Abstract
Long noncoding RNAs (lncRNAs) participate in and regulate the biological process of colorectal cancer (CRC) progression. Our previous research identified differentially expressed lncRNAs in 10 CRC tissues and 10 matched nontumor tissues by next-generation sequencing (NGS). In this study, we identified an lncRNA, FEZF1 antisense RNA 1 (FEZF1-AS1), and further explored its function and mechanism in CRC. We verified that FEZF1-AS1 is highly expressed in CRC tissues and cell lines. Through functional experiments, we found that reduced levels of FEZF1-AS1 significantly suppressed CRC cell migration, invasion, and proliferation and inhibited tumor growth in vivo. Mechanistically, we discovered that reduced levels of the lncRNA FEZF1-AS1 inhibited the activation of epithelial–mesenchymal transition (EMT); the overexpression of orthodenticle homeobox 1 (OTX1) partially rescued the FEZF1-AS1-induced inhibition of protein expression. It indicated that FEZF1-AS1 may play a role in the occurrence and development of CRC by regulating the FEZF1-AS1/OTX1/EMT pathway. Furthermore, it was reported that FEZF1-AS1 is located in both the nucleus and cytoplasm of HCT116 cells. Dual-luciferase reporter assays verified that FEZF1-AS1 directly binds miR-30a-5p and negatively regulated each other. Further, we showed that 5′-nucleotidase ecto (NT5E) is a direct target of miR-30a-5p, and the inhibition of miR-30a-5p expression partially rescued the inhibitory effect of FEZF1-AS1 on NT5E. Our results indicated that the mechanism by which FEZF1-AS1 positively regulates the expression of NT5E is through sponging miR-30a-5p. Our study demonstrated that lncRNA FEZF1-AS1 is involved in the development of CRC and may serve as a diagnostic and therapeutic target for CRC patients.
Key words: Colorectal cancer (CRC), FEZF1 antisense RNA 1 (FEZF1-AS1), miR-30a-5p, 5′-Nucleotidase ecto (NT5E), Orthodenticle homeobox 1 (OTX1), Epithelial–mesenchymal transition (EMT)
INTRODUCTION
Colorectal cancer (CRC) is one of the most common malignant tumors, with mortality rates second highest in the world in 20181. The incidence and mortality of CRC are increasing annually in China2. Its high degree of malignancy, rapid proliferation, significant tumor heterogeneity, and lack of specificity for chemoradiotherapy can lead to the chemoradiotherapy tolerance in CRC3. However, the mechanisms of CRC are still unclear, and it is important to find new ways of treatment and to explore effective therapeutic targets for CRC.
Long noncoding RNAs (lncRNAs) are transcripts exceeding 200 nucleotides in length that have no capability of encoding proteins4. Recently, it was found that lncRNAs participate in the development of cancer and may serve as a diagnostic target and potential drug therapeutic target for human cancers5–7. Numerous studies have indicated that many lncRNAs are aberrantly expressed in human cancer, such as HOTAIR, SChLAP1, GCLnc1, DANCR, and SNHG58–12.
FEZF1 antisense RNA 1 (FEZF1-AS1) plays an important role in the pathogenesis of most tumors13–15. Our previous research found by next-generation sequencing (NGS) assay that the lncRNA FEZF1-AS1 was differentially expressed between 10 CRC tissues and 10 matched nontumor tissues16. In this study, we investigated the role and potential mechanism of FEZF1-AS1 in CRC.
MATERIALS AND METHODS
Patients and Specimens
A total of 90 paired CRC tissues and matched nontumor tissues were obtained from patients who underwent surgery at the Fourth Hospital of Hebei Medical University between 2015 and 2017. All the specimens were diagnosed as CRC by pathological examination. Informed consent was obtained from every patient for the use of the specimens, and approval was obtained from the Medical Ethics Committee of Hebei Medical University Fourth Affiliated Hospital and Hebei Provincial Tumor Hospital. No patients received any form of chemoradiotherapy before surgery, and the clinical data were complete.
Cell Culture and Transfection
The CRC cell lines SW480 and HCT116 were obtained from the GeneChem Company (Shanghai, P.R. China). The CRC cell lines HT29, DLD-1, and RKO and human embryonic kidney (HEK293T) cells were obtained from the State Key Laboratory of Molecular Oncology, Cancer Hospital, Chinese Academy of Medical Sciences and Peking Union Medical College. SW480 cells were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA), and the other four cell lines were cultured in RPMI-1640 (Thermo Fisher Scientific) medium supplemented with 100 U/ml penicillin, 100 mg/ml streptomycin (Invitrogen, Waltham, MA, USA), and 10% fetal bovine serum (Biological Industries, Kibbutz Beit Haemek, Israel). All the CRC cells were incubated at 37°C in a 5% CO2 humidified atmosphere. Cells were transiently transfected with si-FEZF1-AS1 (GenePharma, Shanghai, P.R. China), orthodenticle homeobox 1 (OTX1) overexpression plasmid (Generay, Shanghai, P.R. China), and miR-30a-5p mimics and inhibitors (RiboBio, Guangzhou, P.R. China) using Lipofectamine 2000 (Invitrogen).
RNA Isolation and qRT-PCR Analysis
The total RNA was extracted from the cells or tissues with TRIzol solution (Invitrogen). Reverse transcription was conducted with a Transcriptor cDNA Synth kit (Roche, Basel, Switzerland) according to the instructions of the manufacturer. Quantitative real-time (qRT)-PCR was performed using TransStart Top Green qPCR SuperMix (TransGen, Beijing, P.R. China) with a quantitative Real-Time PCR system (ABI 7500; Shanghai Simenbio Technology Ltd., P.R. China). All the primers used in the experiment are listed in Table 1. The expression levels of the target genes were normalized to those of GAPDH or U6 and analyzed using the 2-ΔΔCt method.
Table 1.
Sense and Antisense Primers for Real-Time Reverse Transcription Polymerase Chain Reaction
| Genes/Primers | Sequences (5′–3′) |
|---|---|
| FEZF1-AS1 | |
| Forward | GTTCTGTGTTTGTGGTTTTGT |
| Reverse | AGGTTGCTTGTGGTGTGA |
| OTX1 | |
| Forward | TCTTTCATTCCTGGGCTCACA |
| Reverse | AACTCTGCCGACTGCTTGGAT |
| HOCX11 | |
| Forward | GCGGAAAGAAGACGAGTTAGAAA |
| Reverse | CTGACGGACCGACAAGTGAAA |
| LEMED1 | |
| Forward | CCTTCGCTGTCATCACTGTCC |
| Reverse | CTGGCCCAATACTACCTTCCA |
| GAPDH | |
| Forward | AATCCCATCACCATCTTCCAG |
| Reverse | CCTTCTCCATGGTGGTGAAGAC |
| miR-30a-5p | |
| Forward | CGCGTGTAAACATCCTCGAC |
| Reverse | AGTGCAGGGTCCGAGGTATT |
| RT primer | GTCGTATCCAGTGCAGGGTCCGAGG |
| TATTCGCACTGGATACGACCTTCCA | |
| miR-4443 | |
| Forward | GCGCGTTGGAGGCGTG |
| Reverse | AGTGCAGGGTCCGAGGTATT |
| RT primer | GTCGTATCCAGTGCAGGGTCCGAGG |
| TATTCGCACTGGATACGACAAAACC | |
| miR-610 | |
| Forward | GCGCGTGAGCTAAATGTGTG |
| Reverse | AGTGCAGGGTCCGAGGTATT |
| RT primer | GTCGTATCCAGTGCAGGGTCCGAGG |
| TATTCGCACTGGATACGACTCCCAG | |
| miR-107 | |
| Forward | GCGAGCAGCATTGTACAGGG |
| Reverse | AGTGCAGGGTCCGAGGTATT |
| RT primer | GTCGTATCCAGTGCAGGGTCCGAGG |
| TATTCGCACTGGATACGACTGATAG | |
| U6 | |
| Forward | GCTTCGGCAGCACATATACTAAAAT |
| Reverse | CGCTTCACGAATTTGCGTGTCAT |
| RT primer | CGCTTCACGAATTTGCGTGTCAT |
Western Blot Analysis
The proteins from transfected cells were extracted in RIPA lysis buffer, and the protein concentration was measured using a BCA protein assay kit (Thermo Fisher Scientific). The proteins were concentrated in 5% stacking gel and separated by 10% SDS-PAGE before being transferred to PVDF membranes for 2 h. The membranes were incubated with antibodies against E-cadherin, N-cadherin, vimentin (Proteintech, Wuhan, P.R. China), OTX1 (Boster, Wuhan, P.R. China) and 5′-nucleotidase ecto (NT5E; Proteintech) at 4°C overnight and secondary antibodies (Cell Signaling Technology, Danvers, MA, USA) for 1 h at room temperature. The experimental results were viewed using the Odyssey system (LI-COR Biosciences, Lincoln, NE, USA).
