Abstract
Laryngeal cancer, one of the most common head and neck malignancies, is an aggressive neoplasm. Increasing evidence has demonstrated that microRNAs (miRNAs) exert important roles in oncogenesis and progression of diverse types of human cancers. miR-632, a tumor-related miRNA, has been reported to be dysregulated and implicated in human malignancies; however, its biological role in laryngeal carcinoma remains to be elucidated. The present study aimed at exploring the role of miR-632 in laryngeal cancer and clarifying the potential molecular mechanisms involved. In the current study, miR-632 was found to be significantly upregulated both in laryngeal cancer tissues and laryngeal cancer cell lines. Functional studies demonstrated that miR-632 accelerated cell proliferation and colony formation, facilitated cell migration and invasion, and enhanced the expression of cell proliferation-associated proteins, cyclin D1 and c-myc. Notably, miR-632 could directly bind to the 3′-untranslated region (3′-UTR) of glycogen synthase kinase 3β (GSK3β) to suppress its expression in laryngeal cancer cells. Mechanical studies revealed that miR-632 promoted laryngeal cancer cell proliferation, migration, and invasion through negative modulation of GSK3β. Pearson’s correlation analysis revealed that miR-632 expression was inversely correlated with GSK3β mRNA expression in laryngeal cancer tissues. Taken together, our findings suggest that miR-632 functions as an oncogene in laryngeal cancer and may be used as a novel therapeutic target for laryngeal cancer.
Key words: miR-632, Cell proliferation, Cell invasion, Laryngeal carcinoma, Glycogen synthase kinase 3β (GSK3β)
INTRODUCTION
Laryngeal squamous cell carcinoma (LSCC), one of the most common head and neck malignancies, originates from the epithelium of the larynx1–3. LSCC is an aggressive neoplasm with a high mortality rate. It was estimated that about 160,000 new cases were diagnosed every year, and a 5-year survival rate was approximately 60%4,5. Surgery or total laryngectomy, followed by chemotherapy or radiotherapy, remains the primary treatment for LSCC6,7. Despite significant advances in the diagnosis and treatment for laryngeal cancer over the past several decades, the long-term prognosis for LSCC patients, especially for advanced stage LSCC patients, is still poor. Hence, it is a critical need to develop effective therapeutic strategies for LSCC and identify sensitive biomarkers for LSCC prognosis.
MicroRNAs (miRNAs) are a large class of small (19∼25 nucleotides) endogenous, noncoding and single-stranded RNA molecules, which exert their functions by sequence-specific base pairing on the 3′-untranslated regions (3′-UTRs) of target messenger RNAs (mRNAs) to inhibit translation8,9. Mounting evidence has demonstrated that the ectopic expression of miRNAs is implicated in the occurrence and development of diverse types of human malignancies and other pathological processes10–12.
Glycogen synthase kinase 3β (GSK3β), a major member of the serine/threonine protein kinase GSK3 family, play crucial roles in various cellular processes, including cell proliferation, cell cycle, cell differentiation, and cell death13,14. Recent studies have revealed that GSK3β is dysregulated and implicated in multiple types of human tumors, including esophageal carcinoma15, hepatic cancer16, pancreatic cancer17, and ovarian cancer18.
miR-632 has been reported to be upregulated in breast cancer and exerts tumor-promoting roles by negative modulation of DNAJB619. However, the role of miR-632 in laryngeal cancer remains unclear. This study aimed to explore the functional role of miR-632 in laryngeal cancer and elucidate the potential molecular mechanisms involved. In the present study, miR-632 was discovered to be remarkably upregulated in both laryngeal cancer tissues and cell lines. Functional studies revealed that miR-632 facilitated laryngeal cancer cell proliferation, colony formation, migration, and invasion. Mechanically, miR-632 exerts tumor-promoting roles in laryngeal cancer partially by negative regulation of GSK3β.
MATERIALS AND METHODS
Patients and Tissue Samples
Laryngeal cancer tissues and corresponding adjacent nontumor tissues (located 5 cm away from laryngeal cancer tissues) were obtained from 10 patients in Renmin Hospital of Wuhan University (Wuhan, P.R. China) from March 2014 to December 2016. All the clinical samples were immediately frozen in liquid nitrogen after being removed from the patients and stored at −80°C for further studies. This study was approved by the ethnics and scientific committee of Wuhan University and complied with the Declaration of Helsinki. All the patients recruited in the present study provided written informed consent.
Cell Culture
Three human laryngeal cancer cell lines (SNU899, TU212, and Hep-2) and one normal human bronchial epithelial cell line BEAS-2B were purchased from Shanghai Cell Bank, Chinese Academy of Sciences (Shanghai, P.R. China). Cells were cultured in RPMI-1640 medium (Gibco, Los Angeles, CA, USA) supplemented with 10% fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin. All the cells were maintained at 37°C in a humidified incubator containing 5% CO2.
RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were extracted from tissues and cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, and quantified using Nanodrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). For microRNA expression analysis, mature miRNA-632 was reverse transcribed using TaqMan MicroRNA Reverse Transcription kit (Applied Biosystems, Foster City, CA, USA). U6 was used an endogenous control to normalize miR-632 expression.
