Abstract
miRNAs play an important role in progression of hepatocellular carcinoma (HCC). In this work, we assessed the function of miR-202 in human HCC and identified BCL2 as its target. We found miR-202 expression was found significantly downregulated, while BCL2 expression was markedly upregulated in HCC tissues and cell lines (HepG2, Hep3B, and HCCLM3). Both miR-202 and BCL2 were closely correlated with major vascular invasion and advanced TNM stage as well as overall survival of HCC patients. Overexpression of miR-202 significantly inhibited cell proliferation, induced apoptosis and cell cycle arrest at the G0/G1 phase, and prevented tumor formation in a xenograft nude mouse model. Further, miR-202 dramatically inhibited migration, invasion, and epithelial–mesenchymal transition. miR-202 bound to the 3′-untranslated region (3′-UTR) of BCL2 mRNA and downregulated the expression level of BCL2 protein. Exogenous BCL2 overexpression weakened the inhibitory effects of miR-202, while inhibition of BCL2 enhanced the inhibitory effects of miR-202. In conclusion, miR-202 serves as a tumor suppressor in HCC progression by downregulating BCL2 expression, indicating miR-202 might be a potential target for HCC.
Key words: miR-202, BCL2, Hepatocellular carcinoma (HCC), Progression
INTRODUCTION
Hepatocellular carcinoma (HCC) is a leading cause of cancer-related deaths in the world1. Chemotherapy is one of the most common treatment methods for patients with advanced HCC2. Although therapeutic strategies, including resection, liver transplantation, image-guided tumor ablation, and transcatheter chemoembolization, have been developed to combat liver cancer, the clinical outcomes remain unsatisfactory due to a lack of sensitive and specific biomarkers for early diagnosis3,4. Therefore, it is necessary to understand occurrence and development of HCC.
miRNAs are small noncoding RNAs [22 nucleotides (nt)] that regulate gene expression through binding to the 3′-untranslated region (3′-UTR) of target genes, thereby leading to degradation of target mRNAs or inhibition of translation5. According to recent studies, miRNAs are now recognized as critical factors for some biological processes of human diseases6,7. Deregulated miRNAs have been referred to as potential biomarkers in different malignancies7. For example, miR-202 affects proliferation, invasion, and migration of some human cancers, including gastric cancer, cervical cancer, and breast cancer8–11. However, the role of miR-202 in the development and progression of HCC is still unknown.
By the aid of the online prediction website TargetScan (http://www.targetscan.org), we preliminarily found that BCL2 may be a potential target gene of miR-202. Therefore, we explored the expressions of miR-202 and BCL2 in HCC tissues and cell lines and analyzed their associations with clinicopathological parameters and prognosis. Mechanistically, we investigated the effect of miR-202 on proliferation, migration, invasion, and epithelial–mesenchymal transition (EMT) in HCC.
MATERIALS AND METHODS
Tissue Collection
The fresh cancer tissues of 60 patients with HCC undergoing curative resection in the Department of Hepatobiliary Surgery of Shandong Provincial Third Hospital (Jinan, Shandong, China) from January 2015 to December 2018 were selected. Of these patients, no patients received adjuvant therapies, including chemo-/radiotherapy prior to surgery. Normal tissues were derived from paracancerous tissues at least 5 cm away from the tumor margin. All tissues were frozen in liquid nitrogen immediately after removal and stored at −80°C. Written informed consent was obtained from all patients. Our study complied with the Declaration of Helsinki and was approved by Shandong Provincial Third Hospital.
Cell Culture
The immortalized liver cell line (LO2) and HCC cell lines (HepG2, Hep3B, and HCCLM3) were obtained from the Institute of Biochemistry and Cell Biology (Chinese Academy of Science, Beijing, China) and maintained in Dulbecco’s modified Eagle’s medium (DMEM) or RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) at 37°C in a humidified atmosphere containing 5% CO2 and 95% air.
Cell Transfection
The siRNA targeting BCL2 (si-BCL2), control siRNA (si-control), BCL2 overexpression plasmid, empty vector, miR-202 mimics, and negative control miRNAs (miR-NC) were obtained from GenePharma (Shanghai, China). Transfection was conducted using Lipofectamine® 3000 reagent (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol reagent (Beyotime, Shanghai, China) was used to extract total RNAs, and then RNAs were reversely transcribed into complementary DNA using a TaqMan® MicroRNA Reverse Transcription Kit (Biosystems, Foster City, CA, USA). The reverse transcription was conducted according to the instructions of the Prime Script™ RT Reagent Kit (Takara, Dalian, China) to quantify levels of related mRNAs. After that, PCR was determined using a SYBR Green kit (Takara). The relative gene expression levels were calculated using the comparative 2−ΔCt method, with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 as an internal standard.
