Abstract
Tamoxifen-induced radioresistance, reported in vitro, might pose a problem for patients who receive neoadjuvant tamoxifen treatment and subsequently receive radiotherapy after surgery. Previous studies suggested that DNA damage repair or cell cycle genes are involved, and could therefore be targeted to preclude the occurrence of cross-resistance. We aimed to characterize the observed cross-resistance by investigating gene expression of DNA damage repair genes and cell cycle genes in estrogen receptor-positive MCF-7 breast cancer cells that were cultured to tamoxifen resistance. RNA sequencing was performed, and expression of genes characteristic for several DNA damage repair pathways was investigated, as well as expression of genes involved in different phases of the cell cycle. The association of differentially expressed genes with outcome after radiotherapy was assessed in silico in a large breast cancer cohort. None of the DNA damage repair pathways showed differential gene expression in tamoxifen-resistant cells compared to wild-type cells. Two DNA damage repair genes were more than two times upregulated (NEIL1 and EME2), and three DNA damage repair genes were more than two times downregulated (PCNA, BRIP1, and BARD1). However, these were not associated with outcome after radiotherapy in the TCGA breast cancer cohort. Genes involved in G1, G1/S, G2, and G2/M phases were lower expressed in tamoxifen-resistant cells compared to wild-type cells. Individual genes that were more than two times upregulated (MAPK13) or downregulated (E2F2, CKS2, GINS2, PCNA, MCM5, and EIF5A2) were not associated with response to radiotherapy in the patient cohort investigated. We assessed the expression of DNA damage repair genes and cell cycle genes in tamoxifen-resistant breast cancer cells. Though several genes in both pathways were differentially expressed, these could not explain the cross-resistance for irradiation in these cells, since no association to response to radiotherapy in the TCGA breast cancer cohort was found.
Key words: Treatment resistant, Tamoxifen treatment, Radiotherapy, DNA damage repair, Cell cycle control
INTRODUCTION
Radiotherapy and hormonal treatment (tamoxifen) are both corner stones of breast cancer treatment, and are successful in a large number of patients. However, when resistance to these treatment modalities occurs, adverse outcomes are likely for the patient. Previously we, and others, showed that breast cancer cells cultured to tamoxifen resistance also acquire radioresistance in vitro1–3. In the classical breast cancer treatment regimen, where tamoxifen is given after radiotherapy following surgery, this poses no problem for patients. However, neoadjuvant endocrine therapy, such as tamoxifen, is increasingly given to breast cancer patients. Endocrine treatment-induced radioresistance could pose a problem for these patients when they receive radiotherapy later in the treatment schedule4. Moreover, patients that receive radiotherapy as treatment for metastasized disease could suffer from reduced efficiency due to earlier adjuvant tamoxifen treatment.
In order to prevent cross-resistance for irradiation in tamoxifen-treated tumors, it is imperative to identify the genes and/or pathways that cause radioresistance in tamoxifen-resistant breast cancer. Several studies have addressed the expression of DNA damage repair genes in tamoxifen-resistant cells. Luzhna et al.2 described that while after irradiation wild-type MCF-7 cells had decreased levels of genes making up the base excision repair (BER), homologous recombination (HR), and mismatch repair (MMR) pathways, these genes were not differentially expressed in tamoxifen-resistant MCF-7 cells after irradiation. Moreover, the tamoxifen-resistant cells displayed more efficient repair of double-strand breaks and were less susceptible to apoptosis. Others found that tamoxifen-resistant MCF-7 cells exhibited increased levels of PARP1 and LIG3, both part of the alternative nonhomologous end joining (NHEJ) pathway, and are dependent on this pathway for repair of double-strand breaks. Significantly more yH2AX foci and a large number of genomic aberrations were present in those resistant cells5. Another study showed that BRCA1 and its associated protein BARD1 are upregulated in tamoxifen-resistant breast cancer cells, which render the cells resistant to DNA-damaging chemotherapy6. Thus, DNA damage repair is reportedly altered in tamoxifen-resistant breast cancer cells, possibly explaining their cross-resistance to radiotherapy.
Next to DNA damage repair genes, differential expression of cell cycle genes might also contribute to the altered radiosensitivity observed in tamoxifen-resistant cells. Tamoxifen treatment decreases the percentage of cells in S phase, while inducing a G1 block7–10. The efficiency of radiotherapy depends on the cell cycle phase cells are in, a phenomenon that has been studied for a long time11. The most sensitive phases of the cell cycle are the G2 and M phases, while cells in the G1 phase are more resistant to irradiation, and cells in the S phase are the most resistant12. Tamoxifen-resistant cells have increased levels of genes regulating the G1/S transition (CCNE1, CDK2, and E2F1), and higher percentages of tamoxifen-resistant cells were present in S phase compared to parental cells13. Also, CCND1 and MYC are reportedly upregulated in tamoxifen resistance, which could lead to G1/S phase blockade14. Higher expression of these genes in tamoxifen-resistant cells could explain the cross-resistance to irradiation.
Here we aim to assess differentially regulated genes in estrogen receptor-positive breast cancer cells cultured to tamoxifen resistance, in order to explain the increased radioresistance observed in these cells. To this end, we analyzed the expression of DNA damage repair genes and cell cycle genes in tamoxifen-resistant breast cancer cells compared to wild-type cells, and validated their predictive power in a large breast cancer cohort in silico.
