Abstract
The coronavirus COVID-19 pandemic is today’s major public health crisis, we have faced since the Second World War. The pandemic is spreading around the globe like a wave, and according to the World Health Organization’s recent report, the number of confirmed cases and deaths are rising rapidly. COVID-19 pandemic has created severe social, economic, and political crises, which in turn will leave long-lasting scars. One of the countermeasures against controlling coronavirus outbreak is specific, accurate, reliable, and rapid detection technique to identify infected patients. The availability and affordability of RT-PCR kits remains a major bottleneck in many countries, while handling COVID-19 outbreak effectively. Recent findings indicate that chest radiography anomalies can characterize patients with COVID-19 infection. In this study, Corona-Nidaan, a lightweight deep convolutional neural network (DCNN), is proposed to detect COVID-19, Pneumonia, and Normal cases from chest X-ray image analysis; without any human intervention. We introduce a simple minority class oversampling method for dealing with imbalanced dataset problem. The impact of transfer learning with pre-trained CNNs on chest X-ray based COVID-19 infection detection is also investigated. Experimental analysis shows that Corona-Nidaan model outperforms prior works and other pre-trained CNN based models. The model achieved 95% accuracy for three-class classification with 94% precision and recall for COVID-19 cases. While studying the performance of various pre-trained models, it is also found that VGG19 outperforms other pre-trained CNN models by achieving 93% accuracy with 87% recall and 93% precision for COVID-19 infection detection. The model is evaluated by screening the COVID-19 infected Indian Patient chest X-ray dataset with good accuracy.
Keywords: Coronavirus, COVID-19, SARS-CoV-2, Chest X-Ray (CXR), Radiology images, Deep learning
Introduction
The outbreak of coronavirus occurred in December 2019, where China reported a cluster of unknown causes of pneumonia cases in the city of Wuhan, Hubei province to the World Health Organisation(WHO) [14, 30, 38]. This SARS-CoV-2 or COVID-19 disease spread rapidly around the world [30, 33] and considering severity, WHO announced COVID-19 as a pandemic. Till date (5th June 2020), a total of 6,675,011 cases of COVID-19 have been reported, including total 391,848 deaths worldwide [10]. Inhaling infected droplets may spread the disease, with an incubation period of between two and fourteen days [29]. People with cough, shortness of breath or difficulty breathing, fever, chills, muscle pain, loss of taste or smell, and sore throat symptoms may have COVID-19 [9, 29]. Other less common symptoms have been reported, such as nausea, vomiting, or diarrhea etc. [9]. Dr. Mike Ryan, Executive Director, WHO Health emergencies said, ”It is important to put this on the table: this virus may become just another endemic virus in our communities, and this virus may never go away” on 14th May 2020 at the Geneva Virtual Press Conference [3]. WHO suggested that rapid testing is one of the effective measures to control the spread of SARS-CoV-2 infection [37]. Currently, Real-time reverse transcription-polymerase chain reaction (RT-PCR) testing technique is used for laboratory diagnosis of COVID-19 [6, 34]; however it suffers with following three issues:
Shortage of RT-PCR kits.
Non-urban community hospitals lack the PCR infrastructure to support high sample throughput.
RT-PCR depends on the existence of detectable SARS-CoV-2 in the collected samples. [34].
Alternatively, it is found that Chest radiography examination can also be used for COVID-19 screening; where X-ray images or computed tomography (CT) images are examined by radiologists to search for visual markers linked with SARS-CoV-2 infection. The major hallmarks of SARS-CoV-2 infection on chest radiography imaging are consolidative pulmonary opacities with more tendency to involve lower lobe(s); bilateral and peripheral ground glass appearance [5]. Pleural effusion is rare in case of COVID infections. Due to low RT-PCR sensitivity (60%-70%), even though negative results found, symptoms can be identified by an analysis of CT images [16]. Use of CT images as a diagnostic modality has following issues:
CT imaging devices are costly and require high level of expertise in handling.
CT imaging devices are not portable; thus, there are higher chances of human to human transmission during patient transport due to lack of personal protective equipment (PPE) kits available with medical staffs [26].
CT imaging takes more time for processing than that of X-ray imaging.
High-quality CT imaging devices may not be available in many hospitals or clinics in non-urban areas, making timely screening of COVID-19 infections difficult.
X-rays, on the other hand, are the most common and easily accessible radiographic examination techniques in the clinical practices and is of great use for low cost and faster screening of COVID-19 infections in the current epidemic situation [25, 36].
In response to the growing coronavirus pandemic situation and the shortage of expert radiologists, an artificial intelligence (AI) based COVID-19 diagnostic system with high sensitivity and specificity; without human intervention is highly desirable for the analysis of radiography imaging features. These AI-powered COVID-19 diagnostic systems can make COVID-19 screening tests cheaper and real-time mass testing effectively. Also, the chances of transmission to the involved technicians will be reduced and the burden on the existing limited health experts or radiologists will also be reduced.
Considering the limitations of the existing RT-PCR and CT based COVID-19 diagnostic techniques, in this work, we propose a low cost, realtime, faster, scalable DCNN model, called Corona-Nidaan for COVID-19 patient screening using chest X-ray samples. The proposed model is end-to-end trained with 20,907 chest X-ray images (including synthetic images) which are collected from the three different open-source datasets [7, 8, 20]. In our proposed model, we use Depth-wise separable convolutional layers instead of traditional 2D convolutional layers in order to make our model lightweight and to reduce the computational complexity. In case of embedded and mobile vision applications with the constrained computational resource requirements, light-weight deep learning models with fewer parameters plays an important role. Our proposed model consists of total of 4.022 million parameters, which is lower than that of 1.0 MobileNet-224 (4.2 million parameters) [13]. This makes our model suitable in case of on-device DNN implementation for COVID-19 screening.
In this research, we also want to answer the following questions: 1) How effective is the transfer-learning with existing pre-trained DCNN models for detecting COVID-19 infection?; 2) Given the X-Ray image of the chest, what is the best way to extract features related to the hallmarks of COVID-19 disease? To find the answers to these questions, we evaluate the performance of five different well-known pre-trained CNN models along with the proposed Corona-Nidaan model. It is found that Corona-Nidaan model outperforms over the transfer-learning models and other state of art works mentioned in recent literature. Proposed Corona-Nidaan model and other implemented transfer-learning based pre-trained models are publicly available at https://github.com/mainak15/Corona-Nidaan.
To summarize, this work has four major contributions:
A novel light weight DCNN model titled Corona-Nidaan is proposed that can learn the hallmarks of SARS-CoV-2 infection from chest X-ray samples and then detect COVID-19 cases within a second, without human intervention.
The efficacy of transfer learning using pre-trained CNNs is investigated on the chest X-ray images for COVID-19 infection detection.
A simple oversampling technique is suggested to overcome the imbalance classification problem.
The efficacy of the Corona-Nidaan model is also validated against the screening of COVID-19 infected Indian Patient chest X-ray dataset.
The remaining paper organized as follows: In Section 2, we review the related work on chest X-Ray based COVID-19 infection detection. In Section 3, we introduce the proposed Corona-Nidaan deep neural network architecture and its design principles along with new minority class oversampling approach. In Section 4, we present the experimental setup, the formation of the ChestX dataset, implementation details, detailed analysis of implemented models, comparison of Corona-Nidaan with other approaches, along with the performance of Corona-Nidaan against Indian COVID-19 patients dataset. Section 5 states the conclusions of this work along with the future work to be established in these directions.
Related work
Recent advancements in the deep learning techniques and the availability of large open-source medical image datasets has enabled creation of deep neural networks to deliver promising results without human intervention in a wide range of medical imaging tasks, such as lung diseases diagnosis from chest X-ray images [25, 36], breast cancer detection [4], pulmonary tuberculosis classification [18], diabetic retinopathy detection [11] and arrhythmia detection [24]. Wang et al. [36] released a new front-view chest X-ray dataset consisting of 108,948 images of 32,717 patients with 8 disease labels. The authors used transfer learning with ImageNet pre-trained CNNs (ResNet-50, GoogLeNet, AlexNet, and VGGNet-16) to detect 8 lung diseases, including pneumonia. Rajpurkar et al. [25] utilized CheXNet for pneumonia detection using the ChestX-ray14 dataset. The dataset contains 112,120 frontal chest X-ray images. The authors achieved the F1 score of value 0.435 which is slightly more than the F1 score value (0.387) which is achieved by using radiologist’s consultation.
