Abstract
The careful evaluation of food is important for survival throughout the animal kingdom, and specialized chemoreceptors have evolved to recognize nutrients, minerals, acids, and many toxins. Vertebrate bitter taste, mediated by the taste receptor type 2 (T2R) family, warns against potentially toxic compounds. During evolution T2R receptors appear first in bony fish, but the functional properties of bony fish T2R receptors are mostly unknown. We performed a phylogenetic analysis showing the “living fossil” coelacanth (Latimeria chalumnae) and zebrafish (Danio rerio) to possess T2R repertoires typical for early-diverged species in the lobe-finned and the ray-finned clade, respectively. Receptors from these two species were selected for heterologous expression assays using a diverse panel of bitter substances. Remarkably, the ligand profile of the most basal coelacanth receptor, T2R01, is identical to that of its ortholog in zebrafish, consistent with functional conservation across >400 Myr of separate evolution. The second coelacanth receptor deorphaned, T2R02, is activated by steroid hormones and bile acids, evolutionary old molecules that are potentially endogenously synthesized agonists for extraoral T2Rs. For zebrafish, we report the presence of both specialized and promiscuous T2R receptors. Moreover, we identified an antagonist for one of the zebrafish receptors suggesting that bitter antagonism contributed to shape this receptor family throughout evolution.
Keywords: bitter taste receptor, bony fish, calcium mobilization assay
Significance
Vertebrate bitter taste is devoted to prevent ingestion of potentially harmful food items and mediated by specialized receptors (T2R), which during evolution first appeared in bony fish. Despite their founding role, the functional properties of bony fish T2Rs are largely unknown and hence, in addition to careful comparative genomic analyses of bony fish and several close relatives, we investigated the agonist profiles of selected coelacanth and zebrafish T2Rs. We demonstrate the functional conservation of T2R1 orthologs in both fish species despite over 400 Myr of separate evolution. Moreover, another ancestral coelacanth receptor recognizes bile acids and steroid hormones, providing a first cue on the putative food selection of this “living fossil.”
Introduction
Bitterness perception in vertebrates is believed to detect and reject potentially toxic compounds and is therefore considered to be important for survival (Behrens and Meyerhof 2018; Mura et al. 2018). On the contrary, mild to moderate bitterness is often tolerated by humans, if associated with food or beverages known to be safe for consumption (Drewnowski and Gomez-Carneros 2000). Moreover, similar to humans, who ingest bitter medicines when sick, some cases of animals seeking bitter plants for medicinal purposes or voluntarily sampling bitter solutions have been documented (Koshimizu et al. 1994; Villalba et al. 2014). Thus, complete bitter rejection represents an extreme case of vertebrates’ consummatory behaviors. Indeed, the fact that the correlation between bitterness and toxicity is rather weak (Glendinning 1994; Pawlik et al. 1995; Nissim et al. 2017) suggests that blunt rejection of bitter tasting food items may not be beneficial for survival under conditions of food scarcity. It should be noted here that the term bitter taste is a human construct, which is frequently used to characterize aversive taste behavior elicited by compounds tasting bitter to humans also in other vertebrates. In the incessant battle between prey and predator, many marine invertebrate organisms have evolved deterrent substances that elicit aversive taste behavior in fish (Pawlik et al. 1995; Marin et al. 1999).
Structurally highly diverse bitter substances are detected in and beyond the oral cavity by taste 2 receptor genes (Tas2r/T2R) (Adler et al. 2000; Chandrashekar et al. 2000; Matsunami et al. 2000), a subgroup of the large G protein-coupled receptor (GPCR) gene family (Fredriksson et al. 2003). Interestingly, the T2R receptor family is the “youngest” among the six major chemosensory receptor families of vertebrates. T2Rs are believed to have originated in bony vertebrates (Grus and Zhang 2009; Sharma et al. 2019), after the divergence of bony fish from cartilaginous fish, but before the split between actinopterygians (ray-finned lineage) and sarcopterygians (lobe-finned lineage), the former giving rise to teleost fish, and the latter to coelacanths, lungfish and tetrapods. In comparison, T1Rs and the four olfactory receptor families have already been found in jawless and/or cartilaginous fish (Grus and Zhang 2009; Shiriagin and Korsching 2019).
T2R genes of many species are under positive selective pressure throughout evolution (Shi et al. 2003), presumably to facilitate the recognition of relevant food sources in ever-changing environments, for different nutritional needs, and varying habitats. The evolution of T2Rs is also highly dynamic on the genomic scale, with frequent gene birth and gene death events resulting in large variations of T2R repertoire sizes within bony vertebrates (Shi et al. 2003), ranging from 0 to 3 receptor genes in dolphins (Jiang et al. 2012), galliform birds (Behrens, Korsching, et al. 2014), and some “modern” teleost fish (Shiriagin and Korsching 2019) to very large numbers in an amphibian species—western clawed frog—(Behrens, Korsching, et al. 2014) and in the “living fossil” coelacanth (Syed and Korsching 2014) (Latimeria chalumnae), the most basal extant species of the lobe-finned lineage. There is a very pronounced difference between the maximal size of T2R repertoires in tetrapods (often above 25 genes) compared with the ray-finned lineage (so far all but one T2R repertoire in the 1–7 gene range [Dong et al. 2009; Li and Zhang 2014; Shiriagin and Korsching 2019]). This difference appears to be a feature of the respective lineages and not related to water-to-land transition, because coelacanth, a lobe-finned fish, also possesses a large T2R repertoire, in fact the largest reported so far (Syed and Korsching 2014). Moreover, ray-finned fish with terrestrial life style such as several mudskipper species exhibit small T2R repertoires similar to other ray-finned fish such as zebrafish (Shiriagin and Korsching 2019) (fig. 1). A potential mechanism for the larger repertoires in the lobe-finned lineage could involve a higher density of class I transposons in T2R gene clusters coupled with positive selection, as has been suggested for coelacanth (Syed and Korsching 2014).
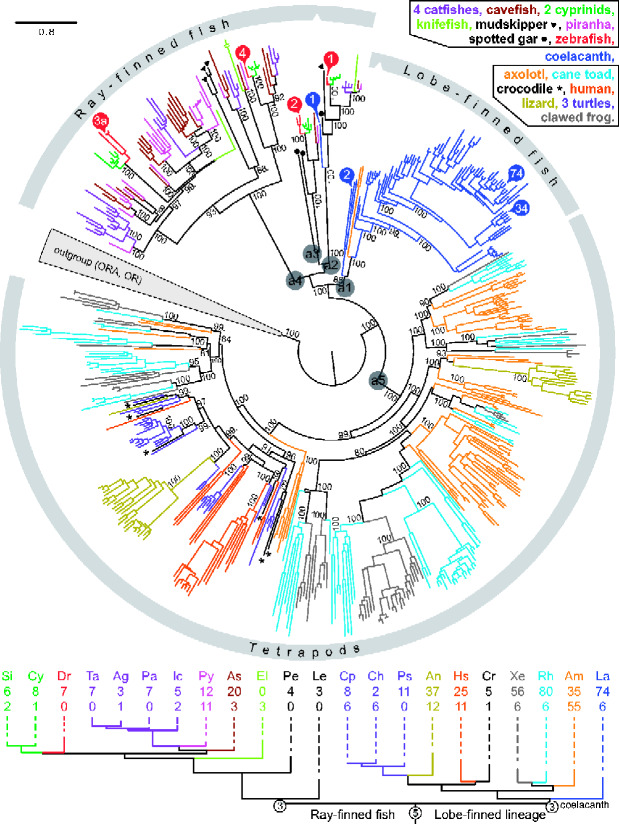
Fig. 1.
Phylogenetic tree visualizing vertebrate T2R evolution. Top panel, complete T2R repertoires from 22 species are shown color coded as indicated. Some colors refer to closely related species as shown. The tree was generated using a maximum-likelihood method, for details see Materials and Methods. Note the high branch support (shown as %) even in basal nodes. Most gene birth events occur late as visualized by a small degree of color mixing within the tree. Note the axolotl gene pair within the large coelacanth clade. Ancestral genes discussed in the main text are indicated by a1–a5. T2Rs selected for deorphanization are indicated by numbers in colored circles, red circles for zebrafish genes, blue for coelacanth, with numbers referring to the gene name, for example number 34 represents T2R34. The evolutionary relationships of the 22 species investigated are approximated in the bottom panel by a phylogenetic tree constructed with the respective species opsin orthologs, for details see Materials and Methods. Species color code same as above. Species are indicated by two letter abbreviations; Dr, Danio rerio, zebrafish; La, Latimeria chalumnae, coelacanth; for other names see supplementary figure S1, Supplementary Material online. The number of putatively functional T2Rs is given below the species acronym, second row of numbers refers to pseudogenes. Circled numbers refer to estimates for the number of ancestral genes.
How such large differences in T2R repertoire sizes between species are related to the chemical space of tastant molecules (taste space) accessible to the respective species is unknown. We have shown previously that small avian T2R gene repertoires do not per se predict a limited range of detectable bitter compounds because large average tuning breadths can compensate to some extent for low gene numbers (Behrens, Korsching, et al. 2014). In contrast, the knowledge about possible functions of T2Rs in bony fish is extremely limited, and nothing is known about the general tuning widths and specificities of bony fish T2Rs. So far, only a single orthologous pair of T2Rs from zebrafish (Danio rerio) and medaka (Oryzias latipes) has been shown to be expressed in taste buds and deorphaned with the synthetic bitter substance denatonium benzoate which elicits an aversive response in zebrafish (Oike et al. 2007). Nothing is known about coelacanth T2R ligands.
To understand the role of taste in the evolution of terrestrial life, it would be essential to know if and how bitter agonist profiles of coelacanth T2Rs are different from those of tetrapods on one hand and ray-finned fishes on the other hand. To what extent are there common characteristics of aquatic T2Rs (coelacanth vs. e.g., zebrafish)? Are coelacanth T2R characteristics carried onto land? Of particular interest are the response profiles of basal T2Rs within the coelacanth T2R clade, which may be expected to yield insights into the evolutionary origin of taste perception. In particular, it is an open question whether the oral function of T2R as bitter taste receptors or the extraoral function as endogenous metabolite sensor (Behrens and Meyerhof 2018) is the evolutionary most ancient function of T2Rs. To investigate these questions, we have cloned the two most basal coelacanth T2R genes as well as two genes representing two major clades within the large gene expansion observed in this species and subjected them to functional heterologous expression assays. We contrast these findings with an examination of ligands for four zebrafish T2R genes, one for each of the four subclades, thus representing the full-sequence divergence of the T2R repertoire.