For mRNA expression analysis, the first strand was synthesized using TaqMan High-Capacity cDNA Reverse Transcription (Applied Biosystems). GAPDH was used as an internal control to normalize the expression of GSK3β mRNA. qRT-PCR was performed on an Applied Biosystems 7500 Fast Real-Time PCR system. The specific primer sequences (Invitrogen) were as follows: GSK3β, 5′-ATTACGGGACCCAAATGTCA-3′ (forward) and 5′-TGCAGAAGCAGCATTATTGG-3′ (reverse); GAPDH, 5′-CACCCACTCCTCCACCTTTG-3′ (forward) and 5′-CCACCACCCTGTTGCTGTAG-3′ (reverse); miR-632, 5′-GACGGGAGGCGGAGCGGGGA-3′ (forward) and 5′-TCCCCGCTCCGCCTCCCGTC-3′ (reverse); U6, 5′-CTCGCTTCGGCAGCACA-3′ (forward) and 5′-AACGCTTCACGAATTTGCGT-3′ (reverse). The relative expression level was calculated using the 2−ΔΔCt method.
Cell Transfection
Cell transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocols. Cells were transfected with miR-632 mimics or miR-632 inhibitor. The siRNA against GSK3β was designed and synthesized by Shanghai GenePharma Co., Ltd. (Shanghai, P.R. China). The sequences of si-GSK3β were as follows: 5′-CGAUUACACGUCUAGUAUA (sense) and UAACAAUCUAUUUACACCC-3′ (antisense). Cells were harvested 48 h posttransfection for further experiments.
Cell Viability Assay
Cell proliferation was evaluated using MTT Cell Proliferation and Cytotoxicity Assay kit (Sigma-Aldrich) according to the manufacturer’s protocol. Briefly, Hep-2 cells were seeded into 96-well plates at a density of 2∼3 × 103 cells/well after transfection. Cells were then incubated at 37°C for different periods of time (0, 24, 48, and 72 h). Subsequently, the culture medium was removed and replaced with 100 μl of sterile MTT (0.5 mg/ml; Sigma-Aldrich). After incubation at 37°C for another 4 h, MTT solution was removed and replaced with 150 μl of DMSO (4%; Sigma-Aldrich). Following incubation with DMSO for 15 min, the absorbance was measured at 560 nm via a microplate reader (Bio-Tek Instruments, Winooski, VT, USA).
Colony Formation Assay
Colony formation assays were performed to assess the clonogenic capacity of Hep-2 cells after transfection. In brief, 400 cells from each treatment were seeded into six-well plates and allowed to grow for 2 weeks. Crystal violet (2%; Sigma-Aldrich) was applied to stain the formed colonies. The number of colonies was then counted under an invert microscope (Olympus, Tokyo, Japan). The experiments were performed in triplicate.
Wound Healing Assay
Hep-2 cells were seeded in six-well culture plates to grow into a monolayer. The cell monolayer was scraped using yellow pipette tips and washed twice with culture medium to form a wound. Following further incubation in the culture medium for 24 h, closure of the scratch was observed using an inverted microscope (Olympus). Cells were observed and photos taken at 0 and 24 h after wounding under an inverted microscope. The cell-free area at 24 h after scratching and the original denuded area were measured using the ImageJ software (National Institute of Health, Bethesda, MD, USA).
Transwell Invasion Assay
For Transwell invasion analysis, 2 × 104 Hep-2 cells were seeded into the upper chamber coated with Matrigel (Sigma-Aldrich) after transfection. The cells were plated in the culture medium without serum, and medium containing 10% FBS in the lower chamber served as chemoattractant. After incubation for 24 h at 37°C, cells that did not invade through the pore of the filter were carefully wiped off with cotton wool. Then the invasive cells were fixed with 95% ethanol (Sangon, Shanghai, P.R. China), stained with 0.5% crystal violet (Sigma-Aldrich), and counted under an inverted microscope (Olympus).
Luciferase Reporter Assays
Wild-type GSK3β 3′-UTR fragments and mutant GSK3β 3′UTR fragments were inserted into pmirGLO reporter vectors (Promega, Madison, WI, USA). Cells were cotransfected with miR-632 mimics and wild-type GSK3β or mutant GSK3β by Lipofectamine 2000 (Invitrogen). The Dual-Luciferase Reporter kit (Promega) was applied to detect the relative luciferase activity 48 h posttransfection. Results were presented as the ratio of Renilla luciferase activity to firefly luciferase activity.
Western Blot Analysis
Proteins were extracted from cells using the protein extraction reagent (Takara, Dalian, P.R. China). The BCA Protein Assay kit (Takara) was applied to detect the concentrations of the extracted proteins. The extracts were separated on 10% SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) microporous membranes (Dupont NEN, Boston, MA, USA). The PVDF membranes were blocked with phosphate-buffered saline (PBS) containing 0.1% Tween-20 (PBST) and 5% (w/v) nonfat milk for 1 h at room temperature. Following washing three times with PBST, the PVDF membranes were probed with corresponding antibodies overnight at 4°C. Anti-GSK3β (ab205710) and anti-GAPDH (ab8245) were purchased from Abcam (Cambridge, MA, USA) and used at the following dilutions: anti-GSK3β (1:1,000) and anti-GAPDH (1:3,000). After the PVDF membranes were washed again with PBST, horseradish peroxidase (HRP)-labeled IgG was added at 1:5,000 dilution and incubated at room temperature for 1 h. The blots were developed using ECL Western blotting reagents.
Statistical Analysis
Data were expressed as mean ± standard deviation (SD) from three separate experiments. Statistical analysis was performed using SPSS 18.0 (SPSS, Chicago, IL, USA). Correlation between miR-632 expression and GSK3β mRNA expression in laryngeal cancer tissues was evaluated using Pearson’s correlation analysis. Two-tailed Student’s t-test was applied to compare the differences between two groups, and one-way analysis of variance (ANOVA) followed by Dunnett’s multiple comparison was employed to compare the differences among three independent groups. A value of p < 0.05 was considered statistically significant.