Recently, several deep learning approaches are suggested by researchers to screen and diagnose coronavirus-infected patients using chest X-ray images. Wang and Lin et al. [35] proposed COVID-Net, a deep learning model, and trained the model with the COVIDx dataset consisting of 13,800 frontal-view chest X-ray images, extracted from total 13,725 patient cases. The dataset consists of 183 COVID-19 samples, 8,066 normal samples, and 5,538 pneumonia samples are obtained from the three different open-access datasets. Although the authors achieved 92.6% test accuracy in three-class classification, it is found that the model exhibits high false negative rate for COVID-19 class and large number of trainable parameters. Hemdan et al. [12] employed transfer learning with ImageNet pre-trained CNNs for COVID-19 detection and reported good performance of VGG19 and DenseNet121 compared to other pre-trained CNN models. However, we found that these CNN models are fine-tuned with only 25 COVID-19 and 25 normal X-ray samples. Ozturk et al. [22] proposed DarkCovidNet model based on DarkNet architecture, and achieved 87.02% test accuracy in the three-class classification. DarkCovidNet model consists of 17 convolutional layers, and the model is trained on minimal COVID-19 samples. Due to the use of under-sampling techniques, it is quite possible that the DarkCovidNet model miss the important signatures of normal and pneumonia class. Mangal et al. [19] presented the CovidAID model that achieves 90.5% accuracy with a 100% recall for COVID-19 screening. To tackle the imbalanced classification problem due to limited COVID samples, the authors considered a random subset of pneumonia data in each batch, while training the model. However, the model suffers with many false negative cases for the normal class. Apostolopoulos et al. [1] analyzed the performance of the transfer learning with pre-trained CNNs. The authors achieved 93.48% and 98.75% classification accuracy in three and two class classifications, respectively, against a dataset consisting of 700 pneumonia, 224 Covid-19, and 504 normal chest X-ray samples. Basu et al. [2] introduced the domain extension transfer learning with a 12 layered CNN model to achieve 95.3% accuracy supporting four-class classification into categories, Covid-19, normal, other diseases, and pneumonia using limited number of samples from each class. Using fixed size filter may not capture multilevel features from the X-ray images; which is essential for generalization capability of any CNN model. Also, detecting COVID-19 becomes a complex learning problem as in many cases COVID patterns may mimic non-COVID pneumonia cases; and hence, end to end training systems can solve such complex problems better than suggested Domain Extension Transfer Learning (DETL) in their paper. Oh et al. [21] employed FC-DenseNet103 to extract the lung segments from the pre-processed X-ray images. From the extracted lung contour, the authors generated random patches. Each patch is fed into ResNet-18 CNN (which is pre-trained on ImageNet) to train and classify Tuberculosis, Normal, Bacterial Pneumonia, Viral Pneumonia, and COVID-19 infections. This model with 11.6 million trainable parameters has achieved 91.9% accuracy. However, the authors trained the model with a minimal number of samples. Khan et al. [17] used 284 COVID-19, 310 Normal, 330 Bacterial Pneumonia, 327 Viral Pneumonia X-ray samples to train the model namely CoroNet (Xception) and achieved 89.6% accuracy for three-class classification. The computational complexity of the model is high, and it is found that the model some times mis-classifies Pneumonia cases as Normal. Perumal et al. [23] utilized the extracted Haralick features from both X-ray and CT images to trained VGG-16 CNN, and obtained 93% accuracy. The model sometimes mis-classifies COVID-19 as viral pneumonia, viral pneumonia as COVID-19, and Normal as Bacterial Pneumonia. Although the authors claimed it easy to use pre-defined CNN models to solve COVID-19 detection problem, such predefined model suffers with large number of trainable parameters. Also, the suggested model by the authors requires manual pre-processing and feature generation from input X-ray images before feeding it to the CNN model.
Many of these works achieved promising results in the detection of COVID-19 infected patients using chest X-ray images. However, most of these models are trained with a limited number of samples (pneumonia and normal X-ray images) to overcome the class balancing problem of the dataset, which results in loss of essential information of the majority classes (pneumonia and normal X-ray images). Also, the total number of trainable parameters of the stated models is too large to use in the embedded and mobile vision applications. On-device DNN implementation for COVID-19 screening may reduce diagnosis cost and time.
In this work, we propose Corona-Nidaan DNN model for COVID-19 patient screening using chest X-ray image analysis along with a new minority class oversampling approach to deal with imbalance classification problem. A detailed experiment with five different pre-trained CNNs is carried out to compare the performance of our model. We analysed that both approaches can achieve comparable performance, but the performance of the Corona-Nidaan model is better in terms of accuracy and model complexity. We trained and tested our model with 245 COVID-19, 8,066 Normal, 5,551 Pneumonia Chest X-ray images, and validated our findings by medical experts of Sardar Vallabhbhai Patel COVID Hospital, New Delhi, India. Corona-Nidaan model can be used as an on-device DNN for COVID-19 screening due to it’s lower complexity than that of 1.0 MobileNet-224. Through the empirical study, we found that our model does not suffer from too many false positive and false negative results.
Proposed approach
In this section, we explain the architecture of proposed Corona-Nidaan Model along with the Minority Class Oversampling Approach.
Proposed Corona-Nidaan model architecture
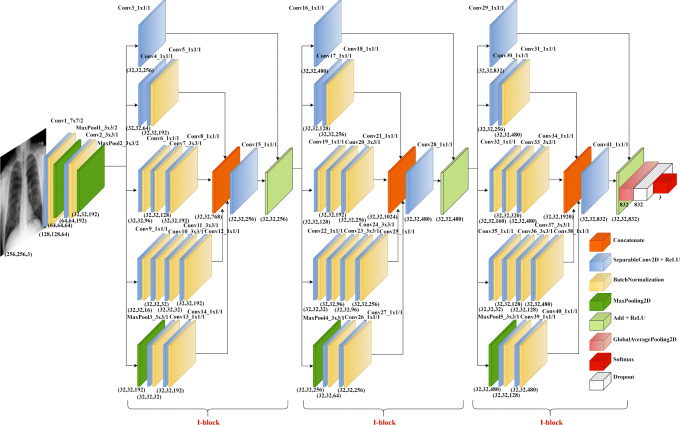
Here, we propose Corona-Nidaan, a novel lightweight deep neural network architecture, mainly inspired by the architectural designs of InceptionV3, InceptionResNetV2, and MobileNetV2. The overall architecture of the model shown in Fig. 1.
Fig. 1.
The overall architecture of the proposed model
The following design principles are taken into account in designing proposed model.
DP1: In a CNN model, the application of multiple different sized convolution filters to the same input can extract multi-level feature representations at the same time and hence, improves the overall performance of the model, minimizing over-fitting and computational costs.
DP2: The introduction of residual connection in the model reduces the Vanishing Gradient effect and accelerates the model’s training speed.
DP3: Introduction of Depth-wise separable convolutions significantly reduces model parameters and extra computation overhead.
DP4: Addition of batch normalization layers to the network has benefits, such as speed up network training, reduce the difficulty of initial starting weights, and introduce additional network regularization.
DP5: The use of Global Average Pooling before the softmax layer is better than the fully connected layer as it reduces the number of trainable parameters from the network.
The proposed model consists of total 91 layers, with forty-one depth-wise separable convolutional layers, thirty-two batch normalization layers, five max-pooling layers, three concatenate layers, three add layers, three activation layers, one global average pooling layer, one dropout layer, one input, and one softmax layer. Instead of the traditional 2D convolutional layer, we used the deep-wise separable convolutional layer in our network, driven by the MobileNetV2 philosophy (refer design principle: DP3). We used ReLU activation and the same padding in all depth-wise separable convolutional layers. Batch normalization is used after activation in the depth-wise separable convolutional layer to accelerate training and improve the generalization error of our model as opposed to the InceptionResNetV2, which use batch normalization before activation in each convolutional layer. The first convolutional layer applies 64 kernels of size (7 × 7) with stride 2 to the input image of dimension (256 × 256 × 3). The output of this convolutional layer is given to the first batch normalization layer and the first max pooling layer with pool size (3 × 3) and stride 2. The second convolutional layer consists of 192 kernels of size (3 × 3) and stride 1 connected to the first max-pooling layer’s output. The second batch normalization layer and the second max pooling layer with pool size (3 × 3) and stride 2 take the second convolutional layer’s output. The output of the second max-pooling layer is given to the next stage of network, which is made up of three consecutive I-blocks, as in Fig. 1. Each I-block follows the five design principles stated earlier in order to improve performance of the final model. The detailed architectural design of a single I-block is as shown in Fig. 2.
Fig. 2.