Immunohistochemistry
Immunohistochemical staining was used to quantify protein expression. The tumor tissues were fixed with 10% formalin and embedded in paraffin. After that, the tissues were dewaxed and dehydrated, antigens were retrieved with dilute gastric enzymes, and endogenous peroxidase activity was blocked with normal goat serum. The specimens were then incubated with primary antibodies against OTX1 and NT5E at 4°C overnight. After washing with PBS three times, the secondary antibody (Servicebio, Wuhan, P.R. China) was added to the sections dropwise at room temperature for 50 min. A DAB solution was used to stain the sections, which were observed under a microscope. To quantify the expression of the proteins, we graded the staining intensity and extent of the immunoreactivity. Five high-power visual fields (400×) were randomly selected from each section. The staining intensity was graded as follows: no staining (0), light brown (1), brown (2), and dark brown (3). The average percentage of positively stained cells was calculated and scored as follows: ≤25% (0), 25%–50% (1), 50%–75% (2), and ≥75% (3). The expression of OTX1 and NT5E in tumor tissues was determined by the staining intensity score and the percentage of positive cells.
Cell Proliferation Assay
After 24 h of transfection with si-FEZF1-AS1 and negative control (NC) siRNA, 2,000 cells/well were cultured with six replicates/group in 96-well plates. Cell proliferation was tested by MTS assay. The proliferation of transfected cells was detected after 0, 24, 48, 72, and 96 h. At each time point, 15 μl of MTS reagent (Promega, Madison, WI, USA) was added to each well for 2 h at 37°C. The absorbance was detected at 492 nm using a microplate reader.
Transwell Migration Assay
In Transwell migration assays, 1.5 × 105 HCT116 cells and SW480 cells were cultured in a Transwell chamber (24-well insert; 8-mm pore size; Corning Coastar, Cambridge, MA, USA). The cells were cultured in medium without fetal bovine serum, and 0.6 ml of cell medium with 20% fetal bovine serum was added to the lower 24-well plates as a chemoattractant. After approximately 24 h, the cells had moved from the upper surface to the lower surface of the membrane and were stained with crystal violet. The number of migrated cells was counted in five randomly selected fields under a microscope.
Cell Invasion Assay
In an invasion assay, a film of Matrigel (Beyotime Biotechnology, Shanghai, P.R. China) was used to cover the bottom surface of the Transwell chamber. A total of 1.8 × 105 HCT116 and SW480 cells were cultured in the Transwell chamber. The cells were cultured in medium without fetal bovine serum, and 0.6 ml of medium with 20% fetal bovine serum was added to the lower 24-well plates as a chemoattractant. After approximately 48 h, the cells had moved from the upper surface to the lower surface of the membrane and were stained with crystal violet. The number of invaded cells was counted in five randomly selected fields under a microscope.
Wound Healing Experiments
The cells were used to inoculate six-well plates. When the transfected cells reached 80% confluence, wounds were made by scraping a pipette tip through each culture plate. To ensure that wound healing was estimating in the same region, the wounded area on the well was marked. The cells were cultured in medium without fetal bovine serum. The migration of the cells was observed under a microscope at the time the scratch was made and after 24 h and 48 h.
Clonogenic Assay
A total of 2,000 transfected HCT116 or SW480 cells/well were cultured in six-well plates for 10 or 14 days. The cells were fixed with 4% paraformaldehyde for 10 min and stained by crystal violet. Cell colony-forming units were counted under a microscope.
Animal Experiments
Four- to 6-week-old male BALB/c athymic nude mice were acquired from the Beijing Vital River Laboratory Animal Technology Company (Beijing, P.R. China). To evaluate the effect of FEZF1-AS1 on nude mice, we constructed stable FEZF1-AS1 knockdown HCT116 cells. A total of 1 × 107 stable FEZF1-AS1 downregulated or control HCT116 cells were injected into all mice on the right side of the ventral subcutaneous layer. Tumor weights and volumes were determined (V = 0.5 × length × width2) with a Vernier caliper every 3 days. After 30 days, the nude mice were sacrificed, and tumor tissues were removed for qRT-PCR and immunohistochemical analysis.
Luciferase Assay
293T cells were cotransfected with the pGL3-FEZF1-AS1-WT, pGL3-NT5E-WT, or pGL3-NT5E-MUT reporter plasmids and miR-30a-5p mimics in 24-well plates. Forty-eight hours after transfection, luciferase activities were measured using the Dual-Luciferase Reporter Assay system (Promega) according to the instruction manual.
Statistical Analysis
All the experimental data were analyzed using SPSS version 21.0 software. The data are expressed as the mean ± standard deviation from three repeated experiments. Student’s t-test and one-way analysis of variance were used for comparison. A chi-square test was performed to analyze categorical variables. Spearman’s rank correlation analysis and linear correlation analysis were used to analyze the correlation between two groups. The difference was considered statistically significant with a value of p < 0.05.
RESULTS
The Overexpression of FEZF1-AS1 in Human CRC Tissues and Cell Lines
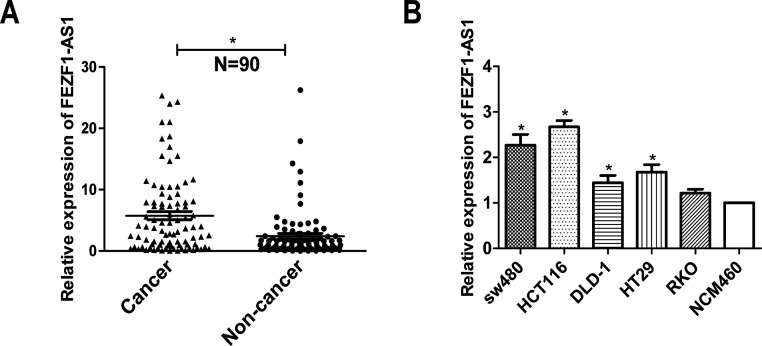
lncRNA FEZF1-AS1 was selected among the differentially expressed lncRNAs identified by NGS, and the expression of FEZF1-AS1 was verified to be overexpressed in CRC compared to that in nontumor tissues16. To confirm whether FEZF1-AS1 regulated the development of human CRC, we first tested the expression of FEZF1-AS1 in CRC tissues from patients by qRT-PCR (N = 90). As shown in Figure 1A, the transcript level of FEZF1-AS1 was significantly upregulated in CRC tissues compared with that in the corresponding nontumor colorectal tissues. Additionally, qRT-PCR was performed to detect the relative expression of FEZF1-AS1 in the SW480, HCT116, HT29, DLD-1, and RKO cell lines. The relative expression levels of FEZF1-AS1 in SW480, HCT116, DLD-1, and HT29 cells were higher than those in NCM460 cells and have statistical difference (Fig. 1B). We analyzed the relationship between FEZF1-AS1 and clinical pathological parameters and found that the increased expression of FEZF1-AS1 was significantly associated with T stage and TNM stage (Table 2). These results indicated that a high level of FEZF1-AS1 expression might play an important role in colorectal cancer.
