Research into the identification of biomarkers for gut health and ways to modulate the microbiota composition and activity to improve health has put Akkermansia muciniphila in the spotlight. As a mucin degrader, A. muciniphila colonizes the interesting but not fully described host-glycan degradation niche. Much of research concerning A. muciniphila has been done, but little is known about its behavior in the complex microbial ecosystem in the colon, about the potential role of mucins to influence A. muciniphila behavior, and the impact of its probiotic administration on the microbial ecosystem.
KEYWORDS: SHIME, colon, gastrointestinal tract, gut, host glycan, microbiome, microbiota, mucus, prebiotic, probiotic
ABSTRACT
Akkermansia muciniphila is an abundantly present commensal mucin-degrading gut bacterium (1 to 4%) that is widely distributed among healthy individuals. It has been positioned as a health biomarker and is currently being explored as a biotherapeutic agent and next-generation probiotic. Preliminary and ongoing research is mostly based on in vivo mouse models and human intervention trials. While these allow the assessment of physiologically relevant endpoints, the analysis of fecal samples presents limitations with respect to the in-depth mechanistic characterization of Akkermansia′s effects at the level of the microbiome. We aimed to evaluate the effect of A. muciniphila treatment on the endogenous community from four different donors in a validated, controlled in vitro model of the gut microbial ecosystem (SHIME). Taking into account the nutritional specificity of A. muciniphila and the prebiotic-like action of mucins in the colon environment, the interplay between mucin, A. muciniphila, and the endogenous community was investigated. The effects on the microbial community composition and functionality of A. muciniphila supplementation without mucin were limited, whereas mucin addition successfully induced compositional and metabolic changes in the gut microbiota. Indeed, mucin addition resulted in significantly higher acetate, propionate, and butyrate production for all four donors and the increase of several bacteria, including A. muciniphila, Ruminococcus, Clostridium cluster XIVa, and Lachnospiraceae. This study revealed that the supplementation of A. muciniphila together with mucin limited the observed prebiotic-like effect of mucin in inducing compositional changes.
IMPORTANCE Research into the identification of biomarkers for gut health and ways to modulate the microbiota composition and activity to improve health has put Akkermansia muciniphila in the spotlight. As a mucin degrader, A. muciniphila colonizes the interesting but not fully described host-glycan degradation niche. Much research concerning A. muciniphila has been done, but little is known about its behavior in the complex microbial ecosystem in the colon, the potential of mucins to influence A. muciniphila behavior, and the impact of its probiotic administration on the microbial ecosystem. This study aimed at investigating the impact of A. muciniphila administration on the endogenous community while also taking into account its nutritional specificity. As such, the effect of A. muciniphila administration was investigated with and without addition of mucin. This allowed us to elucidate the importance of the presence of mucin to modulate the efficiency of probiotic supplementation with A. muciniphila.
INTRODUCTION
Akkermansia muciniphila was isolated from human feces as a mucin-degrading bacterium in 2004 (1). Since its discovery A. muciniphila has gained attention in scientific studies, as its abundance is inversely correlated with disorders such as inflammatory bowel disease (IBD), obesity, autism, appendicitis, and diabetes (2–9). A study with obese mice showed A. muciniphila exerting therapeutic effects, as its supplementation reversed high-fat-diet-induced insulin resistance, dyslipidemia, metabolic endotoxemia, and fat mass gain (10). Plovier et al. (11) showed that pasteurization of A. muciniphila before treatment enhanced its beneficial impact and that the beneficial effects were, at least partly, due to a specific outer membrane protein (Amuc_1100). An ongoing clinical study by Université Catholique de Louvain is investigating the effects associated with the administration of A. muciniphila on the metabolic disorders related to overweight and obesity in humans. Preliminary results demonstrated that daily oral administration of A. muciniphila was safe and well tolerated and after 3 months improved several metabolic parameters, such as insulin sensitivity (12).
A. muciniphila has been referred to as a possible next-generation probiotic (13–15), a broad term that conforms to the normal definition of a probiotic and comprises microorganisms with potential health benefits but which do not necessarily have a qualified presumption of safety (QPS) or generally regarded as safe (GRAS) status. Some of these next-generation probiotics are likely to be used in a pharmaceutical context, which makes them fit well within the emerging concept of live biotherapeutic products: “a biological product that: (1) contains live organisms, such as bacteria; (2) is applicable to the prevention, treatment, or cure of a disease or condition of human beings; and (3) is not a vaccine” (16). Since there is as yet no consensus on the correct terminology, we consider A. muciniphila to be a live biotherapeutic product, thereby avoiding confusion with established probiotic products. A. muciniphila’s mode of action may consist of direct interaction with the host—for example, through the Amuc_1100 protein (11)—and/or indirect interplay with the established microbial community.
This established community, together with the high turnover in the gastrointestinal tract, however, presents a challenge for the stable introduction and maintenance of live biotherapeutics. In that respect, the availability of nutrients, selectively sustaining the growth of a biotherapeutic agent, could be an important factor in determining the success rate of future therapies. In the case of A. muciniphila, the host glycan mucin has been identified as a major determinant of its colonization capacity (17, 18). In the Simulator of the Human Intestinal Microbial Ecosystem (SHIME), a dynamic model of the colonic microbial ecosystem, mucin deprivation and supplementation were shown to specifically affect A. muciniphila abundance, more than any other species present (19, 20).
This reflects the superior ability of A. muciniphila to use up to 85% of the complex mucin structure. This is composed of O-glycosylated and, to a lesser extent, N-glycosylated protein backbones, with chains of 2 to 12 monosaccharides, mostly galactose, fucose, N-acetylgalactosamine, N-acetylglucosamine, mannose, and sialic acid (21, 22). In vivo mouse trials have demonstrated A. muciniphila’s efficient degradation of mucins (18, 23). It possesses an entire repertoire of enzymes with both extracellular and intracellular activity (21). A study of its genome showed the presence of genes encoding 61 proteins predicted to be involved in mucin degradation (11% of all proteins). Mucin degradation by A. muciniphila leads to the release of oligosaccharides and their subsequent fermentation into acetate and propionate, both of which can stimulate microbial metabolic interactions, as well as a host response (1, 24). Other bacteria in close proximity could profit from the mucolytic activity by using the oligosaccharides and acetate for growth and metabolic conversions, such as butyrate production (25, 26). It has been hypothesized that the presence and activity of these cross-feeding bacteria coexisting with A. muciniphila at the mucus layer might provide additional resistance against colonization by pathogens and could impact the host response due to their proximity to the epithelial cells (13, 14).
Only a few other intestinal species have the enzymatic capacity for initiating partial or full mucin degradation, including Bacteroides thetaiotaomicron, Ruminococcus gnavus, Ruminococcus torques, and Bifidobacterium bifidum (2, 27–29). Considering the limited number of species that can degrade the complex mucin structure and the described health effect conferred by its degradation, mucins fit the definition of prebiotic substances, “substrates that are selectively utilized by host microorganisms conferring a health benefit” (14, 30, 31). As mucin glycans constitute 80% of the dry weight of the mucus layer covering the intestinal epithelium and are present in the luminal content as a consequence of the continuous mucus desquamation, the human body has even been described as producing its own prebiotic (32–34). Mucin thus plays an important role in the interaction between A. muciniphila, the microbial community, and the host.
Considering the ongoing studies and future perspective for A. muciniphila as a biotherapeutic agent, we investigated the effect of A. muciniphila administration on the endogenous community. For this purpose, the in vitro SHIME model was used, with colon compartments separately inoculated with the microbiota from four human donors. Taking into account its nutritional specificity, supplementation of A. muciniphila was investigated with and without addition of mucin as its functional niche. This allowed us to elucidate the importance of the presence of mucin to modulate the engraftment success of supplemented A. muciniphila. The interplay between A. muciniphila, mucin, and the microbial community may also confer community resilience in a scenario of antibiotic-induced community disruption. As a final objective, we therefore also evaluated the impact of an antibiotic pulse on the dynamics and recovery of Akkermansia- or mucin-amended microbial ecosystems.
