Abstract
It has been reported that kindlin-3 expression is closely associated with progression of many cancers and microRNA (miRNA) processing. However, the effects and precise mechanisms of kindlin-3 in acute myeloid leukemia (AML) have not been well clarified. Our study aimed to explore the interaction between kindlin-3 and miR-4792 in AML. In our study, we found that the expression of kindlin-3 was dramatically increased in AML samples and cell lines, and the miR-4792 level was significantly downregulated. Interestingly, the low miR-4792 level was closely associated with upregulated kindlin-3 expression in AML samples. Moreover, introduction of miR-4792 dramatically suppressed proliferation and invasion and induced apoptosis of AML cells. We demonstrated that miR-4792 could directly target kindlin-3 by using both bioinformatics analysis and luciferase reporter assay. In addition, kindlin-3 silencing had similar effects with miR-4792 overexpression on AML cells. Overexpression of kindlin-3 in AML cells partially reversed the inhibitory effects of miR-4792 mimic. miR-4792 inhibited cell proliferation and invasion and induced apoptosis of AML cells by directly downregulating kindlin-3 expression, and miR-4792 targeting kindlin-3 was responsible for the regulation of the proliferation, invasion, and apoptosis of AML cells.
Key words: Acute myeloid leukemia (AML), MicroRNA-4792, Kindlin-3, Proliferation, Invasion, Apoptosis
INTRODUCTION
Acute myeloid leukemia (AML) is characterized by maturation arrest of myeloid progenitor cells at different stages, malignant proliferation, and impaired apoptosis of leukemic cells. It is a heterogeneous disease that causes an overall decrease in the human hematopoietic system1. China ranks third worldwide in the incidence of AML and has the highest levels of mortality in young patients with AML2,3. Therefore, there is an urgent need to develop new therapies for AML. However, the pathogenesis of AML is still not fully understood and thus hampers the development of new treatments. Hence, it is urgently needed to illustrate the molecular pathogenesis mechanism responsible for leukemogenesis in AML and find new therapeutic strategies.
Kindlin-3 is expressed in hematopoietic, endothelial cells, leukocytes, and platelets4–6. In addition to leukocyte migration, endothelial tube formation, and platelet aggregation, kindlin-3 is involved in osteoclast-mediated bone resorption7. Recently, kindlin-3 was found to be important in cancer progression, although its role as a promoter or suppresser of cancer metastasis remains controversial8,9. However, the expression of kindlin-3 and its functional roles in AML are still unclear.
MicroRNAs (miRNAs) are a class of small, highly conserved noncoding RNA molecules that regulate gene expression at the posttranscriptional level10. Aberrant expression of miRNAs has been regarded as a new type of “oncomiRs” or “tumor suppressors,” which play critical roles in the progression of a number of cancers including AML11–14. Zhu et al. reported that miR-9 acted as a tumor suppressor and inhibited cell proliferation, migration, and invasion in AML by directly targeting CX chemokine receptor 415. Chen et al. demonstrated that introduction of miR-628 restrained the proliferation of AML cells by directly targeting insulin-like growth factor type 1 receptor (IGF-1R)16. Ding et al. showed that miR-130a was aberrantly overexpressed in adult AML17. High miR-338 level was closely associated with poor prognosis in AML patients undergoing chemotherapy18.
In this study, we for the first time found that kindlin-3 expression was markedly upregulated in AML tissues and cell lines. However, until now the functional roles of kindlin-3 in AML have remained unclear. Furthermore, we confirmed that kindlin-3 was one of the direct targets of miR-4792 in AML. For further study, we also confirmed significant downregulation of miR-4792 in AML tissues and cells. Upregulation of miR-4792 suppressed the proliferation and invasion and induced apoptosis of AML cells. And overexpression of kindlin-3 blocked the effects of miR-4792. Therefore, our outcomes showed critical roles for miR-4792/indlin-3 axis in the pathogenesis of AML and suggested its possible application in AML treatment.
MATERIALS AND METHODS
Patients and Samples
Bone marrow specimens were obtained from 30 patients with AML and 30 healthy controls from Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, between March 2017 and September 2018. None of the patients had received chemotherapy, radiotherapy, targeted therapy, or hematopoietic stem cell transplantation before bone marrow aspiration. This study was approved by the Ethics Committee Hospital of Tongji Hospital (ethic number: 20170220) and was performed in accordance with the Declaration of Helsinki and the guidelines of the Ethics Committee of Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology. All patients signed the informed consent form.
Cell Culture
TF-1a, HL-60, THP-1, NB4, and Kg1a AML cell lines and the human normal stromal cells HS-5 were obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA). All cells were cultured in Roswell Park Memorial Institute (RPMI)-1640 medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Gibco), 2 mM l-glutamine, and 100 U/ml streptomycin plus 100 U/ml penicillin (Gibco) at 37°C with 5% CO2.
Transient Transfection
The miR-4792 mimics, miR-negative control (miR-NC), siRNA for kindlin-3 (si-kindlin-3), and siRNA-negative control (si-NC) were synthesized and purified by Gene-Pharma (Shanghai, China). The kindlin-3 overexpression plasmid was generated by inserting kindlin-3 complementary DNA (cDNA) into a pcDNA3.1 vector. This plasmid was sequenced confirmed by Gene-Pharma. miR-4792 mimics, miR-NC, si-NC, and si-kindlin-3 were transfected by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocols. Total RNA and protein were collected 48 h after transfection.
RNA Extraction and Real-Time Quantitative Polymerase Chain Reaction (PCR)
Total RNA was extracted from cell lines and clinical samples by using TRIzol reagent (Invitrogen) according to the operating instructions. RNA was quantified by using ultraviolet (UV) absorbencies at 260 and 280 nm (A260/280). Subsequently, the RNA was reverse transcribed into cDNA using the reverse transcription system (Thermo Scientific, Waltham, MA, USA). The level of miR-4792 was detected by the ABI PRISM 7500 Sequence Detection System (ABI) using the TaqMan MicroRNA assay kits (Applied Biosystems, Foster City, CA, USA). U6 small nuclear RNA (snRNA) was used as the control. The mRNA expressions of kindlin-1, kindlin-2, kindlin-3, proliferating cell nuclear antigen (PCNA), CDK4, cyclin E1, p27, matrix metalloproteinase-2 (MMP-2), MMP-9, and tissue inhibitor of metalloproteinases 1 (TIMP-1) were also analyzed by SYBR Green and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The judgment of primer sequences’ specificity was based on dissociation curve; 2−ΔΔCt (cycle threshold) was used to calculate the relative gene expression levels. Prime sequences are shown in Table 1.
Table 1.