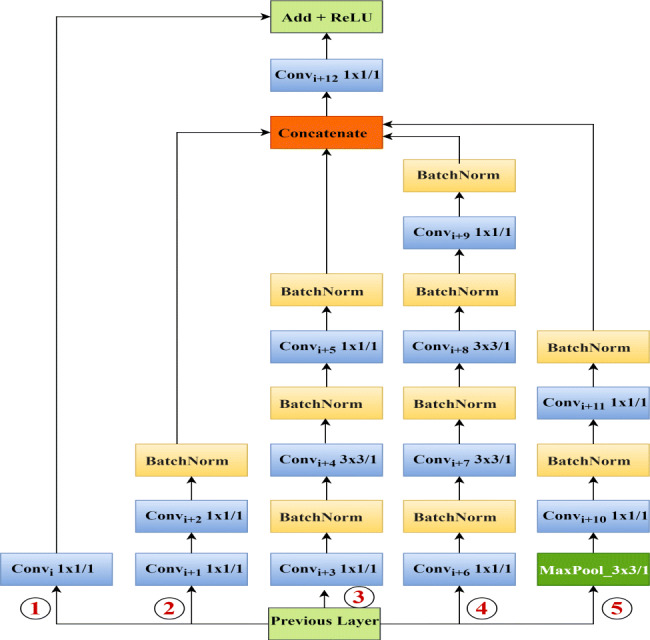
I-blocks of the proposed model
The I-block consists of thirteen depth-wise separable convolutional layers, ten batch normalization layers, one max-pooling layer, one concatenate layer, one add layer, and one activation layer. This block has five parallel paths, acting as multiple convolution kernels applied with some pooling to the output of the previous layer or activation at the same time (refer design principle: DP1, DP2 and DP4). Third and fourth paths perform (1 × 1) convolution at the initial stage to reduce dimension and computational costs, before some costly convolutions. The initial (1 × 1) convolution in the second path and the (3 × 3) convolutions in the third and fourth paths extract multi-level representations of features (as in design principle: DP1). All paths, except the first path, use the last (1 × 1) convolution for dimension expansion. As a result, we get the same feature map dimension as the output dimension of the previous layer or the activation. The max-pooling layer of the fifth layer helps to extract low-level features from the input. The output feature maps of the second, third, fourth, and fifth paths are then concatenated. The concatenate layer output is then fed to a convolutional layer with kernel size (1 × 1) for feature dimension reduction. The path one acts as a shortcut connection in the I-block, i.e., the output of the previous layer added to the stacked layers’ output, which enables residual learning (refer design principle: DP2). We use a (1 × 1) convolutional layer in path one to match with the output dimension of the convolutional layer after the concatenate layer. Finally, the path one convolutional layer output added to the output of the convolutional layer after the concatenate layer. ReLU activation is used in the final activation layer of our I-block. The architecture of each I-block can be represented as in (1).
| 1 |
Where, , , , and denotes five parallel paths of the I-block; whereas i, x, b, w represents convolutional layer’s index, input feature map, biases and weights respectively. The depth-wise separable convolutional layer, max-pooling layer, concatenate layer, add layer, batch normalization, feature map addition, feature map concatenation and ReLU are indicated using fC, fm, fCon, fA, fb, ⊕, ⊗ and σ respectively.
The global average pooling layer takes the output of the third I-block as input (as in design principle: DP5). After the global average pooling layer, we used a dropout layer to avoid overfitting. The dropout layer’s output is then fed to the three-way softmax layer to produce a probability distribution of three class labels. Our proposed model consists of a total of 4,021,974 parameters.
Proposed minority class oversampling approach
In most of the published dataset, it is found that the COVID-19 class has a deficient number of samples compared to the other two classes. This type of problem with the distribution of training data is known as the imbalanced class distribution. It is difficult for any machine learning or deep learning model to obtain optimized results using imbalanced training data. The model will not be capable of learning the characteristics of the minority class because the number of observations is deficient. Three of the most popular approaches to address this problem are 1) undersampling, 2) oversampling, and 3) synthetic sampling. Through experimental analysis using approach one and two, we observed that the random deletion of samples from the majority class loses essential information and, on the other hand, random over-sampling for the minority class (copies of samples already available) leads to overfitting. Using synthetic images from existing samples of the minority class as well as standard image data augmentation techniques did not improve proposed model’s performance. So, we come up with a two-phase oversampling approach. In Phase one, we generated five images (blur, sharp, blue channel, green channel and red channel) using the Algorithm 1 for each sample of the minority class (i.e. COVID-19).

Figure 3 shows the distribution and statistical parameters of original and over-sampled COVID-19 images. It can be seen that, generated over-sampled images using Algorithm 1 follows the same distribution characteristics as that of original COVID-19 images and hence capable of handling dataset imbalance problem effectively.
Fig. 3.
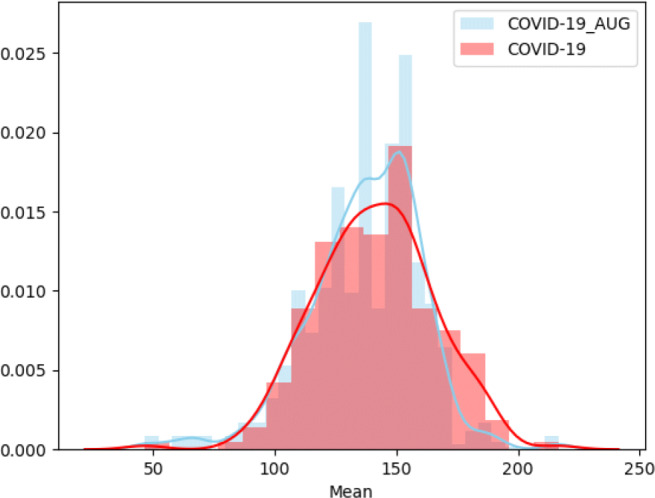
Distribution of means of original and over-sampled COVID-19 images
In phase two, standard data augmentation techniques applied to each sample generated by the Algorithm 1 to generate more sample representations for minority class i.e. COVID cases.
Experiments and results
The experiments are carried out on a laptop running Windows 10 with Intel Core i5-8300H CPU 2.30 GHz, 8 GB memory, NVIDIA GeForce GTX 1050 Ti, Dedicated GPU memory 4.0 GB, CUDA v10.0.130 Tool kit and CuDNN v7.6.5.
Dataset
We trained, validated, and tested the proposed Corona-Nidaan model on the ChestX dataset. The dataset is formed by combining the three different open access chest X-ray datasets 1) RSNA Pneumonia Detection Challenge dataset [20], 2) Fig. 1 COVID-19 Chest X-ray Dataset Initiative [7] and 3) COVID-19 Image Data Collection [8] . There are currently 218 COVID-19 and 33 pneumonia samples in the COVID-19 Image Data Collection dataset, and 27 COVID-19 samples are in Fig. 1 COVID-19 Chest X-ray Dataset Initiative. The RSNA Pneumonia Detection Challenge dataset comprises 8,066 normal samples and 5,518 pneumonia samples. After combining all the samples from the three different datasets, our ChestX dataset consisted of 13,862 samples of which 245 samples belong to the COVID-19 class, 5,551 samples belong to the pneumonia class, and 8,066 samples belong to the normal class (examples of x-ray samples are shown in Fig. 4).
Fig. 4.
Examples of Chest X-ray samples taken from ChestX dataset.
The sample distribution shows that the COVID-19 class has a deficient number of samples compared to the other two classes. Hence, two-phase oversampling approach is used. In Phase one, we generated five images (blur, sharp, blue channel, green channel and red channel) using the Algorithm 1 for each sample of the minority class (i.e. COVID-19).
In phase two, standard data augmentation techniques applied to each sample generated by the Algorithm 1 with parameters set as: horizontal flip=True, rotation range= 10, zoom range= 0.2, height shift range= 0.1, brightness range=(0.9, 1.1) and width shift range= 0.1. The total number of training samples for the minority class (COVID-19) become 7,490 after the two-phase oversampling, in which 1,070 and 6,420 (214 × 5 × 6) samples are produced by phase one and two respectively. The distribution of the final chest X-ray image for training and testing summarized in Table 1.
Table 1.
Chest X-ray image’s distribution for training and testing
| COVID-19 | Normal | Pneumonia | Total | |
|---|---|---|---|---|
| Train | 7,490 | 7,966 | 5,451 | 20,907 |
| Test | 31 | 100 | 100 | 231 |
Implementation details
Proposed Corona-Nidaan DNN is trained end to end using the ChestX dataset. We conducted a detailed experiment with five different pre-trained CNNs to ensure the usefulness of transfer learning for COVID-19 cases detection from the chest X-ray images. We implemented Corona-Nidaan model and other transfer learning models using Python 3.7.7, OpenCV 4.1.1, and the Keras 2.2.4 API, TensorFlow-GPU v1.14.0 backend. We have set the weights for equally penalized under or over-represented classes in training sets in order to overcome the imbalanced classification problem. The implementation details of both Corona-Nidaan model as well as other transfer learning models (based on five different pre-trained CNNs) are explained as:
Corona-Nidaan model
The proposed Corona-Nidaan model is end to end trained on the ChestX dataset and optimized using Adam optimizer. The X-ray images of the ChestX dataset are resized to (256 × 256 × 3). Each pixel value of the images is rescaled with a 1./255 factor. We used depth-wise separable convolutional instead of traditional 2D convolutional to reduce the number of multiplications. In our proposed model, all the convolutional layers activated by ReLU activation, and the final prediction layer activated by softmax activation. We employed glorot uniform kernel initializer, same padding, and bias initialized with a constant value of 0.2 for all depth-wise separable convolutional layers.
