Extracellular vesicles (EVs) are cellular membrane-derived nanosized particles that are produced by most cells in all three kingdoms of life. They play a pivotal role in cell-cell communication through their ability to transport bioactive molecules from donor to recipient cells.
KEYWORDS: EV, NF-κB, anti-inflammatory, comparative proteomics, growth conditions, immunomodulation, membrane vesicle, protein-protein interactions
ABSTRACT
Propionibacterium freudenreichii is a probiotic Gram-positive bacterium with promising immunomodulatory properties. It modulates regulatory cytokines and mitigates the inflammatory response in vitro and in vivo. These properties were initially attributed to specific bacterial surface proteins. Recently, we showed that extracellular vesicles (EVs) produced by P. freudenreichii CIRM-BIA129 mimic the immunomodulatory features of parent cells in vitro (i.e., modulating NF-κB transcription factor activity and interleukin-8 release), which underlies the role of EVs as mediators of the probiotic effects of the bacterium. The modulation of EV properties, and particularly of those with potential therapeutic applications, such as the EVs produced by the probiotic P. freudenreichii, is one of the challenges in the field to achieve efficient yields with the desired optimal functionality. Here, we evaluated whether the culture medium in which the bacteria are grown could be used as a lever to modulate the protein content and, hence, the properties of P. freudenreichii CIRM-BIA129 EVs. The physical, biochemical, and functional properties of EVs produced from cells cultivated on laboratory yeast extract lactate (YEL) medium and cow milk ultrafiltrate (UF) medium were compared. UF-derived EVs were more abundant and smaller in diameter, and they displayed more intense anti-inflammatory activity than YEL-derived EVs. Furthermore, the growth media modulated EV content in terms of both the identities and abundances of their protein cargos, suggesting different patterns of interaction with the host. Proteins involved in amino acid metabolism and central carbon metabolism were modulated, as were the key surface proteins mediating host-propionibacterium interactions.
IMPORTANCE Extracellular vesicles (EVs) are cellular membrane-derived nanosized particles that are produced by most cells in all three kingdoms of life. They play a pivotal role in cell-cell communication through their ability to transport bioactive molecules from donor to recipient cells. Bacterial EVs are important factors in host-microbe interactions. Recently, we have shown that EVs produced by the probiotic P. freudenreichii exhibited immunomodulatory properties. We evaluate here the impact of environmental conditions, notably culture media, on P. freudenreichii EV production and function. We show that EVs display considerable differences in protein cargo and immunomodulation depending on the culture medium used. This work offers new perspectives for the development of probiotic EV-based molecular delivery systems and reinforces the optimization of growth conditions as a tool to modulate the potential therapeutic applications of EVs.
INTRODUCTION
Probiotic organisms are increasingly being used in medical and technological contexts because of their health benefits (1, 2). Among these organisms, dairy bacteria such as Propionibacterium freudenreichii are of particular value because of their long-term safe consumption and economic interest (3, 4). P. freudenreichii is a Gram-positive bacterium that is used traditionally as a Swiss-type cheese starter (5) but has also been studied with respect to the production of vitamin B12 (6, 7) and organic acids (8, 9). Furthermore, this bacterium can survive for a considerable period in vitro and under cheese-making conditions (10). It can also survive and adapt metabolically to animal and human gastrointestinal tracts (11–13) and has been identified in the fecal samples from a discrete cohort of human preterm breastfed infants (14, 15).
P. freudenreichii has long been studied for its probiotic properties, such as modulating the composition of the microbiota, antitumor activity, and immunomodulation (3). This species mainly modulates the composition of the microbiota through a bifidogenic effect, promoted by abundantly produced metabolites such as 1,4-dihydroxy-2-naphthoic acid (DHNA) and 2-amino-3-carboxy-1,4-naphthoquinone (ACNQ) (16–19). Its antitumor activity is linked to the production of short-chain fatty acids such as propionate and acetate (20–24). Regarding immunomodulation, some proteins in this species, and particularly surface layer protein B (SlpB), have been associated with a reduction in the proinflammatory cytokines released and/or an increase in the release of anti-inflammatory cytokines both in vitro and in vivo (19, 25–29).
In addition to bacterial surface exposure, we recently showed that some immunomodulatory P. freudenreichii proteins are found associated with extracellular vesicles (EVs) (30). EVs are spherical nanometric structures produced by cells in the three domains of life (31). They are composed of lipid bilayers and an internal functional cargo, implicated in several biological processes that include interactions with host cells (31–34). The production of EVs has been demonstrated in several probiotic bacteria species (35–41). In the case of P. freudenreichii, EVs produced by the CIRM-BIA129 strain had a proteome predicted to interact with key protein components of the human inflammatory response, which was reinforced by the ability of EVs to reduce both the activity of the transcription factor NF-κB and interleukin-8 (IL-8) release in an intestinal epithelial cell model (30). The modulation exerted by ultrafiltrate (UF)-derived EVs occurred when the inflammatory response was induced by bacterial lipopolysaccharide (LPS) but not by other inducers, such as tumor necrosis factor alpha (TNF-α) and IL-1β. In addition, the EV-borne SlpB protein was shown to play a role in this immunomodulation, because EVs secreted by a ΔslpB mutant displayed reduced anti-inflammatory activity (30).
As well as their functional role in modulating interactions with the host, bacterial EVs are themselves modulated by specific conditions. In other words, environmental shifts impact the production and functional properties of EVs (42). For example, exposure to antibiotics induces vesiculation in several Staphylococcus aureus strains (43) and modifies the protein content of EVs secreted by Acinetobacter baumannii DU202 (44) and Campylobacter jejuni 81-176 (45). Iron-limiting conditions were shown to be associated with quantitative changes to the EV proteome of pathogenic and probiotic Escherichia coli strains (46, 47), of Helicobacter pylori 60190 (48), and of Mycobacterium tuberculosis H37Rv (49). Salt stress was also shown to be linked to drastic changes in the protein content and proinflammatory activity of EVs secreted by Listeria monocytogenes 10403S (50, 51). The EV proteome or its activity also changed in line with growth phases in Bacillus subtilis 168 (52), Rhizobium etli CE3 (53), Pseudomonas aeruginosa PAO1 (54), and H. pylori 26695 (55) and with the composition of media in Gallibacterium anatis 12656-12 (56) and Pseudomonas putida KT2440 (57). The bacterial properties of EVs were also affected by pH shifts (58) and exposure to epibromohydrin (59) or cannabidiol (60) and to other environmental conditions (61).
The impact of environmental conditions on EV production and function still remains poorly documented in probiotic bacteria, although it may represent a tool to modulate the properties of EVs and have potential therapeutic applications. Therefore, our aim was to investigate whether different growth conditions impact the production of P. freudenreichii-derived EVs as well as modulate their properties. In the present study, we have shown that P. freudenreichii CIRM-BIA129 cultured in milk UF or yeast extract-lactate (YEL) growth medium produced EVs with distinct physicochemical, biochemical, and functional properties. In particular, UF- and YEL-derived EVs displayed considerable differences in protein cargo and immunomodulation. This study is a comparative analysis of the properties of the EVs produced by P. freudenreichii under various growth conditions and will contribute to understanding how probiotic traits are affected by different contexts. It also suggests interventional opportunities for the engineering of EV content and activity, enabling improvements to their potential technological and therapeutic roles.
RESULTS
The production of EVs by P. freudenreichii is dependent on growth conditions.
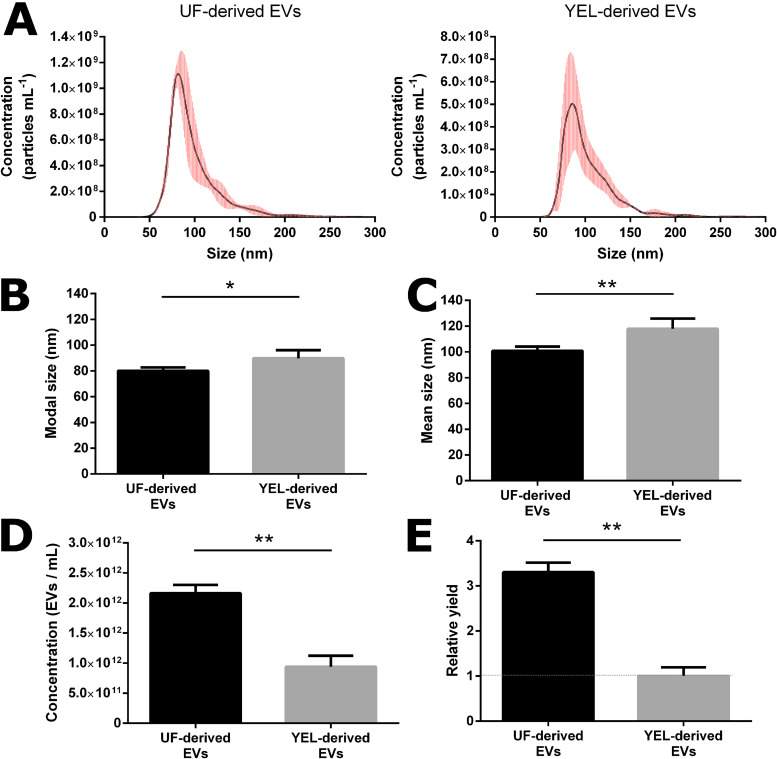
During this work, we tried to determine whether environmental conditions, i.e., different growth media, affected the production of EVs by P. freudenreichii CIRM-BIA129. For that purpose, the sizes and concentrations of EVs purified from UF and YEL culture media were evaluated. Both conditions yielded EVs with a monodisperse size distribution, although YEL-derived EVs were less abundant than UF-derived EVs (Fig. 1A). They also displayed differences in diameter: UF-derived EVs had a modal size of 80.06 ± 2.606 nm and a mean size of 100.9 ± 3.204 nm, whereas YEL-derived EVs were significantly larger (P < 0.05), with a modal size of 89.72 ± 6.324 nm and a mean size of 117.9 ± 7.856 nm (Fig. 1B and C). The total concentration of EVs was significantly higher (P < 0.01) from UF (2.164 × 1012 ± 1.383 × 1011 EVs ml−1) than from YEL (9.394 × 1011 ± 1.846 × 1011 EVs ml−1) medium (Fig. 1D). Finally, the EV relative yield of CIRM-BIA129 (i.e., the amount of recovered EVs normalized by the amount of bacterial cells at sampling time) was more than three times higher from UF than from YEL medium (Fig. 1E).
