Abstract
Osteosarcoma (OS), the most common bone cancer, causes high morbidity in children and young adults. TRIM46 is a member of the family of tripartite motif (TRIM)-containing proteins that serve as important regulators of tumorigenesis. Here we investigate the possible role of TRIM46 in OS and the underlying molecular mechanism. We report an increase in the expression of TRIM46 in OS and its association with tumor size, Enneking’s stage, and patient prognosis. TRIM46 knockdown inhibits OS cell viability and cell cycle progression and induces apoptosis, while TRIM46 overexpression exerts inverse effects, which are inhibited by peroxisome proliferator-activated receptor alpha (PPARα) overexpression and the nuclear factor kappa B (NF-κB) inhibitor, pyrrolidine dithiocarbamate (PDTC). Furthermore, TRIM46 negatively regulates PPARα expression via ubiquitination-mediated protein degradation and modification. PPARα overexpression also inactivates NF-κB signaling and NF-κB promoter activity in OS cells overexpressing TRIM46. Moreover, TRIM46 knockdown inhibits tumor growth and induces apoptosis of OS cells in vivo. TRIM46 acts as an oncogene in OS by interacting with and ubiquitinating PPARα, resulting in the activation of NF-κB signaling pathway. Thus, TRIM46 may be a potential biomarker of carcinogenesis.
Key words: Osteosarcoma (OS), TRIM46, Ubiquitination, NF-κB, PPARα
INTRODUCTION
Osteosarcoma (OS) is the most common primary bone malignancy diagnosed in children and young adults, characterized with the invasion and destruction of the bone and adjacent soft tissues, fatigue, and joint pain1–3. Although new treatment strategies have been recently proposed, the overall survival rate of patients with OS is limited to approximately 60% because of its propensity to lung metastasis, high resistance to chemotherapy, and advanced grade at diagnosis4. Efforts have been directed to elucidate the mechanism underlying the pathogenesis of OS and for the development of new therapeutic strategies. Loss of normal cell cycle control and resistance to apoptosis, which contribute to the uncontrolled growth of cells, are the two hallmarks of human cancers that have posed a huge challenge to cancer treatment5,6. Hence, identification of molecules or signaling pathways contributing to the regulation of cell cycle progression and apoptosis may help develop better treatment strategies and determine the molecular prognostic factor for OS.
Clinical evidence suggests that the dysregulation in the ubiquitin-mediated degradation of oncogene or tumor suppressor that contributes to cell growth, cell cycle, and apoptosis may be associated with the etiology of cancer7,8. E3 ubiquitin ligase involved in the regulation of oncogene or tumor suppressor has attracted attention among cancer researchers. Most of the tripartite motif (TRIM)-containing proteins serve as E3 ubiquitin ligases and carry three zinc-binding domains, including a RING finger, one or two B-box, and a coiled-coil region. These proteins are implicated in oncogenic processes, including cell cycle, apoptosis, and migration7,9,10. TRIM46 is a member of the TRIM family with E3 ligase activity and is associated with small cell lung carcinoma (SCLC) and paraneoplastic neurological syndrome11. The expression of TRIM46 is downregulated in non-small cell lung cancer (NSCLC)10, and TRIM46 regulates the proliferation and metastasis of breast cancer cells both in vivo and in vitro9. We analyzed the publicly available expression data and found TRIM46 expression upregulation in OS, suggestive of its important role in cancer development. However, its function in the regulation of OS cell viability, cell cycle progression, and apoptosis and the underlying molecular mechanism have not been completely understood.
Peroxisome proliferator-activated receptor (PPAR) is a ligand-activated transcription factor with many biological functions, including carcinogenesis. PPARα is one of the three subunits of the PPAR (PPARα, PPARγ, and PPARδ) involved in the regulation of inflammation, chemoresistance, proliferation, and apoptosis of cancer cells by inhibiting the nuclear factor kappa B (NF-κB) signaling pathway12–14. NF-κB family comprises five related transcription factors that regulate gene transcription under various physiological conditions. TRIM21 activates the NF-κB signaling pathway by inducing IKKβ ubiquitination15 and increases the proliferation and chemoresistance of OS cells8. TRIM14 downregulation results in the inhibition of OS cell growth and promotes their apoptosis through NF-κB signaling pathway16. Furthermore, TRIM23 regulates PPARγ protein stability through atypical ubiquitin conjugation to PPARγ17. We therefore speculate that TRIM46 may induce PPARα ubiquitination, resulting in the activation of NF-κB signaling pathway.
In the present study, we investigated the role(s) and molecular mechanism of TRIM46 in regulating OS cell viability, cell cycle progression, and apoptosis. We observed increased TRIM46 expression in OS and its correlation with tumor size, Enneking’s stage, and patient prognosis. TRIM46 promoted cell growth and inhibited apoptosis of OS by activating NF-κB signaling pathway through the ubiquitination of PPARα. Our findings suggest that TRIM46 is a potential oncogene in OS.
MATERIALS AND METHODS
Bioinformatic Analysis
The transcription profiles by of the microarray of bone specimens (n = 14) from OS patients and their normal bone counterpart controls (n = 4) were obtained from ArrayExpress (http://www.ebi.ac.uk/arrayexpress; E-MEXP-3628)18. Gene set enrichment analysis (GSEA) was performed to identify the significantly enriched biological pathways between OS specimens with high and low TRIM46 expression derived from the E-MEXP-3628 dataset.
Clinical Specimens
In the present study, 101 patients with OS and 15 patients with bone cysts were enrolled between March 2009 and August 2014. Patients with a recent history of other cancers or those that had recurrent or primary OS and received chemotherapy or radiation therapy before surgical operation were excluded from the study. Bone cysts or OS tissues were collected from the participants during routine surgery at the Shanghai Tenth People’s Hospital. Twenty-five OS specimens and 15 bone cysts were obtained for real-time quantitative polymerase chain reaction (qPCR) and Western blot assays. In addition, another 76 OS specimens were collected for immunohistochemistry (IHC). Patients’ clinical characteristics, including age, gender, tumor size, local recurrence, Enneking’s stage, anatomic location, and prognosis, were collected for statistical analysis. The study was approved by the Ethics Committee of Shanghai Tenth People’s Hospital, and all participants provided written informed consents.
IHC
IHC staining for TRIM46 and PPARα proteins was performed for 76 OS specimens following the standard protocol using anti-TRIM46 (Proteintech, Philadelphia, PA, USA) and anti-PPARα (Abcam, Cambridge, MA, USA) antibody, respectively. Patients with at least 25% positively stained tumor cells were grouped into the high expression group, while those with less than 25% positively stained tumor cells were classified into the low expression group.
Cell Culture
HOS, MG63, SAOS2, and U2OS human OS cell lines and the human osteoblastic cell line hFOB1.19 were purchased from Cell Collection of Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Lonza, Walkersville, MD, USA) supplemented with 10% (v/v) fetal bovine serum (Gibco, Carlsbad, CA, USA), 4.5% (w/v) glucose (Gibco), 100 U/ml penicillin, and 100 mg/ml streptomycin (Gibco) and maintained in a humidified 5% CO2 incubator at 37°C.