RESULTS
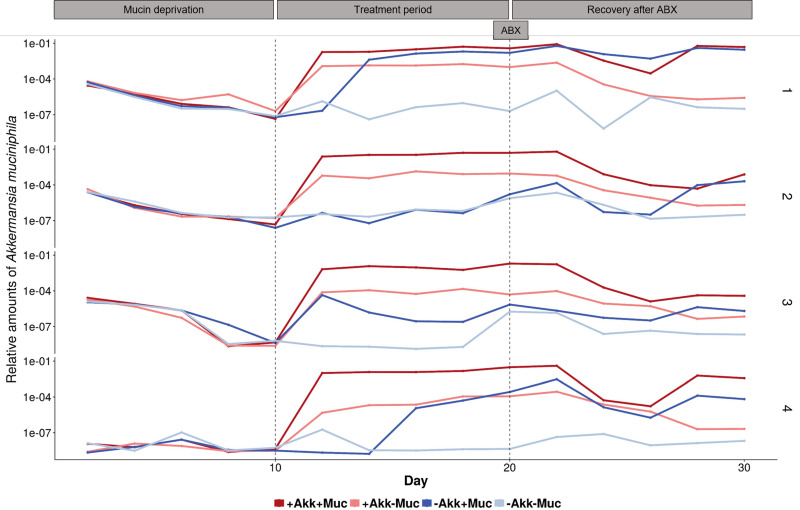
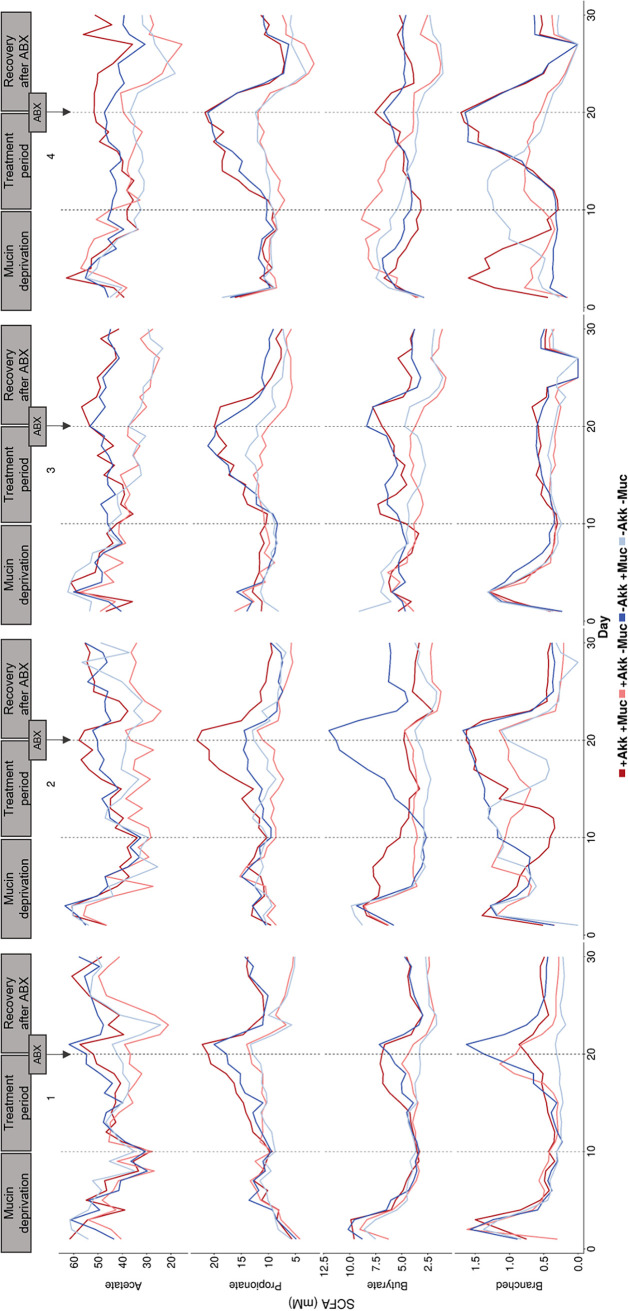
During the 10-day mucin deprivation period, mucin-free medium was fed to the SHIME system, creating mucin-deprived microbial communities, separately derived from the fecal microbiota of four different donors. These different communities were characterized by a profound decrease in A. muciniphila abundance (Fig. 1) and similar short-chain fatty acid (SCFA) profiles (Fig. 2) with 36.3 ± 6.8 mM acetate, 9.4 ± 0.8 mM propionate, and 4.2 ± 1.4 mM butyrate (n = 16). The initial A. muciniphila abundance of around 0.01% in donors 1, 2, and 3 decreased 1,000-fold for donors 1 and 2 upon mucin deprivation and was close to the quantification limit in donor 3 (Fig. 1). In the case of donor 4, A. muciniphila abundance levels remained close to the quantification limit during the entire mucin deprivation period (Fig. 1).
FIG 1.
Log (base 10) scaled relative abundance of A. muciniphila over total bacteria, measured with qPCR. Colon vessels were inoculated with fecal samples of donors 1 to 4. From days 0 to 10, mucin-free feed was added. From days 10 to 20, different treatments were imposed; vessels were treated with either A. muciniphila (+Akk–Muc), mucin (–Akk+Muc) (4 g liter−1), or a combination of both (+Akk+Muc) or received no treatment (–Akk–Muc). At day 20, all vessels were treated with an antibiotic mix (ABX), after which A. muciniphila treatments, in contrast to the mucin treatments, were discontinued.
FIG 2.
Short-chain fatty acid concentration (mM) measured in the colon vessels inoculated with fecal samples of donors 1 to 4. From days 0 to 10, mucin-free feed was administered. From days 10 to 20 onwards, different treatments were applied; vessels were treated with either A. muciniphila (+Akk–Muc), mucin (–Akk+Muc) (4 g liter−1), a combination of both (+Akk+Muc), or neither (–Akk–Muc). At day 20, all vessels were treated with an antibiotic mix (ABX), after which A. muciniphila treatments, in contrast to the mucin treatments, were discontinued.
Effect of treatment on community functionality and A. muciniphila abundance.
From day 10 onward, A. muciniphila and/or mucin was added to the SHIME. A. muciniphila concentrations could be affected by addition of exogenous A. muciniphila and by addition of mucin as its functional niche (Fig. 1). We found the combination of exogenous A. muciniphila with mucin to yield the highest levels of A. muciniphila. This coincided with a sharp increase in propionate for all 4 donors and in acetate and butyrate, albeit to a lesser extent (Fig. 2). Addition of exogenous A. muciniphila in the absence of mucin also resulted in high A. muciniphila levels. However, this addition of A. muciniphila alone did not come with significant shifts in functionality in terms of SCFA production, with A. muciniphila addition contributing to only 0.5% of the variation in SCFA concentrations (P = 0.11) (Fig. 2 and 3).
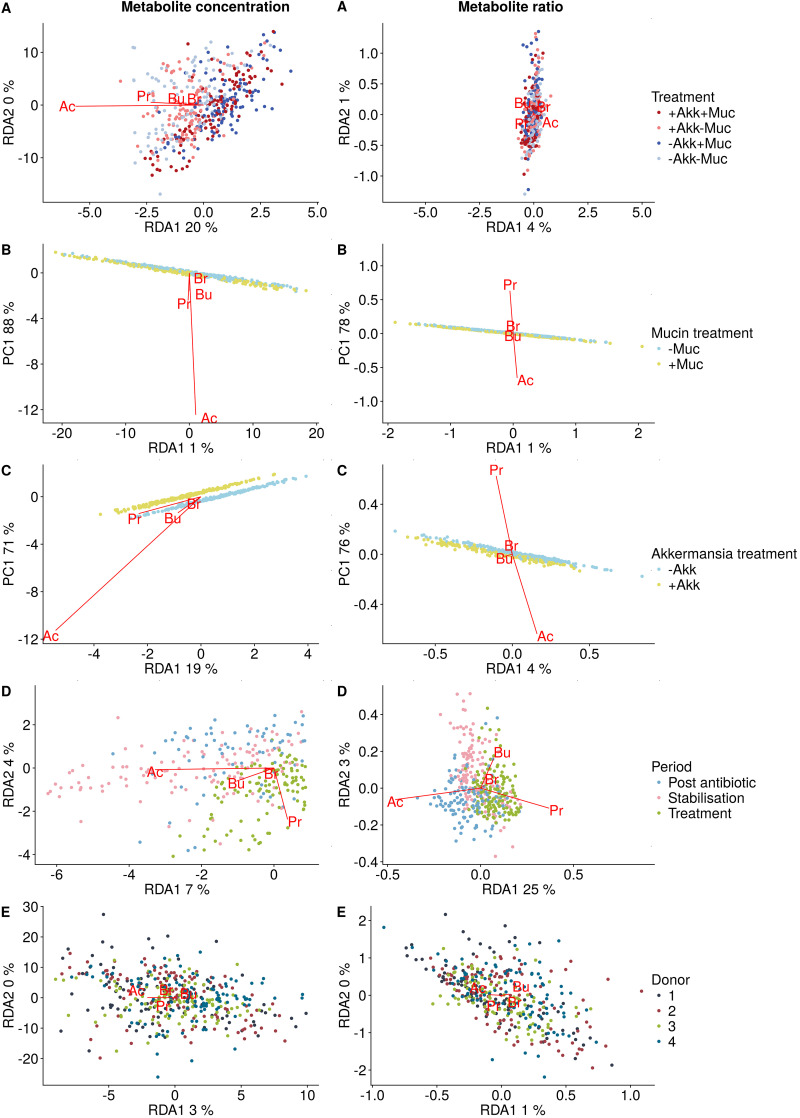
FIG 3.
Partial redundancy analysis correlation triplot with the response variables (Ac, acetate; Pr, propionate; Bu, butyrate; Br, branched SCFA) indicated in red and the different factors represented in the legends.
Mucin seemed to be the most important driver of altered functionality, as partial redundancy analysis indicated that mucin treatment accounted for 17% of the observed variation in SCFA concentrations (P = 0.001) (Fig. 3). Even in the absence of exogenous A. muciniphila, addition of the host glycan displayed a high 10 mM increase in propionate concentrations for all donors (Fig. 2). This propionate increase was less pronounced for donor 2, yet this was compensated for by a high 10 mM increase in butyrate for the mucin amendment alone. Interestingly, the endogenous A. muciniphila in this donor did not respond to the mucin treatment (Fig. 1).
