Abstract
Purpose
Osteonecrosis of the femoral head is a common disease of the hip that leads to severe pain or joint disability. We aimed to identify potential differentially expressed miRNAs and mRNAs in osteonecrosis of the femoral head.
Methods
The data of miRNA and mRNA were firstly downloaded from the database. Secondly, the regulatory network of miRNAs–mRNAs was constructed, followed by function annotation of mRNAs. Thirdly, an in vitro experiment was applied to validate the expression of miRNAs and targeted mRNAs. Finally, GSE123568 dataset was used for electronic validation and diagnostic analysis of targeted mRNAs.
Results
Several regulatory interaction pairs between miRNA and mRNAs were identified, such as hsa-miR-378c-WNT3A/DACT1/CSF1, hsa-let-7a-5p-RCAN2/IL9R, hsa-miR-28-5p-RELA, hsa-miR-3200-5p-RELN, and hsa-miR-532-5p-CLDN18/CLDN10. Interestingly, CLDN10, CLDN18, CSF1, DACT1, IL9R, RCAN2, RELN, and WNT3A had the diagnostic value for osteonecrosis of the femoral head. Wnt signaling pathway (involved WNT3A), chemokine signaling pathway (involved RELA), focal adhesion and ECM-receptor interaction (involved RELN), cell adhesion molecules (CAMs) (involved CLDN18 and CLDN10), cytokine–cytokine receptor interaction, and hematopoietic cell lineage (involved CSF1 and IL9R) were identified.
Conclusion
The identified differentially expressed miRNAs and mRNAs may be involved in the pathology of osteonecrosis of the femoral head.
Keywords: osteonecrosis of the femoral head, miRNAs, mRNAs, signaling pathway
Introduction
Osteonecrosis of the femoral head is a disabling and progressive chronic disease, which leads to femoral head collapse and further total hip arthroplasty.1,2 It is estimated that the age range of about 75% of patients is from 30–60 years old.3 Pain is one of the common clinical symptoms of osteonecrosis of the femoral head.4 However, most patients with a lesion less than 30% of the femoral head are initially asymptomatic.5 Malizos6,7 and Zalavras and Lieberman found that osteocytes death and bone marrow cells was the main characteristic of the early stages of osteonecrosis of the femoral head. In the next moment, the repair reaction of necrotic bone is initiated. During this process, the imbalance between bone resorption and bone reformation leads to structural damage of the femoral head, among which there is a significant degeneration and cracking of the hip articular cartilage, which accelerates the development of osteonecrosis of the femoral head.8,9
The pathogenic mechanism of osteonecrosis of the femoral head is complex. Kerachian et al10 found that local microvascular thrombosis resulted in decreasing blood flow in the femoral head. It was pointed out that fibroblast growth factor 2 (FGF2), insulin-like growth factor 1 (IGF1), SRY-box transcription factor 9 (SoX9), and collagen type ii α1 affected the pathogenesis of osteonecrosis of the femoral head.11 In addition, several risks, such as the trauma, steroids, smoking, alcoholism, irradiation/chemotherapy, clotting disturbances, hyperlipidemia, hyperviscosity, autoimmune diseases, Legg-Calve-Perthes diseases, and genetic factors are possible causes of osteonecrosis of the femoral head.12–22
The incidence of osteonecrosis of the femoral head is on the rise, in spite of various research efforts and trials. Therefore, understanding the pathophysiology of the disease and its progression is urgently needed. Interestingly, miRNAs play roles in targeting mRNAs, and regulate diverse biological processes in bones, such as osteoblasts, osteoclasts differentiation, and bone programming.23–26 In this study, we performed the differentially expression analysis of miRNA and mRNA in osteonecrosis of the femoral head, which may provide a novel field in understanding the pathological mechanism of osteonecrosis of the femoral head.
Materials and Methods
Datasets
The miRNA and mRNA expression profile was downloaded from the Gene Expression Omnibus database (GEO) dataset. The keywords of ((“femur head” [MeSH Terms] OR femoral head [All Fields]) AND (“necrosis” [MeSH Terms] OR necrosis [All Fields])) AND “gse” [Filter] was used to retrieve related datasets. According to above screening criteria, one miRNA dataset (GSE89587, involving ten cases and seven normal controls) and one mRNA dataset (GSE74089, involving four cases and four normal controls) were finally selected.
Identification of Differentially Expressed miRNAs and mRNAs
Firstly, the raw data of miRNAs and mRNAs was preprocessed as follows: the probes corresponding to multiple miRNAs/mRNAs were removed; and the miRNAs/mRNAs corresponding to multiple probes were left with only the one with the highest average expression. In this study, the identification method of differentially expressed miRNAs and mRNAs was referred to previous studies.27,28 The screening criteria of differentially expressed miRNAs and mRNAs was, respectively, FDR<0.05, |log2FC|>3, and FDR<0.05, |log2FC|>1.
Correlation Analysis Between miRNAs and mRNAs
In this study, miRWalk (http://mirwalk.umm.uni-heidelberg.de/) was used to predict targeted mRNAs of miRNAs. The establishment of a miRNA-target regulatory network was visualized using Cytoscape software.
Functional Enrichment of mRNAs
In order to understand the biological function of the targeted differentially expressed mRNAs of differentially expressed miRNAs, we performed functional analysis via GeneCodis3 software. FDR<0.05 was set as the criterion for selecting significantly enriched functional terms.
In vitro Validation of miRNAs and Targeted mRNAs
In total, six patients with osteonecrosis of the femoral head and seven normal controls were enrolled. All patients had not taken corticosteroids or medications for nearly a month. In addition, patients older than 80 years or without incomplete clinical information were excluded. Normal controls were matched by gender and age of the case group. Healthy individuals with a history of osteonecrosis of the femoral head and suffering from bone metabolic disorders (such as osteoporosis) were excluded. Ethical approval was obtained from the ethics committee of Honghui Hospital Xian Jiao Tong University Health Science Center (No.201904006). Those included provided informed written consent. This study was carried out in accordance with the Declaration of Helsinki.
Total RNA of the blood samples was extracted and synthesized cDNA by FastQuant Reverse Transcriptase (TIANGEN). Then real-time PCR was performed in an ABI 7300 Real-time PCR system with SYBR® Green PCR Master Mix (Applied Biosystems). Has-U6 was used for the internal reference of miRNA. ACTB and GAPDH were used for the internal reference of mRNA.
Electronic Validation and Diagnostic Analysis of Targeted mRNAs in GSE123568 Dataset
The GSE123568 dataset (peripheral blood sample) involved 30 patients with osteonecrosis of the femoral head and 10 normal controls, and was used for electronic validation and ROC analysis of targeted mRNAs. The expression result of these mRNAs was shown by box plots.