In this study, the hyperparameters (shown in Table 2) of the model are tuned using a manual search technique. We performed experimental analysis using different optimization algorithms such as Adam, AdaGrad, Stochastic Gradient Descent with Momentum, and RMSProp. We found that our proposed model with Adam Optimizer performs well on the train and test sets. We used 0.001 as an initial learning rate, 300 epochs, and eight batch size during training. If no improvement is observed in validation accuracy for two consecutive epochs, then the learning rate is reduced by 0.5 factors by our algorithm. To reduce overfitting, 40% dropout is applied to the dropout layer after the global average pooling layer, and we adopted an early stop. For multi-class classification, the categorical cross-entropy loss function enhanced during training.
Table 2.
Corona-Nidaan DCNN input dimension, optimization algorithm and hyperparameters
| Input dimension | Epochs | Optimizer | Initial-learning-rate | ReduceLROnPlateau | Batch size | Early stop |
|---|---|---|---|---|---|---|
| 256 × 256 × 3 | 300 | Adam | 0.001 | Yes(0.5 factor) | 8 | 10 patience |
Transfer learning based models
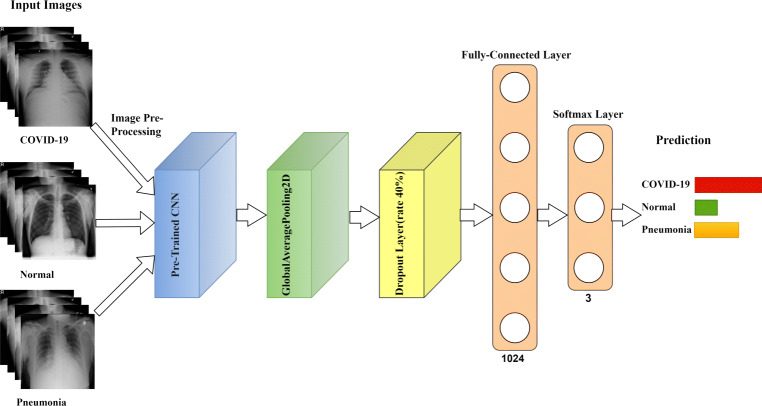
The implemented transfer learning models, are divided into two parts 1) Convolutional base and 2) Classifier. The convolutional base is used as a spatial feature extractor and classifier predicts the class label based on features extracted by CNN. The overall architecture of these models is shown as in Fig. 5.
Fig. 5.
The overall architecture of the transfer learning model
In this experiment, DenseNet201 [15], InceptionResNetV2 [32], MobileNetV2 [27], VGG19 [28] and InceptionV3 [31] are employed as convolutional base of the model. All the CNNs are pre-trained on ImageNet. The final softmax layer is removed from all the pre-trained CNNs as we want to extract features, and not predictions. The ChestX dataset is small and different from the pre-trained model’s dataset; in this scenario, freezing the lower level layers and training the higher-level layer technique works well. The non-trainable layers and the input X-ray image dimensions of the respective pre-trained CNNs shown in Table 3.
Table 3.
Pre-trained CNNs input dimension, non-trainable layers, optimization algorithm and hyperparameters
| CNNs | Input dimension | Non-trainable layers | Optimizer | Learning-rate | ReduceLROnPlateau | Batch size |
|---|---|---|---|---|---|---|
| MobileNetV2 | 224 × 224 × 3 | 100 | Adam | 0.001 | Yes | 32 |
| VGG19 | 224 × 224 × 3 | 15 | SGD | 0.0001 | No | 32 |
| InceptionResNetV2 | 299 × 299 × 3 | 618 | SGD | 0.0001 | No | 32 |
| InceptionV3 | 299 × 299 × 3 | 249 | Adam | 0.001 | Yes | 32 |
| DenseNet-201 | 224 × 224 × 3 | 481 | Adam | 0.001 | Yes | 8 |
The classifier consists of a global average pooling and a fully connected layer with the ReLU activation function, followed by a softmax layer. The manual search method is employed to find the model’s hyperparameters and the most efficient optimization algorithm. Table 3 illustrates the model’s hyperparameter and optimization algorithm based on the pre-trained CNNs. The fully connected layer and the softmax layer consists of 1024 and 3 neurons, respectively. We adopted he-uniform kernel initializer at the fully connected layer. We set the learning rate to 0.0001 value and the batch size to the value of 32 during the training phase of the pre-trained VGG19 and InceptionResNetV2 model. In case of other pre-trained CNNs, we set an initial learning rate to 0.001 value and then reduced it by 0.5 factor, if the validation accuracy does not improve after two epochs. The batch size of the model is set to 8 in case of pre-trained DenseNet201 model, otherwise it is set to 32 for pre-trained MobileNetV2 and InceptionV3 models. The maximum epoch set at 300 for training the model. To overcome overfitting problem, 1) A dropout layer with a dropout rate of 0.4 is added after the global average pooling layer; 2) An early stop with 10 epochs; and 3) The L2 kernel and the bias regularizer is applied in a fully connected layer with a weight decay of 0.001. For disease prediction from X-ray samples, the categorical cross-entropy loss function is enhanced. The model is optimized by using SGD Optimizer in case of pre-trained VGG19 and InceptionResNetV2 models. Adam Optimizer is used in case of other pre-trained CNNs during training.
Detailed analysis of implemented models
In this section, we present results and analysis of the both methods, 1) transfer learning with pre-trained CNNs and; 2) End to end trained Corona-Nidaan model on the ChestX dataset. To investigate the performance of any model, we computed the f1-score, recall (sensitivity), and precision (positive predictive value) for each class on the test dataset. In the case of an imbalanced dataset, only the accuracy metric does not reflect the model’s performance. For example, if the majority class has 98 samples, then the model achieves 98% accuracy. However, the model is not able to detect 2% of minority class samples. To evaluate the overall efficiency of any model, we calculated the accuracy, the macro average, and the weighted average. To visualize the performance of these models, we plotted the confusion matrix.
Transfer learning based models
In this section, we present our results and analysis for the implemented transfer learning based pre-trained models on the ChestX test dataset.
MobileNetV2:
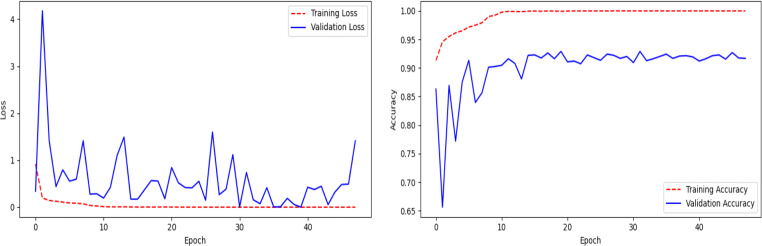
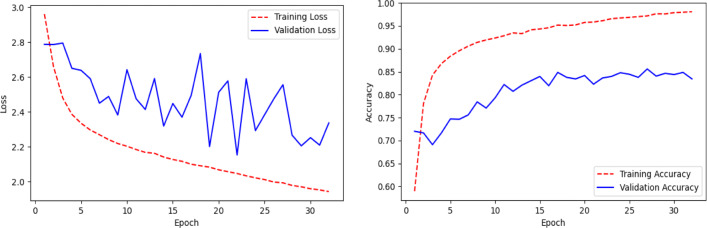
The performance of pre-trained MobileNetV2 (Fig. 5) on the ChestX test dataset is shown in Table 4 and the confusion matrix in Fig. 6. It can be observed that the model achieved 85% on the test dataset, but the recall value for COVID-19 class is only 32%, which means that the model is very picky. The model mis-classifies many COVID-19 cases as normal or pneumonia cases. The line plots of categorical cross-entropy loss and accuracy over training epochs of the model have shown in Fig. 7. The loss plot shows that the training loss remains flat after the 10th epoch, regardless of training, means under-fitting condition, and the model is unable to learn the training dataset well.
Table 4.
Performance of the model (Fig. 5) on the ChestX test dataset using pre-trained MobileNetV2
| Precision | Recall | F1-Score | |
|---|---|---|---|
| COVID-19 | 1.00 | 0.32 | 0.49 |
| Normal | 0.84 | 0.99 | 0.91 |
| Pneumonia | 0.84 | 0.87 | 0.86 |
| Accuracy | 0.85 | ||
| Macro avg | 0.89 | 0.73 | 0.75 |
| Weighted avg | 0.86 | 0.85 | 0.83 |
Fig. 6.
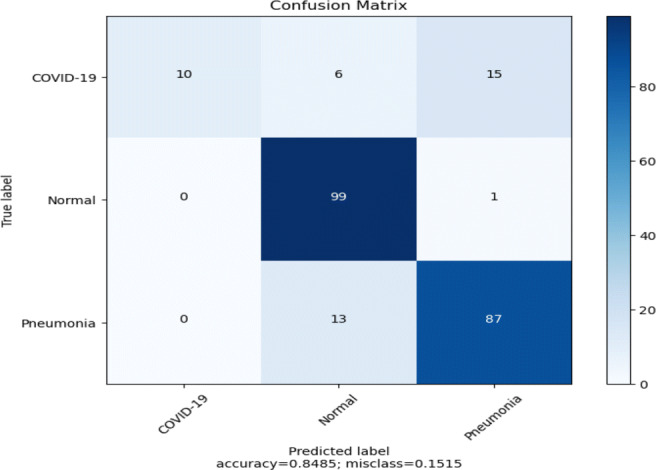
Confusion matrix of the model (Fig. 5) using pre-trained MobileNetV2
Fig. 7.