Cell Transfection
Short hairpin RNA (shRNA) specifically targeting TRIM46 and scramble shRNA were provided by Invitrogen (Grand Island, NY, USA) and inserted into pLKO.1-Puro lentivirus for constructing a TRIM46 knockdown vector. Full-length TRIM46 and PPARα were inserted into the pLVX-Puro lentivirus to obtain TRIM46 and PPARα expression vectors. 293T cells were seeded in six-well plates and transfected with pLKO.1-Puro-shTRIM46 (shTRIM46), pLVX-Puro-TRIM46 (ovTRIM46), pLVX-Puro-PPARα (ovPPARα), or pLKO.1-Puro-scramble shRNA (shNC) and blank pLVX-Puro (vector) as a negative control using Lipofectamine 2000 (Invitrogen). At 48 h after transfection, SAOS2 and HOS cells were transduced with the lentivirus for TRIM46 expression knockdown, while U2OS cells were transduced with the lentivirus-expressing TRIM46 and/or PPARα.
Cell Viability
SAOS2, HOS, and U2OS cells were seeded in a 96-well plate and incubated with the Cell Counting Kit-8 (CCK-8) solution (Dojindo Molecular Technologies, Gaithersburg, MD, USA) to detect viability at 0, 12, 24, and 48 h. Absorbance was measured at 450-nm wavelength.
Cell Cycle and Apoptosis
For cell cycle detection, SAOS2, HOS, and U2OS cells were seeded in a six-well plate and incubated with propidium iodide (PI; BioVision Inc., Mountain View, CA, USA) in the dark for 1 h at 25°C. For cell cycle detection, SAOS2, HOS, and U2OS cells were seeded in six-well plates and stained with annexin V–fluorescein isothiocyanate (FITC) and PI (BioVision Inc.). FACScan flow cytometry (Becton Dickinson, Franklin Lakes, NJ, USA) using Cell Quest software (Becton Dickinson) was used for analysis.
Coimmunoprecipitation (Co-IP) Assay
Cell lysates were prepared from SAOS2 cells using radioimmunoprecipitation assay (RIPA) buffer and incubated with anti-TRIM46 (Biorbyt, Cambridge, UK), anti-PPARα (Abcam), or control immunoglobulin G (IgG) (Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 2 h at 4°C and then with protein A/G Plus agarose (Santa Cruz Biotechnology) for 2 h at 4°C. The immunoprecipitated proteins were analyzed by Western blotting.
Ubiquitination Assay
Cell lysates prepared from SAOS2 cells that were transduced with the lentivirus for TRIM46 knockdown or control lentivirus (shTRIM46 or shNC) were incubated with anti-PPARα (Abcam) or control IgG antibody (Santa Cruz Biotechnology). Immunoprecipitated complexes were detected using anti-ubiquitin (Ub) antibody (Abcam) and standard Western blotting.
Dual-Luciferase Reporter Assay
The full-length NF-κB promoter was inserted into pGL3 vector (Promega, Madison, WI, USA). SAOS2 and HOS cells transduced with the lentivirus for TRIM46 knockdown and U2OS cells transduced with the lentivirus expressing TRIM46 or PPARα with or without pyrrolidine dithiocarbamate (PDTC) were transfected with the pGL3-NF-κB promoter (Sigma-Aldrich, St. Louis, MO, USA). Luciferase activity was detected using a dual-luciferase reporter kit (Promega).
Real-Time qPCR (RT-qPCR)
Total RNA was isolated and collected from the bone specimens derived from patients with OS and normal controls as well as OS cells using TRIzol reagent (Takara, Tokyo, Japan) in accordance with the manufacturer’s instruction. cDNA was synthesized using the iScript cDNA kit (Bio-Rad Laboratories, Hercules, CA, USA) from 1 μg of RNA per sample. qPCR was conducted with the TaqMan Fast Advanced Master Mix (Applied Biosystems, Austin, TX, USA). The mRNA level of genes was normalized to the internal control GAPDH and calculated using the 2−ΔΔCt method.
Western Blot Analysis
Total protein was isolated from the bone specimens of patients with OS and normal controls as well as OS cells using RIPA lysis buffer (Santa Cruz Biotechnology). The proteins were separated on a sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto a polyvinylidene fluoride (PVDF) membrane (Millipore, Billerica, MA, USA). The antibodies used for immunoblotting were as follows: anti-TRIM46, anti-PPARα, and anti-cleaved caspase 3 from Abcam, anti-cyclin D1, anti-p-NF-κB, anti-NF-κB, and anti-glyceraldehyde 3-phsopahte dehydrogenase (GAPDH) from Cell Signaling Technology (Danvers, MA, USA). Horseradish peroxidase (HRP)-conjugated secondary antibodies (Beyotime Biotechnology, Shanghai, China) were used, and immunoreactive bands were determined using an enhanced chemiluminescence system (ECL; Thermo Fisher Scientific, Rockford, IL, USA).
Subcutaneous Xenograft Experiment
SAOS2 and HOS cells stably transduced with the lentivirus for TRIM46 knockdown or control (shTRIM46 or shNC) were subcutaneously injected into the right flank of 6-week-old male nude mice (n = 6 peer group). On day 33 after inoculation, the tumors were collected, photographed, weighed, and analyzed by IHC, terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining, and Western blotting. All animal studies were approved by Shanghai Tenth People’s Hospital Ethics Committee.
Statistical Analysis
All results are reported as means ± standard deviation (SD). GraphPad Prism 5.0 software (GraphPad Software Inc., San Diego, CA, USA) with Student’s t-test or analysis of variance (ANOVA) followed by Dunnett’s multiple comparison test was used for data analysis. A value of p < 0.05 indicates significant difference.