FIG 1.
Characteristics of P. freudenreichii-secreted EVs under different growth conditions. (A) Size distribution of EVs derived from UF (left) and YEL (right) culture media. (B and C) Mode (B) and mean (C) diametric sizes of EVs produced from UF and YEL culture media. (D) Total concentrations of EVs produced from UF and YEL culture media. (E) Relative yields of EVs (normalized by the amount of bacterial cells in each culture medium). Size and concentration measurements were acquired using nanoparticle tracking analysis (NTA). Data are expressed as means ± standard deviations of values obtained from at least three independent biological replicates. Asterisks indicate statistical significance as evaluated by the Mann-Whitney test: **, P ≤ 0.01; *, P ≤ 0.05.
The biological activity of P. freudenreichii-derived EVs is dependent on growth conditions.
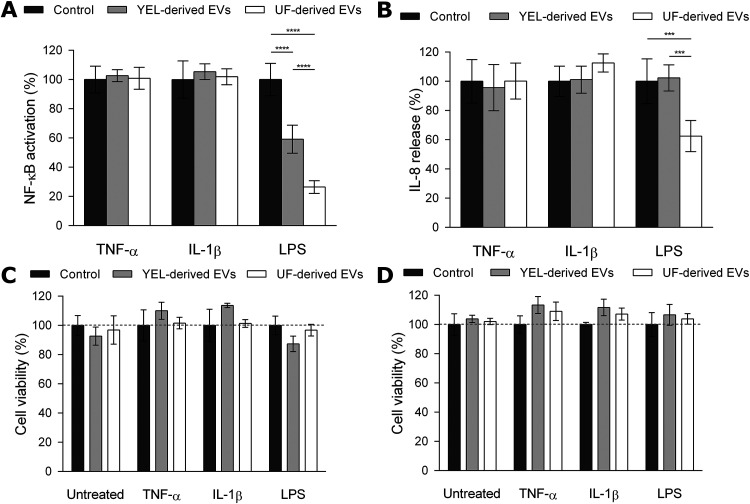
EVs derived from the two growth conditions were also evaluated in terms of their biological activity in vitro, i.e., NF-κB activation and IL-8 release. UF-derived P. freudenreichii CIRM-BIA129 EVs had previously been shown to exert an immunomodulatory effect on HT-29 human intestinal epithelial cells via the NF-κB pathway, i.e., modulation of transcription factor NF-κB activity and IL-8 release (30). To determine whether growth conditions also modulate EV activity, we compared the ability of the EVs produced from UF and YEL media to modulate the regulatory activity of the NF-κB transcription factor and the release of IL-8 from human intestinal epithelial cells. For that purpose, NF-κB/SEAP HT-29 cells, which are designed to measure NF-κB regulatory activity (62), and HT-29 parental cells were induced to an inflammatory state by different proinflammatory inducers and treated with EV preparations. Whatever the growth medium, EVs exerted an inhibitory effect on the regulatory activity of the NF-κB transcription factor only when the inflammation pathway was induced by LPS (Fig. 2A). Nevertheless, EV-mediated reduction of the LPS-induced proinflammatory effect was growth medium dependent. Indeed, the anti-inflammatory activity of EVs was significantly more intense with UF-derived than with YEL-derived EVs (P < 0.0001) (Fig. 2A). The specific LPS-induced anti-inflammatory effect of EVs was confirmed by the evaluation of IL-8 release from the HT-29 parental cell line. However, in this case, only UF-derived EVs were able to reduce LPS-induced IL-8 release (Fig. 2B). Finally, UF- and YEL-derived EVs had no cytotoxic effect on the two HT-29 cell lines, indicating that reductions in NF-κB activation and IL-8 release were not associated with cell death (Fig. 2C and D).
FIG 2.
P. freudenreichii-secreted EVs play different biological roles depending on growth conditions. (A) Percent NF-κB transcription factor activity in HT-29/kb-seap-25 cells left untreated or treated with the inflammatory inducer TNF-α (1 ng ml−1), IL-1β (1 ng ml−1), or LPS (1 ng ml−1) in the presence or absence of UF- and YEL-derived EVs (1.0 × 109 EVs ml−1). The values are normalized by the control conditions (control TBS buffer or stimulation by the inducer in the absence of EVs). (B) Percentage of IL-8 released by HT-29 intestinal epithelial cells after stimulation by LPS (1 ng ml−1), TNF-α (1 ng ml−1), or IL-1β (1 ng ml−1) inducer in the presence or absence of UF- and YEL-derived EVs (1.0 × 109 EVs ml−1). The values are normalized to the control conditions (control TBS buffer or stimulation by inducer in the absence of EVs). (C and D) Percent viability of HT-29/kb-seap-25 reporter (left) and HT-29 parental (right) cells before or after stimulation by LPS (1 ng ml−1), TNF-α (1 ng ml−1), or IL-1β (1 ng ml−1) inducer in the presence or absence of UF- and YEL-derived EVs (1.0 × 109 EVs ml−1). The values are normalized by the control conditions (control TBS buffer or stimulation by an inducer in the absence of EVs). Data are expressed as mean ± standard deviation of values obtained from at least three independent biological replicates. Asterisks indicate statistical significance as evaluated by two-way ANOVA with Tukey's multiple-comparison test: ****, P ≤ 0.0001; ***, P ≤ 0.001.
EVs produced under different growth conditions contain shared and exclusive proteins.
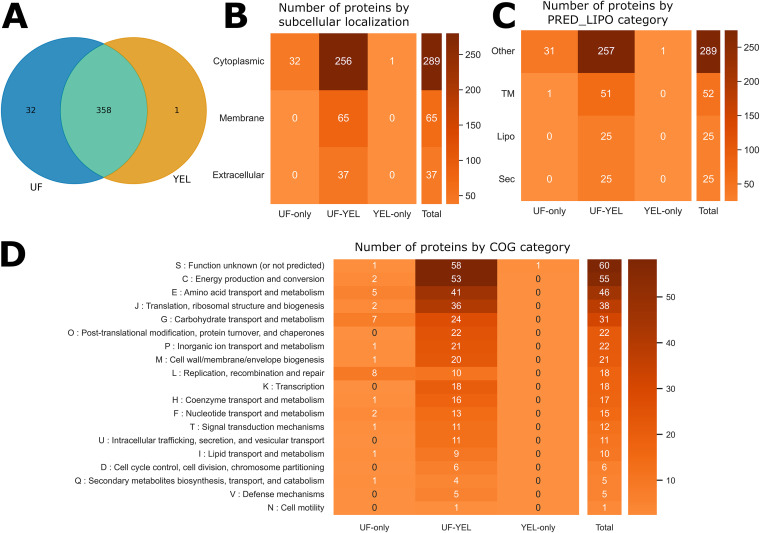
To identify potential factors involved in the differences in biological activity between UF- and YEL-derived EVs, we characterized the protein content of EVs produced under the two growth conditions (see Table S4 in the supplemental material). Qualitative proteomics analysis enabled the identification of 391 proteins, 32 of which were exclusively detected in UF-derived EVs, one was exclusively detected in YEL-derived EVs, and 358 were common to both growth conditions (Fig. 3A). Concerning subcellular localization, all exclusive proteins (UF and YEL exclusive) were predicted to be cytoplasmic. The shared proteins were mainly cytoplasmic (n = 256), but some were also predicted to be membrane proteins (n = 65) or extracellular proteins (n = 37) (Fig. 3B). Predicted lipoproteins were only identified among proteins found under both conditions (n = 25) (Fig. 3C). The frequencies of Clusters of Orthologous Groups (COG) categories were well distributed among shared proteins, mainly being related to metabolism and information processing. UF-exclusive proteins were mainly linked to carbohydrate or amino acid metabolism and DNA processing, while the only YEL-exclusive protein had an unknown function (Fig. 3D).
FIG 3.
Qualitative proteomics can reveal differences in EV protein content as a function of growth conditions. (A) Venn diagram showing the number of shared and specific proteins for EVs under the two growth conditions. (B to D) Heatmaps showing the distribution of proteins as a function of growth condition categories and functional predictions. Columns show growth condition categories (UF-only, proteins identified exclusively in UF-derived EVs; YEL-only, proteins identified exclusively in YEL-derived EVs; UF-YEL, proteins identified under both growth conditions). Rows show subcellular localization prediction (cytoplasmic, membrane, or extracellular) (B), PRED-LIPO prediction (Sec, secretion signal peptide; Lipo, lipoprotein signal peptide; TM, transmembrane; Other, no signals found) (C), and COG functional categories (D).
A comparison of growth conditions reveals several differentially abundant EV proteins.