Line plots of categorical cross-entropy loss and accuracy over training epochs of the model (Fig. 5) using pre-trained MobileNetV2
VGG19:
Table 5 shows the performance of pre-trained VGG19 model (Fig. 5) on the ChestX dataset and Fig. 8 represents the confusion matrix of the model. It is clear from the experimental results that the overall performance of the model is good. The model obtained a test accuracy of 93%, and the train and validation loss and accuracy are very close to each other, showing that the model is not over-fitted (see Fig. 9). The recall and precision values of all three classes are quite impressive, which means low false negative, and positive prediction rates. In this model, the COVID-19 class achieves a precision and recall value of 93% and 87%, respectively. From the confusion matrix, it is clear that the model mis-classifies very few samples. The model correctly predicts 27 samples as COVID-19, 96 as Normal, and 91 as Pneumonia.
Table 5.
Performance of the model (Fig. 5) on the ChestX test dataset using pre-trained VGG19
| Precision | Recall | F1-Score | |
|---|---|---|---|
| COVID-19 | 0.93 | 0.87 | 0.90 |
| Normal | 0.91 | 0.96 | 0.93 |
| Pneumonia | 0.95 | 0.91 | 0.93 |
| Accuracy | 0.93 | ||
| Macro avg | 0.93 | 0.91 | 0.92 |
| Weighted avg | 0.93 | 0.93 | 0.93 |
Fig. 8.
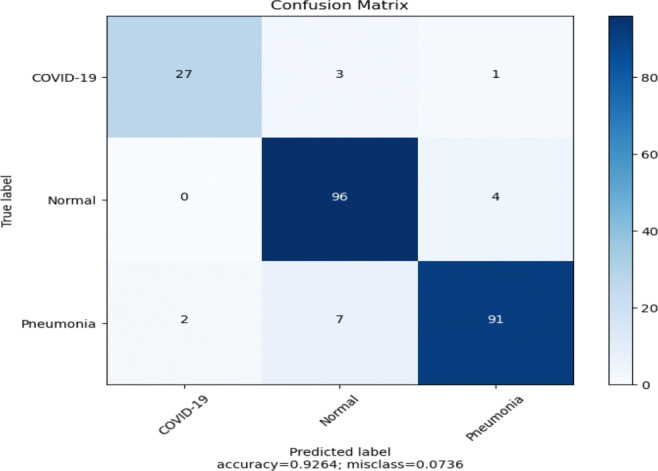
Confusion matrix of the model (Fig. 5) using pre-trained VGG19
Fig. 9.
Line plots of categorical cross-entropy loss and accuracy over training epochs of the model (Fig. 5) using pre-trained VGG19
InceptionResNetV2
The pre-trained InceptionResNetV2 model (Fig. 5) achieved 80% test accuracy with 39% and 55% recall and precision, respectively, for the COVID-19 class, as shown in Table 6 along with low recall (79%) rate for Pneumonia class. The model predicts around 10 false-positive and 19 false-negative cases for the COVID-19 class (Refer: Fig. 10). It can be observed from the loss and accuracy plots of the model as in Fig. 11, that there is no indication of over-fitting or under-fitting.
Table 6.
Performance of the model (Fig. 5) on the ChestX test dataset using pre-trained InceptionResNetV2
| Precision | Recall | F1-Score | |
|---|---|---|---|
| COVID-19 | 0.55 | 0.39 | 0.45 |
| Normal | 0.82 | 0.94 | 0.88 |
| Pneumonia | 0.83 | 0.79 | 0.81 |
| Accuracy | 0.80 | ||
| Macro avg | 0.73 | 0.71 | 0.71 |
| Weighted avg | 0.79 | 0.80 | 0.79 |
Fig. 10.
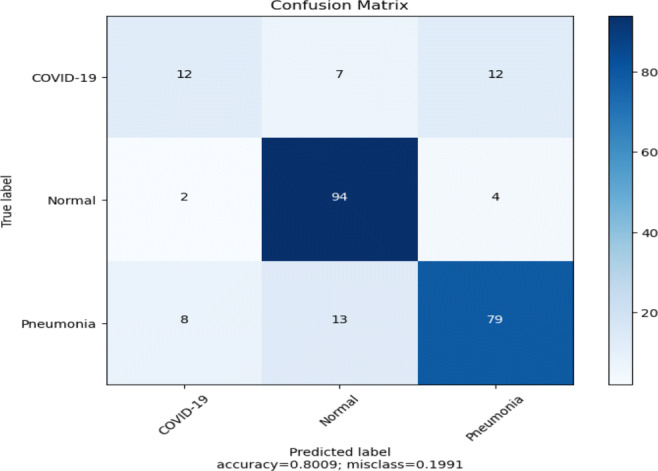
Confusion matrix of the model (Fig. 5) using pre-trained InceptionResNetV2
Fig. 11.
Line plots of categorical cross-entropy loss and accuracy over training epochs of the model (Fig. 5) using pre-trained InceptionResNetV2
InceptionV3
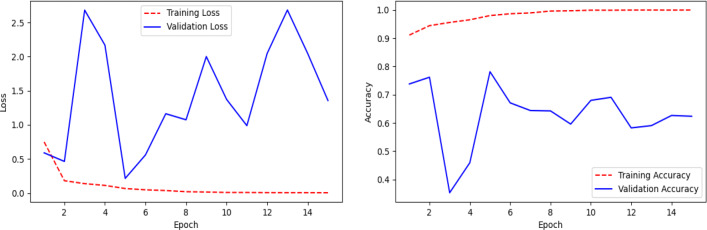
Model performance (Fig. 5) using pre-trained InceptionV3 is summarized in Table 7. This model achieves an accuracy of 77% on the test dataset. From Table 7, it can be seen that the model mis-classifies almost 35% of Pneumonia cases during testing. The precision value of the COVID-19 class is only 48%. The confusion matrix (Fig. 12) shows that Pneumonia and the COVID-19 classifications suffer from high false-negative and false-positive results. The loss and accuracy plots of the model is shown in Fig. 13. The loss plot tells that the validation accuracy curve is decreased to the 2nd epoch and then start to increase again, which means that the model is over-fitted on the training data.
Table 7.
Performance of the model (Fig. 5) on the ChestX test dataset using pre-trained InceptionV3
| Precision | Recall | F1-Score | |
|---|---|---|---|
| COVID-19 | 0.48 | 0.94 | 0.63 |
| Normal | 0.83 | 0.85 | 0.84 |
| Pneumonia | 0.96 | 0.65 | 0.77 |
| Accuracy | 0.77 | ||
| Macro avg | 0.75 | 0.81 | 0.75 |
| Weighted avg | 0.84 | 0.77 | 0.78 |
Fig. 12.
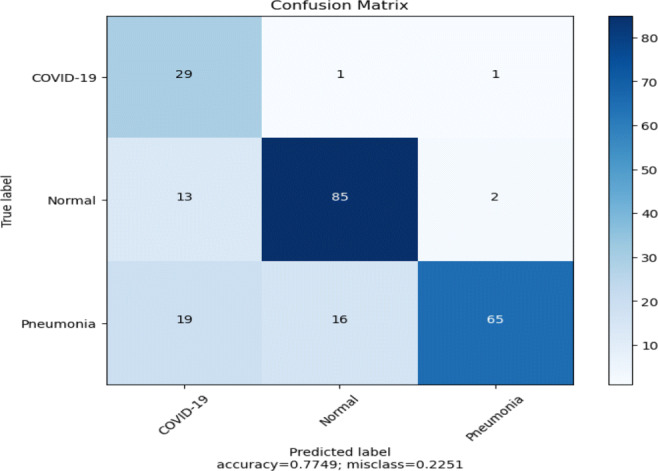
Confusion matrix of the model (Fig. 5) using pre-trained InceptionV3
Fig. 13.
Line plots of categorical cross-entropy loss and accuracy over training epochs of the model (Fig. 5) using pre-trained InceptionV3
DenseNet201
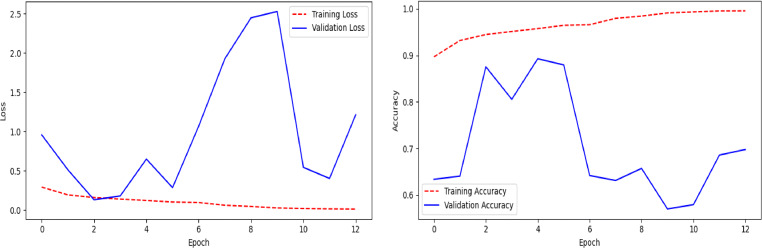
Pretrained DenseNet201 model (Fig. 5) achieved an overall test accuracy of 84% as shown in Table 8.The recall value of the COVID-19 class is very low, i.e., only 42%. We can notice that a lot of COVID-19 cases are misclassified as Normal or Pneumonia, as shown in the confusion matrix in Fig. 14. The model exhibits large false-negative results in case of COVID-19 class. The loss and accuracy plots shown in Fig. 15 reflects the overfitting condition against training data.
