Abstract
Ticks require bacterial symbionts for the provision of necessary compounds that are absent in their hematophagous diet. Such symbionts are frequently vertically transmitted and, most commonly, belong to the Coxiella genus, which also includes the human pathogen Coxiella burnetii. This genus can be divided in four main clades, presenting partial but incomplete cocladogenesis with the tick hosts. Here, we report the genome sequence of a novel Coxiella, endosymbiont of the African tick Amblyomma nuttalli, and the ensuing comparative analyses. Its size (∼1 Mb) is intermediate between symbionts of Rhipicephalus species and other Amblyomma species. Phylogenetic analyses show that the novel sequence is the first genome of the B clade, the only one for which no genomes were previously available. Accordingly, it allows to draw an enhanced scenario of the evolution of the genus, one of parallel genome reduction of different endosymbiont lineages, which are now at different stages of reduction from a more versatile ancestor. Gene content comparison allows to infer that the ancestor could be reminiscent of C. burnetii. Interestingly, the convergent loss of mismatch repair could have been a major driver of such reductive evolution. Predicted metabolic profiles are rather homogenous among Coxiella endosymbionts, in particular vitamin biosynthesis, consistently with a host-supportive role. Concurrently, similarities among Coxiella endosymbionts according to host genus and despite phylogenetic unrelatedness hint at possible host-dependent effects.
Keywords: Amblyomma nuttalli, Coxiella-like endosymbiont, genome reduction, phylogeny, comparative genomics, symbiont evolution
Significance
The genus Coxiella includes the pathogen Coxiella burnetii and widespread nutritional mutualists in ticks. Current knowledge on their evolution is hampered by the limited genomic resources available. Here, we provide the first genome sequence of a Coxiella endosymbiont of clade B, the only clade for which none was available. These data allow to infer an evolutionary scenario of parallel genome reduction among Coxiella endosymbionts, with similar constraints, leading to selective retention of biosynthetic pathways beneficial for the host. The combined predicted functional capabilities of the symbionts appear to be a subset of those of C. burnetii. Accordingly, this pathogen could be closer to an ancestral state of the endosymbionts, rather than being derived from an endosymbiotic ancestor, as previously hypothesized.
Introduction
Mutualistic associations with bacteria are widespread and can allow eukaryotes to colonize novel ecological niches (Bennett and Moran 2015). In arthropods, intracellular and maternally transmitted bacterial mutualists are particularly common (Douglas 1998). A typical role of these endosymbionts is providing essential nutrients that are absent in the host's diet (Sandström and Moran 1999). The classical example is that of sap-feeding insects, such as aphids, acquiring essential amino acids, and vitamins from intracellular bacteria (Wernegreen 2012).
Similarly, blood-feeding arthropods also rely on an incomplete source of nutrients and harbor bacterial mutualists. For instance, “Candidatus Riesia” bacteria supply their host, the human lice, with several compounds, in particular B vitamins (Sasaki-Fukatsu et al. 2006; Boyd et al. 2017). A better understanding of these symbioses and their role can foster the development of control strategies for hematophagous arthropod vectors of diseases (Zindel et al. 2011). Ticks in particular are extensively studied due to their prominent role of disease vectors for humans and domestic animals (Dantas-Torres et al. 2012). The most important pathogens vectored by ticks include Borrelia, the tick-borne encephalitis virus, Coxiella burnetii, and multiple Rickettsiales bacteria (Kernif et al. 2016).
Most tick species present at least one bacterial endosymbiont, and many species more than one (Cafiso et al. 2016; Moutailler et al. 2016; Duron et al. 2017). Transmission of symbionts is mainly dependent on maternal inheritance through transovarial transmission, but horizontal transfer, possibly during cofeeding, also plays a role. Indeed, closely related tick species, and even different individuals of the same species, may harbor different sets of bacterial symbionts (Duron et al. 2017).