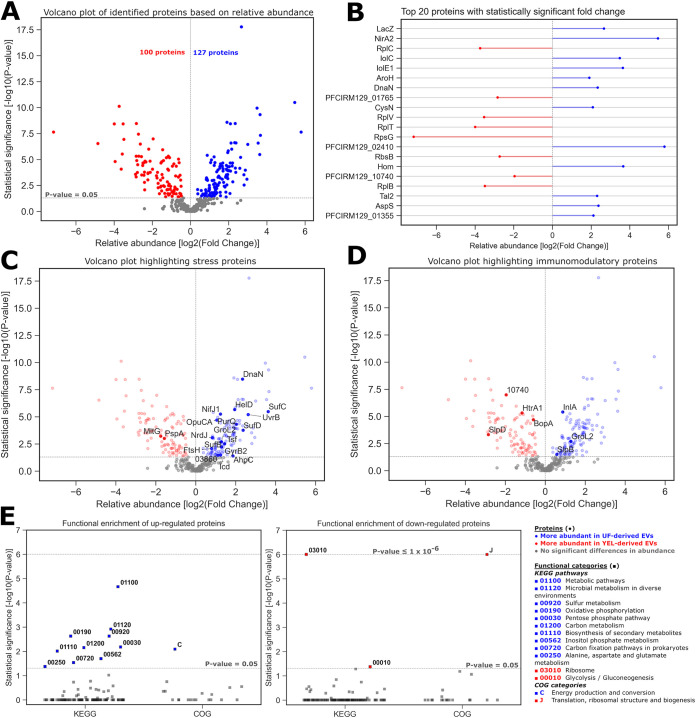
Qualitative proteomic analysis pinpointed proteins specific to YEL- and UF-derived EVs. Further, to better grasp the impact of growth conditions on EV content, the relative abundance of the proteins was analyzed under each condition. Quantitative proteomics showed that the relative abundance of EV proteins could vary as a function of culture medium (Fig. S1). Among the total of 391 identified proteins, the abundances of 164 proteins did not differ between YEL- and UF-derived EVs (P > 0.05), 127 proteins were significantly more abundant in UF-derived EVs (P ≤ 0.05, log[fold change] > 0), and 100 proteins were significantly more abundant in YEL-derived EVs (P ≤ 0.05, log[fold change] < 0) (Fig. 4A). Among the proteins more abundant in UF-derived EVs, the most significant included those related to carbohydrate (LacZ, IolC, and IolE1), amino acid (AroH and Hom), and energy (NirA2) metabolism. Among the proteins more abundant in YEL-derived EVs, the most significant were mainly ribosomal proteins (RplC, RplV, RplT, RplB, and RpsG). Differentially abundant proteins under each condition also included those of unknown function or poorly characterized (PFCIRM129_01355, PFCIRM129_01765, PFCIRM129_02410, and PFCIRM129_10740) (Fig. 4B). Interestingly, among EV proteins undergoing significant changes to their abundance as a function of the culture medium, we identified several that had reportedly been associated with a stress response (Fig. 4C) or immunomodulatory properties (Fig. 4D) in P. freudenreichii or other bacterial species. Functional enrichment analysis (Tables S5 and S6) showed that proteins that were more abundant in UF-derived EVs were mainly related to energy, carbohydrate, amino acid, and sulfur metabolism KEGG pathways, together with COG category C (energy production and conversion). On the other hand, proteins more abundant in YEL-derived EVs were mainly related to glycolysis/gluconeogenesis and ribosome KEGG pathways, together with COG category J (translation, ribosomal structure, and biogenesis) (Fig. 4E).
FIG 4.
EVs derived from different growth conditions carry proteins with different relative abundances. (A) Volcano plot showing the relative abundance of proteins from UF-derived EVs versus YEL-derived EVs (fold change UF/YEL) as well as the significance of their fold changes. The results are presented according to a logarithmic scale, with positive fold change ratios shown in blue, negative fold change ratios shown in red, and nonsignificant ratios shown in gray. The horizontal dotted line shows the threshold of significance, and the vertical line indicates a logarithmic fold change of zero. (B) Relative abundance of proteins with the highest significant UF/YEL fold changes. (C and D) Volcano plots highlighting subsets of proteins reportedly associated with a stress response (C) or immunomodulation (D) in P. freudenreichii or other bacterial strains. (E) Enrichment analysis of KEGG pathways and COG categories for the upregulated (left) and downregulated (right) proteins. Significant results are shown in blue for categories enriched in upregulated proteins, in red for categories enriched in downregulated proteins, and in gray for categories that are not statistically enriched. The lower dotted line represents the minimal threshold of significance, and the higher dotted line indicates a cap for categories with P values of 1 × 10−6 or lower.
Predictions of interactions highlight the relevant bacterial proteins potentially implicated in immunomodulation.
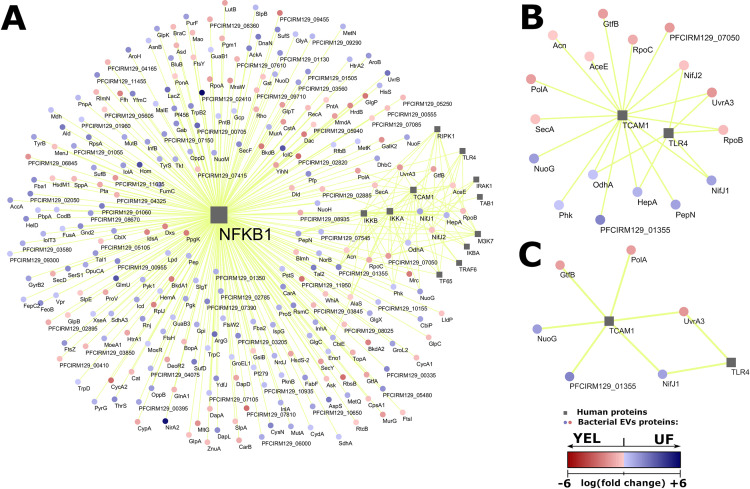
The qualitative and quantitative differences in the protein content of EVs revealed factors that might vary with growth conditions. Our next aim was to investigate whether this variation, when translated into different patterns of interaction, might offer insights into the mechanisms underlying the differential activity of EVs. Therefore, 101 human proteins from the NF-κB pathway and 391 bacterial proteins found in EVs were submitted to the prediction of protein-protein interactions. A total of 893 interactions were considered to be valid (minimum score of 0.9765), involving 49 human and 266 bacterial proteins (Table S7). Next, we performed progressive filtering procedures to select meaningful interactions according to experimental criteria. First, in view of the fact that the anti-inflammatory activity of EVs was LPS specific, we filtered out interactions involving human proteins that were not related to the LPS-induced part of the NF-κB pathway, which resulted in a subnetwork of 357 interactions (Fig. 5A). A further filtering was then performed to retain only human proteins that were specific to the LPS-induced NF-κB pathway, i.e., not related to TNF-α or IL-1β induction. This resulted in 23 interactions, involving just two human proteins: TCAM1 and TLR4 (Fig. 5B). A final filtering was performed in bacterial proteins to retain only those whose differential expression was considered significant (P ≤ 0.05) according to quantitative proteomics (Fig. 5C). Therefore, TLR4 and TCAM1 human proteins were identified within the NF-κB signaling pathway as potential targets for immunomodulation. Moreover, novel bacterial proteins were suggested to be responsible for the differential intensity of the anti-inflammatory response resulting from various growth conditions: pyruvate synthase/pyruvate-flavodoxin oxidoreductase (NifJ1), the NADH-quinone oxidoreductase chain G (NuoG), an uncharacterized protein (PFCIRM129_01355), the GtfB glycosyltransferase (GtfB), a putative DNA polymerase I (PolA), and UvrABC system protein A (UvrA3).
FIG 5.
Prediction of protein-protein interactions highlights relevant bacterial proteins. (A to C) A network representation of predicted interactions filtered by human proteins that correspond to the LPS-induced part of the NF-κB pathway (A), by uman proteins involved in the LPS-induced NF-κB pathway but not in induction by TNF-α or IL-1β (B), and by bacterial proteins whose abundance was considered significant by quantitative proteomic analysis (C). Human proteins are represented by gray squares, and bacterial proteins are represented by red or blue circles. Predictions of interactions are represented by yellow lines connecting squares and circles. The relative abundance of bacterial proteins is seen according to the scale in the bottom right corner (logarithm of fold change), where positive values (blue) indicate a higher abundance in UF-derived EVs and negative values (red) indicate a higher abundance in YEL-derived EVs.
DISCUSSION
Several studies have demonstrated that the production, properties, and roles of bacterial EVs are affected by environmental conditions. Thus, monitoring the environmental conditions under which the bacteria are grown offers a tool to modulate the production and properties of EVs and may have some therapeutic applications (42). We recently reported the production of EVs by the probiotic P. freudenreichii CIRM-BIA129, and our study included their physicochemical and functional characterization (30). In particular, we showed that they displayed anti-inflammatory effects throughout modulation of the regulatory activity of the NF-κB transcription factor and IL-8 release in human epithelial cell models. However, whether the production and immunomodulatory effects of P. freudenreichii-secreted EVs could be modulated by environmental conditions still needed to be determined.
