Type 1 interferons (IFN-1) are critical host responses to viral infection. However, the contribution of IFN-1 responses to control of viruses in specific cell and tissue types is not fully defined.
KEYWORDS: dissemination, interferon, mouse, pathogenesis, reovirus
ABSTRACT
Mammalian orthoreovirus (reovirus) spreads from the site of infection to every organ system in the body via the blood. However, mechanisms that underlie reovirus hematogenous spread remain undefined. Nonstructural protein σ1s is a critical determinant of reovirus bloodstream dissemination that is required for efficient viral replication in many types of cultured cells. Here, we used the specificity of the σ1s protein for promoting hematogenous spread as a platform to uncover a role for lymphatic type 1 interferon (IFN-1) responses in limiting reovirus systemic dissemination. We found that replication of a σ1s-deficient reovirus was restored to wild-type levels in cells with defective interferon-α receptor (IFNAR1) signaling. Reovirus spreads systemically following oral inoculation of neonatal mice, whereas the σ1s-null virus remains localized to the intestine. We found that σ1s enables reovirus spread in the presence of a functional IFN-1 response, as the σ1s-deficient reovirus disseminated comparably to wild-type virus in IFNAR1−/− mice. Lymphatics are hypothesized to mediate reovirus spread from the intestine to the bloodstream. IFNAR1 deletion from cells expressing lymphatic vessel endothelium receptor 1 (LYVE-1), a marker for lymphatic endothelial cells, enabled the σ1s-deficient reovirus to disseminate systemically. Together, our findings indicate that IFN-1 responses in lymphatics limit reovirus dissemination. Our data further suggest that the lymphatics are an important conduit for reovirus hematogenous spread.
IMPORTANCE Type 1 interferons (IFN-1) are critical host responses to viral infection. However, the contribution of IFN-1 responses to control of viruses in specific cell and tissue types is not fully defined. Here, we identify IFN-1 responses in lymphatics as important for limiting reovirus dissemination. We found that nonstructural protein σ1s enhances reovirus resistance to IFN-1 responses, as a reovirus mutant lacking σ1s was more sensitive to IFN-1 than wild-type virus. In neonatal mice, σ1s is required for reovirus systemic spread. We used tissue-specific IFNAR1 deletion in combination with the IFN-1-sensitive σ1s-null reovirus as a tool to test how IFN-1 responses in lymphatics affect reovirus systemic spread. Deletion of IFNAR1 in lymphatic cells using Cre-lox technology enabled dissemination of the IFN-1-sensitive σ1s-deficient reovirus. Together, our results indicate that IFN-1 responses in lymphatics are critical for controlling reovirus systemic spread.
INTRODUCTION
Systemic dissemination is a fundamental step in viral pathogenesis. To spread within the host, viruses need to replicate in multiple cell and tissue types. Viruses also must overcome a variety of physical and physiological barriers, including host antiviral defenses. Mammalian orthoreovirus (reovirus) is a member of the Reoviridae family of nonenveloped, double-stranded RNA (dsRNA) viruses that infects its hosts via respiratory or enteric routes. Following replication at the portal of entry, reovirus traffics to secondary organs and tissues, including the heart and central nervous system (CNS) (1, 2). Reoviruses primarily disseminate via the blood, although serotype 3 (T3) reoviruses can also spread by neural routes (3). In the intestine, reovirus infects intestinal epithelial cells (IECs) and Peyer’s patch (PP) cells (4–6) and is hypothesized to traffic through the mesenteric lymph node (MLN) to the blood via the lymphatics (7). However, the functional route of reovirus systemic spread is not defined.
Reovirus dissemination is influenced by a combination of host and viral factors (8). A key host determinant of reovirus spread is junctional adhesion molecule A (JAM-A), a cell surface receptor for reovirus (9, 10). JAM-A is a tight junction protein that promotes polarization and barrier formation by epithelial and endothelial cells and also is expressed on monocytes, lymphocytes, dendritic cells, and platelets, where it aids in cell migration and extravasation (11–14). JAM-A is dispensable for reovirus replication in the intestine but required for hematogenous spread (10). JAM-A on endothelial cells is required for establishment of viremia as well as egress of reovirus from the bloodstream into organs (15).
Reovirus nonstructural protein σ1s is also required for reovirus systemic spread (16). Like JAM-A, σ1s is dispensable for reovirus replication in the intestine but is essential for spread through the blood (16, 17). The σ1s protein is not needed for reovirus to traffic from the PP to the MLN (16). However, σ1s is required for reovirus replication in the MLN, which is hypothesized to facilitate viral spread through intestinal lymphatics to the bloodstream for the establishment of viremia and systemic dissemination (17). In culture, σ1s enhances reovirus replication in numerous cell lines, including simian virus 40 (SV40) immortalized endothelial cells (SVECs) and murine embryonic fibroblasts (MEFs) (16–19). In these cell lines, σ1s functions as a replication accessory factor that promotes reovirus protein synthesis (19). Therefore, σ1s may promote efficient viral replication in cells that are required for reovirus dissemination.
Type 1 interferons (IFN-1) are critical for host control of viral infections (20). IFN-1 (IFN-α/β) is produced in response to viruses and signals in an autocrine or paracrine manner to induce expression of hundreds of IFN-stimulated genes (ISGs) that function to limit viral replication (20–22). Adult mice are normally refractory to reovirus disease (23, 24). However, reovirus infection is lethal in adult mice that lack interferon-α receptor subunit 1 (IFNAR1) and cannot respond to IFN-1 (25). In adult IFNAR1−/− mice, IFNAR1 expression on hematopoietic cells is required for protection from reovirus (25). Neonatal IFNAR1−/− mice also succumb more rapidly to reovirus and have higher viral loads than wild-type (WT) mice (24, 26). Although IFN-1 is an important host response against reoviruses in vivo, the cell types responsible for IFN-1-mediated protection against reoviruses are not defined.
Here, we used tissue-specific IFNAR1 deletion in combination with the IFN-1-sensitive reovirus as a tool to identify a role for lymphatics in reovirus dissemination. We found that σ1s is a viral determinant of reovirus resistance to IFN-1 responses in cultured cells and in vivo, as σ1s-deficient reovirus disseminates efficiently in IFNAR1−/− mice. Using Cre-lox technology, we found that the IFN-1-sensitive σ1s-deficient reovirus disseminated in mice with lymphatic endothelial cell-specific deletion of IFNAR1. Together, our results indicate that IFN-1 responses in lymphatics are a critical barrier that reovirus must overcome to spread systemically.
RESULTS
σ1s facilitates reovirus replication in the presence of IFN-1 responses.