RESULTS
TRIM46 Expression Is Increased in OS Tissues and Associated With OS Patient Prognosis
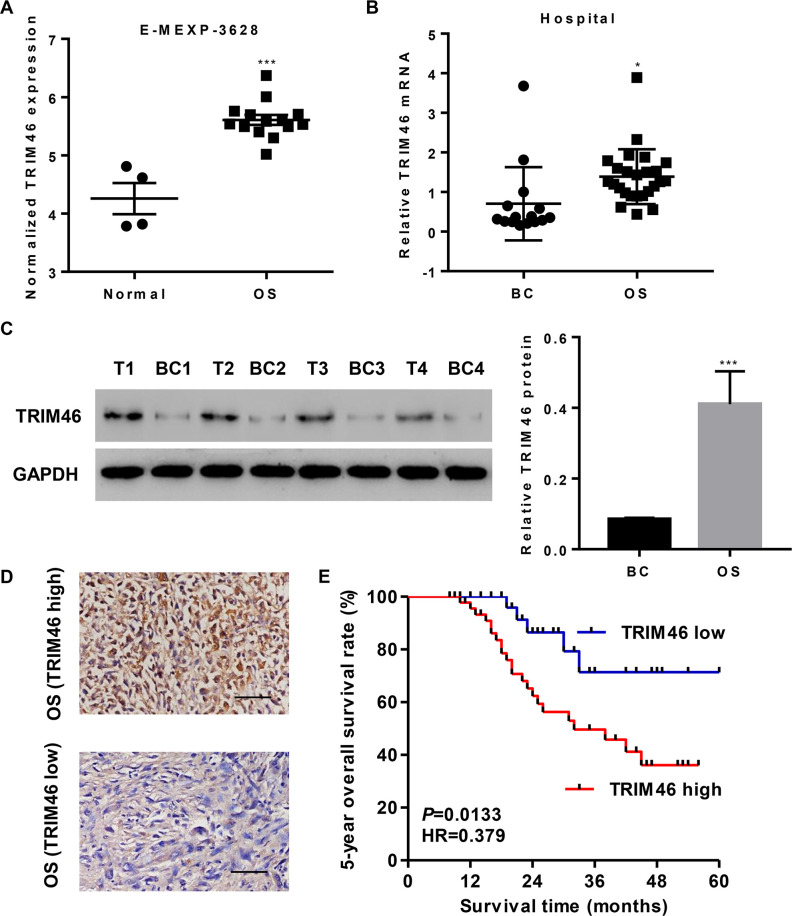
We analyzed the data from ArrayExpress (Access id: E-MEXP-3628) to investigate the differentially expressed genes between patients with OS and their normal bone counterparts. As a result, we found that the expression of TRIM46 was higher in OS tissues than in normal bone tissues (Fig. 1A). To confirm the TRIM46 expression pattern in OS, we examined TRIM46 mRNA level in OS tissues (n = 25) and normal bone tissues (n = 15) and found it to be markedly upregulated in OS tissues (Fig. 1B). Similar results were observed with the Western blot analysis (Fig. 1C).
Figure 1.
TRIM46 expression is upregulated in osteosarcoma (OS) tissues and associated with survival time. (A) TRIM46 expression in OS and their normal bone counterpart tissues based on the E-MEXP-3628 dataset. (B) TRIM46 expression in OS tissues and bone cysts (BC) from our independent hospital cohort 1 was analyzed by quantitative polymerase chain reaction (qPCR). (C) Representative TRIM46 expression in OS tissues (T1, T2, T3, and T4) and bone cysts (BC1, BC2, BC3, and BC4) from our independent hospital cohort 1, as analyzed by Western blotting. (D) Immunohistochemistry (IHC) staining for TRIM46 in OS tissues from our independent hospital cohort 2. Scale bar: 50 μm. (E) Survival analysis of patients from our independent hospital cohort 2. *p < 0.05, ***p < 0.001 compared with normal or BC.
Another 76 OS specimens were divided into two groups based on IHC staining results (Fig. 1D), namely TRIM46 high-expression group with more than 25% positively stained tumor cells (n = 47) and TRIM46 low-expression group (n = 29). Kaplan–Meier analysis and log-rank test showed that the 5-year overall survival rate of patients from the TRIM46 high-expression group was markedly lower than that of patients from the TRIM46 low-expression group (Fig. 1E). Chi-square test indicated that the expression of TRIM46 correlated with tumor size and Enneking’s stage but not with age, gender, local recurrence, and anatomic location (Table 1).
Table 1.
Relationship Between Expression Level of TRIM46 and Clinical Characteristics in Osteosarcoma
| Clinicopathological Parameter | TRIM46 | p Value | |
|---|---|---|---|
| Low (n = 29) | High (n = 47) | ||
| Age | 0.8663 | ||
| <14 | 13 | 22 | |
| ≥14 | 16 | 25 | |
| Gender | 0.7377 | ||
| Female | 10 | 18 | |
| Male | 19 | 29 | |
| Tumor size (cm) | 0.0010 | ||
| <5 | 17 | 101 | |
| ≥5 | 12 | 37 | |
| Local recurrence | 0.4180 | ||
| Yes | 2 | 6 | |
| No | 27 | 41 | |
| Enneking’s stage | 0.0096 | ||
| IIA | 12 | 7 | |
| IIB | 17 | 40 | |
| Anatomic location | 0.9501 | ||
| Femur | 16 | 25 | |
| Tibia | 8 | 13 | |
| Humerus | 3 | 4 | |
| Others | 1 | 5 | |
Differences between groups were done by the chi-square test.
We next performed a univariate analysis of the prognostic factors of overall survival with the Cox regression model (Table 2). TRIM46 level [p = 0.009, hazard ratio (HR) = 0.643, 95% confidence interval (CI) = 0.460–0.886], Enneking’s stage (p = 0.009, HR = 1.549, 95% CI = 1.156–2.118), and local recurrence (p = 0.014, HR = 0.362, 95% CI = 0.103–0.877) were independent prognostic indicators of patients with OS. We also performed a multivariate analysis with Cox regression model (Table 2) and found that TRIM46 level (p = 0.002, HR = 0.592, 95% CI = 0.423–0.810) was an independent prognostic factor. Together, these data demonstrate that TRIM46 may serve as a prognostic factor and that high TRIM46 expression was associated with poor overall survival.
Table 2.
Univariate and Multivariate Analysis of Overall Survival in Patients With Osteosarcoma
| Variables | Univariate Analysis | Multivariate Analysis | ||
|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | |
| Age (<14 vs. ≥14) | 1.125 (0.797–1.585) | 0.490 | ||
| Gender (female vs. male) | 1.182 (0.828–1.639) | 0.333 | ||
| Tumor size (cm) (<5 vs. ≥5) | 0.964 (0.649–1.347) | 0.838 | ||
| Local recurrence (yes vs. no) | 0.362 (0.103–0.877) | 0.014 | ||
| Enneking’s stage (IIA vs. IIB) | 1.549 (1.156–2.118) | 0.009 | ||
| Anatomic location (femur, tibia, humerus vs. others) | 0.893 (0.632–1.808) | 0.687 | ||
| TRIM46 expression (high vs. low) | 0.643 (0.460–0.886) | 0.009 | 0.592 (0.423–0.810) | 0.002 |
TRIM46 Expression Is Upregulated in OS Cell Lines and Promotes Cell Viability
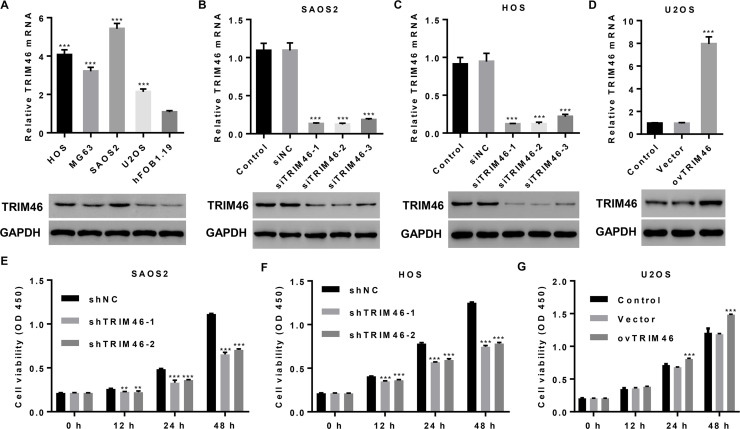
To validate the role of TRIM46 in OS cell viability, we measured TRIM46 mRNA and protein levels in OS cell lines and found them to be significantly higher in four OS cell lines than in hFOB1.19 cells. In particular, TRIM46 level was the highest in SAOS2 and HOS cells and the lowest in U2OS cells (Fig. 2A). We then transduced pLKO.1-Puro-shTRIM46 (shTRIM46) or pLKO.1-Puro-scramble shRNA (shNC) into SAOS2 and HOS cells and pLVX-Puro-TRIM46 (ovTRIM46) or blank pLVX-Puro (vector) into U2OS cells (Fig. 2B–D). The results of the CCK-8 assay demonstrate that TRIM46 knockdown markedly suppressed the viability of the SAOS2 and HOS cells at 12, 24, and 48 h compared with shNC treatment (Fig. 2E and F). On the contrary, TRIM46 overexpression significantly promoted the viability of U2OS cells at 24 and 48 h as compared with the control treatment (Fig. 2G).