Moreover, we wished to establish whether the large coelacanth T2R repertoire (80 genes) was typical for earlier-derived species of the lobe-finned lineage. Similarly, we investigated how representative the zebrafish T2R repertoire (seven genes) is for earlier-derived species in the ray-finned lineage, because a recent report (Shiriagin and Korsching 2019) showed a comparatively large T2R repertoire of 24 genes for Mexican cavefish (Astyanax mexicanus). The Mexican cavefish is an earlier-derived teleost fish as well, in comparison to neoteleost fish such as stickleback, medaka, and puffer fish (Betancur et al. 2013). We have mined several genomes from early-derived tetrapods and from teleost fish species that are phylogenetically close to both zebrafish and Mexican cavefish. We report that both coelacanth and zebrafish T2R repertoires exhibit typical repertoire size and divergence for early-derived members of the lobe-finned and ray-finned lineage, respectively, making them well suited to compare bitter agonist profiles between these two lineages, and in particular between the orthologous receptors, zebrafish T2R1 and coelacanth T2R01.
Results
Largely Species-Specific Gene Expansions Give Rise to Huge Amphibian T2R Families Rivaling the Size of the Coelacanth T2R Repertoire
We have delineated the T2R repertoires of two amphibian and three reptilian species, and find that both amphibian species (Ambystoma mexicanum, axolotl, and Rhinella marina, cane toad) possess T2R repertoires rivaling, and in the case of cane toad, slightly exceeding the coelacanth repertoire of intact genes (fig. 1 and supplementary fig. S1, files S1 and S2, Supplementary Material online). Axolotl exhibits an extremely high frequency of pseudogenes (61%) compared with both coelacanth (8% [Syed and Korsching 2014]) and cane toad (7%). Currently it is unclear whether this high percentage of pseudogenes reflects genome sequence quality or indeed an on-going major downsizing of the axolotl T2R repertoire. Our results show the previously published T2R repertoire (including both intact genes and pseudogenes) of the clawed frog (Xenopus tropicalis, 62 genes [Behrens, Korsching, et al. 2014]) to be in the lower range of amphibian repertoires. Except for lizard, the reptilian T2R repertoires are much smaller, ranging between 6 (Crocodylus porosus, salt water crocodile) and 8–14 genes (Chelonia mydas, green sea turtle; Chrysemys picta belli, western painted turtle) (see fig. 1 and supplementary fig. S1, files S1 and S2, Supplementary Material online). This range is similar to that reported for several avian species (Behrens, Korsching, et al. 2014). The phylogenetic tree suggests most of these differences to result from late gene birth events within species, because many species-specific subclades with up to 30 genes are present (fig. 1). Still, there are several higher-order subnodes that encompass mixed amphibian species, suggesting that the most recent common ancestor (MRCA) of amphibians already possessed several ancestral genes, consistent with the conclusions of an earlier study (Behrens, Korsching, et al. 2014). Taken together, very large T2R repertoires seem to be frequent in early-derived aquatic species of the lobe-finned lineage (coelacanth and amphibians), whereas later-derived species in this lineage (reptiles and birds) exhibit more modest family sizes in general (except for lizard).
Importantly, we observe two axolotl genes within the large coelacanth expansion, but no ortholog in the two frog species examined, consistent with a loss of the corresponding ancestral gene (a1 in fig. 1) in frogs, but not salamanders (axolotl). Because orthologs of ancestral gene a1 also have not been found in any reptilian genome, a second loss in the MRCA of reptiles has to be posited. Judging from the topology of the phylogenetic tree and the maximal branch support of the mixed amphibian/coelacanth clade (fig. 1), the origin of the a1 clade appears to be at least in the MRCA of the lobe-finned lineage. To validate the placement of the two axolotl genes within the a1 clade, we have searched for orthologs of axolotl T2R1 in all amphibian genomes currently available, which include two caecilians, an earlier-derived group of amphibians. We observed a close ortholog of axolotl T2R1 only in one of the caecilian species, Microcaecilia unicolor, which exhibits 43% identity at the amino acid level and presents a sister gene to axolotl T2R1 in phylogenetic analysis (supplementary fig. S2, Supplementary Material online). No coelacanth T2R01 ortholog was found in any amphibian genome, including the caecilians. Next, we compared the genomic environment of axolotl T2R1 and caecilian T2R1. We observed clear synteny between the genomic environment of axolotl T2R1 and the caecilian T2R1, despite the occurrence of four inversions within a 21-gene segment (supplementary fig. S3, Supplementary Material online), supporting the caecilian T2R1 as true ortholog of axolotl T2R1 (axolotl T2R2 is adjacent to T2R1 and a close ortholog, that is, most likely resulted from a species-specific gene duplication). Interestingly, the T2R gene is flanked by two inversion points. We then compared the genomic environment of caecilian T2R1 with that of all coelacanth T2R genes, and found (only) two coelacanth genes with any synteny, T2R01 and T2R04. As expected, synteny is less pronounced due to the much greater evolutionary distance. Three inversions, two translocations and several insertions/losses of genes are observed in the coelacanth contig containing T2R01 and T2R04 relative to the syntenic caecilian genomic sequence (supplementary fig. S3, Supplementary Material online). Nevertheless, 13 of in total 20 neighboring genes on the coelacanth contig are also neighbors for the caecilian T2R1, confirming the synteny, and thus validating the phylogenetic position of axolotl and caecilian T2R1 within the ancestral clade a1 (fig. 1, supplementary figs. S2 and S3, Supplementary Material online).
No synteny was found between the genomic environment of the most basal coelacanth T2R, T2R01 and any of the seven zebrafish T2Rs, including the direct ortholog Dr-T2R1, presumably due to the much larger evolutionary distance. Nevertheless, the topological position of coelacanth T2R01 with the ray-finned T2R1 clade is supported by a very long common branch reflecting a long evolutionary history of the ancestral gene before the split into ray-finned and lobe-finned lineage. Furthermore, branch support is maximal and consistent with an earlier study with less species (Syed and Korsching 2014). We conclude that the ancestral gene for T2R01 (a2, fig. 1) originated in the MRCA of the ray-finned and lobe-finned lineage, that is, close to the origin of bony vertebrates. Thus, we selected this coelacanth gene, plus the most basal gene in the mixed axolotl/coelacanth clade (a1, fig. 1), T2R02, for deorphanization, together with two genes deep inside the coelacanth-specific T2R expansion (fig. 1), the latter in an attempt to understand the tuning of species-specific T2R receptors.
The Zebrafish T2R Repertoire Is Representative for Early-Derived Teleost Fish
All newly analyzed teleost species, except the two cyprinids, possess a single T2R1 gene (fig. 1, supplementary table S1, Supplementary Material online), suggesting a high degree of conservation for this gene, consistent with a previous study with fewer species (Shiriagin and Korsching 2019). Both cyprinid species have recently undergone polyploidization—carp (Xu et al. 2014); Sinocyclocheilus grahami (Yang et al. 2016), thus the duplication in cyprinid T2R1 genes (fig. 1, supplementary fig. S1, Supplementary Material online) is due to polyploidization of the whole genome, and not indicative of the evolutionary dynamic of T2R1 itself.
With respect to repertoire size, the two other cyprinid fish show a very similar T2R repertoire compared with zebrafish, suggesting that the zebrafish T2R repertoire of seven genes is typical for cyprinids (fig. 1, supplementary file S2, Supplementary Material online). The slightly higher gene number of eight and nine genes, respectively, seems due to the recent polyploidization of those species (Xu et al. 2014; Yang et al. 2016). We also analyzed the available genomes for three neighboring phylogenetic orders, which together with the cyprinids constitute a larger group of early-derived teleosts (Otophysa). Four catfish species (order Siluriformes) possess 4–7 T2R genes per species (fig. 1, supplementary file S2, Supplementary Material online), again very similar to the zebrafish T2R repertoire. Interestingly, electric knifefish, a member of another neighboring order (Gymnotiformes), only has three pseudogenes, but not a single intact T2R gene, not even the slowly evolving and highly conserved T2R1 gene (fig. 1, supplementary file S2, Supplementary Material online). It is conceivable that electroreception might de-emphasize other sensory modalities, which could lead to loss of the respective receptor genes. The third order, Characiformes, contains Mexican cavefish with its unusually large T2R repertoire of 24 genes (Shiriagin and Korsching 2019). We report here that the genome of piranha, another Characiformes fish, has a similarly large repertoire of 23 genes, often as direct orthologs of the corresponding cavefish genes, but sometimes also in independent gene expansions (fig. 1). These large T2R repertoire sizes appear to be a specialized development within this particular order (Characiformes), whereas the much smaller zebrafish T2R repertoire size appears typical both for early-derived teleosts (Otophysa) and neoteleosts, cf. (Shiriagin and Korsching 2019). Furthermore, a high frequency of pseudogenes in the piranha T2R repertoire (48%) may show an instability of this enlarged repertoire, although technical reasons cannot be excluded at this point. It is noteworthy that for all species newly examined here, ancestral clades a2 and a3 show no local gene duplications at all, whereas the evolution in ancestral clade a4 is somewhat more dynamic (fig. 1), consistent with an earlier study with less species (Shiriagin and Korsching 2019). Taken together, zebrafish appears to be well suited to understand ligand spectra of a characteristic teleost T2R repertoire. To obtain a representative picture of the taste space accessible to zebrafish, we have selected four of in total seven zebrafish T2R for deorphanization attempts, using one gene from each of four subclades (fig. 1).