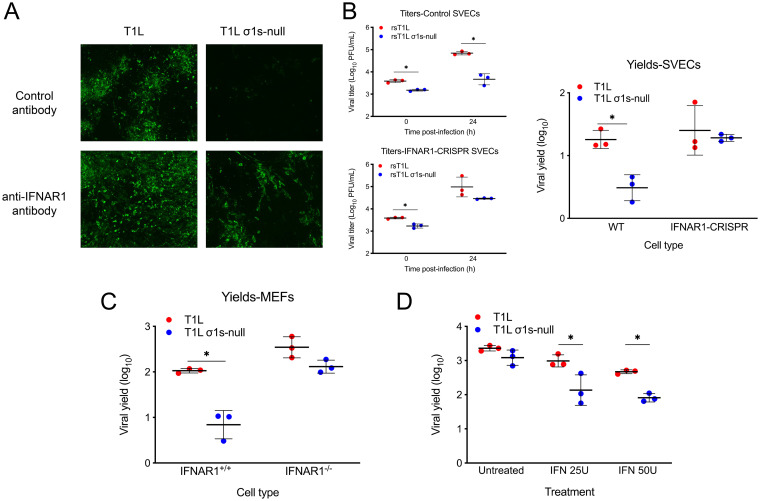
Previous work indicates that σ1s does not affect the induction of IFN-1 responses to reovirus (19). To further explore the relationship between σ1s and IFN-1, we tested whether σ1s promotes reovirus replication in cells when IFN-1 signaling was blocked. Murine SV40 immortalized endothelial cells (SVEC4-10; SVECs), a mouse lymphatic endothelial cell line, were infected with the T1L or T1L σ1s-null strain in the presence of isotype control or anti-IFNAR1 antibodies, and viral spread in culture was assessed over a 7-day period (Fig. 1A). T1L produced large foci and spread throughout the culture in the presence of the control antibody. In contrast, the T1L σ1s-null strain was limited to individual cells with few apparent multicellular foci. Treatment with anti-IFNAR1 antibodies enhanced spread of the T1L and T1L σ1s-null strains. To quantitatively assess the effect of IFN-1 responses on viral replication, we measured replication in wild-type SVECs and SVECs with IFNAR1 deleted using CRISPR-Cas9 editing (IFNAR1-CRISPR SVECs). Consistent with our previous work (19), the T1L strain produced approximately 10-fold more virus than the T1L σ1s-null strain in wild-type SVECs. In contrast, T1L and T1L σ1s-null strains replicated to equivalent levels in IFNAR1-CRISPR SVECs (Fig. 1B). Similarly, T1L replicated to significantly higher levels than the T1L σ1s-null strain in IFNAR1+/+ MEFs, but T1L and T1L σ1s-null strains produced comparable yields in IFNAR1−/− MEFs (Fig. 1C). These results indicate that σ1s facilitates efficient reovirus replication in the presence of IFN-1 responses.
FIG 1.
The σ1s protein facilitates reovirus replication in the presence of IFN-1 responses. (A) SVECs were infected with the T1L or T1L σ1s-null strain at an MOI of 10 PFU/cell and treated with control or anti-IFNAR1 antibodies. At 7 days, infected cells were identified via indirect immunofluorescence using reovirus polyclonal antiserum. (B) Wild-type or IFNAR1-CRISPR SVECs were infected with the T1L or T1L σ1s-null strain at an MOI of 1 PFU/cell. Viral titers were determined at 0 and 24 h. Results are presented as (left) mean viral titers or (right) mean viral yields from three independent experiments. (C) Wild-type (IFNAR1+/+) or IFNAR1−/− MEFs were infected with the T1L or T1L σ1s-null strain at an MOI of 1 PFU/cell. Viral titers were determined at 0 and 24 h, and results are presented as the mean viral yield from three independent experiments. (D) L929 cells were left untreated or treated with 25 or 50 units of recombinant IFN-α/β for 6 h prior to infection with the T1L or T1L σ1s-null strain at an MOI of 1 PFU/cell. Viral titers were determined at 0 and 24 h, and results are presented as the mean viral yield. Error bars represent standard deviation (SD). *, P < 0.05 as determined by Student's t test.
To determine whether σ1s enhances reovirus sensitivity to IFN-1, replication of T1L and T1L σ1s-null strains was measured in L929 cells treated with recombinant IFN-β prior to infection (Fig. 1D) (27). Unlike SVECs or MEFs, σ1s is not required for reovirus replication in L929 cells and allows assessment of the relationship between σ1s and IFN-1 independent of σ1s effects on viral replication (19, 28). Consistent with previous work, T1L and T1L σ1s-null strains replicated equivalently in untreated L929 cells. However, T1L replication was modestly impaired by IFN-β, and the reduction in T1L σ1s-null strain yields were significantly more pronounced. Together, these findings indicate that σ1s enhances reovirus resistance to IFN-1.
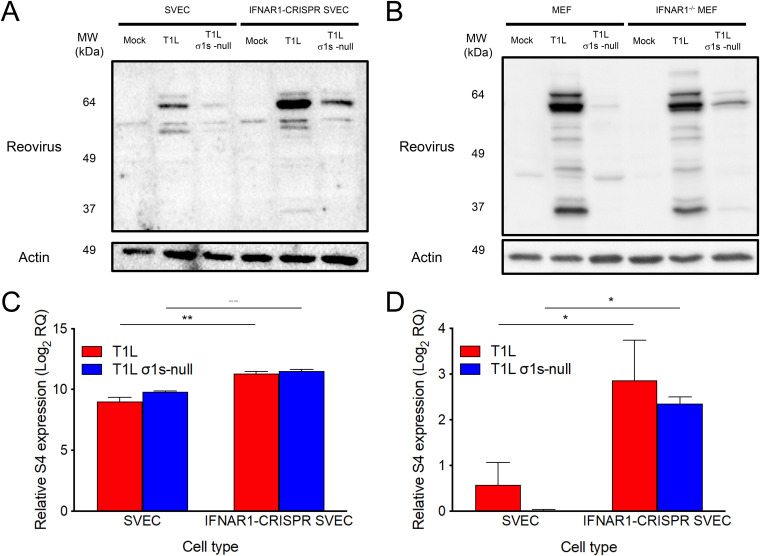
The σ1s protein enhances reovirus replication in SVECs and MEFs by promoting viral protein production (19). Inhibition of viral protein synthesis is a key mechanism by which IFN-1 responses combat viral infections (20). To determine whether σ1s allows reovirus to overcome IFN-1-mediated protein synthesis inhibition, viral protein production was assessed in wild-type and IFNAR1-CRISPR SVECs (Fig. 2A) and IFNAR1+/+ and IFNAR1−/− MEFs (Fig. 2B). Consistent with previous results (19), T1L produced more viral protein than the T1L σ1s-null strain in wild-type SVECs and MEFs. Although both viruses produced more protein in IFNAR1-CRISPR SVECs than in wild-type cells, protein expression by the T1L σ1s-null strain remained substantially lower than that of T1L in both cell types. No difference in T1L protein levels was observed between wild-type and IFNAR1−/− MEFs. However, the T1L σ1s-null strain produced more protein in IFNAR1−/− MEFs than wild-type MEFs. These data indicate that σ1s does not directly counteract the inhibition of reovirus protein synthesis caused by IFN-1.
FIG 2.
The σ1s protein does not counteract IFN-1-mediated inhibition of reovirus protein synthesis. Wild-type or IFNAR1-CRISPR SVECs (A) or wild-type (IFNAR1+/+) or IFNAR1−/− MEFs (B) were infected with the T1L or T1L σ1s-null strain at an MOI of 10 PFU/cell. At 18 h, whole-cell lysates were collected and separated by SDS-PAGE. Reovirus proteins and β-actin were detected by Western blot. (C and D) WT or IFNAR1-CRISPR SVECs were infected with the T1L or T1L σ1s-null strain at an MOI of 10 PFU/cell. At 0 or 18 h, total RNA was collected and relative expression of mRNA (C) or negative-sense RNA (D) was determined compared to 0 h. The RQ of positive- or negative-sense RNA was determined with reference to the quantity at the 0 h. The RQ of reovirus mRNA was determined by subtracting the RQ of negative-sense viral RNA (representing genomic RNA production) from the RQ of positive-sense RNA. Data are presented as the mean log2 RQ from three independent experiments. Error bars indicate SD. *, P < 0.05; **, P < 0.005 as determined by Student's t test.