Figure 2.
TRIM46 expression is upregulated in OS cell lines and promotes cell viability. (A–D) TRIM46 expression in four OS cell lines, hFOB1.19 cells, SAOS2, and HOS cells transduced with a lentivirus for silencing TRIM46 expression or U2OS cells transduced with a lentivirus expressing TRIM46 was detected by qPCR (upper) and Western blotting (lower). (E–G) Cell viability was detected at 0, 12, 24, and 48 h using the Cell Counting Kit-8 (CCK-8) assay after lentiviral transduction in SAOS2, HOS, and U2OS cells. **p < 0.01, ***p < 0.001 compared with hFOB1.19, shNC, or vector group.
TRIM46 Induces the Growth and Suppresses Apoptosis of OS Cells
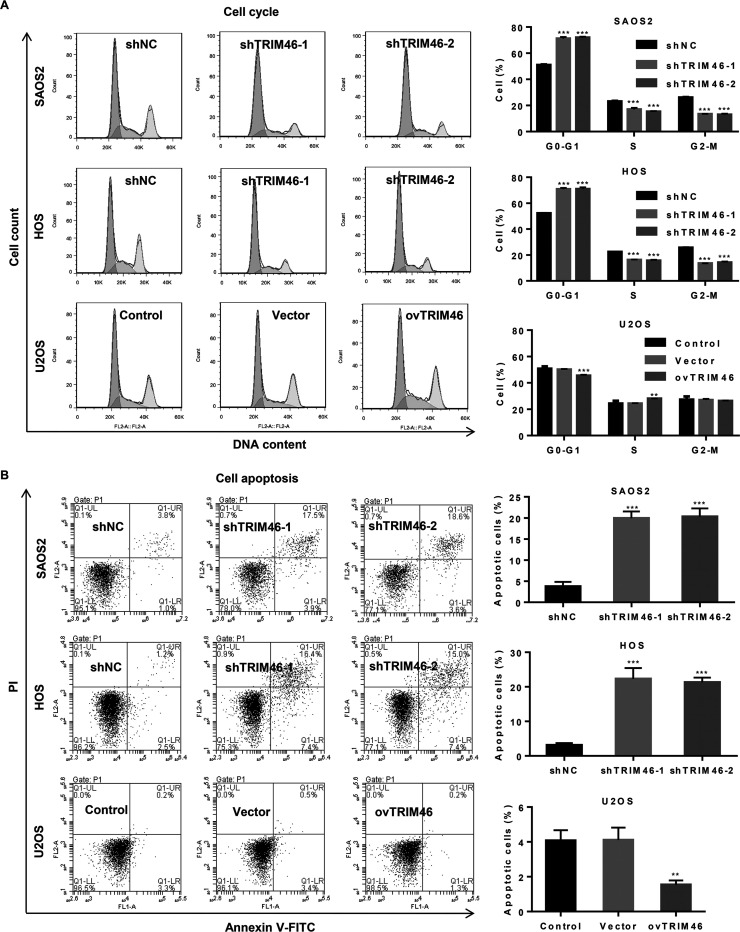
Having confirmed the regulatory effect of TRIM46 on the viability of OS cells, we investigated whether TRIM46 could also be involved in the regulation of apoptosis and cell cycle progression in OS. TRIM46 expression knockdown in SAOS2 and HOS cells significantly increased the G0/G1 phase fraction and decreased the cells in the S and G2/M phase compared with shNC treatment. On the contrary, TRIM46 overexpression in U2OS cells significantly decreased the cells in G0/G1 phase and increased those in the S phase as compared with the control treatment (Fig. 3A). Moreover, TRIM46 knockdown markedly induced the apoptosis of SAOS2 and HOS cells compared with shNC treatment, while TRIM46 overexpression markedly decreased the apoptosis of U2OS cells compared with the vector treatment (Fig. 3B). These results suggest that TRIM46 enhances the growth of OS cells.
Figure 3.
TRIM46 induces OS cell cycle progression and suppresses apoptosis. (A) Cell cycle progression and (B) apoptosis were analyzed using flow cytometry in SAOS2 and HOS cells transduced with a lentivirus for silencing TRIM46 expression or U2OS cells transduced with a lentivirus expressing TRIM46. **p < 0.01, ***p < 0.001 compared with shNC or vector.