RESULTS
miR-632 Is Significantly Upregulated in Laryngeal Cancer Tissues and Cell Lines
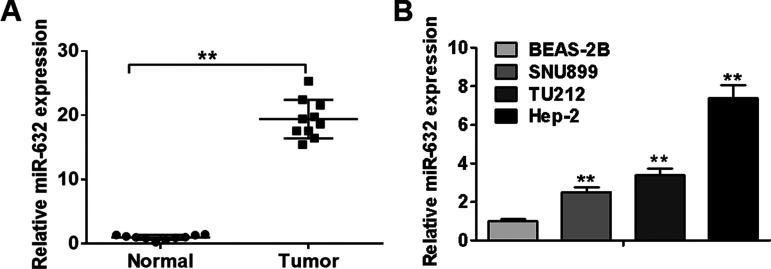
Given that the biological role of miR-632 in laryngeal cancer remains to be elucidated, we initially carried out qRT-PCR analysis to detect its expression levels in 10 pairs of laryngeal cancer tissues and corresponding pericarcinomatous tissues. As illustrated in Figure 1A, laryngeal cancer tissues displayed higher expression levels of miR-632 than adjacent noncancerous tissues. Consistently, miR-632 was observed to be significantly upregulated in laryngeal cancer cell lines (SNU899, TU212, and Hep-2) compared with normal bronchial epithelial cell line BEAS-2B (Fig. 1B). Hep-2 cells (highest endogenous miR-632 expression) were selected for subsequent studies. Taken together, these findings reveal that miR-632 is significantly upregulated in laryngeal cancer tissues and cell lines.
Figure 1.
miR-632 is significantly upregulated in laryngeal cancer tissues and cell lines. (A) Relative expression levels of miR-632 in 10 pairs of laryngeal cancer tissues and adjacent noncancerous tissues were measured using quantitative real-time PCR (qRT-PCR) analysis. (B) Relative expression levels of miR-632 in normal bronchial epithelial cell line BEAS-2B and three laryngeal cancer cell lines (SNU899, TU212, and Hep-2) were identified by qRT-PCR analysis. **p < 0.01.
miR-632 Accelerates Laryngeal Cancer Cell Proliferation and Colony Formation
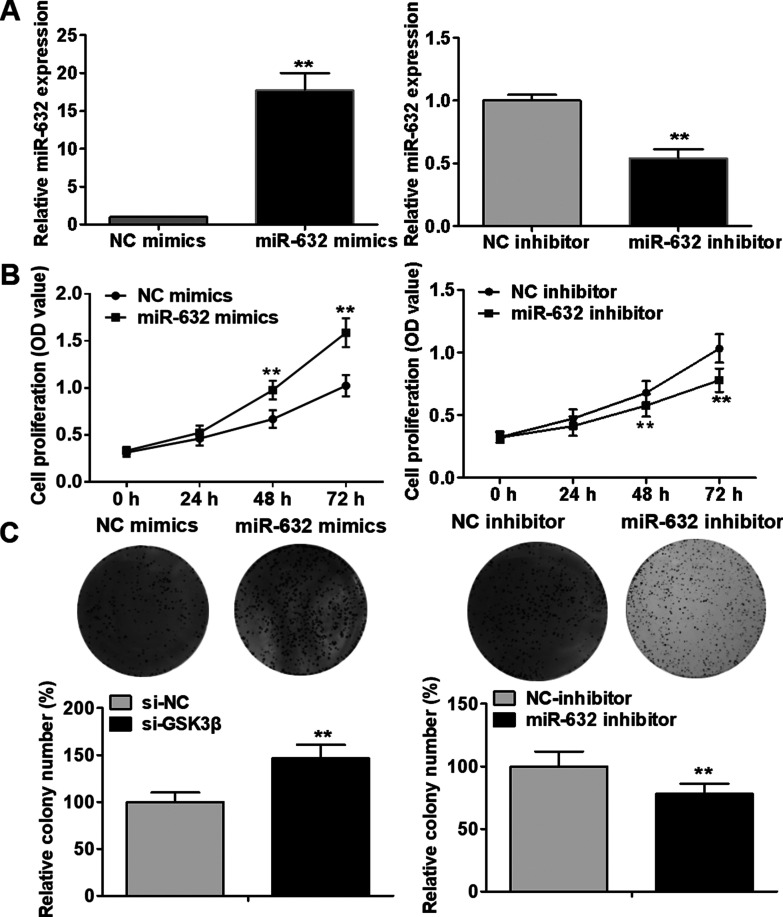
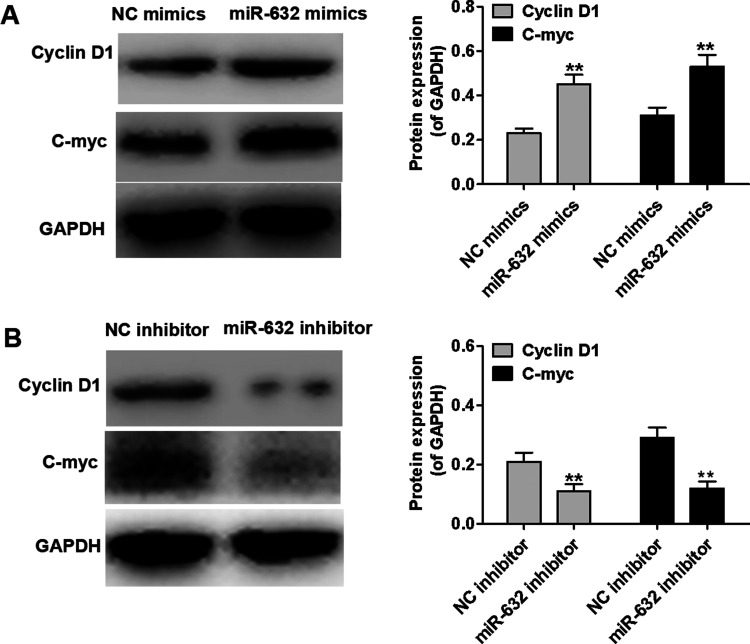
To investigate the potential biological role of miR-632 in laryngeal cancer, Hep-2 cells were transfected with miR-632 mimics or miR-632 inhibitor. The transfection efficacy was evaluated by qRT-PCR analysis (Fig. 2A). As evident from MTT assays, Hep-2 cell proliferation was notably facilitated by miR-632 mimics compared with the negative control group, whereas miR-632 inhibitor markedly repressed Hep-2 cell proliferation (Fig. 2B). As illustrated in Figure 2C, the clonogenic capability of Hep-2 cells was significantly strengthened by miR-632 overexpression compared with the negative control group; however, miR-632 inhibitor dramatically suppressed the clonogenic formation ability. In addition, Western blot analysis was performed to evaluate the effect of miR-632 on the expression of cell proliferation-associated proteins in Hep-2 cells. miR-632 overexpression was discovered to significantly upregulate the expression of cyclin D1 and c-myc, whereas miR-632 inhibitor notably inhibited the expression of cyclin D1 and c-myc (Fig. 3A and B). These findings indicate that miR-632 facilitates laryngeal cancer cell proliferation and colony formation.