Results
Expression Pattern of miRNA and mRNA
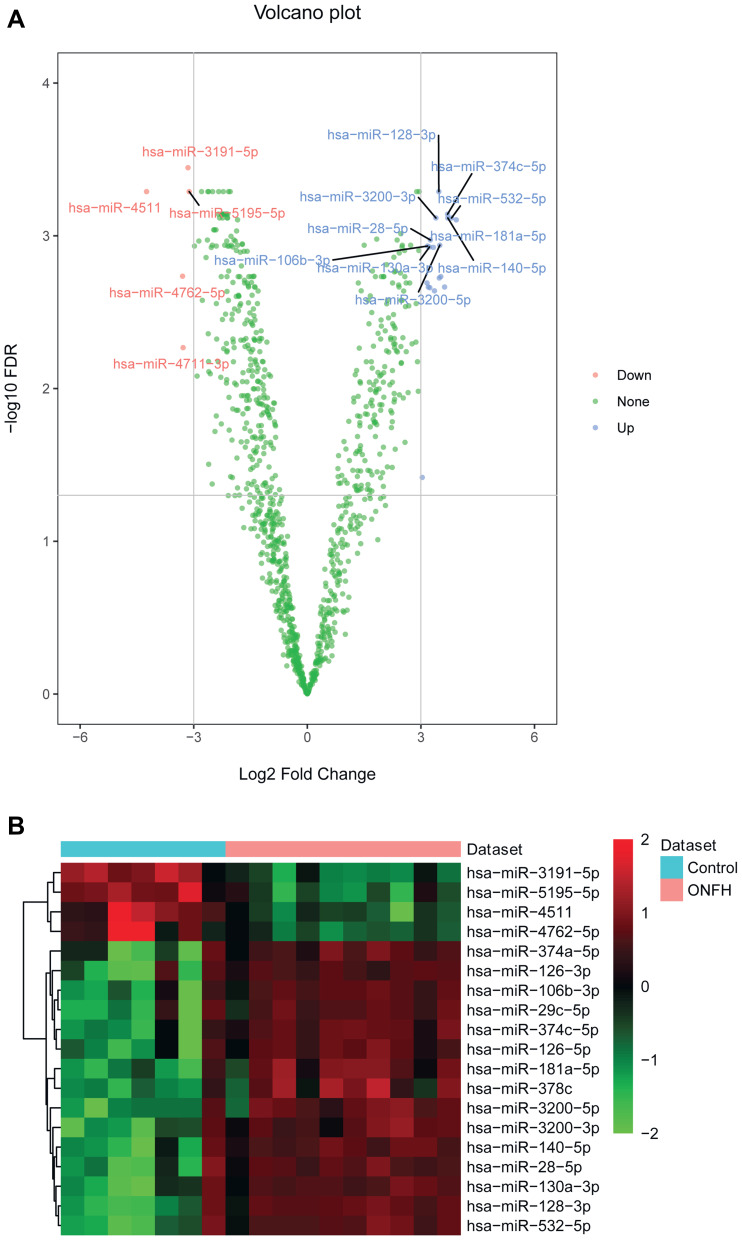
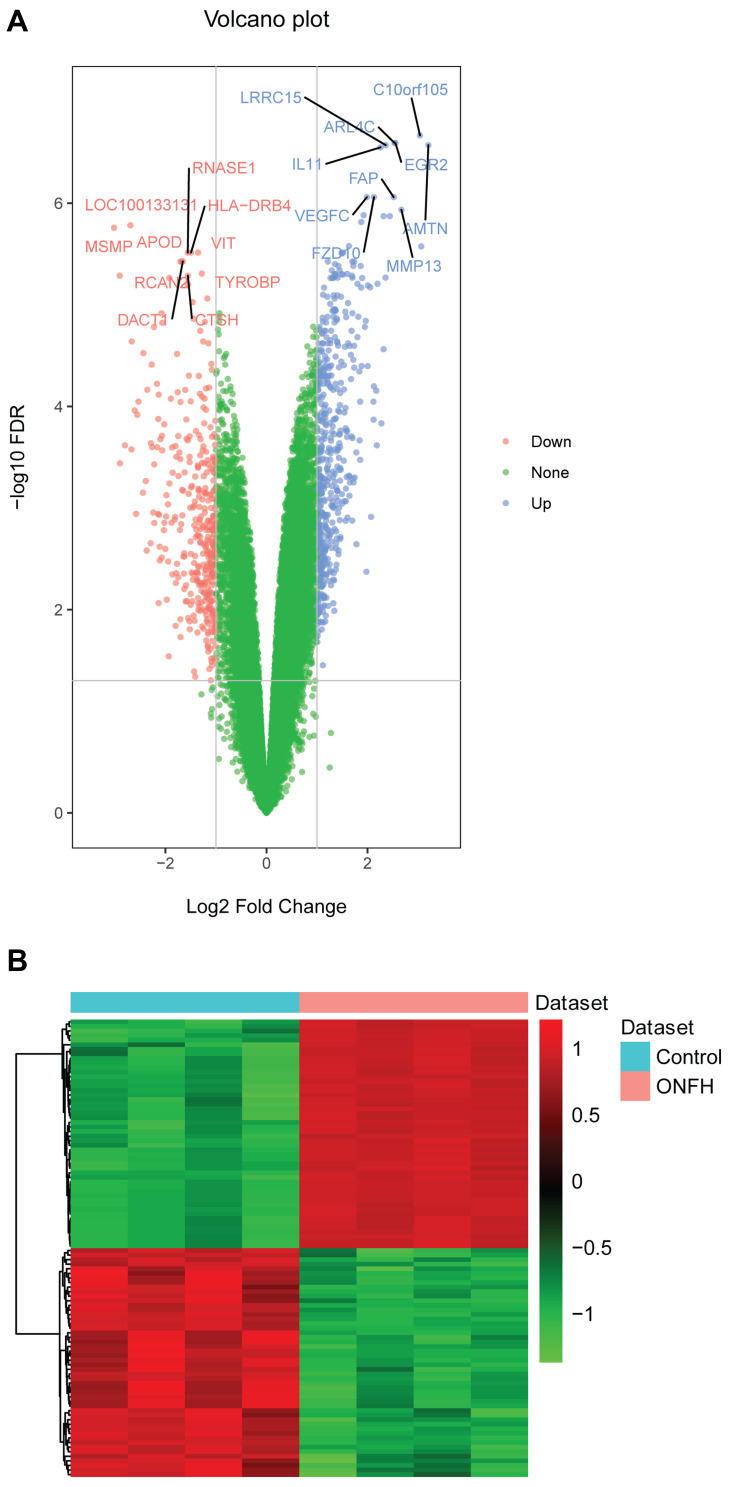
There were 24 differentially expressed miRNAs (supplementary Table 1), and 901 differentially expressed mRNAs (supplementary Table 2) were identified. All differentially expressed miRNAs and the top 20 differentially expressed mRNAs are shown in Tables 1 and 2, respectively. The volcano plot and heat map of all miRNAs and top 100 mRNAs are shown in Figures 1 and 2, respectively.
Table 1.
All Differentially Expressed miRNAs in Osteonecrosis of the Femoral Head
| Symbol | LogFC | P-value | FDR | Up/Down |
|---|---|---|---|---|
| hsa-miR-3191-5p | −3.15065 | 3.62E-07 | 0.000358 | Down |
| hsa-miR-4511 | −4.24635 | 5.12E-06 | 0.000514 | Down |
| hsa-miR-5195-5p | −3.1202 | 5.88E-06 | 0.000514 | Down |
| hsa-miR-128-3p | 3.473807 | 7.27E-06 | 0.000514 | Up |
| hsa-miR-374c-5p | 3.70425 | 1.30E-05 | 0.000722 | Up |
| hsa-miR-532-5p | 3.811221 | 2.07E-05 | 0.000764 | Up |
| hsa-miR-140-5p | 3.711132 | 2.09E-05 | 0.000764 | Up |
| hsa-miR-3200-3p | 3.396543 | 2.39E-05 | 0.000764 | Up |
| hsa-miR-181a-5p | 3.934233 | 2.62E-05 | 0.000786 | Up |
| hsa-miR-28-5p | 3.249402 | 4.52E-05 | 0.001064 | Up |
| hsa-miR-3200-5p | 3.499118 | 6.03E-05 | 0.001152 | Up |
| hsa-miR-106b-3p | 3.17598 | 7.23E-05 | 0.001167 | Up |
| hsa-miR-130a-3p | 3.227388 | 7.73E-05 | 0.001184 | Up |
| hsa-miR-126-5p | 3.329919 | 7.94E-05 | 0.00119 | Up |
| hsa-miR-4762-5p | −3.29506 | 0.000158 | 0.001839 | Down |
| hsa-miR-378c | 3.525841 | 0.000171 | 0.00185 | Up |
| hsa-miR-374a-5p | 3.48526 | 0.000189 | 0.001889 | Up |
| hsa-miR-29c-5p | 3.161635 | 0.000221 | 0.002036 | Up |
| hsa-miR-126-3p | 3.196734 | 0.000246 | 0.002164 | Up |
| hsa-let-7a-5p | 3.626154 | 0.000248 | 0.002164 | Up |
| hsa-miR-339-3p | 3.235942 | 0.000261 | 0.002187 | Up |
| hsa-miR-301a-3p | 3.358697 | 0.000278 | 0.002292 | Up |
| hsa-miR-4711-3p | −3.28102 | 0.001108 | 0.005396 | Down |
| hsa-miR-141-3p | 3.03568 | 0.016527 | 0.03819 | Up |
Abbreviations: FC, fold change; FDR, false discovery rate.
Table 2.
Top 20 Differentially Expressed mRNAs in Osteonecrosis of the Femoral Head
| Symbol | LogFC | P-value | FDR | Up/Down |
|---|---|---|---|---|
| C10orf105 | 3.03737 | 9.90E-12 | 2.15E-07 | Up |
| ARL4C | 2.533696 | 2.73E-11 | 2.55E-07 | Up |
| EGR2 | 2.551482 | 3.52E-11 | 2.55E-07 | Up |
| LRRC15 | 2.360616 | 5.31E-11 | 2.68E-07 | Up |
| AMTN | 3.203584 | 6.15E-11 | 2.68E-07 | Up |
| IL11 | 2.257176 | 7.78E-11 | 2.82E-07 | Up |
| FAP | 2.516977 | 2.84E-10 | 8.71E-07 | Up |
| VEGFC | 1.98741 | 3.44E-10 | 8.71E-07 | Up |
| FZD10 | 2.128076 | 3.60E-10 | 8.71E-07 | Up |
| MMP13 | 2.670397 | 5.32E-10 | 1.16E-06 | Up |
| MSMP | −3.01757 | 1.28E-09 | 1.74E-06 | Down |
| VIT | −1.35337 | 3.11E-09 | 3.06E-06 | Down |
| HLA-DRB4 | −1.49327 | 3.17E-09 | 3.06E-06 | Down |
| RNASE1 | −1.55781 | 3.23E-09 | 3.06E-06 | Down |
| RCAN2 | −1.70034 | 4.41E-09 | 3.75E-06 | Down |
| DACT1 | −1.65655 | 5.18E-09 | 3.75E-06 | Down |
| APOD | −1.67708 | 5.18E-09 | 3.75E-06 | Down |
| TYROBP | −1.27919 | 8.36E-09 | 4.92E-06 | Down |
| CTSH | −1.55528 | 1.03E-08 | 5.16E-06 | Down |
| HLA-DRA | −1.92425 | 1.35E-08 | 5.44E-06 | Down |
Abbreviations: FC, fold change; FDR, false discovery rate.