Here, we compared the properties of P. freudenreichii EVs purified from two growth media that are routinely used for P. freudenreichii cultures: cow’s milk ultrafiltrate (UF) medium and yeast extract-lactate (YEL) medium (29, 63, 64). YEL is the gold-standard laboratory medium for propionibacteria. This medium, in addition to yeast extract (providing the necessary growth factors) and sodium lactate (the preferred carbon and energy source), contains peptone (as a nitrogen source) (65). This medium was developed to mimic the growth conditions of propionibacteria in Swiss-type cheeses after fermentation of the cheese curd by lactic acid bacteria. We later developed a milk UF medium to mimic the growth of propionibacteria in fermented milk (63, 66). This medium represents the aqueous phase of cow milk, added with casein peptone and sodium lactate. UF and YEL were chosen to compare EV properties because they differentially impact the physiology of the bacterium, notably its growth parameters (biomass and generation time), the pH of the extracellular medium at the end of stationary phase, and cell viability after stress challenges (63, 64).
We demonstrated that culture media affect the physical, biochemical, and biological properties of P. freudenreichii-secreted EVs. Notably, compared to EVs recovered from YEL cultures, markedly more were recovered from UF cultures. In some bacterial species, an increase in EV production has been linked to an adaptation to stressful conditions (67–69). In the case of P. freudenreichii, UF could be considered a more stressful growth condition than YEL, since the bacterium accumulated higher proportions of trehalose during growth in UF than in YEL (64), with this sugar being involved in the response to stress, notably during acid adaptation and osmoadaptation (64, 70, 71). Moreover, some of the proteins that we found more abundant in UF than YEL-derived EVs were associated in the parent cells with the adaptation of the bacterium to stress (Fig. 4C). Among these, chaperonin GroL2 (GroL2), pyruvate synthase/pyruvate-flavodoxin oxidoreductase (NifJ1), and alkyl hydroperoxide reductase protein C22 (AhpC) were notably associated with the response to acid stress in this bacterium (72). Taken together, these results suggested that EV production in P. freudenreichii is affected by stress conditions and showed that the control of growth conditions might offer a lever to modulate EV production in this probiotic bacterium.
As mentioned above, in addition to impacting the physical properties of EVs, the culture medium also modulates EV content in terms of both the identities and abundances of its protein cargo. When the protein contents were compared, modest differences were observed between YEL- and UF-derived EVs. Indeed, more than 90% (n = 358) of EV proteins were found to be common to the YEL and UF media. Only one protein with an unknown function was specific to YEL, and 32 of the predicted cytoplasmic proteins were found to be exclusive to UF-derived EVs. This notably included several proteins with functions assigned to the COG categories of “replication, recombination, and repair” (L, n = 8), “carbohydrate transport and metabolism” (G, n = 7), and “amino acid transport and metabolism” (E, n = 5). The most important impact of the culture medium concerned the abundance of proteins packed into EVs. The abundance of approximately 60% of EV proteins changed significantly as a function of culture medium. In the case of YEL-derived EVs, the more abundant proteins were mainly related to ribosomes, glycolysis, and gluconeogenesis, which may have been linked to the higher growth rate of P. freudenreichii in YEL medium (63, 64). In contrast, UF-derived EVs were more enriched in proteins related to pentose phosphate, sulfur, secondary metabolites, inositol phosphate, amino acids, and energy metabolism, which are probably related to stress responses. Although the rules for the cargo selection and sorting into bacterial EVs remain elusive, especially in Gram-positive bacteria (73), it is often thought that the content of EVs reflects the physiology, metabolic status, and biological properties of the cells producing them (32, 52). Therefore, modulation of both the presence and abundance of proteins in EVs as a function of culture medium might be related to the level of their synthesis by cells. Indeed, it seems consistent that the relative abundance of a protein in a whole cell would affect its availability to be packed into EVs in the absence of, or in addition to, a selective process (34, 74). Accordingly, several proteins associated with the adaptation of the bacterium to UF or YEL medium were identified as being more abundant in UF- or YEL-derived EVs, respectively. For example, some of the proteins specifically present or found to be more abundant in UF-derived EVs were involved in the assimilation of the main carbon sources of UF, including beta-galactosidase (LacZ) (which hydrolyzes lactose into glucose and galactose) and inositol assimilation proteins (IolE1, IolC, IolT3, and IolA). However, one cannot exclude that, in addition to the level of synthesis by cells, some of these proteins are also purposely packed into EVs to perform specific functions, such as host cell interactions (75). To date, the molecular mechanisms that drive the recruitment of proteins into bacterial EVs have remained unclear. Determining which proteins are selected, and how, is of crucial value to identifying those purposely packed into EVs, notably to better understand their biological roles. Recently, we showed that abundance, charge, and subcellular localization could influence the protein availability of the vesicle cargo in S. aureus (74). Whether these rules are also applicable to the EVs secreted by P. freudenreichii still needs to be investigated, but our data may offer an additional opportunity to uncover the mechanisms governing the selection of proteins into EVs.
Finally, the culture medium in which bacteria are grown also modulates the biological activity of CIRM-BIA129 EVs. Indeed, UF- and YEL-derived EVs presented remarkable differences in terms of immunomodulation. We showed that EVs from both origins reduced the activity of NF-κB, a key mediator of inflammatory responses, but at different intensities. This reduction occurred specifically when inflammation was induced by bacterial LPS and was achieved in roughly 74% for UF-derived EVs but only 41% for YEL-derived EVs. This differential EV-mediated modulation of NF-κB activity has profound consequences on the cellular inflammatory response. Indeed, LPS-induced IL-8 release by HT-29 cells was significantly reduced by UF-derived EVs but not by YEL-derived EVs. To date, the bacterial effectors involved in the immunomodulatory activity of P. freudenreichii-secreted EVs are still unknown. We recently showed that SlpB participates in EV-mediated anti-inflammatory effects in vitro, since EVs derived from a ΔslpB mutant displayed a partial reduction of NF-κB activation compared to wild-type-derived EVs (30). However, it is not known whether the role of SlpB is direct or indirect. Among P. freudenreichii-associated immunomodulatory proteins, SlpB, internaline A (InlA), and the chaperonin GroL2 were more abundant in UF- than in YEL-derived EVs. In contrast, surface-layer protein D (SlpD), the solute binding protein of the ABC transport system (BopA), trypsin-like serine protease (HtrA1), and a protein of unknown function, PFCIRM129_10740, were more abundant in YEL-derived EVs. The differences in abundance of at least one of these recognized immunomodulatory proteins may explain the differential anti-inflammatory activity seen for UF- and YEL-derived EVs (27–29, 76).
One can also suppose that other differentially abundant proteins are implicated in the differential activity of EVs triggered by culture media. These include those bacterial proteins predicted to interact with the human proteins Toll-like receptor 4 (TLR4) and TIR domain-containing adapter molecule 1 (TICAM-1), which are of particular interest. Indeed, these two proteins are involved in the NF-κB signaling pathway induced by LPS but not by TNF-α or IL-1β; therefore, they are good candidates as molecular targets for immunomodulation mediated by P. freudenreichii-derived EVs with LPS-specific induction. Consequently, the abundances of their potential bacterial interacting partners might also explain the differences in EV activity as a function of culture medium. Among the predicted TLR4- and TICAM1-interacting bacterial proteins, six displayed a different abundance pattern between UF- and YEL-derived EVs. These included NifJ1, the NADH-quinone oxidoreductase chain G (NuoG), and an uncharacterized protein (PFCIRM129_01355), which were more abundant in UF-derived EVs, and GtfB glycosyltransferase (GtfB), a putative DNA polymerase I (PolA), and the UvrABC system protein A (UvrA3), which were more abundant in YEL-derived EVs. These proteins have not previously been characterized as being immunomodulatory, but our in silico network predictions suggest they can interact with human proteins with key roles in the inflammatory response; their abundances also could explain why UF- and YEL-derived EVs display distinct patterns of biological activity. Whether these proteins as well as other differentially abundant immunomodulatory proteins participate in the modulation of EV biological activity as a function of culture medium certainly deserves further investigation.
To sum up, our study showed that the probiotic P. freudenreichii CIRM-BIA129 produced EVs with distinct properties depending on the culture medium in which the bacterium was grown and reinforced the importance of environmental conditions to the properties of EVs. Although little is known about the mechanisms that underpin the modulation of Gram-positive bacterial EV production, content, and activity, they respond definitively to environmental stimuli in the case of P. freudenreichii. Specifically, in the context of probiotic bacteria, the optimization of medium composition might represent a tool to improve the beneficial activity of EVs and enable the development of EV-based therapeutic applications, such as functional foods with improved properties or pure EV formulations. Comprehensive studies might elucidate the relationships between specific medium components and the vesicular export of immunomodulatory proteins, enabling an improvement to the anti-inflammatory properties of EVs.
MATERIALS AND METHODS
Bacterial cultures.
P. freudenreichii CIRM-BIA129 (equivalent to the ITG P20 strain, provided by CNIEL) was supplied, stored, and maintained by the CIRM-BIA Biological Resource Center (Centre International de Ressources Microbiennes-Bactéries d'Intérêt Alimentaire, INRAE, Rennes, France). The bacteria were grown under two conditions: in yeast extract-lactate medium (YEL) or cow milk ultrafiltrate medium (UF), which was further supplemented with 100 mM sodium lactate and 5 g liter−1 casein hydrolysate, as previously described (63, 65). For both conditions, incubation was at 30°C, without agitation, until the start of stationary phase (2 × 109 bacteria ml−1 for UF and 3 × 109 bacteria ml−1 for YEL), as reported elsewhere (63). The culture starting volume was 500 ml for each biological replicate.
Purification of EVs.