Heterologous Expression
In order to facilitate functional characterization of coelacanth and zebrafish T2Rs, the successful expression of the corresponding proteins in heterologous cells is imperative. We therefore utilized the hsv-epitope fused to the carboxyl end of the receptors for immunocytochemical detection in HEK 293T-Gα16gust44 cells, transiently transfected with the coelacanth bitter taste receptor constructs lcT2R01, lcT2R02, lcT2R34, and lcT2R74 as well as the zebrafish constructs drT2R1-4. The localization of the T2Rs in the cells was visualized using a mouse anti-hsv antiserum and the corresponding fluorescently labeled secondary antibody anti-mouseAlexa488 (green) together with a cell surface labeling procedure using biotinylated concanavalin A in combination with streptavidin-Alexa633 for fluorescence detection (red) and a fluorescent counterstaining of the cells’ nuclei with DAPI (blue). As seen in supplementary figure S4, Supplementary Material online, confocal laser scanning microscopy resulted in all cases in the detection of readily visible receptor proteins in a fraction of transfected cells. The specificity of the staining procedure was demonstrated by the absence of green signals in cells transfected with an empty expression vector. Counting the nuclear DAPI signals reflecting all cells and the green fluorescence signals indicative of cells expressing the receptors at detectable levels revealed that the fraction of cells expressing coelacanth T2Rs ranges from 22–49%, whereas for the zebrafish T2Rs the expression frequency was between 10% and 48%. Hence, the successful expression in heterologous cells allowed the next step in our study, the functional screening with bitter compounds.
For the screening of bony fish T2Rs, we selected 90 substances from our library of bitter compounds including a large variety of synthetic (21 compounds) as well as natural chemicals (69 compounds) with different structures (see supplementary table S2, Supplementary Material online). The majority of the natural compounds were plant metabolites (47 compounds), some represented animal (8 compounds) or bacterial (5 compounds) metabolites and some co-occur in plants as well as in animals (9 compounds). A considerable number of the natural bitter substances are present in terrestrial as well as in aquatic systems (22 compounds), whereas the majority occur dominantly in terrestrial systems (47 compounds). The bitter substances were applied in two concentrations, one maximum concentration (listed in supplementary table S2, Supplementary Material online) and one-third of that. The maximum concentration was chosen based on previous experiments (Meyerhof et al. 2010; Thalmann et al. 2013; Behrens, Korsching, et al. 2014; Lossow et al. 2016; Risso et al. 2017) taking substance solubilities as well as receptor dependence of compound-induced fluorescence changes into account. HEK 293T-Gα16gust44 cells transiently transfected with one of the four coelacanth or four zebrafish T2Rs were screened using an automated fluorometric imaging plate reader (FLIPRtetra), which automatically applied compounds dissolved in buffer to the transfected cells loaded with the calcium-sensitive dye Fluo4-am and measured the resulting fluorescence changes. For negative controls, cells transfected with an empty expression vector were treated identically. Compound-receptor combinations showing fluorescence changes exceeding those observed in identically treated empty vector-transfected cells were considered as candidate hits (not shown) and selected for further functional experiments. The verification/falsification of putative agonists selected for further screening resulted in the identification of 29 substances acting as agonist of at least one of the bony fish T2Rs (see table 1 for an overview of the identified receptor-agonist combinations resulting in receptor activations).
Table 1.
Response Profiles of Deorphaned Coelacanth and Zebrafish T2Rs with Bitter Compounds
| Substance | Source | Coelacanth |
Zebrafish |
||||
|---|---|---|---|---|---|---|---|
| T2R01 | T2R02 | T2R1 | T2R2 | T2R3a | T2R4 | ||
| Acetaminophen | S | • | |||||
| Amarogentin | N, P, T | • | |||||
| Andrographolide | N, P, T | • | |||||
| Androsterone | N, Ap, TA | • | |||||
| Benzoate, sodium salt | N, Pa, TA | • | • | • | |||
| Chenodesoxycholate, sodium salt | N, A, TA | • | |||||
| Chloramphenicol | N, B, TA | • | |||||
| Chloroquine | S | • | • | • | |||
| Colchicine | N, P, T | • | |||||
| Coumarin | N, P, TA | • | |||||
| Denatonium benzoate | S | • | • | • | |||
| Denatonium saccharide | S | • | |||||
| Desoxycholate, sodium salt | N, A, TA | • | |||||
| Dimethyl sulfoxide (DMSO) | Sn, TA | • | |||||
| (-)-Epicatechin | N, P, T | • | |||||
| Epigallocatechin gallate | N, P, T | • | |||||
| Glycocholate, sodium salt | N, A, TA | • | |||||
| Papaverine | N, P, T | • | |||||
| 1,10-Phenanthroline | S | • | |||||
| Progesterone | N, Ap, TA | • | |||||
| 6-n-propyl-2-thiouracil (PROP) | S | • | |||||
| Quassin | N, P, T | • | |||||
| Quinine sulphate | N, P, T | • | |||||
| Santonin | N, P, T | • | |||||
| Strychnine | N, P, T | • | |||||
| Taurocholic acid | N, A, TA | • | |||||
| Taurolithocholic acid | N, A, TA | • | • | • | |||
| Umbelliferone | N, P, TA | • | |||||
| Xanthotoxin | N, P, T | • | |||||
Note.—First group of symbols, natural compound (N), synthetic compound (S); second group of symbols, plant (P), animal (A), bacterial (B) metabolite; third group of symbols, terrestrial (T), aquatic (A) occurrence. Small letters indicate minor sources.
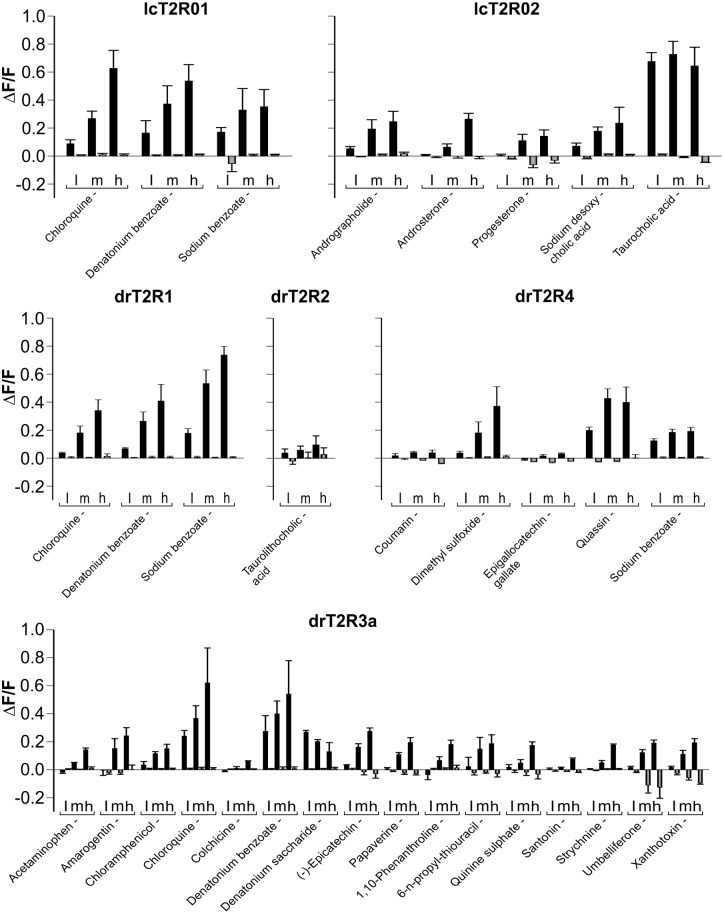
In our screening of coelacanth bitter taste receptors, we identified three compounds activating lcT2R01 and eight substances activating lcT2R02, whereas for the two receptors lcT2R34 and lcT2R74, no agonists were identified. Hence, with responses to 3% and 8% of the screened substances lcT2R01 and lcT2R02 can be considered as narrowly and intermediately tuned receptors, respectively. The zebrafish drT2R2 responded only to a single compound and therefore represents a narrowly tuned receptor, whereas for drT2R3a 16 agonists (18%) could be identified and thus this receptor exhibits intermediate to broad tuning width. Moreover, six substances activated drT2R4 (7%) and three drT2R1 (3%) indicating intermediate and narrow tuning breadths, respectively.
We wished to investigate whether fish taste ligands might tend to be more hydrophilic than ligands of terrestrial species, due to the aquatic habitat and the presence of taste buds on the exterior body surface (Yasuoka and Abe 2009) of fish. Therefore, we compared the available partition coefficients, a value that indicates the hydrophilic/hydrophobic nature of a substance, of identified fish T2R agonists with those of the compounds that did not activate fish T2Rs. However, no such tendency was observed, on the contrary the average hydrophobicity of identified agonists was significantly higher than that of compounds not activating any of the fish receptors (partition coefficient log P = 2.2 ± 1.4 SD vs. 0.8 ± 2.2 SD, respectively).