Figure 2.
miR-632 accelerates laryngeal cancer cell proliferation and colony formation. (A) miR-632 expression levels in Hep-2 cells were identified using qRT-PCR after transfection with miR-632 mimics or miR-632 inhibitor. (B) Hep-2 cell proliferation was assessed using MTT assays after transfection with miR-632 mimics or miR-632 inhibitor. (C) Clonogenic capability of Hep-2 cells was evaluated using colony formation assays after transfection with miR-632 mimics or miR-632 inhibitor. **p < 0.01.
Figure 3.
miR-632 suppresses the expression of cell proliferation-associated proteins in Hep-2 cells. (A) Expression levels of cyclin D1 and c-myc in Hep-2 cells were determined using Western blot after transfection with NC-mimics or miR-632 mimics. (B) Expression levels of cyclin D1 and c-myc in Hep-2 cells were identified using Western blot after transfection with NC-inhibitor or miR-632 inhibitor. **p < 0.01.
miR-632 Promotes Cell Laryngeal Cancer Cell Migration and Invasion
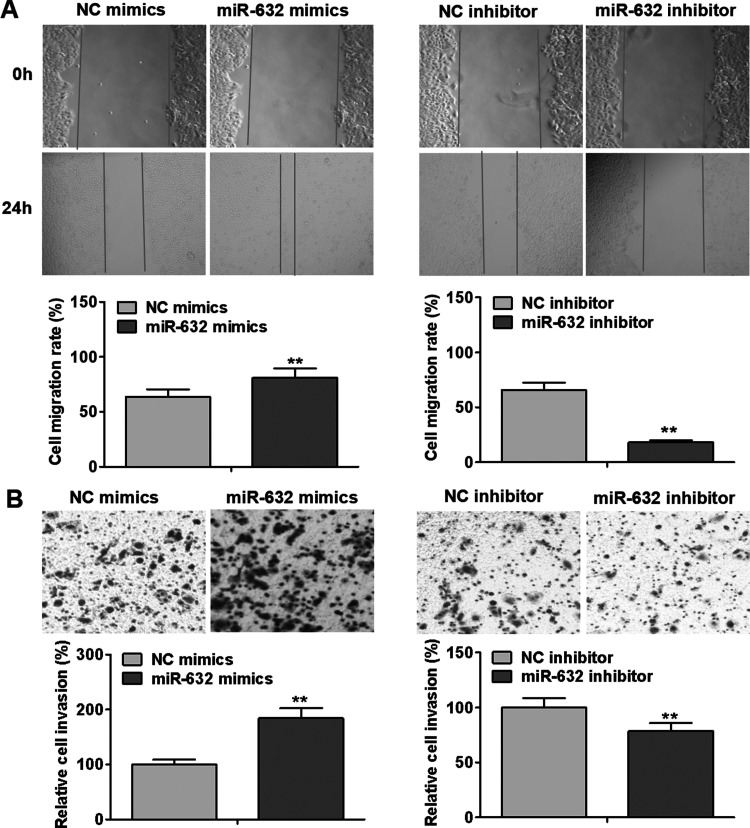
To investigate whether miR-632 influences the motility of laryngeal cancer cells, we assessed the migrative and invasive abilities of Hep-2 cells after transfection with miR-632 mimics or miR-632 inhibitor. As presented in Figure 4A, miR-632 mimics significantly enhanced the migrative capability of Hep-2 cells in comparison with the negative control group, whereas miR-632 inhibitor markedly slowed down Hep-2 cell migration. As evident from Transwell invasion assays, Hep-2 cell invasion was notably promoted by miR-632 overexpression compared with the negative control group, whereas miR-632 inhibitor remarkably repressed the invasion of Hep-2 cells (Fig. 4B). These results suggest that miR-632 accelerates laryngeal cancer migration and invasion.
Figure 4.
miR-632 promotes laryngeal cancer cell migration and invasion. (A) Hep-2 cell migration was examined by wound healing assays after transfection with miR-632 mimics or miR-632 inhibitor. (B) Hep-2 cell invasion was identified using Transwell invasion assays after transfection with miR-632 mimics or miR-632 inhibitor. **p < 0.01.