Figure 1.
The overexpression of FEZF1 antisense RNA (FEZF1-AS1) in human colorectal cancer (CRC) tissues and cell lines. (A) The expression of FEZF1-AS1 in CRC tissues and corresponding nontumor colorectal tissues was analyzed by quantitative real-time (qRT)-PCR (N = 90). (B) qRT-PCR analysis indicated that the expression of FEZF1-AS1 was higher in the CRC cell lines than that in the NCM460 normal colonic epithelial cell line. *p < 0.05.
Table 2.
Correlation of the Expression of FEZF1-AS1 in Colorectal Cancer (CRC) With Clinicopathologic Characteristics
| Feature | lncRNA FEZF1-AS1 | χ2 | p Value | |
|---|---|---|---|---|
| Low | High | |||
| Age | 0.413 | 0.520 | ||
| <60 | 20 | 17 | ||
| ≥60 | 25 | 28 | ||
| Gender | 1.169 | 0.280 | ||
| Male | 30 | 25 | ||
| Female | 15 | 20 | ||
| Tumor size (cm) | 2.915 | 0.138 | ||
| <5 | 17 | 24 | ||
| ≥5 | 28 | 21 | ||
| Pathological differentiation | 0.494 | 0.482 | ||
| Well and moderate | 42 | 39 | ||
| Poor | 3 | 6 | ||
| T stage | 5.789 | 0.016* | ||
| T1 + T2 | 22 | 11 | ||
| T3 + T4 | 23 | 34 | ||
| Tumor embolus | 1.216 | 0.270 | ||
| Positive | 6 | 10 | ||
| Negative | 39 | 35 | ||
| TNM stage | 7.647 | 0.006* | ||
| I + II | 26 | 13 | ||
| III + IV | 19 | 32 | ||
p < 0.05.
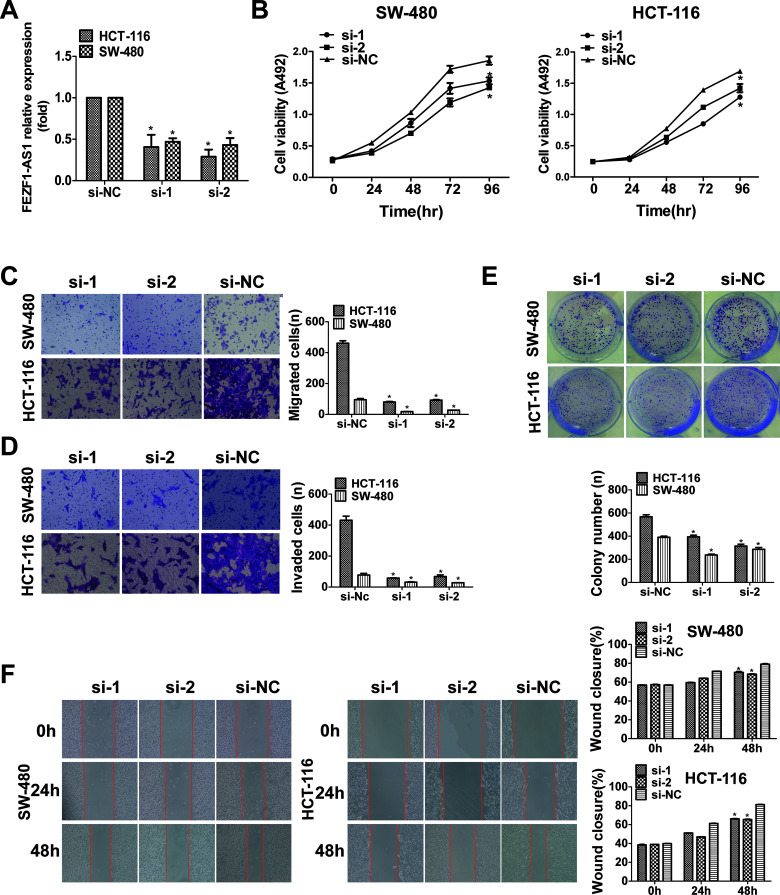
Silencing FEZF1-AS1 Suppressed CRC Cell Proliferation, Clonogenicity, Invasion, and Migration
To determine the function of FEZF1-AS1, we transfected HCT116 and SW480 cells with FEZF1-AS1 siRNA. Compared to its expression in the NC group, the expression level of FEZF1-AS1 was significantly decreased (Fig. 2A). Furthermore, the transfection of FEZF1-AS1 siRNA significantly inhibited the proliferation capacity of CRC cells compared with the NC group, but there was no significant difference between the si-1 and si-2 groups. (Fig. 2B). Transwell (Fig. 2C and D) and wound healing (Fig. 2F) assays demonstrated that the silencing of FEZF1-AS1 reduced the invasion and migration capability of CRC cells. In addition, downregulation of FEZF1-AS1 significantly inhibited the colony formation activity of CRC cells (Fig. 2E). In conclusion, these data clearly revealed that FEZF1-AS1 plays a crucial role in the formation and growth of colorectal cancer.
Figure 2.
Silencing FEZF1-AS1 suppressed CRC cell proliferation, clonogenicity, invasion, and migration. (A) The expression of FEZF1-AS1 was detected by qRT-PCR in HCT116 and SW480 cells transfected with si-FEZF1-AS1. (B) The proliferation of HCT116 and SW480 cells following FEZF1-AS1 knockdown was detected by MTS assay. Transwell assays assessed cell migration and invasion (C, D), clone formation assays assessed colony formation (E), and wound healing assays assessed cell migration (F) in the si-FEZF1-AS1 group and the negative control group, which were compared. The numbers of proliferating, invading, and migrating cells are shown in the right-hand histogram. *p < 0.05.