TRIM46 Induces OS Cell Growth and Suppresses Apoptosis Through PPARα Ubiquitination
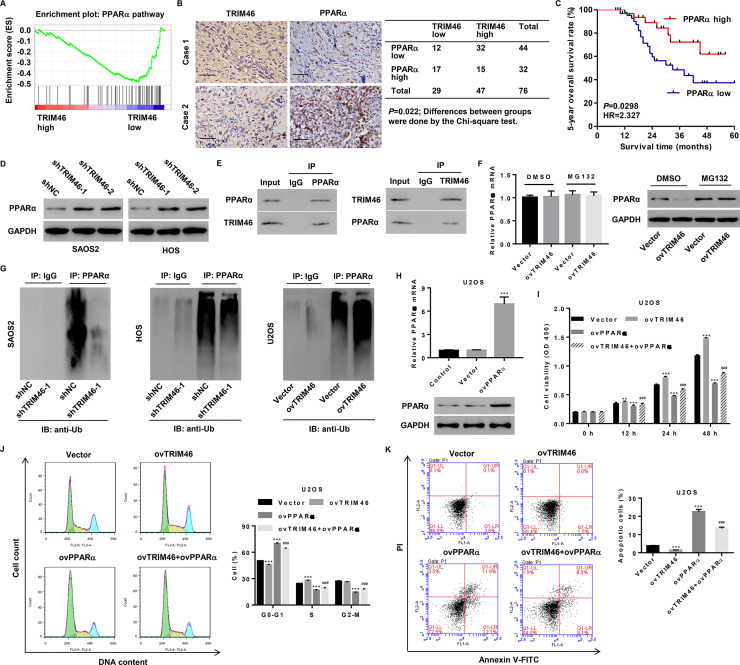
The mechanism underlying the TRIM46-mediated increase in cell growth and suppression of cellular apoptosis remains unknown. To explore the TRIM46-related signaling pathways in an unbiased manner, GSEA based on the ArrayExpress dataset was performed and PPARα pathway was identified. This pathway plays a role in the inhibition of tumorigenesis and uncontrolled cell proliferation and induction of apoptosis4 and was associated with lower TRIM46 expression (Fig. 4A). In addition, we also conducted IHC staining on OS tissues and scored TRIM46 and PPARα expression (Fig. 4B). Among 76 cases successfully stained for TRIM46 and PPARα, TRIM46 low and TRIM46 high groups showed significantly different TRIM46 staining intensity; 17/29 (58.6%) of TRIM46 low and 15/47 (31.9%) of TRIM46 high specimens showed high PPARα expression. The difference was significant (Fig. 4B), indicating a negative correlation between TRIM46 and PPARα expression. Kaplan–Meier analysis and log-rank test showed that the 5-year overall survival rate of patients with low PPARα expression was markedly lower than that of patients with high PPARα expression (Fig. 4C). The TRIM46-mediated negative regulation of PPARα was also observed in SAOS2 and HOS cells following TRIM46 knockdown (Fig. 4D). Furthermore, we performed Co-IP assay and found that TRIM46 interacted with PPARα and that the TRIM46 overexpression-mediated decrease in PPARα protein expression but not its mRNA expression was significantly inhibited by the proteasome inhibitor, MG132 (Fig. 4E and F). We therefore investigated whether TRIM46 affected PPARα ubiquitination and found that TRIM46 knockdown significantly inhibited PPARα ubiquitination in SAOS2 and HOS cells, while TRIM46 overexpression significantly promoted PPARα ubiquitination in U2OS cells (Fig. 4G). These results indicate that TRIM46 inhibits PPARα expression through its posttranslational modification.
Figure 4.
TRIM46 induces ubiquitination of peroxisome proliferator-activated receptor alpha (PPARα) and regulates cell viability, cell cycle, and apoptosis of OS through PPARα. (A) GSEA for the comparison between TRIM46 lower-expression group (blue) and TRIM46 higher-expression group (red) of OS patients from the E-MEXP-3628 dataset. Enrichment plots are shown for a set of activated genes involved in the PPARα pathway. (B) IHC staining and correlation analysis of TRIM46 and PPARα in OS tissues. Scale bar: 50 μm. (C) Survival analysis of patients from our independent hospital cohort 2. (D) PPARα expression in SAOS2 and HOS cells transduced with a lentivirus for silencing TRIM46 expression was analyzed by Western blotting. (E) The interaction between TRIM46 and PPARα in SAOS2 cells was assayed by coimmunoprecipitation (Co-IP). (F) PPARα expression in U2OS cells transduced with a lentivirus expressing TRIM46 and treated with 10 μM MG132 by qPCR (left) and Western blotting (right). (G) Effect of TRIM46 on the ubiquitination of PPARα in SAOS2, HOS, and U2OS cells. (H) PPARα expression in U2OS cells transduced with a lentivirus expressing PPARα was detected by qPCR (upper) and Western blotting (lower). (I) Cell viability, (J) cell cycle progression, and (K) cell apoptosis of U2OS cells transduced with the lentivirus expressing TRIM46 and PPARα. **p < 0.01, ***p < 0.001 compared with vector. ###p < 0.001 compared with the lentivirus expressing TRIM46 group (ovTRIM46).
Having reported the TRIM46-mediated regulation of PPARα expression in OS, we investigated whether PPARα is involved in mediating the effects of TRIM46 on cell viability, cell cycle progression, and apoptosis. U2OS cells transduced with pLVX-Puro-PPARα (ovPPARα) showed an increase in PPARα expression level compared with those from the vector control group (Fig. 4H). CCK-8 assay showed that PPARα overexpression in U2OS cells inhibited the effect of TRIM46 overexpression on cell viability (Fig. 4I). Flow cytometry analysis demonstrated that PPARα overexpression in U2OS cells significantly inhibited cellular apoptosis and cell cycle progression induced by TRIM46 overexpression (Fig. 4J and K). These results indicate that TRIM46 regulates the growth and apoptosis of OS cells through the ubiquitination of PPARα.
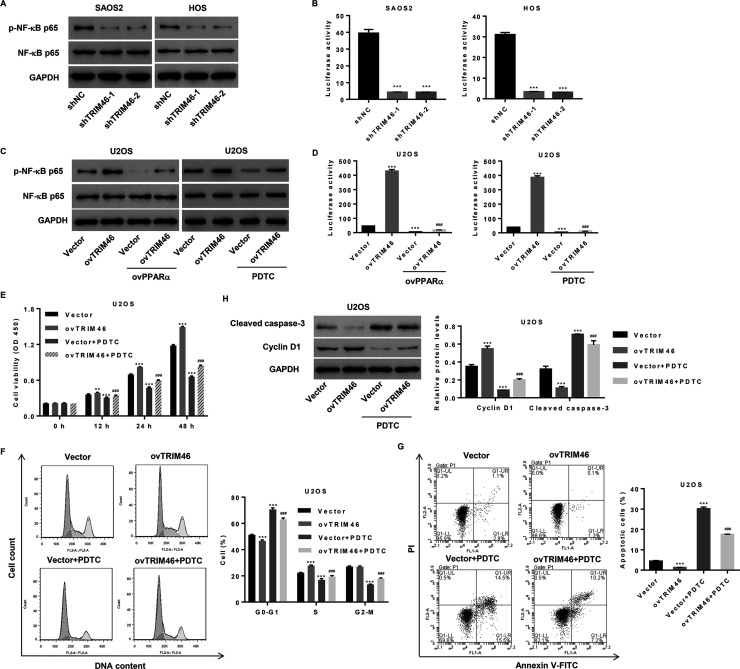
NF-κB Signaling Is Activated by TRIM46 Through PPARα and May Be Involved in Mediating the Effects of TRIM46 on Viability, Cell Cycle Progression, and Apoptosis of OS Cells
TRIM46 induces ubiquitination of PPARα, which has been reported to be associated with the inhibition of NF-κB signaling pathway14. We verified the importance of this TRIM46-regulated pathway in OS cells. As shown in Figure 5A and B, TRIM46 knockdown in SAOS2 and HOS cells significantly decreased NF-κB activation and promoter activity. Moreover, the increase in NF-κB activation and promoter activity induced by TRIM46 overexpression in U2OS cells was significantly downregulated following PPARα overexpression or NF-κB inhibitor PDTC treatment (Fig. 5C and D).
