Abstract
Background
Overexpression of epidermal growth factor receptor (EGFR), and downstream pathway activation appears to be a common oncogenic driver in the majority of head and neck squamous cell cancers (HNSCCs); yet targeting EGFR for the treatment of HNSCC has met with limited success. Apart from the anti-EGFR antibody cetuximab, no small molecule EGFR/tyrosine kinase inhibitors (TKIs) have progressed to routine clinical use. The aim of this study was to determine factors contributing to the lack of response to TKIs and identify alternative therapeutic vulnerabilities.
Methods
Genomic and transcriptomic sequencing, high-throughput compound screens, overexpression and siRNA knockdown, western blot, in vivo xenograft studies.
Findings
We derived three pairs of isogenic gefitinib (TKI)-sensitive and resistant patient-derived HNSCC cell lines. Genomic sequencing of gefitinib-resistant cell lines identified a lack of activating and resistance-associated EGFR mutations. Instead, transcriptomic sequencing showed upregulated EMT gene signature in the gefitinib-resistant cells with a corresponding increase in their migratory phenotype. Additionally, the resistant cell displayed reduced growth rate. Surprisingly, while gefitinib-resistant cells were independent of EGFR for survival, they nonetheless displayed activation of downstream ERK and AKT signalling. High-throughput screening (HTS) of druggable, small molecule libraries revealed that the gefitinib-resistant cells were particularly sensitive to inhibitors of genes involved in cell cycle and mitosis, such as Aurora kinase inhibitors (AKIs), cyclin-dependent kinase (CDK) inhibitors, and microtubule inhibitors. Notably our results showed that in the EGFR inhibited state, Aurora kinases are essential for cell survival.
Interpretation
Our study demonstrates that in the absence of activating EGFR mutations, HNSCCs may gain resistance to gefitinib through decreased cell proliferation, which makes them exceptionally vulnerable to cell-cycle inhibitors.
Funding
Agency for Science, Technology, and Research (A*STAR), National Medical Research Council (NMRC), and the National Institutes of Health (NIH)/National Cancer Institute (NCI).
Keywords: Head and neck squamous cell carcinoma, Gefitinib resistance, Aurora kinase inhibition, EGFR T790M negative, Chemical genetics
Research in context.
Evidence before this study
The epidermal growth factor receptor (EGFR) is commonly overexpressed in head and neck squamous cell carcinoma (HNSCC) and has been associated with poor prognosis. While the anti-EGFR antibody cetuximab has been approved for locally advanced or metastatic HNSCC in combination with chemotherapy or radiotherapy, small molecule EGFR inhibitors have met with little success. Investigation of resistance to anti-EGFR therapy in HNSCCs has thus been mainly focused on resistance to cetuximab. Some mechanisms of resistance to anti-EGFR therapy include HER2/HER3 activation, MET overexpression, and epithelial to mesenchymal transition (EMT).
Added value of this study
In this study, we show that in HNSCCs, the evolution of gefitinib resistance is not driven by significant genomic alterations. More importantly, EGFR remains wild-type in the gefitinib-resistant HNSCCs, which is in contrast with TKI-resistance in non-small cell lung carcinoma (NSCLC), where it is predominantly driven by the gain of activating mutations, such as T790M in EGFR. Instead, our study shows that gefitinib-resistant HNSCC cells no longer depend on EGFR signalling for survival. These resistant cells also display decreased rates of proliferation thereby making them vulnerable to inhibitors of cell division such as Aurora kinase inhibitors (AKIs), cyclin dependent kinase (CDK) inhibitors, and microtubule inhibitors. The results from our study also indicate that in the EGFR inhibited state, Aurora kinases are essential for cell survival.
Implications of all the available evidence
Our results demonstrate the potential clinical utility of inhibitors of cell division to target HNSCCs that display resistance to EGFR-directed TKIs.
Alt-text: Unlabelled box
1. Introduction
Head and neck squamous cell carcinomas (HNSCCs) represent the most common malignancy arising from the larynx, pharynx, oral cavity, nasal cavity, and paranasal sinuses [1]. Treatment modalities for HNSCC include surgery or radiotherapy for early stage (I/II) patients while patients with locoregionally advanced (stage II/IV) disease undergo multimodal treatment involving combinations of surgery, radiotherapy, cisplatin-based chemoradiotherapy [2]. However conventional radiotherapy and chemotherapy are often associated with adverse side effects and reduced quality of life for patients [2]. Additionally, recurrent or metastatic disease occur in majority of HNSCC patients (more than 65%) despite treatment [1].