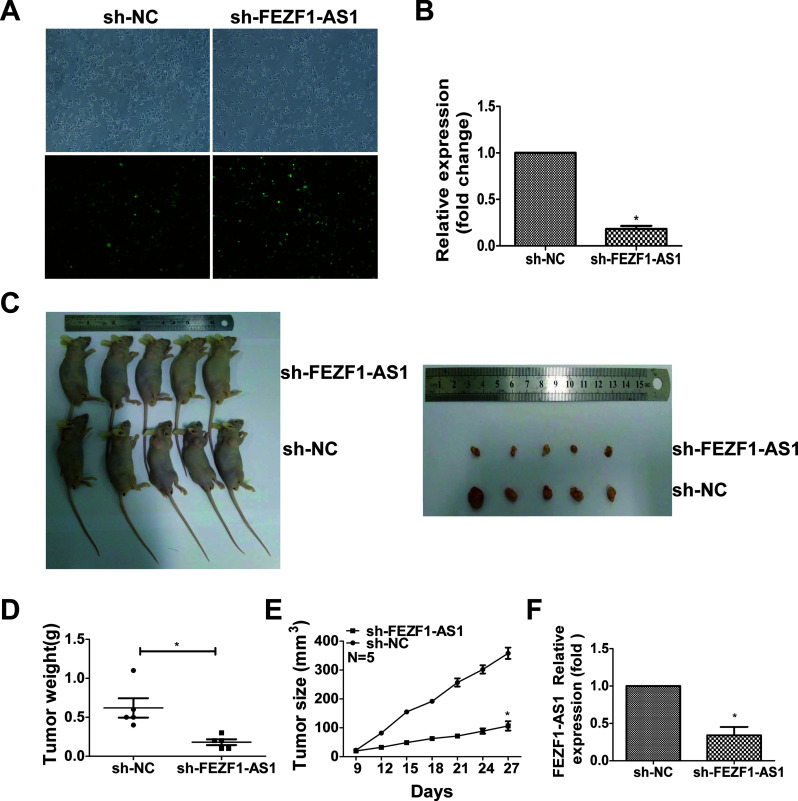
Silencing FEZF1-AS1 Suppressed the Growth of Transplanted Tumors In Vivo
To verify the effect of FEZF1-AS1 on the tumorigenesis of CRC cells in vivo, we established sh-FEZF1-AS1-HCT116 cells, in which FEZF1-AS1 was stably knocked down in HCT116 cells (Fig. 3A and B). Then sh-FEZF1-AS1-HCT116 or sh-nc-HCT116 cells were injected into the subcutaneous layer of nude mice. After 30 days, tumor growth in the sh-FEZF1-AS1-HCT116 group was much slower than that in the sh-nc-HCT116 group (Fig. 3C). The weights and sizes of tumors from the sh-FEZF1-AS1-HCT116 group were smaller than those in tumors from the NC group (Fig. 3D and E). qRT-PCR analysis found that the expression level of FEZF1-AS1 in sh-FEZF1-AS1-HCT116 xenografts was lower than that in the sh-nc-HCT116 xenografts (Fig. 3F). In conclusion, the downregulation of FEZF1-AS1 suppressed the growth of tumors in vivo.
Figure 3.
Silencing of FEZF1-AS1 suppressed the growth of transplanted tumors in vivo. Detection of stable FEZF1-AS1 expressed at low levels in HCT116 colorectal cancer cells by immunofluorescence microscopy (A) and qRT-PCR (B) after infection with lentivirus carrying the full-length sequence of human FEZF1-AS1. (C) The growth of sh-FEZF1-AS1-transfected HCT116 cells and sh-nc-transfected HCT116 cells injected into the subcutaneous layer of nude mice. (D) Tumor weights and (E) sizes were measured. (F) The expression of FEZF1-AS1 in nude mouse tissues from different groups was analyzed by qRT-PCR. *p < 0.05.
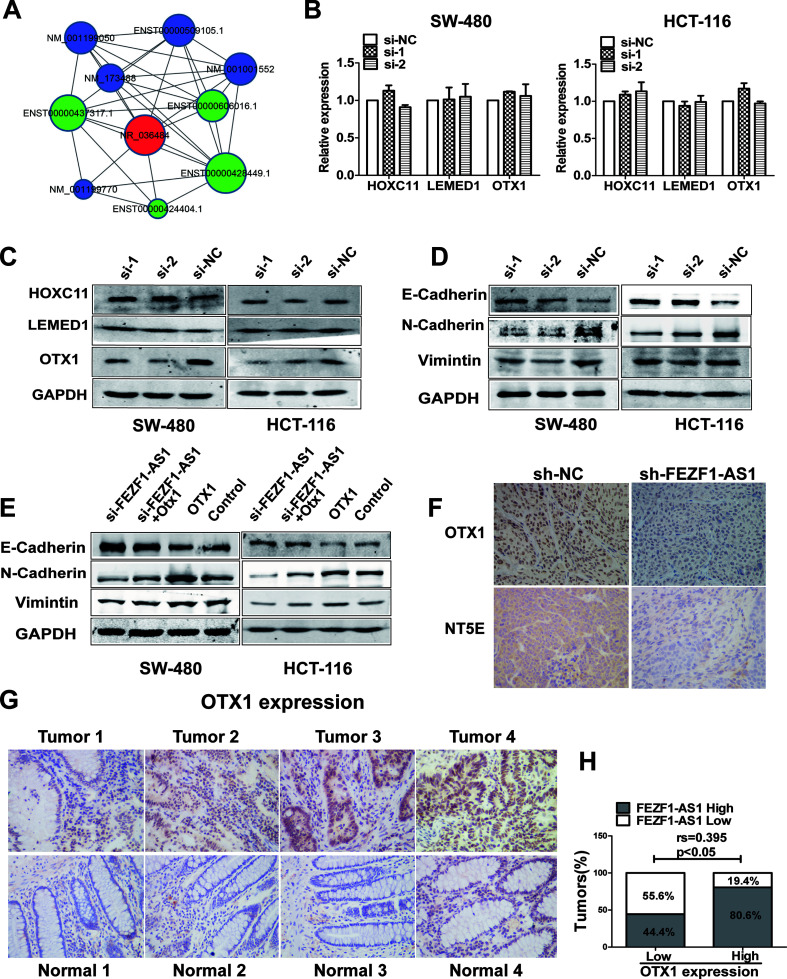
FEZF1-AS1 Was Associated With the OTX1 Protein, and the Silencing of FEZF1-AS1 Inhibited the OTX1/EMT Signaling Pathway
Gene coexpression networks were positively or negatively correlated with FEZF1-AS1 (NR-036484) in colorectal cancers based on NGS results (Fig. 4A). Among these genes, three genes, NM_014212 (HOXC11), NM_001001552 (LEMED1), and NM_001199770 (OTX1), were chosen for verification. As shown by qRT-PCR, there was no significant difference in the expression of these three proteins after FEZF1-AS1 knockdown in HCT116 and SW480 cells (Fig. 4B). However, Western blotting showed that silencing FEZF1-AS1 suppressed the expression of OTX1 (Fig. 4C). Therefore, we chose OTX1 for further experiments.
Figure 4.
FEZF1-AS1 was associated with the orthodenticle homeobox 1 (OTX1) protein, and the silencing of FEZF1-AS1 inhibited the OTX1/epithelial–mesenchymal transition (EMT) signaling pathway. (A) Expression network of genes positively or negatively related to FEZF1-AS1 (NR-036484) in colorectal cancers. Three genes positively related to FEZF1-AS1 expression were verified by qRT-PCR (B) and Western blot (C). (D) The expression of E-cadherin, N-cadherin, and vimentin in CRC cells transfected with si-FEZF1-AS1 was detected by Western blot. (E) The expression of E-cadherin, N-cadherin, and vimentin in CRC cells transfected with an OTX1 overexpression plasmid or cotransfected with si-FEZF1-AS1 and an OTX1 overexpression plasmid was detected by Western blot. (F) Representative immunohistochemical images indicating the expression of OTX1 and 5′-nucleotidase ecto (NT5E) in two groups of nude mouse tumors. (G) The expression of OTX1 in CRC tissues and normal tissues was detected by immunohistochemistry. (H) Spearman’s rank correlation analysis showed a positive correlation between OTX1 and FEZF1-AS1 in CRC tissues.