While we had expected A. muciniphila to completely wash out from the SHIME reactor when no exogenous A. muciniphila was added and no mucin was supplemented as its nutritional preference, endogenous A. muciniphila levels never disappeared (Fig. 1).
Effect of treatment on community composition.
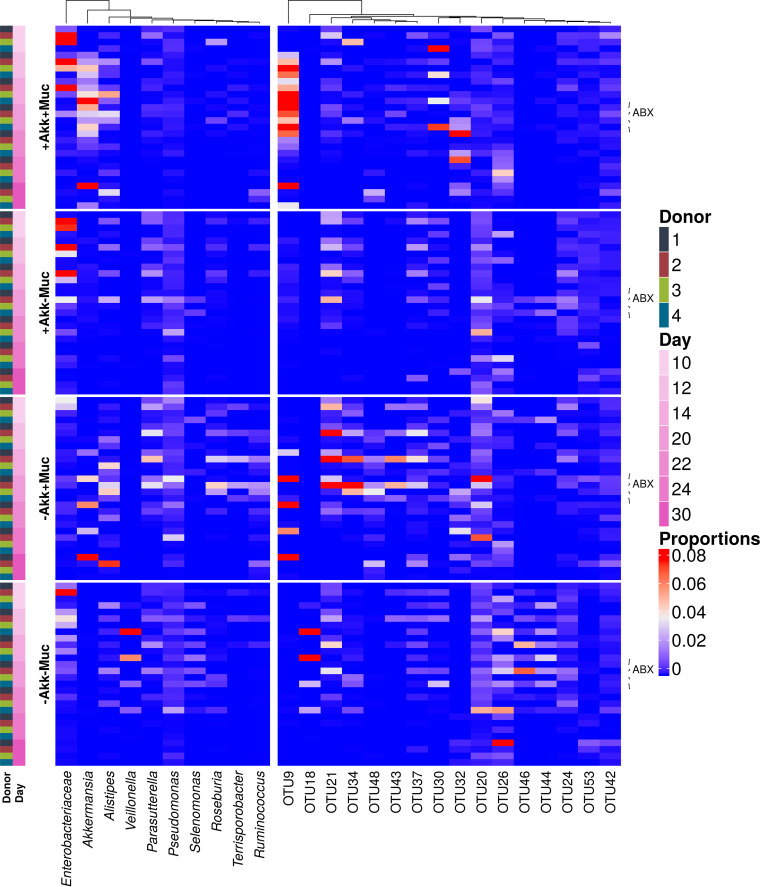
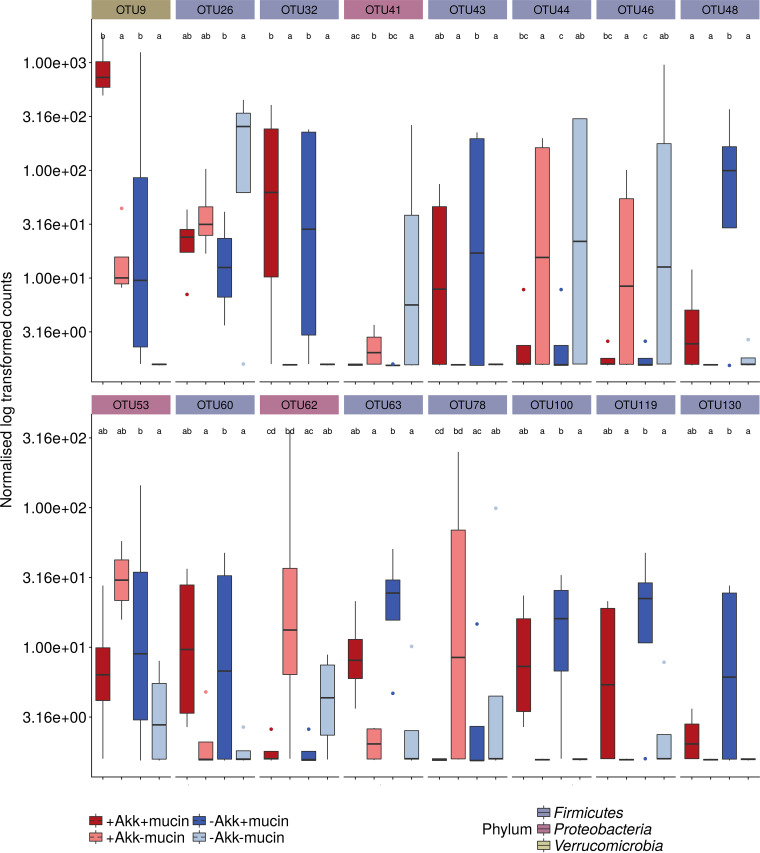
Besides A. muciniphila (OTU9), other members of the microbial community were affected by the different treatment combinations. An unsupervised principal-coordinate ordination displayed no clear donor- or treatment-dependent clustering, illustrating the individuality of the response to mucin and A. muciniphila (Fig. 4). To quantify and distinguish between donor and treatment effects a partial distance-based redundancy analysis (RDA) was performed, showing that the effects of mucin (8%) and A. muciniphila (7%) were limited and nonsignificant (see Table S2 in the supplemental material). In order to select the taxonomic entities which were most discriminative for each of the different treatments, a sparse partial least squares discriminant analysis (sPLS-DA) was performed. The final model, retaining only the 75 most predictive operational taxonomic units (OTUs), showed a clustering by treatment (three-dimensional [3D] plot is shown in Fig. S2). Clostridium cluster XIVa OTU26 and Veillonella OTUs 44 and 46 were characteristic of the control treatment without mucin or A. muciniphila (Fig. 5). The addition of A. muciniphila had little effect on the microbial community, whereas mucin supplementation resulted in proportional increases of A. muciniphila (OTU9), OTU20, OTU21, OTU24, OTU48, OTU43, OTU32, and OTU37. Interestingly, coadministration of A. muciniphila restricted the effect of mucin on these OTUs and amplified the A. muciniphila upsurge (Fig. 5). In the presence of added A. muciniphila (+Akk+Muc [i.e., treatment with both A. muciniphila and mucin] versus +Akk–Muc [i.e., treatment with A. muciniphila alone]), fewer OTUs were significantly affected, amounting to OTU9 and OTU32 increasing in abundance and OTU62 (corresponding phylogenetically to Enterobacteriaceae [∼Enterobacteriaceae]) and OTU78 (∼Lachnospiraceae) decreasing in abundance (Fig. 5). This was also reflected at the genus level (Fig. 5) and in the DESeq2 analysis, comparing the different conditions after 10 days of treatment (Fig. 6). Mucin treatment (–Akk+Muc [i.e., treatment with mucin alone] versus –Akk–Muc [i.e., no treatment]) significantly stimulated A. muciniphila (OTU9), OTU32 to OTU63 (∼Clostridium cluster XIVa), and OTU48 (∼Ruminococcus torques) (Fig. 6). OTU46 (∼Veilonellaceae), OTU41 (∼Enterobacter), and OTU26 (∼Clostridium cluster XIVa), on the other hand, were characteristic of mucin-deprived communities (Fig. 6).
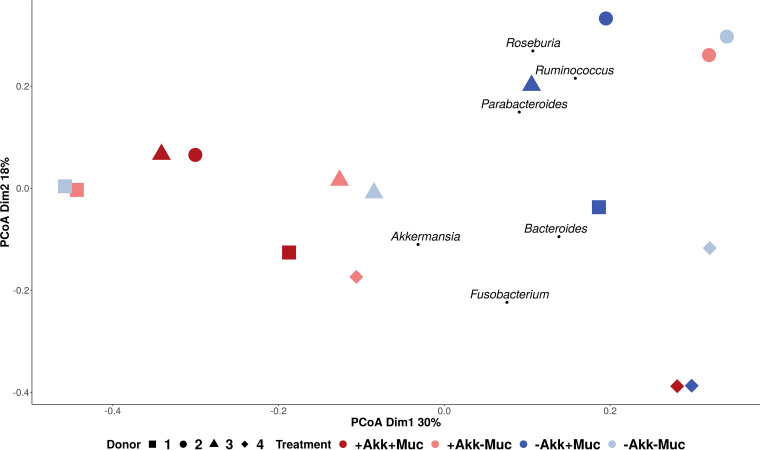
FIG 4.
A PCoA biplot revealed the effect of treatment (colors) on the bacterial communities of the different donors (shapes) comparing days 10 and 20 (size). Samples from donors 1, 2, and 3 with mucin clustered together, independent from A. muciniphila treatment. Samples from donors 1, 2, and 3 without mucin also clustered together, independent from A. muciniphila treatment. Weighted average scores of genera characteristic of treatments were a posteriori projected.
FIG 5.
Heatmap representation of the most predictive genera (left side) and OTUs (right side) for the different treatments as determined by sPLS-DA regression analysis.
FIG 6.
Boxplots of OTUs that were significantly different in abundance between treatments (day 20) across all four donors as determined by DESeq2 analysis (α = 0.05). The color of the boxplots represents the different treatments, and facet labels are colored according to the phylum-level classification. Letter codes show significance.
In line with the sPLS-DA, the effect of A. muciniphila supplementation on the community was very limited, with only 0.33% of the community at the OTU level significantly affected. A. muciniphila adversely affected OTU48 (∼R. torques) abundances in the presence of mucin (+Akk+Muc) and OTU41 (∼Enterobacter) in the treatment without mucin (+Akk–Muc).