The application of targeted therapy poses an attractive strategy for the treatment of HNSCC. However, this has not come into fruition in the vast majority of cases. Indeed, there has been significant interest in the use of epidermal growth factor receptor (EGFR) targeted therapies for HNSCC. EGFR is a receptor tyrosine kinase (RTK) belonging to the ErbB family of proteins. Other members of the ErbB family include HER2, HER3, and HER4. Activation of EGFR occurs upon ligand binding to the extracellular domain, resulting in homodimerization or heterodimerization with other members of the ErbB family and subsequent phosphorylation of its intracellular domains. This in turn activates various downstream signalling pathways such as the RAS-RAF-MEK-ERK and the PI3K-AKTmTOR pathways [3]. Notably, EGFR and its downstream signalling effectors are upregulated in about 90% of HNSCC tumours [4,5]. In fact, high expression of EGFR has been reported to be associated with poor prognosis and decreased overall survival [6], [7], [8]. Thus far, the only EGFR targeted therapy that has been approved for HNSCC is the EGFR monoclonal antibody cetuximab [9]. Numerous clinical trials have also explored the use of EGFR tyrosine kinase inhibitors (TKIs), including gefitinib, erlotinib and afatinib, for patients with locoregionally advanced, metastatic, and/or recurrent disease [10], [11], [12], [13]. However, response to these TKIs have been dismally low, with less than 10% of patients showing any tumour regression with no significantly benefits to patient outcomes. Hence, none of the EGFR TKIs have been approved for clinical use on HNSCC patients.
A reason for the lack of success of EGFR TKIs in HNSCC is the lack of biomarkers that predict clinical response. Unlike in non-small cell lung cancer (NSCLC), whereby activating mutations in the EGFR kinase domain improved clinical responses to EGFR TKIs [14,15], such mutations are rare in HNSCC [16], [17], [18]. Additionally, these tumours may also have intrinsic or primary resistance to TKIs or acquire these during drug-induced tumour evolution. Furthermore, the EGFR signalling pathway can cross-talk with multiple other signal transduction pathways such as MET [19] and IGF1R-signalling [20]. The complexity of EGFR signalling combined with the lack of predictive EGFR mutations in HNSCC patients presents a major challenge for the identification of strategies to overcome EGFR-TKI resistance and thereby improve the response against EGFR-TKIs.
Therefore, in this study we sought to understand the mechanism of TKI-resistance in HNSCC through a chemical genetics approach. We generated three pairs of isogenic patient-derived, gefitinib-sensitive and resistant HNSCC cell lines in vitro, and employed in-depth functional genomic characterisation together with high-throughput chemical screens. These approaches allowed us to not only explore the mechanistic basis for drug-insensitivity in the gefitinib-resistant setting, but also uncover alternative therapeutic vulnerabilities in the TKI-resistant HNSCC cell lines. Altogether the results demonstrate that in the absence of any activating EGFR mutations (T790M), HNSCC cells adopt a strategy of reduced cell proliferation to gain resistance to EGFR TKIs, thus gaining an alternative vulnerability to drugs that target cell proliferation.
2. Methods
2.1. Compounds
Gefitinib (cat# 13166) was purchased from Cayman Chemical. Afatinib (cat# S1011), AZD9291 (osimertinib, cat# S7297), BGB324 (R428, cat# S2841), TAK-901 (cat# S2718), as well as the kinase inhibitor (cat# L1200) and anti-cancer (cat# L3000) compound libraries were all purchased from Selleckchem. All drug stock solutions were prepared in dimethylsulfoxide (DMSO) and stored at −20 °C. Gefitinib used for in vivo studies were clinical grade tablets for human dosage while TAK-901 (cat# HY-12201) used for in vivo studies was purchased from MedChemExpress.
2.2. Cell culture
HN19, HN64, and HN90 cells were derived and validated as previously described [21,22]. No further validation of the cell lines was carried out in this study. All HNSCC cells were cultured in RPMI 1640 medium (Hyclone, cat# SH30027.01) supplemented with 10% FBS (Hyclone, cat# SV30160.03) and 100 U/mL penicillin-streptomycin (Gibco, cat# 15140122). HEK293T cells were cultured in DMEM medium (Gibco, cat# 11965084) supplemented with 15% FBS. Cells were grown in a humidified incubator at 37 °C with 5% CO2 and were routinely tested for mycoplasma contamination using MycoAlert mycoplasma detection kit (Lonza, cat# LT07-118).