Sequences of Primers for qRT-PCR
| Gene | Primer Sequence |
|---|---|
| Kindlin-1 | F: 5′-GCGTTGACCATCCCAATGAAG-3′ |
| R: 5′-ACCAAAGAGCAAAGTCTGACC-3′ | |
| Kindlin-2 | F: 5′-TCTGACCATGCTCTCTGGTG-3′ |
| R: 5′-AGTTCTTCGGGGTGTCTGATA-3′ | |
| Kindlin-3 | F: 5′-GGACTACATCGACTCGTCATGG-3′ |
| R: 5′-CTCCACAATCTTCAGGAGCAC-3′ | |
| PCNA | F: 5′-CCTGCTGGGATATTAGCTCCA-3′ |
| R: 5′-CAGCGGTAGGTGTCGAAGC-3′ | |
| CDK4 | F: 5′-GGGGACCTAGAGCAACTTACT-3′ |
| R: 5′-CAGCGCAGTCCTTCCAAAT-3′ | |
| Cyclin E1 | F: 5′-AAGGAGCGGGACACCATGA-3′ |
| R: 5′-ACGGTCACGTTTGCCTTCC-3′ | |
| p27 | F: 5′-AACGTGCGAGTGTCTAACGG-3′ |
| R: 5′-CCCTCTAGGGGTTTGTGATTCT-3′ | |
| MMP-2 | F: 5′-CTGCGGTTTTCTCGAATCCA-3′ |
| R: 5′-GGGTATCCATCGCCATGCT-3′ | |
| MMP-9 | F: 5′-CCCTGGAGACCTGAGAACCA-3′ |
| R: 5′- CCACCCGAGTGTAACCATAGC-3′ | |
| TIMP-1 | F: 5′-CTTCTGCAATTCCGACCTCGT-3′ |
| R: 5′-ACGCTGGTATAAGGTGGTCTG-3′ | |
| miR-4792 | F: 5′-CGGTGAGCGCTCGCTGG-3′ |
| F: 5′-GAACATGTCTGCGTATCTC-3′ | |
| U6 | F: 5′-CTCGCTTCGGCAGCACA-3′ |
| F: 5′-AACGCTTCACGAATTTGCGT-3′ | |
| GAPDH | F: 5′-GAGTCAACGGATTTGGTCGTATTG-3′ |
| R: 5′-CCTGGAAGATGGTGATGGGATT-3′ |
Cell Proliferation Assay
To study the effect of miR-4792 on proliferation of AML cells, HL-60 and Kg1a cells (104/well) were seeded in a 96-well plate and incubated overnight in complete medium. After removing the medium, cells were transfected with miR-4792 mimic or miR-NC for 48 h. The BrdU (colorimetric) Kit (Millipore, Boston, MA, USA) was used to assess the cell proliferation according to the manufacturer’s protocols.
Annexin V–Fluorescein Isothiocyanate (FITC)/Propidium Iodide (PI) Analysis
Cell apoptosis was detected by Annexin-V–FITC/PI Kit (Beijing Biosea Biotechnology, Beijing, China) according to the manuals. Briefly, the cells (100,000 cells/well) were seeded in a six-well plate. Treated cells were washed twice with cold phosphate-buffered saline (PBS; Gibco) and resuspended in buffer. The adherent and floating cells were combined and treated according to the manufacturer’s instruction and measured with a flow cytometer (Beckman Coulter, Brea, CA, USA) to differentiate apoptotic cells (annexin V positive and PI negative) from necrotic cells (annexin V and PI positive).
Apoptosis Detection
Cell apoptosis was also determined using a Cell Death Detection ELISAPLUS Kit (Cat. No. 11774425001; Roche Diagnostics, Basel, Switzerland) that measures cytoplasmic DNA–histone complexes generated during apoptotic DNA fragmentation. Cell apoptosis detection was performed according to the manufacturer’s protocol and monitored at 405 nm.
Caspase 3 Activity Assay
A caspase 3 fluorescent assay kit (Nanjing KeyGen Biotech. Co. Ltd., Nanjing, China) was used to detect caspase activity. Briefly, cells were lysed using the lysis buffer provided by the kit and centrifuged at 10,000 × g for 1 min at 4°C. Supernatants were collected; equal amounts (30 μg) of protein were reacted with the synthetic fluorescent substrates, which were provided by the kit, at 37°C for 90 min, and fluorescence was measured at 405 nm using a microplate reader.
In Vitro Invasion Assay
After treatment, the cells were plated onto the 24-well upper chamber with a membrane that was pretreated with Matrigel (100 μg per well; BD Biosciences, San Jose, CA, USA). In the lower portion of the chamber, we added fresh medium with 10% FBS. After the cells were incubated for 24 h at 37°C, we carefully removed the cells in the upper chamber. Invaded cells were fixed with 4% formaldehyde, stained with 0.5% crystal violet, and counted under a microscope (Nikon, Tokyo, Japan). To quantify the number of invasive cells, the washing solution was examined at 540 nm. All assays were independently repeated three times.
Protein Extraction and Western Blot Analysis
Transfected cells were solubilized with radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime Biotechnology, Shanghai, China) containing protease inhibitors (Millipore). The concentration of protein was measured by using a BCA protein assay kit (Beyotime Biotechnology). Equal amounts (50 μg) of protein were separated with 10% SDS-PAGE and transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were then blocked with 5% nonfat milk in TBST for 1 h at room temperature, followed by incubation with primary antibodies of kindlin-3 (ab68040), MMP-2 (ab97779), MMP-9 (ab58803), TIMP-1 (ab211926) (Abcam, Cambridge, UK), PCNA (#2586), CDK4 (#12790), cyclin D1 (#2922), and p27 (#3688) (Cell Signaling Technology Inc., Danvers, MA, USA) overnight at 4°C. Subsequently, the membranes were washed with TBST three times and probed with the corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies (Cell Signaling Technology Inc.) for 2 h at room temperature. ECL reagent (Pierce, Rockford, IL, USA) was used to detect the signals on the membranes.
Luciferase Reporter Assay
The luciferase reporter vectors [pGL3-kindlin-3-3′-UTR wild type (WT) and pGL3-kindlin-3-3′-UTR mutant (MUT)] were synthesized by Gene-Pharma. Cells were seeded into 24-well plates and transfected with pGL3-kindlin-3-3′-UTR WT or pGL3-kindlin-3-3′-UTR MUT, along with miR-4792 mimics or niR-NC using Lipofectamine 3000 (Invitrogen), following the manufacturer’s instructions. After transfection for 48 h, luciferase reporter assays were performed with Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA). The relative firefly luciferase activities were measured by normalizing to Renilla luciferase activities.
Statistical Analysis
The data are expressed as the mean ± standard error of the mean (SEM). The number of independent experiments is represented by n. Correlations between miR-4792 and kindlin-3 mRNA levels were analyzed using Pearson’s correlation coefficient. Multiple comparisons were performed using one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. Other comparisons were analyzed using two-tailed Student’s t-test. A value of p < 0.05 was considered statistically significant.