Table 8.
Performance of the model (Fig. 5) on the ChestX test dataset using pre-trained DenseNet201
| Precision | Recall | F1-Score | |
|---|---|---|---|
| COVID-19 | 0.81 | 0.42 | 0.55 |
| Normal | 0.85 | 0.96 | 0.90 |
| Pneumonia | 0.84 | 0.86 | 0.85 |
| Accuracy | 0.84 | ||
| Macro avg | 0.84 | 0.75 | 0.77 |
| Weighted avg | 0.84 | 0.84 | 0.83 |
Fig. 14.
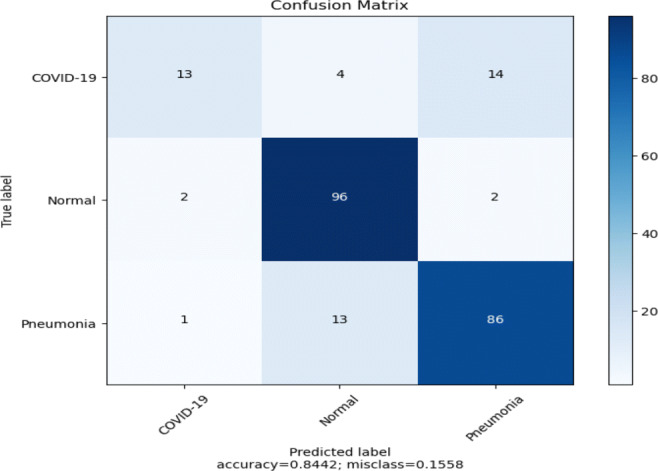
Confusion matrix of the model (Fig. 5) using pre-trained DenseNet201
Fig. 15.
Line plots of categorical cross-entropy loss and accuracy over training epochs of the model (Fig. 5) using pre-trained DenseNet201
Corona-Nidaan model
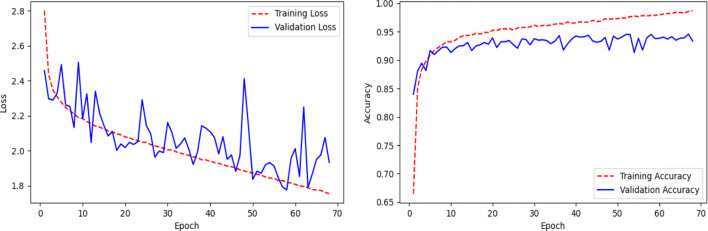
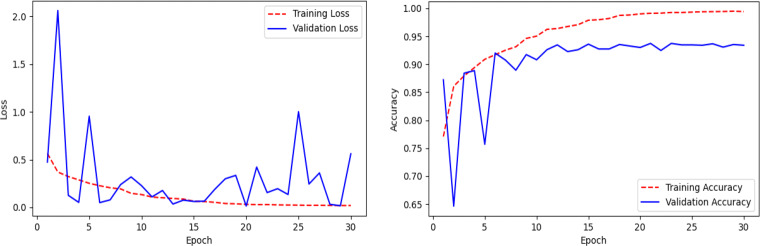
Here, we present the results and analysis of the Corona-Nidaan model on the ChestX test dataset. The f1-score, precision, and recall for each class and the overall test accuracy is shown in Table 9. Corona-Nidaan achieved 1) 95% overall test accuracy; 2) 94% precision and recall value for the COVID-19 class classification; 3) 93% precision and 98% recall for the Normal class classification; and 4) 97% precision and 92% recall for the Pneumonia class classification. The confusion matrix in Fig. 16 shows that the model does not suffer from too many false-negative and false-positive results. That is an excellent indication because high false-negative and false-positive results increase the burden on public health services due to the requirement for additional PCR testing. The loss and accuracy plots indicate that the training process converged well as in Fig. 17.
Table 9.
Performance of Corona-Nidaan on the ChestX test dataset
| Precision | Recall | F1-Score | |
|---|---|---|---|
| COVID-19 | 0.94 | 0.94 | 0.94 |
| Normal | 0.93 | 0.98 | 0.96 |
| Pneumonia | 0.97 | 0.92 | 0.94 |
| Accuracy | 0.95 | ||
| Macro avg | 0.95 | 0.95 | 0.95 |
| Weighted avg | 0.95 | 0.95 | 0.95 |
Fig. 16.
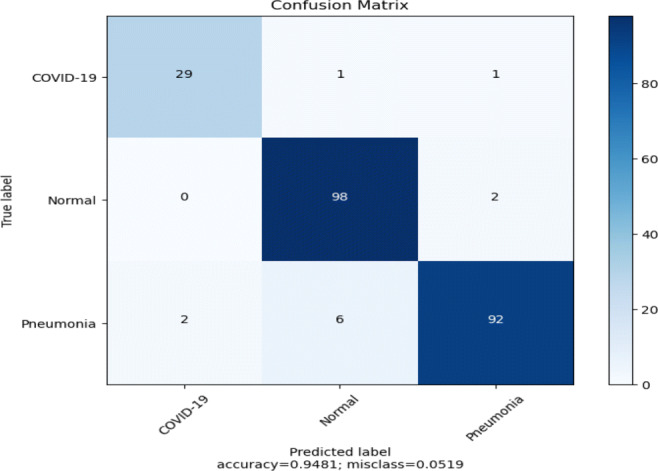
Confusion matrix for Corona-Nidaan on the ChestX test dataset
Fig. 17.
Line plots of categorical cross-entropy loss and accuracy over training epochs of the Corona-Nidaan model
Comparison of Corona-Nidaan with the transfer learning models
It can be seen from the results (Refer Table 10) that the performance of the Corona-Nidaan is always better than that of other pre-trained models. During experimentation, it is found that pre-trained VGG19 model outperformed other pre-trained CNNs. Hence, in case if the dataset is small, pre-trained VGG19 model can be used instead of other pre-trained models.
Table 10.
Comparison of Corona-Nidaan with the transfer learning model
| Params (M) | Accuracy | |
|---|---|---|
| MobileNetV2 | 3.572 | 85% |
| VGG19 | 20.55 | 93% |
| InceptionResNetV2 | 55.91 | 80% |
| InceptionV3 | 23.90 | 77% |
| DenseNet201 | 20.29 | 84% |
| Corona-Nidaan | 4.022 | 95% |
Comparison of Corona-Nidaan with other published approaches
In this section, we compared the Corona-Nidaan model with other published approaches (shown in Table 11). The proposed Corona-Nidaan model achieves 95% accuracy on the ChestX dataset and outperforms other previously published approaches. The dataset comprised of total 13862 chest X-ray images which are collected from the three different open-source image repository [7, 8, 20]. The studies mentioned in this section have used the same COVID-19 samples from COVID-19 Image Data Collection [8]. Wang and Lin et al. [35] proposed the COVID-Net deep learning model and achieved 92.6% test accuracy in three-class classifications. Their model is trained and tested with the same number of samples collected from the same data sources. The overall performance of the model is good. However, COVID-Net suffers from false-negative results for the COVID-19 class. The COVID-Net consists of 117.4 million parameters, which is 29 times larger than that of Corona-Nidaan model. The precision and recall value of COVID-Net for each class type is lower than that of our proposed model. Hemdan et al. [12] achieved 90% accuracy by utilizing transfer learning with seven pre-trained CNNs. The authors claimed that VGG19 and DenseNet201 performed well compared to the other five pre-trained models. However, in case of these CNN models, the f1-score for COVID-19 and Normal class calculated is 91% and 89% respectively, where it is 94% and 96% respectively in case of our model. Ozturk et al. [22] used a deep convolutional neural network, DarkCovidNet, and achieved 87.02% test accuracy in three-class classifications. The model is trained using 500 pneumonia, 500 normal, and 125 COVID-19 chest X-ray samples. The sensitivity, precision, and f1-score of DarkCovidNet model stated for three-class classifications are 85.35%, 89.96%, and 87.37%, respectively, which are lower than that of Corona-Nidaan model (shown in Table 9).
Table 11.