2.3. Derivation of gefitinib-resistant cell lines
Gefitinib-resistant HN19-GR, HN64-GR, and HN90-GR cell lines were derived by chronic dosing of the corresponding parental cell line with increasing concentrations of gefitinib till a final concentration of 6.4 µM. The GR cells were subsequently maintained at 6.4 µM gefitinib.
2.4. POLARIS sequencing
DNA from HN19, HN60, HN90 and their corresponding GR cells was extracted using DNeasy blood and tissue kit (Qiagen, cat# 69504) and following the manufacturer's instructions. The DNA was then subjected to POLARIS XPLORA cancer panel (https://www.a-star.edu.sg/polaris/) (Table S1) sequencing assay to identify SNVs in 740 cancer-associated genes. The full lists of SNVs detected can be found in Table S2.
2.5. RNA sequencing
RNA from HN19, HN60, HN90 and their corresponding GR cells was extracted using RNeasy plus mini kit (Qiagen, cat# 74136) and following the manufacturer's instructions. RNA quality was checked using the RNA 6000 nano kit (Agilent, cat# 5067-1511) and RNA with RIN values of more than 9.5 were used for RNA sequencing. RNA sequencing was carried out by Theragen Etex Bio Institute. Libraries were prepared with Illumina TruSeq stranded kit for paired end sequencing. Generated libraries were run on an Illumina HiSeq4K system and 30 million raw reads were collated per sample. Reads were mapped to the human genome (GRCh37) followed by analysis for differential expression using Cuffdiff (Table S3). Gene sets in the Molecular Signatures Database (MSigDB) were used with gene set enrichment analysis (GSEA) software [23] to identify classes of genes that are over-represented in the resistant cell lines treated with the gefitinib as compared to the sensitive cell lines (Tables S4–S6). Gene sets with absolute normalized enrichment score greater than 1.5 and FDR q value less than 0.25 in all the three cell lines were reported. The RNA sequencing datasets generated during the course of this study have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE157374.
2.6. Chemical compound library screen and analysis
Cells were plated at 2500 cells per well in 50 µL of growth medium in white, flat bottom 384-well plates (Corning, cat# 3570) and allowed to attach overnight at 37 °C, 5% CO2. The next day, cells were treated with the kinase inhibitor (Selleckchem, cat# L1200) and anti-cancer (Selleckchem, cat# L3000) compound libraries at a final concentration of 1 µM, 1% DMSO. After 72 h, viability of the treated cells was determined using CellTiter-Glo luminescent cell viability assay (Promega, cat# G7573) according the manufacturer's protocol. Luminescence signals were measured using Tecan Infinite M1000 microplate reader.
Analysis of the chemical screen was carried out by calculating the median luminescent signal of DMSO control in each plate. The %inhibition score of each compound was then determined using the following formula:
Compounds for which the %inhibition score is greater than 50 were identified as hits. The complete list of compounds tested and their %inhibition scores against each cell line can be found in Table S7. Target enrichment p values for each cell line were calculated using Fisher's exact test. The FDR p values can be found in Table S8.
2.7. Western blot
Cells were washed twice with ice cold PBS on ice and lysed on ice in RIPA buffer (Thermo Fisher Scientific, cat# 89900) supplemented with complete protease inhibitor cocktail (Roche, cat# 11697498001) and PhosSTOP phosphatase inhibitor cocktail (Roche, cat# 4906837001). Protein concentration in lysates were determined using BCA protein assay kit (Pierce, cat# 23225). 30–50 µg of protein lysate per sample was resolved on Tris-Glycine SDS-polyacrylamide gels and blotted onto PVDF membranes (Millipore, cat# IPVH00010). The membranes were blocked with Intercept® (TBS) blocking buffer (Li-COR, cat# 927-60001), following which the membranes were incubated with primary antibodies in Intercept® T20 (TBS) antibody diluent (Li-COR, cat# 927-65001) overnight at 4 °C. The list of primary and secondary antibodies used and their dilutions can be found in Table S9. Signals were detected on Li-COR Odyssey CLx imaging system.