We next quantified viral RNA to determine the effect of IFN-1 responses on viral RNA synthesis by T1L and T1L σ1s-null strains. Consistent with previous results (19), T1L and T1L σ1s-null S4 mRNA levels (Fig. 2C) were comparable in wild-type SVECs (log2 9.0 and log2 9.8, respectively). In contrast, T1L and T1L σ1s-null strains produced more S4 mRNA in IFNAR1-CRISPR SVECs (log2 11.3 and log2 11.5, respectively). Negative-sense RNA was detected in wild-type SVECs infected with T1L (Fig. 2D), but the T1L σ1s-null strain produced little, if any, negative-sense RNA (Fig. 2D). Negative-sense RNA levels increased in IFNAR1-CRISPR SVECs for T1L and T1L σ1s-null strains. These data are consistent with previous findings that σ1s is dispensable for reovirus RNA synthesis (19). These results indicate that while IFN-1 responses limit reovirus RNA synthesis, σ1s does not specifically modulate antiviral responses that prevent viral RNA accumulation.
The σ1s protein facilitates reovirus dissemination in the face of IFN-1 responses in vivo.
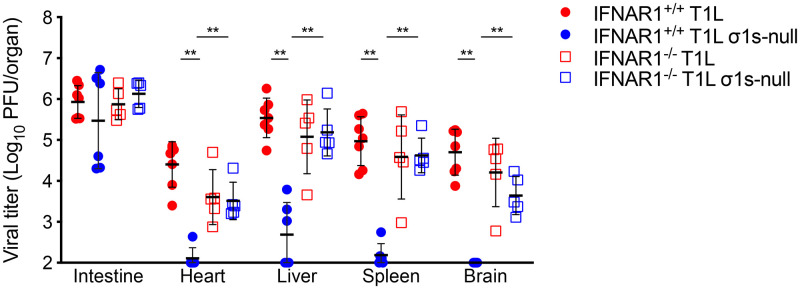
Our data and published studies (29) indicate that σ1s contributes to reovirus IFN-1 resistance in cultured cells. To determine whether σ1s is required for reovirus to overcome IFN-1 responses in vivo, we assessed T1L and T1L σ1s-null strains spread in wild-type and IFNAR1−/− mice (Fig. 3). T1L replicated in the intestine and spread systemically in wild-type mice. Consistent with previous findings (16), the T1L σ1s-null strain produced viral titers similar to those of T1L in the intestine of wild-type mice but T1L σ1s-null titers in target organs (brain, heart, liver, and spleen) were near or below the limit of detection. As in wild-type mice, T1L and T1L σ1s-null strains produced comparable intestinal titers in IFNAR1−/− mice. However, equivalent titers of T1L and T1L σ1s-null strains were recovered from peripheral sites in IFNAR1−/− mice. These data indicate that the IFN-1 response acts as a barrier to reovirus hematogenous dissemination. These findings further suggest that σ1s is required for reovirus to spread systemically in the presence of IFN-1 responses.
FIG 3.
The σ1s protein is required for reovirus dissemination in the presence of IFN-1 responses. Three- to four-day-old wild-type (IFNAR1+/+) or IFNAR1−/− neonatal mice were infected orally with 104 PFU of the T1L or T1L σ1s-null strain. At 4 days, the indicated organs were resected and homogenized, and viral titer was determined by plaque assay. Error bars represent SD. **, P < 0.005 as determined by Mann-Whitney test.
The σ1s protein is required for efficient reovirus replication in lymphatic endothelial cells.
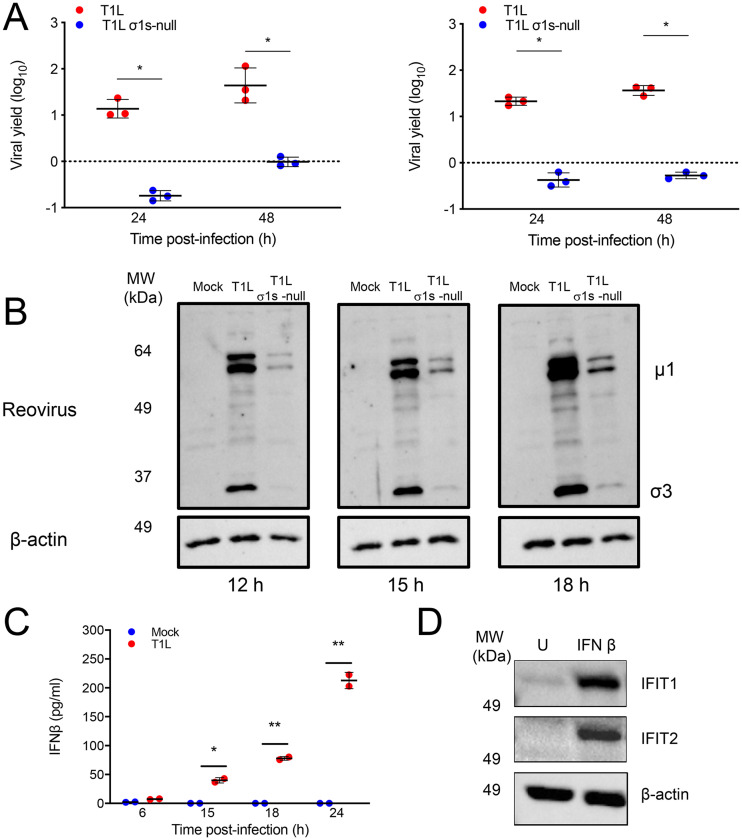
Reovirus is hypothesized to spread via the lymphatics, which are largely formed from lymphatic endothelial cells (LECs) (30). Previous work revealed that σ1s was required for efficient reovirus replication in SVECs, an immortalized lymphatic endothelial cell line (19). To determine whether σ1s is required for reovirus replication in primary LECs, we quantified T1L and T1L σ1s-null progeny yields produced by LECs derived from C57BL/6 mice (Fig. 4A). We found that T1L generated significantly higher progeny yields than the T1L σ1s-null strain in primary LECs at both multiplicity of infection (MOI) values tested. In cells where σ1s is required for reovirus replication, σ1s also mediates optimal viral protein production (19). In primary LECs, we observed that differential replication of wild-type and σ1s-deficient viruses correlated with differences in viral protein production, as T1L produced substantially more viral protein than the T1L σ1s-null strain (Fig. 4B). Together, these data indicate that σ1s promotes reovirus replication in primary LECs.
FIG 4.
The σ1s protein facilitates reovirus protein synthesis and replication in primary lymphatic endothelial cells. (A) Primary LECs from C57BL/6 mice were infected with the T1L or T1L σ1s-null strain at MOIs of 1 (left) or 10 (right) PFU/cell. At 0, 24, and 48 h, viral titers were determined by plaque assay. Results are presented as the mean viral yield from two independent experiments. Error bars represent SD. *, P < 0.05 as determined by Student's t test. (B) Primary LECs were mock infected or infected with the T1L or T1L σ1s-null strain at an MOI of 5 PFU/cell. At the indicated times, whole-cell lysates were collected and separated by SDS-PAGE. Reovirus proteins and β-actin were detected by Western blot. (C) Primary LECs were mock infected or infected with the T1L strain at an MOI of 100 PFU/cell. At the indicated times, IFN-β levels in the supernatants were determined by enzyme-linked immunosorbent assay (ELISA). (D) Primary LECs were treated with 200 U IFN-β or left untreated. At 6 h, whole-cell lysates were collected and separated by SDS-PAGE. IFIT1, IFIT2, and β-actin were detected by Western blot.