Figure 1.
The volcano plot and heat map of all differentially expressed miRNAs in osteonecrosis of the femoral head. (A) The volcano plot of all differentially expressed miRNAs. The X and Y axis represents Log2 Fold Change and –log10 FDR, respectively. Blue and red represents up-regulated and down-regulated miRNAs, respectively. (B) The heat map of all differentially expressed miRNAs.
Abbreviation: ONFH, osteonecrosis of the femoral head.
Figure 2.
The volcano plot and heat map of the top 100 differentially expressed mRNAs in osteonecrosis of the femoral head. (A) The volcano plot of the top 100 differentially expressed mRNAs. The X- and Y-axes represent Log2 Fold Change and –log10 FDR, respectively. Blue and red represent up-regulated and down-regulated mRNAs, respectively. (B) The heat map of the top 100 differentially expressed mRNAs.
Abbreviation: ONFH, osteonecrosis of the femoral head.
Network of miRNAs–mRNAs
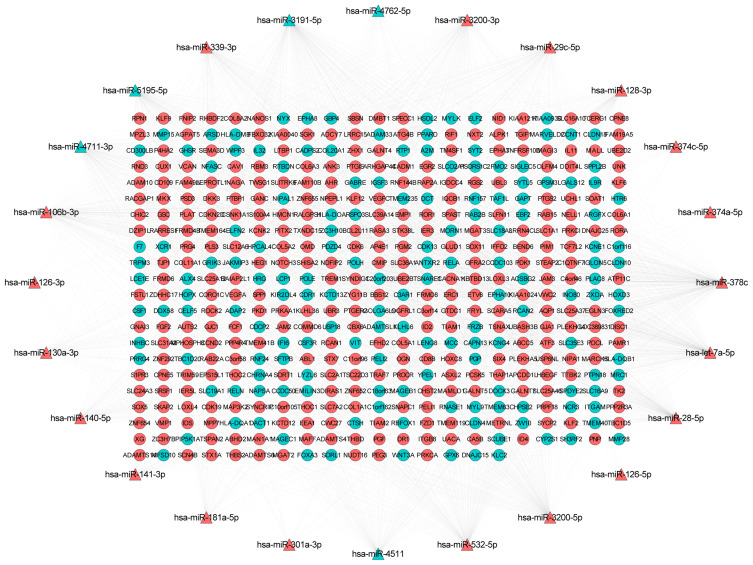
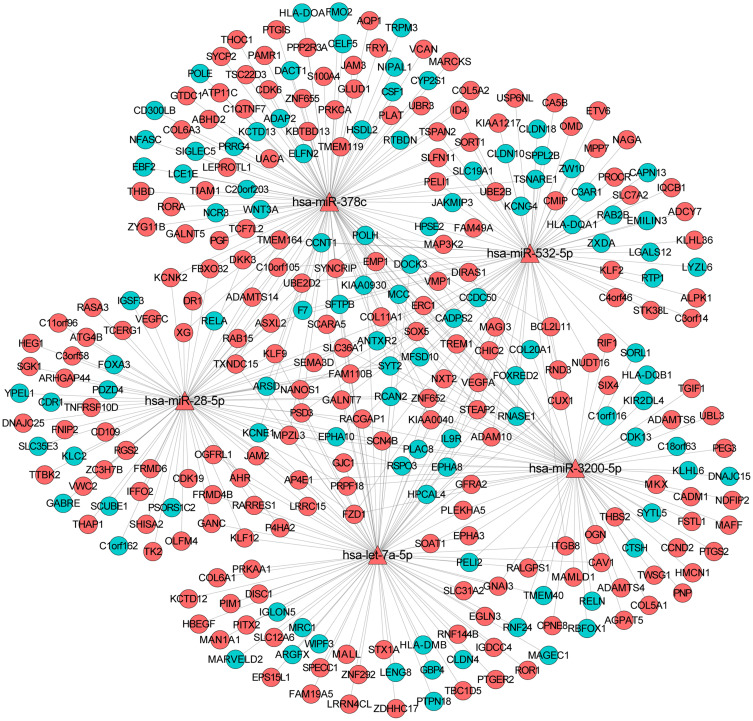
Depending on the targeted analysis, 2,137 miRNA–mRNA pairs (involving 24 miRNA and 457 mRNA) were identified (supplementary Table 3). The established regulatory network of miRNA–targeted mRNA is illustrated in Figure 3. In the network, there were 481 nodes and 1,205 edges. The top 10 differentially expressed miRNAs that targeted the most differentially expressed mRNAs were hsa-miR-378c, hsa-miR-3191-5p, hsa-let-7a-5p, hsa-miR-28-5p, hsa-miR-3200-5p, hsa-miR-532-5p, hsa-miR-106b-3p, hsa-miR-339-3p, hsa-miR-5195-5p, and hsa-miR-3200-3p. In addition, the sub-network of miRNA-target mRNAs between hsa-miR-378c, hsa-let-7a-5p, hsa-miR-28-5p, hsa-miR-3200-5p, hsa-miR-532-5p, and their targeted mRNAs are shown in Figure 4. In addition, we used the TargetScan (http://www.targetscan.org/vert_72/) software to further validate the targeted relationship between miRNA and mRNA, such as hsa-miR-378c-DACT1 and hsa-miR-28-5p-RELA (supplementary Figure 1).
Figure 3.
The network of miRNA-target mRNAs between 24 miRNAs and 457 mRNAs in osteonecrosis of the femoral head. The triangle and circule represent the differentially expressed miRNAs and targeted differentially expressed mRNAs, respectively. The red and green color represent up-regulation and down-regulation, respectively.
Figure 4.
The sub-network of miRNA-target mRNAs between hsa-miR-378c, hsa-let-7a-5p, hsa-miR-28-5p, hsa-miR-3200-5p, hsa-miR-532-5p, and their targeted mRNAs in osteonecrosis of the femoral head. The triangle and circule represent the differentially expressed miRNAs and targeted differentially expressed mRNAs, respectively. The red and green color represent up-regulation and down-regulation, respectively.
Functional Analysis of Targeted mRNAs
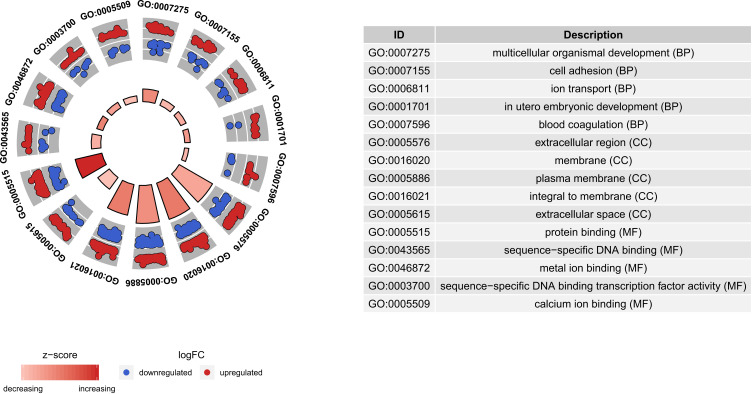
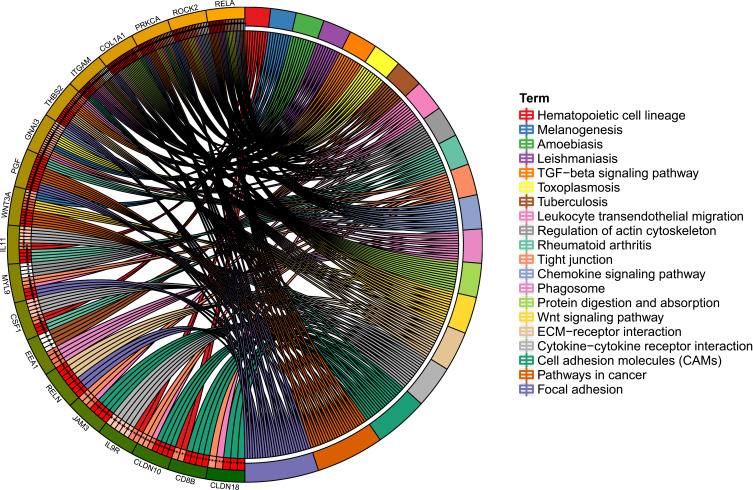
GO and KEGG analysis of targeted mRNAs is shown in supplementary Table 4. The top five significant enrichment GO terms and all KEGG terms are presented in Figures 5 and 6, respectively. Total KEGG terms involving targeted differentially expressed mRNAs are shown in Table 3. In the KEGG terms, we found seven valuable signaling pathways including the Wnt signaling pathway (involved WNT3A), chemokine signaling pathway (involved RELA), focal adhesion and ECM-receptor interaction (involved RELN), cell adhesion molecules (CAMs) (involved CLDN18 and CLDN10), cytokine–cytokine receptor interaction, and hematopoietic cell lineage (involving CSF1 and IL9R).