The DESeq2 procedure resulted in few OTUs whose abundance was significantly changed by the treatments across all four donors. These interindividual differences are also apparent from the principal-coordinate analyses (PCoA) at the genus level after treatment (day 20) (Fig. 4). For donors 1 to 3, different clusters were distinguished in response to the treatments. Communities after treatment with mucin (without A. muciniphila) are characterized by the presence of Ruminococcus, Roseburia, and Parabacteroides and clustered separately from communities treated with both mucin and A. muciniphila. Those without mucin, on the other hand, clustered according to donor, independent from A. muciniphila treatment. Addition of A. muciniphila thus impacted mucin′s effect on the community composition, whereas A. muciniphila had no effect without mucin. Samples from donor 4 clustered separately, partly due to the higher relative abundance of Fusobacterium spp., and showed a different response to the treatments, with the addition of A. muciniphila affecting community composition when no mucin was added, but not in combination with mucin supplementation.
Effect of antibiotic pulse.
After 10 days of treatment (day 20), an antibiotic pulse, containing ciprofloxacin, tetracycline, and amoxicillin, was applied to the colon vessels, after which A. muciniphila treatment ceased but mucin treatment continued. The effect of this antibiotic disturbance was followed up to investigate whether preceding treatment with mucin and/or A. muciniphila would have protective effects. At the functional level, no protective effects were observed, as the drop in SCFA production after the antibiotic pulse resulted in more similar SCFA profiles across treatments (Fig. 2). The decrease in propionate and butyrate after the antibiotic pulse was significantly larger (P < 0.05) in the presence of mucin, offsetting the initial positive effects of the addition of mucin. Acetate almost fully recovered to the levels found before antibiotic disturbance within 10 days. Propionate and butyrate levels remained significantly lower throughout the antibiotic wash-out period (P < 0.01). Four to six days after the disturbance, A. muciniphila abundance was lowest, after which it increased again in conditions with mucin. After 10 days of recovery, a clear, although not significant, difference in A. muciniphila abundances between conditions with and without mucin was visible (Fig. 1). Also, at the community level, no protective effects were observed from the treatments, and the disturbance persisted after 10 days. The antibiotic pulse marginally reduced the total bacterial count (Fig. S3) and affected the same genera, such as Roseburia, Bifidobacterium, Alistipes, Butyricicoccus, Enterobacteriaceae, etc., independent of the preceding treatment, as determined by DESeq analysis (Fig. S4). Alpha diversity was significantly reduced after antibiotic treatment and did not recover within 10 days (P < 0.01) (Fig. S5).
The case of donor 2.
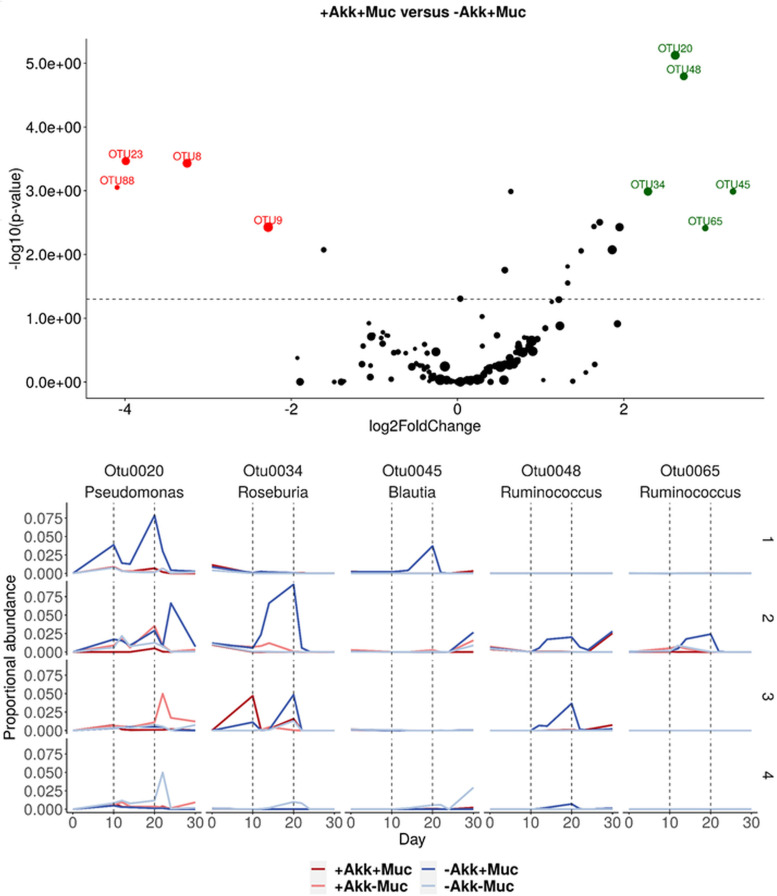
An interesting effect of the treatments and antibiotic disturbance was observed for donor 2. Endogenous A. muciniphila did not increase with mucin addition prior to antibiotics but suddenly responded to mucin after the antibiotic disturbance (Fig. 1). Interestingly, this mucin treatment, without a response of A. muciniphila, caused an increase in butyrate significantly larger than in any other donor or for any other treatment and induced no response in propionate (Fig. 2). A detailed inspection of the time course of the relative abundances from species that were significantly affected solely by the mucin treatment revealed an interesting response of Ruminococcus species OTU48 and OTU65, together with butyrate-producing Roseburia OTU34, to the mucin treatment in donor 2 (Fig. 7). These species might be involved in the observed difference in butyrate between those two treatments (Fig. 1). Ruminococcus species OTU48 and OTU65 responded to mucin treatment without A. muciniphila supplementation, but not to other treatments, together with butyrate-producing Roseburia species (OTU34), which increased greatly. After antibiotic disturbance, OTU65 and OTU34 did not recover, whereas OTU48 did. OTU34 and OTU48 displayed similar responses to mucin treatment in donor 3, which was characterized by a less pronounced A. muciniphila response (Fig. 1 and 7).
FIG 7.
(Top) Volcano plot showing results from the DESeq2 analysis between treatments +Akk+Muc and –Akk+Muc in donor 2. Green and red dots represent OTUs that are more abundant in –Akk+Muc and +Akk+Muc, respectively, and the size indicates the relative abundance of the OTU in the community. (Bottom) Relative abundance of OTUs stimulated by mucin treatment –Akk+Muc in donor 2. As a comparison, relative abundances for the other donors are shown too.
DISCUSSION
In this study, we hypothesized that the combination of the candidate live biotherapeutic A. muciniphila with mucin as its functional niche may yield increased survival and activity for A. muciniphila and subsequently bring about prebiotic effects to the community. Being a specialist mucin degrader, A. muciniphila would use the mucin, produce acetate and propionate, and release mucin-derived oligosaccharides and thus have a greater impact on the community composition and functionality, for example, by stimulating cross-feeding on acetate by butyrate-producing species (25, 26).
Our initial hypothesis, stating that joint supplementation of A. muciniphila and mucin more effectively induces cross-feeding of, for instance, butyrate, compared to mucin alone, does not seem to hold. Mucin addition had the largest impact on microbial community composition and functionality. Mucin-enriched communities, without addition of exogenous A. muciniphila, were characterized by higher endogenous Akkermansia, Roseburia, Ruminococcus, and Parabacteroides proportions. Similar community shifts upon mucin addition were observed in previous studies (19, 20). Mucin addition resulted in significant increases in acetate, propionate, and butyrate production (P < 0.01) for all donors. This increase was independent of A. muciniphila addition, except for donor 2, where the butyrate increase was three times higher in the absence of A. muciniphila supplementation. Endogenous A. muciniphila, although present, did not increase upon mucin supplementation in this donor. The highest increase in butyrate was thus induced by mucin at low A. muciniphila abundance. OTU32, belonging to the butyrate-producing genus Roseburia, was specifically increased by mucin treatment in donor 2 and to a lesser extent in donor 3. However, no Roseburia species have been identified to degrade mucin. Butyrate production would thus be the result of cross-feeding, not with A. muciniphila but with, for example, OTU48 and OTU65, both belonging to Ruminococcus and increased by the mucin treatment. Species like R. gnavus and R. torques are known mucin degraders and might thus deliver acetate and mucin-derived oligosaccharides to Roseburia and other butyrate-producing species (2, 27, 28, 35). It is a possibility that this cross-feeding consortium prevented endogenous A. muciniphila from benefitting from the mucin. In support of this hypothesis, we observed that A. muciniphila abundance increased upon mucin treatment after community disruption by antibiotics, together with OTU48 (∼R. torques), while OTU32 (∼Roseburia) and OTU65 (∼Ruminococcus) did not recover. These hypotheses, however, need to be substantiated with further experiments before real conclusions can be drawn.