RESULTS
High Expression of Kindlin-3 Correlates With Low Level of miR-4792 in AML Samples and Cells
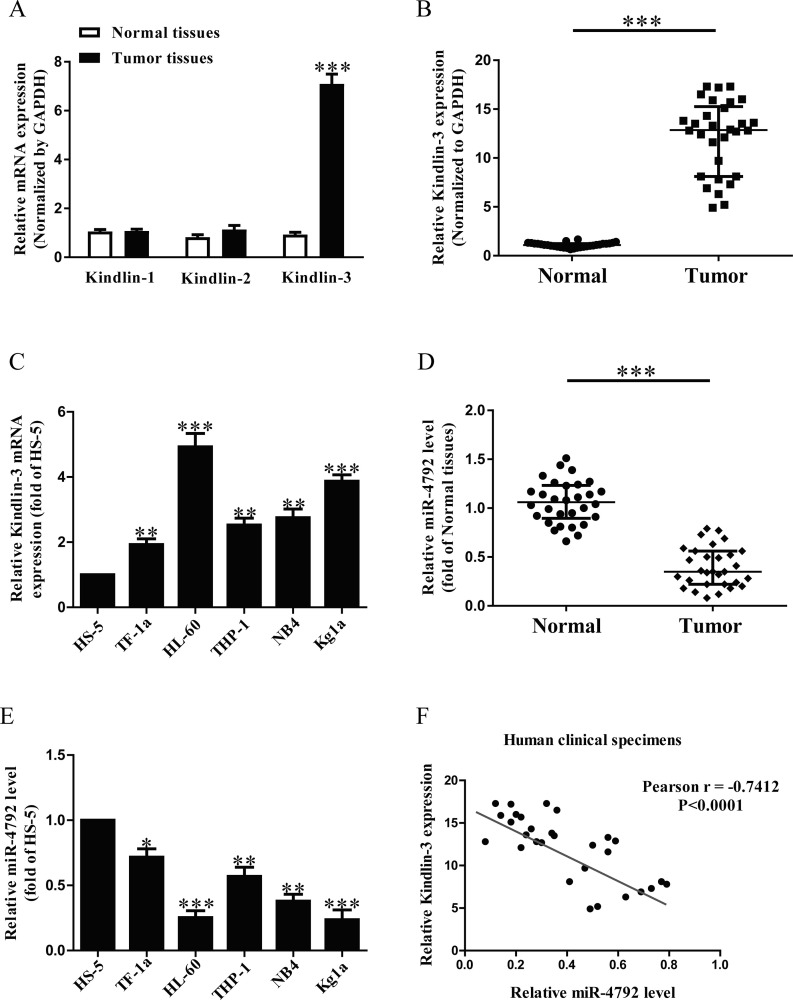
It has been reported that kindlins, including kindlin-1, kindlin-2, and kindlin-3, are closely associated with cancers. However, which one plays critical roles in AML is still unknown. In this study, kindlin-1, kindlin-2, and kindlin-3 expressions were detected by quantitative reverse transcription PCR (qRT-PCR) assays in bone marrow specimens derived from six patients with AML and six healthy controls. The data indicated that the mRNA expression of kindlin-3 was higher than that in kindlin-1 and kindlin-2 in AML samples compared with the adjacent samples (Fig. 1A). To further confirm the expression of kindlin-3, we detected kindlin-3 expression in bone marrow specimens derived from 30 patients with AML and 30 healthy controls. As had been expected, kindlin-3 mRNA expression was significantly increased in AML samples compared with the adjacent samples (Fig. 1B). Furthermore, we also determined the mRNA level of kindlin-3 in five AML cell lines such as TF-1a, HL-60, Kg1a, NB4, and THP-1, and the human normal stromal cells HS-5. Compared with HS-5, the mRNA expression of kindlin-3 in HL-60 cells was higher than that in the other four AML cell lines (Fig. 1C). For further study, the online database TargetScan 7.2 predicted that miR-4792 might directly target kindlin-3. Furthermore, our data confirmed that the miR-4792 level in the AML samples was markedly lower than that in the adjacent samples (Fig. 1D). To support this result, we also demonstrated that the miR-4792 level was lower in HL-60 cells compared with that in the other four AML cell lines, as shown in Figure 1E. Therefore, HL-60 cells were used in the following experiments. To study whether kindlin-3 expression was closely associated with miR-4792 in AML, Pearson’s correlation analysis revealed a significant inverse correlation between kindlin-3 and miR-4792 in AML tissues (Fig. 1F).
Figure 1.
The expressions of kindlin-3 and microRNA-4792 (miR-4792) in acute myeloid leukemia (AML) samples and cell lines. (A) Quantitative reverse transcription polymerase chain reaction (qRT-PCR) analysis of kindlin-1, kindlin-2, and kindlin-3 expressions in AML samples and adjacent normal samples (n = 6). Transcript levels were normalized by glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression. (B) qRT-PCR analysis of kindlin-3 expressions in AML samples and adjacent normal samples (n = 30). Transcript levels were normalized by GAPDH expression. (C) Relative kindlin-3 expression analyzed by qRT-PCR in five AML cell lines were normalized with GAPDH (n = 6). (D) qRT-PCR analysis of miR-4792 level in AML samples and adjacent normal samples (n = 30). Transcript levels were normalized by U6 level. (E) Relative miR-4792 level analyzed by qRT-PCR in five AML cell lines were normalized with U6 (n = 6). (F) Pearson’s correlation analysis of the relative expression levels of miR-4792 and the relative kindlin-3 mRNA levels in AML samples (n = 30). All data are presented as mean ± standard error of the mean (SEM). *p < 0.05, **p < 0.01, ***p < 0.001 versus normal samples or HS-5.
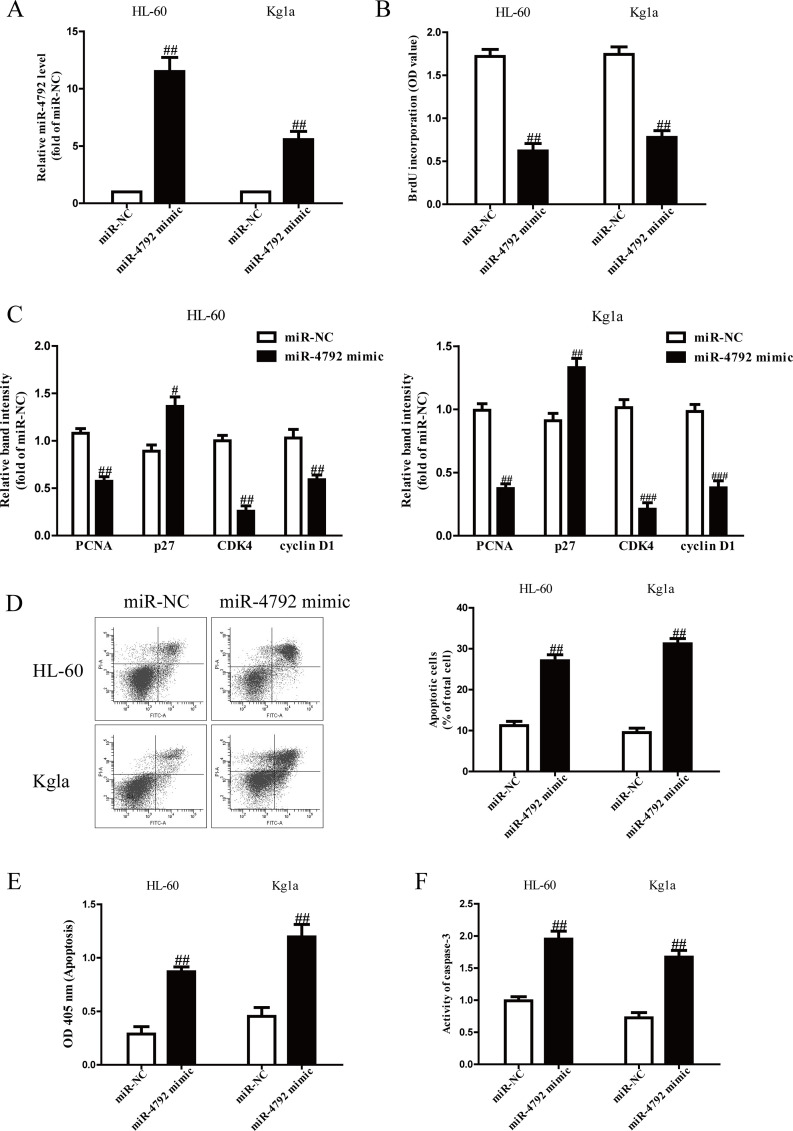
miR-4792 Inhibited Cell Proliferation and Induced Cell Apoptosis in AML Cells
The qRT-PCR analysis confirmed that the miR-4792 level was significantly increased in miR-4792 mimic compared to the miR-NC group (Fig. 2A). To investigate the effect of miR-4792 on AML cell proliferation, the BrdU-enzyme-linked immunosorbent assay (ELISA) assay showed that introduction of miR-4792 markedly suppressed the proliferation of both HL-60 and Kg1a cells (Fig. 2B). To further confirm the above results, we explored the effects of miR-4792 on several genes involved in cell proliferation and cell cycle. Overexpression of miR-4792 reduced the mRNA expressions of PCNA, CDK4, and cyclin D1 and enhanced the mRNA expressions of p27 in HL-60 and Kg1a cells (Fig. 2C). Next, FCM and ELISA assays revealed that overexpression of miR-4792 dramatically induced apoptosis of HL-60 and Kg1a cells (Fig. 2D and E). Finally, to confirm the above apoptosis results, we detected the activity of caspase 3. The activity of caspase 3 was significantly increased by miR-4792 mimic (Fig. 2F).