Figure 5.
Top five significantly enriched GO terms of targeted differentially expressed mRNAs in osteonecrosis of the femoral head. The z-score clustering in the GO terms of targeted differentially expressed mRNA is shown below. The red and blue color represent up-regulated and down-regulated mRNA, respectively.
Abbreviations: BP, biological process; CC, cellular component; MF, molecular function; FC, fold change.
Figure 6.
KEGG signaling pathways of targeted differentially expressed mRNAs in osteonecrosis of the femoral head. Different colors represent different signaling pathways; mRNA outside the circle represents the enriched one of mRNAs in the particular signaling pathway.
Table 3.
Total KEGG Terms Involved Targeted Differentially Expressed mRNAs in Osteonecrosis of the Femoral Head
| Terms | Count | P-value | FDR | mRNAs |
|---|---|---|---|---|
| Focal adhesion | 19 | 2.09E-11 | 3.15E-09 | RELN,PGF,MYLK,ROCK2,CCND2,SPP1,PRKCA,COL5A2,CAV1,COL1A1,THBS2,VEGFC,COL5A1,ITGB8,COL6A1,VEGFA,MYL9,COL11A1,COL6A3 |
| Pathways in cancer | 16 | 7.87E-06 | 0.00017 | CDK6,PPARD,EGLN3,FGF2,PGF,ABL1,WNT3A,CSF3R,PRKCA,FZD1,VEGFC,TCF7L2,RELA,SLC2A1,VEGFA,PTGS2 |
| Cell adhesion molecules (CAMs) | 15 | 1.25E-10 | 9.43E-09 | JAM3,JAM2,HLA-DOA,NFASC,ITGAM,CD8B,CADM1,HLA-DQA1,CLDN18,CLDN10,HLA-DQB1,ITGB8,HLA-DMB,VCAN,CLDN4 |
| Tight junction | 11 | 1.35E-06 | 4.07E-05 | JAM3,JAM2,TJP1,GNAI3,PRKCA,MAGI3,CLDN18,CLDN10,PRKCI,MYL9,CLDN4 |
| ECM-receptor interaction | 10 | 1.70E-07 | 8.54E-06 | RELN,SPP1,COL5A2,COL1A1,THBS2,COL5A1,ITGB8,COL6A1,COL11A1,COL6A3 |
| Leukocyte transendothelial migration | 10 | 2.71E-06 | 6.83E-05 | JAM3,JAM2,ROCK2,ITGAM,GNAI3,PRKCA,CLDN18,CLDN10,MYL9,CLDN4 |
| Cytokine-cytokine receptor interaction | 9 | 0.00791727 | 0.031461 | TNFRSF10D,IL11,INHBC,CSF3R,CSF1,XCR1,VEGFC,VEGFA,IL9R |
| Protein digestion and absorption | 9 | 9.24E-07 | 3.49E-05 | SLC36A1,SLC1A1,COL5A2,COL1A1,COL5A1,COL6A1,COL11A1,COL6A3,SLC16A10 |
| Regulation of actin cytoskeleton | 9 | 0.00196338 | 0.01098 | FGF2,TIAM1,MYLK,ROCK2,ITGAM,PIP5K1A,TIAM2,ITGB8,MYL9 |
| Phagosome | 9 | 9.15E-05 | 0.001256 | HLA-DOA,ITGAM,HLA-DQA1,THBS2,STX7,EEA1,HLA-DQB1,MRC1,HLA-DMB |
| Tuberculosis | 8 | 0.00214245 | 0.011554 | HLA-DOA,ITGAM,HLA-DQA1,RELA,EEA1,HLA-DQB1,MRC1,HLA-DMB |
| Wnt signaling pathway | 8 | 0.000859587 | 0.006181 | PPARD,ROCK2,CCND2,WNT3A,PRKCA,FZD1,TCF7L2,CSNK1A1 |
| Rheumatoid arthritis | 8 | 1.58E-05 | 0.000299 | PGF,IL11,HLA-DOA,CSF1,HLA-DQA1,HLA-DQB1,VEGFA,HLA-DMB |
| Chemokine signaling pathway | 7 | 0.0121892 | 0.044892 | TIAM1,ROCK2,ADCY7,GNAI3,XCR1,TIAM2,RELA |
| TGF-beta signaling pathway | 7 | 0.000108744 | 0.001368 | ROCK2,INHBC,PITX2,LTBP1,ID4,THBS2,ID2 |
| Toxoplasmosis | 7 | 0.00128054 | 0.007734 | HLA-DOA,GNAI3,HLA-DQA1,RELA,HLA-DQB1,PDK1,HLA-DMB |
| Melanogenesis | 7 | 0.000330342 | 0.003118 | WNT3A,ADCY7,GNAI3,PRKCA,FZD1,TCF7L2,DCT |
| Leishmaniasis | 7 | 2.67E-05 | 0.000448 | HLA-DOA,ITGAM,HLA-DQA1,RELA,HLA-DQB1,HLA-DMB,PTGS2 |
| Amoebiasis | 7 | 0.000421534 | 0.003744 | ITGAM,PRKCA,COL5A2,COL1A1,COL5A1,RELA,COL11A1 |
| Hepatitis C | 6 | 0.00865149 | 0.033497 | DDX58,CLDN18,CLDN10,RELA,PDK1,CLDN4 |
| Lysosome | 6 | 0.00552762 | 0.023848 | NAPSA,NAGA,CTSH,SORT1,AP4E1,IDS |
| Axon guidance | 6 | 0.00722661 | 0.029492 | EPHA8,ABL1,ROCK2,GNAI3,SEMA3D,EPHA3 |
| Hematopoietic cell lineage | 6 | 0.000824081 | 0.006222 | IL11,ITGAM,CSF3R,CSF1,CD8B,IL9R |
| Gap junction | 6 | 0.00111969 | 0.007045 | TJP1,ADCY7,GNAI3,PRKCA,MAP3K2,GJA1 |
| Epithelial cell signaling in Helicobacter pylori infection | 6 | 0.000260077 | 0.002805 | JAM3,JAM2,TJP1,ADAM10,HBEGF,RELA |
| Staphylococcus aureus infection | 6 | 3.08E-05 | 0.000466 | HLA-DOA,ITGAM,HLA-DQA1,C3AR1,HLA-DQB1,HLA-DMB |
| Viral myocarditis | 6 | 0.000185105 | 0.00215 | ABL1,HLA-DOA,CAV1,HLA-DQA1,HLA-DQB1,HLA-DMB |
| Complement and coagulation cascades | 5 | 0.00169723 | 0.009857 | THBD,A2M,PLAT,C3AR1,F7 |
| Gastric acid secretion | 5 | 0.00266688 | 0.01299 | MYLK,ADCY7,GNAI3,PRKCA,KCNK2 |
| Renal cell carcinoma | 5 | 0.00235692 | 0.011863 | EGLN3,PGF,VEGFC,SLC2A1,VEGFA |
| Pancreatic cancer | 5 | 0.00235692 | 0.011863 | CDK6,PGF,VEGFC,RELA,VEGFA |
| Antigen processing and presentation | 5 | 0.000864725 | 0.005935 | HLA-DOA,CD8B,HLA-DQA1,HLA-DQB1,HLA-DMB |
| Autoimmune thyroid disease | 4 | 0.00292128 | 0.013367 | HLA-DOA,HLA-DQA1,HLA-DQB1,HLA-DMB |
| Type I diabetes mellitus | 4 | 0.00101917 | 0.006691 | HLA-DOA,HLA-DQA1,HLA-DQB1,HLA-DMB |
| Intestinal immune network for IgA production | 4 | 0.0026893 | 0.01269 | HLA-DOA,HLA-DQA1,HLA-DQB1,HLA-DMB |
| N-Glycan biosynthesis | 4 | 0.00398592 | 0.017702 | RPN1,MAN1A1,MGAT2,MGAT3 |
| Shigellosis | 4 | 0.00867634 | 0.032753 | ABL1,ROCK2,UBE2D2,RELA |
| Acute myeloid leukemia | 4 | 0.006843 | 0.028703 | PPARD,TCF7L2,RELA,PIM1 |
| Allograft rejection | 4 | 0.000549845 | 0.004613 | HLA-DOA,HLA-DQA1,HLA-DQB1,HLA-DMB |
| Graft-versus-host disease | 4 | 0.000627861 | 0.00499 | HLA-DOA,HLA-DQA1,HLA-DQB1,HLA-DMB |
| Asthma | 4 | 0.000305331 | 0.003074 | HLA-DOA,HLA-DQA1,HLA-DQB1,HLA-DMB |
Abbreviation: FDR, false discovery rate.