Lymphatics facilitate reovirus dissemination.
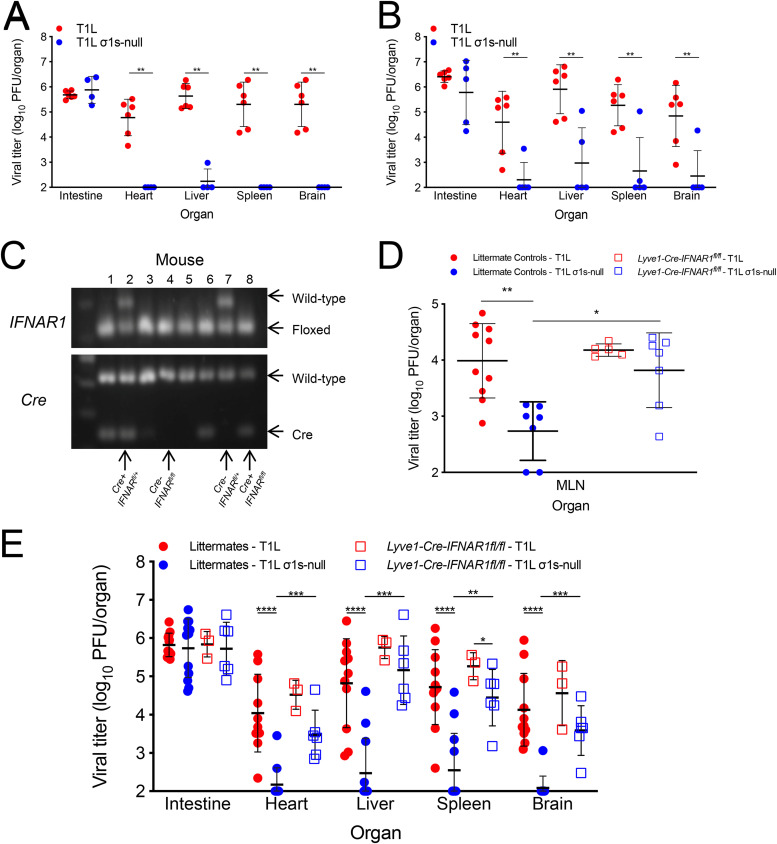
If lymphatics function as conduits for reovirus dissemination, we hypothesized that ablating IFN-1 responses specifically in LECs would enable dissemination of σ1s-deficient reovirus. To test this hypothesis, we used the IFN-1-sensitive σ1s-null reovirus in combination with lymphatic-specific deletion of IFNAR1. We first confirmed that primary LECs secrete IFN-β in response to reovirus infection (Fig. 4C) and produce ISGs following IFN-1 treatment (Fig. 4D). To generate lymphatic-specific IFNAR1 deletion mice, IFNAR1fl/fl mice were crossed with Lyve1-Cre mice (31, 32). Lymphatic vessel endothelium receptor 1 (LYVE-1) is a receptor for hyaluronan that promotes LEC proliferation (33, 34). LYVE-1 is a commonly used marker for the lymphatic endothelium but is also expressed on liver sinusoid, some tissue-resident macrophages, and a subset of hematopoietic stem cells (31, 35–37). T1L disseminated in the parental IFNAR1fl/fl (Fig. 5A) and Lyve1-Cre (Fig. 5B) strains, similar to results obtained with C57BL/6 mice (16). In contrast, the T1L σ1s-null strain did not spread efficiently in either parental mouse strain.
FIG 5.
IFN-1 responses in lymphatics limit reovirus dissemination. IFNAR1fl/fl (A) or Lyve1-Cre (B) mice were orally infected with 104 PFU T1L or T1L σ1s-null virus. At 4 days, the indicated organs were resected and homogenized, and viral titer was determined by plaque assay. (C) The genotype of transgenic mice was determined by performing PCR analysis of chromosomal DNA using primers specific for IFNAR1 and Cre. The floxed IFNAR1 alleles and Cre were differentiated based on migration of the PCR products in 1% agarose gel and stained with ethidium bromide (EtBr). (D and E) Littermate control (IFNAR1fl/fl, IFNAR1+/fl, Lyve1-Cre-IFNAR1+/fl) or Lyve1-Cre-IFNAR1fl/fl mice were orally infected with 104 PFU T1L or T1L σ1s-null virus. At 4 days, MLNs (D) or the indicated organs (E) were resected and homogenized, and viral titer was determined by plaque assay. Error bars represent SD. *, P < 0.05; **, P < 0.01; ***, P < 0.0005; ****, P < 0.0001 as determined by Mann-Whitney test.
The F1 progeny resulting from crossing IFNAR1fl/fl and Lyve1-Cre strains were bred to IFNAR1fl/fl mice. The resulting progeny (Lyve1-Cre-IFNAR1+/fl, IFNAR1+/fl, IFNAR1fl/fl littermate controls, and Lyve1-Cre-IFNAR1fl/fl LEC IFNAR1 deletions) were infected with the T1L or T1L σ1s-null strain, and at 4 days, viral tissue titers were determined. Mice were genotyped by PCR at the time of harvest to determine their Cre and IFNAR1 status (Fig. 5C). Following oral infection, we found that T1L titers were substantially higher on day 4 than the T1L σ1s-null strain in the MLNs of littermate control animals (Fig. 5D). In contrast, T1L and T1L σ1s-null strains produced comparable titers in MLNs from Lyve1-Cre-IFNAR1fl/fl mice. These data indicate that IFN-1 responses in the lymphatics impair spread of σ1s-deficient reovirus to the MLN.
We next assessed reovirus spread in mice with lymphatic IFNAR1 deletion. In littermate control mice, T1L and T1L σ1s-null strains produced comparable titers in the intestine, but only T1L had high titers in peripheral organs. T1L σ1s-null titers in organs from littermate control mice were near or below the level of detection (Fig. 5E). These data are consistent with observations in wild-type, IFNAR1fl/fl, and Lyve1-Cre mice that σ1s is required for efficient reovirus dissemination. In contrast, T1L and T1L σ1s-null strains produced largely comparable titers in all organs of Lyve1-Cre-IFNAR1fl/fl mice. Thus, deletion of IFNAR1 in lymphatics enables dissemination of σ1s-deficient reovirus. Together, these results indicate that lymphatic IFN-1 responses are critical for controlling reovirus dissemination.
DISCUSSION
Here, we identified a role for lymphatic IFN-1 responses in controlling hematogenous reovirus dissemination. In the intestine, reovirus is transcytosed by microfold cells (M-cells) in the gut-associated lymphoid tissue (GALT) where it infects the basolateral surface of IECs (8). Replication in IECs mediates reovirus release into the stool for spread to future hosts (8, 38, 39). To disseminate systemically, reovirus is taken up by cells in the Peyer’s patch (4, 10, 16) and hypothesized to traffic through the MLN via the lymphatics and then to the bloodstream (8). However, the operant route of reovirus dissemination is not known. Our data provide support for the hypothesis that the lymphatics function in hematogenous reovirus dissemination, as IFNAR1 deletion in LYVE-1-expressing cells allowed spread of the IFN-1-senstive σ1s-deficient reovirus.