Figure 2.
Effects of miR-4792 on the proliferation and apoptosis in AML cells. HL-60 and Kg1a cells were transfected with miR-4792 mimic or miR-NC for 48 h. (A) The level of miR-4792 was determined by qRT-PCR assay. (B) Cell proliferation was assessed by BrdU-enzyme-linked immunosorbent assay (ELISA) assay. (C) The mRNA expressions of proliferating cell nuclear antigen (PCNA), CDK4, cyclin D1, and p27 were determined by qRT-PCR assay. (D, E) Cell apoptosis was measured by flow cytometric analysis of cells labeled with annexin V/propidium iodide (PI) double staining and nucleosomal degradation, respectively. (F) The activities of caspase 3 were determined by caspase 3 activity detection assay. All data are presented as mean ± SEM, n = 6. #p < 0.05, ##p < 0.01, ###p < 0.001 versus miR-NC.
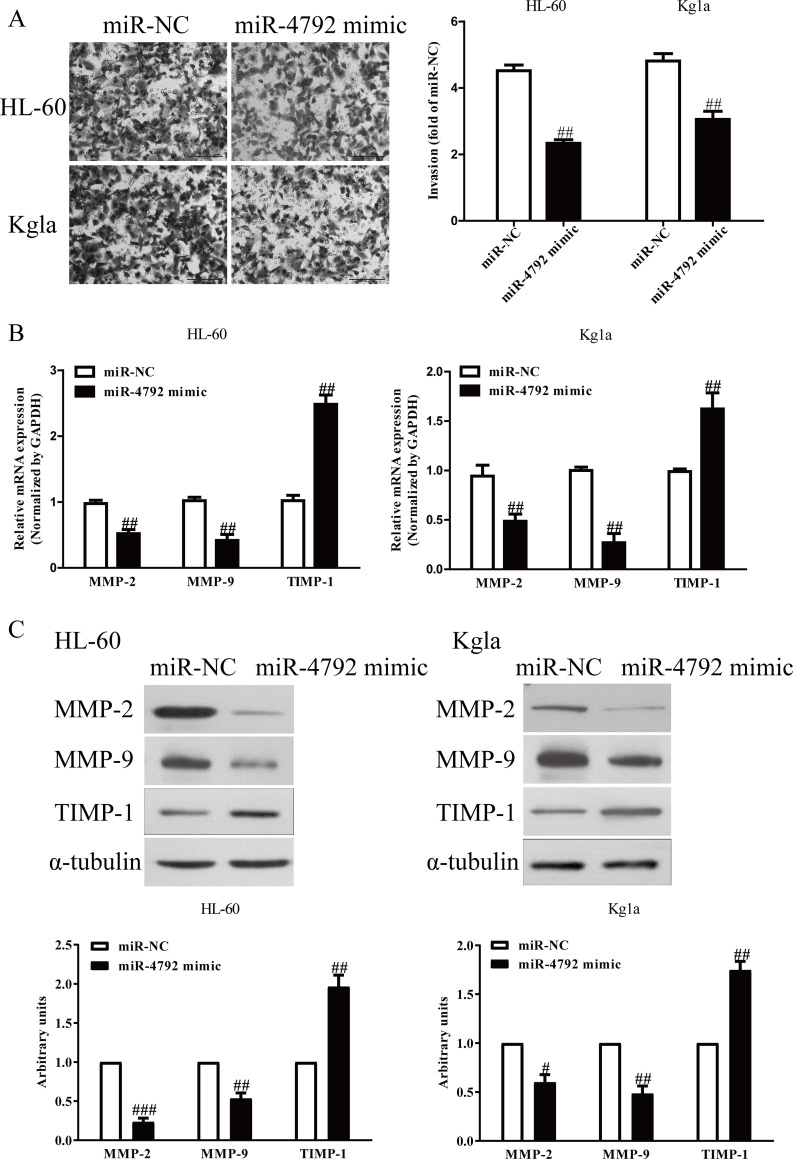
The Effects of miR-4792 on the Invasion in AML Cells
To study the effects of miR-4792 on invasion of AML cells, we assessed the invasive ability of AML cells by using Transwell assay after transfection with miR-4792 mimic or miR-NC. The miR-4792 mimic group exhibited that the number of invading AML cells was significantly reduced compared to the miR-NC group (Fig. 3A). Next, the expressions of MMP-2, MMP-9, and TIMP-1 were determined. Our qRT-PCR and Western blot assays demonstrated that both mRNA and protein expressions of MMP-2 and MMP-9 were markedly decreased, and TIMP-1 expression was significantly increased by overexpression of miR-4792 in HL-60 and Kg1a cells (Fig. 3B and C).
Figure 3.
The effects of miR-4792 on invasion and related molecules in AML cells. HL-60 and Kg1a cells were transfected with miR-4792 mimic or miR-NC for 48 h. (A) The invasion was assessed by Transwell assay. (B, C) Both mRNA and protein expressions of matrix metalloproteinase-2 (MMP-2), MMP-9, and tissue inhibitor of metalloproteinases 1 (TIMP-1) were determined by qRT-PCR and Western blot, respectively. All data are presented as mean ± SEM, n = 6. #p < 0.05, ##p < 0.01, ###p < 0.001 versus miR-NC.