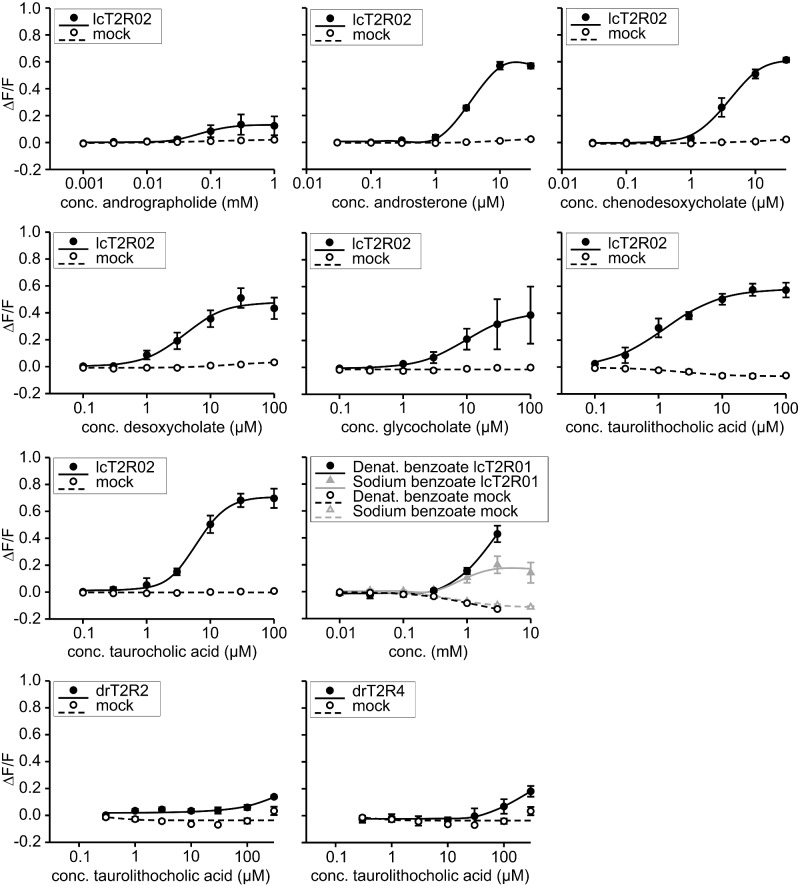
The verification/falsification of candidate agonists was done using three different concentrations of the putative agonists (fig. 2) and/or by obtaining dose–response curves (fig. 3). As shown in figure 2 coelacanth bitter taste receptor lcT2R01 responded with pronounced amplitudes to the substances chloroquine, denatonium benzoate, and sodium benzoate. The dose–response curves established with the two compounds denatonium benzoate and sodium benzoate (fig. 3) show that the benzoate anion common to both compounds might represent the relevant agonist as indicated by the same threshold concentrations of 1 mM denatonium benzoate and sodium benzoate, respectively. The receptor lcT2R02 exerted a strong bias for cholesterol-derived agonists, both steroid hormones (androsterone and progesterone) and bile acids (chenodesoxycholic acid, desoxycholic acid, glycocholic acid, taurocholic acid, taurolithocholic acid). The only exception was andrographolide, which was also found to activate lcT2R02 (figs. 2 and 3). For a better comparison of the potencies and efficacies of these chemically related agonists, we monitored the full dose–response relationships of all substances except progesterone, which exhibited limited efficacy (fig. 3). We identified the bile acid taurolithocholic acid as being the most potent lcT2R02 agonist, followed by androsterone, chenodesoxycholic acid, and desoxycholic acid with approximately similar potencies, whereas taurocholic acid and glycocholic acid trailed the set of cholesterol-based agonists. Andrographolide represented the least potent agonist.
Fig. 2.
Activation profiles of coelacanth and zebrafish bitter taste receptors. The cDNA constructs of the bitter taste receptors of coelacanth (lcT2R#) and zebrafish (drT2R#) were transiently transfected into HEK 293T-Gα16gust44 cells and subsequently subjected to calcium mobilization assays using an automated fluorometric imaging plate reader (FLIPRtetra). Bitter substances that were considered as potential activators during the first round of screening were applied in three different concentrations (low [l], medium [m], high [h]; the high concentration corresponds to the maximal concentration given in supplementary table S1, Supplementary Material online, medium and low concentrations represent each 3-fold dilution steps of test compounds) to the corresponding receptor transfected cells and the changes in fluorescence (y axis, ΔF/F) were monitored (means ± SD, n = 4). Only compound-receptor pairs resulting in statistically significant (Student’s t-test, P ≤ 0.05) fluorescence changes in transfected cells with at least one of the tested concentrations were judged positive. The receptors and the corresponding identified agonists are indicated.
Fig. 3.
Dose–response curves of selected agonist-receptor pairs. The cDNA constructs of the bitter taste receptors of coelacanth (lcT2R#) and zebrafish (drT2R#) were transiently transfected into HEK 293T-Gα16gust44 cells and subsequently subjected to calcium mobilization assays using an automated fluorometric imaging plate reader (FLIPRtetra). Cells transfected with an empty expression vector served as negative controls (mock). The relative changes in fluorescence (ΔF/F) are plotted on the y axis (means ± SD, n = 4), the applied compound concentrations on the logarithmically scaled x axis.
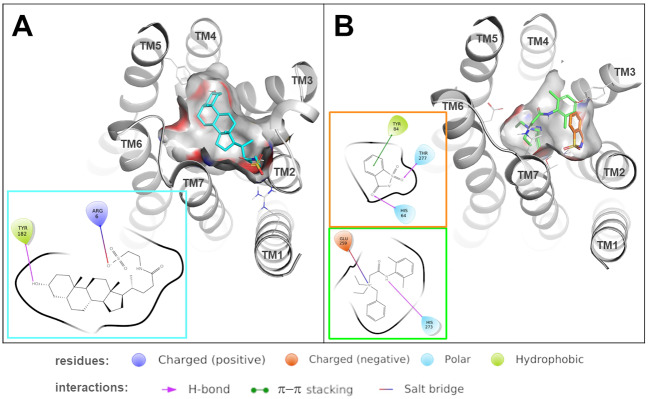
The strong bias of lcT2R02 for rather large steran-based agonists suggested that these types of ligands should fit well into the binding pocket of the receptor. In order to provide insights into the modes of interactions of cholesterol-derived compounds with lcT2R02, we performed docking simulations of taurolithocholic acid, the most potent identified lcT2R02 agonist. The results indicate that the agonist establishes a salt bridge with Arg6, which enters in the binding site from transmembrane (TM) 1, and with Tyr182 (TM5), whereas the hydrophobic portion of the ligand is accommodated in the core of the binding site (fig. 5). The contact between Tyr182 of the receptor with the hydroxyl group at the carbon atom C3 of taurolithocholic acid, which is also present in all the other identified cholesterol-derived agonists of lcT2R02, highlights the importance of this contact for ligand selectivity. This is supported by the fact that progesterone, which exhibits an oxo-group at this position instead, failed to activate the receptor. Moreover, the basic nature of the receptor’s Arg6, which establishes contact with a negatively charged group of taurolithocholic acid, could represent another specificity-determining factor as all other steran-based agonists possess negatively charged terminal groups including androsterone’s C17 oxo-group.
Fig. 5.
Predicted binding modes of ligands in lcT2R01 and drT2R3a. Predicted binding modes of taurolithocholic acid (colored in cyan) into lcT2R02 (A) and of denatonium (colored in green) and saccharine (colored in orange) into drT2R3a (B). 3D representations show how the ligand is accommodated in the binding site, ligand–receptor interactions (including π–π stacking interactions, salt bridges and hydrogen bonds) are detailed in the 2D diagrams.
Also zebrafish drT2R2 showed small but significant responses to its single agonist taurocholic acid (figs. 2 and 3). By far the most broadly tuned bony fish bitter taste receptor in our screening was zebrafish drT2R3a, which responded to 6 synthetic as well as 10 natural bitter substances. Also drT2R4 responded to synthetic (dimethyl sulfoxide, sodium benzoate) and natural (coumarin, epigallocatechin gallate, quassin) bitter substances, with coumarin and epigallocatechin gallate eliciting only small but significant signal amplitudes (fig. 2). Similar to drT2R2, drT2R4 was activated by taurolithocholic acid (fig. 3). Finally, drT2R1 showed responses to chloroquine, denatonium benzoate, and sodium benzoate (fig. 2). Remarkably, this is a complete functional overlap with its coelacanth ortholog lcT2R01, separated by over 400 Myr of evolution. The response to denatonium benzoate confirms a previous report (Oike et al. 2007), but the response to benzoate and the absence of response to denatonium saccharide suggest that benzoate, not denatonium is the active compound in denatonium benzoate.
In the course of the functional experiments with the zebrafish receptor drT2R3a, we observed a peculiar response upon stimulation with denatonium saccharide, a chemical composed of the denatonium cation and the saccharide anion. In contrast to denatonium benzoate responses which increased with increasing concentrations, the denatonium saccharide activation decreased with increasing concentrations (see fig. 2). We reasoned that this response pattern might indicate an inhibitory effect of saccharin on denatonium-induced activation of drT2R3a and therefore performed inhibition experiments (fig. 4). The determination of the dose–response relationship of drT2R3a with denatonium benzoate revealed an EC50 concentration of 0.23 ± 0.02 mM. Stimulation of drT2R3a with 1 mM denatonium benzoate and increasing concentrations of saccharin indeed demonstrated that saccharin is able to block denatonium benzoate induced responses of drT2R3a almost completely. The saccharin concentration resulting in the half-maximal inhibition of denatonium benzoate responses (IC50) was 0.92 ± 0.28 mM. Saccharin alone elicited no drT2R3a responses and hence, partial agonism could be excluded to be responsible for the saccharin-dependent drop in denatonium responses.
Fig. 4.
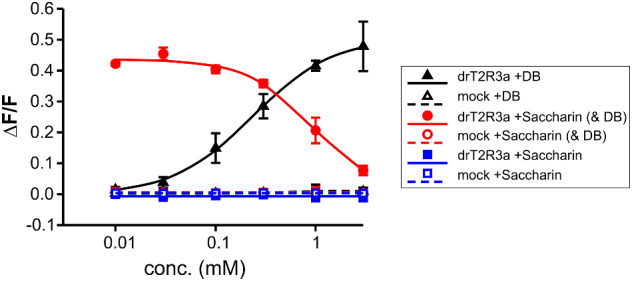
Inhibition of denatonium responses of drT2R3a by saccharin. The cDNA of the bitter taste receptor drT2R3a was transiently transfected into HEK 293T-Gα16gust44 cells and subsequently subjected to calcium mobilization assays using an automated fluorometric imaging plate reader (FLIPRtetra). Cells transfected with an empty expression vector served as negative controls (mock). The responses of drT2R3a to denatonium benzoate (DB, black curves), saccharin (blue curves), and 1 mM denatonium benzoate and increasing saccharin concentrations (red curves) are shown. The relative changes in fluorescence (ΔF/F) are plotted on the y axis (means ± SD, n = 4), the applied compound concentrations on the logarithmically scaled x axis.