MATERIALS AND METHODS
Cell Culture
The culturing of estrogen receptor-positive MCF-7 cells (LCG Standards, Teddington, UK) was described previously, including the number of passages and authentication of the cell lines3. Tamoxifen-resistant cells were acquired by culturing MCF-7 cells with 4-hydroxytamoxifen (#H7904; Sigma-Aldrich, St. Louis, MO, USA), increasing the dose weekly up to 10 μM15.
RNA Isolation
The Total RNA Purification kit (Norgen Biotek Corp., Thorold, ON, Canada) was used to isolate RNA. On column DNase treatment (RNase-Free DNase Set; #79254; Qiagen, Hilden, Germany) was performed, all according to the manufacturer’s instructions.
RNA Sequencing
RNA sequencing was performed on wild-type MCF-7 and tamoxifen-resistant MCF-7 cells as described previously3.
Patients: TCGA Database
Data from the Cancer Genome Atlas (TCGA) project’s breast cancer cohort (https://portal.gdc.cancer.gov/projects/TCGA-BRCA) were analyzed for expression of genes of interest and associated to patient outcome after radiotherapy. Data were accessed and processed with the University of California at Santa Cruz Xena Browser at http://xena.ucsc.edu/. Node-negative and metastasis-free patients were selected and grouped into radiotherapy-treated (n = 209) and nonradiotherapy-treated (n = 194) patients. Relapse-free survival was assessed.
Statistics
Mean deviation from 1 was calculated for the genes in different DNA damage repair pathways or cell cycle phase with a Student’s t-test. Xena Browser TCGA data were imported into SPSS (SPSS Inc., Chicago, IL, USA), and Breslow p values at median cutoff were calculated for all genes of interest.
RESULTS
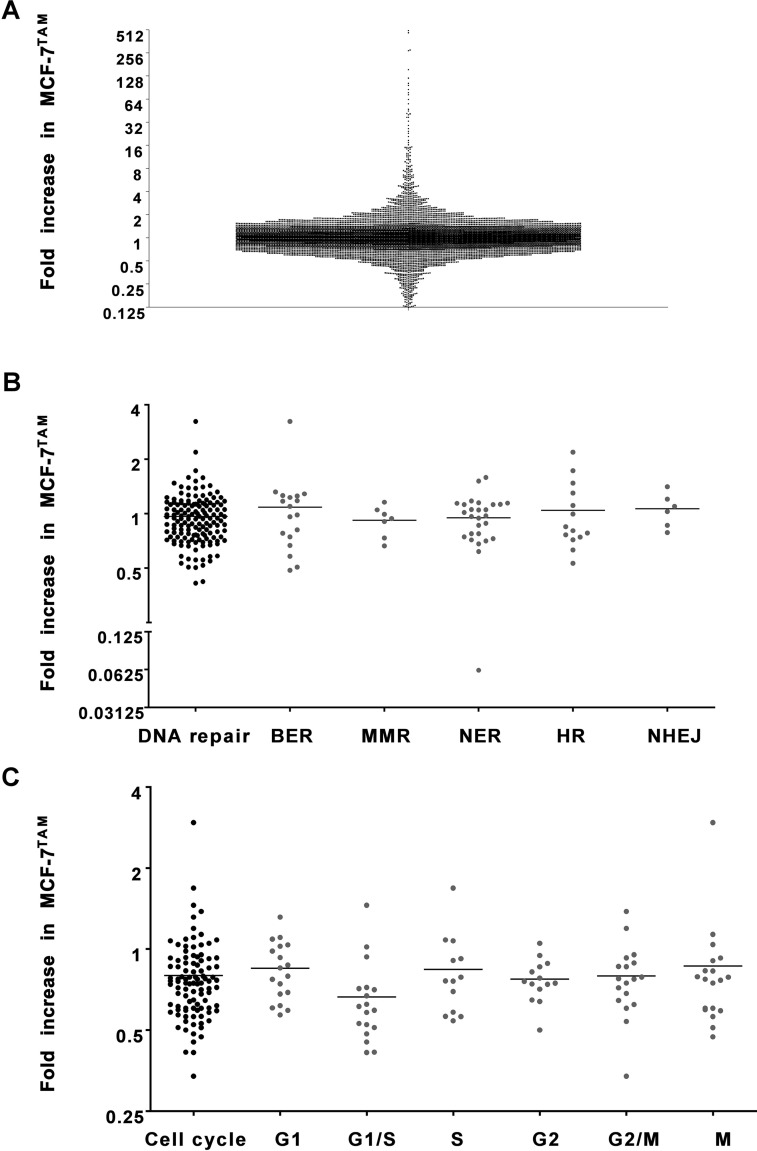
RNA sequencing of tamoxifen-resistant cells (MCF-7TAM) and MCF-7 wild-type cells (MCF-7WT) was performed. For further analyses, only protein-coding genes were included that had more than 10 reads in either the wild-type cells or tamoxifen-resistant cells. The relative expression of these genes in tamoxifen-resistant cells compared to wild-type cells was assessed (Fig. 1A). Many genes were not differentially expressed in tamoxifen-resistant cells compared to wild-type cells. However, 487 genes were more than two times increased, and 493 genes were more than two times decreased. The genes with the highest change in expression were 500 times increased, or over 6,000 times decreased in tamoxifen-resistant cells compared to wild-type cells.
Figure 1.