Bacterial cultures (500 ml) were centrifuged at room temperature (6,000 × g, 15 min) and the supernatants filtered through 0.22-µm top filters (Nalgene, Thermo Scientific). The supernatants were then concentrated 1,000-fold using 100-kDa ultrafiltration units (Amicon, Merck Millipore) by successive centrifugations (2,500 × g). The concentrated suspension of EVs was recovered in TBS buffer (Tris-buffered saline; 150 mM NaCl, 50 mM Tris-Cl, pH 7.5) and further purified by size exclusion chromatography (qEV original, 70 nm; iZON) as recommended by the manufacturer (30, 77). The resulting fractions containing EVs in TBS buffer were pooled (1.5 ml) and further concentrated to achieve approximately 50 µl using 10-kDa centrifugal filter units (Amicon, Merck Millipore). The aliquots were stored at −20°C or used immediately.
Characterization of EVs.
The characterization of EVs in terms of size and concentration was performed by nanoparticle tracking analysis using a NanoSight NS300 (Malvern Panalytical), equipped with a sCMOS camera, Blue488 laser, and NTA 3.3 Dev Build 3.3.104 software. The experiments were conducted at 25°C, with a camera level of 15 and a syringe pump speed of 50. Concentration, focus, and other parameters were adjusted accordingly for each experimental group. A detection threshold of 3 was employed to determine valid particles. EV homogeneity and integrity were also confirmed by negative staining electron microscopy (data not shown).
Evaluation of EV activity in vitro.
Two HT-29 human colon adenocarcinoma cell lines were employed to characterize the activity of EVs in vitro, as previously reported (30). Briefly, the parental HT-29 intestinal epithelial cells (ATCC HTB-38) were used to measure IL-8 release (IL-8/CXCL8 DuoSet; R&D Systems), and the lineage transfected with the secreted alkaline phosphatase (SEAP) reporter system (HT-29/kb-seap-25) was used to monitor NF-κB activity (Quanti-Blue reagent; Invivogen) (62). HT-29/kb-seap-25 cells were cultured in RPMI-glutamine medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum (Corning), 1% nonessential amino acids, 1% sodium pyruvate, 1% HEPES buffer (Thermofisher Scientific), and 1% penicillin-streptomycin (Lonza) (62). The parental HT-29 cells were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM) (Dominique Dutscher) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin (76). The cells were treated with TBS buffer as a control or the EV preparations (1.0 × 109 EVs ml−1) purified from UF or YEL cultures. To induce inflammation, the cells were also treated with TNF-α (1 ng ml−1; PeproTech), IL-1β (1 ng ml−1; Invivogen), LPS from Escherichia coli O111:B4 (1 ng ml−1; Sigma-Aldrich), or the TBS control. After seeding in 96-well plates (3 × 104 cells/well) and stimulation with the samples, the cells were incubated at 37°C, under 5% CO2, for 24 h. Cell confluence was verified under the microscope before and after stimulation. Cell viability after stimulation was investigated using the CellTiter 96 AQueous one-solution cell proliferation assay (MTS; Promega). Absorbance was measured with a Xenius (SAFAS Monaco) microplate reader at 655 nm for the SEAP (NF-κB) activity assay, 450 nm for the IL-8 enzyme-linked immunosorbent assay, and 490 nm for the MTS cell viability assay.
Identification and quantification of proteins using mass spectrometry.
One microgram of EV was dissolved in SDS and subjected to SDS-PAGE for a short period, allowing the entry of the protein into 2 to 3 mm of separating gel. The gel pieces were subjected to in-gel trypsinolysis followed by peptide extraction, as described previously (30, 72, 78). After digestion, the peptides were stored at −20°C until further analysis. Nano-liquid chromatography tandem mass spectrometry (LC-MS/MS) experiments were performed as previously described (30, 79). The peptides were identified from MS/MS spectra using X!TandemPipeline software (80), and searches were performed against the genome sequence of P. freudenreichii CIRM-BIA129 (GenBank accession no. NZ_HG975455). The database search parameters were specified as the following: trypsin cleavage was used, and peptide mass tolerance was set at 10 ppm for MS and 0.05 Da for MS/MS. Methionine oxidation was selected as a variable modification. For each peptide identified, an E value lower than 0.05 was considered a prerequisite for validation. A minimum of two peptides per protein was imposed, resulting in a false discovery rate (FDR) of <0.15% for protein identification. Each peptide identified by tandem mass spectrometry was quantified using the free MassChroQ software (81) before data treatment and statistical analysis with R software (R 3.4; Project for Statistical Computing). A specific R package, called MassChroqR (v0.4.3), was used to automatically filter dubious peptides and group the peptide quantification data into proteins. Two different and complementary analytical methods were used, based on peak counting or XIC (extracted ion current). For peak counting, variance analysis was performed on proteins with a minimum peak ratio of 1.5 between both culture conditions. Proteins with an adjusted P value of <0.05 were considered significantly different. For XIC-based quantifications, normalization was performed to take account of possible global quantitative variations between LC-MS runs. Peptides shared between different proteins were automatically excluded from the data set, as were peptides present in fewer than two of the three biological replicates. Missing data were then imputed from a linear regression based on other peptide intensities for the same protein. Analysis of variance was used to determine proteins whose abundance differed significantly between growth conditions.
Proteomic analysis.
Protein subcellular localizations were predicted by Cello2GO (82); lipoproteins were predicted by PRED-LIPO (83) and Clusters of Orthologous Groups (COG) categories, and KEGG Pathways were predicted by eggNOG-mapper v2 (84, 85). The Venn diagram, heatmaps, and volcano plot were conceived using Python’s Matplotlib-venn 0.11.5, Seaborn 0.10.0, and Bioinfokit 0.7 packages, respectively.
Functional enrichment analysis.
For functional enrichment analysis, the lists of up- and downregulated proteins were submitted to the g:Profiler web server (86, 87), together with an ortholog-based Gene Matrix Transposed (GMT) file (see Files S1 and S2 in the supplemental material) constructed from KEGG pathways and COG whole-proteome annotation categories obtained from eggNOG-mapper v2 (84, 85). Protein lists were ordered by decreasing relative level of abundance, and a significance threshold (adjusted P value) of 0.05 was adopted. For visualization, adjusted P values of 1 × 10−6 or lower were capped to this value.
Prediction of protein-protein interactions.
Human and bacterial protein sequences were retrieved from the UniProt and NCBI databases, respectively. Bacterial sequences identified in EVs derived from at least one of the growth conditions, as well as the human sequences corresponding to KEGG’s NF-κB pathway, were submitted to the InterSPPI web server for the prediction of interactions (88). Only interactions with a score of 0.9765 or higher were considered valid, with a specificity of 0.99. These interactions were gradually filtered so that human counterparts corresponded to, or were specific to, the LPS-induced portion of the NF-κB pathway, and/or bacterial proteins displayed a significant fold change in their expression according to quantitative proteomics. The lists of human proteins used for filtering are available in Tables S1 to S3. Predicted interactions and protein annotations were processed using the Pandas Python package (89). Visualization of the interaction network was achieved using Cytoscape software (90).
Statistical analysis.
All experiments were performed independently and at least in triplicate. The results are presented as means ± standard deviations. Absorbance measurements were normalized to the control conditions. The differences between experimental groups were analyzed using the Mann-Whitney test or two-way analysis of variance (ANOVA) followed by Tukey’s multiple-comparison test. Statistical analysis was performed using GraphPad Prism (GraphPad Software, San Diego, California, USA).
Data availability.
The mass spectrometry proteomics data can be found at https://doi.org/10.15454/Q6PPXY.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Ludovica Marinelli and Nicolas Lapaque (INRAE, AgroParisTech, University Paris-Saclay, Micalis Institute, Jouy-en-Josas, France) for their technical advice on the NF-κB reporter cell line. We also thank Victoria Hawken for English language editing of the manuscript. We thank the CNIEL (Centre National Interprofessionnel de l'Economie Laitière) for providing the ITG P20 strain (alias CIRM-BIA129) of P. freudenreichii.
V.R.R., G.J., Y.L.L., V.A.C.A., and E.G. conceived and designed the experiments. V.R.R., V.B.-B., J.J., and B.S.R.L. performed the experiments. V.R.R., A.N., J.J., and E.G. analyzed the data. F.L.R.C., A.N., J.J., V.B.-B., G.J., and E.G. gave practical suggestions regarding performance of the experiments. V.A.C.A., Y.L.L., and E.G. contributed to funding acquisition. V.R.R. and E.G. wrote the original draft. All authors contributed to data interpretation, drafting the manuscript, critically revising the manuscript, and approving its final version.