In contrast to the mucin treatment, the addition of A. muciniphila hardly affected the community (Fig. S6 to S9). This is not surprising, as prebiotic treatments (such as mucin) generally induce more significant changes in community composition compared to probiotic treatments (such as A. muciniphila) (36). When comparing the effect of A. muciniphila with and without mucin at the community level, only two species were significantly increased by the combined supplementation of A. muciniphila and mucin; these were A. muciniphila for obvious reasons and a Clostridium cluster XIVa species (OTU32) (Fig. S7). It is not clear whether the latter would benefit from putative cross-feeding interactions. Ruminococcus torques, a known mucin degrader (2), on the other hand, was significantly decreased by coadministered A. muciniphila and mucin compared to supplementation with only mucin. This suggests that coadministration of 8 log units of A. muciniphila gives an initial numerical advantage over other species, resulting in a more efficient occupation of the mucin-degradation niche, thereby outcompeting endogenous community members, such as R. torques. In contrast, if mucin is administered alone, the endogenous microbiota can probably compete more efficiently with the endogenous A. muciniphila, eventually resulting in a bigger community change.
However, the combined addition of A. muciniphila and mucin may still provide a protective advantage in case of an acute stress. We chose antibiotic administration as a relevant stress factor for the gut microbiota, and the mix of amoxicillin, tetracycline, and ciprofloxacin was previously found to cause microbial dysbiosis (37). Antibiotic disruption of the microbial community 10 days after the mucin and/or Akkermansia treatment caused a profound decrease in SCFA production, in line with results from in vivo and in vitro studies (37–39), although the proportional SCFA profiles did not alter. Mucin-supplemented communities retained higher SCFA levels than mucin-depleted communities, as before the antibiotic treatment. In addition, community composition was heavily affected, and it did not recover within the 10-day recovery period; however A. muciniphila levels did recover faster in mucin-rich communities. Thus, although no general protective effects from mucin and/or Akkermansia treatment at the community level were observed, mucin-supplemented communities did benefit from the presence of mucin.
In contrast to previous studies, A. muciniphila was not washed out of the system, even when no mucin was added to the feed during 30 days (–Akk –Muc) (19, 20). Its abundance decreased due to the mucin deprivation in the first 10 to 14 days but stabilized afterwards. Plovier et al. (11) previously obtained dense A. muciniphila cultures on a mucin-free medium containing peptone, glucose, N-acetylglucosamine, and threonine. All compounds of this mucin-free medium were also present in our mucin-free SHIME feed, possibly explaining why A. muciniphila did not completely disappear.
The ability to control the supply of mucin glycan (by varying the concentration but not the structure) necessitated the use of in vitro models. Besides, the nutritional role of mucins cannot be separated from the protective role of the mucus layer in vivo, and attachment to the mucus layer or the antimicrobial peptides in the mucus layer would have confounding effects. The in vitro model (SHIME) used in this research thus provided essential advantages to study the impact of mucin degradation on the community. However, there still remain many aspects of the mucin degradation niche that should be addressed in future research, such as variability in mucin structure and supply influenced by interindividual variability, host health, and the cross talk with the microbial community.
To conclude, this in vitro study with four donors revealed that the presence or absence of mucin as a functional niche has a far greater effect than A. muciniphila on the gut microbiota. The joint supplementation of A. muciniphila with mucin limited the prebiotic-like effect that was observed for mucin in inducing compositional and metabolic changes. While cross-feeding on mucin has been shown for butyrate-producing bacteria and A. muciniphila in coculture experiments (25), A. muciniphila does not seem to enhance cross-feeding in a complex microbial background. Addition of both mucin and A. muciniphila might lead to A. muciniphila dominating the mucin degradation niche, while addition of mucin alone leads to involvement of several bacteria, including A. muciniphila, Ruminococcus, Clostridium cluster XIVa, and Lachnospiraceae. When aiming at the modulation of (mucus-associated) microbiota, stimulation of endogenous A. muciniphila might thus be more successful than its administration as a live biotherapeutic product.
MATERIALS AND METHODS
Chemicals, growth media, and bacterial strains.
Chemicals were obtained from Sigma (Bornem, Belgium), unless stated otherwise. Akkermansia muciniphila (DSMZ 22959, type strain) was cultured in reinforced clostridial medium (RCM) with 4 g liter−1 of partially purified porcine gastric mucin type III for 24 h prior to the daily treatment of the colon vessels (days 10 to 20). After 24 h of growth, the pure culture was washed with anaerobic phosphate-buffered saline (PBS) (0.8 g liter−1 NaCl and 0.2 g liter−1 KCl) in an anaerobic (10% CO2 and 90% N2) workstation (GP Campus, Jacomex, Dagneux, France). Using flow cytometry, the A. muciniphila concentration was quantified and was standardized to 2.5 · 108 ± 5 · 107 cells ml−1 before supplementation (10 ml) to the SHIME colon compartments (40). This daily dose of an average of 2.5 · 109 cells of A. muciniphila was based on other in vitro SHIME experiments and in vivo studies of the effect of bacterial supplementation/probiotics (11, 12, 41, 42). Samples of the pure A. muciniphila cultures were diluted in a filter-sterilized phosphate-buffered solution to obtain cell numbers within the detection range (103 to 106 cells/ml). Next, the samples were stained with SYBR green I (10,000× diluted from stock; Invitrogen) and incubated for 13 min at 37°C before measurement. The flow cytometer (BD Accuri C6 flow cytometer; BD, Erembodegem, Belgium) was equipped with a 488-nm solid-state laser, and Milli-Q water was used as the sheath fluid. Cell counts were done by measuring the number of particles in a set volume, and quality control of cell counting was done with standardized beads. The background was monitored by measuring a filtered sample, diluted identically to the test samples. Each sample was performed in triplicate (40).
The nutritional medium for the SHIME consisted of (in g liter−1) Arabic gum (1.0), starch (4.0) (Anco, Roeselare, Belgium), xylan (1.0), pectin (2.0), d-(+)-glucose (0.4), yeast extract (3.0) (Oxoid Ltd., Basingstoke, Hampshire, UK), peptone (1.0) (Oxoid Ltd., Basingstoke, Hampshire, UK), and commercial pig gastric mucin type II (4.0). The monosaccharide composition of mucin was (in grams per 100 g dry matter) l-arabinose (0.05), d-xylose (0.04), d-mannose (0.28), d-galactose (7.47), and d-glucose (1.95). This medium was autoclaved and acidified with 37% HCl to pH 2.0. The pancreatic juice contained (in g liter −1) NaHCO3 (12.5), bile salts (6.0) (Difco, Bierbeek, Belgium), and pancreatin (0.9).
Long-term dynamic gut model for the luminal colon microbiota (SHIME).
The dynamic in vitro SHIME model (ProDigest, Ghent University, Ghent, Belgium) was used to study the impact of supplementation of live A. muciniphila, with or without the presence of a host glycan degradation niche, in different microbial communities. The model is described by Van Herreweghen et al. (19); the model consists of multiple compartments that simulate the stomach, the small intestine, and the colon regions (43). Each anaerobic compartment was continuously stirred at 37°C and flushed with N2 (15 min/day) to ensure anaerobic conditions after sampling. On day 0, the colon compartments were filled with nutritional medium and inoculated with 40 ml of 20% (wt/vol) fecal slurry. Following an overnight static incubation of the colon compartments (16 h), the stomach and small intestine compartments operated on the fill-and-draw principle, with peristaltic pumps adding nutritional medium and pancreatic juice three times a day and gradually emptying the small intestine compartment into the colon compartments after gastrointestinal digestion. The volume in the colon compartments was kept constant by the simultaneous fluid flow into and out of compartments (44). Samples were taken from the vessels daily before new feed entered the colon compartments.
Fecal samples were collected from healthy donors between the ages of 25 and 35 and prepared within 1 h according to standard procedures (45) prior to inoculation.
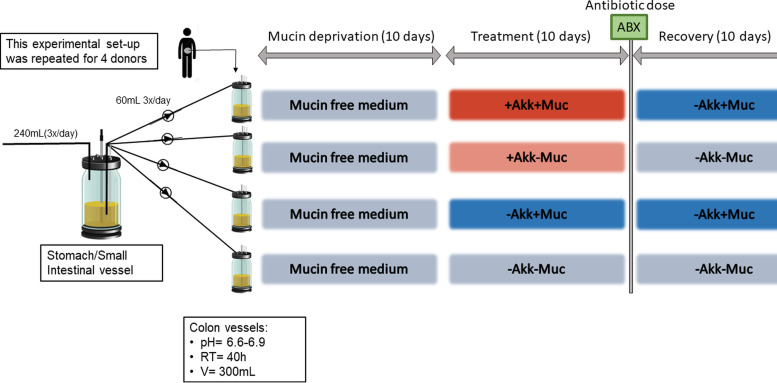
The setup for this experiment is shown in Fig. 8. Fecal suspension from 4 donors was used to inoculate the colon vessels (4 colon vessels/donor), with a retention time (RT) of 40 h and a pH between 6.6 and 6.9 (distal colon pH). During the mucin deprivation period (days 0 to 10), a mucin-free nutritional medium was fed to the colon vessels. From days 10 to 20 onward (treatment period), the 4 following treatments were applied to the 4 colon vessels/donor: +Akk+Muc, where A. muciniphila was administered daily to the colon vessels after sampling and mucin (4 g liter−1) was added to the feed; +Akk–Muc, where only A. muciniphila was added; –Akk+Muc, where only mucin (4 g liter−1) was added; and –Akk–Muc, which is identical to the medium provided during the mucin deprivation period. After this 10-day treatment period, an antibiotic mix, containing ciprofloxacin, amoxicillin, and tetracycline at, respectively, 40, 40, and 10 mg liter−1 final colonic concentrations, was supplemented directly into every colon vessel to induce acute stress (37).