In vitro Validation
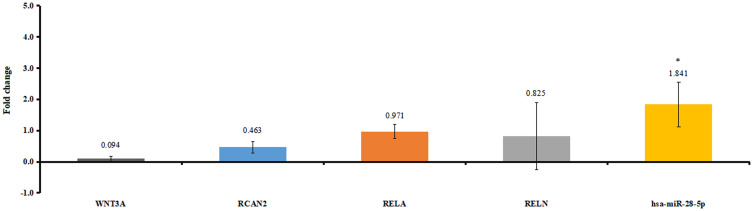
Six patients with osteonecrosis of the femoral head and seven normal controls were incorporated in our study. Clinical information of these individuals is shown in Table 4. As mentioned above, hsa-miR-28-5p was one of the top 10 differentially expressed miRNAs that targeted the most differentially expressed mRNAs. WNT3A, RELA, and RELN were involved in KEGG pathways. RCAN2 was in the top 20 down-regulated mRNAs. We selected hsa-miR-28-5p, WNT3A, RCAN2, RELA, and RELN for validation (Figure 7). The relative expression of hsa-miR-28-5p was significantly up-regulated, the relative expression of WNT3A, RCAN2, RELA, and RELN was down-regulated in patients with osteonecrosis of the femoral head. The validated result was in line with the bioinformatics analysis.
Table 4.
Clinical Information of Enrolled Individuals in vitro Validation
| Group | Gender | Age | Weight | Pain | Function | Malformation | Joint Activities | Cartilage Injury of Hip Joint | ARCO Stage |
|---|---|---|---|---|---|---|---|---|---|
| NC | Male | 53 | 77 | No | Good | No | Normal | No | No |
| Male | 62 | 68 | No | Good | No | Normal | No | No | |
| Female | 58 | 63 | No | Good | No | Normal | No | No | |
| Female | 58 | 63 | No | Good | No | Normal | No | No | |
| Male | 51 | 70 | No | Good | No | Normal | No | No | |
| Female | 63 | 55 | No | Good | No | Normal | No | No | |
| Female | 49 | 62 | No | Good | No | Normal | No | No | |
| Case | Male | 50 | 80 | Yes | Limp | Shortening | Adduction abduction limited and buckling 90° | Yes | IV |
| Male | 39 | 72 | Yes | Limp | Shortening | Adduction abduction limited and buckling 90° | Yes | IV | |
| Male | 46 | 66 | Yes | Limp | Shortening | Adduction abduction limited and buckling 80° | Yes | IV | |
| Female | 35 | 62 | Yes | Limp | Shortening | Adduction abduction limited and buckling 90° | Yes | IV | |
| Female | 58 | 67 | Yes | Limp | Shortening | Adduction abduction limited and buckling 90° | Yes | IV | |
| Male | 20 | 65 | Yes | Limp | Shortening | Adduction abduction limited and buckling 90° | Yes | IV |
Abbreviation: NC, normal controls.
Figure 7.
The in vitro validation of differentially expressed miRNAs and targeted differentially expressed mRNAs. Fold change >1 and fold change <1 represent up-regulation and down-regulation, respectively. *P<0.05.
Expression Validation and Diagnostic Analysis of Targeted mRNAs
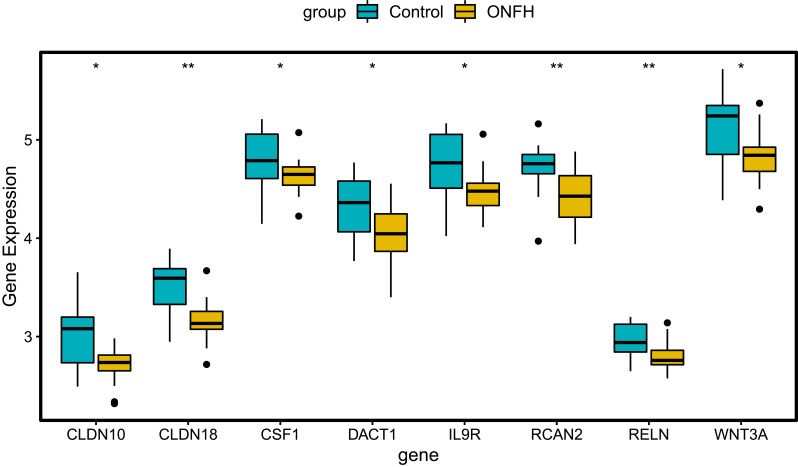
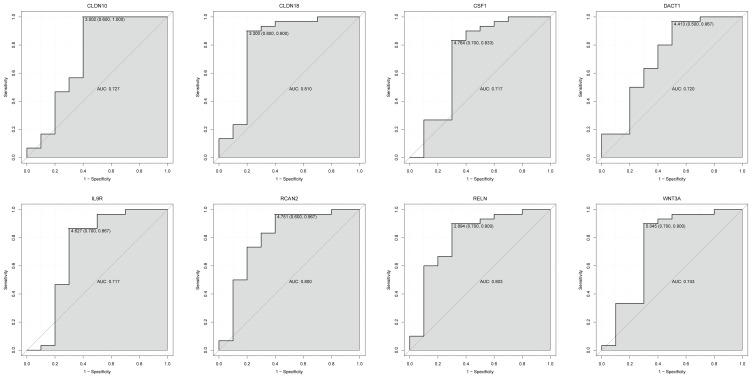
The GSE123568 dataset was firstly utilized to validate the expression of CLDN10, CLDN18, CSF1, DACT1, IL9R, RCAN2, RELN, and WNT3A (Figure 8). The expression of these mRNAs was all significantly down-regulated, which is in line with the bioinformatics analysis. In addition, the ROC curve analysis was performed to assess the diagnosis ability of these mRNAs in the GSE123568 dataset (Figure 9). The AUC of these mRNAs was more than 0.7, which suggested that they had a diagnostic value for osteonecrosis of the femoral head.
Figure 8.
Expression box plots of CLDN10, CLDN18, CSF1, DACT1, IL9R, RCAN2, RELN, and WNT3A in the GSE123568 dataset. *P<0.05, **P<0.01.
Abbreviation: ONFH, osteonecrosis of the femoral head.
Figure 9.
The ROC curves of CLDN10, CLDN18, CSF1, DACT1, IL9R, RCAN2, RELN, and WNT3A between osteonecrosis of the femoral head and normal controls. The ROC curves were used to show the diagnostic ability of these mRNAs with 1-specificity and sensitivity.
Discussion
Hsa-miR-378c, a hypoxia-regulated miRNA, is reported to be down-regulated in rheumatoid arthritis and osteosarcomas.29,30 The level of hsa-miR-378c was increased in osteonecrosis of the femoral head in this study. Moreover, three down-regulated mRNAs including Wnt family member 3A (WNT3A), dishevelled binding antagonist of beta catenin 1 (DACT1), and colony stimulating factor 1 (CSF1) were all regulated by hsa-miR-378c. Significantly, WNT3A, DACT1, and CSF1 had a diagnostic value for osteonecrosis of the femoral head. WNT3A can induce chondrocytes proliferation and alter the extracellular matrix synthesis function of the chondrocytes.31 In addition, WNT3A and hypoxia could act together to promote angiogenesis by regulating cell death.32 It is noted that liposome-reconstituted recombinant human WNT3A protein has been used to treat osteonecrosis defect.33 The expression of DACT1 is found in primary chondrocytes and vascular endothelial cells.34,35 Käkönen and Mundy36 found that CSF-1 could interact with osteoblast to regulate the RANK–RANKL pathway to stimulate osteoclast precursors, ultimately leading to osteolysis. Additionally, CSF1, combined with VEGF-A, induces angiogenesis and recruitment of pericyte to neovessels.37 Our result suggested that WNT3A, DACT1, and CSF1 may play roles in bone remodeling and angiogenesis under the regulation of hsa-miR-378c in the process of osteonecrosis of the femoral head.
Hsa-let-7a-5p is down-regulated in osteogenic differentiation, while up-regulated during osteoclastogenesis, which indicates the role of hsa-let-7a-5p in the bone imbalance.38–40 Under mechanical tension, hsa-let-7a-5p is remarkably increased in cartilage endplate chondrocytes.41 In this study, we found that hsa-let-7a-5p was up-regulated in osteonecrosis of the femoral head. Furthermore, a regulator of calcineurin 2 (RCAN2) and interleukin 9 receptor (IL9R) were down-regulated and targeted by hsa-let-7a-5p. It is worth mentioning that RCAN2 and IL9R could be considered as diagnostic biomarkers for osteonecrosis of the femoral head. RCAN2, an angiogenesis related gene, is transcribed activation by vascular endothelial growth factor (VEGF).42–45 The expression of RCAN2 is negatively correlated with cartilage proliferation and differentiation.46 It is reported that disruption of RCAN2 could lead to reducing bone mass, which is related to increased osteoclast function and reduced osteoblast function.47 IL9R is associated with hematopoietic cell lineage.48 Our result indicated that hsa-let-7a-5p and its target (RCAN2 and IL9R) could be involved in osteonecrosis of the femoral head.