This work received financial support from INRAE (Rennes, France) and Institut Agro (Rennes, France). V.R.R. and B.S.R.L. were supported by the International Cooperation Program CAPES/COFECUB at the Federal University of Minas Gerais funded by CAPES–the Brazilian Federal Agency for the Support and Evaluation of Graduate Education of the Brazilian Ministry of Education (number 99999.000058/2017-03 and 88887.179897/2018-00, respectively).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Ozen M, Dinleyici EC. 2015. The history of probiotics: the untold story. Benef Microbes 6:159–165. doi: 10.3920/BM2014.0103. [DOI] [PubMed] [Google Scholar]
- 2.Lebeer S, Bron PA, Marco ML, Van Pijkeren J-P, O'Connell Motherway M, Hill C, Pot B, Roos S, Klaenhammer T. 2018. Identification of probiotic effector molecules: present state and future perspectives. Curr Opin Biotechnol 49:217–223. doi: 10.1016/j.copbio.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Rabah H, Rosa do Carmo F, Jan G. 2017. Dairy propionibacteria: versatile probiotics. Microorganisms 5:24. doi: 10.3390/microorganisms5020024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaucher F, Bonnassie S, Rabah H, Marchand P, Blanc P, Jeantet R, Jan G. 2019. Review: adaptation of beneficial propionibacteria, lactobacilli, and bifidobacteria improves tolerance toward technological and digestive stresses. Front Microbiol 10:841. doi: 10.3389/fmicb.2019.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ojala T, Laine PKS, Ahlroos T, Tanskanen J, Pitkänen S, Salusjärvi T, Kankainen M, Tynkkynen S, Paulin L, Auvinen P. 2017. Functional genomics provides insights into the role of Propionibacterium freudenreichii ssp. shermanii JS in cheese ripening. Int J Food Microbiol 241:39–48. doi: 10.1016/j.ijfoodmicro.2016.09.022. [DOI] [PubMed] [Google Scholar]
- 6.Deptula P, Chamlagain B, Edelmann M, Sangsuwan P, Nyman TA, Savijoki K, Piironen V, Varmanen P. 2017. Food-like growth conditions support production of active vitamin B12 by Propionibacterium freudenreichii 2067 without DMBI, the lower ligand base, or cobalt supplementation. Front Microbiol 8:368. doi: 10.3389/fmicb.2017.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamlagain B, Sugito TA, Deptula P, Edelmann M, Kariluoto S, Varmanen P, Piironen V. 2018. In situ production of active vitamin B12 in cereal matrices using Propionibacterium freudenreichii. Food Sci Nutr 6:67–76. doi: 10.1002/fsn3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pillai VV, Prakash G, Lali AM. 2018. Growth engineering of Propionibacterium freudenreichii shermanii for organic acids and other value-added products formation. Prep Biochem Biotechnol 48:6–12. doi: 10.1080/10826068.2017.1381619. [DOI] [PubMed] [Google Scholar]
- 9.Piwowarek K, Lipińska E, Hać-Szymańczuk E, Bzducha-Wróbel A, Synowiec A. 2018. Research on the ability of propionic acid and vitamin B12 biosynthesis by Propionibacterium freudenreichii strain T82. Antonie Van Leeuwenhoek 111:921–932. doi: 10.1007/s10482-017-0991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aburjaile FF, Rohmer M, Parrinello H, Maillard MB, Beaucher E, Henry G, Nicolas A, Madec MN, Thierry A, Parayre S, Deutsch SM, Cocaign-Bousquet M, Miyoshi A, Azevedo V, Le Loir Y, Falentin H. 2016. Adaptation of Propionibacterium freudenreichii to long-term survival under gradual nutritional shortage. BMC Genomics 17:1007. doi: 10.1186/s12864-016-3367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hervé C, Fondrevez M, Chéron A, Barloy-Hubler F, Jan G. 2007. Transcarboxylase mRNA: a marker which evidences P freudenreichii survival and metabolic activity during its transit in the human gut. Int J Food Microbiol 113:303–314. doi: 10.1016/j.ijfoodmicro.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Saraoui T, Parayre S, Guernec G, Loux V, Montfort J, Cam AL, Boudry G, Jan G, Falentin H. 2013. A unique in vivo experimental approach reveals metabolic adaptation of the probiotic Propionibacterium freudenreichii to the colon environment. BMC Genomics 14:911. doi: 10.1186/1471-2164-14-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lan A, Bruneau A, Philippe C, Rochet V, Rouault A, Hervé C, Roland N, Rabot S, Jan G, Herve C, Roland N, Rabot S, Jan G. 2007. Survival and metabolic activity of selected strains of Propionibacterium freudenreichii in the gastrointestinal tract of human microbiota-associated rats. Br J Nutr 97:714–724. doi: 10.1017/S0007114507433001. [DOI] [PubMed] [Google Scholar]
- 14.Colliou N, Ge Y, Sahay B, Gong M, Zadeh M, Owen JL, Neu J, Farmerie WG, Alonzo F, Liu K, Jones DP, Li S, Mohamadzadeh M. 2017. Commensal Propionibacterium strain UF1 mitigates intestinal inflammation via Th17 cell regulation. J Clin Investig 127:3970–3986. doi: 10.1172/JCI95376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang HY, Chen JH, Chang JH, Lin HC, Lin CY, Peng CC. 2017. Multiple strains probiotics appear to be the most effective probiotics in the prevention of necrotizing enterocolitis and mortality: an updated meta-analysis. PLoS One 12:e0171579. doi: 10.1371/journal.pone.0171579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mori H, Sato Y, Taketomo N, Kamiyama T, Yoshiyama Y, Meguro S, Sato H, Kaneko T. 1997. Isolation and structural identification of bifidogenic growth stimulator produced by Propionibacterium freudenreichii. J Dairy Sci 80:1959–1964. doi: 10.3168/jds.S0022-0302(97)76138-1. [DOI] [PubMed] [Google Scholar]
- 17.Isawa K, Hojo K, Yoda N, Kamiyama T, Makino S, Saito M, Sugano H, Mizoguchi C, Kurama S, Shibasaki M, Endo N, Sato Y. 2002. Isolation and identification of a new bifidogenic growth stimulator produced by Propionibacterium freudenreichii ET-3. Biosci Biotechnol Biochem 66:679–681. doi: 10.1271/bbb.66.679. [DOI] [PubMed] [Google Scholar]
- 18.Okada Y, Tsuzuki Y, Narimatsu K, Sato H, Ueda T, Hozumi H, Sato S, Hokari R, Kurihara C, Komoto S, Watanabe C, Tomita K, Kawaguchi A, Nagao S, Miura S. 2013. 1,4-Dihydroxy-2-naphthoic acid from Propionibacterium freudenreichii reduces inflammation in interleukin-10-deficient mice with colitis by suppressing macrophage-derived proinflammatory cytokines. J Leukoc Biol 94:473–480. doi: 10.1189/jlb.0212104. [DOI] [PubMed] [Google Scholar]
- 19.Rabah H, Ferret-Bernard S, Huang S, Le Normand L, Cousin FJ, Gaucher F, Jeantet R, Boudry G, Jan G. 2018. The cheese matrix modulates the immunomodulatory properties of Propionibacterium freudenreichii CIRM-BIA 129 in healthy piglets. Front Microbiol 9:2584. doi: 10.3389/fmicb.2018.02584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cousin FJ, Jouan-Lanhouet S, Théret N, Brenner C, Jouan E, Le Moigne-Muller G, Dimanche-Boitrel MT, Jan G. 2016. The probiotic Propionibacterium freudenreichii as a new adjuvant for TRAIL-based therapy in colorectal cancer. Oncotarget 7:7161–7178. doi: 10.18632/oncotarget.6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cousin FJ, Jouan-Lanhouet S, Dimanche-Boitrel MT, Corcos L, Jan G. 2012. Milk fermented by Propionibacterium freudenreichii induces apoptosis of HGT-1 human gastric cancer cells. PLoS One 7:e31892. doi: 10.1371/journal.pone.0031892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan G, Belzacq A-SS, Haouzi D, Rouault A, Métivier D, Kroemer G, Brenner C. 2002. Propionibacteria induce apoptosis of colorectal carcinoma cells via short-chain fatty acids acting on mitochondria. Cell Death Differ 9:179–188. doi: 10.1038/sj.cdd.4400935. [DOI] [PubMed] [Google Scholar]
- 23.Lan A, Lagadic-Gossmann D, Lemaire C, Brenner C, Jan G. 2007. Acidic extracellular pH shifts colorectal cancer cell death from apoptosis to necrosis upon exposure to propionate and acetate, major end-products of the human probiotic propionibacteria. Apoptosis 12:573–591. doi: 10.1007/s10495-006-0010-3. [DOI] [PubMed] [Google Scholar]
- 24.Lan A, Bruneau A, Bensaada M, Philippe C, Bellaud P, Rabot S, Jan G. 2008. Increased induction of apoptosis by Propionibacterium freudenreichii TL133 in colonic mucosal crypts of human microbiota-associated rats treated with 1,2-dimethylhydrazine. Br J Nutr 100:1251–1259. doi: 10.1017/S0007114508978284. [DOI] [PubMed] [Google Scholar]