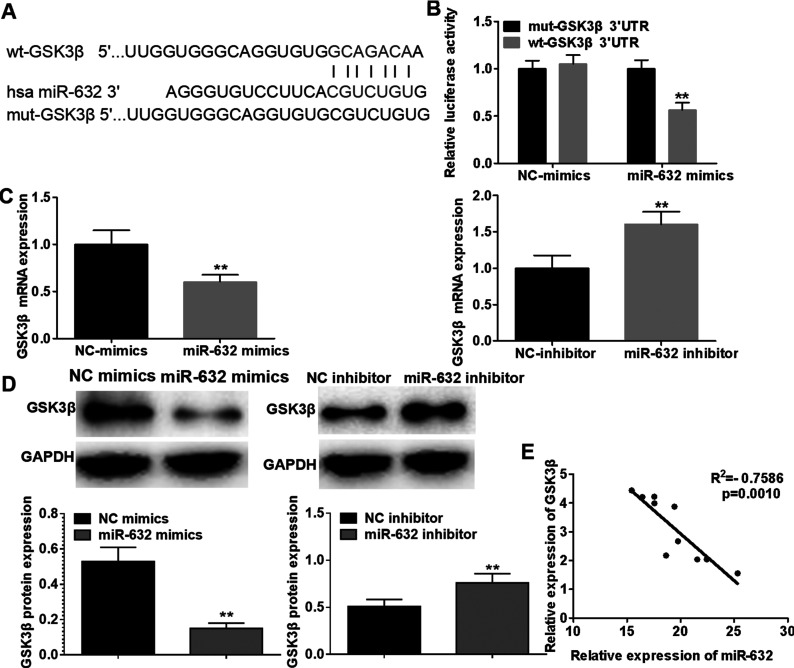
GSK3β Is a Downstream Target of miR-632
To elucidate the potential molecular mechanism by which miR-632 promotes laryngeal cancer cell proliferation, migration, and invasion, TargetScan algorithm was performed to predict the potential targets of miR-632. GSK3β, frequently reported to be involved in pathogenesis and development of a variety of human malignancies, was selected as a candidate target of miR-632 (Fig. 5A). To verify whether GSK3β is regulated by direct binding of miR-632 to its 3′-UTR, luciferase reporter assays were conducted. We constructed luciferase reporter vectors containing the predicted miR-632 targeting sequences (wild-type GSK3β 3′-UTR) or its corresponding mutant fragments (mutant GSK3β 3′-UTR). The results revealed that cotransfection of miR-632 mimics and wild-type GSK3β 3′-UTR notably decreased the activity of luciferase, while cotransfection of miR-608 mimics and mutant GSK3β 3′-UTR failed to reduce the activity of luciferase (Fig. 5B). Furthermore, qRT-PCR analysis and Western blotting analysis demonstrated that GSK3β was remarkably downregulated in Hep-2 cells transfected with miR-632 mimics compared with the negative control group, whereas miR-632 inhibitor dramatically upregulated the mRNA and protein expression levels of GSK3β (Fig. 5C and D). Additionally, Pearson’s correlation analysis demonstrated that miR-632 expression was negatively correlated with GSK3β mRNA expression in laryngeal cancer tissues (Fig. 5E). Taken together, our results suggest that miR-632 directly binds to GSK3β 3′-UTR to suppress its expression.
Figure 5.
Glycogen synthase kinase 3β (GSK3β) is a downstream target of miR-632. (A) A putative binding site of miR-632 in the 3′-untranslated region (3′-UTR) of GSK3β was predicted by TargetScan online software. (B) Luciferase activity of reporter vectors carrying wild-type or mutant GSK3β 3′-UTR in Hep-2 cells was detected in the presence of miR-632. (C) GSK3β mRNA expression in Hep-2 cells was examined using qRT-PCR after transfection with miR-632 mimics or miR-632 inhibitor. (D) Protein expression of GSK3β in Hep-2 cells was measured by Western blotting analysis after transfection with miR-632 mimics or miR-632 inhibitor. (E) Correlation between miR-632 expression and GSK3β mRNA expression in laryngeal cancer tissues was determined by Pearson’s correlation analysis. **p < 0.01.
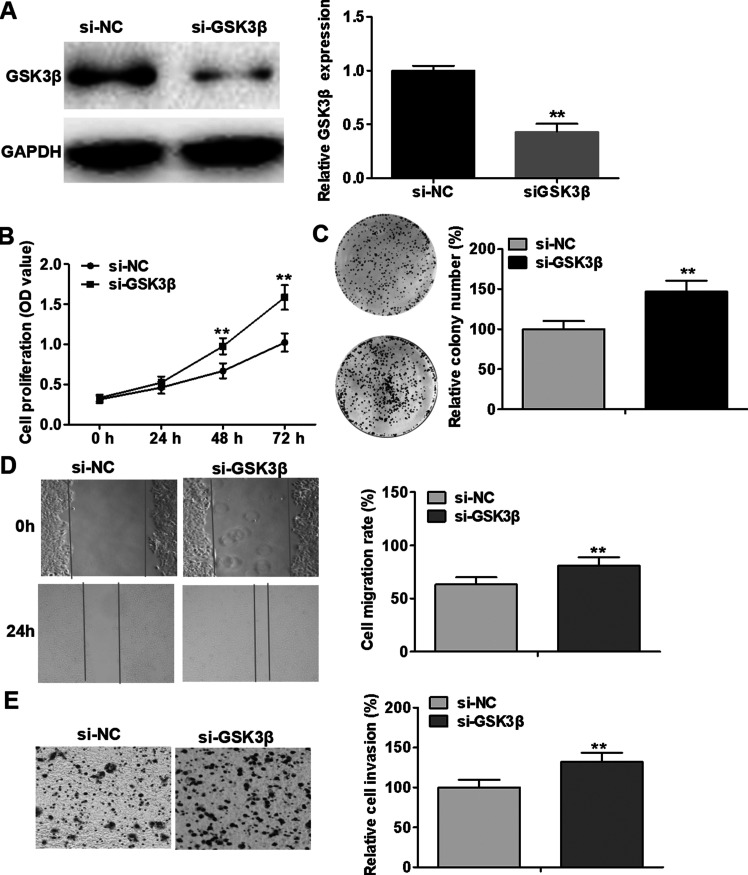
Downregulation of GSK3β Has Similar Effects With Overexpression of miR-632 in Laryngeal Cancer Cells
Given that GSK3β is closely associated with oncogenesis and progression of a wide range of human tumors, we downregulate the expression of GSK3β in Hep-2 cells, with a view to further investigating the functional connection between miR-632 and GSK3β. The interference efficiency was assessed using Western blotting analysis (Fig. 6A). As illustrated in Figure 6B–E, knockdown of GSK3β notably facilitated Hep-2 cell proliferation, colony formation, migration, and invasion compared with the negative control treatment. These results demonstrate that downregulation of GSK3β has similar effects with overexpression of miR-632 in laryngeal cancer cells.