Expression of DNA damage repair genes and cell cycle genes in tamoxifen-resistant breast cancer cells. The expression of (A) all genes, (B) DNA damage repair genes, and (C) cell cycle genes in tamoxifen-resistant MCF-7 cells compared to wild-type cells. For each gene, the fold change in expression levels in MCF-7TAM compared to MCF-7WT is shown (based on normalized read counts), as measured by RNA sequencing.
Expression of DNA Damage Repair Genes in Tamoxifen-Resistant Breast Cancer Cells
We assessed the expression of genes that are known to be involved in various types of DNA damage repair (https://www.mdanderson.org/documents/Labs/Wood-Laboratory/human-dna-repair-genes.html 16,17); 148 of these genes that were protein coding and passed our read–count threshold were further analyzed (Fig. 1B). The genes of interest were subdivided in the following groups: base excision repair (BER), mismatch repair (MMR), nucleotide excision repair (NER), homologous recombination (HR), and nonhomologous end joining (NEHJ) (Fig. 1B). None of the groups were differentially expressed in tamoxifen-resistant cells compared to the wild-type cell lines (two-sided t-test compared to mean = 1; BER: p = 0.5483, MMR: p = 0.2744, NER: p = 0.4092, HR: p = 0.7267, NHEJ: p = 0.5107).
Out of these 148 genes, we identified individual genes with a minimum twofold change in expression (Table 1). Only two DNA damage repair genes (NEIL1 and EME2) were upregulated in tamoxifen-resistant cells, while two other DNA damage repair genes (BRIP1 and PCNA) were downregulated.
Table 1.
Differential Expression of DNA Damage Repair Genes in Tamoxifen-Resistant Breast Cancer Cells
| Gene Name | Gene Function | Fold Change MCF-7TAM |
|---|---|---|
| NEIL1 | DNA glycosylase, initiation of base excision repair | 3.24 |
| EME2 | Endonuclease, homologous recombination | 2.19 |
| BRIP1 | Helicase, BRCA1 interacting | 0.42 |
| PCNA | DNA polymerase cofactor, nonhomologous end joining | 0.41 |
| LIG3 | DNA ligase, base excision repair | 1.25 |
| PARP1 | Base excision repair | 1.18 |
| BRCA1 | Homologous recombination | 0.63 |
| BARD1 | BRCA1 interacting | 0.42 |
List of the differentially expressed DNA damage repair genes in MCF-7TAM and genes that had previously been found to be differentially expressed in tamoxifen-resistant breast cancer cells5,6. For each gene, the fold change in expression levels in MCF-7TAM compared to MCF-7WT is shown (based on normalized read counts), as measured by RNA sequencing.
We further included genes that were shown to be differentially expressed in tamoxifen-resistant MCF-7 cells according to the literature: LIG3, PARP1, BRCA1, and BARD1 5,6 (Table 1). Of these, only BARD1 had a more than twofold decreased expression in tamoxifen-resistant MCF-7 cells compared to MCF-7 wild-type cells. In contrast, BARD1 has previously been found to exhibit increased expression in tamoxifen-resistant MCF-7 cells6.
Expression of Cell Cycle Genes in Tamoxifen-Resistant Breast Cancer Cells
Besides DNA damage repair genes, differential expression of cell cycle genes in tamoxifen-resistant breast cancer cells might also contribute to radioresistance12. One hundred and twenty genes characteristic of six different phases in the cell cycle (20 each for G1, G1/S, S, G2, G2/M, and M)18 were analyzed for their expression in the tamoxifen-resistant cells. After selecting for genes that were protein coding and that had more than 10 reads in either the wild-type cells or tamoxifen-resistant cells, 100 genes were left (G1: 18, G1/S: 18, S: 13, G2: 14, G2/M: 18, M: 19) (Fig. 1C). Gene expression was plotted as fold increase in tamoxifen-resistant MCF-7 cells compared to wild-type MCF-7 cells. Four of the groups showed differential expression in tamoxifen-resistant cells compared to the wild-type cells (two-sided t-test compared to mean = 1; G1: p = 0.0072, G1/S: p < 0.0001, S: p = 0.0907, G2: p < 0.0001, G2/M: p = 0.0017, M: p = 0.2806).
We also analyzed the individual genes for those that had at least two times increased expression or decreased expression in tamoxifen-resistant cells compared to wild-type cells (Table 2). Only one gene was more than two times upregulated (MAPK13). Six genes were downregulated in tamoxifen-resistant cells: E2F2, CKS2, GINS2, PCNA, MCM5, and EIF5A2.
Table 2.
Differential Expression of Cell Cycle Genes in Tamoxifen-Resistant Breast Cancer Cells
| Gene Name | Gene Function | Cell Cycle Phase | Fold Change MCF-7TAM |
|---|---|---|---|
| MAPK13 | Cellular stress-induced signaling protein | M | 2.95 |
| E2F2 | Cell cycle control | G1/S | 0.48 |
| CKS2 | Maintenance of cell wall integrity | M | 0.47 |
| GINS2 | Initiation of DNA replication | G1/S | 0.45 |
| PCNA | DNA polymerase cofactor | G1/S | 0.41 |
| MCM5 | Initiation of DNA replication | G1/S | 0.41 |
| EIF5A2 | Cell cycle control | G2/M | 0.34 |
| CCNE1 | CDK2 regulator | G1 | 0.87 |
| CDK2 | Initiation of DNA synthesis | G1/S | 0.71 |
| E2F1 | Cell cycle control | G1/S | 0.59 |
| MYC | Cell cycle progression | 0.33 | |
| CCND1 | CDK4/6 regulator | G1/S | 0.31 |
List of the differentially expressed cell cycle genes in MCF-7TAM. For each gene, the fold change in expression levels in MCF-7TAM compared to MCF-7WT is shown (based on normalized read counts), as measured by RNA sequencing.