OTX1 is highly expressed in many cancers and involved in the development of cancers. A recent report found that OTX1 regulated epithelial–mesenchymal transition (EMT) to promote CRC progression17. Then we found that FEZF1-AS1 knockdown downregulated the EMT markers vimentin and N-cadherin, whereas it upregulated the expression of the epithelial marker E-cadherin (Fig. 4D). Furthermore, the inhibited EMT markers caused by FEZF1-AS1 knockdown were partially rescued by the upregulated expression of OTX1 in HCT116 and SW480 cells (Fig. 4E). Immunohistochemical analysis of animal tissues indicated that the expression levels of OTX1 and NT5E was significantly reduced in the sh-FEZF1-AS1-HCT116 group compared with those in the sh-nc-HCT116 group (Fig. 4F). Moreover, an immunohistochemical assay was used to detect the expression of OTX1 in 40 clinical CRC tissues and their normal counterparts (Table 3). The OTX1 expression levels in CRC tissues were higher than those in the corresponding nontumor colorectal tissues (Fig. 4G). As mentioned above, the expression of FEZF1-AS1 was increased in CRC tissues compared to its expression in corresponding normal colorectal tissues. In addition, Spearman’s rank correlation analysis showed that there was a positive correlation between OTX1 and FEZF1-AS1 (Fig. 4H). These results indicated that FEZF1-AS1 exerts its function in CRC cells through the FEZF1-AS1/OTX1/EMT signaling pathway.
Table 3.
OTX1 Protein Expression in Human Colorectal Noncancer Specimens and CRC
| Type of Tissues | Total (N) | Negative (−/+) | Positive (++/+++) | p (χ2) |
|---|---|---|---|---|
| Cancer | 40 | 9 | 31 | |
| Noncancer | 40 | 27 | 13 | 16.364* |
p < 0.05.
The Reciprocal Repression of FEZF1-AS1 and miR-30a-5p and miR-30a-5p Repression Promoted CRC Cell Proliferation and Migration
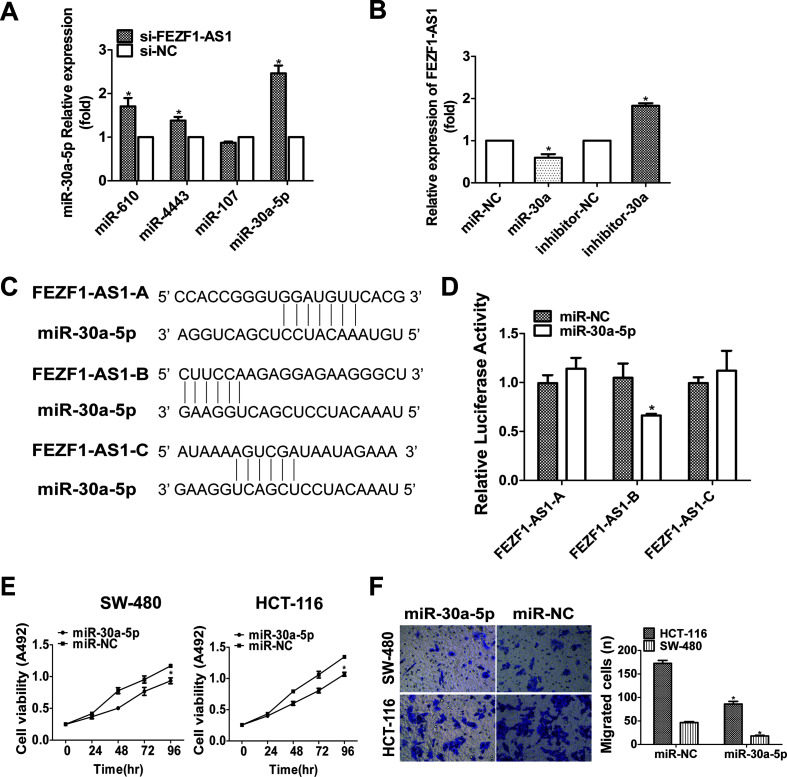
There are a variety of mechanisms by which lncRNAs work. Many studies have found that the function of lncRNAs is also related to the expression of miRNAs. FEZF1-AS1 was demonstrated in CRC cells by nucleoplasm separation technology, and FEZF1-AS1 located in both the nucleus and cytoplasm of HCT116 cells18. Therefore, we speculated that FEZF1-AS1 may play a role as a competing endogenous RNA (ceRNA). First, we predicted numerous miRNAs that could be bound to FEZF1-AS1 by using the bioinformatics tool StarBase. We chose four possible miRNAs: miR-610, miR-4443, miR-107, and miR-30a-5p. We examined the expression of these four miRNAs by qRT-PCR after FEZF1-AS1 knockdown in HCT116 cells. There was a significant negative correlation between miR-30a-5p and FEZF1-AS1 expression (Fig. 5A). Therefore, we chose miR-30a-5p for subsequent experiments. To determine whether miR-30a-5p negatively regulates FEZF1-AS1, miR-30a-5p mimics/inhibitors were transfected into HCT116 cells. The overexpression of miR-30a-5p suppressed the expression of FEZF1-AS1, and an miR-30a-5p inhibitor increased the expression of FEZF1-AS1 (Fig. 5B). The three putative complementary sequences between miR-30a-5p and FEZF1-AS1 are illustrated in Figure 5C. A dual-luciferase reporter assay showed that an miR-30a-5p mimics inhibited luciferase activity when it was cotransfected with pGL3-FEZF1-AS1-fragment-2, but not with the other two fragments (Fig. 5D). This suggested that fragment 2 of FEZF1-AS1 may be a target site of miR-30a-5p. These experimental results indicated a negative correlation between FEZF1-AS1 and miR-30a-5p.
Figure 5.
The reciprocal repression of FEZF1-AS1 and miR-30a-5p and miR-30a-5p repression promoted CRC cell proliferation and migration. (A) The expression of miR-610, miR-4443, miR-107, and miR-30a-5p was detected by qRT-PCR after FEZF1-AS1 knockdown in HCT116 cells. (B) qRT-PCR analysis of FEZF1-AS1 expression in HCT116 cells transfected with miR-NC, miR-30a-5p mimic, miR-NC inhibitor, or miR-30a-5p inhibitor. (C) Bioinformatics was used to predict miR-30a-5p binding sites in FEZF1-AS1. (D) The indicated constructs were transfected into human embryonic kidney (HEK293T) cells and cotransfected with miR-30a-5p mimics, and then the relative luciferase activity was detected. (E) The effect of the overexpression of miR-30a-5p on the proliferation of HCT116 and SW480 cells was detected by MTS assay. (F) The effect of the overexpression of miR-30a-5p on the migration of HCT116 and SW480 cells was detected by Transwell assay. *p < 0.05.
To determine the function of miR-30a-5p in CRC cells, we transfected HCT116 and SW480 cells with miR-30a-5p mimics. Compared to proliferation and migration in the NC group, the overexpression of miR-30a-5p significantly inhibited the proliferation (Fig. 5E) and migration (Fig. 5F) of HCT116 and SW480 cells.
The Expression of miR-30a-5p Was Decreased in Human CRC Tissues, and NT5E Was the Target of miR-30a-5p
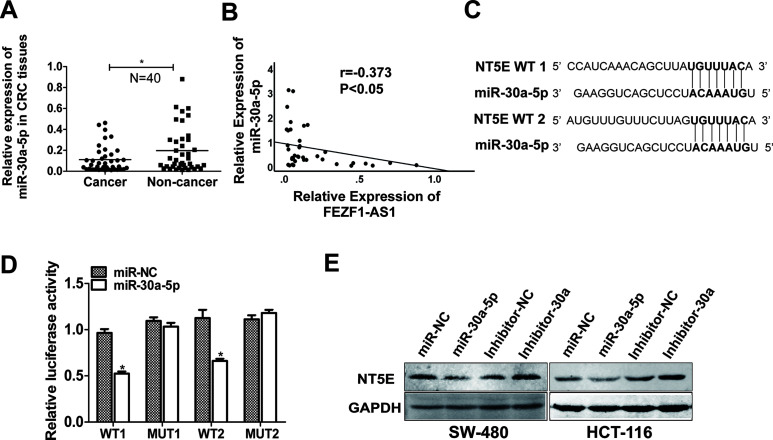
We tested the expression of miR-30a-5p in CRC tissues from patients by qRT-PCR (N = 40). As shown in Figure 6A, the transcript level of miR-30a-5p was significantly downregulated in CRC tissues compared with that in the corresponding normal colorectal tissues. In addition, linear correlation analysis showed a negative correlation between FEZF1-AS1 and miR-30a-5p in paired colorectal samples (Fig. 6B). These results indicated that miR-30a-5p is negatively correlated with FEZF1-AS1 in CRC tissues.