Experimental evidence indicates a major role of such symbionts in tick physiology, as their depletion resulted in impaired growth and reproduction in multiple species belonging to different genera (Zhong et al. 2007; Guizzo et al. 2017; Ben-Yosef et al. 2020). Interestingly, in Ornithodoros moubata such developmental defects were rescued by supplementation with B vitamins (Duron et al. 2018). Collectively, these data suggest a common role of different, phylogenetically unrelated, symbionts as B vitamins providers. Other parallel roles have been hypothesized, including protection from pathogens, provision of energy, support for feeding, protection against oxidative and osmotic stress, and waste molecule recycling (Buysse et al. 2019; Olivieri et al. 2019).
Most of characterized tick symbionts are affiliated to Coxiella (Gammaproteobacteria) (Duron et al. 2017), and will now be abbreviated as CEs (Coxiella endosymbionts). Besides many CEs of ticks, this genus includes the tick-borne pathogen C. burnetii, causative agent of the Q-fever (Angelakis and Raoult 2010). It has been hypothesized that the latter arose from a CE ancestor, but the origin of its virulence is unclear (Duron et al. 2015).
Currently, a total of seven genomes of CEs are available (Gottlieb et al. 2015; Smith et al. 2015; Guizzo et al. 2017; Ramaiah and Dasch 2018; Tsementzi et al. 2018). These present the hallmarks of genome reduction in bacterial symbionts (McCutcheon and Moran 2012), but at different stages. CEs of Rhipicephalus spp. have comparatively larger genomes (1.2–1.7 Mb) with a higher number of pseudogenes (Gottlieb et al. 2015; Guizzo et al. 2017; Ramaiah and Dasch 2018; Tsementzi et al. 2018), characteristics of relatively recent symbioses with ongoing genome reduction. Conversely, the CEs of Amblyomma ticks have more streamlined genomes (0.6–0.7 Mb), with high gene density and low number of mobile elements (Smith et al. 2015), typical of a later stage of symbiosis.
Nevertheless, all CEs possess the pathways for production of many B vitamins, consistently with the hypothesized role and the general trend observed in nutritional symbionts, which retain some host-supportive pathways even in case of severe genome reduction (Nakabachi et al. 2006; López-Madrigal et al. 2011).
Four phylogenetic clades (A–D) were identified in the genus Coxiella, exhibiting only partial congruence with their hosts’ phylogeny (partial cocladogenesis). Clade C displays a good degree of cocladogenesis with Rhipicephalus hosts, whereas CEs of two unrelated clades (B and D) were found in Amblyomma hosts (Duron et al. 2015). Interestingly, the known clade B Amblyomma hosts came from the African continent, whereas clade D hosts are American (Duron et al. 2015; Binetruy et al. 2020). Currently, genomes of representatives of three of the four Coxiella clades have been sequenced, that is, CEs from clades C and D, and C. burnetii from clade A.
Here, we sequenced the genome of a novel CE of Amblyomma nuttalli belonging to the fourth clade (B), providing a basis for comparative analyses aimed to improve the understanding of the diversity and evolution of CEs.
Materials and Methods
An adult female of A. nuttalli was collected from a white rhinoceros in the Masai Mara National Reserve, Kenya in February 2016. The tick was morphologically identified following standard taxonomic keys (Theiler and Salisbury 1959) and subjected to DNA extraction, using NucleoSpin Tissue Kit (Macherey Nagel, Duren, Germany), according to the manufacturer’s instructions. DNA was subjected to Illumina HiSeq X by Admera Health (South Plainfield, NJ, USA) using a Nextera XT library, obtaining 2,73,511,224 150-nt paired-end reads.
The reads were assembled using SPAdes (3.6.0), and subjected to a modified version of the blobology pipeline (Kumar et al. 2013), in order to select only the symbiont sequences (for a complete description of the process, see Castelli et al. [2019]). Briefly, we selected contigs with a log10 coverage >2.5, extracted and reassembled separately the reads mapping on those contigs (Langmead and Salzberg 2012), and extensively revised manually the results. Finally, in order to close the assembly, PCR reactions were performed with primers designed in proximity of contig ends, product were sequenced and results processed, as described previously (Castelli et al. 2019),
The completeness level of the genome was confronted with all published CE genomes using BUSCO, using the Gammaproteobacteria lineage data set (Seppey et al. 2019).