Figure 5.
TRIM46 activates nuclear factor kappa B (NF-κB) signaling and regulates NF-κB transcription via PPARα. (A) Expression of p-NF-κBp65 and NF-κBp65 and (B) activity of NF-κB promoter in SAOS2 and HOS cells transduced with a lentivirus for silencing TRIM46 expression, as detected by Western blotting and luciferase reporter assay. (C) Expression of p-NF-κBp65 and NF-κBp65 and (D) activity of the NF-κB promoter in U2OS cells transduced with a lentivirus expressing TRIM46 and PPARα or treated with 50 μM PDTC, as detected by Western blotting and luciferase reporter assay. (E) Cell viability, (F) cell cycle progression, (G) cell apoptosis, and (H) expression of cleaved caspase 3 and cyclin D1 in U2OS cells transduced with the lentivirus expressing TRIM46 and/or treated with PDTC. **p < 0.01, ***p < 0.001 as compared with shNC or vector. ###p < 0.001 compared with the lentivirus expressing TRIM46 (ovTRIM46).
To explore the role of NF-κB signaling pathway in mediating the effects of TRIM46 on cell viability, cell cycle progression, and apoptosis, U2OS cells were transduced with pLVX-Puro-TRIM46 (ovTRIM46) or blank pLVX-Puro (vector) with or without PDTC treatment. As shown in Figure 5E–G, the increase in cell growth and decrease in cell apoptosis induced by TRIM46 overexpression were inhibited by PDTC. We analyzed the levels of cell cycle- and apoptosis-associated proteins, cyclin D1 and caspase 3, by Western blotting and found that cyclin D1 expression increased but that of cleaved caspase 3 decreased after TRIM46 overexpression, and these effects were inhibited by PDTC (Fig. 5H). Thus, the NF-κB signaling pathway contributes to the TRIM46-mediated effects on cell viability, cell cycle, and apoptosis in OS.
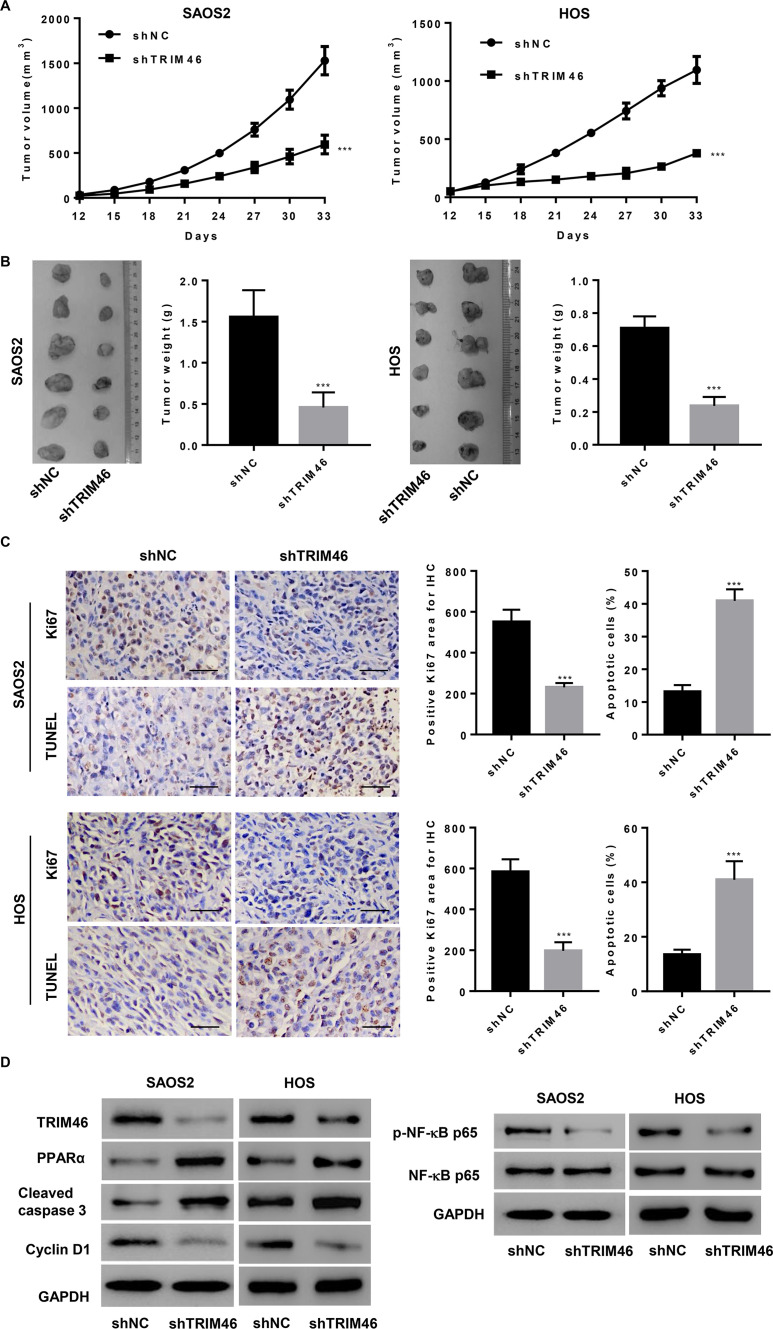
TRIM46 Induces OS Cell Growth and Suppresses Apoptosis In Vivo
Next, we investigated if TRIM46 knockdown in OS cells could reduce tumor growth in vivo. SAOS2 and HOS cells transduced with pLKO.1-Puro-shTRIM46 (shTRIM46) or pLKO.1-Puro-scramble shRNA (shNC) were subcutaneously injected into nude mice, and tumor volume was determined for 33 days. As shown in Figure 6A, shTRIM46-treated tumors grew much slower than shNC-treated tumors in mice. After 33 days of inoculation, the mice were killed; tumor weights of shTRIM46-treated mice were significantly lower than those of shNC-treated mice (Fig. 6B). The shTRIM46-treated mice also showed reduced Ki-67 expression and increased cell apoptosis as compared with shNC-treated mice, as evident through IHC and TUNEL staining, respectively (Fig. 6C). Moreover, the xenograft from shTRIM46-treated mice showed a decrease in the expression of TRIM46, cyclin D1, and p-NF-κB and an increase in the expression of PPARα and cleaved caspase-3 (Fig. 6D). These results suggest that TRIM46 downregulation inhibits tumor growth in vivo.
Figure 6.