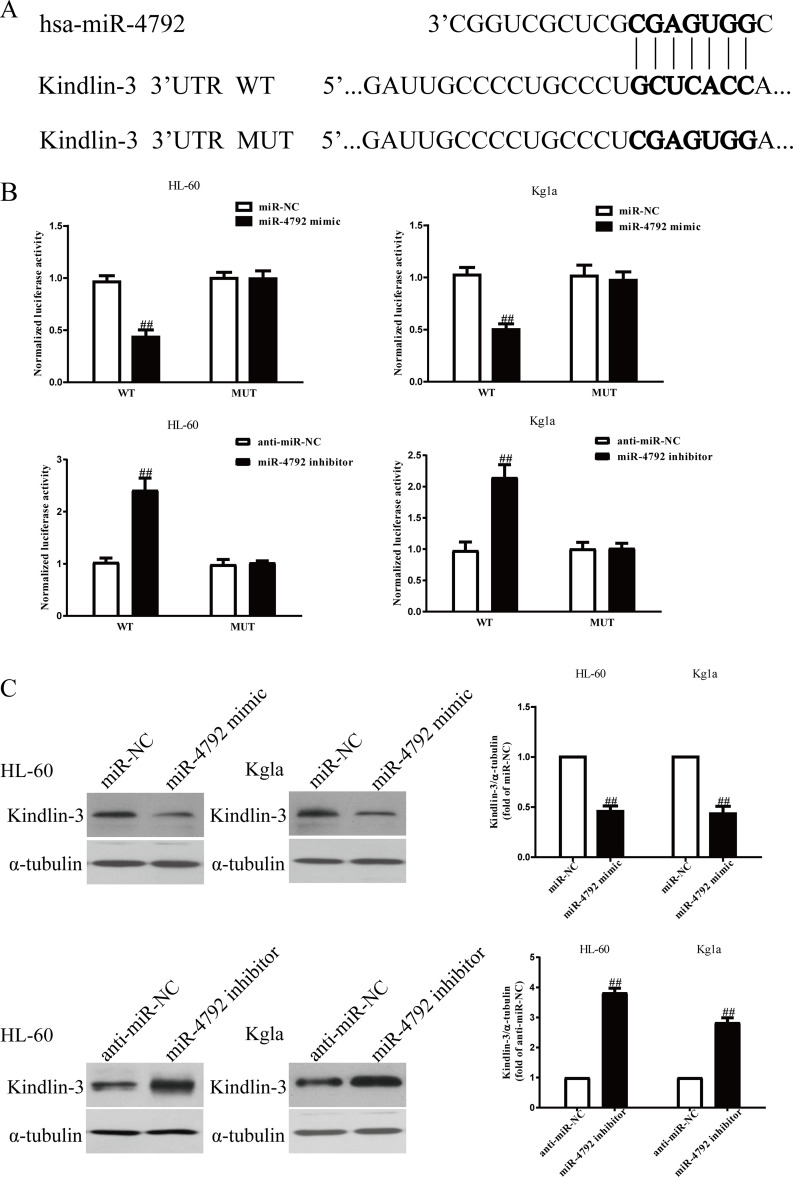
miR-4792 Directly Targeted Kindlin-3 3′-Untranslated Region (3′-UTR)
According to the online database microRNA.org, we identified an miR-4792 binding site in the 3′-UTR of kindlin-3 (Fig. 4A). To validate whether kindlin-3 is a direct target of miR-4792, luciferase plasmids containing the potential kindlin-3 miR-4792 binding sites (WT) or a mutated kindlin-3 3′-UTR were constructed (Fig. 4A). Overexpression of miR-4792 inhibited WT kindlin-3 reporter activity but not the activity of the mutated reporter construct in HL-60 and Kg1a cells, demonstrating that miR-4792 could specifically target the kindlin-3 3′-UTR by binding to the seed sequence (Fig. 4B). Downregulation of miR-4792 enhanced WT kindlin-3 reporter activity but not the activity of the mutated reporter construct in HL-60 and Kg1a cells (Fig. 4B). Next, we confirmed the results at protein level. Introduction of miR-4792 could significantly decrease the expression of kindlin-3 in HL-60 and Kg1a cells (Fig. 4C), whereas knockdown of miR-4792 increased kindlin-3 expression (Fig. 4C). These data indicated that miR-4792 directly regulated kindlin-3 expression in AML cells through 3′-UTR sequence binding.
Figure 4.
Kindlin-3 was a direct target of miR-4792. HL-60 and Kg1a cells were transfected with miR-4792 mimic or miR-NC for 48 h. (A) Schematic representation of kindlin-3 3′-UTRs showing putative miRNA target site. (B) The analysis of the relative luciferase activities of kindlin-3-WT and kindlin-3-MUT. (C) The protein expression of kindlin-3 was determined by Western blot assay. All data are presented as mean ± SEM, n = 6. ##p < 0.01 versus miR-NC or anti-miR-NC.
Knockdown of Kindlin-3 Inhibited the Proliferation, Invasion, and Induced Apoptosis of AML Cells
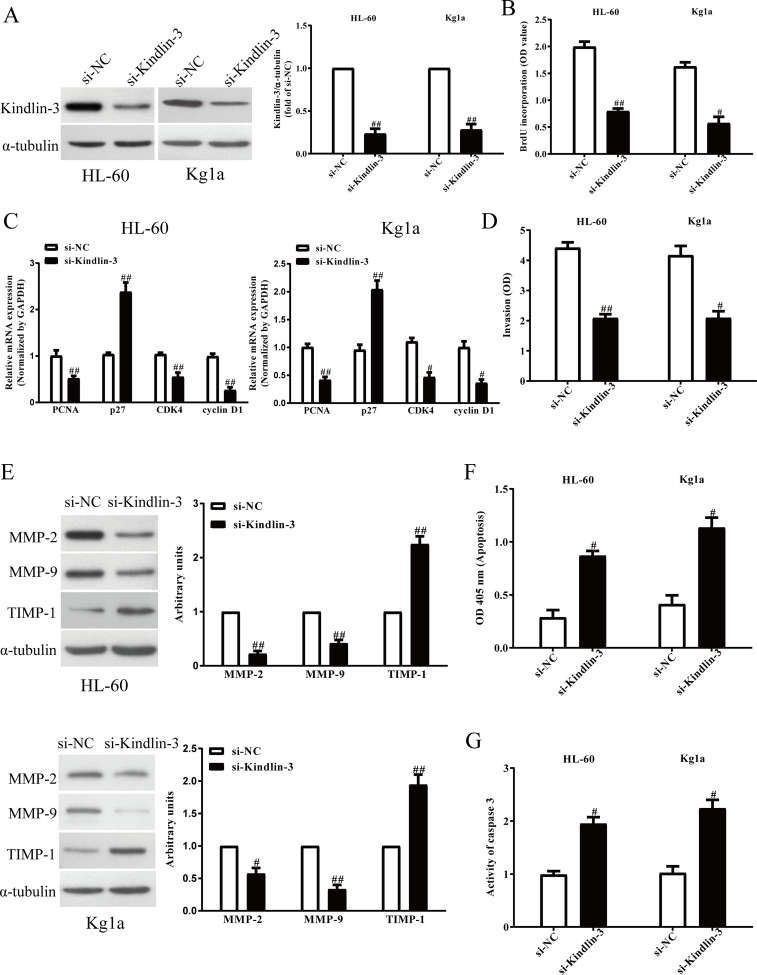
To study the effects of kindlin-3 on AML cells, cell proliferation, invasion, and apoptosis were estimated in HL-60 and Kg1a cells after transfection with si-NC or si-kindlin-3 for 48 h. Western blot analysis showed that kindlin-3 expression was significantly decreased in HL-60 and Kg1a transfected with si-kindlin-3 for 48 h compared to the si-NC group (Fig. 5A). The BrdU-ELISA assay indicated that knockdown of kindlin-3 could significantly suppress the proliferation of AML cells (Fig. 5B), and qRT-PCR assay showed that downregulation of kindlin-3 decreased the mRNA levels of PCNA, CDK4, and cyclin D1 and increased the mRNA levels of p27 (Fig. 5C). Furthermore, both Transwell and Western blot assays suggested that decreased kindlin-3 expression inhibited invasive ability of HL-60 and Kg1a cells (Fig. 5D), dramatically downregulated the expressions of MMP-2 and MMP-9 (Fig. 5E), and upregulated TIMP-1 expression (Fig. 5E). Finally, knockdown of kindlin-3 resulted in inducing apoptosis (Fig. 5F) in HL-60 and Kg1a cells and increasing the activity of caspase 3 (Fig. 5G). Consequently, kindlin-3 silencing had similar effects with miR-4792 overexpression on AML cells.