Genome annotation was performed using Prokka 1.11 (Seemann 2014) and manually curated by inspecting blastp hits of predicted ORFs on NCBI nr, Uniprot, and Legionellales sequences.
ISEScan (Xie and Tang 2017) and ISfinder (Siguier et al. 2006) were used to identify insertion sequences and PHASTER (Arndt et al. 2016) for prophages. Pseudogene prediction on the novel genome, all published CE genomes, and representative C. burnetii genomes (table 1) was performed using Pseudofinder (Syberg-Olsen et al. 2020).
Table 1.
List of Genomes Included in the Analyses, with Their Accession Numbers, and for Coxiella Total Genome Size, Coding DNA Size, and GC Content
| Organism | Accession | Genome Length (bp) | Coding DNA (bp) | CG Content |
|---|---|---|---|---|
| Coxiella burnetii RSA 493 | GCA_000007765.2 | 2,032,807 | 1601033 | 42.6% |
| Coxiella burnetii Dugway 5J108-111 | GCA_000017105.1 | 2,212,937 | 1769726 | 42.4% |
| Coxiella endosymbiont of Amblyomma nuttalli — | 1,003,026 | 660534 | 35.9% | |
| Coxiella endosymbiont of Amblyomma americanum 1 | GCA_000815025.1 | 656,901 | 568842 | 34.6% |
| Coxiella endosymbiont of Amblyomma americanum 2 | GCA_002850495.1 | 656,933 | 566434 | 34.6% |
| Coxiella endosymbiont of Amblyomma sculptum | GCA_009883795.1 | 622,921 | 539589 | 38.1% |
| Coxiella endosymbiont of Rhipicephalus microplus 1 | GCA_002871095.1 | 1,194,772 | 683985 | 32.6% |
| Coxiella endosymbiont of Rhipicephalus microplus 2 | GCA_002930125.1 | 1,296,467 | 623102 | 31.7% |
| Coxiella endosymbiont of Rhipicephalus sanguineus | GCA_002804145.1 | 1,715,759 | 907053 | 38.0% |
| Coxiella endosymbiont of Rhipicephalus turanicus | GCA_001077715.1 | 1,733,840 | 917489 | 38.2% |
| Aquicella siphonis | GCA_902459485.1 | |||
| “Candidatus Berkiella cookevillensis” | GCA_001431315.1 | |||
| Legionella pneumophila | GCA_000008485.1 | |||
| Environmental Coxiellaceae bacterium | GCA_001795425.1 | |||
| Rickettsiella grylli | GCA_000168295.1 | |||
| Rickettsiella isopodorum | GCA_001881495.1 | |||
| Rickettsiella viridis | GCA_003966755.1 | |||
| Tatlockia micdadei | GCA_000953635.1 |
Note.—The newly obtained genome is highlighted in bold.
COGs were predicted on the same data set using the NCBI pipeline (Galperin et al. 2015) on validated genes (i.e., ORFs excluding predicted pseudogenes). COG repertoires were used for comparative analyses. Metabolic pathways were manually reconstructed employing the BioCyc database reference (Karp 2019).
Two data sets were used for phylogeny. The first one involved a wide taxonomic sampling, analyzed through MLST (multilocus sequence typing) as in Duron et al. (2015), thus employing five genes and 96 organisms (published data set plus all available CEs).
The second set was analyzed by using a phylogenomic approach, and included the previous selection of Coxiella genomes, a representative selection of Coxiellaceae, including 1 MAG (metagenome assembled genome), and two other Legionellales as outgroup table 1. Using OrthoFinder (2.3.3) (Emms and Kelly 2019), 213 single copy conserved orthologs were identified.