Genes upregulated in tamoxifen-resistant cells, according to literature13,14, were also investigated here. CCNE1, CDK2, and E2F1 were marginally (13–41%) downregulated in tamoxifen-resistant cells, but less than our threshold of twofold change. MYC and CCND1 had a decreased expression more than twofold, contrary to the increase that was reported previously14.
Association of In Vitro Differentially Expressed Genes in a Breast Cancer Patient Cohort
Thus, only a limited number of DNA damage repair and cell cycle control genes exhibit differential expression in tamoxifen-resistant breast cancer cells. To assess the clinical association of the DNA damage repair genes and cell cycle genes with radioresistance, we analyzed data generated by the TCGA Research Network (https://cancergenome.nih.gov/) for relations between the expression of genes of interest and outcome after radiotherapy. Patients with T1–4, N0, M0 tumors were selected, and the cohort was divided in patients that did or did not receive radiotherapy as part of their primary treatment. Patients were dichotomized by the median value of each of the investigated genes. Of the five differentially expressed DNA damage repair genes, none was significantly associated with outcome in patients who received radiotherapy as part of their primary treatment (Table 3). Only expression of EME2 was specifically associated with poor outcome in patients who did not receive radiotherapy, but not in patients that had received radiotherapy.
Table 3.
Association of Differentially Expressed DNA Damage Genes and Cell Cycle Genes With Outcome After Radiotherapy
| Gene Name | TCGA: RT− | TCGA: RT+ |
|---|---|---|
| DNA damage repair | ||
| Increased | ||
| EME2 | 0.03 | 0.37 |
| NEIL1 | 0.44 | 0.10 |
| Decreased | ||
| PCNA * | 0.56 | 0.38 |
| BRIP1 | 0.19 | 0.30 |
| BARD1 | 0.78 | 0.32 |
| Cell cycle | ||
| Increased | ||
| MAPK12 | 0.31 | 0.16 |
| Decreased | ||
| E2F2 | 0.74 | 0.47 |
| CKS2 | 0.62 | 0.77 |
| GINS2 | 0.40 | 0.26 |
| PCNA * | 0.56 | 0.38 |
| MCM5 | 0.67 | 0.33 |
| EIF5A2 | 0.37 | 0.89 |
| CCND1 | 0.28 | 0.68 |
| MYC | 0.15 | 0.08 |
For each of the DNA damage genes and cell cycle genes that were differentially expressed in MCF-7TAM, the p values of the association with relapse-free survival in patients treated with or without radiotherapy in the TCGA breast cancer cohort are depicted, based on the Breslow test.
PCNA is both a DNA damage repair and a cell cycle gene.
The cell cycle genes that were differentially expressed in tamoxifen-resistant breast cancer cells were also examined for an association between their expression and outcome after radiotherapy in the TCGA cohort. None of the more than two times upregulated or downregulated genes were associated with outcome in patients either treated with or without radiotherapy (Table 3).
Therefore, we conclude that none of the differentially expressed genes (DNA damage repair or cell cycle related) are associated with radioresistance in the breast cancer patient cohort investigated.
DISCUSSION
Here we aimed to identify possible mechanisms of radioresistance in acquired tamoxifen-resistant breast cancer cells by investigating the expression of DNA damage repair genes and cell cycle genes in breast cancer cells cultured to tamoxifen resistance. Increased expression of genes that stimulate DNA damage repair could mean that these are responsible for radioresistance observed in these cells. Conversely, decreased expression of genes that inhibit DNA damage repair could also lead to radioresistance. Moreover, increased expression of cell cycle genes that correspond to radioresistant parts of the cell cycle could explain radioresistance observed in tamoxifen-resistant cells. Interestingly, cross-resistance for irradiation was previously observed in hormone treatment-insensitive prostate cancer cells. These show increased radioresistance and upregulated genes involved in cell cycle arrest and DNA damage repair, suggesting common mechanisms might be involved in various hormone-sensitive cancers19.
None of the known pathways involved in DNA damage repair (BER, MMR, NER, HR, or NEHJ) were as a whole differentially expressed in the tamoxifen-resistant cells. Luzhna et al. also investigated the expression of genes in DNA repair pathways in tamoxifen resistance, albeit after irradiation, and found that genes in these pathways are not up- or downregulated as well2. Interestingly, proteins involved in DNA damage repair are poor prognostic factors in estrogen receptor-positive breast cancer patients treated with endocrine therapy17. Therefore, differential expression of DNA damage repair genes could merely be associated with the occurrence of tamoxifen resistance, and not specifically with the radioresistant phenotype. This paper also showed that estrogen receptor-positive breast cancer patients have increased levels of damaging mutations in NER, BER, and NHEJ genes17.