To investigate the molecular basis for the inhibition of denatonium responses by saccharin, we again performed in silico studies, this time using a model of drT2R3a and docking experiments with denatonium and saccharin (fig. 5). These studies revealed that the agonist denatonium may bind to the drT2R3a by establishing a salt bridge with Glu259 (TM6) and a hydrogen bond with His273 (TM7). Interestingly, the antagonist saccharin is predicted to be accommodated by a subpocket formed by TMs 7-2-3: specifically, it forms hydrogen bonds with His64 (TM2) and Thr277 (TM7) and π–π stacking interaction between the mesomeric ring system of Tyr84 (TM3) and the benzene ring of saccharin. Therefore, our docking analysis suggests that the two ligands interact with a different pattern of residues in the binding site, and the binding of saccharin may compete with denatonium. As saccharin is able to bind to drT2R3a, but not to activate the receptor as shown by our screening results, the simultaneous presence of both molecules at the receptor results in inhibition, because the receptor’s binding cavity would only allow accommodation of one ligand at a time.
Discussion
Our phylogenetic analysis of early-derived tetrapods showed large T2R repertoires in two amphibian species, similar in size to the coelacanth repertoire. Both because of this similarity and because coelacanth is the earliest-diverging extant member of the lobe-finned lineage, we consider coelacanth as a foundational stepping stone to understand the evolution of bitter taste. Similarly, our analysis of early-derived teleosts showed the zebrafish T2R repertoire as typical for this group of bony fish, and together with the importance of zebrafish as a vertebrate model system suggested zebrafish as suitable representative to elucidate early-derived teleost fish bitter sensation. The comparison of both species allows insights in the evolution of agonist profiles within large (coelacanth, 74 intact T2R genes [Syed and Korsching 2014]) versus small (zebrafish, 7 intact genes [Shiriagin and Korsching 2019]) T2R repertoires in bony fish. Previously, no functional studies had been performed on any bony fish T2Rs except a preliminary study that identified a single agonist for a zebrafish receptor and its medaka ortholog (Oike et al. 2007). This agonist was confirmed in our study.
All four zebrafish T2Rs studied could be deorphaned. For drT2R2 a single agonist was identified, suggesting this receptor to be highly selective. drT2R1 with 3 agonists appears to be narrowly tuned as well, whereas drT2R4 (6 agonists) and drT2R3a (16 agonists) can be considered as intermediately and intermediately to broadly tuned receptors, respectively. This range of specificities is actually broader than previously observed in tetrapods with small T2R gene repertoires such as chicken or turkey, which only possess T2Rs with broad agonist profiles (Behrens, Korsching, et al. 2014). All four zebrafish T2R together responded to about one-quarter of the test compounds, which is a surprisingly high fraction when considering that the compound library is certainly biased by human bitter taste perception (Meyerhof et al. 2010). In nature, zebrafish exhibit an omnivorous lifestyle (Spence et al. 2008). Its varied natural habitats are river basins widely distributed in and around India, where it feeds on small worms, crustaceans, insect larvae, but also phytoplankton (Spence et al. 2008). It is tempting to speculate that the existence of “generalist” receptors such as the drT2R3a, which recognizes a large variety of chemically different molecules, represents an evolutionary advantage allowing the survival of species in such varied habitats.
As drT2R1 is activated by benzoate in the same concentration range as denatonium benzoate, but not by denatonium saccharide, we conclude that rather the benzoate anion is responsible for the observed response. Denatonium, an intensely bitter compound for humans who possess narrowly tuned receptors for it (Meyerhof et al. 2010), is detected by the broadly tuned drT2R3a in zebrafish. We wish to emphasize that the agonist range of drT2R1 is identical to that of coelacanth lcT2R01, its direct ortholog, consistent with a conservation of the agonist profile across over 400 Myr (for an alignment of the amino acid sequences of lcT2R01 and drT2R1, see supplementary fig. S5, Supplementary Material online). Although we cannot exclude that these identical agonist profiles might have arisen from convergent evolution, it appears more likely that they reflect an ancestral pattern. Such high conservation is an extremely atypical feature for bitter taste receptors—even in the human versus mouse comparison of several species-specific changes in agonist profiles of one-to-one orthologous T2Rs are seen (Lossow et al. 2016)—but parallels the atypically slow evolution and low evolutionary dynamics of this clade of T2R receptors, which exhibits nearly no gene birth and death events (Shiriagin and Korsching 2019, this study). It is tempting to speculate that this observed ligand profile for drT2R1 and lcT2R01 may be close to the original ligand profile at the birth of the T2R family. This would also imply that the ancestral receptor might have been narrowly tuned as well, and that broadly tuned receptors may constitute a later evolutionary achievement. To understand the ecological function of this ancestral receptor will however require a better understanding of the natural ligands—so far only synthetic compounds could be identified (chloroquine and benzoate). Besides the narrowly tuned lcT2R01, our screening of four coelacanth T2Rs showed the second most basal receptor within the coelacanth T2R repertoire, lcT2R02 to exhibit intermediate tuning width, whereas we did not find any agonists for the later-derived receptors lcT2R34 and lcT2R74. Although we cannot exclude technical issues, this circumstance might reflect an extraordinarily high selectivity of the two receptors. At the very least, no broadly tuned T2R receptor was found in coelacanth, in contrast to zebrafish. If the entire coelacanth bitter taste receptor repertoire might not include any broadly tuned receptor, the recognizable chemical space would be correspondingly limited.
Only few studies addressed the habitat and feeding behavior of coelacanths so far (Fricke and Plante 1988; Fricke et al. 1991), however, from the stomach content of few captured specimen it is beyond doubt that coelacanth is a carnivore feeding on a considerable array of prey (Uyeno and Tsutsumi 1991). In contrast to carnivores, herbivores and omnivores might be expected to require a larger taste space because toxic bitter compounds mainly occur in plant materials. A previously suggested correlation of T2R repertoire size with feeding habit (high in herbivores, low in carnivores) (Li and Zhang 2014) is broken by the large T2R family in coelacanth (Syed and Korsching 2014), but this conundrum might be solved by considering taste space as the relevant criterion instead of just counting receptors. A large family of specialized receptors might still cover a more restricted chemical space than an intermediate-sized family with some broadly tuned receptors. For example, three zebra finch T2R receptors together respond to slightly less chemicals than the single orthologous chicken T2R receptor (Behrens, Korsching, et al. 2014). The assumption that “carnivores require fewer T2R genes” (Li and Zhang 2014) may have to be modified to “carnivores require a reduced bitter chemical space” regardless of the T2R gene numbers. However it is also possible that the marine environment presents particular challenges for fish preying on invertebrates, many of which contain taste-aversive and partially toxic compounds (Pawlik et al. 1995; Marin et al. 1999). To gain a deeper understanding of the relationship between the ecological constraints and the taste space covered by the T2R family it will be necessary to identify the natural ligand spectra of these molecules and in particular for coelacanth to obtain a much deeper sampling of co-occurring potential prey species and feeding habits.
Intriguingly, the second most basal coelacanth receptor, lcT2R02, nearly exclusively responded to agonists exhibiting a steran scaffold (steroid hormones and bile acids), indicating a more restricted chemical space than the sheer number of agonists would imply (a similar situation has been found for the two human TAS2Rs, TAS2R38 and TAS2R16 [Meyerhof et al. 2010]). Steroid hormones and bile acids are compounds that are almost exclusively synthesized in animals and nearly absent in plants. Because of the profound and conserved effects of steroids in vertebrates, it would be advantageous for a predator to avoid ingesting tissues with a high steroid content. This consideration does not seem to apply for bile acids, because these molecules would not necessarily affect the predator’s physiology when ingested. However, bile acids are produced in the liver, which in some species produce or store powerful toxins such as tetrodotoxin in fugu (Halstead 1978). Recognizing bile acids may serve as a mere indicator for potentially toxic liver tissue. In fact, the white-spotted puffer Arothron hispidus, a toxic member of the family tetraodontidae (puffer fish), occurs in waters of the Comores, where coelacanth live (Smith and Heemstra 1986). Alternatively, because T2Rs have been identified in numerous non-gustatory tissues in a variety of mammals (for a recent review see Behrens, Prandi, et al. 2017) one could speculate that steroid hormones and bile acids are endogenous agonists for extra-oral T2Rs in tissues such as heart (Foster et al. 2014), brain (Dehkordi et al. 2012), and thyroid gland (Clark et al. 2015) that are not directly accessible to tastants or other xenobiotics. Indeed, zebrafish which occur in a completely different fresh water habitat also possess two bile acid-sensitive receptors.
To date, the evolutionary sequence with which taste receptors started to acquire physiological functions is unknown. A number of extant taste receptor clades exhibit pseudogenization events affecting one or even multiple taste qualities without apparent negative physiological consequences (Li et al. 2005; Zhao et al. 2010; Jiang et al. 2012) suggesting a dominant and perhaps primordial taste function of these receptors, because extraoral functions have been associated with a number of crucial physiological roles in fertility, innate immunity, etc. In other words, if extraoral functions would be dominant, one would expect severe phenotypes after pseudogenization. However, the existence of extraoral taste receptors as well as endogenously produced activators may hint at a different sequence. Future research is required to solve this conundrum.
Somewhat surprisingly, we found that the discovered fish T2R agonists exhibited a higher partition coefficient (log P = 2.2 ± 1.4) than the compounds not activating any of the fish T2Rs (log P = 0.8 ± 2.2) suggesting that bitter activators in aquatic environments are not more hydrophilic compared with ligands for terrestrial T2Rs. Therefore, fish T2R ligand requirements are not systematically different from terrestrial animals, which may have enabled conquering terrestrial habitats. This is quite different from the massive restructuring which became necessary in the olfactory system during the water-to-land transition (Korsching 2016). A structural similarity of aquatic and terrestrial T2Rs is also supported by our modeling study for drT2R3a which shows binding pockets with comparable location to those assigned previously to human and chicken bitter taste receptors (Brockhoff et al. 2010; Born et al. 2013; Marchiori et al. 2013; Di Pizio et al. 2017; Nowak et al. 2018). Moreover, several receptor positions located in the binding pockets of the fish T2Rs agree with positions that have been shown in detailed structure-function studies of human TAS2Rs to be involved in agonist interactions. According to the Balesteros–Weinstein numbering system (Ballesteros and Weinstein 1995) position 5.38, Y182 in lcT2R02, corresponds to L178 in TAS2R14 (Di Pizio et al. 2020) and S175 in TAS2R46 (Sandal et al. 2015). Similarly, for the receptor drT2R3a position 6.54, E259, corresponds to S244 in TAS2R46 (Sandal et al. 2015), position 7.35, H273, corresponds to I262 in TAS2R14 (Di Pizio et al. 2020) and F261 in TAS2R46 (Sandal et al. 2015), and position 7.39, T277, corresponds to E265 in TAS2R46 (Brockhoff et al. 2010), T266 in TAS2R10 (Born et al. 2013), and Q266 in TAS2R14 (Nowak et al. 2018), all of which were shown to contribute to agonist interactions in human bitter taste receptors.