Figure 6.
The expression of miR-30a-5p was decreased in human CRC tissues, and NT5E was the target of miR-30a-5p. (A) The expression of miR-30a-5p in CRC tissues and corresponding nontumor colorectal tissues was analyzed by qRT-PCR (N = 40). (B) Linear correlation analysis showed a significant negative correlation between FEZF1-AS1 and miR-30a-5p in paired colorectal samples. (C) Bioinformatics was used to predict miR-30a-5p binding sites in NT5E. (D) NT5E-WT or mutant reporter vectors were transfected into HEK293T cells and cotransfected with miR-30a-5p mimics, and then the relative luciferase activity was detected. (E) Western blot analysis of NT5E expression in HCT116 cells transfected with miR-NC, miR-30a-5p mimic, miR-NC inhibitor, and miR-30a-5p inhibitor. *p < 0.05.
We used three algorithms, TargetScan, TargetMiner, and miRDB, to identify putative cotarget genes of miR-30a-5p. We predicted that NT5E was a target of miR-30a-5p. Then we predicted that the binding site of miR-30a-5p was in the 3′-UTR region of NT5E (Fig. 6C). A dual-luciferase reporter assay was used to verify our predictions, and miR-30a-5p mimics inhibited luciferase activity when cotransfected with NT5E-3′-UTR-WT. Mutation of the NT5E-3′-UTR substantially eliminated this inhibition (Fig. 6D). In addition, Western blot analysis showed that the expression of NT5E was downregulated by miR-30a-5p mimics, while the expression of NT5E was upregulated by an miR-30a-5p inhibitor (Fig. 6E). These results indicated that NT5E is a direct target of miR-30a-5p.
The FEZF1-AS1/miR-30a-5p Axis Regulates CRC Cell Migration and Proliferation by Targeting NT5E
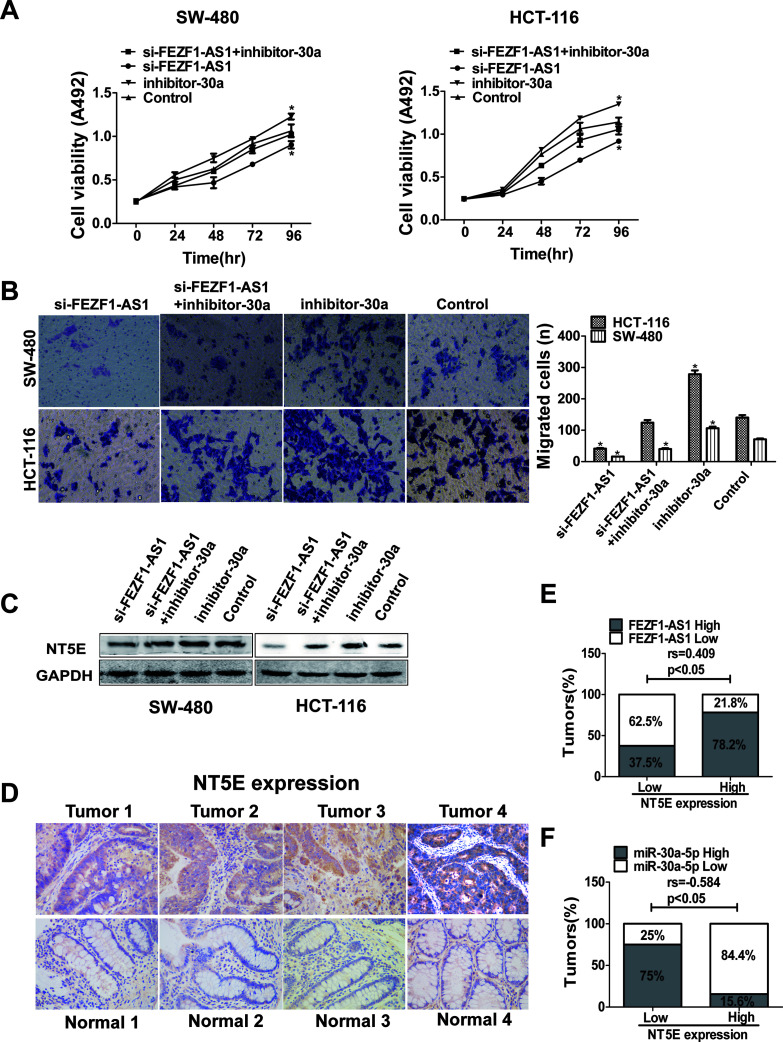
To verify whether the mutual inhibition of FEZF1-AS1 and miR-30a-5p plays a role in CRC cells, we cotransfected si-FEZF1-AS1 and miR-30a-5p inhibitor into HCT116 and SW480 cells. The inhibition of cell proliferation and migration caused by FEZF1-AS1 knockdown was partially rescued when HCT116 and SW480 cells were cotransfected with miR-30a-5p inhibitor and si-FEZF1-AS1 (Fig. 7A and B). Western blotting was used to explore whether FEZF1-AS1 regulates NT5E expression by interacting with miR-30a-5p. NT5E protein expression was inhibited by FEZF1-AS1 knockdown and was rescued by the inhibition of miR-30a-5p in HCT116 and SW480 cells (Fig. 7C). Therefore, our results indicated that FEZF1-AS1 regulates NT5E expression through interacting with miR-30a-5p.
Figure 7.
The lncRNA FEZF1-AS1/miR-30a-5p axis regulates CRC cell migration and proliferation by targeting NT5E. (A) The proliferation of HCT116 and SW480 cells was detected by MTS assay 96 h after transfection. (B) Cell migration was detected by Transwell assay 24 h after transfection. (C) The expression of NT5E in CRC cells transfected with miR-30a-5p inhibitor or cotransfected with si-FEZF1-AS1 and miR-30a-5p inhibitor was detected by Western blot. (D) The expression of NT5E was detected by immunohistochemistry in CRC tissues and normal tissues. (E) Spearman’s rank correlation analysis showed a positive correlation between NT5E and FEZF1-AS1, whereas there was a negative correlation between NT5E and miR-30a-5p (F). *p < 0.05.
NT5E is highly expressed in many cancers and involved in the development of cancers, including CRC19. We used an immunohistochemical assay to verify the expression of NT5E in 40 pairs of clinical CRC tissues and their normal counterparts (Table 4). The NT5E expression level was higher in CRC tissues than in corresponding normal colorectal tissues (Fig. 7D). Spearman’s rank correlation analysis showed a positive correlation between NT5E and FEZF1-AS1 (Fig. 7E), whereas there was a negative correlation between NT5E and miR-30a-5p (Fig. 7F) in paired colorectal samples. These results suggested that the FEZF1-AS1/miR-30a-5p/NT5E axis plays an important role in CRC cell migration and proliferation.
Table 4.
NT5E Protein Expression in Human Colorectal Noncancer Specimens and CRC
| Type of Tissues | Total (N) | Negative (−/+) | Positive (++/+++) | p (χ2) |
|---|---|---|---|---|
| Cancer | 40 | 8 | 32 | |
| Noncancer | 40 | 29 | 11 | 22.175* |
p < 0.05.