Figure 6.
Downregulation of GSK3β has similar effects with overexpression of miR-632 in laryngeal cancer cells. (A) Hep-2 cells were transfected with si-GSK3β to achieve the knockdown of GSK3β, as validated by Western blotting analysis. (B) Hep-2 cell viability was examined by MTT assays after transfection with si-NC or si-GSK3β. (C) Clonogenic ability of Hep-2 cells was evaluated by colony formation assays after transfection with si-NC or si-GSK3β. (D) Hep-2 cell migration was identified via wound healing assays after transfection with si-NC or si-GSK3β. (E) Hep-2 cell invasion was assessed using Transwell invasion assays after transfection with si-NC or si-GSK3β. **p < 0.01.
Downregulation of GSK3β Has Opposite Effects With Inhibition of miR-632 in Laryngeal Cancer Cells
To further explore the functional connection between miR-632 and GSK3β, Hep-2 cells were cotransfected with miR-632 inhibitor and si-GSK3β. miR-632 inhibitor was observed to remarkably suppress Hep-2 cell proliferation, colony formation, migration, and invasion. In addition, the inhibitory effects of miR-632 on Hep-2 cell proliferation, colony formation, migration, and invasion were partially alleviated by the knockdown of GSK3β (Fig. 7A–D). Taken together, our data suggest that the suppressing effects of miR-632 in laryngeal cancer cells were partially mediated by GSK3β.
Figure 7.
Downregulation of GSK3β has opposite effects to inhibition of miR-632 in laryngeal cancer cells. (A) Hep-2 cell viability was measured using MTT assays after cotransfection with miR-632 inhibitor and si-GSK3β. (B) Clonogenic capability of Hep-2 cells was identified by colony formation assays after cotransfection with miR-632 inhibitor and si-GSK3β. (C) Hep-2 cell migration was assessed by wound healing assays after cotransfection with miR-632 inhibitor and si-GSK3β after cotransfection with miR-632 inhibitor and si-GSK3β. (D) Hep-2 cell invasion was evaluated via Transwell invasion assays after cotransfection with miR-632 inhibitor and si-GSK3β. **p < 0.01 versus NC-inhibitor + si-NC group; ##p < 0.01 versus miR-632 inhibitor + si-NC group.
DISCUSSION
Laryngeal cancer, an aggressive malignancy, has the second highest morbidity among all the head and neck carcinomas20. Laryngeal cancer has brought immense economic pressures and high health risks to the people worldwide. Even though great progress in diagnosis and treatment of laryngeal cancer has been achieved, the long-term prognosis remains poor. Evidence is accumulating that miRNAs function as important regulator in tumorigenesis and progression of laryngeal carcinoma21–24. miR-632, a novel cancer-related miRNA, has been reported to promote cell proliferation and invasion in breast cancer by downregulating DNAJB619. However, the biological role of miR-632 in laryngeal cancer is poorly understood.
In the present study, we initially examined the expression levels of miR-632 in 10 pairs of laryngeal cancer tissues and corresponding adjacent noncancerous tissues, and found that miR-632 was significantly upregulated in laryngeal cancer tissues. Consistently, miR-632 was observed to be notably upregulated in laryngeal cancer cell lines. To get a better understanding of the biological role of miR-632 in laryngeal cancer, functional studies were subsequently conducted. The results of functional studies demonstrated that miR-632 accelerated laryngeal cancer cell proliferation and colony formation, facilitated cell migration and invasion, and enhanced the expression of cell proliferation-associated proteins, cyclin D1 and c-myc. It is well acknowledged that cyclin D1 and c-myc are two important proteins that positively regulate cell proliferation25,26. Our findings suggest that miR-632 may serve as a tumor promoter in laryngeal cancer.
To explore the potential molecular mechanisms by which miR-632 promotes laryngeal cancer cell proliferation, migration, and invasion, we performed bioinformatics analysis using TargetScan online software. GSK3β, frequently reported to be implicated in a variety of human cancers, was selected as a candidate target of miR-632. Dual-luciferase reporter assays confirmed GSK3β as a direct target of miR-632. Previous studies have showed that aberrant expression of GSK3β is implicated in diverse human malignancies, including esophageal cancer15, ovarian cancer18, non-small cell lung cancer27, hepatocellular carcinoma28, breast cancer29, melanoma30, and nasopharyngeal carcinoma31. GSK3β is an important point of convergence for cellular signaling pathways involved in oncogenesis and progression of human cancers. It is documented that GSK3β can phosphorylate some key proteins (e.g., c-jun, c-myc, and β-catenin) involved in cancer-related pathways, thereby inducing ubiquitin-mediated degradation and inhibiting cancer-associated signal pathways32–34.