Deregulation of hsa-miR-28-5p is related to rheumatic diseases, such as axial spondyloarthritis and rheumatoid arthritis.49,50 Herein, we first found that hsa-miR-28-5p was up-regulated in osteonecrosis of the femoral head. In addition, the down-regulated RELA proto-oncogene, NF-kB subunit (RELA, also called p65) was one of the targets of hsa-miR-28-5p. It is reported that RELA is the most potent transcriptional factor of hypoxic induction factor 2 (HIF2) that regulates chondrocyte differentiation and cartilage degradation.51 Lacking or deletion of RELA inhibits the expression of cartilage catabolic factors such as matrix metalloproteinases 9 (MMP9), SRY-box transcription factor 9 (SOX9), nitric oxide synthase 2 (NOS2), and cyclooxygenase 2 (COX2) in chondrocytes, which results in reduced bone loss and accelerated cartilage degeneration.52–57 In addition, RELA is regarded as an angiogenesis modulating agent.58 It is suggested that RELA may be involved in cartilage degeneration of osteonecrosis of the femoral head under that regulation of hsa-miR-28-5p.
Previous studies on hsa-miR-3200-5p in orthopedic disease are very rare, and only a recent report showed significantly higher expression of hsa-miR-3200-5p in the osteosarcoma.59 In our study, we found that the expression level of hsa-miR-3200-5p was increased in osteonecrosis of the femoral head. Moreover, down-regulated reelin (RELN) was targeted by hsa-miR-3200-5p. It is noted that RELN had a diagnostic value for osteonecrosis of the femoral head. RELN, expressed in osteoblast lineage cells, is considered as a stromal cell-specific and hematopoietic cell-lineage marker.60 Clinically, RELN is a potential molecular target candidate for diagnosis and therapy of rheumatoid arthritis.61 Our result indicated that hsa-miR-3200-5p and RELN may play a critical role in osteonecrosis of the femoral head.
Hsa-miR-532-5p plays roles in the regulation of the adaptation to hypoxia in endothelial cells.62 Hsa-miR-532-5p is differentially expressed in chondrocytes from distinct regions of developing human cartilage.63 In the present study, we found that hsa-miR-532-5p was up-regulated in osteonecrosis of the femoral head. Furthermore, claudin 18 (CLDN18) and claudin 10 (CLDN10) were down-regulated and regulated by hsa-miR-532-5p. In addition, CLDN18 and CLDN10 were associated with disease diagnosis. Elevated expression of CLDN18 is found in osteoblasts.64,65 Knock-out of CLDN18 leads to reduced bone mass from hyperactive osteoclasts.64 The expression of CLDN10 is increased in osteosarcoma osteoblast cells.66 Our result suggested that CLDN18 and CLDN10 could be associated with bone loss under the regulation of hsa-miR-532-5p in osteonecrosis of the femoral head.
Based on KEGG analysis, we found seven valuable signaling pathways including the Wnt signaling pathway (involved WNT3A), chemokine signaling pathway (involved RELA), focal adhesion and ECM-receptor interaction (involved RELN), cell adhesion molecules (CAMs) (involved CLDN18 and CLDN10), cytokine–cytokine receptor interaction, and hematopoietic cell lineage (involved CSF1 and IL9R) in osteonecrosis of the femoral head. The Wnt/β-catenin pathway induces VEGF to promote neovascularization.67,68 In addition, the signaling pathway plays a key role in regulating chondrocyte proliferation. It is found that the Wnt/β-catenin pathway is involved in the process of cartilage damage.69 It is noted that the Wnt/β-catenin pathway is associated with the pathogenesis of early stage femoral head osteonecrosis via regulating of transcriptional regulator Myc-like (c-Myc) that affects the cell apoptosis and cell cycle.70 Chemokines are involved in angiogenesis and wound healing. It is reported that chemokines secreted from chondrocytes alter functional abilities of subchondral bone osteoblasts.71 The chemokine signaling pathway is significantly enriched in immature articular cartilage after osteonecrosis of the femoral head.72 Focal adhesion, involved in cell growth, shape, and movement, attach chondrocytes to the pericellular cartilage matrix and link to intracellular organelles. It has been shown that focal adhesion is a remarkably enriched biological pathway in the immature articular cartilage after osteonecrosis of the femoral head.72 Cell adhesion molecules, such as cadherins, selectins, and immunoglobulin superfamily proteins, are associated with angiogenesis.73–75 In addition, cell adhesion molecules play a vital role in regulating cartilage matrix turnover.76,77 ECM-receptor interaction is associated with angiogenesis, chondrogenesis, and cartilage degeneration.78 It is found that ECM-receptor interaction is significantly enriched in hip cartilage with osteonecrosis of the femoral head.79 It has been identified that cytokine–cytokine receptor interaction is one of the most dramatically important pathways in osteonecrosis of the femoral head.72,80 Osteoclasts originated from hematopoietic stem cells are involved in maintaining bone integrity. Normal femoral head shows trabecular bones surrounded by bone marrow endowed with hematopoietic cells. Infiltration of hematopoietic cell to the ischemic area plays a significant role in regulating ischemia-induced angiogenesis.81
In conclusion, the epigenetic modifications of hsa-miR-378c-WNT3A/DACT1/CSF1, hsa-let-7a-5p-RCAN2/IL9R, hsa-miR-28-5p-RELA, hsa-miR-3200-5p-RELN, and hsa-miR-532-5p-CLDN18/CLDN10, and seven signaling pathways (Wnt signaling pathway, chemokine signaling pathway, focal adhesion, cell adhesion molecules (CAMs), ECM-receptor interaction, cytokine-cytokine receptor interaction, and hematopoietic cell lineage) may be involved in osteonecrosis of the femoral head. In addition, CLDN10, CLDN18, CSF1, DACT1, IL9R, RCAN2, RELN, and WNT3A had a diagnostic value for osteonecrosis of the femoral head. However, there are limitations of our study. Firstly, the deeper mechanism study of identified differentially expressed miRNAs, mRNAs, and relevant downstream molecules in the disease is further needed in animal models. Secondly, the regulatory relationship between identified miRNAs and targeted mRNAs is not investigated. Further in vitro experiment, such as luciferase reporter gene assay is needed in the further study.
Acknowledgment
This study was funded by the Department of Science and Technology of Shanxi Provincial of “The mechanism of Tongluoshenggu Decoction in the treatment of steroid-induced femoral head necrosis based on autophagy mediated by PI3K/Akt/mTOR signaling pathway” (2020SF-287).
Abbreviations
CLDN10, claudin 10; CLDN18, claudin 18; CSF1, colony stimulating factor 1; COX2, cyclooxygenase 2; DACT1, dishevelled binding antagonist of beta catenin 1; RELN, reelin; CAMs, cell adhesion molecules; FGF2, fibroblast growth factor 2; GEO, Gene Expression Omnibus database; HIF2, hypoxic induction factor 2; IGF1, insulin-like growth factor 1; IL9R, interleukin 9 receptor; MMP9, matrix metalloproteinases 9; NOS2, nitric oxide synthase 2; RCAN2, regulator of calcineurin 2; SOX9, SRY-box transcription factor 9; VEGF, vascular endothelial growth factor; WNT3A, Wnt family member 3A.
Disclosure
No competing financial interests exist and the authors report no conflicts of interest for this work.