Comparison of Corona-Nidaan with other previously published approaches developed using chest X-ray images
| Study | Number of Samples | Methods | Accuracy | Params (in million) | Remarks |
|---|---|---|---|---|---|
| Wang and Lin et al. [35] | 183 COVID-19 8,066 Normal 5,538 Pneumonia | COVID-Net b | 92.6% | 117.4 | Model suffers from false-negative results for COVID-19 cases and consists of more number of trainable parameters. |
| Hemdan et al. [12] | 25 COVID-19 25 Normal | COVIDX-Net(VGG-19) a | 90% | 20.55 | 93% of accuracy found on our dataset. |
| Ozturk et al. [22] | 125 COVID-19 500 Normal 500 Pneumonia | DarkCovidNet a | 87.02% | 1.16 | Model suffers from false-positive and false-negative results. Under-sampling technique loses important details of pneumonia and normal classes. 71% of accuracy found on our dataset. |
| Mangal et al. [19] | 115 COVID-19 1,341 Normal 3,867 Pneumonia | CovidAID c | 90.5% | – | Model suffers from false-negative cases in case of normal class. |
| Apostolopoulos et al. [1] | 224 COVID-19 504 Normal 700 Pneumonia | VGG-19 a | 93.48% | 20.55 | 93% accuracy found on our dataset. |
| Basu et al. [2] | 225 COVID-19 350 Normal 322 Pneumonia 50 Other-disease | 12 layer CNN c | 95.3% | – | The proposed model trained on very limited Normal and Pneumonia samples. |
| Oh et al. [21] | 180 COVID-19 191 Normal 54 Bacterial Pneumonia 57 Tuberculosis 20 Viral Pneumonia | FC-DenseNet103 + ResNet-18 b | 91.9% | 11.6 | Model is trained with very limited number of samples, suffers from false positive and negative results. More number of trainable parameters. |
| Khan et al. [17] | 284 COVID-19 310 Normal 330 Bacterial Pneumonia 327 Viral Pneumonia | CoroNet (Xception) c | 89.6% | 33 | More number of trainable parameters, trained on limited number of training samples, Model mis-classifies many Pneumonia cases as Normal. |
| Perumal et al. [23] | 205 COVID-19(X-ray) 1,349 Normal 2,538 Bacterial Pneumonia 202 COVID-19(CT) 1,345 Viral pneumonia | VGG-16 c | 93% | – | Model sometimes mis-classifies COVID-19, viral pneumonia, and Normal cases. More number of trainable parameters as VGG-16 used. Manual pre-processing and feature generation used. |
| ours | 245 COVID-19 8,066 Normal 5,551 Pneumonia | Corona-Nidaan | 95% | 4.02 | End to end learning approach, low mis-classification rates, lightweight, and less number of trainable parameters. |
a Implemented, trained and tested against our dataset
b Tested on the same samples as we did
c Reported accuracy by the author in their work
From the confusion matrix for three class classifications, it is clear that the DarkCovidNet model suffers from false-positive and false-negative results. DarkCovidNet used only 500 samples for each pneumonia and normal class, using under-sampling technique to avoid the problem of imbalanced classification, however, such random deletion of samples from the majority class loses important class information. For COVID-19 screening, Mangal et al. [19] used transfer learning and obtained 90.5% accuracy. They used 115 COVID-19, 1,341 normal, and 3,867 pneumonia chest X-ray samples to train the CovidAID model. The model’s overall performance is impressive, especially for COVID-19 cases with a sensitivity of 100 %. However, the model suffers with the same problem of getting many false-negative cases for the normal class. It is important to minimize false negative and false positive rates for initial screening techniques. Apostolopoulos et al. [1] adopted the same transfer learning technique as Hemdan et al. [12] and also observed that the performance of VGG19 is better than other pre-trained CNNs. They achieved 93.48% three-class classification accuracy on the test dataset. Basu et al. [2] achieved 95.3% using 12 layers of CNN against a dataset consisting of 225 COVID-19, 50 Other-disease, 322 Pneumonia, and 350 Normal samples. It is found that the authors used five-fold cross-validation to measure the performance of their model, and the model is trained on minimal Normal and Pneumonia samples. Oh et al. [21] used 57 Tuberculosis, 191 Normal, 54 Bacterial Pneumonia, 20 Viral Pneumonia, and 180 COVID-19 X-ray images to train proposed patch-based CNN. The authors stated 91.9% test accuracy when tested on the same samples, however, the model suffers from false positive and negative results. The number of trainable parameters in their model is as twice as that of our model. The precision value of COVID-19 (76.9%), Pneumonia (90.3%) infections, and the sensitivity of Normal class (90%) is lower. Khan et al. [17] used a limited number of training samples to train CoroNet (Xception pre-trained on ImageNet) and achieved 89.6% three-class classification accuracy. The number of trainable parameters of their model is eight times larger compared to Corona-Nidaan and the model also mis-classifies many Pneumonia cases as Normal cases. Perumal et al. [23] used 205 COVID-19 (X-ray), 1,349 Normal, 2,538 Bacterial Pneumonia, 202 COVID-19(CT) and 1,345 Viral pneumonia samples to train VGG-16 CNN and achieved 93% accuracy. The model sometimes misclassifies COVID-19 as viral pneumonia, viral pneumonia as COVID-19, and Normal as Bacterial Pneumonia. The computational complexity of the proposed model is also enormous.
Study of Corona-Nidaan performance against Indian COVID-19 patient dataset
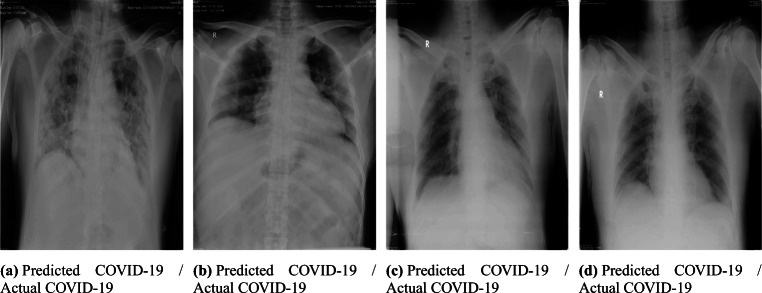
In this study, we tested the performance of Corona-Nidaan on Indian patient chest X-ray samples. We consulted medical experts from the Pune region, India, and Sardar Vallabhbhai Patel COVID Hospital, New Delhi, India and validated the performance of our model against almost 1000 X-ray samples belonging to two different classes (Normal, COVID-19). These chest X-ray images’ assessment results by Corona-Nidaan model against confirmed COVID-19 cases is shown in Fig 18. The model is validated for the screening of COVID-19 infected patients by the medical experts. The remarks of the end-users (Medical Experts) on the performance of Corona-Nidaan model are as under:
The overall performance of the model on Indian patient’s chest X-ray images is good, and the prediction results are convincing, especially in the present pandemic.
This low-cost tool can differentiate COVID-19 infection, Pneumonia, and Normal cases from chest X-ray images without human intervention.
In exceptional cases with the early-stage symptoms, COVID-19 X-ray images may be misclassified by the model as Pneumonia or Normal. The early features of COVID-19 infection may mimick other non-COVID pneumonia features. Hence, to overcome this problem, the model needs to train with more samples of Chest X-ray images.
The model does not suffer from high false negative and positive results, which indicates the suitability of the model for medical imaging classification tasks. This is in alignment with aim of this model, wherein we do not want to miss any COVID case in the current CORONA pandemic.
The information generated as a result of the X-ray image analysis can be used as a screening test for rapid and mass testing.
Fig. 18.
Chest X-ray images diagnosis by the proposed Corona-Nidaan model against confirmed COVID-19 cases
Conclusion
In this study, Corona-Nidaan, a lightweight deep convolutional neural network, is introduced for COVID-19 cases screening using the chest X-ray samples. A simple oversampling method to tackle the imbalance classification problem is also suggested in this work. The following conclusions can be drawn based on the results of this study: (1) Our proposed Corona-Nidaan model achieved 95% test accuracy with 94% recall and precision value for multi-class classification. (2) The Corona-Nidaan model does not suffer from too many false positive and false negative results, which is an excellent indicator because it reflects our model’s reliability. (3) Three stacked I-blocks are capable of capturing critical COVID-19 features for better classification. (4) Transfer learning can be used for medical imaging tasks when the data set is small, and it found that VGG19 outperformed other pre-trained CNNs.
In our future work, the proposed model will be enhanced to perform severity level analysis by collecting serial X-ray images of COVID-19 infected with local radiologists and hospitals’ help, as the major drawback in such research work is the availability of minimal open-source COVID-19 chest X-ray samples.
Acknowledgements
This research is supported by the Defence Institute of Advanced Technology, DRDO Lab, Ministry of Defence, India, and the Indian National Academy of Engineering. The authors would like to thank Sardar Vallabhbhai Patel COVID Hospital, New Delhi, India, to facilitate validation of Corona-Nidaan against the Indian Patient X-Ray samples. The authors would like to thank NVIDIA for the GPU grant for carrying out deep learning based research work.