Western Blot
The total protein was extracted from cells using radioimmunoprecipitation assay (RIPA) lysis buffer (Abcam, Cambridge, MA, USA). The protein concentration was determined using a BCA kit (Beyotime). Following separation of the protein via sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), a total of 30 μg of protein was transferred onto a polyvinylidene fluoride (PVDF) membrane. The membranes were blocked in 5% skimmed milk in TBS-Tween for 1 h at room temperature and incubated with primary antibodies (1:1,000; Abcam) overnight at 4°C, and washed with phosphate-buffered saline (PBS) with Tween 20 three times, followed by appropriate horseradish peroxidase-conjugated secondary antibody for 1 h. Membranes were developed in enhanced chemiluminescence reagent (Pierce, Rockford, IL, USA). GAPDH was used as the loading control.
Cell Proliferation Assay
Cells were plated at a density of 5 × 103 cells per well in 96-well plates with serum-free medium and cultured for 24 h. Then the medium was changed to normal medium containing 10% FBS and incubated for 24, 48, or 72 h. Subsequently, 10 μl of cell counting kit-8 (CCK-8) reagent (6 mg/ml) (Dojindo, Kumamoto, Japan) was added to each well and cultured for 4 h. The optical density (OD) was measured at 450 nm using a microplate reader (Bio-Tek, Winooski, VT, USA).
Cell Cycle Assay
Briefly, cells were collected and washed with PBS, and then fixed with 70% cold ethanol at −20°C for 2 h. Then cells were passed through Falcon Filters (70 μm) so that samples were monodispersed, which were incubated with RNase A and stained with propidium iodide (PI) at room temperature for 30 min (Cell Cycle Detection Kit; BD Pharmingen, San Diego, CA, USA). Flow cytometry analysis was carried out with a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA).
Cell Apoptosis Assay
Cell apoptosis assay was performed using Annexin-V–fluorescein isothiocyanate (FITC)/PI (PI/FITC) Apoptosis Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. In brief, cells were resuspended in binding buffer and then incubated with 5 μl of annexinV–FITC reagent and 5 μl of PI in the dark for 15 min. The samples were analyzed by flow cytometry with BD FACSCalibur (BD Biosciences, San Jose, CA, USA).
Transwell Assay
Cells (3 × 104) maintained in 20% Matrigel (no Matrigel for migration assay) in serum-free medium were seeded into the upper chambers with filters (8.0-μm membrane pores), and complete medium with 10% FBS supplementation was added to the lower chamber. After incubation for 24 h, cells migrated into the lower chambers. To quantify the cell number in the lower chambers, cells were fixed and stained with 0.5% crystal violet and then counted manually.
Dual-Luciferase Reporter Assay
The possible candidate targets of miR-202 were predicted by TargetScan (http://www.targetscan.org). The 3′-UTR of the human BCL2 gene with the predicted miR-202 binding site was amplified and subcloned to a pmirGLO vector (Promega Corporation, Fitchburg, WI, USA). The BCL2 3′-UTR with mutant binding site was also generated through a KOD-Plus-Mutagenesis Kit. HEK-293T cells were transfected with the wild-type (WT) or mutant (MUT) 3′-UTR vectors and miR-202 mimics or miR-NC. Luciferase activity was detected by the Dual-Luciferase Reporter Assay System.
Induction of Subcutaneous Tumors in Nude Mice
All animals were maintained and used in accordance with the guidelines of the Animal Center of Shandong University. Briefly, HepG2 cells (2 × 106/100 μl PBS) were subcutaneously injected into the nude mice (n = 5 mice per group; Shandong University Animal Center, Shandong, China). Tumor growth in nude mice was monitored every week. The tumor volume was calculated using the formula V = 0.5 × length × width2. Finally, mice were euthanized with an intraperitoneal injection of pentobarbital sodium (200 mg/kg) at the end of the experiment, and then tumors were collected for the determination of weight and volume. qRT-PCR and Western blotting were employed to detect the abundance of Ki-67 and BCL2.
Statistical Analysis
Three independent experiments were performed for each data point. The statistical results were represented as mean ± standard deviation (SD). Statistical analysis was conducted with GraphPad Prism 7 using Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test as appropriate. Survival curves were calculated using Kaplan–Meier and log-rank tests. A value of p < 0.05 was considered to be statistically significant.
RESULTS
Expressions of miR-202 and BCL2 in HCC Samples and Cells
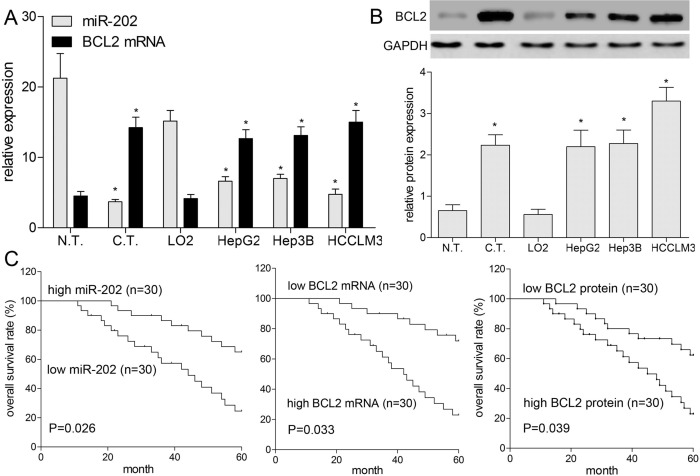
In this work, miR-202 and BCL2 expressions were analyzed in 60 pairs of HCC tissues and their matched adjacent normal liver tissues using qRT-PCR and Western blotting, respectively. As shown in Figure 1A, compared with paracancerous tissues, miR-202 expression was obviously lower, whereas the expression of BCL2 mRNA was significantly higher in 60 cases of HCC tissues (p < 0.05). In addition, BCL2 protein level was also higher in 60 cases of HCC tissues compared with that in paracancerous tissues (p < 0.05) (Fig. 1B). Furthermore, we detected the expressions miR-202 and BCL2 in the normal LO2 and three HCC cell lines (HepG2, Hep3B, and HCCLM3). The results showed that miR-202 expression was lower in HepG2, Hep3B, and HCCLM3 cells (p < 0.05) (Fig. 1A), while the expressions of BCL2 mRNA and protein were significantly higher in HCC cell lines as compared with those in LO2 cells (Fig. 1A and B), which indicated both miR-202 and BCL2 were implicated into progression of HCC.