DISCUSSION
CRC is one of the most common causes of cancer-related death. Recently, genomic studies have found that lncRNAs are key regulatory factors of gene expression and cells20. lncRNAs have emerged as key molecules in CRC (e.g., the lncRNA CCAL, which is an important regulator of CRC). The overexpression of CCAL was associated with a low overall survival rate and predicted a poor response to adjuvant chemotherapy in patients with CRC21. The lncRNA HOXB-AS3 encodes a peptide that suppresses CRC growth22. High expression levels of the lncRNAs CCAT1 and CCAT2 are closely related to poor recurrence-free survival and overall survival, and these two lncRNAs are important prognostic biomarkers in CRC23. However, the potential molecular mechanisms of lncRNAs in CRC are not yet clear. The lncRNA FEZF1-AS1 participates in the progression of colorectal carcinoma, lung adenocarcinoma, gastric cancer, and pancreatic ductal adenocarcinoma and is highly expressed in these cancers24–27. In this study, we selected lncRNA FEZF1-AS1 from the results of NGS16 and verified that FEZF1-AS1 was highly expressed in CRC tissues and cell lines. Furthermore, clinical data demonstrated that the expression of FEZF1-AS1 is positively correlated with T stage and TNM stage. Through functional experiments, we found that reduced levels of FEZF1-AS1 significantly suppressed CRC cell migration, invasion, and proliferation and inhibited tumor growth in a CRC nude mouse model. These results revealed the participation of FEZF1-AS1 in the development of CRC.
OTX1 acts as a transcription factor and may play a role in the development of the brain and sensory organs. Recently, OTX1 was found to participate in several tumors28–30. In our study, our data also indicated that the OTX1 expression level in CRC tissues was higher than that in the corresponding nontumor colorectal tissues. As shown by qRT-PCR, there was no significant difference in the expression of OTX1 after FEZF1-AS1 knockdown. However, Western blotting showed that the expression of OTX1 was decreased after FEZF1-AS1 knockdown. The results indicated that knockdown of FEZF1-AS1 did not affect the transcriptional level and posttranscriptional degradation level of OTX1, but affected its protein translation level or protein degradation level. Yu et al. reported that OTX1 participated in human colon tumorigenesis through EMT17. Nieto et al. suggested that EMT played a major role in all stages of tumorigenesis and development, such as tumor proliferation, invasion, diffusion, and colonization31. We found that the expression of the EMT markers vimentin and N-cadherin was decreased, and the expression of the epithelial marker E-cadherin was upregulated after FEZF1-AS1 knockdown. Next, cotransfection with si-FEZF1-AS1 and pCDNA-OTX1 partially rescued the FEZF1-AS1-induced inhibition of protein expression. Taken together, these results indicated that FEZF1-AS1 might affect the growth and metastasis of CRC cells through the OTX1/EMT pathway, and FEZF1-AS1 might be a target for the diagnosis and therapy of CRC.
The ceRNA mechanism is a regulatory model of the interactions between RNAs. Increasing evidence suggests that ceRNAs are a kind of RNA with miRNA binding sites that can competitively bind miRNA to inhibit its regulation of target genes. For instance, Hmga2 functions as a competing endogenous RNA to promote lung carcinogenesis32. The lncRNA ADNCR suppresses adipogenic differentiation by targeting miR-20433. The lncRNA CCR492 functions as a let-7 miRNA that competes with endogenous RNA to regulate c-Myc expression34. Previous articles found that miR-30a-5p was downregulated in CRC tissues35,36. Our data also suggested that miR-30a-5p was downregulated in CRC tissues, which decreased the proliferation and migration of CRC cells. Whether FEZF1-AS1 and miR-30a-5p directly bind requires further investigation. In this study, FEZF1-AS1 located in both the nucleus and cytoplasm of HCT116 cells, and FEZF1-AS1 and miR-30a-5p negatively regulated each other. Dual-luciferase reporter assays verified that FEZF1-AS1 directly binds miR-30a-5p. Zhu et al. found that miR-30a was involved in the development of nonsmall cell lung cancer by regulating the expression of NT5E37. We hypothesized that FEZF1-AS1 regulated NT5E by sponging miR-30a-5p. Our luciferase reporter assays found that NT5E is a direct target of miR-30a-5p. Furthermore, the expression of NT5E was downregulated by miR-30a-5p mimics and upregulated by an miR-30a-5p inhibitor. We proved that the inhibition of miR-30a-5p expression partially rescued the inhibitory effect of FEZF1-AS1 on NT5E. These results suggest that FEZF1-AS1 functions as a ceRNA to regulate NT5E.
In summary, our data showed that FEZF1-AS1 may play a carcinogenic role in the occurrence and development of CRC by regulating the FEZF1-AS1/OTX1/EMT and FEZF1-AS1/miR-30a-5p/NT5E pathways. We suggest that FEZF1-AS1 is a promising therapeutic target for the treatment of CRC.
ACKNOWLEDGMENTS
Funding information: National Natural Science Foundation of China, Grant No: 81772550; Health and Family Planning Commission of Hebei Province, Grant No: 220180584; Key project of Hebei Province Health and Family Planning Commission, Grant No: G201735. J.L. carried out the experiments and interpreted the manuscript. L.M.Z. and G.Y.W. designed the experiments. M.L, X.H.H., and G.B. collected and analyzed the data. J.L. and J.C. wrote the manuscript, and all the authors reviewed the final manuscript.
Footnotes
The authors declare no conflicts of interest.