Global and conditional deletion of JAM-A revealed that endothelial cells, but not hematopoietic cells, mediate establishment of reovirus viremia and egress from the blood into tissues (10, 15). It is possible that σ1s promotes reovirus replication in LECs that line lymphatic vessels and lymph nodes to provide a reservoir that seeds virus for trafficking through the lymphatics to the blood. Consistent with this hypothesis, we found that σ1s was required for reovirus protein expression and replication in primary LECs (Fig. 4). LYVE-1 is predominantly expressed on LECs but also on a small subset of fetal and adult hematopoietic stem cells, liver sinusoidal endothelial cells, and adult tissue-resident macrophages (31, 35–37). Conditional expression of JAM-A on hematopoietic cells is insufficient to restore reovirus hematogenous spread in JAM-A-deficient mice, indicating that hematopoietic cells do not mediate reovirus dissemination (15). Liver sinusoidal epithelial cells are not reported to harbor reovirus (40). In the liver, reovirus is taken up by Kupffer cells and is detected in hepatocytes (40). Treatment with silica dioxide or carrageenan to prevent macrophage uptake reduced reovirus levels in bile following intravenous inoculation (40). In contrast, carrageenan increased viral blood titers, indicating that macrophages restrict systemic spread when virus is administered intravenously (40). However, the role tissue-specific macrophages, including Kupfer cells, play in reovirus dissemination remains to be determined.
It is also possible that loss of IFN-1 signaling in LYVE-1-positive cells increases the permeability of the lymphatic endothelium, thereby allowing reovirus to escape the lymphatic vessels. IFN-1 controls LEC expansion in response to viral infection (41) and also modulates vascular endothelial barrier function, particularly at the blood-brain barrier (42, 43). Lack of IFN-1 signaling in the lymphatic endothelium could allow the σ1s-null virus to leak into the lymphatic vessels. Loss of IFN-1 signaling in the LECs also could alter the transport dynamics of the lymphatics. In the skin, IFN-1 signaling blocks fluid transport to the regional lymph node and limits poxvirus dissemination (44). If IFN-1 has similar effects on the dynamics of gut lymphatics, removing IFNAR1 from LECs could prevent the interruption of lymphatic flow intended to impede viral spread.
Why σ1s is dispensable for reovirus replication in the intestine remains an open question. One possibility is that reovirus replication in the intestine is largely controlled by interferon λ (IFN-λ) as opposed to IFN-1 (38, 39, 45). Like IFN-1, IFN-λ provokes ISG expression, but IFN-1 induces ISGs with more rapid kinetics and to a greater magnitude than IFN-λ (46). IFN-λ limits reovirus replication in the intestine, as mice lacking IFNLR1 or IFN-λ2/3 have elevated reovirus IEC infection and shedding (38, 39, 47). We found no difference in viral intestinal titers between wild-type and σ1s-deficient viruses in wild-type or IFNAR1-knockout mice. These data are consistent with IFN-λ as the primary means of controlling reovirus replication in the intestine. It is possible that σ1s is more important for resisting IFN-1 than IFN-λ responses due to the lower potency of IFN-λ compared to that of IFN-1. However, the relationship between σ1s and IFN-λ remains unexplored.
Like most viruses, reovirus activates cellular mechanisms that function to impair viral protein synthesis, including the dsRNA-dependent protein kinase (PKR) that phosphorylates α subunit of eukaryotic initiation factor 2 (eIF2α) to block translation (48–50) and the 2′-5′ oligoadenylate synthase (OAS)-RNase L system that degrades viral RNA (48). Reovirus must produce viral proteins in the face of host translational shutoff in order to replicate efficiently. It is also hypothesized that reovirus benefits from host shutoff, as viral replication is decreased in MEFs lacking PKR or expressing a constitutively active form of eIF2α (50). Reovirus uses multiple mechanisms to evade host translational shutoff, including outer capsid protein σ3 binding dsRNA to blunt PKR activation (51) and IFN-1 signaling (52). Nonstructural protein σNS also facilitates escape from host shutoff by mediating dissolution of stress granules (53). However, reovirus has other means to circumvent host translational arrest. It is possible that σ1s promotes reovirus protein expression by counteracting the function of one or more ISGs that block host translation (20, 54). Although σ1s is required for reovirus replication in the presence of IFN-1 responses, σ1s does not function as a classical IFN-1 antagonist (19), as IFN-1 secretion, IFNAR signaling, and ISG induction are comparable between wild-type and σ1s-deficient viruses (19). We observed that viral protein expression by the σ1s-deficient virus is only partially restored in the absence of IFNAR1 signaling. This result suggests that σ1s promotes reovirus protein expression via an IFN-1-independent mechanism.
The σ1s protein is required for systemic reovirus spread (16, 17). Here, we found that σ1s is important for reovirus resistance to IFN-1 in cell culture and in vivo. We used the IFN-1-sensitive σ1s-deficient reovirus in combination with tissue-specific deletion of IFNAR1 in lymphatic endothelial cells to identify a role for IFN-1 responses in lymphatics in controlling reovirus spread. Together, our findings provide new insight into mechanisms that control reovirus dissemination and further define how reovirus spreads from mucosal sites of infection to target organs and tissues.
MATERIALS AND METHODS
Cells and viruses.
Murine L929 fibroblasts were maintained in Joklik’s modified Eagle medium (JMEM; Sigma) supplemented with 5% heat-inactivated fetal bovine serum (FBS; Invitrogen), 2 mM l-glutamine (Invitrogen), 100 U/ml penicillin-100 μg/ml streptomycin (Invitrogen), and 250 ng/ml amphotericin B (Sigma). SV40 immortalized endothelial cells (SVECs; ATCC), C57BL/6 murine embryonic fibroblasts (MEFs), IFNAR1−/− MEFs, and human embryonic kidney 293 cells (HEK293T) were maintained in Dulbecco’s modified Eagle medium (DMEM; Invitrogen) supplemented to contain 10% heat-inactivated FBS and 2 mM l-glutamine. SVECs lacking the IFN-α/β receptor (IFNAR1-CRISPR) were maintained in the same medium as SVECs with 2 μg/ml puromycin (Gibco). Primary LECs (Cell Biologics) were maintained in endothelial cell medium (Cell Biologics) supplemented to contain 5% FBS, 2 mM l-glutamine, antibiotic-antimycotic solution (100 U/ml penicillin-100 U/ml streptomycin-50 ng/ml amphotericin B), and each of the following according to the manufacturer’s instructions: vascular endothelial growth factor (VEGF), heparin, endothelial cell growth supplement (ECGS), epidermal growth factor (EGF), and hydrocortisone.
Reoviruses were generated using plasmid-based reverse genetics as described (8, 16, 17, 55, 56). Purified reovirus stocks were obtained from second- or third-passage L929 cell lysates from twice-plaque-purified reovirus (57). Vertrel was used to extract reovirus particles, which were separated on a 1.2- to 1.4-g/cm3 CsCl density gradient and exhaustively dialyzed in virion storage buffer (150 mM NaCl, 15 mM MgCl2, 10 mM Tris-HCl, pH 7.8). The titers of the viral stocks were determined by plaque assay on L929 cells (58).
CRISPR-Cas9 deletion of IFNAR1.
IFNAR1 was deleted in SVECs using the CRISPR-Cas9 system (59). The plentiCRISPRV.2 (Addgene) was digested with BsmBI and ligated with guide RNA sequences specific for IFNAR1, (IFNAR1+) 5′-CACCGGCTCGCTGTCTGTGGCGCGG-3′ and (IFNAR1−) 5′-AAACCCGCGCCCACGACAGCGAGCC-3′. The cloned plasmids were transfected in to HEK293T cells in combination with pSPAX2 and pCMV-G plasmids using Lipofectamine 2000 (Invitrogen). Supernatants were collected at 24 and 48 h posttransfection, passed through 0.45-μm syringe filters, and applied to SVECs in 6-well plates (∼50% confluent). At 48 h posttransduction, puromycin (Invitrogen; 2 μg/ml) was added to the medium. Puromycin-selected SVECs were tested for IFNAR1 deletion by treatment with IFN-β (PBL) followed by reverse transcriptase quantitative PCR (RT-qPCR) to measure ISG expression.