TRIM46 promotes OS cell viability and inhibits cell apoptosis in vivo. SAOS2 and HOS cells transduced with a lentivirus to silence TRIM46 expression injected in a xenograft nude mouse model showed attenuated tumor growth (n = 6 per group). (A) Tumor volume was evaluated every 3 days for 33 days. (B) At day 33, images of xenograft tumors were obtained (left), and the tumor weights were plotted (right). (C) Xenograft tumors subjected to Ki-67 immunostaining and terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining. Scale bar: 50 μm. (D) Expression of TRIM46, PPARα, cleaved caspase 3, cyclin D1, p-NF-κBp65, and NF-κBp65 in xenograft tumors was detected by Western blotting. ***p < 0.001 compared with shNC group.
DISCUSSION
In this study, we demonstrate the upregulated expression of TRIM46 in OS tissues based on the microarray data from ArrayExpress. We provide the evidence for the important effects of TRIM46 on the regulation of OS cell viability, cell cycle progression, and apoptosis and propose a mechanism that involves PPARα/NF-κB.
In recent years, many studies have shown that TRIM family proteins such as TRIM219, TRIM218, TRIM1416, TRIM4420, TRIM5921, and TRIM6622 are known to be dysregulated and involved in the pathogenesis of OS. However, the biologic functions of most TRIM proteins in OS, including TRIM46, have not been well elucidated. TRIM46 is essential for uniform axonal microtubule orientation and axon specification and neuronal polarity23, and its expression is dysregulated in lung cancer10,11. Further, it is known to be involved in breast cancer progression9. We found that TRIM46 expression was upregulated in OS tissues, consistent with the finding in SCLC tissues11 but not in NSCLC tissues10, suggesting that the TRIM46 expression profile varies with different cancers and even in the same cancer with different histological types. Considering the correlation between TRIM46 expression and clinical characteristics of patients with OS, TRIM14 and TRIM66 expression correlated with tumor stage, histological grade, lung metastasis, local recurrence, and survival time in human OS22,24. Similar correlation between TRIM46 and poor prognosis was also observed, while local recurrence was not associated with TRIM46. Lung metastasis was not analyzed, as the patients enrolled were diagnosed with Enneking’s stage IIA and IIB without metastasis. Moreover, in addition to local recurrence and Enneking’s stage, TRIM46 was also an independent prognostic factor. These data indicate that TRIM46 expression could serve as a novel prognostic factor in patients with OS.
Loss of normal cell cycle control and resistance to apoptosis are the two hallmarks of human cancers that have posed a challenge to cancer treatment5,6. TRIM2 downregulation inhibited OS cell viability and promoted apoptosis both in vitro and in vivo19. Further, TRIM14 silencing increased OS cell proliferation and cell cycle progression in vitro and inhibited tumor growth in vivo, while its overexpression demonstrated inverse effects24. TRIM66 knockdown in OS cells decreased cell proliferation and increased apoptosis and cell cycle arrest22. These data suggest that TRIM proteins are of great importance in the regulation of OS cell growth, apoptosis, and cell cycle progression. In line with these previous studies, our data confirm the proproliferative and antiapoptotic properties of TRIM46 in OS. The tumor inhibition effect of TRIM46 knockdown was also observed in nude mice. The nuclear proliferation antigen Ki-67 is a tumor growth marker and its expression correlates with tumor size, lymph node, histological grade, and metastasis in OS. Thus, Ki-67 may contribute to OS progression25, and inhibition of Ki-67 expression was found to result in human hepatocellular carcinoma (HCC) HepG2 cell cycle arrest and apoptosis26. In addition to the decreased tumor volume and weight, decreased Ki-67 expression and increased TUNEL-positive staining were also observed in nude mice lacking TRIM46 expression. These data suggest that TRIM46 promotes OS progression both in vivo and in vitro.
PPARα activation inhibits cell proliferation and cell cycle progression in HCC and lung cancer14,27 but exerts opposite effects in glioma28 and renal cancer13. Our bioinformatic analysis shows that TRIM46 expression was significantly associated with the PPARα signaling pathway, while IHC and Western blotting results revealed the negative correlation between TRIM46 and PPARα expression in OS tissues and cells. TRIM23, like TRIM46, is a member of the TRIM family, and ubiquitinates PPARγ17. Here we explored whether TRIM46 regulates PPARα protein stability in OS cells via its E3 ubiquitin ligase activity and found that TRIM46 interacts with PPARα and inhibits its expression in a ubiquitin proteasome-dependent manner. Moreover, antiproliferative and proapoptotic effects of PPARα in OS cells overexpressing TRIM46 were observed. As PPARα is capable of inhibiting tumor progression by suppressing NF-κB activation13,14, we further investigated the role of this pathway in TRIM46-induced OS cell behavior. As expected, TRIM46 expression positively correlated with NF-κB activation and its promoter activity, which was inhibited by PPARα overexpression or PDTC treatment. The increase in cell viability and decrease in apoptosis and cell cycle arrest induced by TRIM46 overexpression were significantly inhibited by the suppression of NF-κB activation. Previous studies have reported that PPARα or inhibition of NF-κB activation results in the suppression of cyclin D1 expression and promotes caspase 3 activation in HCC14,29, which was partly similar to our findings. Thus, TRIM46 may regulate OS progression by activating NF-κB through the ubiquitination of PPARα.
In summary, our study confirms the expression pattern of TRIM46 in OS and demonstrates the increase in OS cell viability and inhibition of apoptosis and cell cycle arrest following TRIM46 ectopic expression via NF-κB signaling activation. TRIM46 promotes NF-κB activation and its promoter activity by the ubiquitination of PPARα.
ACKNOWLEDGMENT
This work was supported by the China national natural surface project (81572632 and 8187100734).