Figure 1.
Expression profile and significance of miR-202 and BCL2 were analyzed in hepatocellular carcinoma (HCC) tissues and cells. (A) The expressions of miR-202 and BCL2 mRNA were detected by quantitative real-time polymerase chain reaction (qRT-PCR) in 60 cases of HCC tissues and four kinds of cell lines (HepG2, Hep3B, HCCLM3, and LO2). (B) The BCL2 protein expression was detected by Western blotting. (C) Kaplan–Meier analysis was used to assess overall survival (OS) of patients with HCC. *p < 0.05 by two-way analysis of variance (ANOVA) or Student’s t-test. N.T., normal tissues; C.T., cancer tissues.
Relationship of miR-202 and BCL2 With Clinicopathology and Prognosis of HCC
We analyzed the correlations among miR-202, BCL2, and a series of clinicopathologic indicators, including age, gender, vascular invasion, tumor size, and TNM stage (Table 1). The expressions of miR-202, BCL2 mRNA, and protein were found to be significantly correlated with major vascular invasion (p = 0.038, 0.029, and 0.031, respectively) and TNM stage (p = 0.012, 0.013, and 0.005, respectively). Therefore, we further determined that downregulated miR-202 and upregulated BCL2 were involved in the progression of HCC. As illustrated in Figure 1C, Kaplan–Meier analysis showed that patients with low expression of miR-202 had a worse overall survival (OS) than patients with high expression of miR-202. In addition, a worse OS was also associated with high expression of BCL2 mRNA or protein (p < 0.05) (Fig. 1C). Multivariate survival analysis showed that miR-202 (p = 0.001), BCL2 (p = 0.028 for mRNA, and 0.002 for protein), TNM stage (p = 0.020), and major vascular invasion (p = 0.022) were independent prognostic factors for patients with HCC (Table 2).
Table 1.
Association of miR-202 and BCL2 With Clinicopathological Parameters of Hepatocellular Carcinoma (HCC)
| Parameters | Case | miR-202 | p Value | BCL2 mRNA | p Value | BCL2 Protein | p Value |
|---|---|---|---|---|---|---|---|
| Age | 0.354 | 0.546 | 0.657 | ||||
| <55 | 28 | 4.81 ± 0.99 | 13.16 ± 3.11 | 2.03 ± 0.44 | |||
| ≥55 | 32 | 4.23 ± 1.03 | 14.74 ± 2.99 | 2.35 ± 0.38 | |||
| Gender | 0.197 | 0.335 | 0.391 | ||||
| Male | 38 | 4.69 ± 1.02 | 13.77 ± 3.25 | 2.39 ± 0.36 | |||
| Female | 22 | 2.78 ± 1.53 | 14.45 ± 2.98 | 1.89 ± 0.29 | |||
| Tumor size | 0.295 | 0.199 | 0.365 | ||||
| <5 cm | 27 | 5.01 ± 1.33 | 12.82 ± 3.24 | 1.95 ± 0.26 | |||
| ≥5 cm | 33 | 4.08 ± 1.12 | 14.98 ± 3.46 | 2.42 ± 0.34 | |||
| Tumor number | 0.426 | 0.116 | 0.422 | ||||
| Single | 38 | 4.73 ± 1.22 | 13.33 ± 3.64 | 2.33 ± 0.35 | |||
| Multiple | 22 | 4.10 ± 1.34 | 15.14 ± 4.14 | 1.98 ± 0.37 | |||
| Cirrhosis | 0.107 | 0.136 | 0.144 | ||||
| No | 19 | 3.92 ± 2.09 | 12.55 ± 3.24 | 1.65 ± 0.25 | |||
| Yes | 41 | 4.77 ± 1.25 | 14.67 ± 2.79 | 2.46 ± 0.33 | |||
| Major vascular invasion | 0.038* | 0.029* | 0.031* | ||||
| No | 21 | 6.01 ± 1.15 | 10.61 ± 3.43 | 0.91 ± 0.35 | |||
| Yes | 39 | 3.68 ± 0.65 | 15.84 ± 2.46 | 2.89 ± 0.89 | |||
| TNM stage | 0.012* | 0.013* | 0.005* | ||||
| I–II | 25 | 6.11 ± 1.27 | 11.16 ± 3.09 | 1.01 ± 0.98 | |||
| III–IV | 35 | 3.36 ± 1.01 | 16.03 ± 2.96 | 3.00 ± 1.11 |
Statistically significant.