FIG 8.
Experimental setup of the SHIME experiment. Akk, A. muciniphila; Muc, mucin; RT, retention time.
Samples were taken daily for SCFA analysis, as described previously (46), and every 2 days for DNA extraction (47), followed by 16S rRNA gene amplicon sequencing (Illumina MiSeq) (48) and A. muciniphila quantitative PCR (qPCR) quantification (49).
Microbial community analysis.
DNA extraction was performed by a combination of chemical and mechanical lysis through a bead-beating step as reported by Geirnaert et al. (50). As starting material, the pellet obtained after centrifuging 1 ml of luminal sample at 5,000 × g for 10 min was used. The DNA quality was verified on a 1.5% (wt/vol) agarose gel.
The total bacterial and Akkermansia-specific 16S rRNA gene copy number was quantified by qPCR on 100- and 10-fold-diluted DNA extracts, respectively, using a StepOnePlus real-time PCR system (Applied Biosystems, Carlsbad, CA) as described in reference 19. The total bacterial 16S rRNA genes and the species-specific 16S rRNA genes of A. muciniphila were quantified by qPCR on 100- and 10-fold-diluted DNA extracts, respectively, using a StepOnePlus real-time PCR system (Applied Biosystems, Carlsbad, CA). Primers for total bacteria (338F [ACTCCTACGGGAGGCAGCAG] and 518R [ATTACCGCGGCTGCTGG]) were used with the following cycling program: 3 min at 95°C, followed by 40 cycles of 1 min at 95°C, 40 s at 56°C, and 40 s at 72°C (51). A. muciniphila-specific primers (AM1 [GAGCACGTGAAGGTGGGGAC] and AM2 [CCTTGCGGTTGGCTTCAGAT]) were used with the following cycling program: 5 min at 95°C, followed by 40 cycles of 15 s at 95°C, 40 s at 60°C, and 30 s at 72°C and a final extension at 72°C for 5 min (49). The results of the A. muciniphila qPCR are shown as relative abundance, normalized to the total bacterial count. The qPCR mix consisted of 14.19 μl sterile nuclease-free water (Sigma-Aldrich, St. Louis, MO, USA) and 2.5 μl Taq buffer (10×, with KCl) containing 0.025 units of recombinant Taq DNA polymerase μl−1, 0.2 mM deoxynucleoside triphosphate (dNTP) mix, 1.5 mM MgCl2 (Fermentas Molecular Biology Tools, Waltham, MA, USA), 0.2 μM primer F, 0.2 μM primer R, 0.05 μg μl−1 bovine serum albumin (BSA) (Roche Applied Science, Penzberg, Germany), and 0.125 μl 20 × SYBR green (1:500 diluted from a 10,000 × SYBR green I nucleic acid stain concentrate in DMSO; Sigma-Aldrich, St. Louis, MO, USA). For each sample, 5 μl diluted DNA extract was added to 20 μl PCR mix in technical triplicate in a qPCR plate, and for each qPCR assay, standard curves were created by a 10-fold dilution series of DNA of a plasmid containing the targeted 16S rRNA gene fragment.
At various time points during the experiment, the bacterial community was assessed using amplicon sequencing of the 16S rRNA gene (48). DNA samples were sent to LGC Genomics (Teddington, Middlesex, UK) for library preparation and sequencing on an Illumina MiSeq platform, as described by De Paepe et al. (48). The V3-V4 region of the 16S rRNA gene was amplified by PCR using primers (341F [CCTACGGGNGGCWGCAG] and 785R [GACTACHVGGGTATCTAAKCC]) obtained from Klindworth et al. (52), with a slight modification to the reverse primer by introducing another degenerated position (K) to make it more universal. The mothur software package v.1.39.5 and its guidelines were used to process the amplicon data generated by LGC Genomics, as described in detail by De Paepe et al. (48).
Statistical analysis.
All statistical analyses were performed in R v.3.4.3.
Functional data. Nonparametric, rank-based longitudinal data analysis of the SCFA production (measured acetate, propionate, butyrate, and branched SCFA concentrations) over time was conducted using the R package nparLD (nparLD_2.1). Wald- and analysis of variance (ANOVA)-type statistics were used to assess the significance of the combined mucin and A. muciniphila treatment as a function of time (f1-ld-f1 design). A significant time effect was observed, which was expected, as the treatment was applied after an initial stabilization period of 10 days and the system was disturbed after 20 days by an antibiotic pulse. The longitudinal data analysis was therefore repeated on the data subsets (stabilization, treatment prior to antibiotic pulse, and treatment post-antibiotic pulse). The relative treatment effects obtained by nparLD were verified by a partial redundancy analysis, followed by a PCA (vegan 2.4-4 package).
Acetate, propionate, butyrate, and branched SCFA levels were modeled as a function of the treatment (with A. muciniphila and/or mucin), conditional on the period (stabilization, treatment prior to antibiotic pulse, and treatment post-antibiotic pulse), and interindividual differences (factor donor). Similarly, donor and period were considered main effects, conditional on the other factors. Permutation tests were applied to assess the statistical significance of the global model and the individual canonical axes (53). The RDA results were plotted in a type II scaling correlation triplot, displaying the constrained canonical (labeled RDA1/2) and, in the case of the A. muciniphila or mucin effect, the first unconstrained residual (labeled PC1) axis. Both axes were annotated with the proportional eigenvalues representing their contribution to the total (both constrained and unconstrained) variance. The coordinates of the sites were derived from the weighted sums of the scores of the response variables. Next to the absolute metabolite concentrations, the relative proportion of the metabolites is an important marker. Therefore, the above-outlined procedure was repeated using the metabolite ratios. Additionally, in order to assess if significant interactions occurred between the explanatory variables, a global RDA was performed based on a regression model including interaction terms in addition to each of the main effects.
Microbial community data. To visualize differences in microbial community composition between donors, treatments, and antibiotic responses, ordination and clustering techniques were applied. For these purposes, the shared file was further processed to remove OTUs with too low an abundance according to the arbitrary cutoffs described by McMurdie and Holmes (54). An OTU is defined in the manuscript as a collection of sequences that are found to be more than 97% similar to one another in the V3-V4 region of their 16S rRNA gene after applying hierarchical clustering (55–58). To deal with differences in sampling depth, proportional data transformed on the common scale to the lowest number of reads was used (54). A table with the most abundant OTUs classified to the species level using both the RDP Seqmatch tool and NCBI BLAST is given in Table S1.
Principal-coordinate analysis (PCoA; package stats) was conducted based on the abundance-based Jaccard dissimilarity matrix (vegan package; visualized with ggplot2) (59–62). This procedure was repeated at the OTU and genus levels, focusing on the comparison between the donors and between the applied treatments. At the genus level, weighted averages of genus abundances were a posteriori added to the ordination plot using the wascores function in vegan (61). Donor and treatment both influenced the grouping of samples, which was further explored using a partial distance-based redundancy analysis at the species level (db RDA) (63). The scores obtained by a PCoA were modeled as a function of the treatment, with the effects of the interindividual variability and treatment period being parceled out using the capscale function of the vegan package (vegan_2.4-4) (61, 62). Interpretation of the results was preceded by a permutation test of the db RDA results to confirm that a relationship exists between the response data and the exploratory variables. Using the same principle, the significance of the first two constrained axes was evaluated. The constrained fraction of the variance, explained by the exploratory variables, was adjusted by applying a subtractive procedure (64, 65). The fraction of the variance explained by the exploratory variables and its significance are given in Table S2.
In a next step, sparse partial least squares discriminant analysis (sPLS-DA) (mixOmics_6.3.1) was performed to select the taxonomic features most predictive of the treatment (+Akk+Muc, –Akk+Muc, +Akk–Muc, and –Akk–Muc) (Fig. 1). Here, too, a factorial response variable was created, indicating the treatment condition of each sample. The filtered proportional OTU-level abundances were used as predictors. The number of components and OTUs or genera to include in the sPLS-DA model were assessed based on the classification error rates obtained after a 5-fold cross-validation. The final sPLS-DA model, with an optimum of 3 components, was displayed (Fig. S2), and the proportional abundances of the most predictive and most abundant OTUs and genera were represented in a heatmap (Fig. 5).
Finally, in order to find statistically significant differences in the species- and genus-level abundances between the different treatments, the DESeq package was applied on the filtered, unnormalized data at the end of the treatment period (day 20) (α = 0.05) as suggested by McMurdie and Holmes (54) and Love et al. (66). The factors “treatment” and “donor” were used in the design formula, and the effect of the treatment was determined by a likelihood ratio test on the difference in deviance between a full- and reduced-model formula. An empirical Bayes shrinkage correction was employed for low counts (66). Pairwise significant differences were obtained using Wald tests, specifying all pairwise combinations of treatments as the contrast argument. Results from the pairwise comparisons were visualized in a volcano plot, showing the −log10 adjusted P value as a function of the shrunken log2 fold change. Species with an absolute shrunken log2 fold change exceeding 2 were annotated in the plot (67). The most pronounced significant differences at the species level were shown in side-by-side boxplots comparing the normalized counts (plus a 0.5 pseudocount) during treatments.