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–92. [DOI] [PubMed] [Google Scholar]
- 2. Berner K, Hall KS. Prognostic factors and treatment results of high-grade osteosarcoma in norway: A scope beyond the “classical” patient. Sarcoma 2015;2015:516843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sanchez-Pareja A, Larousserie F, Boudabbous S, Beaulieu JY, Mach N, Saiji E, Rougemont AL. Giant cell tumor of bone with pseudosarcomatous changes leading to premature denosumab therapy interruption: A case report with review of the literature. Int J Surg Pathol. 2016;24(4):366–72. [DOI] [PubMed] [Google Scholar]
- 4. Wagner ER, He BC, Chen L, Zuo GW, Zhang W, Shi Q, Luo Q, Luo X, Liu B, Luo J, Rastegar F, He CJ, Hu Y, Boody B, Luu HH, He TC, Deng ZL, Haydon RC. Therapeutic implications of PPARgamma in human osteosarcoma. PPAR Res. 2010;2010:956427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang L, Yang L, Lu Y, Chen Y, Liu T, Peng Y, Zhou Y, Cao Y, Bi Z, Liu T, Liu Z, Shan H. Osthole induces cell cycle arrest and inhibits migration and invasion via PTEN/Akt pathways in osteosarcoma. Cell Physiol Biochem. 2016;38(6):2173–82. [DOI] [PubMed] [Google Scholar]
- 6. Zhou Y, Zhao RH, Tseng KF, Li KP, Lu ZG, Liu Y, Han K, Gan ZH, Lin SC, Hu HY, Min DL. Sirolimus induces apoptosis and reverses multidrug resistance in human osteosarcoma cells in vitro via increasing microRNA-34b expression. Acta Pharmacol Sin. 2016;37(4):519–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer 2011;11(11):792–804. [DOI] [PubMed] [Google Scholar]
- 8. Zeng QZ, Liu WT, Lu JL, Liu XH, Zhang YF, Liu LX, Gao XJ. YWHAZ binds to TRIM21 but is not involved in TRIM21-stimulated osteosarcoma cell proliferation. Biomed Environ Sci. 2018;31(3):186–96. [DOI] [PubMed] [Google Scholar]
- 9. Zhang L, Li X, Dong W, Sun C, Guo D, Zhang L. Mmu-miR-1894-3p inhibits cell proliferation and migration of breast cancer cells by targeting Trim46. Int J Mol Sci. 2016;17(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhan W, Han T, Zhang C, Xie C, Gan M, Deng K, Fu M, Wang JB. TRIM59 Promotes the proliferation and migration of non-small cell lung cancer cells by upregulating cell cycle related proteins. PLoS One 2015;10(11):e0142596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. van Coevorden-Hameete MH, van Beuningen SFB, Perrenoud M, Will LM, Hulsenboom E, Demonet JF, Sabater L, Kros JM, Verschuuren J, Titulaer MJ, de Graaff , Sillevis Smitt PAE, Hoogenraad CC. Antibodies to TRIM46 are associated with paraneoplastic neurological syndromes. Ann Clin Transl Neurol. 2017;4(9):680–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huang D, Zhao Q, Liu H, Guo Y, Xu H. PPAR-alpha agonist WY-14643 inhibits LPS-induced inflammation in synovial fibroblasts via NF-kB pathway. J Mol Neurosci. 2016;59(4):544–53. [DOI] [PubMed] [Google Scholar]
- 13. Aimudula A, Nasier H, Yang Y, Zhang R, Lu P, Hao J, Bao Y. PPARα mediates sunitinib resistance via NF-κB activation in clear cell renal cell carcinoma. Int J Clin Exp Pathol. 2018;11(5):2389–400. [PMC free article] [PubMed] [Google Scholar]
- 14. Zhang N, Chu ES, Zhang J, Li X, Liang Q, Chen J, Chen M, Teoh N, Farrell G, Sung JJ, Yu J. Peroxisome proliferator activated receptor alpha inhibits hepatocarcinogenesis through mediating NF-kappaB signaling pathway. Oncotarget 2014;5(18):8330–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wada K, Niida M, Tanaka M, Kamitani T. Ro52-mediated monoubiquitination of IKK{beta} down-regulates NF-{kappa}B signalling. J Biochem. 2009;146(6):821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li YJ, Zhang GP, Zhao F, Li RQ, Liu SJ, Zhao ZR, Wang X. Target therapy of TRIM-14 inhibits osteosarcoma aggressiveness through the nuclear factor-kappaB signaling pathway. Exp Ther Med. 2018;15(3):2365–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe M, Takahashi H, Saeki Y, Ozaki T, Itoh S, Suzuki M, Mizushima W, Tanaka K, Hatakeyama S. The E3 ubiquitin ligase TRIM23 regulates adipocyte differentiation via stabilization of the adipogenic activator PPARgamma. Elife 2015;4:e05615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72(7):1865–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qin Y, Ye J, Zhao F, Hu S, Wang S. TRIM2 regulates the development and metastasis of tumorous cells of osteosarcoma. Int J Oncol. 2018;53(4):1643–56. [DOI] [PubMed] [Google Scholar]
- 20. Wang H, Fang ZL, Zhang GH, Ma X. TRIM44, a crucial target of miR-410, functions as a potential oncogene in osteosarcoma. Onco Targets Ther. 2018;11:3637–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang J, Xing D, Li Z, Shen J, Zhao H, Li S. TRIM59 is upregulated and promotes cell proliferation and migration in human osteosarcoma. Mol Med Rep. 2016;13(6):5200–6. [DOI] [PubMed] [Google Scholar]
- 22. Chen Y, Guo Y, Yang H, Shi G, Xu G, Shi J, Yin N, Chen D. TRIM66 overexpresssion contributes to osteosarcoma carcinogenesis and indicates poor survival outcome. Oncotarget 2015;6(27):23708–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Beuningen SFB, Will L, Harterink M, Chazeau A, van Battum EY, Frias CP, Franker MAM, Katrukha EA, Stucchi R, Vocking K, Antunes AT, Slenders L, Doulkeridou S, Sillevis Smitt P, Altelaar AFM, Post JA, Akhmanova A, Pasterkamp RJ, Kapitein LC, de Graaff , Hoogenraad CC. TRIM46 controls neuronal polarity and axon specification by driving the formation of parallel microtubule arrays. Neuron 2015;88(6):1208–26. [DOI] [PubMed] [Google Scholar]
- 24. Xu G, Guo Y, Xu D, Wang Y, Shen Y, Wang F, Lv Y, Song F, Jiang D, Zhang Y, Lou Y, Meng Y, Yang Y, Kang Y. TRIM14 regulates cell proliferation and invasion in osteosarcoma via promotion of the AKT signaling pathway. Sci Rep. 2017;7:42411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mardanpour K, Rahbar M, Mardanpour S. Coexistence of HER2, Ki67, and p53 in osteosarcoma: A strong prognostic factor. N Am J Med Sci. 2016;8(5):210–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lima KG, Krause GC, da Silva EFG, Xavier LL, Martins LAM, Alice LM, da Luz LB, Gassen RB, Filippi-Chiela EC, Haute GV, Garcia MCR, Funchal GA, Pedrazza L, Reghelin CK, de Oliveira JR. Octyl gallate reduces ATP levels and Ki67 expression leading HepG2 cells to cell cycle arrest and mitochondria-mediated apoptosis. Toxicol In Vitro 2018;48:11–25. [DOI] [PubMed] [Google Scholar]
- 27. Liang H, Kowalczyk P, Junco JJ, Klug-De Santiago HL, Malik G, Wei SJ, Slaga TJ. Differential effects on lung cancer cell proliferation by agonists of glucocorticoid and PPARalpha receptors. Mol Carcinog. 2014;53(9):753–63. [DOI] [PubMed] [Google Scholar]
- 28. Haynes HR, Scott HL, Killick-Cole CL, Shaw G, Brend T, Hares KM, Redondo J, Kemp KC, Ballesteros LS, Herman A. shRNA-mediated PPARα knockdown in human glioma stem cells reduces in vitro proliferation and inhibits orthotopic xenograft tumour growth. J Pathol. 2019;247(4):422–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ren K, Li Z, Li Y, Zhang W, Han X. Sulforaphene enhances radiosensitivity of hepatocellular carcinoma through suppression of the NF-kappaB pathway. J Biochem Mol Toxicol. 2017;31(8). [DOI] [PubMed] [Google Scholar]