References
- 1.Aaron RK, Ciombor DM. Coagulopathies and osteonecrosis. Curr Opin Orthop. 2001;12(5):378–383. doi: 10.1097/00001433-200110000-00003 [DOI] [Google Scholar]
- 2.Chandler FA, Peltier LF. Coronary disease of the hip. Clin Orthop Relat Res. 2001;386:7–10. doi: 10.1097/00003086-200105000-00002 [DOI] [PubMed] [Google Scholar]
- 3.Candinas R, Jakob M, Buckingham TA, Mattmann H, Amann FW. Vibration, acceleration, gravitation, and movement: activity controlled rate adaptive pacing during treadmill exercise testing and daily life activities. Pacing Clin Electrophysiol. 1997;20(7):1777–1786. doi: 10.1111/j.1540-8159.1997.tb03566.x [DOI] [PubMed] [Google Scholar]
- 4.Shah KN, Racine J, Jones LC, Aaron RK. Pathophysiology and risk factors for osteonecrosis. Curr Rev Musculoskelet Med. 2015;8:201–209. doi: 10.1007/s12178-015-9277-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pouya F, Kerachian MA. Avascular necrosis of the femoral head: are any genes involved? Arch Bone Jt Surg. 2015;3:149–155. [PMC free article] [PubMed] [Google Scholar]
- 6.Malizos KN, Karantanas AH, Varitimidis SE, Dailiana ZH, Bargiotas K, Maris T. Osteonecrosis of the femoral head: etiology, imaging and treatment. Eur J Radiol. 2007;63:0–28. [DOI] [PubMed] [Google Scholar]
- 7.Zalavras CG, Lieberman JR. Osteonecrosis of the femoral head: evaluation and treatment. J Am Acad Orthop Surg. 2014;22(7):455–464. doi: 10.5435/JAAOS-22-07-455 [DOI] [PubMed] [Google Scholar]
- 8.Mccarthy J, Puri L, Barsoum W, Lee JA, Laker M, Cooke P. Articular cartilage changes in avascular necrosis: an arthroscopic evaluation. Clin Orthop Relat Res. 2003;406:64. doi: 10.1097/00003086-200301000-00011 [DOI] [PubMed] [Google Scholar]
- 9.Magnussen RA, Guilak F, Vail TP. Articular cartilage degeneration in post-collapse osteonecrosis of the femoral head. J Bone Joint Surg Am. 2005;87:1272–1277. [DOI] [PubMed] [Google Scholar]
- 10.Kerachian MA, Harvey EJ, Cournoyer D, Chow TY, Seguin C. Avascular necrosis of the femoral head: vascular hypotheses. Endothelium. 2006;13:237–244. doi: 10.1080/10623320600904211 [DOI] [PubMed] [Google Scholar]
- 11.Huang G, Zhao G, Xia J, et al. FGF2 and FAM201A affect the development of osteonecrosis of the femoral head after femoral neck fracture. Gene. 2018;652:39–47. doi: 10.1016/j.gene.2018.01.090 [DOI] [PubMed] [Google Scholar]
- 12.Nobillot R, Le Parc JM, Benoit J, Paolaggi JB. Idiopathic osteonecrosis of the hip in twins. Ann Rheum Dis. 1994;53:702. doi: 10.1136/ard.53.10.702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyamoto Y, Matsuda T, Kitoh H, et al. A recurrent mutation in type II collagen gene causes Legg-Calvé-Perthes disease in a Japanese family. Hum Genet. 2007;121:625–629. [DOI] [PubMed] [Google Scholar]
- 14.Chen WM, Liu Y-F, Lin M-W, et al. Autosomal dominant avascular necrosis of femoral head in two taiwanese pedigrees and linkage to chromosome 12q13. Am J Hum Genet. 2004;75:310–317. doi: 10.1086/422702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu YF, Chen W-M, Lin Y-F, et al. Type II collagen gene variants and inherited osteonecrosis of the femoral head. N Engl J Med. 2005;352:2294–2301. doi: 10.1056/NEJMoa042480 [DOI] [PubMed] [Google Scholar]
- 16.Larson E, Jones LC, Goodman SB, Koo KH, Cui Q. Early-stage osteonecrosis of the femoral head: where are we and where are we going in year 2018? Int Orthop. 2018;42:1–6. doi: 10.1007/s00264-018-3917-8 [DOI] [PubMed] [Google Scholar]
- 17.Arlet J. Nontraumatic avascular necrosis of the femoral head. Past Present Future. 1992;277:12–21. [PubMed] [Google Scholar]
- 18.Desforges JF, Mankin HJ. Nontraumatic necrosis of bone (Osteonecrosis). N Engl J Med. 1992;326:1473–1479. doi: 10.1056/NEJM199205283262206 [DOI] [PubMed] [Google Scholar]
- 19.Hamadeh IS, Ngwa BA, Gong Y. Drug induced osteonecrosis of the jaw. Cancer Treat Rev. 2015;41(5):455–464. doi: 10.1016/j.ctrv.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 20.Fang B, Li Y, Chen C, et al. Huo Xue Tong Luo capsule ameliorates osteonecrosis of femoral head through inhibiting lncRNA-Miat. J Ethnopharmacol. 2019;238:111862. doi: 10.1016/j.jep.2019.111862 [DOI] [PubMed] [Google Scholar]
- 21.Kamiya N, Yamaguchi R, Adapala NS, et al. Legg-calve-perthes disease produces chronic hip synovitis and elevation of interleukin-6 in the synovial fluid. J Bone Miner Res. 2015;30(6):1009–1013. doi: 10.1002/jbmr.2435 [DOI] [PubMed] [Google Scholar]
- 22.Hyman JE, Trupia EP, Wright ML, et al. Interobserver and intraobserver reliability of the modified Waldenstrom classification system for staging of Legg-Calve-Perthes disease. J Bone Joint Surg Am. 2015;97:643–650. doi: 10.2106/JBJS.N.00887 [DOI] [PubMed] [Google Scholar]
- 23.Eskildsen T, Taipaleenmäki H, Stenvang J, Abdallah M. Basem, Ditzel and Nicholas, MicroRNA-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc Natl Acad Sci. 2011;108:6139–6144. doi: 10.1073/pnas.1016758108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh T, Nozawa Y, Akao Y. MicroRNA-141 and −200a are involved in bone morphogenetic protein-2-induced mouse pre-osteoblast differentiation by targeting distal-less homeobox 5. J Biol Chem. 2009;284(29):19272–19279. doi: 10.1074/jbc.M109.014001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Zhao L, Xing L, et al. MicroRNA-204 regulates Runx2 protein expression and mesenchymal progenitor cell differentiation. Stem Cells (Dayton, Ohio). 2010;28(2):357–364. doi: 10.1002/stem.288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuang W, Zheng L, Xu X, et al. Dysregulation of the miR-146a-Smad4 axis impairs osteogenesis of bone mesenchymal stem cells under inflammation. Bone Res. 2017;5(1):1–9. doi: 10.1038/boneres.2017.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reiner-Benaim A. FDR control by the BH procedure for two-sided correlated tests with implications to gene expression data analysis. Biom J. 2007;49:107–126. doi: 10.1002/bimj.200510313 [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc. 1995;57:289–300. [Google Scholar]
- 29.Chen Y-J, Chang W-A, Hsu Y-L, Chen C, Kuo P-L. Deduction of novel genes potentially involved in osteoblasts of rheumatoid arthritis using next-generation sequencing and bioinformatic approaches. Int J Mol Sci. 2017;18(11):2396. doi: 10.3390/ijms18112396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao H, Li M, Lihua L, et al. MiR-133b is down-regulated in human osteosarcoma and inhibits osteosarcoma cells proliferation, migration and invasion, and promotes apoptosis. PLoS One. 2013;8(12):e83571. doi: 10.1371/journal.pone.0083571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nalesso G, Sherwood J, Bertrand J, et al. WNT-3A modulates articular chondrocyte phenotype by activating both canonical and noncanonical pathways. J Cell Biol. 2011;193:551–564. doi: 10.1083/jcb.201011051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fustermatanzo A, Manferrari G, Marchetti B, Pluchino S. Wnt3a promotes pro-angiogenic features in macrophages in vitro: implications for stroke pathology. Exp Biol Med. 2018;243:22–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salmon B, Liu B, Shen E, et al. WNT-activated bone grafts repair osteonecrotic lesions in aged animals. Sci Rep. 2017;7(1):14254. doi: 10.1038/s41598-017-14395-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.James CG, Ulici V, Tuckermann J, Underhill TM, Beier F. Expression profiling of Dexamethasone-treated primary chondrocytes identifies targets of glucocorticoid signalling in endochondral bone development. BMC Genom. 2007;8:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee YW, Eum SY, Chen KC, Hennig B, Toborek M. Gene expression profile in interleukin-4-stimulated human vascular endothelial cells. Mol Med. 2004;10:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Käkönen SM, Mundy GR. Mechanisms of osteolytic bone metastases in breast carcinoma. Cancer. 2003;97:834–839. doi: 10.1002/cncr.11132 [DOI] [PubMed] [Google Scholar]
- 37.Hsu C-W, Poché RA, Saik JE, et al. Improved angiogenesis in response to localized delivery of macrophage-recruiting molecules. PLoS One. 2015;10:e0131643. doi: 10.1371/journal.pone.0131643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Yu X, Yu Y, et al. MicroRNA expression analysis during FK506-induced osteogenic differentiation in rat bone marrow stromal cells. Mol Med Rep. 2017;16:581–590. doi: 10.3892/mmr.2017.6655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–3657. doi: 10.1182/blood-2010-10-311415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kagiya T, Nakamura S. Expression profiling of microRNAs in RAW264.7 cells treated with a combination of tumor necrosis factor alpha and RANKL during osteoclast differentiation. J Periodontal Res. 2013;48:373–385. doi: 10.1111/jre.12017 [DOI] [PubMed] [Google Scholar]
- 41.Feng C, Liu M, Fan X, Yang M, Zhou Y. Intermittent cyclic mechanical tension altered the microRNA expression profile of human cartilage endplate chondrocytes. Mol Med Rep. 2018;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang S, Li X, Parra M, Verdin E, Bassel-Duby R, Olson EN. Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc Natl Acad Sci U S A. 2008;105:7738–7743. doi: 10.1073/pnas.0802857105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ginnan R, Sun LY, Schwarz JJ, Singer HA. MEF2 is regulated by CaMKIIdelta2 and a HDAC4-HDAC5 heterodimer in vascular smooth muscle cells. Biochem J. 2012;444:105–114. doi: 10.1042/BJ20120152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin L, Zhao D, Liu X, et al. Down syndrome candidate region 1 isoform 1 mediates angiogenesis through the calcineurin-NFAT pathway. Mol Cancer Res. 2006;4:811–820. doi: 10.1158/1541-7786.MCR-06-0126 [DOI] [PubMed] [Google Scholar]