The specific DNA damage repair genes that were upregulated or downregulated in tamoxifen-resistant cells in our current study cannot explain radioresistance observed in those cells. Although NEIL1 and EME2 (both >2-fold increased RNA expression in tamoxifen resistance) are both stimulators of DNA damage repair20,21, we did not find an association of their gene expression with outcome after radiotherapy in the TCGA patient cohort, meaning that the gene is not likely to induce radioresistance in patients. BRIP1 and PCNA were downregulated in tamoxifen-resistant breast cancer cells. Since these are both stimulators of DNA damage repair as well22,23, their decreased expression does not explain radioresistance observed in these cells. BRIP1 was associated with the repair of DNA double strand breaks, as evidenced by assessment of γH2AX foci, after chemotherapy24. Its lower expression in the tamoxifen-resistant cells in this study is therefore unclear.
Interestingly, PCNA is involved in DNA damage repair as well as in cell cycle regulation. In a study with radiotherapy-treated oral cancer patients, low expression of PCNA was associated with a better patient survival25, which indeed points toward an opposite role for PCNA in radioresistance than its expression in tamoxifen-resistant cells in this study suggests. However, in pancreatic tumor cells, PCNA was increased in cells treated with rapamycin, which was associated with decreased radioresistance26; in another study PCNA inhibition increased the number of double-strand breaks after treatment with DNA-damaging chemo agent cisplatin and therefore sensitized cells to this treatment27. PCNA may therefore have a dual role, either promoting or inhibiting DNA damage repair, and this may differ in different cancer models.
A BRCA1-interacting protein, BARD1, also showed decreased gene expression in tamoxifen-resistant cells, as well as BRCA1 itself, which had a slightly decreased expression, contrary to previous reports6. We did not observe an increased expression of LIG3 or PARP1, as Tobin et al. did5. This shows that even though some aspects of tamoxifen resistance are consistent when creating tamoxifen-resistant cells from wild-type MCF-7 cells, heterogeneity remains an important issue.
Cells are relatively resistant to irradiation in G1 and S phases, since DNA damage repair genes are highly expressed in those phases to guarantee correct DNA replication. We found a decrease in genes related to G1 and G1/S phases, which is not in line with the radioresistant phenotype of these cells. The only increased gene was MAPK13. Although it is present in the M phase, a relative radiosensitive phase, it has previously been associated with paclitaxel resistance in breast cancer28, and it is expressed in radioresistant gynecological cancer stem cells29. One of the genes with decreased expression (E2F2) is a mediator of apoptosis that is induced after DNA damage30,31. Its decreased expression could lead to a decrease in radiation-induced apoptosis, and therefore contributes to radioresistance in tamoxifen-resistant cells. CKS2, which slows down the cell cycle in order to allow repair of DNA damage32, is associated with enhanced sensitivity to different chemotherapeutic agents when overexpressed33, which could correspond to enhanced radioresistance in tamoxifen-resistant breast cancer cells where it is downregulated. However, in another study high expression levels of CKS2 were associated with decreased overall survival in breast cancer patients, which is contrary to these data34. MCM5 and GINS2 showed decreased expression in the tamoxifen-resistant cells. These genes are essential for DNA replication35. MCM5 is associated with worse outcome in patients when highly present in breast cancer patients36. GINS2 was previously associated with tamoxifen resistance when higher expressed in breast cancer patients, in contrast to what we found37. In another study, GINS2 knockdown was found to induce apoptosis38. These observations are contrary to the implications that our findings here have. EIF5A2 was earlier found to be upregulated in radioresistant colorectal cancer cell lines39, contrary to our findings, and high levels of EIF5A2 were associated with poor outcome after chemoradiotherapy in nasopharyngeal carcinoma patients40. In breast cancer high expression levels of EIF5A2 were associated with chemoresistance as well41. Finally, CCND1, a regulator of G1/S transition42, and MYC were also downregulated in tamoxifen-resistant breast cancer cells. CCND1 overexpression was previously associated with tamoxifen resistance43,44, while it was also associated with increased radiosensitivity in MCF-7 cells45. The latter is in line with our data. However, CCND1 knockdown in prostate cancer cells sensitized them to irradiation46. Downregulation of MYC has previously been shown to impair cell cycle progression47. High expression levels of MYC were present in radioresistant breast cancer cells48 and similarly in docetaxel-resistant lung cancer cells that were cross-resistant to irradiation49.
Many of the genes that were differentially expressed in breast cancer cells are associated with radiosensitivity. However, we did not find an association between the expression of the genes that were differentially expressed in tamoxifen-resistant breast cancer cells and outcome after radiotherapy in a breast cancer patient cohort. In some cases, our findings were contradictory to what others found in terms of association with radiosensitivity. This can partly be explained by the fact that we found these genes to be differentially expressed after chronic tamoxifen treatment. This change in expression could be an effect of this treatment alone and not involved in the cross-resistance for radiotherapy we observed. Therefore, the effect of these genes on radiosensitivity should indeed be further investigated in tamoxifen-resistant cells.
Other pathways involved in radioresistance ought to be investigated for their role in the cross-resistance observed in tamoxifen-resistant cells, such as hypoxia, one of the three crucial factors in radioresistance, next to proliferation and DNA damage repair50. Tamoxifen has been shown to induce hypoxia in MCF-7 xenografts51. Therefore, the role hypoxia-induced genes play in radioresistance in tamoxifen-resistant breast cancer should be further investigated. Moreover, in vitro studies may lack information that is crucial for therapy outcome in patients. Other cells in the tumor microenvironment can also contribute to responses to treatment. Another related factor is neovascularization, which is known to codetermine the response to cancer treatment52. Therefore, we wanted to confirm the results in clinical datasets, to establish correlations of DNA damage repair genes and cell cycle genes on patient outcome. Finally, the results obtained from retrospective clinical studies may be biased by the fact that patients who have been treated with radiotherapy differ from patients who have not been thus treated. Therefore, more clinical data are necessary before final conclusions about the role of these DNA damage repair genes and cell cycle genes can be drawn.