To further determine the functional correlation between miR-632 and GSK3β, we downregulated the expression of GSK3β in laryngeal cancer cells. Knockdown of GSK3β was observed to have similar effects on cell proliferation, colony formation, cell migration, and invasion with overexpression of miR-632 in laryngeal cancer cells. Furthermore, knockdown of GSK3β was found to partially reverse the suppressive effects of miR-632 inhibitor on laryngeal cancer cells. These results suggest that the promoting effects of miR-632 on cell proliferation, migration, and invasion are mediated by GSK3β in laryngeal cancer cells.
In conclusion, miR-632 is significantly upregulated in laryngeal cancer tissues and cell line, and exerts tumor-promoting roles in laryngeal cancer via targeting GSK3β. Our study provides new insights into understanding the molecular mechanisms underlying laryngeal cancer oncogenesis and development. Thus, miR-632 may be used to be a novel prognostic marker and a potential therapeutic target for laryngeal cancer.
ACKNOWLEDGMENTS
We thank the reviewers for the helpful comments on this manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Liu Y, Liu J, Wang L, Yang X, Liu X. MicroRNA195 inhibits cell proliferation, migration and invasion in laryngeal squamous cell carcinoma by targeting ROCK1. Mol Med Rep. 2017;16(5):7154–62. [DOI] [PubMed] [Google Scholar]
- 2. Wu Q, Zhuang K, Li H. PAQR3 plays a suppressive role in laryngeal squamous cell carcinoma. Tumour Biol. 2016;37(1):561–5. [DOI] [PubMed] [Google Scholar]
- 3. Wu CP, Zhou L, Gong HL, Du HD, Tian J, Sun S, Li JY. Establishment and characterization of a novel HPV-negative laryngeal squamous cell carcinoma cell line, FD-LSC-1, with missense and nonsense mutations of TP53 in the DNA-binding domain. Cancer Lett. 2014;342(1):92–103. [DOI] [PubMed] [Google Scholar]
- 4. Marsit CJ, Posner MR, Mcclean MD, Kelsey KT. Hypermethylation of E-cadherin is an independent predictor of improved survival in head and neck squamous cell carcinoma. Cancer 2008;113(7):1566–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Langevin SM, Stone RA, Bunker CH, Lyons-Weiler MA, Laframboise WA, Kelly L, Seethala RR, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation is associated with poorer overall survival in patients with squamous cell carcinoma of the head and neck. Cancer 2011;117(7):1454–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Strojan P, Haigentz MJ, Bradford CR, Wolf GT, Hartl DM, Langendijk JA, Rinaldo A, Eisbruch A, Mendenhall WM, Forastiere AA, Takes RP, Ferlito A. Chemoradiotherapy vs. total laryngectomy for primary treatment of advanced laryngeal squamous cell carcinoma. Oral Oncol. 2013;49(4):283–6. [DOI] [PubMed] [Google Scholar]
- 7. Haigentz MJ, Silver CE, Hartl DM, Takes RP, Rodrigo JP, Robbins KT, Rinaldo A, Ferlito A. Chemotherapy regimens and treatment protocols for laryngeal cancer. Expert Opinion Pharmacother. 2010;11(8):1305–16. [DOI] [PubMed] [Google Scholar]
- 8. Wang X, Liu XS. Systematic curation of miR-base annotation using integrated small RNA high-throughput sequencing data for C. elegans and Drosophila. Front Genet. 2011;2:25–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y, Nie Y, Cao J, Tu S, Lin Y, Du Y, Li Y. G-A variant in miR-200c binding site of EFNA1 alters susceptibility to gastric cancer. Mol Carcinog. 2014;53(3):219–29. [DOI] [PubMed] [Google Scholar]
- 10. Wang S, Guo D, Li C. Downregulation of miRNA-26b inhibits cancer proliferation of laryngeal carcinoma through autophagy by targeting ULK2 and inactivation of the PTEN/AKT pathway. Oncol Rep. 2017;38(3):1679–87. [DOI] [PubMed] [Google Scholar]
- 11. Yao XD, Li P, Wang JS. MicroRNA differential expression spectrum and microRNA-125a-5p inhibition of laryngeal cancer cell proliferation. Exp Ther Med. 2017;14(2):1699–705. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12. Saito K, Inagaki K, Kamimoto T, Ito Y, Sugita T, Nakajo S, Ishikura T, Hanaoka H, Okubo K, Onozaki T, Zama T. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS One 2013;8(8):71480–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lee HC, Tsai JN, Liao PY, Tsai WY, Lin KY, Chuang CC, Sun CK, Chang WC, Tsai HJ. Glycogen synthase kinase 3α and 3β have distinct functions during cardiogenesis of zebrafish embryo. BMC Dev Biol. 2007;7(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shin SH, Lee EJ, Chun J, Hyun S, Kim YI, Kang SS. The nuclear localization of glycogen synthase kinase 3β is required its putative PY-nuclear localization sequences. Mol Cells 2012;34(4):375–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu K, Luo Y, Tian H, Yu KZ, He JX, Shen WY. The tumor suppressor LKB1 antagonizes WNT signaling pathway through modulating GSK3beta activity in cell growth of esophageal carcinoma. Tumour Biol. 2014;35(2):995–1002. [DOI] [PubMed] [Google Scholar]