2.8. Immunofluorescence
Cells were seeded into black, clear, flat bottom 96-well plates (Corning, cat# 3904) at 6000 cells per well and allowed to proliferate for 2 days at 37 °C, 5% CO2. The cells were washed once with PBS and fixed for 20 min at room temperature with 4% PFA. After which the cells were permeabilised with 0.1% Triton X-100 (Sigma, cat# T8787) in PBS for 15 min at room temperature and blocked with 3% BSA in PBS for 1 h at room temperature. The cells were incubated overnight at 4 °C with primary antibodies in blocking buffer followed by incubation with secondary antibodies and DAPI (Thermo Fisher Scientific, cat# 62248, 1:10,000) in PBS for 1 h at room temperature. Primary antibody used was anti-N-Cadherin (Cell Signalling Technology, cat# 13116, RRID: AB_2687616, 1:500). Signals were detected using Alexa Fluor 488 goat anti-rabbit antibody (Invitrogen, cat# A-11034, RRID: AB_2576217, 1:5000) and images were captured using Opera Phenix high content screening (HCS) system from Perkin Elmer.
2.9. Immunofluorescence dose response
Cells were seeded into black, clear, flat bottom 96-well plates at 4500 cells per well and allowed to attach overnight at 37 °C, 5% CO2. The next day, serially diluted TAK-901 in DMSO were added to the appropriate wells. DMSO was also added to control wells to a final concentration of 0.1%. At 6 h, 16 h, and 24 h after addition of TAK-901, the cells were washed and fixed with 4% PFA for 20 min at room temperature. The cells were then permeabilised with 0.1% Triton X-100, blocked with 3% BSA, and incubated overnight at 4 °C with p-Histone H3 (S10) antibody (Cell Signalling Technology, cat# 9701, RRID: AB_331535, 1:500). Signals were detected using Alexa Fluor 488 goat anti-rabbit antibody (1:1000) and images were captured using Operetta HCS from Perkin Elmer.
Columbus image analysis software from Perkin Elmer was used to identify and quantify the total p-Histone H3 staining intensity of each well. The positive intensities from the treated wells at each time point were then normalised against the average positive intensity of DMSO treated wells at that time point. The normalised values were plotted against the respective TAK-901 concentrations using GraphPad Prism. Dose response EC50 values were then obtained using 3-parameter non-linear regression.
2.10. Scratch wound assay
Cells were seeded into 2-well silicone insert (Ibidi, cat# 80209) at a density in which the cells are 100% confluent 24 h later. The next day, cells were treated with mitomycin C (MedKoo Biosciences, cat# 100630) at 10 μg/mL for 2 h. After 2 h, the insert was removed and the cells were washed with PBS 3 times before replacing with growth medium. The size of the wounds at this time (0 h), and at different time intervals, were monitored using either the EVOS cell imaging system or the Incucyte® S3 Live-Cell Analysis System. The percentage wound healing at the various time points (i.e., x h) was calculated based on the equation below:
2.11. Spheroid formation assay
Cells were seeded at 1600 cells per well in 100 μL of spheroid medium in 96-well ultra-low attachment round bottom plates (Corning, cat# 7007) and allowed to grow for 4 days at 37 °C, 5% CO2. Spheroid medium consists of DMEM/F12 (Gibco, cat# 11320) supplemented with B27 (minus vitamin A, Gibco, cat# 12587), 100 U/mL penicillin-streptomycin (Gibco, cat# 15140122), 20 ng/mL recombinant EGF (Gibco, cat# PHG0313), and 10 ng/mL recombinant FGF (Gibco, cat# PHG0023).
2.12. Growth assay
Cells were plated at 3000 cells per well in 200 µL of growth medium in white, clear, flat bottom 96-well plates and allowed to proliferate at 37 °C, 5% CO2. The next day (0 h) and subsequently at 24 h intervals, the viability of the cells was determined using CellTiter-Glo luminescent cell viability assay.
2.13. Cell cycle analysis
Cells were harvested by trypsinisation and fixed with ice cold 70% ethanol at 4 °C for at least 24 h. The fixed cells were washed twice with PBS, and stained with a solution containing 0.1% Triton X-100, 0.2 mg/mL RNase A (Invitrogen, cat# 12091021), and 20 µg/mL propidium iodide (Sigma, cat# P4864) for 30 min at room temperature. Prior to FACS analysis the cells were strained through a 35 µm cell strainer (Falcon, cat# 352235). The cells were analysed using BD FACSJazzTM cell sorter.