Then, for the two sets, respectively the nucleotide and protein sequences of each single gene were aligned separately using Muscle (Edgar 2004), polished with Gblocks (Talavera and Castresana 2007), and finally concatenated (3,118 and 59,256 total positions, respectively).
For each set, we inferred the best model (GTR + I + G and LG + I + G, respectively) using modeltest-ng 0.1.3 (Darriba et al. 2019), built a maximum likelihood tree with RAxML 8.2.4 (Stamatakis 2014) with 1,000 bootstrap pseudoreplicates.
Results and Discussion
The obtained genome assembly of the CE of A. nuttalli has a total length of 1,003,026 bp and a GC content of 35.9%. BUSCO completeness score was 79.2%, similar to other CEs. A total of 764 genes were found, including 44 RNA genes (including 38 tRNAs and 3 rRNAs). Among these, we identified 702 functional CDSs and 18 pseudogenes, accounting for a total of 660,777 bp (65.8%) coding (including structural RNA genes). Neither prophages nor ISs were found.
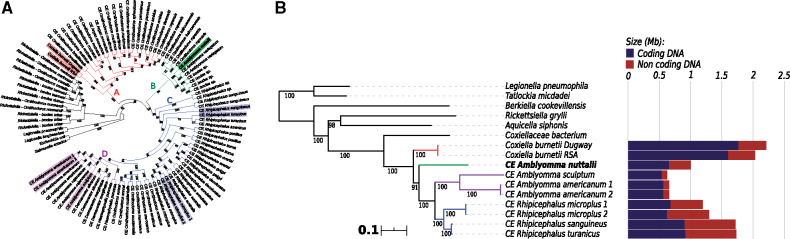
MLST phylogeny provided an overall consistent topology with most previous studies (Duron et al. 2015; Gottlieb et al. 2015), in particular for the major Coxiella clades and their relationships (clade A earliest divergent, clade B sister group of clades C + D), with moderate to high support (fig. 1A). Consistently with what could be expected based on geographical origin, the CE of A. nuttalli lies in the clade B, composed by CEs of African ticks.
Fig. 1.
(A) Maximum likelihood MLST phylogenetic tree of Coxiella and other Legionellales, and (B) maximum likelihood phylogenomic tree of Coxiellaceae. In (A, B), the four major Coxiella clades are evidenced by different colors, numbers on branches stand for bootstrap support after 1,000 pseudoreplicates, CE stands for Coxiella endosymbiont, and respective host organisms are indicated. In (A), symbionts with available genomes are highlighted. In (B), scale bar stands for estimated proportional sequence divergence, and the bar plot on the right shows the respective coding (blue) and noncoding (red) genome sizes.
For the available Coxiella genomes, phylogenomic analysis showed the same relations of the MLST phylogeny (fig. 1B). Interestingly, branch lengths are proportional to the degree of genome reduction within the Coxiella genus table 1, consistently with previous analyses (Duron et al. 2015; Gottlieb et al. 2015). This would indicate a higher evolutionary rate in smaller genomes, as predicted for obligate symbionts by genome reduction models (McCutcheon and Moran 2012).
Specifically, C. burnetii, a pathogen capable of living in different environments and hosts (Angelakis and Raoult 2010), presents the largest genome (2 Mb) and, with a high coding density (79.9%), the highest amount of coding DNA (1.8 Mb). All the host-restricted CEs have smaller genomes, with similar sizes within each clade: 1.2–1.7 Mb for CEs of Rhipicephalus (clade C), 1.0 Mb for the novel CE of A. nuttalli (clade B), and 0.6–0.7 Mb for CEs of other Amblyomma species (clade D). However, the degree of genome reduction does not correlate with the phylogenetic branching pattern between clades, in particular the CEs with more reduced genomes (clades B, D) do not form a single monophyletic group (fig. 1B). Accordingly, considering also the novel CE of A. nuttalli, a scenario with parallel independent genome reduction in genus Coxiella (at least for monophyletic CEs of B–D clades together) appears plausible.