Table 2.
Multivariate Cox Regression Analysis of Overall Survival of HCC Patients
| Parameters | Hazard Ratio | 95% CI | p Value |
|---|---|---|---|
| Major vascular invasion | 1.55 | 1.07–2.26 | 0.022 |
| TNM stage | 1.51 | 1.07–2.15 | 0.020 |
| miR-202 | 2.48 | 1.77–3.49 | 0.001 |
| BCL2 mRNA | 1.78 | 1.06–3.01 | 0.028 |
| BCL2 protein | 1.78 | 1.23–2.57 | 0.002 |
miR-202 Represses HCC Cell Growth and Promotes Apoptosis
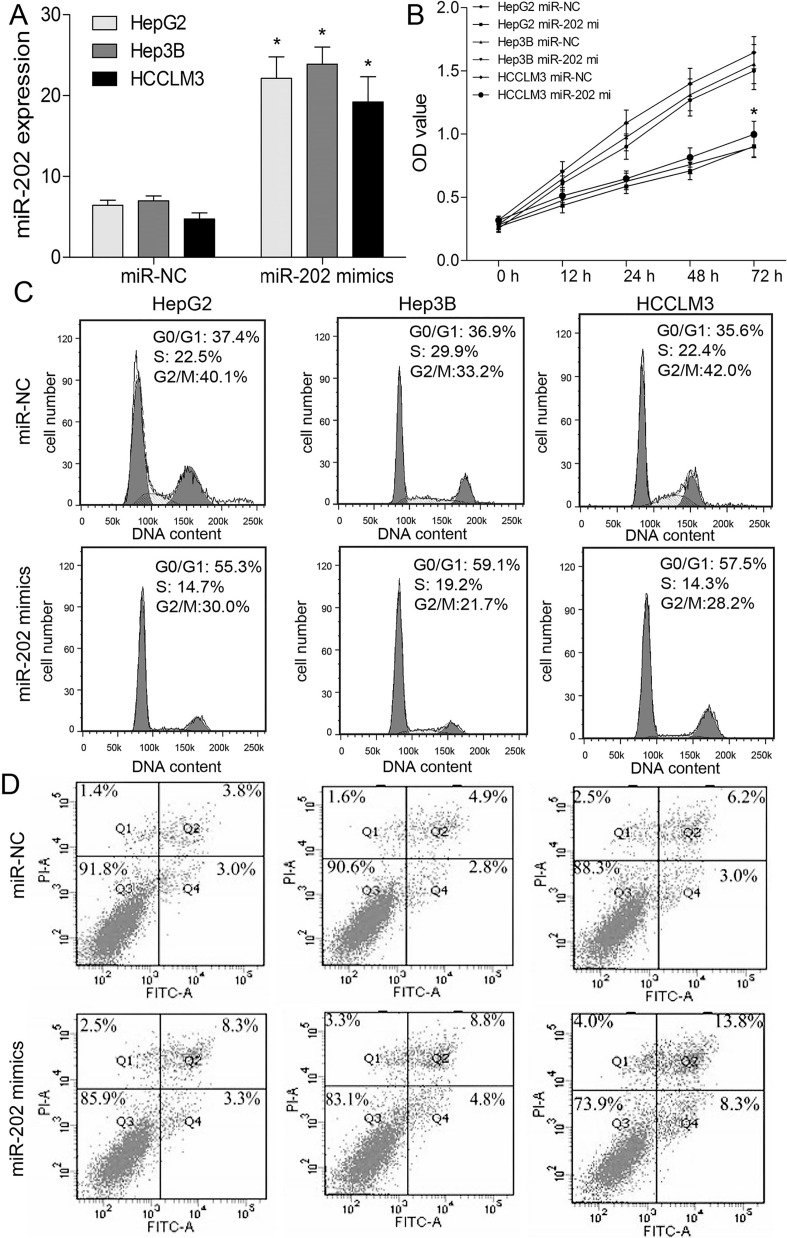
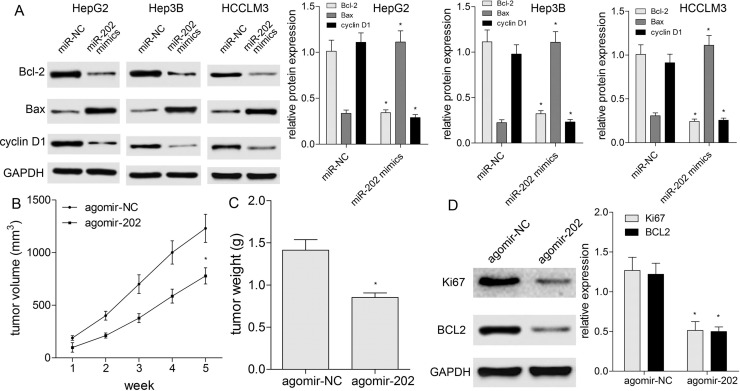
To further figure out the role of miR-202 in proliferation, cell cycle, and apoptosis of HCC cells, transfection of miR-202 mimics, CCK-8, and flow cytometric analysis were conducted. At 48 h posttransfection, miR-202 was markedly upregulated compared with miR-NC (Fig. 2A). The CCK-8 assay showed miR-202 overexpression significantly inhibited proliferation of HepG2, Hep3B, and HCCLM3 cells (p < 0.05) (Fig. 2B). Cell cycle analysis by flow cytometry showed that the cell populations in the G0/G1 phase were increased, while the cell populations in the S and G2/M phases were decreased in HCC cells transfected with miR-202 mimics (p < 0.05) (Fig. 2C). Additionally, apoptosis assay revealed that overexpression of miR-202 induced apoptosis of HCC cells (p < 0.05) (Fig. 2D). Based on molecular level, we detected the cell cycle- and apoptosis-associated proteins, including cyclin D1, Bax, and Bcl-2. Western blotting revealed that miR-202 repressed the expressions of cyclin D1 and Bcl-2 proteins, and enhanced Bax protein expression (p < 0.05) (Fig. 3A). To validate the tumor-suppressive role of miR-202 in HCC in vivo, HepG2 cells were subcutaneously implanted into nude mice to establish a HepG2 xenograft model. We noticed obviously smaller tumor volume (p < 0.05) (Fig. 3B) and lower tumor weight (p < 0.05) (Fig. 3C) in miR-202-overexpressing mice than agomir-NC. Moreover, Western blotting displayed lower Ki-67 and BCL2 expressions in miR-202-overexpressing nude mice compared with agomir-NC, indicating miR-202 repressed in vivo xenografted tumor growth (p < 0.05) (Fig. 3D).