- 25.Foligné B, Deutsch S-M, Breton J, Cousin FJ, Dewulf J, Samson M, Pot B, Jan G. 2010. Promising immunomodulatory effects of selected strains of dairy propionibacteria as evidenced in vitro and in vivo. Appl Environ Microbiol 76:8259–8264. doi: 10.1128/AEM.01976-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma S, Yeom J, Lim Y-HH. 2020. Dairy Propionibacterium freudenreichii ameliorates acute colitis by stimulating MUC2 expression in intestinal goblet cell in a DSS-induced colitis rat model. Sci Rep 10:5523. doi: 10.1038/s41598-020-62497-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Do Carmo FLR, Rabah H, Cordeiro BF, da Silva SH, Pessoa RM, Fernandes SOA, Cardoso VN, Gagnaire V, Deplanche M, Savassi B, Figueiroa A, Oliveira ER, Fonseca CC, Queiroz MIA, Rodrigues NM, de Cicco Sandes SH, Nunes ÁC, Lemos L, de Lima Alves J, Faria AMC, Ferreira Ê, Le Loir Y, Jan G, Azevedo V. 2019. Probiotic Propionibacterium freudenreichii requires SlpB protein to mitigate mucositis induced by chemotherapy. Oncotarget 10:7198–7219. doi: 10.18632/oncotarget.27319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Le Maréchal C, Peton V, Plé C, Vroland C, Jardin J, Briard-Bion V, Durant G, Chuat V, Loux V, Foligné B, Deutsch SM, Falentin H, Jan G. 2015. Surface proteins of Propionibacterium freudenreichii are involved in its anti-inflammatory properties. J Proteomics 113:447–461. doi: 10.1016/j.jprot.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 29.Deutsch S-MM, Mariadassou M, Nicolas P, Parayre S, Le Guellec R, Chuat V, Peton V, Le Maréchal C, Burati J, Loux V, Briard-Bion V, Jardin J, Plé C, Foligné B, Jan G, Falentin H. 2017. Identification of proteins involved in the anti-inflammatory properties of Propionibacterium freudenreichii by means of a multi-strain study. Sci Rep 7:46409. doi: 10.1038/srep46409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de R Rodovalho V, da Luz BSR, Rabah H, do Carmo FLR, Folador EL, Nicolas A, Jardin J, Briard-Bion V, Blottière H, Lapaque N, Jan G, Le Loir Y, de Carvalho Azevedo VA, Guédon E. 2020. Extracellular vesicles produced by the probiotic Propionibacterium freudenreichii CIRM-BIA 129 mitigate inflammation by modulating the NF-κB pathway. Front Microbiol 11:1544. doi: 10.3389/fmicb.2020.01544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Woith E, Fuhrmann G, Melzig MF. 2019. Extracellular vesicles—connecting kingdoms. Int J Mol Sci 20:5695. doi: 10.3390/ijms20225695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gill S, Catchpole R, Forterre P. 2019. Extracellular membrane vesicles in the three domains of life and beyond. FEMS Microbiol Rev 43:273–303. doi: 10.1093/femsre/fuy042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu Y, Defourny KAYY, Smid EJ, Abee T. 2018. Gram-positive bacterial extracellular vesicles and their impact on health and disease. Front Microbiol 9:1–8. doi: 10.3389/fmicb.2018.01502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyofuku M, Nomura N, Eberl L. 2019. Types and origins of bacterial membrane vesicles. Nat Rev Microbiol 17:13–24. doi: 10.1038/s41579-018-0112-2. [DOI] [PubMed] [Google Scholar]
- 35.Li M, Lee K, Hsu M, Nau G, Mylonakis E, Ramratnam B. 2017. Lactobacillus-derived extracellular vesicles enhance host immune responses against vancomycin-resistant enterococci. BMC Microbiol 17:66. doi: 10.1186/s12866-017-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alvarez C-S, Giménez R, Cañas M-A, Vera R, Díaz-Garrido N, Badia J, Baldomà L. 2019. Extracellular vesicles and soluble factors secreted by Escherichia coli Nissle 1917 and ECOR63 protect against enteropathogenic E. coli-induced intestinal epithelial barrier dysfunction. BMC Microbiol 19:166. doi: 10.1186/s12866-019-1534-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim M-H, Choi SJ, Choi H-I, Choi J-P, Park H-K, Kim EK, Kim M-J, Moon BS, Min T, Rho M, Cho Y-J, Yang S, Kim Y-K, Kim Y-Y, Pyun BY. 2018. Lactobacillus plantarum-derived extracellular vesicles protect atopic dermatitis induced by Staphylococcus aureus-derived extracellular vesicles. Allergy Asthma Immunol Res 10:516–532. doi: 10.4168/aair.2018.10.5.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cañas M-A, Fábrega M-J, Giménez R, Badia J, Baldomà L. 2018. Outer membrane vesicles from probiotic and commensal Escherichia coli activate NOD1-mediated immune responses in intestinal epithelial cells. Front Microbiol 9:498. doi: 10.3389/fmicb.2018.00498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Behzadi E, Mahmoodzadeh Hosseini H, Imani Fooladi AA. 2017. The inhibitory impacts of Lactobacillus rhamnosus GG-derived extracellular vesicles on the growth of hepatic cancer cells. Microb Pathog 110:1–6. doi: 10.1016/j.micpath.2017.06.016. [DOI] [PubMed] [Google Scholar]
- 40.Grande R, Celia C, Mincione G, Stringaro A, Di Marzio L, Colone M, Di Marcantonio MC, Savino L, Puca V, Santoliquido R, Locatelli M, Muraro R, Hall-Stoodley L, Stoodley P. 2017. Detection and physicochemical characterization of membrane vesicles (MVs) of Lactobacillus reuteri DSM 17938. Front Microbiol 8:1040. doi: 10.3389/fmicb.2017.01040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Choi JH, Moon CM, Shin T-S, Kim EK, McDowell A, Jo M-K, Joo YH, Kim S-E, Jung H-K, Shim K-N, Jung S-A, Kim Y-K. 2020. Lactobacillus paracasei-derived extracellular vesicles attenuate the intestinal inflammatory response by augmenting the endoplasmic reticulum stress pathway. Exp Mol Med 52:423–437. doi: 10.1038/s12276-019-0359-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orench-Rivera N, Kuehn MJ. 2016. Environmentally controlled bacterial vesicle-mediated export. Cell Microbiol 18:1525–1536. doi: 10.1111/cmi.12676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreoni F, Toyofuku M, Menzi C, Kalawong R, Mairpady Shambat S, François P, Zinkernagel AS, Eberl L. 2018. Antibiotics stimulate formation of vesicles in Staphylococcus aureus in both phage-dependent and -independent fashions and via different routes. Antimicrob Agents Chemother 63:e01439-18. doi: 10.1128/AAC.01439-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun SH, Park EC, Lee SY, Lee H, Choi CW, Yi YS, Ro HJ, Lee JC, Jun S, Kim HY, Kim GH, Kim SI. 2018. Antibiotic treatment modulates protein components of cytotoxic outer membrane vesicles of multidrug-resistant clinical strain, Acinetobacter baumannii DU202. Clin Proteom 15:11. doi: 10.1186/s12014-018-9204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Godlewska R, Klim J, Dębski J, Wyszyńska A, Łasica A. 2019. Influence of environmental and genetic factors on proteomic profiling of outer membrane vesicles from Campylobacter jejuni. Pol J Microbiol 68:255–261. doi: 10.33073/pjm-2019-027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hong J, Dauros-Singorenko P, Whitcombe A, Payne L, Blenkiron C, Phillips A, Swift S. 2019. Analysis of the Escherichia coli extracellular vesicle proteome identifies markers of purity and culture conditions. J Extracell Vesicles 8:1632099. doi: 10.1080/20013078.2019.1632099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chan KW, Shone C, Hesp JR. 2017. Antibiotics and iron-limiting conditions and their effect on the production and composition of outer membrane vesicles secreted from clinical isolates of extraintestinal pathogenic E. coli. Prot Clin Appl 11:1600091. doi: 10.1002/prca.201600091. [DOI] [PubMed] [Google Scholar]
- 48.Keenan JI, Allardyce RA. 2000. Iron influences the expression of Helicobacter pylori outer membrane vesicle-associated virulence factors. Eur J Gastroenterol Hepatol 12:1267–1273. doi: 10.1097/00042737-200012120-00002. [DOI] [PubMed] [Google Scholar]
- 49.Prados-Rosales R, Weinrick BC, Piqué DG, Jacobs WR, Casadevall A, Rodriguez GM. 2014. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol 196:1250–1256. doi: 10.1128/JB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee T, Jun SH, Choi CW, Kim S, Lee JC, Shin JH. 2018. Salt stress affects global protein expression profiles of extracellular membrane-derived vesicles of Listeria monocytogenes. Microb Pathog 115:272–279. doi: 10.1016/j.micpath.2017.12.071. [DOI] [PubMed] [Google Scholar]
- 51.Jun SH, Lee T, Lee JC, Shin JH. 2019. Different epithelial cell response to membrane vesicles produced by Listeria monocytogenes cultured with or without salt stress. Microb Pathog 133:103554. doi: 10.1016/j.micpath.2019.103554. [DOI] [PubMed] [Google Scholar]
- 52.Kim Y, Edwards N, Fenselau C. 2016. Extracellular vesicle proteomes reflect developmental phases of Bacillus subtilis. Clin Proteom 13:6. doi: 10.1186/s12014-016-9107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taboada H, Meneses N, Dunn MF, Vargas-Lagunas C, Buchs N, Castro-Mondragón JA, Heller M, Encarnación S. 2019. Proteins in the periplasmic space and outer membrane vesicles of Rhizobium etli CE3 grown in minimal medium are largely distinct and change with growth phase. Microbiology (Reading) 165:638–650. doi: 10.1099/mic.0.000720. [DOI] [PubMed] [Google Scholar]
- 54.Tashiro Y, Ichikawa S, Shimizu M, Toyofuku M, Takaya N, Nakajima-Kambe T, Uchiyama H, Nomura N. 2010. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in Pseudomonas aeruginosa. Appl Environ Microbiol 76:3732–3739. doi: 10.1128/AEM.02794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zavan L, Bitto NJ, Johnston EL, Greening DW, Kaparakis-Liaskos M. 2019. Helicobacter pylori growth stage determines the size, protein composition, and preferential cargo packaging of outer membrane vesicles. Proteomics 19:e1800209. doi: 10.1002/pmic.201970004. [DOI] [PubMed] [Google Scholar]
- 56.Bager RJ, Persson G, Nesta B, Soriani M, Serino L, Jeppsson M, Nielsen TK, Bojesen AM. 2013. Outer membrane vesicles reflect environmental cues in Gallibacterium anatis. Vet Microbiol 167:565–572. doi: 10.1016/j.vetmic.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Choi CW, Park EC, Yun SH, Lee SY, Lee YG, Hong Y, Park KR, Kim SH, Kim GH, Kim SI. 2014. Proteomic characterization of the outer membrane vesicle of Pseudomonas putida KT2440. J Proteome Res 13:4298–4309. doi: 10.1021/pr500411d. [DOI] [PubMed] [Google Scholar]