Biographies
Mainak Chakraborty
is presently pursuing his Ph.D. from Defence Institute of Advanced Technology (DIAT), an autonomous institute under the Ministry of Defence, Girinagar, Pune, INDIA. He is working as an Assistant Professor in the Department of Computer Science & Engineering at Dream Institute of Technology, Kolkata, INDIA, since 2011. He received his M.Tech and B.Tech degrees in Computer Science & Engineering and Information Technology from the Maulana Abul Kalam Azad University of Technology, West Bengal, formerly known as West Bengal University of Technology, in 2011 and 2008, respectively. He is a recipient of AICTE-INAE Teachers Research Fellowship from The Indian National Academy of Engineering (INAE). He won Smart India Hackathon-2019 and 2nd runner up in Smart India Hackathon-2017. His research areas are Deep Learning, Machine Learning, Natural Language Processing, Human Activity Detection, and Medical Image Analysis.

Dr. Sunita Vikrant Dhavale
presently associated with Defence Institute of Advanced Technology (DIAT), an autonomous institute under Ministry of Defence, Pune, as an Assistant Professor in Department of Computer Engineering. She received her M.E. and Ph.D. degrees in Computer Science from Pune University in 2009 and DIAT University in 2015, respectively. She is a recipient of Atal Tinkering Lab Mentorship, ISACA Academic Advocate Membership, EC-Council’s CEH-v8, IETE M. N. Saha Memorial Award, and Outstanding Woman Achiever Award from Venus International Foundation. She was selected as one of the top performers in four weeks AICTE approved Faculty Development Program on ICT tools by IIT, Bombay, in September 2016. She has more than 20 publications in International Journals, International Conference Proceedings, and Book chapter. Her research areas are computer vision, deep learning, and cybersecurity. She is a member of many professional bodies, including IEEE, AAAI, ACM, ISTE, IETE, IAENG, and ISACA.

Dr. Jitendra Ingole
(MD Internal Medicine) works as a Professor of Internal Medicine at Smt Kashibai Navale Medical College, Pune. He has many publications to his credit and author of a book. He has been invited as a guest speaker at many institutes and is also a postgraduate guide for MD Internal Medicine. His areas of interest are Nutrition, application of technology for patient betterment.

Code Availability
Compliance with Ethical Standards
Conflict of interests
The authors declare that they have no conflict of interest.
Footnotes
This article belongs to the Topical Collection: Artificial Intelligence Applications for COVID-19, Detection, Control, Prediction, and Diagnosis
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Mainak Chakraborty, Email: mainak.mail@gmail.com.
Sunita Vikrant Dhavale, Email: sunitadhavale@gmail.com.
Jitendra Ingole, Email: jitendra.ingole@gmail.com.
References
- 1.Apostolopoulos ID, Mpesiana TA (2020) Covid-19: automatic detection from x-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med, p 1 [DOI] [PMC free article] [PubMed]
- 2.Basu S, Mitra S (2020) Deep learning for screening covid-19 using chest x-ray images. arXiv:200410507
- 3.BBC (2020) Coronavirus may never go away, world health organization warns. https://www.bbc.com/news/world-52643682
- 4.Bejnordi BE, Veta M, Van Diest PJ, Van Ginneken B, Karssemeijer N, Litjens G, Van Der Laak JA, Hermsen M, Manson QF, Balkenhol M, et al. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. Jama. 2017;318(22):2199–2210. doi: 10.1001/jama.2017.14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bernheim A, Mei X, Huang M, Yang Y, Fayad ZA, Zhang N, Diao K, Lin B, Zhu X, Li K, et al (2020) Chest ct findings in coronavirus disease-19 (covid-19): relationship to duration of infection. Radiology p 200463 [DOI] [PMC free article] [PubMed]
- 6.Chan JFW, Yip CCY, To KKW, Tang THC, Wong SCY, Leung KH, Fung AYF, Ng ACK, Zou Z, Tsoi HW, et al. (2020) Improved molecular diagnosis of covid-19 by the novel, highly sensitive and specific covid-19-rdrp/hel real-time reverse transcription-pcr assay validated in vitro and with clinical specimens. J Clin Microbiol 58(5) [DOI] [PMC free article] [PubMed]
- 7.Chung A (2020) Figure 1 COVID-19 chest x-ray data initiative
- 8.Cohen JP, Morrison P, Dao L (2020) Covid-19 image data collection. arXiv:200311597https://github.com/ieee8023/covid-chestxray-dataset
- 9.For Disease Control C Prevention (2020) Symptoms of coronavirus. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html
- 10.Dong E, Du H, Gardner L (2020) An interactive web-based dashboard to track covid-19 in real time. The Lancet infectious diseases [DOI] [PMC free article] [PubMed]
- 11.Gulshan V, Peng L, Coram M, Stumpe MC, Wu D, Narayanaswamy A, Venugopalan S, Widner K, Madams T, Cuadros J, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. Jama. 2016;316(22):2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 12.Hemdan EED, Shouman MA, Karar ME (2020) Covidx-net: a framework of deep learning classifiers to diagnose covid-19 in x-ray images. arXiv:200311055
- 13.Howard AG, Zhu M, Chen B, Kalenichenko D, Wang W, Weyand T, Andreetto M, Adam H (2017) Mobilenets: efficient convolutional neural networks for mobile vision applications. arXiv:170404861
- 14.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, et al. Clinical features of patients infected with 2019 novel coronavirus in wuhan, china. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
- 16.Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH (2020) Essentials for radiologists on covid-19: an update—radiology scientific expert panel [DOI] [PMC free article] [PubMed]
- 17.Khan AI, Shah JL, Bhat MM (2020) Coronet: a deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Comput Meth Prog Biomed p 105581 [DOI] [PMC free article] [PubMed]
- 18.Lakhani P, Sundaram B. Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology. 2017;284(2):574–582. doi: 10.1148/radiol.2017162326. [DOI] [PubMed] [Google Scholar]
- 19.Mangal A, Kalia S, Rajgopal H, Rangarajan K, Namboodiri V, Banerjee S, Arora C (2020) Covidaid: Covid-19 detection using chest x-ray. arXiv:200409803
- 20.Of North America RS (2019) Rsna pneumonia detection challenge dataset. https://www.kaggle.com/c/rsna-pneumonia-detection-challenge/data
- 21.Oh Y, Park S, Ye JC (2020) Deep learning covid-19 features on cxr using limited training data sets. IEEE Trans Med Imaging [DOI] [PubMed]
- 22.Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Acharya UR (2020) Automated detection of covid-19 cases using deep neural networks with x-ray images. Comput Biol Med p 103792 [DOI] [PMC free article] [PubMed]
- 23.Perumal V, Narayanan V, Rajasekar SJS (2020) Detection of COVID-19 using CXR and CT images using Transfer Learning and Haralick features. Appl Intell pp 1–18, 10.1007/s10489-020-01831-z [DOI] [PMC free article] [PubMed]
- 24.Rajpurkar P, Hannun AY, Haghpanahi M, Bourn C, Ng AY (2017a) Cardiologist-level arrhythmia detection with convolutional neural networks. arXiv:170701836
- 25.Rajpurkar P, Irvin J, Zhu K, Yang B, Mehta H, Duan T, Ding D, Bagul A, Langlotz C, Shpanskaya K, et al. (2017b) Chexnet: radiologist-level pneumonia detection on chest x-rays with deep learning. arXiv:171105225
- 26.Rubin GD, Ryerson CJ, Haramati LB, Sverzellati N, Kanne JP, Raoof S, Schluger NW, Volpi A, Yim JJ, Martin IB, et al. (2020) The role of chest imaging in patient management during the covid-19 pandemic: a multinational consensus statement from the fleischner society. Chest [DOI] [PMC free article] [PubMed]
- 27.Sandler M, Howard A, Zhu M, Zhmoginov A, Chen LC (2018) Mobilenetv2: Inverted residuals and linear bottlenecks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4510–4520
- 28.Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv:14091556
- 29.Singhal T (2020) A review of coronavirus disease-2019 (covid-19). The Indian Journal of Pediatrics pp 1–6 [DOI] [PMC free article] [PubMed]
- 30.Sohrabi C, Alsafi Z, O’Neill N, Khan M, Kerwan A, Al-Jabir A, Iosifidis C, Agha R (2020) World health organization declares global emergency: A review of the 2019 novel coronavirus (covid-19). Int J Surg [DOI] [PMC free article] [PubMed]
- 31.Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
- 32.Szegedy C, Ioffe S, Vanhoucke V, Alemi AA (2017) Inception-v4 inception-resnet and the impact of residual connections on learning. In: Thirty-First AAAI conference on artificial intelligence
- 33.Tandon PN, et al. Covid-19: impact on health of people & wealth of nations. Indian J Med Res. 2020;151(2):121. doi: 10.4103/ijmr.IJMR_664_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VY, Chen H, Mubareka S, Gubbay J, Chan WC (2020) Diagnosing covid-19: the disease and tools for detection. ACS nano [DOI] [PubMed]
- 35.Wang L, Wong A (2020) Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest radiography images. arXiv:200309871 [DOI] [PMC free article] [PubMed]
- 36.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM (2017) Chestx-ray8: hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2097–2106
- 37.WHO (2020) Who director-general’s opening remarks at the media briefing on covid-19 - 16 march 2020 https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---16-march-2020
- 38.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in china. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.