To assess the effect of the antibiotic pulse on the microbial community, the DESeq package was again applied. The effect of the antibiotic pulse was determined by a likelihood ratio test on the difference in deviance between a full- and reduced-model formula. An empirical Bayes shrinkage correction was employed for low counts (66). Results from the pairwise comparisons, comparing values obtained before and after the antibiotic pulse for each treatment, were visualized in a volcano plot showing the −log10 adjusted P value as a function of the shrunken log2 fold change. Genera with an absolute shrunken log2 fold change exceeding 1 were annotated in the plot (67) (Fig. S4). Also, alpha diversity was calculated using the Shannon coefficient (vegan package) and visualized in Fig. S5.
Data availability.
The sequencing data have been submitted to the NCBI (National Center for Biotechnology Information) database under accession number SRP126579.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the Agency for Innovation by Science and Technology (grant number 131774), the Special Research Fund (BOF) Concerted Research Actions (GOA, BOF17/GOA/032), and UGent-VUB ALO2015 for funding this research.
We thank Jana De Bodt and Chloë Rotsaert for the technical support.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Derrien M, Vaughan EE, Plugge CM, de Vos WM. 2004. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol 54:1469–1476. doi: 10.1099/ijs.0.02873-0. [DOI] [PubMed] [Google Scholar]
- 2.Png CW, Linden SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, McGuckin MA, Florin THJ. 2010. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol 105:2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 3.Santacruz A, Collado MC, Garcia-Valdes L, Segura MT, Martin-Lagos JA, Anjos T, Marti-Romero M, Lopez RM, Florido J, Campoy C, Sanz Y. 2010. Gut microbiota composition is associated with body weight, weight gain and biochemical parameters in pregnant women. Br J Nutr 104:83–92. doi: 10.1017/S0007114510000176. [DOI] [PubMed] [Google Scholar]
- 4.Swidsinski A, Dorffel Y, Loening-Baucke V, Theissig F, Ruckert JC, Ismail M, Rau WA, Gaschler D, Weizenegger M, Kuhn S, Schilling J, Dorffel WV. 2011. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 60:34–40. doi: 10.1136/gut.2009.191320. [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Christophersen CT, Sorich MJ, Gerber JP, Angley MT, Conlon MA. 2011. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol 77:6718–6721. doi: 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. 2009. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earley H, Lennon G, Balfe A, Coffey JC, Winter DC, O’Connell PR. 2019. The abundance of Akkermansia muciniphila and its relationship with sulphated colonic mucins in health and ulcerative colitis. Sci Rep 9:15683. doi: 10.1038/s41598-019-51878-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dao MC, Belda E, Prifti E, Everard A, Kayser BD, Bouillot JL, Chevallier JM, Pons N, Le Chatelier E, Ehrlich SD, Dore J, Aron-Wisnewsky J, Zucker JD, Cani PD, Clement K. 2019. Akkermansia muciniphila abundance is lower in severe obesity, but its increased level after bariatric surgery is not associated with metabolic health improvement. Am J Physiol Endocrinol Metab 317:E446–E459. doi: 10.1152/ajpendo.00140.2019. [DOI] [PubMed] [Google Scholar]
- 9.Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, Kayser BD, Levenez F, Chilloux J, Hoyles L, Consortium MI-O, Dumas ME, Rizkalla SW, Dore J, Cani PD, Clement K. MICRO-Obes Consortium. 2016. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut 65:426–436. doi: 10.1136/gutjnl-2014-308778. [DOI] [PubMed] [Google Scholar]
- 10.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, Guiot Y, Derrien M, Muccioli GG, Delzenne NM, de Vos WM, Cani PD. 2013. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A 110:9066–9071. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plovier H, Everard A, Druart C, Depommier C, Van Hul M, Geurts L, Chilloux J, Ottman N, Duparc T, Lichtenstein L, Myridakis A, Delzenne NM, Klievink J, Bhattacharjee A, van der Ark KCH, Aalvink S, Martinez LO, Dumas ME, Maiter D, Loumaye A, Hermans MP, Thissen JP, Belzer C, de Vos WM, Cani PD. 2017. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat Med 23:107–113. doi: 10.1038/nm.4236. [DOI] [PubMed] [Google Scholar]
- 12.Depommier C, Everard A, Druart C, Plovier H, Van Hul M, Vieira-Silva S, Falony G, Raes J, Maiter D, Delzenne NM, de Barsy M, Loumaye A, Hermans MP, Thissen JP, de Vos WM, Cani PD. 2019. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat Med 25:1096–1103. doi: 10.1038/s41591-019-0495-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belzer C, de Vos WM. 2012. Microbes inside—from diversity to function: the case of Akkermansia. ISME J 6:1449–1458. doi: 10.1038/ismej.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cani PD, de Vos WM. 2017. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol 8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou K 2017. Strategies to promote abundance of Akkermansia muciniphila, an emerging probiotics in the gut, evidence from dietary intervention studies. J Funct Foods 33:194–201. doi: 10.1016/j.jff.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O’Toole PW, Marchesi JR, Hill C. 2017. Next-generation probiotics: the spectrum from probiotics to live biotherapeutics. Nat Microbiol 2:17057. doi: 10.1038/nmicrobiol.2017.57. [DOI] [PubMed] [Google Scholar]
- 17.Ottman N, Davids M, Suarez-Diez M, Boeren S, Schaap PJ, Martins Dos Santos VAP, Smidt H, Belzer C, de Vos WM. 2017. Genome-scale model and omics analysis of metabolic capacities of Akkermansia muciniphila reveal a preferential mucin-degrading lifestyle. Appl Environ Microbiol 83:e01014-17. doi: 10.1128/AEM.01014-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berry D, Stecher B, Schintlmeister A, Reichert J, Brugiroux S, Wild B, Wanek W, Richter A, Rauch I, Decker T, Loy A, Wagner M. 2013. Host-compound foraging by intestinal microbiota revealed by single-cell stable isotope probing. Proc Natl Acad Sci U S A 110:4720–4725. doi: 10.1073/pnas.1219247110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Herreweghen F, De Paepe K, Roume H, Kerckhof FM, Van de Wiele T. 2018. Mucin degradation niche as a driver of microbiome composition and Akkermansia muciniphila abundance in a dynamic gut model is donor independent. FEMS Microbiol Ecol 94:fiy186. doi: 10.1093/femsec/fiy186. [DOI] [PubMed] [Google Scholar]
- 20.Van Herreweghen F, Van den Abbeele P, De Mulder T, De Weirdt R, Geirnaert A, Hernandez-Sanabria E, Vilchez-Vargas R, Jauregui R, Pieper DH, Belzer C, De Vos WM, Van de Wiele T. 2017. In vitro colonisation of the distal colon by Akkermansia muciniphila is largely mucin and pH dependent. Benef Microbes 8:81–96. doi: 10.3920/BM2016.0013. [DOI] [PubMed] [Google Scholar]
- 21.Derrien M 2007. Mucin utilisation and host interactions of the novel intestinal microbe A. muciniphila. PhD thesis Wageningen University, Wageningen, The Netherlands. [Google Scholar]
- 22.Lai SK, Wang YY, Wirtz D, Hanes J. 2009. Micro- and macrorheology of mucus. Adv Drug Deliv Rev 61:86–100. doi: 10.1016/j.addr.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Passel MW, Kant R, Zoetendal EG, Plugge CM, Derrien M, Malfatti SA, Chain PS, Woyke T, Palva A, de Vos WM, Smidt H. 2011. The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. doi: 10.1371/journal.pone.0016876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reunanen J, Kainulainen V, Huuskonen L, Ottman N, Belzer C, Huhtinen H, de Vos WM, Satokari R. 2015. Akkermansia muciniphila adheres to enterocytes and strengthens the integrity of the epithelial cell layer. Appl Environ Microbiol 81:3655–3662. doi: 10.1128/AEM.04050-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belzer C, Chia LW, Aalvink S, Chamlagain B, Piironen V, Knol J, de Vos WM. 2017. Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and vitamin B12 production by intestinal symbionts. mBio 8:e00770-17. doi: 10.1128/mBio.00770-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chia LW, Hornung BVH, Aalvink S, Schaap PJ, de Vos WM, Knol J, Belzer C. 2018. Deciphering the trophic interaction between Akkermansia muciniphila and the butyrogenic gut commensal Anaerostipes caccae using a metatranscriptomic approach. Antonie Van Leeuwenhoek 111:859–873. doi: 10.1007/s10482-018-1040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoskins LC, Agustines M, Mckee WB, Boulding ET, Kriaris M, Niedermeyer G. 1985. Mucin degradation in human-colon ecosystems: isolation and properties of fecal strains that degrade ABH blood group antigens and oligosaccharides from mucin glycoproteins. J Clin Invest 75:944–953. doi: 10.1172/JCI111795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. 2013. A refined palate: bacterial consumption of host glycans in the gut. Glycobiology 23:1038–1046. doi: 10.1093/glycob/cwt040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martens EC, Chiang HC, Gordon JI. 2008. Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4:447–457. doi: 10.1016/j.chom.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gibson GR, Hutkins R, Sanders ME, Prescott SL, Reimer RA, Salminen SJ, Scott K, Stanton C, Swanson KS, Cani PD, Verbeke K, Reid G. 2017. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat Rev Gastroenterol Hepatol 14:491–502. doi: 10.1038/nrgastro.2017.75. [DOI] [PubMed] [Google Scholar]