2.14. Dose response studies
Cells were plated at 4500 cells per well in 200 µL of growth medium in white, clear, flat bottom 96-well plates (Corning, cat# 3903) and allowed to attach overnight at 37 °C, 5% CO2. The next day, serially diluted compounds in DMSO were added to the appropriate wells. DMSO was also added to control wells to a final concentration of 0.5%. Each concentration was tested in triplicate. After 72 h, viability of the treated cells was determined using CellTiter-Glo luminescent cell viability assay.
For analysis, the average luminescent signal of DMSO control in each plate was calculated. The percentage viability of each treated well was then determined using the following formula:
The %viability values were plotted against the respective compound concentrations using GraphPad Prism. Dose response EC50 values were then obtained using 4-parameter non-linear regression.
For combination studies, synergy scores were calculated using SynergyFinger [24].
2.15. Apoptosis assay
Cells were plated at 4500 cells per well in 100 µL of growth medium in white, clear, flat bottom 96-well plates (Corning, cat# 3903) and allowed to attach overnight at 37 °C, 5% CO2. The next day, serially diluted compounds in DMSO were added to the appropriate wells. DMSO was also added to control wells to a final concentration of 0.5%. Each concentration was tested in triplicate. After 24 h, viability of the treated cells was determined using Caspase Glo 3/7 assay (Promega, cat# G8093) according the manufacturer's protocol. Luminescence signals were measured using Perkin Elmer EnSpire multimode microplate reader.
For analysis, the average luminescent signal of DMSO control in each plate was calculated. The fold change caspase 3/7 activation was calculated using the formula below:
The fold change values were plotted against the respective compound concentrations using GraphPad Prism. Dose response EC50 values were then obtained using 4-parameter non-linear regression.
2.16. siRNA knockdown and dose response
Pre-designed Silencer Select siRNAs (Thermo Fisher Scientific) were used for knockdown of target genes. EGFR siRNA IDs: s563, s564, s565, AXL siRNA IDs: s1845, s1846, s1847, and negative control No. 1 siRNA (cat# 4390843) were used. Unless otherwise stated, pooled siRNAs were used. For siRNA knockdown dose response studies, 20 µL of OptiMEM (Gibco, cat# 31985070) containing a complex of siRNA and 0.2 µL of Lipofectamine 3000 (Invitrogen, cat# L3000150) was dispensed into each well of white, clear, flat bottom 96-well plates. 3500 cells per well in 100 µL of antibiotic-free RPMI medium supplemented with 12% FBS were then added to the respective wells and incubated at 37 °C, 5% CO2 for 2 days. Following which the medium was replaced with growth medium containing serially diluted compounds at 0.5% DMSO and the cells were further cultured for another 3 days. The viability of the treated cells was determined using CellTiter-Glo luminescent cell viability assay. The percentage viability values were plotted against the respective compound concentrations using GraphPad Prism. Dose response EC50 values were then obtained using 4-parameter non-linear regression.
2.17. siRNA knockdown effects on cell viability and caspase 3/7 activation
Pre-designed Silencer Select siRNAs (Thermo Fisher Scientific) were used for knockdown of target genes. AURKA siRNA IDs: s195, s196, s197, AURKB siRNA IDs: s17611, s17612, s17613, and negative control No. 1 siRNA (cat# 4390843) were used. Unless otherwise stated, pooled siRNAs were used. For siRNA knockdown, 20 µL of OptiMEM (Gibco, cat# 31985070) containing a complex of siRNA and 0.2 µL of Lipofectamine 3000 (Invitrogen, cat# L3000150) was dispensed into each well of white, clear, flat bottom 96-well plates. 4500 cells per well in 100 µL of antibiotic-free RPMI medium supplemented with 12% FBS were then added to the respective wells and incubated at 37 °C, 5% CO2. At the respective time points, the viability of the cells was determined using CellTiter-Glo luminescent cell viability assay and caspase 3/7 activation was determined using Caspase Glo 3/7 assay. The effects of the siRNAs at the respective time points were normalized against the effects of the negative control siRNA.