Interestingly, most size variation resides in the noncoding genome (from 88 kb to 816 kb), whereas the length of the functional coding genome is overall less variable in CEs, ranging from 540 kb of CE of Amblyomma sculptum (Clade D), to 917 kb of CE of Rhipicephalus turanicus (Clade C). These features as well are consistent with a recent and still ongoing parallel genome reduction of CEs under similar constraints, possibly due to an equivalent role for the host. Accordingly, the CE of A. nuttalli has traits of a relatively long-term obligate symbiont, at an intermediate stage among CEs for its genome size and coding density (table 1), and having no predicted mobile elements.
The functional capabilities, as represented by COG repertoires, are consistent with the observed pattern of genome reduction (supplementary fig. 1A, Supplementary Material online). Coxiella burnetii is the richest in COGs for all functional categories. CEs have lower COGs numbers, roughly proportional to the respective coding genomes. Many core functions are highly conserved, such as translation machinery (J), coenzyme (H), and nucleotide (F) metabolism, energy production (C), protein modification and chaperones (O), lipid synthesis (I), cell cycle regulation (D). This is consistent with their expected major role for bacterial survival and/or host-support (coenzymes). On the other side, all CEs are more pronouncedly depleted in accessory and regulative functions, including poorly characterized ones (R and S), signal transduction (T), secondary metabolite metabolism (Q), cell motility (N), secretion systems (U), extracellular and defense structures (W and V). Such functionalities are probably less important in strictly host-associated bacteria. Notable is the case of type IV secretion, probably ancestral in Legionellales (Hugoson et al. 2019) and an important virulence factor in C. burnetii (Luedtke et al. 2017), but absent in all CEs. Some functions display gradients of conservation along the genome size, for example, membrane structure biogenesis (M), which correlates with decrease in lipopolysaccharide complexity, whereas peptidoglycan synthesis is conserved.
Such scenario is reflected at the level of single COGs (supplementary fig. 1B, Supplementary Material online), as C. burnetii (clade A) presents the highest number of unique COGs. CEs of all clades are a substantial subset of C. burnetii, which lacks only 60 of the 1,207 total COGs in the data set. Similar observations can be drawn among progressively more reduced CE clades, with the CE of A. nuttalli at an intermediate level between clade B and D CEs.
Few lineage-specific peculiarities were found, and the functional significance of many of these is unclear, for example, the A. nuttalli-specific COGs display redundant or poorly characterized functions (supplementary table 1, Supplementary Material online). However, some relevant variability was also observed (supplementary table 2, Supplementary Material online). For instance, the symbiont of A. nuttalli, while retaining the predicted capability to perform glycolysis, Krebs cycle and oxidative phosphorylation, does not possess a conventional citrate synthase, differently from other CEs. Instead, it presents a 2-methylcitrate synthase, which may also catalyze the same reaction (Patton et al. 1993). As C. burnetii has both genes, this partial redundancy could have been ancestral, and independently lost in different CE lineages.
Similarly to other Coxiella, although biosynthetic abilities for amino acids are scarce in the CE of A. nuttalli, those for vitamins and cofactors (H) are, as expected, abundant and highly conserved. These include in particular genes for the synthesis for riboflavin (B2), pantothenate (B5) and its derivative CoA, pyridoxine (B6), folic acid (B9), and biotin. For biotin (and lipid) synthesis, missing FabI functionality is possibly replaced by FabV (Massengo-Tiassé and Cronan 2008). Similarly to the other CEs of Amblyomma, it also displays complete biosynthetic pathways for thiamine (B1) and NAD (B3). Interestingly, despite overall larger functional capabilities, symbionts of Rhipicephalus retain only partial pathways. Such differences can be possibly explained by the presence of not yet identified transporters and/or noncanonical enzymes (Gottlieb et al. 2015), or by different, not yet clarified, metabolic requirements of the tick hosts.
A similar scenario may hold for nucleotide metabolism (F), which is also more reduced in the CEs of Rhipicephalus, lacking the initial path for the synthesis of purines.