Figure 2.
miR-202 inhibits the growth of HCC cells. (A) The expression level of miR-202 was detected by qRT-PCR in HepG2, Hep3B, and HCCLM3 cells transfected with miR-202 mimics or miR-NC. (B) Cell viability was analyzed by cell counting kit-8 (CCK-8) assay. (C) Cell cycle was determined by flow cytometry. (D) The apoptosis was analyzed by flow cytometry. *p < 0.05 by two-way ANOVA or Student’s t-test. miR-202 mi, miR-202 mimics.
Figure 3.
miR-202 affects apoptotic proteins and xenografted tumor growth. (A) Western blotting was used to detect cell cycle- and apoptosis-related biomarkers (cyclin D1, Bax, and Bcl-2) in HCC cells with miR-202 mimics or miR-NC. Protein expression was quantified by densitometry analysis using ImageJ and normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) level. (B) Size of xenografted tumors (n = 5 mice per group) was measured every week. (C) Weight of xenografted tumors (n = 5 mice per group) was summarized as a bar graph. (D) Western blotting was used to detect Ki-67 and BCL2 expressions in xenografted tumor tissues (n = 5 mice per group). Protein expression was quantified by densitometry analysis using ImageJ and normalized by GAPDH level. *p < 0.05 by two-way ANOVA or Student’s t-test.
miR-202 Represses Cell Migration, Invasion, and EMT
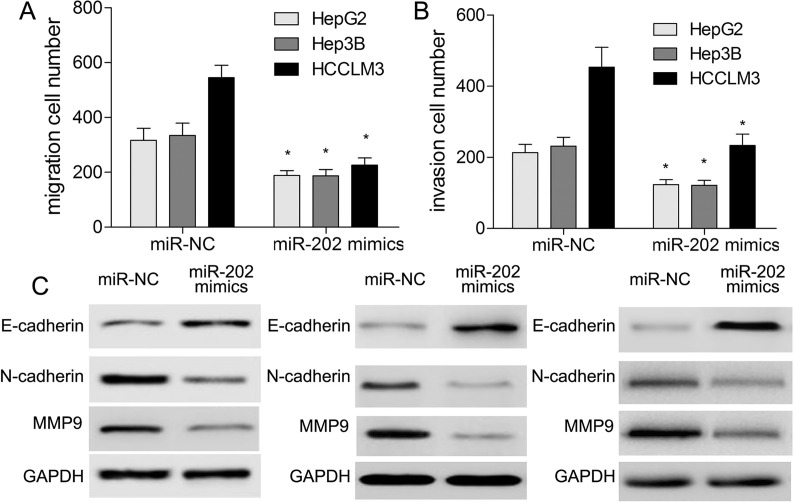
To understand the effect of miR-202 on migration, invasion, and EMT of HCC cells, we conducted Transwell assay and Western blotting. The results showed that in comparison with the miR-NC group, overexpression of miR-202 led to inhibition of migration and invasion capacities in HCC cells (p < 0.05) (Fig. 4A and B). Western blotting analysis suggested that E-cadherin expression was significantly increased, while the expressions of N-cadherin and matrix metalloproteinase 9 (MMP9) were significantly decreased following miR-202 overexpression (p < 0.05) (Fig. 4C), indicating that miR-202 may affect major vascular invasion of HCC cells.