Viral replication assays.
Monolayers of cells in 24-well plates (1 × 105 cells/well) were infected with the T1L or T1L σ1s-null strain at an MOI of 1 PFU/cell at 4°C for 1 h. Cells were washed twice with cold phosphate-buffered saline with calcium and magnesium (PBS+/+; Invitrogen), and fresh medium was added. Infected cells were freeze-thawed twice at the indicated times in the figure legends, and viral titers were determined by plaque assay on L929 cells (58). Viral yields were calculated using the following formula: log10 yieldtx = log10(PFU/ml)tx – log10(PFU/ml)t0, where tx is the time postinfection.
Immunoblotting.
Monolayers of cells in 6-well plates (1 × 106 cells/well) were mock infected or infected with reovirus or treated with recombinant IFN-β (PBL Assay Science) as indicated in the figure legends. Whole-cell lysates were collected in radioimmunoprecipitation assay (RIPA) buffer (20 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% sodium dodecyl sulfate, 1% desoxycholate, and 1/100 IGEPAL [NP-40]) at the indicated times. Total protein in each sample was quantified using the DC protein assay (Bio-Rad), and 10 μg protein was separated by 10% SDS-PAGE. Proteins were transferred to a nitrocellulose membrane and incubated in blocking buffer (5% milk in 1× Tris-buffered saline [TBS] with 0.05% Tween 20 [TBS-T]) for 1 h. Membranes were incubated in blocking buffer containing reovirus-specific rabbit polyclonal antiserum (1:2,000 dilution), IFIT1 antibody (Abcam 111821; 1:1,000 dilution), or IFIT2 antibody (Thermo Fisher PA3-834; 1:1,000 dilution) overnight at 4°C. Membranes were washed three times with TBS-T followed by incubation in blocking buffer containing horseradish peroxidase-conjugated goat anti-rabbit IgG (Jackson Immunolabs; 1:2,000 dilution) for 1 h with rocking. Following three TBS-T washes, proteins were detected using SuperSignal West chemiluminescent substrate (Thermo Fisher) and imaged using a ChemiDoc imaging system (Bio-Rad). Blots were stripped for reprobing by washing membranes three times with TBS followed by incubation in Restore Western blot stripping buffer (Thermo Scientific) for 15 min at room temperature (RT). Following three washes in TBS, membranes were blocked as described above, and β-actin was detected using mouse β-acting-specific monoclonal antibody (Sigma; 1:10,000 dilution) and peroxidase-conjugated goat anti-mouse IgG (Jackson Immunolabs; 1:2,000 dilution).
RT-qPCR.
Monolayers of cells in 6-well plates (1 × 106 cells/well) were infected with the T1L or T1L σ1s-null strain at an MOI of 10 PFU/cell. Total RNA was collected using the RNeasy Plus kit (Qiagen). Reovirus S4 RNA was quantified using the TaqMan fast virus one-step master mix (Applied Biosystems), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was detected as an endogenous control using the predeveloped TaqMan assay reagent kit for mouse GAPDH (Applied Biosystems) as described previously (19). The relative quantity (RQ) of reovirus positive- or negative-sense RNA was quantified using t = 0 h postinfection as the reference sample. The ΔΔCT was calculated for each sample using the following formula: ΔΔCT = (unknown CTtx – GAPDH CTtx) – (unknown CTt0 – GAPDH CTt0) where tx = time postinfection. The ΔΔCT was then used to calculate RQ using the following formula: RQ = 2−ΔΔCT.
Mouse experiments.
Animal husbandry, housing, and experiments were performed according to the guidelines of the Division of Laboratory Animal Medicine (DLAM) at University of Arkansas for Medical Sciences (UAMS). C57BL/6 (JAX stock number 000664), C57BL/6 IFNAR1−/− (JAX stock number 028288), C57BL/6 IFNAR1fl/fl (JAX stock number 028256), and C57BL/6 Lyve1-Cre (JAX stock number 012601) mice were obtained from Jackson Laboratory. IFNAR1fl/fl and Lyve1-Cre mice were crossed to obtain IFNAR1+/fl/Lyve1-Cre heterozygous mice. IFNAR1+/fl/LYVE1-Cre heterozygous mice were crossed to IFNAR1fl/fl mice for experiments, yielding litters of IFNAR1fl/fl, IFNAR1+/fl, IFNAR1+/fl/Lyve1-Cre, and IFNAR1fl/fl/Lyve1-Cre mice. Three- to four-day-old mice were infected orally with 104 PFU T1L or T1L σ1s-null strain diluted in PBS as previously described (10, 60). At 4 days postinfection, organs were resected and homogenized, and viral titer was determined by plaque assay. Infected mice were genotyped after experiments using the KAPA HotStart mouse genotyping kit (KAPA Biosystems) and primers for the floxed IFNAR1 allele and the Lyve1-Cre gene from the Jackson Laboratory website.
Statistics.
Differences in viral replication were determined by an unpaired Student's t test. Differences in viral titer from mouse experiments were determined by Mann-Whitney test. Statistical tests were performed using Prism software (GraphPad Software, Inc.). P values of <0.05 were considered significant.
ACKNOWLEDGMENTS
This research was supported by Public Health Award K22 AI90497 (K.W.B.) and R01 AI118801 (K.W.B.). Additional support was provided by the Center for Microbial Pathogenesis and Host Inflammatory Response (P20 GM103625).
The pMSCV-puro plasmid and EcoPak cells were provided by Craig Forrest (UAMS).
We thank Pranav Danthi and Craig Forrest for critical readings of the manuscript.