Consistently with the symbiotic condition (McCutcheon and Moran 2012), CEs are depleted in DNA repair abilities (L), with lineage-specific features (supplementary table 2, Supplementary Material online). For example both CEs of R. microplus are devoid of the RecFOR pathway and RecA, involved in homologous recombination (Kuzminov 1999). Interestingly, the MutSL pathway is fully absent in the smaller genomes of CEs of Amblyomma of clades B and D, but complete in C. burnetii (clade A) and in most members of clade C. Among those, the exception is the CE of R. microplus “2,” found to have a full mutL gene, but a truncated mutS pseudogene, probably retaining only partial or no functionality. Parsimoniously, we can identify at least three multiple convergent losses of this pathway among CEs (complete in the clades B and D, and still ongoing within clade C). Considering the strong correlation with the degree of genome reduction, it is reasonable to hypothesize that this loss may have had a major evolutionary impact, possibly directly causing increased mutation rates (Schofield and Hsieh 2003), and eventually resulting in accelerated and more pronounced genome reduction. Thus, the seminal speculations by Gottlieb et al. (2015) on a smaller data set are reinforced. This effect would be particularly evident from the differences in coding size and functional categories among the two closely related CEs of R. microplus (fig. 1B).
Conclusions
The novel sequence of CE of A. nuttalli expands the available diversity of Coxiella genomes, being the first obtained from clade B. Despite reduced genome size, biosynthetic pathways for vitamins appear to be conserved, as in other CEs, supporting a role of CEs in dietary supplementation of these compounds to the hosts. At same time, some variations were found in vitamin and purine synthesis, possibly dependent on the host species.
Combining phylogenetic and genomic data, an evolutionary scenario of parallel genome reduction with analogous constraints among CEs (clades B–D) can be hypothesized. Consistently with previous observations (Gottlieb et al. 2015), the convergent loss of MutSL could have been a driver of such reduction, with the CE of A. nuttalli representing an intermediate level between clade C and D, and all CEs substantially being a subset of C. burnetii. Accordingly, and differently from previous views (Duron et al. 2015), CEs could have evolved from a more versatile C. burnetii-like ancestor, analogously to other unrelated symbionts (Taylor et al. 2005; Gerhart et al. 2016). In-depth evolutionary genomic analyses, in particular involving novel sequences of representatives of clade A other than C. burnetii, may provide further insights.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
This study was supported by the Human Frontier Science Programme (Grant No. RGY0075/2017) to D.S. and by the Italian Ministry of Education, University and Research (MIUR): Dipartimenti di Eccellenza Programme (2018–2022)—Department of Biology and Biotechnology “L. Spallanzani,” University of Pavia to D.S.
Data Availability
The data underlying this article are available in the NCBI GenBank Database at ncbi.nlm.nih.gov/, and can be accessed with CP064834 (CE genome sequence) and with SRR12168527 (total reads in SRA).