REFERENCES
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–32. [DOI] [PubMed] [Google Scholar]
- 3. Dulal S, Keku TO. Gut microbiome and colorectal adenomas. Cancer J. 2014;20(3):225–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tsai MC, Spitale RC, Chang HY. Long intergenic noncoding RNAs: New links in cancer progression. Cancer Res. 2011;71(1):3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. [DOI] [PubMed] [Google Scholar]
- 6. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. 2015;21(11):1253–61. [DOI] [PubMed] [Google Scholar]
- 7. Lin C, Yang L. Long Noncoding RNA in cancer: Wiring signaling circuitry. Trends Cell Biol. 2018;28(4):287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 2010;464(7291):1071–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Prensner JR, Iyer MK, Sahu A, Asangani IA, Cao Q, Patel L, Vergara IA, Davicioni E, Erho N, Ghadessi M, Jenkins RB, Triche TJ, Malik R, Bedenis R, McGregor N, Ma T, Chen W, Han S, Jing X, Cao X, Wang X, Chandler B, Yan W, Siddiqui J, Kunju LP, Dhanasekaran SM, Pienta KJ, Feng FY, Chinnaiyan AM. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45(11):1392–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sun TT, He J, Liang Q, Ren LL, Yan TT, Yu TC, Tang JY, Bao YJ, Hu Y, Lin Y, Sun D, Chen YX, Hong J, Chen H, Zou W, Fang JY. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6(7):784–801. [DOI] [PubMed] [Google Scholar]
- 11. Yuan SX, Wang J, Yang F, Tao QF, Zhang J, Wang LL, Yang Y, Liu H, Wang ZG, Xu QG, Fan J, Liu L, Sun SH, Zhou WP. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology 2016;63(2):499–511. [DOI] [PubMed] [Google Scholar]
- 12. Zhao L, Han T, Li Y, Sun J, Zhang S, Liu Y, Shan B, Zheng D, Shi J. The lncRNA SNHG5/miR-32 axis regulates gastric cancer cell proliferation and migration by targeting KLF4. FASEB J. 2017;31(3):893–903. [DOI] [PubMed] [Google Scholar]
- 13. Liu YW, Xia R, Lu K, Xie M, Yang F, Sun M, De W, Wang C, Ji G. LincRNA FEZF1-AS1 represses p21 expression to promote gastric cancer proliferation through LSD1-Mediated H3K4me2 demethylation. Mol Cancer 2017;16(1):39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ou ZL, Zhang M, Ji LD, Luo Z. Long noncoding RNA FEZF1-AS1 predicts poor prognosis and modulates pancreatic cancer cell proliferation and invasion through miR-142/HIF-1alpha and miR-133a/EGFR upon hypoxia/normoxia. J Cell Physiol. 2019;1–13. [DOI] [PubMed] [Google Scholar]
- 15. Wang YD, Sun XJ, Yin JJ, Yin M, Wang W, Nie ZQ, Xu J. Long non-coding RNA FEZF1-AS1 promotes cell invasion and epithelial–mesenchymal transition through JAK2/STAT3 signaling pathway in human hepatocellular carcinoma. Biomed Pharmacother. 2018;106:134–41. [DOI] [PubMed] [Google Scholar]
- 16. Li M, Zhao LM, Li SL, Li J, Gao B, Wang FF, Wang SP, Hu XH, Cao J, Wang GY. Differentially expressed lncRNAs and mRNAs identified by NGS analysis in colorectal cancer patients. Cancer Med. 2018;7(9):4650–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu K, Cai XY, Li Q, Yang ZB, Xiong W, Shen T, Wang WY, Li YF. OTX1 promotes colorectal cancer progression through epithelial–mesenchymal transition. Biochem Biophys Res Commun. 2014;444(1):1–5. [DOI] [PubMed] [Google Scholar]
- 18. Bian Z, Zhang J, Li M, Feng Y, Wang X, Zhang J, Yao S, Jin G, Du J, Han W, Yin Y, Huang S, Fei B, Zou J, Huang Z. LncRNA-FEZF1-AS1 promotes tumor proliferation and metastasis in colorectal cancer by regulating PKM2 signaling. Clin Cancer Res. 2018;24(19):4808–19. [DOI] [PubMed] [Google Scholar]
- 19. Wu R, Chen Y, Li F, Li W, Zhou H, Yang Y, Pei Z. Effects of CD73 on human colorectal cancer cell growth in vivo and in vitro. Oncol Rep. 2016;35(3):1750–6. [DOI] [PubMed] [Google Scholar]
- 20. Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007;447(7146):799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ma Y, Yang Y, Wang F, Moyer MP, Wei Q, Zhang P, Yang Z, Liu W, Zhang H, Chen N, Wang H, Wang H, Qin H. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/beta-catenin signalling pathway via suppression of activator protein 2alpha. Gut 2016;65(9):1494–504. [DOI] [PubMed] [Google Scholar]
- 22. Huang JZ, Chen M, Chen, Gao XC, Zhu S, Huang H, Hu M, Zhu H, Yan GR. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell 2017;68(1):171–184.e176. [DOI] [PubMed] [Google Scholar]
- 23. Ozawa T, Matsuyama T, Toiyama Y, Takahashi N, Ishikawa T, Uetake H, Yamada Y, Kusunoki M, Calin G, Goel A. CCAT1 and CCAT2 long noncoding RNAs, located within the 8q.24.21 ‘gene desert’, serve as important prognostic biomarkers in colorectal cancer. Ann Oncol. 2017;28(8):1882–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen N, Guo D, Xu Q, Yang M, Wang D, Peng M, Ding Y, Wang S, Zhou J. Long non-coding RNA FEZF1-AS1 facilitates cell proliferation and migration in colorectal carcinoma. Oncotarget 2016;7(10):11271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jin S, Chen S, Ma Y, Yang B, Liu Y. LincRNA FEZF1-AS1 contributes to the proliferation of LAD cells by silencing p57 expression. Oncotarget 2017;8(61):103004–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wu X, Zhang P, Zhu H, Li S, Chen X, Shi L. Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of gastric cancer and promotes tumorigenesis via activation of Wnt signaling pathway. Biomed Pharmacother. 2017;96:1103–8. [DOI] [PubMed] [Google Scholar]
- 27. Ye H, Zhou Q, Zheng S, Li G, Lin Q, Ye L, Wang Y, Wei L, Zhao X, Li W, Fu Z, Liu Y, Li Z, Chen R. FEZF1-AS1/miR-107/ZNF312B axis facilitates progression and Warburg effect in pancreatic ductal adenocarcinoma. Cell Death Dis. 2018;9(2):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Miao Q. OTX1 contributes to hepatocellular carcinoma progression by regulation of ERK/MAPK pathway. J Korean Med Sci. 2016;31(8):1215–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qin SC, Zhao Z, Sheng JX, Xu XH, Yao J, Lu JJ, Chen B, Zhao GD, Wang XY, Yang YD. Dowregulation of OTX1 attenuates gastric cancer cell proliferation, migration and invasion. Oncol Rep. 2018;40(4):1907–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Terrinoni A, Pagani IS, Zucchi I, Chiaravalli AM, Serra V, Rovera F, Sirchia S, Dionigi G, Miozzo M, Frattini A, Ferrari A, Capella C, Pasquali F, Curto FL, Albertini A, Melino G, Porta G. OTX1 expression in breast cancer is regulated by p53. Oncogene 2011;30(27):3096–103. [DOI] [PubMed] [Google Scholar]
- 31. Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166(1):21–45. [DOI] [PubMed] [Google Scholar]
- 32. Kumar MS, Armenteros-Monterroso E, East P, Chakravorty P, Matthews N, Winslow MM, Downward J. HMGA2 functions as a competing endogenous RNA to promote lung cancer progression. Nature 2014;505(7482):212–7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33. Li M, Sun X, Cai H, Sun Y, Plath M, Li C, Lan X, Lei C, Lin F, Bai Y, Chen H. Long non-coding RNA ADNCR suppresses adipogenic differentiation by targeting miR-204. Biochim Biophys Acta 2016;1859(7):871–82. [DOI] [PubMed] [Google Scholar]
- 34. Maldotti M, Incarnato D, Neri F, Krepelova A, Rapelli S, Anselmi F, Parlato C, Basile G, Dettori D, Calogero R, Oliviero S. The long intergenic non-coding RNA CCR492 functions as a let-7 competitive endogenous RNA to regulate c-Myc expression. Biochim Biophys Acta 2016;1859(10):1322–32. [DOI] [PubMed] [Google Scholar]
- 35. Liu M, Huang F, Zhang D, Ju J, Wu XB, Wang Y, Wang Y, Wu Y, Nie M, Li Z, Ma C, Chen X, Zhou JY, Tan R, Yang BL, Zen K, Zhang CY, Chen YG, Zhao Q. Heterochromatin protein HP1gamma promotes colorectal cancer progression and is regulated by miR-30a. Cancer Res. 2015;75(21):4593–604. [DOI] [PubMed] [Google Scholar]
- 36. Wei W, Yang Y, Cai J, Cui K, Li RX, Wang H, Shang X, Wei D. MiR-30a-5p suppresses tumor metastasis of human colorectal cancer by targeting ITGB3. Cell Physiol Biochem. 2016;39(3):1165–76. [DOI] [PubMed] [Google Scholar]
- 37. Zhu J, Zeng Y, Li W, Qin H, Lei Z, Shen D, Gu D, Huang JA, Liu Z. CD73/NT5E is a target of miR-30a-5p and plays an important role in the pathogenesis of non-small cell lung cancer. Mol Cancer 2017;16(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]