A recurring theme in the study of bitter taste receptor/ligand interaction is the occurrence of antagonists. Often the same chemical compound acts as agonist at one receptor and antagonist at another (Slack et al. 2010; Brockhoff et al. 2011), and the same plant can synthesize both bitter antagonists and bitter agonists for the same receptor (Brockhoff et al. 2011). The phenomenon of bitter taste antagonism thus is expected to have greatly impacted the evolution of the bitter taste receptor gene family (Brockhoff et al. 2011). However, there is no obvious evolutionary benefit for bitter taste antagonists, as they neither seem to serve the protection of plant or prey, nor the protection of herbivore or predator against toxins. Enlarging the size of the T2R repertoire with corresponding specialization of the receptor genes may have provided the best protection against a complete overlap in receptor activation and inhibition spectra (Brockhoff et al. 2011; Behrens and Meyerhof 2013). The antagonist of drT2R3a identified in this study (saccharin), albeit a synthetic compound, suggests that similar natural compounds which are capable of T2R binding without concomitant activation, might exist for early-derived teleostean T2Rs, and that bitter antagonism might have shaped T2R genes throughout the entire evolution of this gene family. Intriguingly, saccharin was recently shown to antagonize the cyclamate-responsive human TAS2R1 suggesting that this noncaloric synthetic sweetener has a broad inhibitory activity against bitter taste receptors of a variety of species (Behrens, Blank, et al. 2017).
Our phylogenetic studies allow us to infer some aspects of the early evolution of T2Rs both in the lobe-finned and the ray-finned lineage. The T2R repertoires of four catfish, two cyprinids, electric eel, and piranha confirm the previous deduction of four ancestral clades in the ray-finned lineage (and the loss of one of them in teleosts [Shiriagin and Korsching 2019]), with overall minor gene birth and death events except a large gene expansion in piranha, limited to one ancestral clade. This is in stark contrast to the lobe-finned lineage, where we describe two amphibian repertoires (axolotl and cane toad) that exceeded the coelacanth repertoire in number of total genes, the one of cane toad even in number of intact genes.
All T2Rs examined form a single sister clade to the ORA (olfactory receptors related to class A, synonym V1R) clade in the phylogenetic tree, with maximal branch support. Because the ORA clade contains both cartilaginous and bony fish ora genes, this would push the origin of the posited ancestral T2R gene back to the MRCA of at least cartilaginous and bony fish. Because no T2Rs were found in cartilaginous fish despite extensive searches (Grus and Zhang 2009, Sharma et al 2019) this would require to hypothesize a loss of this ancestral gene in cartilaginous fish. However, we cannot exclude a potential distortion of the tree topology due to effects such as incomplete lineage sorting and long branch attraction and a correspondingly younger birth of the first T2R gene inside the bony fish lineage. In the MRCA of all bony vertebrates, the ancestral T2R gene appears to already have undergone gene birth events, because a mixed ray-finned/lobe-finned clade (node a2 in fig. 1) exhibits sister groups that are either ray finned (node a3, a4 in fig. 1) or lobe finned (a1, a5 in fig. 1). These five nodes thus could correspond to five ancestral genes in the MRCA of bony vertebrates, of which two have been lost in the ray-finned lineage (nodes a1, a5) and another 2 in the lobe-finned lineage (nodes a3, a4). Furthermore, the tree topology is consistent with the presence of three ancestral genes in the MRCA of coelacanth and amphibians, one of which has been lost in tetrapods (T2R1, node a2), and one in coelacanth (no ortholog in the large tetrapod T2R clade a5). In other words, most gene duplications leading to the current sets of T2R genes in coelacanth and in amphibia have happened after the separation of the respective lineages.
There is no obvious correlation between T2R repertoire size and habitat, as both large and small repertoires are found in salt water, fresh water, and terrestrial species. For small T2R repertoires: salt water crocodile, and one of the turtle species investigated (green sea turtle) live in salt water, whereas axolotl and the other turtle species examined (western painted turtle) live in fresh water. Examples for completely terrestrial species with small to very small T2R repertoires are chicken and turkey (Behrens, Korsching, et al. 2014). For large repertoires: Coelacanth and axolotl are obligate aquatic (salt water and fresh water, respectively), clawed frog strongly prefers the aquatic habitat, and adult cane toad despite its misleading epithet marina is completely terrestrial. However, amphibians have an obligatory aquatic phase before metamorphosis, so we can state that very large T2R repertoires are only found in species (of the lobe-finned lineage) with at least one obligate aquatic life phase. It will be interesting to see whether this correlation holds up, when the genome of lungfish (the only other extant lobe-finned fish) becomes available.
Our agonist identification studies show extreme conservation of the agonist profile of the most basic T2R receptor across over 400 Myr of evolutionary distance (Betancur et al. 2013). Even in T2R clades which have been differentially retained in teleost fish versus coelacanth we see a range of specificities extending from more specialized to more broadly tuned. The suitability of a set of substances tasting bitter to humans for the deorphanization of both lobe-finned and ray-finned fish T2Rs strongly suggests the overall conservation of taste ligands. No systematic differences to terrestrial tetrapod T2R sensitivities were observed, suggesting that the water-to-land transition did not require major modifications of the bitter taste receptor repertoire after conquering land.
Materials and Methods
Bitter Compounds
Commercially available bitter compounds were ordered with the highest available purity (Sigma-Aldrich, ChromaDex, Apin Chemicals, CPS Chemie, LGC Promochem), absinthin and parthenolide were gifts from G. Appendino, Novara, Italy. A full list of chemicals used for the screening can be found in supplementary table S2, Supplementary Material online.
Sequence Data Mining and Phylogenetic Analysis
T2R genes from 13 species were newly identified in basic local alignment search tool (TBLASTN [Altschul et al. 1997]) searches in published genomes available through the NCBI interface (NCBI BLAST [Johnson et al. 2008]). A set of divergent mouse, zebrafish, and coelacanth T2R was used as queries. Genome assemblies used were as follows: Ageneiosus marmoratus, driftwood catfish, GCA_003347165.1; Ambystoma mexicanum, axolotl, GCA_002915635.2; Chelonia mydas, green sea turtle, GCA_900303285.1; Chrysemys picta bellii, western painted turtle, GCA_000241765.2; Crocodylus porosus, saltwater crocodile, GCA_001723895.1; Cyprinus carpio, common carp, GCA_000951615.2; Electrophorus electricus, knife fish, GCA_003665695.2; Ictalurus punctatus, channel catfish, GCA_001660625.1; Pangasianodon hypophthalmus, iridescent shark (despite its name not a shark, but a catfish), GCA_003671635.1; Pygocentrus nattereri, piranha, GCA_001682695.1; Rhinella marina, cane toad, GCA_900303285.1; Sinocyclocheilus grahami, golden-line barbel, GCA_001515645.1; Tachysurus fulvidraco, yellowhead catfish, GCA_003724035.1. Candidates were evaluated down to an e-value of 10−8 and validated by position in phylogenetic trees containing a reference set of published T2Rs (mouse, zebrafish, and coelacanth), as well as large outgroups from several rhodopsin type GPCRs and the frizzled GPCR family to avoid false-positive and false-negative assignments. In our experience the standard method of accepting the designation of the top blast hits as assignment of the respective candidate sequences is less reliable, in particular for less well annotated regions of the tree of life. All candidate sequences were curated manually, if necessary, to complete 5′ and 3′ regions, using Expasy Translate (https://web.expasy.org/translate/; last accessed September 2018). Sequences with >98% amino acid identity were considered allelic variants. Pseudogenes and intron-containing genes were predicted using Genewise (Madeira et al. 2019) with a closely related T2R sequence as template. The predicted amino acid sequences (supplementary file 2, Supplementary Material online) and their genomic location (supplementary table S1, Supplementary Material online) are given in supplementary data, Supplementary Material online. Sequences were aligned with MAFFT7 (Katoh et al. 2019) and sequence positions with over 90% gaps were removed using Gapstreeze (http://hcv.lanl.gov/content/sequence/GAPSTREEZE/gap.html). A phylogenetic tree was constructed using a modified maximum likelihood method (PhyML-aLRT) with SPR setting for tree optimization and chi square-based aLRT for branch support (Guindon et al. 2010) available online (Dereeper et al. 2008; 2010). SMS (smart model selection) was used with AIC setting (Lefort et al. 2017). Branch support above 80% was considered significant. Trees were drawn using figtree1.43 (http://tree.bio.ed.ac.uk/software/figtree/) and annotated using Adobe Illustrator CS6. Zebrafish and shark ORs and ORAs (Saraiva and Korsching 2007) served as outgroup.
Nomenclature of T2R Genes
For previously known genes, we took the gene names from previous publications as follows: coelacanth T2Rs (Syed and Korsching 2014); Xenopus laevis (Behrens, Korsching, et al. 2014); zebrafish (Shiriagin and Korsching 2019). Newly described genes were named according to closest ortholog named gene, whenever possible, or consecutively according to the position in the phylogenetic tree.