Figure 4.
miR-202 represses migration, invasion, and EMT of HCC cells. Transwell assay was used to determine migration (A) and invasion (B) capacities of HCC cells after transfection of miR-202 mimics. (C) Western blotting was applied to detect the expressions of EMT-associated markers. *p < 0.05 by two-way ANOVA or Student’s t-test.
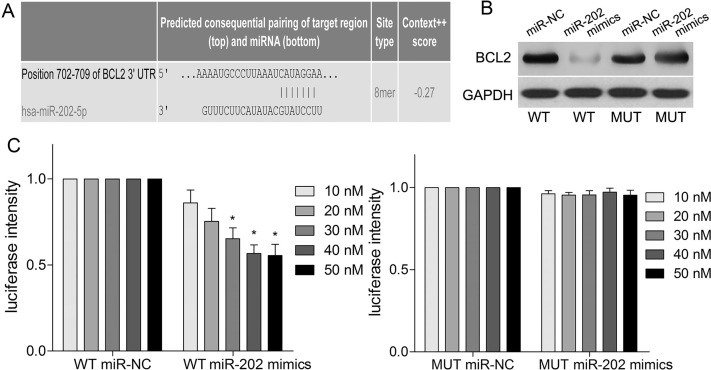
miR-202 Directly Targets BCL2 3′-UTR
Using an online prediction website (http://www.targetscan.org), we demonstrated that the 3′-UTR of BCL2 mRNA may be a binding sequence of miR-202 (Fig. 5A). Further experiments demonstrated miR-202 mimics significantly suppressed the protein level of BCL2 in HEK-293T cells transfected with miR-202 mimics and BCL2 overexpression vector containing WT 3′-UTR compared with MUT 3′-UTR (p < 0.05) (Fig. 5B). Then dual-luciferase reporter assay showed that the luminescence intensity from the wild-type vector carrying the 3′-UTR of BCL2 was significantly reduced after transfection of miR-202 mimics (p < 0.01) (Fig. 5C), while the mutant vector showed no change in luminescence (p > 0.05) (Fig. 5C).
Figure 5.
BCL2 is a direct target of miR-202. (A) The 3′-untranslated region (3′-UTR) of BCL2 was a predicted target sequence of miR-202. (B) Western blotting was used to analyze the expression of BCL2 in HEK-293T cells, which were transfected with miR-202 mimics and 3′-UTR of BCL2. (C) Analysis of relative activities of luciferase reporter containing BCL2-wild type (WT) or BCL2-mutant (MUT). *p < 0.05 by two-way ANOVA or Student’s t-test.
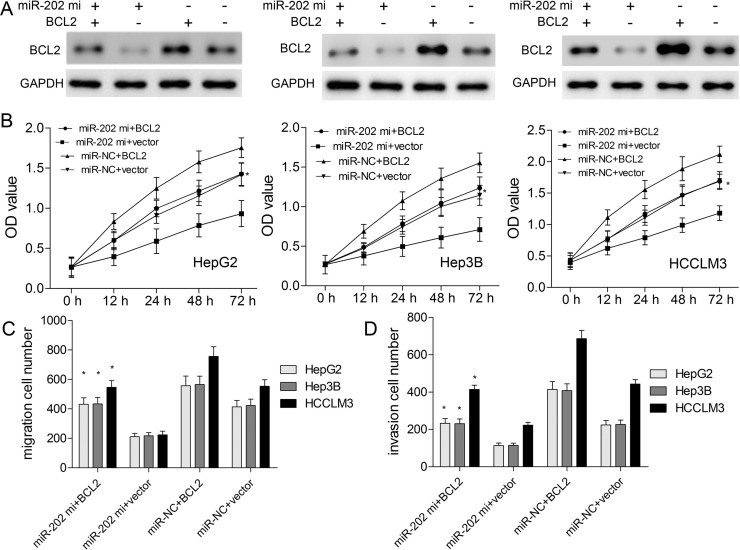
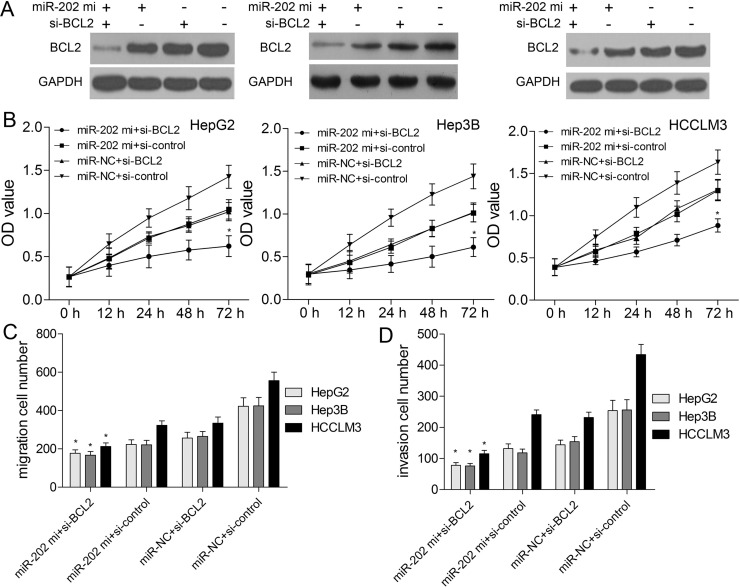
miR-202 Regulates Biological Behaviors of HCC via Targeting BCL2
To verify whether miR-202 could regulate biological behaviors of HCC by negatively regulating BCL2 expression, BCL2 protein expression was upregulated in HepG2, Hep3B, and HCCLM3 cells. BCL2 plasmids without its 3′-UTR were constructed and transfected into HepG2, Hep3B, and HCCLM3 cells to enhance its expression (Fig. 6A). The in vitro results demonstrated that BCL2 overexpression could weaken the inhibitory effects of miR-202 mimics on cell proliferation (Fig. 6B), migration (Fig. 6C), and invasion (Fig. 6D). In addition, BCL2-specific siRNAs were transfected into HCC cells to silence BCL2 protein expression. BCL2 protein expression was significantly downregulated in the BCL2-siRNA group, whereas the expression level of BCL2 in the si-control group did not significantly change (Fig. 7A). Further assays demonstrated that BCL2 downregulation could enhance the inhibitory effects of miR-202 mimics on proliferation (Fig. 7B), migration (Fig. 7C), and invasion (Fig. 7D). Intriguingly, miR-202 overexpression and BCL2 silencing showed the synergistic effects on biological behaviors of HCC cells.