REFERENCES
- 1.Sabin AB. 1959. Reoviruses. Science 130:1387–1389. doi: 10.1126/science.130.3386.1387. [DOI] [PubMed] [Google Scholar]
- 2.Dryden KA, Wang G, Yeager M, Nibert ML, Coombs KM, Furlong DB, Fields BN, Baker TS. 1993. Early steps in reovirus infection are associated with dramatic changes in supramolecular structure and protein conformation: analysis of virions and subviral particles by cryoelectron microscopy and image reconstruction. J Cell Biol 122:1023–1041. doi: 10.1083/jcb.122.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tyler KL, McPhee DA, Fields BN. 1986. Distinct pathways of viral spread in the host determined by reovirus S1 gene segment. Science 233:770–774. doi: 10.1126/science.3016895. [DOI] [PubMed] [Google Scholar]
- 4.Wolf JL, Rubin DH, Finberg R, Kauffman RS, Sharpe AH, Trier JS, Fields BN. 1981. Intestinal M cells: a pathway for entry of reovirus into the host. Science 212:471–472. doi: 10.1126/science.6259737. [DOI] [PubMed] [Google Scholar]
- 5.Wolf JL, Kauffman RS, Finberg R, Dambrauskas R, Fields BN, Trier JS. 1983. Determinants of reovirus interaction with the intestinal M cells and absorptive cells of murine intestine. Gastroenterology 85:291–300. doi: 10.1016/0016-5085(83)90313-X. [DOI] [PubMed] [Google Scholar]
- 6.Fleeton MN, Contractor N, Leon F, Wetzel JD, Dermody TS, Kelsall BL. 2004. Peyer's patch dendritic cells process viral antigen from apoptotic epithelial cells in the intestine of reovirus-infected mice. J Exp Med 200:235–245. doi: 10.1084/jem.20041132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dermody TS, Parker JSL, Sherry B. 2013. Mammalian orthoreovirus, 6th ed. Lippincott Williams & Williams, Philadelphia, PA. [Google Scholar]
- 8.Boehme KW, Lai CM, Dermody TS. 2013. Mechanisms of reovirus bloodstream dissemination. Adv Virus Res 87:1–35. doi: 10.1016/B978-0-12-407698-3.00001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barton ES, Forrest JC, Connolly JL, Chappell JD, Liu Y, Schnell FJ, Nusrat A, Parkos CA, Dermody TS. 2001. Junction adhesion molecule is a receptor for reovirus. Cell 104:441–451. doi: 10.1016/S0092-8674(01)00231-8. [DOI] [PubMed] [Google Scholar]
- 10.Antar AA, Konopka JL, Campbell JA, Henry RA, Perdigoto AL, Carter BD, Pozzi A, Abel TW, Dermody TS. 2009. Junctional adhesion molecule-A is required for hematogenous dissemination of reovirus. Cell Host Microbe 5:59–71. doi: 10.1016/j.chom.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams LA, Martin-Padura I, Dejana E, Hogg N, Simmons DL. 1999. Identification and characterisation of human junctional adhesion molecule (JAM). Mol Immunol 36:1175–1188. doi: 10.1016/S0161-5890(99)00122-4. [DOI] [PubMed] [Google Scholar]
- 12.Ebnet K. 2017. Junctional adhesion molecules (JAMs): cell adhesion receptors with pleiotropic functions in cell physiology and development. Physiol Rev 97:1529–1554. doi: 10.1152/physrev.00004.2017. [DOI] [PubMed] [Google Scholar]
- 13.Liu Y, Nusrat A, Schnell FJ, Reaves TA, Walsh S, Pochet M, Parkos CA. 2000. Human junction adhesion molecule regulates tight junction resealing in epithelia. J Cell Sci 113:2363–2374. [DOI] [PubMed] [Google Scholar]
- 14.Martin-Padura I, Lostaglio S, Schneemann M, Williams L, Romano M, Fruscella P, Panzeri C, Stoppacciaro A, Ruco L, Villa A, Simmons D, Dejana E. 1998. Junctional adhesion molecule, a novel member of the immunoglobulin superfamily that distributes at intercellular junctions and modulates monocyte transmigration. J Cell Biol 142:117–127. doi: 10.1083/jcb.142.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai CM, Boehme KW, Pruijssers AJ, Parekh VV, Van Kaer L, Parkos CA, Dermody TS. 2015. Endothelial JAM-A promotes reovirus viremia and bloodstream dissemination. J Infect Dis 211:383–393. doi: 10.1093/infdis/jiu476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boehme KW, Guglielmi KM, Dermody TS. 2009. Reovirus nonstructural protein σ1s is required for establishment of viremia and systemic dissemination. Proc Natl Acad Sci U S A 106:19986–19991. doi: 10.1073/pnas.0907412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehme KW, Frierson JM, Konopka JL, Kobayashi T, Dermody TS. 2011. The reovirus σ1s protein is a determinant of hematogenous but not neural virus dissemination in mice. J Virol 85:11781–11790. doi: 10.1128/JVI.02289-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoyt CC, Richardson-Burns SM, Goody RJ, Robinson BA, Debiasi RL, Tyler KL. 2005. Nonstructural protein σ1s is a determinant of reovirus virulence and influences the kinetics and severity of apoptosis induction in the heart and central nervous system. J Virol 79:2743–2753. doi: 10.1128/JVI.79.5.2743-2753.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phillips MB, Stuart JD, Simon EJ, Boehme KW. 2018. Nonstructural protein σ1s is required for optimal reovirus protein expression. J Virol 92:e02259-17. doi: 10.1128/JVI.02259-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goubau D, Deddouche S, Reis e Sousa C. 2013. Cytosolic sensing of viruses. Immunity 38:855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schlee M. 2013. Master sensors of pathogenic RNA - RIG-I like receptors. Immunobiology 218:1322–1335. doi: 10.1016/j.imbio.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kauffman RS, Wolf JL, Finberg R, Trier JS, Fields BN. 1983. The σ1 protein determines the extent of spread of reovirus from the gastrointestinal tract of mice. Virology 124:403–410. doi: 10.1016/0042-6822(83)90356-2. [DOI] [PubMed] [Google Scholar]
- 24.Wu AG, Pruijssers AJ, Brown JJ, Stencel-Baerenwald JE, Sutherland DM, Iskarpatyoti JA, Dermody TS. 2018. Age-dependent susceptibility to reovirus encephalitis in mice is influenced by maturation of the type-I interferon response. Pediatr Res 83:1057–1066. doi: 10.1038/pr.2018.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson C, Wetzel JD, He J, Mikacenic C, Dermody TS, Kelsall BL. 2007. Type I interferons produced by hematopoietic cells protect mice against lethal infection by mammalian reovirus. J Exp Med 204:1349–1358. doi: 10.1084/jem.20061587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luethy LN, Erickson AK, Jesudhasan PR, Ikizler M, Dermody TS, Pfeiffer JK. 2016. Comparison of three neurotropic viruses reveals differences in viral dissemination to the central nervous system. Virology 487:1–10. doi: 10.1016/j.virol.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs BL, Ferguson RE. 1991. The Lang strain of reovirus serotype 1 and the Dearing strain of reovirus serotype 3 differ in their sensitivities to beta interferon. J Virol 65:5102–5104. doi: 10.1128/JVI.65.9.5102-5104.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodgers SE, Connolly JL, Chappell JD, Dermody TS. 1998. Reovirus growth in cell culture does not require the full complement of viral proteins: identification of a sigma1s-null mutant. J Virol 72:8597–8604. doi: 10.1128/JVI.72.11.8597-8604.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lanoie D, Cote S, Degeorges E, Lemay G. 2019. A single mutation in the mammalian orthoreovirus S1 gene is responsible for increased interferon sensitivity in a virus mutant selected in Vero cells. Virology 528:73–79. doi: 10.1016/j.virol.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 30.Randolph GJ, Ivanov S, Zinselmeyer BH, Scallan JP. 2017. The lymphatic system: integral roles in immunity. Annu Rev Immunol 35:31–52. doi: 10.1146/annurev-immunol-041015-055354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pham TH, Baluk P, Xu Y, Grigorova I, Bankovich AJ, Pappu R, Coughlin SR, McDonald DM, Schwab SR, Cyster JG. 2010. Lymphatic endothelial cell sphingosine kinase activity is required for lymphocyte egress and lymphatic patterning. J Exp Med 207:17–27. doi: 10.1084/jem.20091619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prigge JR, Hoyt TR, Dobrinen E, Capecchi MR, Schmidt EE, Meissner N. 2015. Type I IFNs act upon hematopoietic progenitors to protect and maintain hematopoiesis during Pneumocystis lung infection in mice. J Immunol 195:5347–5357. doi: 10.4049/jimmunol.1501553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu M, Du Y, Liu Y, He Y, Yang C, Wang W, Gao F. 2014. Low molecular weight hyaluronan induces lymphangiogenesis through LYVE-1-mediated signaling pathways. PLoS One 9:e92857. doi: 10.1371/journal.pone.0092857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerji S, Ni J, Wang SX, Clasper S, Su J, Tammi R, Jones M, Jackson DG. 1999. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J Cell Biol 144:789–801. doi: 10.1083/jcb.144.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mouta Carreira C, Nasser SM, di Tomaso E, Padera TP, Boucher Y, Tomarev SI, Jain RK. 2001. LYVE-1 is not restricted to the lymph vessels: expression in normal liver blood sinusoids and down-regulation in human liver cancer and cirrhosis. Cancer Res 61:8079–8084. [PubMed] [Google Scholar]