Figure 5.
The effects of kindlin-3 silencing on cell proliferation, invasion, and epithelial–mesenchymal transition (EMT) in AML cells. HL-60 and Kg1a cells were transfected with si-kindlin-3 or si-NC for 48 h. (A) The protein expressions of kindlin-3 were determined by Western blot assay. (B) Cell proliferation was assessed by BrdU-ELISA assay. (C) The mRNA expressions of PCNA, CDK4, cyclin D1, and p27 were determined by qRT-PCR. (D) The invasion was assessed by Transwell assay. (E) The protein expressions of MMP-2, MMP-9, and TIMP-1 were detected by Western blot assay. (F) Cell apoptosis was measured by nucleosomal degradation by using Roche’s cell death ELISA detection kit. (G) The activities of caspase 3 were determined by caspase 3 activity detection assay. All data are presented as mean ± SEM, n = 6. #p < 0.05, ##p < 0.01 versus si-NC.
Overexpression of Kindlin-3 Markedly Reversed the Effects of miR-4792 Upregulation on the Proliferation, Invasion, and Apoptosis of AML Cells
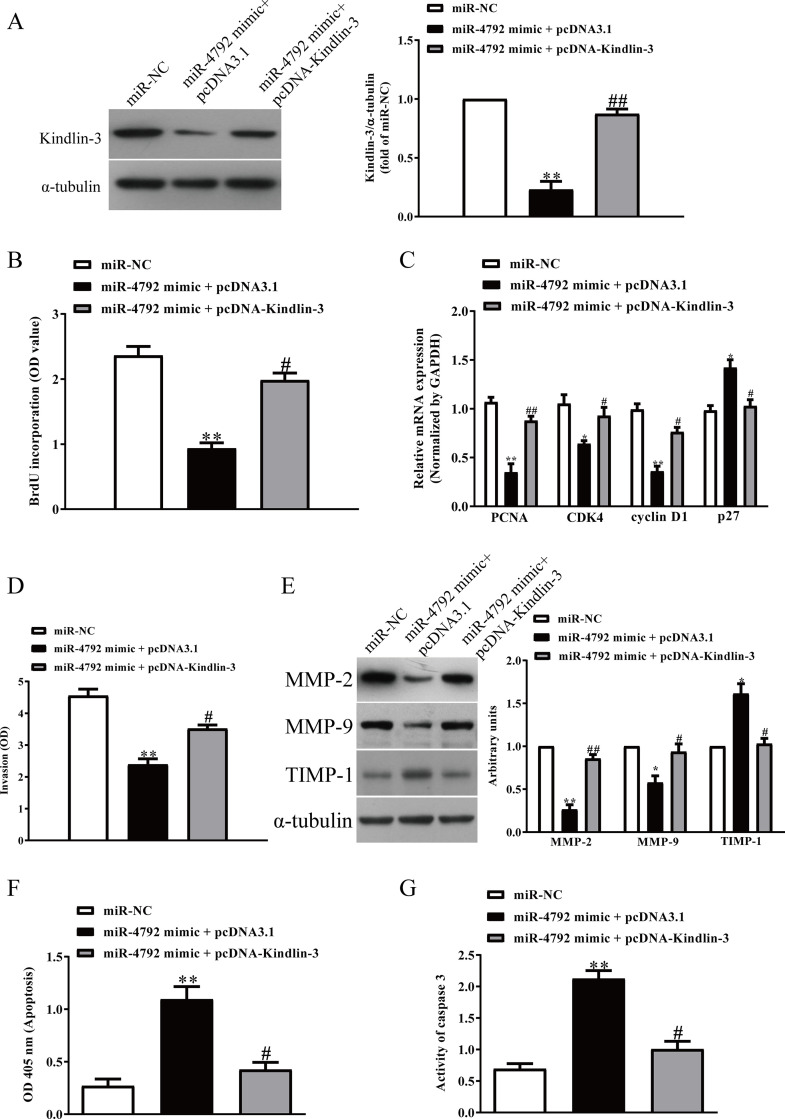
To determine whether miR-4792 targeting kindlin-3 was responsible for regulation of the proliferation, invasion, and apoptosis of AML cells, we constructed an expression vector that encoded the entire kindlin-3 coding sequence but lacked the 3′-UTR. Then we cotransfected miR-NC or miR-4792 mimic with pcDNA3.1 or pcDNA-kindlin-3 into HL-60 cells (Fig. 6A). Cell proliferation assay data showed that concomitant overexpression of miR-4792 and kindlin-3 abrogated the inhibitory effect of miR-4792 mimic (Fig. 6B). The mRNA levels of PCNA, CDK4, and cyclin D1 were increased and the mRNA levels of p27 were decreased in miR-4792-overexpressing AML cells after exogenous upregulation of kindlin-3 (Fig. 6C). Next we found that enhanced kindlin-3 expression partially reversed the inhibitory effect of miR-4792 upregulation on invasion of AML cells (Fig. 6D), upregulated the expressions of MMP-2 and MMP-9 (Fig. 6E), and downregulated TIMP-1 expression compared with the miR-4792 mimic group (Fig. 6E). Furthermore, overexpression of kindlin-3 inhibited cell apoptosis (Fig. 6F) and the activity of caspase 3 in HL-60 cells transfected with miR-4792 mimic (Fig. 6G). Therefore, the inhibitory effects of miR-4792 were partially reversed by kindlin-3 overexpression. Altogether, our findings demonstrated that miR-4792 suppressed cell proliferation, invasion, and induced apoptosis of AML cells by directly decreasing kindlin-3 expression, and miR-4792 targeting kindlin-3 was responsible for regulation of the proliferation, invasion, and apoptosis of AML cells.
Figure 6.
Overexpression of kindlin-3 partially promoted cell proliferation, invasion, and EMT in miR-4792-overexpressing AML cells. HL-60 cells were cotransfected with miR-NC or miR-4792 mimic with pcDNA3.1 or pcDNA-kindlin-3 vector. (A) The protein expression of kindlin-3 was determined by Western blot assay. (B) Cell proliferation was assessed by BrdU-ELISA assay. (C) The mRNA expressions of PCNA, CDK4, cyclin D1, and p27 were determined by qRT-PCR. (D) The invasion was assessed by Transwell assay. (E) The expressions of MMP-2, MMP-9, and TIMP-1 were detected by Western blot assay. (F) Cell apoptosis was measured by nucleosomal degradation by using Roche’s cell death ELISA detection kit. (G) The activities of caspase 3 were determined by caspase 3 activity detection assay. All data are presented as mean ± SEM, n = 6. *p < 0.05, **p < 0.01 versus miR-NC; #p < 0.05, ##p < 0.01 versus miR-4792 mimic + pcDNA3.1.