Generation of cDNA Constructs
The coding sequences of coelacanth bitter taste receptors lcT2R01, lcT2R02, lcT2R34, and lcT2R74 (Syed and Korsching 2014) were synthesized (MWG Eurofins GmbH) and subsequently subcloned into the modified vector pcDNA5FRT PM (Bufe et al. 2002), which contained at the 5′ end of the cDNA integration site sequences coding for the first 45 amino acids of the rat somatostatin receptor subtype 3 (Ammon et al. 2002) serving as export tag to facilitate efficient plasma membrane targeting as well as the herpes simplex virus glycoprotein D epitope at the 3′ end of the integration site for immunocytochemical detection of the receptors. Full-length coding sequences of zebrafish bitter taste receptors drT2R1, drT2R2, drT2R3a, and drT2R4 (sequence information of drT2R2-4 were taken from Dong et al. [2009], the sequence of drT2R1 (zfT2R5) was published by Oike et al. [Oike et al. 2007]) were amplified by PCR from zebrafish genomic DNA available from a previous study (Behrens, Frank, et al. 2014), using the following oligonucleotides (5′ to 3′):
drT2R1_for, AATTGGGAATTCATGAGCACCGACGTTGGG AAC,
drT2R1_rev, TCATCAGCGGCCGCCGACATCCTGTGTCA CAACTG;
drT2R2_for, AATTGGGAATTCATGTCATATCAGTGCAGG ACTC,
drT2R2_rev, TCATCAGCGGCCGCCAACATCCCCTGTCA CAACTGTG;
drT2R3a_for, AATTGGGAATTCATGGGTTTCTTTTTTGTTTA CATTAG,
drT2R3a_rev, TCATCAGCGGCCGCCGGATTTTGGTTTTCT TGTCTTTTTG;
drT2R4_for, AATTGGGAATTCATGGAGCCATGGCTGTAC GC,
drT2R4_rev, TCATCAGCGGCCGCCACCTTTTAGCTTTTTA ATCACACTTAC;
The resulting PCR-amplicons were cloned using EcoRI and NotI into the vector described above. The full cDNA sequences are provided in supplementary file 3, Supplementary Material online.
Immunocytochemistry
For immunolocalization of Coelacanth and zebrafish T2Rs, cDNA constructs were transiently transfected into HEK 293T-Gα16gust44 cells as before (Behrens, Frank, et al. 2014). Briefly, the cells were seeded onto poly-d-lysin-coated glass cover slips in 24-well plates and grown in DMEM supplemented with 10% fetal bovine serum at 37 °C, 5% CO2, 100% air humidity. On the next day, cells were transfected with 800 ng of the cDNA constructs per well using lipofectamine 2000 according to the manufacturer’s protocol. After 24 h, cells were washed twice with warm PBS buffer and placed on ice for 1 h. Plasma membrane staining was initiated by incubation with concanavalin A diluted 1:2,000 in ice cold PBS buffer for 1 h, followed by brief rinses with ice cold PBS buffer to remove excess concanavalin. Fixation of the cells was done using ice cold methanol-acetone (1:1, v/v). After three rinses with PBS for 5 min each at room temperature, cells were incubated for 45 min in PBS supplemented with 5% normal horse serum to prevent unspecific binding of antisera. Next, mouse anti-HSV diluted 1:15,000 with PBS, 5% normal horse serum was applied to the cells for 1 h at room temperature to facilitate detection of the carboxy terminal herpes simplex virus glycoprotein D epitope fused to the receptor proteins. After four 5 min rinses with PBS buffer at room temperature, cells were incubated with anti-mouse Alexa488 (1:2,000) and streptavidin Alexa633 (1:1,000) in PBS, 5% normal horse serum for 1 h at room temperature. Following three PBS washes for 5 min each, 4′,6-diamidine-2′-phenylindole dihydrochloride (DAPI) diluted 1:500 in PBS was added to the cells for 15 min to stain nuclei. Finally, the cells were washed three times with PBS for 5 min each, rinsed once with ddH2O and mounted with Dako mounting medium. Pictures were taken by confocal laser scanning microscopy (Leica TCS SP8) using the following excitation and detection wavelengths: DAPI: 405 nm excitation, 415–440 nm detection; Alexa 488: 488 nm excitation, 500–542 nm detection; Alexa 633: 633 nm excitation, 650–692 nm detection. For determination of the expression rates of the T2R constructs, three representative areas were counted and the number of receptor-expressing cells (green channel) was expressed in percent of the number of total cells (blue channel, DAPI-stained nuclei).
Calcium Mobilization Assays
The screening and subsequent characterization of coelacanth and zebrafish T2Rs was done as before (Behrens, Korsching, et al. 2014; Behrens, Gu, et al. 2017; Risso et al. 2017). Briefly, HEK 293T-Gα16gust44 cells (Behrens et al. 2004; Kuhn et al. 2004) were plated on 96-well plates and transiently transfected with T2R constructs using lipofectamine 2000. After incubation for ∼24 h, cells were loaded with the calcium-sensitive dye Fluo4-am in the presence of 2.5 mM probenecid, washed twice with C1 buffer and placed in a fluorometric imaging plate reader (FLIPRtetra) for automatic application of test compounds and monitoring of fluorescence changes. For the initial screening of candidate compound-receptor pairs, 90 bitter compounds in two different concentrations were applied to cells expressing the individual receptors. The highest concentrations were selected based on previous experiments (Meyerhof et al. 2010; Thalmann et al. 2013; Behrens, Korsching, et al. 2014; Lossow et al. 2016; Risso et al. 2017) taking substance solubilities as well as receptor dependence of compound-induced fluorescence changes into account. In addition, a substance concentration of one-third of the maximum concentration was tested to assess the presumptive potency of the compounds. The initial screening was performed in duplicates for each compound concentration–receptor combination with positive and negative controls included on each 96-well plate. A full list of test substances together with the highest concentrations applied can be found in the supplementary file, Supplementary Material online. All receptor-compound combinations considered potentially positive were subsequently tested in two independent functional experiments using three concentrations of the newly identified agonist in duplicates. Only compound-receptor pairs resulting in statistically significant (Student’s t-test, P ≤ 0.05) fluorescence changes in transfected cells with at least one of the tested concentrations were judged positive. For selected agonists dose–response curves were obtained by two independent experiments performed in duplicates. The graphs were calculated using SigmaPlot software. Similarly, the graph showing the inhibition of sodium saccharide on the denatonium benzoate-stimulated drT2R3a expressing cells was calculated with SigmaPlot.
Molecular Modeling
Structures of denatonium, saccharine, and taurolithocholic acid were built using the Builder tool and prepared for docking through the generation of stereoisomers and protonation states at pH 7 ± 0.5 with LigPrep, as implemented in the Schrödinger Small-Molecule Drug Discovery Suite 2018-3 (Schrödinger, LLC, New York, NY, 2018). Three-dimensional structure models of lcT2R02 and drT2R3a were generated by homology modeling using the structure of the human TAS2R14 receptor model as constructed and refined previously (Thawabteh et al. 2019) as template. Sequence alignment and homology modeling were performed with Prime (Schrödinger Release 2018-3: Prime, Schrödinger, LLC, New York, NY, 2018) (Jacobson et al. 2004). lcT2R02 and drT2R3a have a sequence similarity to TAS2R14 of 40% and 35%, respectively (see supplementary file 4, Supplementary Material online). The Schrödinger Induced-Fit docking protocol was then used to investigate the binding modes of denatonium, saccharine, and taurolithocholic acid into their cognate receptor models (Schrödinger Suite 2018-34 Induced Fit Docking protocol; Glide, Schrödinger, LLC, New York, NY, 2016; Prime, Schrödinger, LLC, New York, NY, 2018) (Sherman et al. 2006).
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Acknowledgments
The author(s) received no financial support for the research and publication of this article.
Data Availability
The data underlying this article are available in the article and in its Supplementary Material online.