CONCLUSIONS
We identified differentially expressed DNA damage repair genes and cell cycle genes in tamoxifen-resistant breast cancer cells. Many of these are related to radiosensitivity according to literature. However, a direct relation between these genes and radioresistance could not be identified, since none of them was associated with outcome after radiotherapy in a breast cancer patient cohort. Thus, changes in DNA damage repair or cell cycle genes do not explain cross-resistance of tamoxifen-resistant breast cancer for radiotherapy and are not likely targets to preclude the occurrence of this cross-resistance.
ACKNOWLEDGMENTS
A.E.M.P. designed the study, developed the methodology, acquired, analyzed, and interpreted the data, and wrote the manuscript. J.B. designed the study, interpreted the data, reviewed and revised the manuscript, and supervised the study. F.C.G.J.S. designed the study, interpreted the data, reviewed and revised the manuscript, and supervised the study. P.N.S. designed the study, developed the methodology, acquired, analyzed, and interpreted the data, reviewed and revised the manuscript, and supervised the study. The datasets used during the current study are available from the corresponding author on request.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Paulsen GH, Strickert T, Marthinsen AB, Lundgren S. Changes in radiation sensitivity and steroid receptor content induced by hormonal agents and ionizing radiation in breast cancer cells in vitro. Acta Oncol. 1996;35(8):1011–9. [DOI] [PubMed] [Google Scholar]
- 2. Luzhna L, Lykkesfeldt AE, Kovalchuk O. Altered radiation responses of breast cancer cells resistant to hormonal therapy. Oncotarget 2015;6(3):1678–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Post AEM, Smid M, Nagelkerke A, Martens JWM, Bussink J, Sweep F, Span PN. Interferon-stimulated genes are involved in cross-resistance to radiotherapy in tamoxifen-resistant breast cancer. Clin Cancer Res. 2018;24(14):3397–408. [DOI] [PubMed] [Google Scholar]
- 4. Guerrero-Zotano AL, Arteaga CL. Neoadjuvant trials in ER(+) breast cancer: A tool for acceleration of drug development and discovery. Cancer Discov. 2017;7(6):561–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tobin LA, Robert C, Nagaria P, Chumsri S, Twaddell W, Ioffe OB, Greco GE, Brodie AH, Tomkinson AE, Rassool FV. Targeting abnormal DNA repair in therapy-resistant breast cancers. Mol Cancer Res. 2012;10(1):96–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhu Y, Liu Y, Zhang C, Chu J, Wu Y, Li Y, Liu J, Li Q, Li S, Shi Q, Jin L, Zhao J, Yin D, Efroni S, Su F, Yao H, Song E, Liu Q. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat Commun. 2018;9(1):1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Osborne CK, Boldt DH, Clark GM, Trent JM. Effects of tamoxifen on human breast cancer cell cycle kinetics: Accumulation of cells in early G1 phase. Cancer Res. 1983;43(8):3583–5. [PubMed] [Google Scholar]
- 8. Osborne CK, Boldt DH, Estrada P. Human breast cancer cell cycle synchronization by estrogens and antiestrogens in culture. Cancer Res. 1984;44(4):1433–9. [PubMed] [Google Scholar]
- 9. Lykkesfeldt AE, Larsen JK, Christensen IJ, Briand P. Effects of the antioestrogen tamoxifen on the cell cycle kinetics of the human breast cancer cell line, MCF-7. Br J Cancer 1984;49(6):717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bruno S, Di Vinci A, Geido E, Giaretti W. Cell cycle synchronization induced by tamoxifen and 17 beta-estradiol on MCF-7 cells using flow cytometry and a monoclonal antibody against bromodeoxyuridine. Breast Cancer Res Treat. 1988;11(3):221–9. [DOI] [PubMed] [Google Scholar]
- 11. Sinclair WK, Morton RA. X-ray sensitivity during the cell generation cycle of cultured Chinese hamster cells. Radiat Res. 1966;29(3):450–74. [PubMed] [Google Scholar]
- 12. Pawlik TM, Keyomarsi K. Role of cell cycle in mediating sensitivity to radiotherapy. Int J Radiat Oncol Biol Phys. 2004;59(4):928–42. [DOI] [PubMed] [Google Scholar]
- 13. Louie MC, McClellan A, Siewit C, Kawabata L. Estrogen receptor regulates E2F1 expression to mediate tamoxifen resistance. Mol Cancer Res. 2010;8(3):343–52. [DOI] [PubMed] [Google Scholar]
- 14. Butt AJ, McNeil CM, Musgrove EA, Sutherland RL. Downstream targets of growth factor and oestrogen signalling and endocrine resistance: The potential roles of c-Myc, cyclin D1 and cyclin E. Endocr Relat Cancer 2005;12(Suppl 1):S47–59. [DOI] [PubMed] [Google Scholar]
- 15. Nagelkerke A, Sieuwerts AM, Bussink J, Sweep FC, Look MP, Foekens JA, Martens JW, Span PN. LAMP3 is involved in tamoxifen resistance in breast cancer cells through the modulation of autophagy. Endocr Relat Cancer 2014;21(1):101–12. [DOI] [PubMed] [Google Scholar]