DISCUSSION
Kindlins are a small family of 4.1-ezrin-radixin-moesin (FERM)-containing cytoplasmic proteins19. Kindlin-3 expression has been reported in endothelial cells, platelets, and hematopoietic cells4–6. Kindlin-3 promotes integrin activation, clustering, and outside-in signaling20. The high expression of kindlin-3 was found in glioblastoma, whereas the low expression of kindlin-3 was reported in melanoma21,22. Intriguingly, kindlin-3 has been demonstrated to act as tumor suppressor or oncogene to regulate cancer cell metastasis21,22. In this study, kindlin-3 expression was highest among the kindlins in AML samples, suggesting that kindlin-3 played a critical role in AML. To test the biological function of kindlin-3 in AML, we found that silencing kindlin-3 expression significantly inhibited the proliferation of AML cells. For further study, we demonstrated that knockdown of kindlin-3 decreased the mRNA levels of PCNA, CDK4, and cyclin D1 and increased the mRNA levels of p27 in AML cells. Invasion is one process of metastasis. In this study, our results demonstrated that knockdown of kindlin-3 significantly inhibited the invasive ability of HL-60 cells compared with the miR-NC group. Moreover, degradation of extracellular matrix (ECM) components by proteolytic enzymes is very important for invasion of cancer cells23, and the MMPs degrading the ECM process is closely associated with invasion, metastasis, and angiogenesis of cancer cells24–26. Particularly, both MMP-2 and MMP-9 are responsible for the invasion and epithelial–mesenchymal transition (EMT) of malignant tumors, by degrading components of the basement membrane25,27,28. Next, TIMPs bind and inhibit enzymatically active MMPs, and the imbalance between MMPs and TIMPs is of critical importance in early events of tumor progression29. Here we found that expressions of MMP-2 and MMP-9 were significantly decreased in HL-60 cells after transfection with si-kindlin-3, whereas TIMP-1 expression was dramatically increased by silencing kindlin-3.
A variety of miRNAs have been known to exhibit aberrant expression in AML30–32. These dysregulated miRNAs perform crucial roles in tumorigenesis and tumor progression of AML and may regulate the major cancer-associated biological traits33–35. Therefore, the comprehensive investigation of the regulatory mechanism underlying miRNA functions in AML occurrence and development is of significant importance for the development of therapeutic strategies to treat patients with AML. To the best of our knowledge, this is the first study to detect the expression level of miR-4792 in AML. In addition, we explored the detailed roles and molecular mechanisms responsible for the action of miR-4792 on AML progression. Circulating miRNAs derived from the tumors can be stably detected in many kinds of biofluids such as plasma, serum, urine, and saliva, which contributes to early detection and diagnosis of multiple types of cancers36. A previous study reported that overexpressed miR-4792 significantly inhibited EMT and invasion of nasopharyngeal carcinoma by directly targeting FOXC137. However, the function of miR-4792 during cancer progression and development is still largely unknown. In this study, we evaluated the effect of miR-4792 in the proliferation and apoptosis of AML cell lines HL-60 and Kg1a and the possible molecular mechanism involved in this process. We found that miR-4792 mimics significantly inhibited the proliferation of HL-60 and Kg1a cells, and simultaneously promoted both cell lines’ apoptosis compared with miR-NC. As we expected, invasion of AML cells was suppressed in the miR-4792 mimic group. Moreover, cell apoptosis results showed that miR-4792 but not miR-NC induced AML cell apoptosis. Altogether, miR-4792 acted as a tumor suppressor to inhibit the proliferation and invasion of AML cells, and induce AML cell apoptosis.
However, no studies have demonstrated the relationship between miR-4792 and kindlin-3 in AML. Based on our findings, we demonstrated that the overexpression of miR-4792 reduced the expression of kindlin-3 and inhibited cancerous signals such as proliferation and invasion and induced apoptosis of AML cells. Such an effect was replicated by the knockdown of kindlin-3 in HL-60 cells. Furthermore, restoration of kindlin-3 reversed the inhibitory effects of miR-4792, indicating that miR-4792 inhibited the proliferation, invasion, and induced apoptosis of AML cells through regulation of kindlin-3, and kindlin-3 might play critical roles in the progression and metastasis of AML.
In conclusion, our results have shown that the expression of kindlin-3 was highest among kindlin-1–3, and the miR-4792 level was dramatically downregulated in AML samples. Overexpression of miR-4792 inhibited proliferation, invasion, and induced apoptosis of AML cells through directly downregulating kindlin-3 expression. Therefore, our study provided functional evidence that fully supported that the miR-4792/kindlin-3 axis was the prognostic factor for AML.
ACKNOWLEDGMENT
The authors declare no conflicts of interest.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. De Kouchkovsky I, Abdul-Hay M. Acute myeloid leukemia: A comprehensive review and 2016 update. Blood Cancer J. 2016;6(7):e441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xu XH, Zhang L, Cao XX, Li J, Zhang W, Zhu TN, Cai HC, Chen M, Han X, Yang C, Han B, Zhang Y, Zhuang JL, Zhou DB, Duan MH. Evaluation of the implementation rate of primary antifungal prophylaxis and the prognosis of invasive fungal disease in acute leukemia patients in China. J Infect Chemother. 2017;23(6):360–7. [DOI] [PubMed] [Google Scholar]
- 3. Danen-van Oorschot AA, Kuipers JE, Arentsen-Peters S, Schotte D, de Haas V, Trka J, Baruchel A, Reinhardt D, Pieters R, Zwaan CM, van den Heuvel-Eibrink MM. Differentially expressed miRNAs in cytogenetic and molecular subtypes of pediatric acute myeloid leukemia. Pediatr Blood Cancer 2012;58(5):715–21. [DOI] [PubMed] [Google Scholar]
- 4. Bialkowska K, Ma YQ, Bledzka K, Sossey-Alaoui K, Izem L, Zhang X, Malinin N, Qin J, Byzova T, Plow EF. The integrin co-activator kindlin-3 is expressed and functional in a non-hematopoietic cell, the endothelial cell. J Biol Chem. 2010;285(24):18640–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moser M, Bauer M, Schmid S, Ruppert R, Schmidt S, Sixt M, Wang HV, Sperandio M, Fässler R. Kindlin-3 is required for beta2 integrin-mediated leukocyte adhesion to endothelial cells. Nat Med. 2009;15(3):300–5. [DOI] [PubMed] [Google Scholar]
- 6. Moser M, Nieswandt B, Ussar S, Pozgajova M, Fassler R. Kindlin-3 is essential for integrin activation and platelet aggregation. Nat Med. 2008;14(3):325–30. [DOI] [PubMed] [Google Scholar]