- 16. Ni CX, Qi Y, Zhang J, Liu Y, Xu WH, Xu J, Hu HG, Wu QY, Wang Y, Zhang JP. WM130 preferentially inhibits hepatic cancer stem-like cells by suppressing AKT/GSK3beta/β-catenin signaling pathway. Oncotarget 2016;7(48):79544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang HW, Liu GH, Liu YQ, Zhao HC, Yang Z, Zhao CL, Zhang XF, Ye H. Over-expression of microRNA-940 promotes cell proliferation by targeting GSK3β and sFRP1 in human pancreatic carcinoma. Biomed Pharmacother. 2016;83:593–601. [DOI] [PubMed] [Google Scholar]
- 18. Cao Q, Lu X, Feng YJ. Glycogen synthase kinase-3β positively regulates the proliferation of human ovarian cancer cells. Cell Res. 2006;16(7):671–7. [DOI] [PubMed] [Google Scholar]
- 19. Mitra A, Rostas JW, Dyess DL, Shevde LA, Samant RS. Micro-RNA-632 downregulates DNAJB6 in breast cancer. Lab Invest. 2012;92(9):1310–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shuang Y, Zhou X, Li C, Huang Y, Zhang L. MicroRNA-503 serves an oncogenic role in laryngeal squamous cell carcinoma via targeting programmed cell death protein 4. Mol Med Rep. 2017;16(4):5249–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Long XB, Sun GB, Hu S, Liang GT, Wang N, Zhang XH, Cao PP, Zhen HT, Cui YH, Liu Z. Let-7a microRNA functions as a potential tumor suppressor in human laryngeal cancer. Oncol Rep. 2009;22(5):1189–95. [DOI] [PubMed] [Google Scholar]
- 22. Sun X, Liu B, Zhao XD, Wang LY, Ji WY. MicroRNA-221 accelerates the proliferation of laryngeal cancer cell line Hep-2 by suppressing Apaf-1. Oncol Rep. 2015;33(3):1221–6. [DOI] [PubMed] [Google Scholar]
- 23. Li M, Chen SM, Chen C, Zhang ZX, Dai MY, Zhang LB, Wang SB, Dai Q, Tao ZZ. microRNA2993p inhibits laryngeal cancer cell growth by targeting human telomerase reverse transcriptase mRNA. Mol Med Rep. 2015;11(6):4645–9. [DOI] [PubMed] [Google Scholar]
- 24. Shen Z, Zhan G, Ye D, Ren Y, Cheng L, Wu Z, Guo J. MicroRNA-34a affects the occurrence of laryngeal squamous cell carcinoma by targeting the antiapoptotic gene survivin. Med Oncol. 2012;29(4):2473–80. [DOI] [PubMed] [Google Scholar]
- 25. Capaldo CT, Koch S, Kwon M, Laur O, Parkos CA, Nusrat A. Tight function zonula occludens-3 regulates cyclin D1-dependent cell proliferation. Mol Bio Cell 2011;22(10):1677–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2α promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell 2007;11(4):335–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zeng J, Liu D, Qiu Z, Huang Y, Chen B, Wang L, Xu H, Huang N, Liu L, Li W. GSK3β overexpression indicates poor prognosis and its inhibition reduces cell proliferation and survival of non-small cell lung cancer cells. PLoS One 2014;9(3):91231–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chua HH, Tsuei DJ, Lee PH, Jeng YM, Lu J, Wu JF, Su DS, Chen YH, Chien CS, Kao PC, Lee CN, Hu RH, Ni YH, Chang MH. RBMY, a novel inhibitor of glycogen synthase kinase 3β, increases tumor stemness and predicts poor prognosis of hepatocellular carcinoma. Hepatology 2015;62(5):1480–96. [DOI] [PubMed] [Google Scholar]
- 29. Panka DJ, Cho DC, Atkins MB, Mier JW. GSK-3β inhibition enhances sorafenib-induced apoptosis in melanoma cell lines. J Biol Chem. 2008;283(2):726–32. [DOI] [PubMed] [Google Scholar]
- 30. Kim DY, Park EY, Chang E, Kang HG, Koo Y, Lee EJ, Ko JY, Kong HK, Chun KH, Park JH. A novel miR-34a target, protein kinase D1, stimulates cancer stemness and drug resistance through GSK3/β-catenin signaling in breast cancer. Oncotarget 2016;7(12):14791–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma R, Wei Y, Huang X, Fu R, Luo X, Zhu X, Lei W, Fang J, Li H, Wen W. Inhibition of GSK 3β activity is associated with excessive EZH2 expression and enhanced tumour invasion in nasopharyngeal carcinoma. PLoS One 2013;8(7):68614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shao J, Teng Y, Padia R, Hong S, Noh H, Xie X, Mumm JS, Dong Z, Ding HF, Cowell J, Kim J, Han J, Huang S. COP1 and GSK3beta cooperate to promote c-Jun degradation and inhibit breast cancer cell tumorigenesis. Neoplasia 2013;15(9):1075–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mao L, Dauchy RT, Blask DE, Slakey LM, Xiang S, Yuan L, Dauchy EM, Shan B, Brainard GC, Hanifin JP, Frasch T, Duplessis TT, Hill SM. Circadian gating of epithelial-to-mesenchymal transition in breast cancer cells via melatonin-regulation of GSK3β. Mol Endocrinol. 2012;26(11):1808–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kang T, Wei Y, Honaker Y, Yamaguchi H, Appella E, Hung MC, Piwnica-Worms H. GSK-3β targets Cdc25A for ubiquitin-mediated proteolysis, and GSK-3β inactivation correlates with Cdc25A overproduction in human cancers. Cancer Cell 2008;13(1):36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]