- 31.Ouwehand AC, Derrien M, de Vos W, Tiihonen K, Rautonen N. 2005. Prebiotics and other microbial substrates for gut functionality. Curr Opin Biotechnol 16:212–217. doi: 10.1016/j.copbio.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Johansson ME, Ambort D, Pelaseyed T, Schutte A, Gustafsson JK, Ermund A, Subramani DB, Holmen-Larsson JM, Thomsson KA, Bergstrom JH, van der Post S, Rodriguez-Pineiro AM, Sjovall H, Backstrom M, Hansson GC. 2011. Composition and functional role of the mucus layers in the intestine. Cell Mol Life Sci 68:3635–3641. doi: 10.1007/s00018-011-0822-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson MEV 2012. Fast renewal of the distal colonic mucus layers by the surface goblet cells as measured by in vivo labeling of mucin glycoproteins. PLoS One 7:e41009. doi: 10.1371/journal.pone.0041009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johansson MEV, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. 2008. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci U S A 105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoskins LC 1993. Mucin degradation in the human gastrointestinal-tract and its significance to enteric microbial ecology. Eur J Gastroenterol Hepatol 5:205–213. doi: 10.1097/00042737-199304000-00004. [DOI] [Google Scholar]
- 36.Ballan R, Battistini C, Xavier-Santos D, Saad SMI. 2020. Interactions of probiotics and prebiotics with the gut microbiota. Prog Mol Biol Transl Sci 171:265–300. doi: 10.1016/bs.pmbts.2020.03.008. [DOI] [PubMed] [Google Scholar]
- 37.Marzorati M, Vilchez-Vargas R, Bussche JV, Truchado P, Jauregui R, El Hage RA, Pieper DH, Vanhaecke L, Van de Wiele T. 2017. High-fiber and high-protein diets shape different gut microbial communities, which ecologically behave similarly under stress conditions, as shown in a gastrointestinal simulator. Mol Nutr Food Res 61. doi: 10.1002/mnfr.201600150. [DOI] [PubMed] [Google Scholar]
- 38.Gustafsson A, Lund-Tonnesen S, Berstad A, Midtvedt T, Norin E. 1998. Faecal short-chain fatty acids in patients with antibiotic-associated diarrhoea, before and after faecal enema treatment. Scand J Gastroenterol 33:721–727. doi: 10.1080/00365529850171666. [DOI] [PubMed] [Google Scholar]
- 39.Van den Abbeele P, Roos S, Eeckhaut V, MacKenzie DA, Derde M, Verstraete W, Marzorati M, Possemiers S, Vanhoecke B, Van Immerseel F, Van de Wiele T. 2012. Incorporating a mucosal environment in a dynamic gut model results in a more representative colonization by lactobacilli. Microb Biotechnol 5:106–115. doi: 10.1111/j.1751-7915.2011.00308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Van Nevel S, Koetzsch S, Weilenmann HU, Boon N, Hammes F. 2013. Routine bacterial analysis with automated flow cytometry. J Microbiol Methods 94:73–76. doi: 10.1016/j.mimet.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 41.Duysburgh C, Van den Abbeele P, Krishnan K, Bayne TF, Marzorati M. 2019. A synbiotic concept containing spore-forming Bacillus strains and a prebiotic fiber blend consistently enhanced metabolic activity by modulation of the gut microbiome in vitro. Int J Pharm X 1:100021. doi: 10.1016/j.ijpx.2019.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geirnaert A, Calatayud M, Grootaert C, Laukens D, Devriese S, Smagghe G, De Vos M, Boon N, Van de Wiele T. 2017. Butyrate-producing bacteria supplemented in vitro to Crohn’s disease patient microbiota increased butyrate production and enhanced intestinal epithelial barrier integrity. Sci Rep 7:11450. doi: 10.1038/s41598-017-11734-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van den Abbeele P, Grootaert C, Marzorati M, Possemiers S, Verstraete W, Gérard P, Rabot S, Bruneau A, El Aidy S, Derrien M, Zoetendal E, Kleerebezem M, Smidt H, Van de Wiele T. 2010. Microbial community development in a dynamic gut model is reproducible, colon-region specific, and selects for Bacteroidetes and Clostridium cluster IX. Appl Environ Microbiol 76:5237–5246. doi: 10.1128/AEM.00759-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Possemiers S, Verthe K, Uyttendaele S, Verstraete W. 2004. PCR-DGGE-based quantification of stability of the microbial community in a simulator of the human intestinal microbial ecosystem. FEMS Microbiol Ecol 49:495–507. doi: 10.1016/j.femsec.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Molly K, Vande Woestyne M, Verstraete W. 1993. Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol 39:254–258. doi: 10.1007/BF00228615. [DOI] [PubMed] [Google Scholar]
- 46.Andersen SJ, Hennebel T, Gildemyn S, Coma M, Desloover J, Berton J, Tsukamoto J, Stevens C, Rabaey K. 2014. Electrolytic membrane extraction enables production of fine chemicals from biorefinery sidestreams. Environ Sci Technol 48:7135–7142. doi: 10.1021/es500483w. [DOI] [PubMed] [Google Scholar]
- 47.Geirnaert A 2015. Probiotic potency of butyrate-producing bacteria for modulating the microbiome and epithelial barrier in inflammatory bowel disease. PhD thesis. Ghent University, Ghent, Belgium. [Google Scholar]
- 48.De Paepe K, Kerckhof FM, Verspreet J, Courtin CM, Van de Wiele T. 2017. Inter-individual differences determine the outcome of wheat bran colonization by the human gut microbiome. Environ Microbiol 19:3251–3267. doi: 10.1111/1462-2920.13819. [DOI] [PubMed] [Google Scholar]
- 49.Collado MC, Derrien M, Isolauri E, de Vos WM, Salminen S. 2007. Intestinal integrity and Akkermansia muciniphila, a mucin-degrading member of the intestinal microbiota present in infants, adults, and the elderly. Appl Environ Microbiol 73:7767–7770. doi: 10.1128/AEM.01477-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geirnaert A, Wang J, Tinck M, Steyaert A, Van den Abbeele P, Eeckhaut V, Vilchez-Vargas R, Falony G, Laukens D, De Vos M, Van Immerseel F, Raes J, Boon N, Van de Wiele T. 2015. Interindividual differences in response to treatment with butyrate-producing Butyricicoccus pullicaecorum 25-3T studied in an in vitro gut model. FEMS Microbiol Ecol 91:fiv054. doi: 10.1093/femsec/fiv054. [DOI] [PubMed] [Google Scholar]
- 51.Ovreas L, Forney L, Daae FL, Torsvik V. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl Environ Microbiol 63:3367–3373. doi: 10.1128/AEM.63.9.3367-3373.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Legendre P, Oksanen J, ter Braak CJF. 2011. Testing the significance of canonical axes in redundancy analysis. Methods in Ecology and Evolution 2:269–277. doi: 10.1111/j.2041-210X.2010.00078.x. [DOI] [Google Scholar]
- 54.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen W, Zhang CK, Cheng YM, Zhang SW, Zhao HY. 2013. A comparison of methods for clustering 16S rRNA sequences into OTUs. PLoS One 8:e70837. doi: 10.1371/journal.pone.0070837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schloss PD, Westcott SL. 2011. Assessing and improving methods used in operational taxonomic unit-based approaches for 16S rRNA gene sequence analysis. Appl Environ Microbiol 77:3219–3226. doi: 10.1128/AEM.02810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X, Cai Y, Sun Y, Knight R, Mai V. 2012. Secondary structure information does not improve OTU assignment for partial 16s rRNA sequences. ISME J 6:1277–1280. doi: 10.1038/ismej.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anderson MJ, Ellingsen KE, McArdle BH. 2006. Multivariate dispersion as a measure of beta diversity. Ecol Lett 9:683–693. doi: 10.1111/j.1461-0248.2006.00926.x. [DOI] [PubMed] [Google Scholar]
- 60.Cox TF 2001. Multidimensional scaling used in multivariate statistical process control. J Appl Stat 28:365–378. doi: 10.1080/02664760120034108. [DOI] [Google Scholar]
- 61.Oksanen JG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens H, Szoecs E, Wagner H. 2016. vegan: community ecology package. R package version 2.4-0. https://CRAN.R-project.org/package=vegan.
- 62.Ramette A 2007. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol 62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vardakou M, Palop CN, Gasson M, Narbad A, Christakopoulos P. 2007. In vitro three-stage continuous fermentation of wheat arabinoxylan fractions and induction of hydrolase activity by the gut microflora. Int J Biol Macromol 41:584–589. doi: 10.1016/j.ijbiomac.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 64.Peres-Neto PR, Legendre P, Dray S, Borcard D. 2006. Variation partitioning of species data matrices: estimation and comparison of fractions. Ecology 87:2614–2625. doi: 10.1890/0012-9658(2006)87[2614:VPOSDM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 65.Borcard D, Gillet F, Legendre P. 2011. Numerical ecology with R. Springer, New York, New York. [Google Scholar]
- 66.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Quackenbush J 2002. Microarray data normalization and transformation. Nat Genet 32:496–501. doi: 10.1038/ng1032. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data have been submitted to the NCBI (National Center for Biotechnology Information) database under accession number SRP126579.