- 7. Schmidt S, Nakchbandi I, Ruppert R, Kawelke N, Hess MW, Pfaller K, Jurdic P, Fässler R, Moser M. Kindlin-3-mediated signaling from multiple integrin classes is required for osteoclast-mediated bone resorption. J Cell Biol. 2011;192(5):883–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sossey-Alaoui K, Pluskota E, Davuluri G, Bialkowska K, Das M, Szpak D, Lindner DJ, Downs-Kelly E, Thompson CL, Plow EF. Kindlin-3 enhances breast cancer progression and metastasis by activating Twist-mediated angiogenesis. FASEB J. 2014;28(5):2260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Djaafri I, Khayati F, Menashi S, Tost J, Podgorniak MP, Sadoux A, Daunay A, Teixeira L, Soulier J, Idbaih A, Setterblad N, Fauvel F, Calvo F, Janin A, Lebbé C, Mourah S. A novel tumor suppressor function of kindlin-3 in solid cancer. Oncotarget 2014;5(19):8970–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wahid F, Shehzad A, Khan T, Kim YY. MicroRNAs: Synthesis, mechanism, function, and recent clinical trials. Biochim Biophys Acta 2010;1803(11):1231–43. [DOI] [PubMed] [Google Scholar]
- 11. Zhao H, Wang D, Du W, Gu D, Yang R. MicroRNA and leukemia: Tiny molecule, great function. Crit Rev Oncol Hematol. 2010;74(3):149–55. [DOI] [PubMed] [Google Scholar]
- 12. Lu YC, Chang JT, Chan EC, Chao YK, Yeh TS, Chen JS, Cheng AJ. miR-196, an emerging cancer biomarker for digestive tract cancers. J Cancer 2016;7(6):650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weiss M, Brandenburg LO, Burchardt M, Stope MB. MicroRNA-1 properties in cancer regulatory networks and tumor biology. Crit Rev Oncol Hematol. 2016;104:71–7. [DOI] [PubMed] [Google Scholar]
- 14. Penna E, Orso F, Taverna D. miR-214 as a key hub that controls cancer networks: Small player, multiple functions. J Invest Dermatol. 2015;135(4):960–9. [DOI] [PubMed] [Google Scholar]
- 15. Zhu B, Xi X, Liu Q, Cheng Y, Yang H. MiR-9 functions as a tumor suppressor in acute myeloid leukemia by targeting CX chemokine receptor 4. Am J Transl Res. 2019;11(6):3384–97. [PMC free article] [PubMed] [Google Scholar]
- 16. Chen L, Jiang X, Chen H, Han Q, Liu C, Sun M. microRNA-628 inhibits the proliferation of acute myeloid leukemia cells by directly targeting IGF-1R. Onco Targets Ther. 2019;12:907–19. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17. Ding C, Chen SN, Macleod RAF, Drexler HG, Nagel S, Wu DP, Sun AN, Dai HP. MiR-130a is aberrantly overexpressed in adult acute myeloid leukemia with t(8;21) and its suppression induces AML cell death. Ups J Med Sci. 2018;123(1):19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu L, Qi J, Gao X, Zhang N, Zhang H, Wang R, Xu L, Yao Y, Niu M, Xu K. High expression of miR-338 is associated with poor prognosis in acute myeloid leukemia undergoing chemotherapy. J Cell Physiol. 2019;234(11):20704–12. [DOI] [PubMed] [Google Scholar]
- 19. Meves A, Stremmel C, Gottschalk K, Fassler R. The Kindlin protein family: New members to the club of focal adhesion proteins. Trends Cell Biol. 2009;19(10):504–13. [DOI] [PubMed] [Google Scholar]
- 20. Xue ZH, Feng C, Liu WL, Tan SM. A role of kindlin-3 in integrin αMß2 outside-in signaling and the Syk-Vav1-Rac1/Cdc42 signaling axis. PloS One 2013;8(2):e56911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu C, Cui C, Liu B, Zou S, Song H, Tian H, Zhao J, Li Y. FERMT3 contributes to glioblastoma cell proliferation and chemoresistance to temozolomide through integrin mediated Wnt signaling. Neurosci Lett. 2017;657:77–83. [DOI] [PubMed] [Google Scholar]
- 22. Feng C, Wee WK, Chen H, Ong LT, Qu J, Tan HF, Tan SM. Expression of kindlin-3 in melanoma cells impedes cell migration and metastasis. Cell Adh Migr. 2017;11(5–6):419–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simpson-Haidaris PJ, Rybarczyk B. The role of fibrinogen as an extracellular matrix protein. Ann NY Acad Sci. 2001;936:406–25. [PubMed] [Google Scholar]
- 24. Bogenrieder T, Herlyn M. Axis of evil: Molecular mechanisms of cancer metastasis. Oncogene 2003;22(42):6524–36. [DOI] [PubMed] [Google Scholar]
- 25. Vihinen P, Kähäri VM. Matrix metalloproteinases in cancer: Prognostic markers and therapeutic targets. Int J Cancer 2002;99(2):157–66. [DOI] [PubMed] [Google Scholar]
- 26. Sounni NE, Janssen M, Foidart JM, Noel A. Membrane type-1 matrix metalloproteinase and TIMP-2 in tumor angiogenesis. Matrix Biol. 2003;22(1):55–61. [DOI] [PubMed] [Google Scholar]
- 27. Hornebeck W, Emonard H, Monboisse JC, Bellon G. Matrix-directed regulation of pericellular proteolysis and tumor progression. Semin Cancer Biol. 2002;12(3):231–41. [DOI] [PubMed] [Google Scholar]
- 28. Klein G, Vellenga E, Fraaije MW, Kamps WA, de Bont ES. The possible role of matrix metalloproteinase (MMP)-2 and MMP-9 in cancer, e.g. acute leukemia. Crit Rev Oncol Hematol. 2004;50(2):87–100. [DOI] [PubMed] [Google Scholar]
- 29. Herszényi L, Hritz I, Lakatos G, Varga MZ, Tulassay Z. The behavior of matrix metalloproteinases and their inhibitors in colorectal cancer. Int J Mol Sci. 2012;13(10):13240–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chang H, Gao Q, Ding W, Qing X. Identification of a robust subpathway-based signature for acute myeloid leukemia prognosis using an miRNA integrated strategy. PLoS One 2018;13(3):e0194245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ma QL, Wang JH, Yang M, Wang HP, Jin J. MiR-362-5p as a novel prognostic predictor of cytogenetically normal acute myeloid leukemia. J Transl Med. 2018;16(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krejcik Z, Belickova M, Hrustincova A, Votavova H, Jonasova A, Cermak J, Dyr JE, Merkerova MD. MicroRNA profiles as predictive markers of response to azacitidine therapy in myelodysplastic syndromes and acute myeloid leukemia. Cancer Biomark. 2018;22(1):101–10. [DOI] [PubMed] [Google Scholar]
- 33. Bi L, Zhou B, Li H, He L, Wang C, Wang Z, Zhu L, Chen M, Gao S. A novel miR-375–HOXB3–CDCA3/DNMT3B regulatory circuitry contributes to leukemogenesis in acute myeloid leukemia. BMC Cancer 2018;18(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Si X, Zhang X, Hao X, Li Y, Chen Z, Ding Y, Shi H, Bai J, Gao Y, Cheng T, Yang FC, Zhou Y. Upregulation of miR-99a is associated with poor prognosis of acute myeloid leukemia and promotes myeloid leukemia cell expansion. Oncotarget 2016;7(47):78095–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang X, Zuo D, Yuan Y, Yang X, Hong Z, Zhang R. MicroRNA-183 promotes cell proliferation via regulating programmed cell death 6 in pediatric acute myeloid leukemia. J Cancer Res Clin Oncol. 2017;143(1):169–80. [DOI] [PubMed] [Google Scholar]
- 36. Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 2008;105(30):10513–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li Y, Chen X. miR-4792 inhibits epithelial–mesenchymal transition and invasion in nasopharyngeal carcinoma by targeting FOXC1. Biochem Biophys Res Commun. 2015;468(4):863–9. [DOI] [PubMed] [Google Scholar]