- 45.Zeng H, Qin L, Zhao D, et al. Orphan nuclear receptor TR3/Nur77 regulates VEGF-A-induced angiogenesis through its transcriptional activity. J Exp Med. 2006;203:719–729. doi: 10.1084/jem.20051523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Katsunori S, Naosuke K, Shunsuke T, et al. Quality evaluation of human bone marrow mesenchymal stem cells for cartilage repair. Stem Cells Int. 2017;1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim JH, Kim K, Kim I, et al. RCANs regulate the convergent roles of NFATc1 in bone homeostasis. Sci Rep. 2016;6:38526. doi: 10.1038/srep38526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luo D, Fan X, Ma C, et al. A study on the effect of neurogenesis and regulation of GSK3beta/PP2A expression in acupuncture treatment of neural functional damage caused by focal ischemia in MCAO rats. Evid Based Complement Alternat Med. 2014;2014:962343. doi: 10.1155/2014/962343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fogel O, Bugge Tinggaard A, Fagny M, et al. Deregulation of microRNA expression in monocytes and CD4+ T lymphocytes from patients with axial spondyloarthritis. Arthritis Res Ther. 2019;21:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maeda Y, Farina NH, Matzelle MM, et al. Synovium-derived microRNAs regulate bone pathways in rheumatoid arthritis. J Bone Miner Res. 2017;32:461–472. doi: 10.1002/jbmr.3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito T, Fukai A, Mabuchi A, et al. Transcriptional regulation of endochondral ossification by HIF-2α during skeletal growth and osteoarthritis development. Nat Med. 2010;16:678–686. [DOI] [PubMed] [Google Scholar]
- 52.Vaira S, Johnson T, Hirbe AC, et al. RelB is the NF-κB subunit downstream of NIK responsible for osteoclast differentiation. Proc Nat Acad Sci USA. 2008;105:3897–3902. doi: 10.1073/pnas.0708576105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vaira S, Alhawagri M, Anwisye I, Kitaura H, Faccio R, Novack DV. RelA/p65 promotes osteoclast differentiation by blocking a RANKL-induced apoptotic JNK pathway in mice. J Clin Invest. 2008;118:2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clohisy JC, Yamanaka Y, Faccio R, Abu-Amer Y. Inhibition of IKK activation, through sequestering NEMO, blocks PMMA-induced osteoclastogenesis and calvarial inflammatory osteolysis. J Orthop Res. 2006;24(7):1358–1365. doi: 10.1002/jor.20184 [DOI] [PubMed] [Google Scholar]
- 55.Kobayashi H, Chang SH, Mori D, et al. Biphasic regulation of chondrocytes by Rela through induction of anti-apoptotic and catabolic target genes. Nat Commun. 2016;7(1):13336. doi: 10.1038/ncomms13336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Caron MM, Emans PJ, Surtel DA, et al. Activation of NF-κB/p65 facilitates early chondrogenic differentiation during endochondral ossification. PLoS One. 2012;7:e33467. doi: 10.1371/journal.pone.0033467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lianxu C, Hongti J, Changlong Y. NF-κBp65-specific siRNA inhibits expression of genes of COX-2, NOS-2 and MMP-9 in rat IL-1β-induced and TNF-α-induced chondrocytes. Osteoarthr Cartil. 2006;14:367–376. doi: 10.1016/j.joca.2005.10.009 [DOI] [PubMed] [Google Scholar]
- 58.Knudsen AR, Kannerup AS, Dich R, et al. Expression of genes involved in rat liver angiogenesis after ischaemia and reperfusion: effects of ischaemic pre- and post-conditioning. HPB. 2010;12:554–560. doi: 10.1111/j.1477-2574.2010.00215.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li G, Li L, Qi S, Wu J, Ming C. MicroRNA-3200-5p promotes osteosarcoma cell invasion via suppression of BRMS1. Mol Cells. 2018;41:523–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Berendam SJ, Koeppel AF, Godfrey NR, et al. Comparative transcriptomic analysis identifies a range of immunologically related functional elaborations of lymph node associated lymphatic and blood endothelial cells. Front Immunol. 2019;10:816. doi: 10.3389/fimmu.2019.00816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.You S, Cho C, Lee I, et al. A systems approach to rheumatoid arthritis. PLoS One. 2012;7(12):e51508. doi: 10.1371/journal.pone.0051508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kochan-Jamrozy K, Króliczewski J, Moszyńska A, et al. miRNA networks modulate human endothelial cell adaptation to cyclic hypoxia. Cell Signal. 2019;54:150–160. doi: 10.1016/j.cellsig.2018.11.020 [DOI] [PubMed] [Google Scholar]
- 63.Audrey MA, Nobish V, Louisa W, et al. Differentially expressed microRNAs in chondrocytes from distinct regions of developing human cartilage. PLoS One. 2013;8:e75012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Linares GR, Brommage R, Powell DR, et al. Claudin 18 is a novel negative regulator of bone resorption and osteoclast differentiation. J Bone Miner Res. 2012;27:1553–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wongdee K, Pandaranandaka J, Teerapornpuntakit J, et al. Osteoblasts express claudins and tight junction-associated proteins. Histochem Cell Biol. 2008;130:79–90. doi: 10.1007/s00418-008-0419-6 [DOI] [PubMed] [Google Scholar]
- 66.Zhang X, Wang X, Wang A, et al. CLDN10 promotes a malignant phenotype of osteosarcoma cells via JAK1/Stat1 signaling. J Cell Commun Signal. 2019;13(3):395–405. doi: 10.1007/s12079-019-00509-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Carsten S, Henrike M, Edward R, Ann B, Thomas F, Kenneth W. Glycogen-Synthase Kinase3beta/beta-catenin axis promotes angiogenesis through activation of vascular endothelial growth factor signaling in endothelial cells. Circ Res. 2005;96:308–318. [DOI] [PubMed] [Google Scholar]
- 68.Kaga S, Zhan L, Altaf E, Maulik N. Glycogen synthase kinase-3β/β-catenin promotes angiogenic and anti-apoptotic signaling through the induction of VEGF, Bcl-2 and survivin expression in rat ischemic preconditioned myocardium. J Mol Cell Cardiol. 2006;40(1):138–147. doi: 10.1016/j.yjmcc.2005.09.009 [DOI] [PubMed] [Google Scholar]
- 69.de Sousa Rabelo F, da Mota LM, Lima RA, et al. The Wnt signaling pathway and rheumatoid arthritis. Autoimmun Rev. 2010;9:207–210. doi: 10.1016/j.autrev.2009.08.003 [DOI] [PubMed] [Google Scholar]
- 70.Zhang C, Zou YL, Ma J, Dang XQ, Wang KZ. Apoptosis associated with Wnt/β-catenin pathway leads to steroid-induced avascular necrosis of femoral head. BMC Musculoskelet Disord. 2015;16. doi: 10.1186/s12891-015-0606-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma AR, Jagga S, Lee SS, Nam JS. Interplay between cartilage and subchondral bone contributing to pathogenesis of osteoarthritis. Int J Mol Sci. 2013;14:19805–19830. doi: 10.3390/ijms141019805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adapala NS, Kim HKW. Comprehensive genome-wide transcriptomic analysis of immature articular cartilage following ischemic osteonecrosis of the femoral head in piglets. PloS one. 2016;11:e0153174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bach TL, Barsigian C, Chalupowicz DG, et al. VE-cadherin mediates endothelial cell capillary tube formation in fibrin and collagen gels1. Exp Cell Res. 1998;238:324–334. doi: 10.1006/excr.1997.3844 [DOI] [PubMed] [Google Scholar]
- 74.Barreiro O, Yanez-Mo M, Serrador JM, et al. Dynamic interaction of VCAM-1 and ICAM-1 with moesin and ezrin in a novel endothelial docking structure for adherent leukocytes. J Cell Biol. 2002;157:1233–1245. doi: 10.1083/jcb.200112126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bazzoni G. The JAM family of junctional adhesion molecules. Curr Opin Cell Biol. 2003;15:525–530. doi: 10.1016/S0955-0674(03)00104-2 [DOI] [PubMed] [Google Scholar]
- 76.Huang D. Extracellular matrix-cell interactions and chondrogenesis. Clin Orthop Relat Res. 1977;123:169–176. [PubMed] [Google Scholar]
- 77.Arner EC, Tortorella MD. Signal transduction through chondrocyte integrin receptors induces matrix metalloproteinase synthesis and synergizes with interleukin-1. Arthritis Rheum. 1995;38:1304–1314. doi: 10.1002/art.1780380919 [DOI] [PubMed] [Google Scholar]
- 78.Hou C, Zhang Z, Zhang Z, et al. Presence and function of microRNA-92a in chondrogenic ATDC5 and adipose-derived mesenchymal stem cells. Mol Med Rep. 2015;12:4877–4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhe L, Lin Y. Identification of potential crucial genes associated with steroid-induced necrosis of femoral head based on gene expression profile. Gene. 2017;627:322. doi: 10.1016/j.gene.2017.05.026 [DOI] [PubMed] [Google Scholar]
- 80.Yuan C, Cai J. Time-series expression profile analysis of fracture healing in young and old mice. Mol Med Rep. 2017;16:4529–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Klotzsche-von Ameln A, Cremer S, Hoffmann J, et al. Endogenous developmental endothelial locus-1 limits ischemia-related angiogenesis by blocking inflammation. Thromb Haemost. 2017;117(6):1150. doi: 10.1160/TH16-05-0354 [DOI] [PMC free article] [PubMed] [Google Scholar]