetic mechanisms leading to overexpression of HMGA proteins in human pituitary adenomas. Front Med. (Lausanne) 2015;2:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, Parlow AF, Visone R, Pierantoni GM, Outwater E, Santoro M, Croce CM, Fusco A. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene 2002;21:3190–8. [DOI] [PubMed] [Google Scholar]
- 8. Fusco A, Fedele M. Roles of HMGA proteins in cancer. Nat Rev Cancer 2007;7:899–910. [DOI] [PubMed] [Google Scholar]
- 9. D’Angelo D, Mussnich P, Sepe R, Raia M, Del Vecchio L, Cappabianca P, Pellecchia S, Petrosino S, Saggio S, Solari D, Fraggetta F, Fusco A. RPSAP52 lncRNA is overexpressed in pituitary tumors and promotes cell proliferation by acting as miRNA sponge for HMGA proteins. J Mol Med. (Berl) 2019;97:1019–32. [DOI] [PubMed] [Google Scholar]
- 10. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 2001;25:402–8. [DOI] [PubMed] [Google Scholar]
- 11. Roberts TC. The microRNA machinery. Adv Exp Med Biol. 2015;887:15–30. [DOI] [PubMed] [Google Scholar]
- 12. Fedele M, Fusco A. Role of the high mobility group A proteins in the regulation of pituitary cell cycle. J Mol Endocrinol. 2010;44:309–18. [DOI] [PubMed] [Google Scholar]
- 13. Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang W, Vreeland AC, Noy N. RNA-binding protein HuR regulates nuclear import of protein. J Cell Sci. 2016;129:4025–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Matoulkova E, Michalova E, Vojtesek B, Hrstka R. The role of the 3’ untranslated region in post-transcriptional regulation of protein expression in mammalian cells. RNA Biol. 2012;9:563–76. [DOI] [PubMed] [Google Scholar]
- 16. Wang W, Furneaux H, Cheng H, Caldwell MC, Hutter D, Liu Y, Holbrook N, Gorospe M. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–9.10629032 [Google Scholar]
- 17. Lafarga V, Cuadrado A, Lopez de Silanes I, Bengoechea R, Fernandez-Capetillo O, Nebreda AR. p38 Mitogen-activated protein kinase- and HuR-dependent stabilization of p21(Cip1) mRNA mediates the G(1)/S checkpoint. Mol Cell Biol. 2009;29:4341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whitmore SA, Settasatian C, Crawford J, Lower KM, McCallum B, Seshadri R, Cornelisse CJ, Moerland EW, Cleton-Jansen AM, Tipping AJ, Mathew CG, Savnio M, Savoia A, Verlander P, Auerbach AD, Van Berkel C, Pronk JC, Doggett NA, Callen DF. Characterization and screening for mutations of the growth arrest-specific 11 (GAS11) and C16orf3 genes at 16q24.3 in breast cancer. Genomics 1998;52:325–31. [DOI] [PubMed] [Google Scholar]
- 19. Evron T, Philipp M, Lu J, Meloni AR, Burkhalter M, Chen W, Caron MG. Growth Arrest Specific 8 (Gas8) and G protein-coupled receptor kinase 2 (GRK2) cooperate in the control of Smoothened signaling. J Biol Chem. 2011;286:27676–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–74. [DOI] [PubMed] [Google Scholar]
- 21. Fedele M, Palmieri D, Fusco A. HMGA2: A pituitary tumour subtype-specific oncogene? Mol Cell Endocrinol. 2010;326:19–24. [DOI] [PubMed] [Google Scholar]
- 22. Fedele M, Pierantoni GM, Visone R, Fusco A. Critical role of the HMGA2 gene in pituitary adenomas. Cell Cycle 2006;5:2045–8. [DOI] [PubMed] [Google Scholar]
- 23. Fedele M, Visone R, De Martino I, Troncone G, Palmieri D, Battista S, Ciarmiello A, Pallante P, Arra C, Melillo RM, Helin K, Croce CM, Fusco A. HMGA2 induces pituitary tumorigenesis by enhancing E2F1 activity. Cancer Cell 2006;9:459–71. [DOI] [PubMed] [Google Scholar]
- 24. Esposito F, De Martino M, D’Angelo D, Mussnich P, Raverot G, Jaffrain-Rea ML, Fraggetta F, Trouillas J, Fusco A. HMGA1-pseudogene expression is induced in human pituitary tumors. Cell Cycle 2015;14:1471–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fedele M, Pentimalli F, Baldassarre G, Battista S, Klein-Szanto AJ, Kenyon L, Visone R, De Martino I, Ciarmiello A, Arra C, Viglietto G, Croce CM, Fusco A. Transgenic mice overexpressing the wild-type form of the HMGA1 gene develop mixed growth hormone/prolactin cell pituitary adenomas and natural killer cell lymphomas. Oncogene 2005;24:3427–35. [DOI] [PubMed] [Google Scholar]
- 26. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, Regev A, Lander ES, Rinn JL. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA 2009;106:11667–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–7. [DOI] [PubMed] [Google Scholar]
- 28. Zeng C, Fukunaga T, Hamada M. Identification and analysis of ribosome-associated lncRNAs using ribosome profiling data. BMC Genomics 2018;19:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. He Z, He J, Liu Z, Xu J, Yi SF, Liu H, Yang J. MAPK11 in breast cancer cells enhances osteoclastogenesis and bone resorption. Biochimie 2014;106:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Browne AJ, Gobel A, Thiele S, Hofbauer LC, Rauner M, Rachner TD. p38 MAPK regulates the Wnt inhibitor Dickkopf-1 in osteotropic prostate cancer cells. Cell Death Dis. 2016;7:e2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Randhawa H, Chikara S, Gehring D, Yildirim T, Menon J, Reindl KM. Overexpression of peptide deformylase in breast, colon, and lung cancers. BMC Cancer 2013;13:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sheth A, Escobar-Alvarez S, Gardner J, Ran L, Heaney ML, Scheinberg DA. Inhibition of human mitochondrial peptide deformylase causes apoptosis in c-myc-overexpressing hematopoietic cancers. Cell Death Dis. 2014;5:e1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen P, Fang X, Xia B, Zhao Y, Li Q, Wu X. Long noncoding RNA LINC00152 promotes cell proliferation through competitively binding endogenous miR-125b with MCL-1 by regulating mitochondrial apoptosis pathways in ovarian cancer. Cancer Med. 2018;7:4530–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li HJ, Sun XM, Li ZK, Yin QW, Pang H, Pan JJ, Li X, Chen W. LncRNA UCA1 promotes mitochondrial function of bladder cancer via the MiR-195/ARL2 signaling pathway. Cell Physiol Biochem. 2017;43:2548–61. [DOI] [PubMed] [Google Scholar]
- 35. Tian T, Lv X, Pan G, Lu Y, Chen W, He W, Lei X, Zhang H, Liu M, Sun S, Ou Z, Lin X, Cai L, He L, Tu Z, Wang X, Tannous BA, Ferrone S, Li J, Fan S. Long noncoding RNA MPRL promotes mitochondrial fission and cisplatin chemosensitivity via disruption of pre-miRNA processing. Clin Cancer Res. 2019;25:3673–88. [DOI] [PMC free article] [PubMed] [Google Scholar]