- 36.Lee LK, Ghorbanian Y, Wang W, Wang Y, Kim YJ, Weissman IL, Inlay MA, Mikkola HKA. 2016. LYVE1 marks the divergence of yolk sac definitive hemogenic endothelium from the primitive erythroid lineage. Cell Rep 17:2286–2298. doi: 10.1016/j.celrep.2016.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schledzewski K, Falkowski M, Moldenhauer G, Metharom P, Kzhyshkowska J, Ganss R, Demory A, Falkowska-Hansen B, Kurzen H, Ugurel S, Geginat G, Arnold B, Goerdt S. 2006. Lymphatic endothelium-specific hyaluronan receptor LYVE-1 is expressed by stabilin-1+, F4/80+, CD11b+ macrophages in malignant tumours and wound healing tissue in vivo and in bone marrow cultures in vitro: implications for the assessment of lymphangiogenesis. J Pathol 209:67–77. doi: 10.1002/path.1942. [DOI] [PubMed] [Google Scholar]
- 38.Baldridge MT, Lee S, Brown JJ, McAllister N, Urbanek K, Dermody TS, Nice TJ, Virgin HW. 2017. Expression of Ifnlr1 on intestinal epithelial cells is critical to the antiviral effects of interferon lambda against norovirus and reovirus. J Virol 91:e02079-16. doi: 10.1128/JVI.02079-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson ST, Kennedy EA, Brigleb PH, Taylor GM, Urbanek K, Bricker TL, Lee S, Shin H, Dermody TS, Boon ACM, Baldridge MT. 2019. Disruption of type III interferon genes Ifnl2 and Ifnl3 recapitulates loss of the type III IFN receptor in the mucosal antiviral response. J Virol 93:e01073-19. doi: 10.1128/JVI.01073-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rubin DH, Costello T, Witzleben CL, Greene MI. 1987. Transport of infectious reovirus into bile: class II major histocompatibility antigen-bearing cells determine reovirus transport. J Virol 61:3222–3226. doi: 10.1128/JVI.61.10.3222-3226.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lucas ED, Finlon JM, Burchill MA, McCarthy MK, Morrison TE, Colpitts TM, Tamburini BAJ. 2018. Type 1 IFN and PD-L1 coordinate lymphatic endothelial cell expansion and contraction during an inflammatory immune response. J Immunol 201:1735–1747. doi: 10.4049/jimmunol.1800271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minagar A, Long A, Ma T, Jackson TH, Kelley RE, Ostanin DV, Sasaki M, Warren AC, Jawahar A, Cappell B, Alexander JS. 2003. Interferon (IFN)-β1a and IFN-β1b block IFN-gamma-induced disintegration of endothelial junction integrity and barrier. Endothelium 10:299–307. doi: 10.1080/10623320390272299. [DOI] [PubMed] [Google Scholar]
- 43.Liu P, Woda M, Ennis FA, Libraty DH. 2009. Dengue virus infection differentially regulates endothelial barrier function over time through type I interferon effects. J Infect Dis 200:191–201. doi: 10.1086/599795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Loo CP, Nelson NA, Lane RS, Booth JL, Loprinzi Hardin SC, Thomas A, Slifka MK, Nolz JC, Lund AW. 2017. Lymphatic vessels balance viral dissemination and immune activation following cutaneous viral infection. Cell Rep 20:3176–3187. doi: 10.1016/j.celrep.2017.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ingle H, Peterson ST, Baldridge MT. 2018. Distinct effects of type I and III interferons on enteric viruses. Viruses 10:46. doi: 10.3390/v10010046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lazear HM, Schoggins JW, Diamond MS. 2019. Shared and distinct functions of type I and type III interferons. Immunity 50:907–923. doi: 10.1016/j.immuni.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahlakoiv T, Hernandez P, Gronke K, Diefenbach A, Staeheli P. 2015. Leukocyte-derived IFN-alpha/beta and epithelial IFN-lambda constitute a compartmentalized mucosal defense system that restricts enteric virus infections. PLoS Pathog 11:e1004782. doi: 10.1371/journal.ppat.1004782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith JA, Schmechel SC, Williams BR, Silverman RH, Schiff LA. 2005. Involvement of the interferon-regulated antiviral proteins PKR and RNase L in reovirus-induced shutoff of cellular translation. J Virol 79:2240–2250. doi: 10.1128/JVI.79.4.2240-2250.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qin Q, Hastings C, Miller CL. 2009. Mammalian orthoreovirus particles induce and are recruited into stress granules at early times postinfection. J Virol 83:11090–11101. doi: 10.1128/JVI.01239-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin Q, Carroll K, Hastings C, Miller CL. 2011. Mammalian orthoreovirus escape from host translational shutoff correlates with stress granule disruption and is independent of eIF2alpha phosphorylation and PKR. J Virol 85:8798–8810. doi: 10.1128/JVI.01831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yue Z, Shatkin AJ. 1997. Double-stranded RNA-dependent protein kinase (PKR) is regulated by reovirus structural proteins. Virology 234:364–371. doi: 10.1006/viro.1997.8664. [DOI] [PubMed] [Google Scholar]
- 52.Roebke KE, Guo Y, Parker JSL, Danthi P. 2020. Reovirus σ3 protein limits interferon expression and cell death induction. J Virol 94:e01485-20. doi: 10.1128/JVI.01485-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choudhury P, Bussiere L, Miller CL. 2017. Mammalian orthoreovirus factories modulate stress granule protein localization by interaction with G3BP1. J Virol 91:e01298-17. doi: 10.1128/JVI.01298-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sharpe AH, Fields BN. 1982. Reovirus inhibition of cellular RNA and protein synthesis: role of the S4 gene. Virology 122:381–391. doi: 10.1016/0042-6822(82)90237-9. [DOI] [PubMed] [Google Scholar]
- 55.Boehme KW, Ikizler M, Kobayashi T, Dermody TS. 2011. Reverse genetics for mammalian reovirus. Methods 55:109–113. doi: 10.1016/j.ymeth.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kobayashi T, Antar AA, Boehme KW, Danthi P, Eby EA, Guglielmi KM, Holm GH, Johnson EM, Maginnis MS, Naik S, Skelton WB, Wetzel JD, Wilson GJ, Chappell JD, Dermody TS. 2007. A plasmid-based reverse genetics system for animal double-stranded RNA viruses. Cell Host Microbe 1:147–157. doi: 10.1016/j.chom.2007.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Furlong DB, Nibert ML, Fields BN. 1988. Sigma 1 protein of mammalian reoviruses extends from the surfaces of viral particles. J Virol 62:246–256. doi: 10.1128/JVI.62.1.246-256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Virgin HW, IV, Bassel-Duby R, Fields BN, Tyler KL. 1988. Antibody protects against lethal infection with the neurally spreading reovirus type 3 (Dearing). J Virol 62:4594–4604. doi: 10.1128/JVI.62.12.4594-4604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanjana NE, Shalem O, Zhang F. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tyler KL, Bronson RT, Byers KB, Fields B. 1985. Molecular basis of viral neurotropism: experimental reovirus infection. Neurology 35:88–92. doi: 10.1212/wnl.35.1.88. [DOI] [PubMed] [Google Scholar]