Figure 6.
BCL2 overexpression weakens the inhibitory effects of miR-202 on HCC cells. (A) Western blotting was used to detect BCL2 protein level after transfection of pcDNA-BCL2. (B) CCK-8 assay was used to measure cell viability. The migration (C) and invasion (D) capacities of HCC cells with miR-202 mimics/miR-NC and pcDNA-BCL2/empty vector were analyzed by Transwell assay. *p < 0.05 by two-way ANOVA or Student’s t-test.
Figure 7.
BCL2 silencing enhances the inhibitory effects of miR-202 on HCC cells. (A) Western blotting was used to detect BCL2 protein expression when HCC cells were transfected with miR-202 mimics and si-BCL2. (B) CCK-8 assay was applied to analyze cell viability. Transwell assay was used to validate migration (C) and invasion (D) capacities of HCC cells after cotransfection of miR-202 mimics and si-BCL2. *p < 0.05 by two-way ANOVA or Student’s t-test.
DISCUSSION
To date, some studies have demonstrated that the expression of miR-202 in cancer tissues varies somewhat depending on the type of cancers, and miR-202 may serve as a biomarker for the diagnosis and prognosis of patients with most cancers5. Many studies have also clearly shown that miR-202 is downregulated in breast cancer and gastric cancer cells8,11. In the present study, we found miR-202 was obviously reduced in HCC tissues and cell lines, whereas BCL2 showed the inverse expression profile. Statistically, miR-202 or BCL2 expression was negatively or positively correlated with vascular invasion, TNM, and poor prognosis. These findings indicated the potential role of miR-202 and BCL2 in progression of HCC.
Jiang et al. reported that miR-202 inhibited cell proliferation and induced cell cycle arrest in the G0/G1 phase and apoptosis12. Meng et al. found that miR-202 may function as a novel tumor suppressor in esophageal squamous cell carcinoma by repressing cell proliferation and migration13. Consistent with the above findings, we identified that miR-202 inhibited proliferation and in vivo xenografted tumor volume and weight. Also, miR-202 caused cell cycle arrest and occurrence of apoptosis, decreased cyclin D1 and Bcl-2 expressions, and increased Bax expression. Additionally, EMT has been reported as an important process in tumorigenesis. When EMT occurs, a polarized epithelial cell converts into a mesenchymal cell, which is characterized by an enhanced migration, invasion, and resistance to apoptosis. In this work, miR-202 also suppressed the expressions of EMT-associated markers to inhibit migration and invasiveness of HCC cells.
According to TargetScan, BCL2 was a potential target gene of miR-202. The BCL2 family serves as a key death factor in an irreversible intrinsic apoptotic event14 and plays an important role in regulating cell viability, determining cell fate and protecting against cellular stress. To this end, the functions of the BCL2 family have been widely elucidated by a number of reports15. Li et al. reported that IL-17A affected autophagic activity of HCC cells by inhibiting degradation of BCL216. Liao et al. found that miR-448 inhibited cell growth by targeting BCL2 in HCC17. Zhang et al. suggested that dioscin suppressed HCC tumor growth by inducing apoptosis and regulating BCL218. Consequently, further studies demonstrated the interaction between miR-202 and BCL2. Exogenous expression of BCL2 weakened the inhibitory effects of miR-202 overexpression on malignant proliferation, migration, and invasion of HCC cells.