- 58.Lynch JB, Schwartzman JA, Bennett BD, McAnulty SJ, Knop M, Nyholm SV, Ruby EG. 2019. Ambient pH alters the protein content of outer membrane vesicles, driving host development in a beneficial symbiosis. J Bacteriol 201:e00319-19. doi: 10.1128/JB.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ballok AE, Filkins LM, Bomberger JM, Stanton BA, O'Toole GA. 2014. Epoxide-mediated differential packaging of cif and other virulence factors into outer membrane vesicles. J Bacteriol 196:3633–3642. doi: 10.1128/JB.01760-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kosgodage US, Matewele P, Awamaria B, Kraev I, Warde P, Mastroianni G, Nunn AV, Guy GW, Bell JD, Inal JM, Lange S. 2019. Cannabidiol is a novel modulator of bacterial membrane vesicles. Front Cell Infect Microbiol 9:324. doi: 10.3389/fcimb.2019.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Adriani R, Mousavi Gargari SL, Nazarian S, Sarvary S, Noroozi N. 2018. Immunogenicity of Vibrio cholerae outer membrane vesicles secreted at various environmental conditions. Vaccine 36:322–330. doi: 10.1016/j.vaccine.2017.09.004. [DOI] [PubMed] [Google Scholar]
- 62.Lakhdari O, Cultrone A, Tap J, Gloux K, Bernard F, Dusko Ehrlich S, Lefèvre F, Doré J, Blottière HM. 2010. Functional metagenomics: a high throughput screening method to decipher microbiota-driven NF-KB modulation in the human gut. PLoS One 5:e13092. doi: 10.1371/journal.pone.0013092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cousin FJ, Louesdon S, Maillard M-B, Parayre S, Falentin H, Deutsch S-M, Boudry G, Jan G. 2012. The first dairy product exclusively fermented by Propionibacterium freudenreichii: a new vector to study probiotic potentialities in vivo. Food Microbiol 32:135–146. doi: 10.1016/j.fm.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 64.Gaucher F, Rabah H, Kponouglo K, Bonnassie S, Pottier S, Dolivet A, Marchand P, Jeantet R, Blanc P, Jan G. 2020. Intracellular osmoprotectant concentrations determine Propionibacterium freudenreichii survival during drying. Appl Microbiol Biotechnol 104:3145–3156. doi: 10.1007/s00253-020-10425-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malik AC, Reinbold GW, Vedamuthu ER. 1968. An evaluation of the taxonomy of Propionibacterium. Can J Microbiol 14:1185–1191. doi: 10.1139/m68-199. [DOI] [PubMed] [Google Scholar]
- 66.Cousin FJ, Foligné B, Deutsch SM, Massart S, Parayre S, Le Loir Y, Boudry G, Jan G. 2012. Assessment of the probiotic potential of a dairy product fermented by Propionibacterium freudenreichii in piglets. J Agric Food Chem 60:7917–7927. doi: 10.1021/jf302245m. [DOI] [PubMed] [Google Scholar]
- 67.Volgers C, Savelkoul PHMM, Stassen FRMM. 2018. Gram-negative bacterial membrane vesicle release in response to the host-environment: different threats, same trick? Crit Rev Microbiol 44:258–273. doi: 10.1080/1040841X.2017.1353949. [DOI] [PubMed] [Google Scholar]
- 68.Gerritzen MJH, Maas RHW, van den Ijssel J, van Keulen L, Martens DE, Wijffels RH, Stork M. 2018. High dissolved oxygen tension triggers outer membrane vesicle formation by Neisseria meningitidis. Microb Cell Fact 17:157. doi: 10.1186/s12934-018-1007-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gerritzen MJH, Martens DE, Uittenbogaard JP, Wijffels RH, Stork M. 2019. Sulfate depletion triggers overproduction of phospholipids and the release of outer membrane vesicles by Neisseria meningitidis. Sci Rep 9:4716. doi: 10.1038/s41598-019-41233-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cardoso FS, Gaspar P, Hugenholtz J, Ramos A, Santos H. 2004. Enhancement of trehalose production in dairy propionibacteria through manipulation of environmental conditions. Int J Food Microbiol 91:195–204. doi: 10.1016/S0168-1605(03)00387-8. [DOI] [PubMed] [Google Scholar]
- 71.Huang S, Rabah H, Jardin J, Briard-Bion V, Parayre S, Maillard M-B, Le Loir Y, Chen XD, Schuck P, Jeantet R, Jan G. 2016. Hyperconcentrated sweet whey, a new culture medium that enhances Propionibacterium freudenreichii stress tolerance. Appl Environ Microbiol 82:4641–4651. doi: 10.1128/AEM.00748-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Leverrier P, Vissers JPC, Rouault A, Boyaval P, Jan G. 2004. Mass spectrometry proteomic analysis of stress adaptation reveals both common and distinct response pathways in Propionibacterium freudenreichii. Arch Microbiol 181:215–230. doi: 10.1007/s00203-003-0646-0. [DOI] [PubMed] [Google Scholar]
- 73.Caruana JC, Walper SA. 2020. Bacterial membrane vesicles as mediators of microbe–microbe and microbe–host community interactions. Front Microbiol 11:432. doi: 10.3389/fmicb.2020.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tartaglia NR, Nicolas A, de R Rodovalho V, da Luz BSR, Briard-Bion V, Krupova Z, Thierry A, Coste F, Burel A, Martin P, Jardin J, Azevedo V, Le Loir Y, Guédon E. 2020. Extracellular vesicles produced by human and animal Staphylococcus aureus strains share a highly conserved core proteome. Sci Rep 10:8467. doi: 10.1038/s41598-020-64952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toyofuku M 2019. Bacterial communication through membrane vesicles. Biosci Biotechnol Biochem 83:1599–1605. doi: 10.1080/09168451.2019.1608809. [DOI] [PubMed] [Google Scholar]
- 76.Do Carmo FLR, Rabah H, Huang S, Gaucher F, Deplanche M, Dutertre S, Jardin J, Le Loir Y, Azevedo V, Jan G. 2017. Propionibacterium freudenreichii surface protein SlpB is involved in adhesion to intestinal HT-29 cells. Front Microbiol 8:1–11. doi: 10.3389/fmicb.2017.01033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Böing AN, van der Pol E, Grootemaat AE, Coumans FAWW, Sturk A, Nieuwland R. 2014. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J Extracell Vesicles 3:23430. doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gagnaire V, Jardin J, Rabah H, Briard-Bion V, Jan G. 2015. Emmental cheese environment enhances Propionibacterium freudenreichii stress tolerance. PLoS One 10:e0135780. doi: 10.1371/journal.pone.0135780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gaucher F, Bonnassie S, Rabah H, Leverrier P, Pottier S, Jardin J, Briard-Bion V, Marchand P, Jeantet R, Blanc P, Jan G. 2020. Data from a proteomic analysis highlight different osmoadaptations in two strain of Propionibacterium freudenreichii. Data Brief 28:104932. doi: 10.1016/j.dib.2019.104932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Langella O, Valot B, Balliau T, Blein-Nicolas M, Bonhomme L, Zivy M. 2017. X!TandemPipeline: a tool to manage sequence redundancy for protein inference and phosphosite identification. J Proteome Res 16:494–503. doi: 10.1021/acs.jproteome.6b00632. [DOI] [PubMed] [Google Scholar]
- 81.Valot B, Langella O, Nano E, Zivy M. 2011. MassChroQ: a versatile tool for mass spectrometry quantification. Proteomics 11:3572–3577. doi: 10.1002/pmic.201100120. [DOI] [PubMed] [Google Scholar]
- 82.Yu CS, Cheng CW, Su WC, Chang KC, Huang SW, Hwang JK, Lu CH. 2014. CELLO2GO: a web server for protein subCELlular lOcalization prediction with functional gene ontology annotation. PLoS One 9:e99368. doi: 10.1371/journal.pone.0099368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bagos PG, Tsirigos KD, Liakopoulos TD, Hamodrakas SJ. 2008. Prediction of lipoprotein signal peptides in Gram-positive bacteria with a Hidden Markov model. J Proteome Res 7:5082–5093. doi: 10.1021/pr800162c. [DOI] [PubMed] [Google Scholar]
- 84.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, Von Mering C, Bork P. 2019. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314. doi: 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, Bork P. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol 34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, Vilo J. 2019. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res 47:191–198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Reimand J, Isserlin R, Voisin V, Kucera M, Tannus-Lopes C, Rostamianfar A, Wadi L, Meyer M, Wong J, Xu C, Merico D, Bader GD. 2019. Pathway enrichment analysis and visualization of omics data using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat Protoc 14:482–517. doi: 10.1038/s41596-018-0103-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lian X, Yang S, Li H, Fu C, Zhang Z. 2019. Machine-learning-based predictor of human–bacteria protein–protein interactions by incorporating comprehensive host-network properties. J Proteome Res 18:2195–2205. doi: 10.1021/acs.jproteome.9b00074. [DOI] [PubMed] [Google Scholar]
- 89.McKinney W 2010. Data structures for statistical computing in Python, p 56–61. In Proc 9th Python Sci Conf doi: 10.25080/Majora-92bf1922-00a. [DOI] [Google Scholar]
- 90.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data can be found at https://doi.org/10.15454/Q6PPXY.