Literature Cited
- Arndt D, et al. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44(W1):W16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett GM, Moran NA.. 2015. Heritable symbiosis: the advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 112(33):10169–10176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Yosef M, et al. 2020. Coxiella-like endosymbiont of Rhipicephalus sanguineus is required for physiological processes during ontogeny. Front Microbiol. 11:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binetruy F, et al. 2020. Microbial community structure reveals instability of nutritional symbiosis during the evolutionary radiation of Amblyomma ticks. Mol Ecol. 29(5):1016–1029. [DOI] [PubMed] [Google Scholar]
- Boyd BM, et al. 2017. Primates, lice and bacteria: speciation and genome evolution in the symbionts of hominid lice. Mol Biol Evol. 34(7):1743–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buysse M, Plantard O, McCoy KD, Duron O, Menard C.. 2019. Tissue localization of Coxiella-like endosymbionts in three European tick species through fluorescence in situ hybridization. Ticks Tick-Borne Dis. 10(4):798–804. [DOI] [PubMed] [Google Scholar]
- Cafiso A, et al. 2016. Molecular screening for Midichloria in hard and soft ticks reveals variable prevalence levels and bacterial loads in different tick species. Ticks Tick-Borne Dis. 7(6):1186–1192. [DOI] [PubMed] [Google Scholar]
- Castelli M, et al. 2019. Deianiraea, an extracellular bacterium associated with the ciliate Paramecium, suggests an alternative scenario for the evolution of Rickettsiales. ISME J. 13(9):2280–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas-Torres F, Chomel BB, Otranto D.. 2012. Ticks and tick-borne diseases: a one health perspective. Trends Parasitol. 28(10):437–446. [DOI] [PubMed] [Google Scholar]
- Darriba D, et al. 2019. ModelTest-NG: a new and scalable tool for the selection of DNA and protein evolutionary models. Mol Biol Evol. 37(1):291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE. 1998. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 43(1):17–37. [DOI] [PubMed] [Google Scholar]
- Duron O, et al. 2015. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. 11(5):e1004892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duron O, et al. 2017. Evolutionary changes in symbiont community structure in ticks. Mol Ecol. 26(11):2905–2921. [DOI] [PubMed] [Google Scholar]
- Duron O, et al. 2018. Tick-bacteria mutualism depends on B vitamin synthesis pathways. Curr Biol. 28(12):1896–1902.e5. [DOI] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2019. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin MY, , Makarova KS, , Wolf YI, , Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43(Database issue):D261–D269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart JG, Moses AS, Raghavan R.. 2016. A Francisella-like endosymbiont in the Gulf Coast tick evolved from a mammalian pathogen. Sci Rep. 6: Article number: 33670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlieb Y, Lalzar I, Klasson L.. 2015. Distinctive genome reduction rates revealed by genomic analyses of two Coxiella-like endosymbionts in ticks. Genome Biol Evol. 7(6):1779–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizzo MG, et al. 2017. A Coxiella mutualist symbiont is essential to the development of Rhipicephalus microplus. Sci Rep. 7. doi:10.1038/s41598-017-17309-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugoson E, Ammunét T, Guy L.. 2019. Host-adaptation in Legionellales is 2.4 Gya, coincident with eukaryogenesis. bioRxiv 852004. doi:10.1101/852004. [DOI] [PMC free article] [PubMed]
- Karp PD, et al. . 2019. The BioCyc collection of microbial genomes and metabolic pathways. Brief Bioinform. 20(4):1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kernif T, Leulmi H, Raoult D, Parola P.. 2016. Emerging tick-borne bacterial pathogens. Microbiol Spectr. 4(3). doi:10.1128/microbiolspec.EI10-0012-2016. [DOI] [PubMed] [Google Scholar]
- Kumar S, , Jones M, , Koutsovoulos G, , Clarke M, , Blaxter M. 2013. Blobology: exploring raw genome data for contaminants, symbionts and parasites using taxon-annotated GC-coverage plots. Front Genet. 4:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol Mol Biol Rev. 63(4):751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL.. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Madrigal S, Latorre A, Porcar M, Moya A, Gil R.. 2011. Complete genome sequence of “Candidatus Tremblaya princeps” strain PCVAL, an intriguing translational machine below the living-cell status. J Bacteriol. 193(19):5587–5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedtke BE, Mahapatra S, Lutter EI, Shaw EI.. 2017. The Coxiella burnetii type IVB secretion system (T4BSS) component DotA is released/secreted during infection of host cells and during in vitro growth in a T4BSS-dependent manner. Pathog Dis. 75:ftx047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massengo-Tiassé RP, Cronan JE.. 2008. Vibrio cholerae FabV defines a new class of enoyl-acyl carrier protein reductase. J Biol Chem. 283(3):1308–1316. [DOI] [PubMed] [Google Scholar]