Taken together, our research indicated that miR-202 affected proliferation, migration, invasion, and EMT of HCC cells by inhibiting BCL2 expression. Notably, these findings could provide a novel target for deciphering the pathogenesis of HCC and may provide new strategies for the diagnosis and therapy of HCC.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Jindal A, Thadi A, Shailubhai K. Hepatocellular carcinoma: Etiology and current and future drugs. J Clin Exp Hepatol. 2019;9:221–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wang J, Yan C, Fu A. A randomized clinical trial of comprehensive education and care program compared to basic care for reducing anxiety and depression and improving quality of life and survival in patients with hepatocellular carcinoma who underwent surgery. Medicine (Baltimore). 2019;98:e17552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kuwaki K, Nouso K, Miyashita M, Makino Y, Hagihara H, Moriya A, Adachi T, Wada N, Yasunaka Y, Yasunaka T, Takeuchi Y, Onishi H, Nakamura S, Ikeda F, Shiraha H, Takaki A, Okada H. The efficacy and safety of steroids for preventing postembolization syndrome after transcatheter arterial chemoembolization of hepatocellular carcinoma. Acta Med Okayama 2019;73:333–9. [DOI] [PubMed] [Google Scholar]
- 4. Ju JX, Zeng QJ, Xu EJ, He XQ, Tan L, Huang QN, Li K, Zheng RQ. Intraprocedural contrast-enhanced ultrasound-CT/MR fusion imaging assessment in HCC thermal ablation to reduce local tumor progression: Compared with routine contrast-enhanced ultrasound. Int J Hyperthermia 2019;36:785–93. [DOI] [PubMed] [Google Scholar]
- 5. Weidle UH, Schmid D, Birzele F, Brinkmann U. MicroRNAs involved in metastasis of hepatocellular carcinoma: Target candidates, functionality and efficacy in animal models and prognostic relevance. Cancer Genomics Proteomics 2020;17:1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ding L, Gu H, Xiong X, Ao H, Cao J, Lin W, Yu M, Lin J, Cui Q. MicroRNAs involved in carcinogenesis, prognosis, therapeutic resistance and applications in human triple-negative breast cancer. Cells 2019;8 pii: E1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yang X, Pan W, Xu G, Chen L. Mitophagy: A crucial modulator in the pathogenesis of chronic diseases. Clin Chim Acta 2020;502:245–54. [DOI] [PubMed] [Google Scholar]
- 8. Zhao Y, Li C, Wang M, Su L, Qu Y, Li J, Yu B, Yan M, Yu Y, Liu B, Zhu Z. Decrease of miR-202-3p expression, a novel tumor suppressor, in gastric cancer. PLoS One 2013;8:e69756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dou D, Shi YF, Liu Q, Luo J, Liu JX, Liu M, Liu YY, Li YL, Qiu XD, Tan HY. Hsa-miR-202-3p, up-regulated in type 1 gastric neuroendocrine neoplasms, may target DUSP1. World J Gastroenterol. 2018;24:573–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yi Y, Li H, Lv Q, Wu K, Zhang W, Zhang J, Zhu D, Liu Q, Zhang W. miR-202 inhibits the progression of human cervical cancer through inhibition of cyclin D1. Oncotarget 2016;7:72067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu F, Li H, Hu C. MiR-202 inhibits cell proliferation, invasion, and migration in breast cancer by targeting ROCK1 gene. J Cell Biochem. 2019;120:16008–18. [DOI] [PubMed] [Google Scholar]
- 12. Jiang J, Huang J, Wang XR, Quan YH. MicroRNA-202 induces cell cycle arrest and apoptosis in lung cancer cells through targeting cyclin D1. Eur Rev Med Pharmacol Sci. 2016;20:2278–84. [PubMed] [Google Scholar]
- 13. Meng X, Chen X, Lu P, Ma W, Yue D, Song L, Fan Q. MicroRNA-202 inhibits tumor progression by targeting LAMA1 in esophageal squamous cell carcinoma. Biochem Biophys Res Commun. 2016;473:821–7. [DOI] [PubMed] [Google Scholar]
- 14. Montero J, Letai A. Why do BCL-2 inhibitors work and where should we use them in the clinic? Cell Death Differ. 2018;25:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hao W, Luo W, Bai M, Li J, Bai X, Guo J, Wu J, Wang M. MicroRNA-206 inhibited the progression of glioblastoma through BCL-2. J Mol Neurosci. 2016;60:531–8. [DOI] [PubMed] [Google Scholar]
- 16. Li S, Lin Z, Zheng W, Zheng L, Chen X, Yan Z, Cheng Z, Yan H, Zheng C, Guo P. IL-17A inhibits autophagic activity of HCC cells by inhibiting the degradation of Bcl2. Biochem Biophys Res Commun. 2019;509:194–200. [DOI] [PubMed] [Google Scholar]
- 17. Liao ZB, Tan XL, Dong KS, Zhang HW, Chen XP, Chu L, Zhang BX. miRNA-448 inhibits cell growth by targeting BCL-2 in hepatocellular carcinoma. Dig Liver Dis. 2019;51:703–11. [DOI] [PubMed] [Google Scholar]
- 18. Zhang G, Zeng X, Zhang R, Liu J, Zhang W, Zhao Y, Zhang X, Wu Z, Tan Y, Wu Y, Du B. Dioscin suppresses hepatocellular carcinoma tumor growth by inducing apoptosis and regulation of TP53, BAX, BCL2 and cleaved CASP3. Phytomedicine 2016;23:1329–36. [DOI] [PubMed] [Google Scholar]