Supplementary Material
Literature Cited
- Adler E, et al. 2000. A novel family of mammalian taste receptors. Cell 100(6):693–702. [DOI] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammon C, Schafer J, Kreuzer OJ, Meyerhof W.. 2002. Presence of a plasma membrane targeting sequence in the amino-terminal region of the rat somatostatin receptor 3. Arch Physiol Biochem. 110(1–2):137–145. [DOI] [PubMed] [Google Scholar]
- Ballesteros J, Weinstein H.. 1995. Integrated methods for the construction of three-dimensional models of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25:366–428. [Google Scholar]
- Behrens M, Blank K, Meyerhof W.. 2017. Blends of non-caloric sweeteners saccharin and cyclamate show reduced off-taste due to TAS2R bitter receptor inhibition. Cell Chem Biol. 24(10):1199–1204 e1192. [DOI] [PubMed] [Google Scholar]
- Behrens M, et al. 2004. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem Biophys Res Commun. 319(2):479–485. [DOI] [PubMed] [Google Scholar]
- Behrens M, Frank O, et al. 2014. ORA1, a zebrafish olfactory receptor ancestral to all mammalian V1R genes, recognizes 4-hydroxyphenylacetic acid, a putative reproductive pheromone. J Biol Chem. 289(28):19778–19788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Gu M, Fan S, Huang C, Meyerhof W.. 2017. Bitter substances from plants used in traditional Chinese medicine exert biased activation of human bitter taste receptors. Chem Biol Drug Des. 91(2):422–433. [DOI] [PubMed] [Google Scholar]
- Behrens M, Korsching SI, Meyerhof W.. 2014. Tuning properties of avian and frog bitter taste receptors dynamically fit gene repertoire sizes. Mol Biol Evol. 31(12):3216–3227. [DOI] [PubMed] [Google Scholar]
- Behrens M, Meyerhof W.. 2013. Bitter taste receptor research comes of age: from characterization to modulation of TAS2Rs. Semin Cell Dev Biol. 24(3):215–221. [DOI] [PubMed] [Google Scholar]
- Behrens M, Meyerhof W.. 2018. Vertebrate bitter taste receptors: keys for survival in changing environments. J Agric Food Chem. 66(10):2204–2213. [DOI] [PubMed] [Google Scholar]
- Behrens M, Prandi S, Meyerhof W.. 2017. Taste and Smell. Taste receptor gene expression outside the gustatory system In: Krautwurst D, editor. Cham: Springer International Publishing; p. 1–34. [Google Scholar]
- Betancur RR, et al. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. 5:ecurrents.tol.53ba26640df0ccaee75bb165c8c26288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born S, Levit A, Niv MY, Meyerhof W, Behrens M.. 2013. The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J Neurosci. 33(1):201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhoff A, Behrens M, Niv MY, Meyerhof W.. 2010. Structural requirements of bitter taste receptor activation. Proc Natl Acad Sci U S A. 107(24):11110–11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhoff A, et al. 2011. Receptor agonism and antagonism of dietary bitter compounds. J Neurosci. 31(41):14775–14782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W.. 2002. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 32(3):397–401. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, et al. 2000. T2Rs function as bitter taste receptors. Cell 100(6):703–711. [DOI] [PubMed] [Google Scholar]
- Clark AA, et al. 2015. TAS2R bitter taste receptors regulate thyroid function. FASEB J. 29(1):164–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehkordi O, et al. 2012. Neuronal expression of bitter taste receptors and downstream signaling molecules in the rat brainstem. Brain Res. 1475:1–10. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Audic S, Claverie JM, Blanc G.. 2010. BLAST-EXPLORER helps you building datasets for phylogenetic analysis. BMC Evol Biol. 10(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A, et al. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36(Web Server):W465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pizio A, et al. 2017. Ligand binding modes from low resolution GPCR models and mutagenesis: chicken bitter taste receptor as a test-case. Sci Rep. 7(1):8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pizio A, et al. 2020. Rational design of agonists for bitter taste receptor TAS2R14: from modeling to bench and back. Cell Mol Life Sci. 77:531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Jones G, Zhang S.. 2009. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 9(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A, Gomez-Carneros C.. 2000. Bitter taste, phytonutrients, and the consumer: a review. Am J Clin Nutr. 72(6):1424–1435. [DOI] [PubMed] [Google Scholar]
- Foster SR, et al. 2014. Bitter taste receptor agonists elicit G-protein-dependent negative inotropy in the murine heart. FASEB J. 28(10):4497–4508. [DOI] [PubMed] [Google Scholar]
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB.. 2003. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 63(6):1256–1272. [DOI] [PubMed] [Google Scholar]
- Fricke H, Plante R.. 1988. Habitat requirements of the living coelacanth Latimeria chalumnae at grande comore, Indian Ocean. Naturwissenschaften 75(3):149–151. [Google Scholar]
- Fricke H, Schauer J, Hissmann K, Kasang L, Plante R.. 1991. Coelacanth Latimeria chalumnae aggregates in caves: first observations on their resting habitat and social behavior. Environ Biol Fish. 30(3):281–286. [Google Scholar]
- Glendinning JI. 1994. Is the bitter rejection response always adaptive? Physiol. Behav. 56(6):1217–1227. [DOI] [PubMed] [Google Scholar]
- Grus WE, Zhang J.. 2009. Origin of the genetic components of the vomeronasal system in the common ancestor of all extant vertebrates. Mol Biol Evol. 26(2):407–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. [DOI] [PubMed] [Google Scholar]
- Halstead BW. 1978. Poisonous and venomous marine animals of the world. Princeton (NJ: ): The Darwin Press, Inc. [Google Scholar]
- Jacobson MP, et al. 2004. A hierarchical approach to all-atom protein loop prediction. Proteins. 55(2):351–367. [DOI] [PubMed] [Google Scholar]
- Jiang P, et al. 2012. Major taste loss in carnivorous mammals. Proc Natl Acad Sci U S A. 109(13):4956–4961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson M, et al. 2008. NCBI BLAST: a better web interface. Nucleic Acids Res. 36(Web Server):W5–W9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD.. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korsching SI. 2016. Aquatic olfaction In: Zufall F, Munger SD, editors. Chemosensory transduction. London, United Kingdom: Academic Press. p. 81–100. [Google Scholar]
- Koshimizu K, Ohigashi H, Huffman MA.. 1994. Use of Vernonia amygdalina by wild chimpanzee: possible roles of its bitter and related constituents. Physiol Behav. 56(6):1209–1216. [DOI] [PubMed] [Google Scholar]
- Kuhn C, et al. 2004. Bitter taste receptors for saccharin and acesulfame K. J Neurosci. 24(45):10260–10265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort V, Longueville JE, Gascuel O.. 2017. SMS: smart model selection in PhyML. Mol Biol Evol. 34(9):2422–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Zhang J.. 2014. Diet shapes the evolution of the vertebrate bitter taste receptor gene repertoire. Mol Biol Evol. 31(2):303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, et al. 2005. Pseudogenization of a sweet-receptor gene accounts for cats’ indifference toward sugar. PLoS Genet. 1(1):e3–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossow K, et al. 2016. Comprehensive analysis of mouse bitter taste receptors reveals different molecular receptive ranges for orthologous receptors in mice and humans. J Biol Chem. 291(29):15358–15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madeira F, et al. 2019. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 47(W1):W636–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchiori A, et al. 2013. Coarse-grained/molecular mechanics of the TAS2R38 bitter taste receptor: experimentally-validated detailed structural prediction of agonist binding. PLoS One. 8(5):e64675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin A, Alvarez LA, Cimino G, Spinella A.. 1999. Chemical defence in cephalaspidean gastropods: origin, anatomical location and ecological roles. J Mollusc Stud. 65(1):121–131. [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB.. 2000. A family of candidate taste receptors in human and mouse. Nature 404(6778):601–604. [DOI] [PubMed] [Google Scholar]
- Meyerhof W, et al. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses 35(2):157–170. [DOI] [PubMed] [Google Scholar]
- Mura E, Taruno A, Yagi M, Yokota K, Hayashi Y.. 2018. Innate and acquired tolerance to bitter stimuli in mice. PLoS One 13(12):e0210032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissim I, Dagan-Wiener A, Niv MY.. 2017. The taste of toxicity: a quantitative analysis of bitter and toxic molecules. IUBMB Life 69(12):938–946. [DOI] [PubMed] [Google Scholar]
- Nowak S, et al. 2018. Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochim Biophys Acta Gen Subj. 1862(10):2162–2173. [DOI] [PubMed] [Google Scholar]
- Oike H, et al. 2007. Characterization of ligands for fish taste receptors. J Neurosci. 27(21):5584–5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlik J, Chanas B, Toonen R, Fenical W.. 1995. Defenses of Caribbean sponges against predatory reef fish. I. Chemical deterrency. Mar Ecol Prog Ser. 127:183–194. [Google Scholar]
- Risso D, Behrens M, Sainz E, Meyerhof W, Drayna D.. 2017. Probing the evolutionary history of human bitter taste receptor pseudogenes by restoring their function. Mol Biol Evol. 34(7):1587–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandal M, et al. 2015. Evidence for a transient additional ligand binding site in the TAS2R46 bitter taste receptor. J Chem Theory Comput. 11(9):4439–4449. [DOI] [PubMed] [Google Scholar]
- Saraiva LR, Korsching SI.. 2007. A novel olfactory receptor gene family in teleost fish. Genome Res. 17(10):1448–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K, Syed AS, Ferrando S, Mazan S, Korsching SI.. 2019. The chemosensory receptor repertoire of a true shark is dominated by a single olfactory receptor family. Genome Biol Evol. 11(2):398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman W, Day T, Jacobson MP, Friesner RA, Farid R.. 2006. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 49(2):534–553. [DOI] [PubMed] [Google Scholar]
- Shi P, Zhang J, Yang H, Zhang YP.. 2003. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 20(5):805–814. [DOI] [PubMed] [Google Scholar]
- Shiriagin V, Korsching SI.. 2019. Massive expansion of bitter taste receptors in blind cavefish, Astyanax mexicanus. Chem Senses. 44:23–32. [DOI] [PubMed] [Google Scholar]
- Slack JP, et al. 2010. Modulation of bitter taste perception by a small molecule hTAS2R antagonist. Curr Biol. 20(12):1104–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MM, Heemstra PC, 1986. Tetraodontidae In: Smith MM, Heemstra PC. editors. Smiths’ sea fishes. Berlin: Springer; p. 894–903. [Google Scholar]
- Spence R, Gerlach G, Lawrence C, Smith C.. 2008. The behaviour and ecology of the zebrafish, Danio rerio. Biol Rev Camb Philos Soc. 83(1):13–34. [DOI] [PubMed] [Google Scholar]
- Syed AS, Korsching SI.. 2014. Positive Darwinian selection in the singularly large taste receptor gene family of an ‘ancient’ fish, Latimeria chalumnae. BMC Genomics 15:650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalmann S, Behrens M, Meyerhof W.. 2013. Major haplotypes of the human bitter taste receptor TAS2R41 encode functional receptors for chloramphenicol. Biochem Biophys Res Commun. 435(2):267–273. [DOI] [PubMed] [Google Scholar]
- Thawabteh A, et al. 2019. Bitterless guaifenesin prodrugs-design, synthesis, characterization, in vitro kinetics, and bitterness studies. Chem Biol Drug Des. 93(3):262–271. [DOI] [PubMed] [Google Scholar]
- Uyeno T, Tsutsumi T.. 1991. Stomach contents of Latimeria chalumnae and further notes on its feeding habits. Environ Biol Fish. 32(1–4):275–279. [Google Scholar]
- Villalba JJ, Miller J, Ungar ED, Landau SY, Glendinning J.. 2014. Ruminant self-medication against gastrointestinal nematodes: evidence, mechanism, and origins. Parasite 21:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, et al. 2014. Genome sequence and genetic diversity of the common carp, Cyprinus carpio. Nat Genet 46(11):1212–1219. [DOI] [PubMed] [Google Scholar]
- Yang J, et al. 2016. The Sinocyclocheilus cavefish genome provides insights into cave adaptation. BMC Biol. 14(1):1. doi: 10.1186/s12915-015-0223-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuoka A, Abe K.. 2009. Gustation in fish: search for prototype of taste perception. Results Probl Cell Differ. 47:239–255. [DOI] [PubMed] [Google Scholar]
- Zhao H, Yang JR, Xu H, Zhang J.. 2010. Pseudogenization of the umami taste receptor gene Tas1r1 in the giant panda coincided with its dietary switch to bamboo. Mol Biol Evol. 27(12):2669–2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its Supplementary Material online.