- McCutcheon JP, Moran NA.. 2012. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 10(1):13–26. [DOI] [PubMed] [Google Scholar]
- Moutailler S, et al. 2016. Co-infection of ticks: the rule rather than the exception. PLoS Negl Trop Dis. 10:e0004539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, et al. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314(5797):267. [DOI] [PubMed] [Google Scholar]
- Olivieri E, et al. 2019. Tissue tropism and metabolic pathways of Midichloria mitochondrii suggest tissue-specific functions in the symbiosis with Ixodes ricinus. Ticks Tick-Borne Dis. 10(5):1070–1077. [DOI] [PubMed] [Google Scholar]
- Patton AJ, Hough DW, Towner P, Danson MJ.. 1993. Does Escherichia coli possess a second citrate synthase gene? Eur J Biochem. 214(1):75–81. [DOI] [PubMed] [Google Scholar]
- Ramaiah A, Dasch GA.. 2018. Genome sequence of Coxiella-like endosymbiont strain CLE-RmD, a bacterial agent in the cattle tick (Rhipicephalus microplus) Deutsch strain. Genome Announc. 6(13):e00003-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandström J, Moran N.. 1999. How nutritionally imbalanced is phloem sap for aphids? In: Simpson SJ, Mordue AJ, Hardie J, editors. Proceedings of the 10th International Symposium on Insect–Plant Relationships. Dordrecht (Netherlands: ): Series Entomologica Springer Netherlands; p. 203–210. doi:10.1007/978-94-017-1890-5_26. [Google Scholar]
- Sasaki-Fukatsu K, et al. 2006. Symbiotic bacteria associated with stomach discs of human lice. Appl Environ Microbiol. 72(11):7349–7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield MJ, Hsieh P.. 2003. DNA mismatch repair: molecular mechanisms and biological function. Annu Rev Microbiol. 57(1):579–608. [DOI] [PubMed] [Google Scholar]
- Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30(14):2068–2069. [DOI] [PubMed] [Google Scholar]
- Seppey M, Manni M, Zdobnov EM.. 2019. BUSCO: assessing genome assembly and annotation completeness. Methods Mol Biol. 1962:227–245. [DOI] [PubMed] [Google Scholar]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M.. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34(90001):D32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith TA, Driscoll T, Gillespie JJ, Raghavan R.. 2015. A Coxiella-like endosymbiont is a potential vitamin source for the lone star tick. Genome Biol Evol. 7(3):831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syberg-Olsen M, Garber A, Keeling P, McCutcheon J, Husnik F. 2020. Pseudofinder. GitHub repository:. https://github.com/filip-husnik/pseudofinder/. Accessed September 1, 2020.
- Talavera G, Castresana J.. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A.. 2005. Wolbachia bacterial endosymbionts of filarial nematodes. Adv Parasitol. 60:245–284. [DOI] [PubMed] [Google Scholar]
- Theiler G, Salisbury L.. 1959. Ticks in the South African zoological survey collection. Part IX. ‘The Amblyomma marmoreum group’. Onderstepoort J Vet Res. 28(1):77. [Google Scholar]
- Tsementzi D, Gordillo JC, Mahagna M, Gottlieb Y, Konstantinidis KT.. 2018. Comparison of closely related, uncultivated Coxiella tick endosymbiont population genomes reveals clues about the mechanisms of symbiosis. Environ Microbiol. 20(5):1751–1764. [DOI] [PubMed] [Google Scholar]
- Wernegreen JJ. 2012. Endosymbiosis. Curr Biol. 22(14):R555–R561. [DOI] [PubMed] [Google Scholar]
- Xie Z, Tang H.. 2017. ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33(21):3340–3347. [DOI] [PubMed] [Google Scholar]
- Zhong J, Jasinskas A, Barbour AG.. 2007. Antibiotic treatment of the tick vector Amblyomma americanum reduced reproductive fitness. PLoS ONE 2(5):e405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindel R, Gottlieb Y, Aebi A.. 2011. Arthropod symbioses: a neglected parameter in pest- and disease-control programmes. J Appl Ecol. 48(4):864–872. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the NCBI GenBank Database at ncbi.nlm.nih.gov/, and can be accessed with CP064834 (CE genome sequence) and with SRR12168527 (total reads in SRA).