- 16. Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science 2001;291(5507):1284–9. [DOI] [PubMed] [Google Scholar]
- 17. Anurag M, Punturi N, Hoog J, Bainbridge MN, Ellis MJ, Haricharan S. Comprehensive profiling of DNA repair defects in breast cancer identifies a novel class of endocrine therapy resistance drivers. Clin Cancer Res. 2018;24(19):4887–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu Z, Lou H, Xie K, Wang H, Chen N, Aparicio OM, Zhang MQ, Jiang R, Chen T. Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nat Commun. 2017;8(1):22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xie BX, Zhang H, Yu L, Wang J, Pang B, Wu RQ, Qian XL, Li SH, Shi QG, Wang LL, Zhou JG. The radiation response of androgen-refractory prostate cancer cell line C4-2 derived from androgen-sensitive cell line LNCaP. Asian J Androl. 2010;12(3):405–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosenquist TA, Zaika E, Fernandes AS, Zharkov DO, Miller H, Grollman AP. The novel DNA glycosylase, NEIL1, protects mammalian cells from radiation-mediated cell death. DNA Repair (Amst) 2003;2(5):581–91. [DOI] [PubMed] [Google Scholar]
- 21. Fadden AJ, Schalbetter S, Bowles M, Harris R, Lally J, Carr AM, McDonald NQ. A winged helix domain in human MUS81 binds DNA and modulates the endonuclease activity of MUS81 complexes. Nucleic Acids Res. 2013;41(21):9741–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cantor SB, Bell DW, Ganesan S, Kass EM, Drapkin R, Grossman S, Wahrer DC, Sgroi DC, Lane WS, Haber DA, Livingston DM. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 2001;105(1):149–60. [DOI] [PubMed] [Google Scholar]
- 23. Choe KN, Moldovan GL. Forging ahead through darkness: PCNA, still the principal conductor at the replication fork. Mol Cell 2017;65(3):380–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Monteiro LJ, Khongkow P, Kongsema M, Morris JR, Man C, Weekes D, Koo CY, Gomes AR, Pinto PH, Varghese V, Kenny LM, Charles Coombes R, Freire R, Medema RH, Lam EW. The forkhead box M1 protein regulates BRIP1 expression and DNA damage repair in epirubicin treatment. Oncogene 2013;32(39):4634–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mallick S, Agarwal J, Kannan S, Pawar S, Kane S, Teni T. PCNA and anti-apoptotic Mcl-1 proteins predict disease-free survival in oral cancer patients treated with definitive radiotherapy. Oral Oncol. 2010;46(9):688–93. [DOI] [PubMed] [Google Scholar]
- 26. Dai ZJ, Gao J, Kang HF, Ma YG, Ma XB, Lu WF, Lin S, Ma HB, Wang XJ, Wu WY. Targeted inhibition of mammalian target of rapamycin (mTOR) enhances radiosensitivity in pancreatic carcinoma cells. Drug Des Devel Ther. 2013;7:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inoue A, Kikuchi S, Hishiki A, Shao Y, Heath R, Evison BJ, Actis M, Canman CE, Hashimoto H, Fujii N. A small molecule inhibitor of monoubiquitinated proliferating cell nuclear antigen (PCNA) inhibits repair of interstrand DNA cross-link, enhances DNA double strand break, and sensitizes cancer cells to cisplatin. J Biol Chem. 2014;289(10):7109–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chang H, Jeung HC, Jung JJ, Kim TS, Rha SY, Chung HC. Identification of genes associated with chemosensitivity to SAHA/taxane combination treatment in taxane-resistant breast cancer cells. Breast Cancer Res Treat. 2011;125(1):55–63. [DOI] [PubMed] [Google Scholar]
- 29. Yasuda K, Hirohashi Y, Kuroda T, Takaya A, Kubo T, Kanaseki T, Tsukahara T, Hasegawa T, Saito T, Sato N, Torigoe T. MAPK13 is preferentially expressed in gynecological cancer stem cells and has a role in the tumor-initiation. Biochem Biophys Res Commun. 2016;472(4):643–7. [DOI] [PubMed] [Google Scholar]
- 30. Vigo E, Muller H, Prosperini E, Hateboer G, Cartwright P, Moroni MC, Helin K. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol Cell Biol. 1999;19(9):6379–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Martinez LA, Goluszko E, Chen HZ, Leone G, Post S, Lozano G, Chen Z, Chauchereau A. E2F3 is a mediator of DNA damage-induced apoptosis. Mol Cell Biol. 2010;30(2):524–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Frontini M, Kukalev A, Leo E, Ng YM, Cervantes M, Cheng CW, Holic R, Dormann D, Tse E, Pommier Y, Yu V. The CDK subunit CKS2 counteracts CKS1 to control cyclin A/CDK2 activity in maintaining replicative fidelity and neurodevelopment. Dev Cell 2012;23(2):356–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. del Rincon SV, Widschwendter M, Sun D, Ekholm-Reed S, Tat J, Teixeira LK, Ellederova Z, Grolieres E, Reed SI, Spruck C. Cks overexpression enhances chemotherapeutic efficacy by overriding DNA damage checkpoints. Oncogene 2015;34(15):1961–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang J, Xu L, Liu Y, Chen J, Jiang H, Yang S, Tan H. Expression of cyclin kinase subunit 2 in human breast cancer and its prognostic significance. Int J Clin Exp Pathol. 2014;7(12):8593–601. [PMC free article] [PubMed] [Google Scholar]
- 35. Costa A, Ilves I, Tamberg N, Petojevic T, Nogales E, Botchan MR, Berger JM. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat Struct Mol Biol. 2011;18(4):471–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Eissa S, Matboli M, Shehata HH, Essawy NO. MicroRNA-10b and minichromosome maintenance complex component 5 gene as prognostic biomarkers in breast cancer. Tumour Biol. 2015;36(6):4487–94. [DOI] [PubMed] [Google Scholar]
- 37. Zheng M, Zhou Y, Yang X, Tang J, Wei D, Zhang Y, Jiang JL, Chen ZN, Zhu P. High GINS2 transcript level predicts poor prognosis and correlates with high histological grade and endocrine therapy resistance through mammary cancer stem cells in breast cancer patients. Breast Cancer Res Treat. 2014;148(2):423–36. [DOI] [PubMed] [Google Scholar]
- 38. Yan T, Liang W, Jiang E, Ye A, Wu Q, Xi M. GINS2 regulates cell proliferation and apoptosis in human epithelial ovarian cancer. Oncol Lett. 2018;16(2):2591–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ojima E, Inoue Y, Miki C, Mori M, Kusunoki M. Effectiveness of gene expression profiling for response prediction of rectal cancer to preoperative radiotherapy. J Gastroenterol. 2007;42(9):730–6. [DOI] [PubMed] [Google Scholar]
- 40. Huang PY, Zeng TT, Ban X, Li MQ, Zhang BZ, Zhu YH, Hua WF, Mai HQ, Zhang L, Guan XY, Li Y. Expression of EIF5A2 associates with poor survival of nasopharyngeal carcinoma patients treated with induction chemotherapy. BMC Cancer 2016;16:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu Y, Du F, Chen W, Yao M, Lv K, Fu P. EIF5A2 is a novel chemoresistance gene in breast cancer. Breast Cancer 2015;22(6):602–7. [DOI] [PubMed] [Google Scholar]
- 42. Stacey DW. Cyclin D1 serves as a cell cycle regulatory switch in actively proliferating cells. Curr Opin Cell Biol. 2003;15(2):158–63. [DOI] [PubMed] [Google Scholar]
- 43. Stendahl M, Kronblad A, Ryden L, Emdin S, Bengtsson NO, Landberg G. Cyclin D1 overexpression is a negative predictive factor for tamoxifen response in postmenopausal breast cancer patients. Br J Cancer 2004;90(10):1942–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thangavel C, Dean JL, Ertel A, Knudsen KE, Aldaz CM, Witkiewicz AK, Clarke R, Knudsen ES. Therapeutically activating RB: Reestablishing cell cycle control in endocrine therapy-resistant breast cancer. Endocr Relat Cancer 2011;18(3):333–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Coco Martin JM, Balkenende A, Verschoor T, Lallemand F, Michalides R. Cyclin D1 overexpression enhances radiation-induced apoptosis and radiosensitivity in a breast tumor cell line. Cancer Res. 1999;59(5):1134–40. [PubMed] [Google Scholar]
- 46. Marampon F, Gravina G, Ju X, Vetuschi A, Sferra R, Casimiro M, Pompili S, Festuccia C, Colapietro A, Gaudio E, Di Cesare E, Tombolini V, Pestell RG. Cyclin D1 silencing suppresses tumorigenicity, impairs DNA double strand break repair and thus radiosensitizes androgen-independent prostate cancer cells to DNA damage. Oncotarget 2016;7(5):5383–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bretones G, Delgado MD, Leon J. Myc and cell cycle control. Biochim Biophys Acta 2015;1849(5):506–16. [DOI] [PubMed] [Google Scholar]
- 48. Zhang Y, Lai J, Du Z, Gao J, Yang S, Gorityala S, Xiong X, Deng O, Ma Z, Yan C, Susana G, Xu Y, Zhang J. Targeting radioresistant breast cancer cells by single agent HK1 inhibitor via enhancing replication stress. Oncotarget 2016;7(23):34688–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang R, Chen DQ, Huang JY, Zhang K, Feng B, Pan BZ, Chen J, De W, Chen LB. Acquisition of radioresistance in docetaxel-resistant human lung adenocarcinoma cells is linked with dysregulation of miR-451/c-Myc-survivin/rad-51 signaling. Oncotarget 2014;5(15):6113–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bussink J, van der Kogel AJ, Kaanders JH. Activation of the I3-K/AKT pathway and implications for radioresistance mechanisms in head and neck cancer. Lancet Oncol. 2008;9(3):288–96. [DOI] [PubMed] [Google Scholar]
- 51. Evans SM, Koch CJ, Laughlin KM, Jenkins WT, Van Winkle T, Wilson DF. Tamoxifen induces hypoxia inMCF-7 xenografts. Cancer Res. 1997;57(22):5155–61. [PubMed] [Google Scholar]
- 52. Klein D. The tumor vascular endothelium as decision maker in cancer therapy. Front Oncol. 2018;8:367. [DOI] [PMC free article] [PubMed] [Google Scholar]