2.18. RNA extraction and quantitative polymerase chain reaction (qPCR)
RNA was extracted using RNeasy plus mini kit (Qiagen, cat# 74136) and following the manufacturer's instructions. 500 ng of RNA was reverse transcribed into cDNA using SuperScript IV VILO master mix (Invitrogen, cat# 11756050). 10 ng cDNA per reaction qPCR was carried out using KAPA SYBR FAST qPCR Kit (Sigma Aldrich, cat# KK4602) in accordance to the manufacturer's protocol. Relative gene expressions were normalized to GAPDH levels. The sequences of primers used for qPCR can be found in Supplementary Table S10.
2.19. Generation of overexpression construct
pLenti CMV Puro DEST (w118-1) destination clone was a gift from Eric Campeau (Addgene plasmid # 17452, RRID: Addgene_17452). Entry clones for Aurora kinase A (clone ID IOH82199), EGFR variant 1 (clone ID IOH81788), and AXL (clone ID IOH22600) were obtained from the Ultimate ORF LITE library (Invitrogen). The expression construct was obtained through Gateway recombination reaction between the entry and destination clones using LR clonase II enzyme mix (Invitrogen, cat# 11791020) and following the manufacturer's protocol.
2.20. Generation of overexpression cell lines
Expression-ready construct for Aurora kinase B (clone ID 101927034) was obtained from the CCSB-Broad lentiviral expression library. Each overexpression construct, together with ViraSafeTM lentiviral packaging system (Cell Biolabs, cat# VPK-206) were transfected into HEK293T cells using Lipofectamine 3000. After 24 h, the medium was replaced with growth medium of the cells to be transduced and cultured for another 24 h to collect the viral particles. The viral particles were harvested and applied to target cells in the presence of 8 µg/mL of polybrene (Santa Cruz Biotechnology, cat# sc-134220). Cells overexpressing Aurora kinase B were selected with (12 µg/mL) and (6 µg/mL) of Blasticidin S (Gibco, cat# R21001) for HN19 parental and HN19-GR cells respectively. HN19 parental cells overexpressing Aurora kinase A and AXL were selected with (0.6 µg/mL) of Puromycin (Gibco, cat# A1113802) while HN19-GR cells overexpressing Aurora kinase A or EGFR variant 1 were selected with (0.8 µg/mL) of Puromycin.
2.21. In vivo drug treatment
Cells were harvested and resuspended in serum free DMEM/F12 (Gibco, cat# 11320082) medium and mixed with 10% matrigel (Corning, cat# 354234). About 2 million cells were injected subcutaneously into one flank of each female NOD scid gamma (NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ, RRID: IMSR_JAX:005557) mice and allowed to form tumours. When the tumours reached 50–75 mm3 the mice were randomized into treatment and control cohorts and treatment was started. Each cohort included at least 6 mice representing 6 tumours per cohort. Sample size for each drug treatment was calculated by E = Total number of animals − Total number of groups. For each drug treatment, there are 4 groups (control vs treatment, and HN19 vs HN19-GR) of 6 animals per group which gives an E value of 20. Treatment was stopped and tumours harvested before control tumours reached a size of 2000 mm3. Toxicity of drug treatment was monitored by changes in body weights of the mice. Tumour sizes were monitored every other day and volumes were calculated according to the equation below:
Gefitinib was prepared by resuspending a clinical grade tablet (250 mg) in 0.05% Tween-80 (Sigma, cat# P4780) in water to a final concentration of 10 mg/mL. Gefitinib was dosed at 20 mg/kg daily via oral gavage. TAK-901 was prepared by first dissolving the compound in DMSO to a concentration of 100 mg/mL. This solution was further diluted in a 25 mM citrate buffer solution (pH 4) containing 20% (2-hydroxypropyl)-β-cyclodextrin (Sigma, cat# 332607) to a final concentration of 5 mg/mL, 5% DMSO. TAK-901 was dosed at 30 mg/kg every other day via IP. All animal work was carried out at the Biological Resource Centre (BRC), A*STAR under IACUC protocol number 151065.
2.22. Immunohistochemistry
Tumour tissues harvested at the end of the experiment were embedded into paraffin blocks by the Advanced Molecular Pathology Laboratory (AMPL). For immunohistochemistry, 5 µm tumour tissue paraffin sections were cut no longer than two weeks before the assay was performed manually. The tissue sections were deparaffinised in xylene and rehydrated through a series of alcohols to distilled water. Epitope retrieval was performed by incubating in 0.01 M citrate buffer at pH 6.0 in a steamer for 30 min at 80 °C. The slides were pre-treated with 3% hydrogen peroxide for 10 min then blocked with 5% normal goat serum (Cell Signaling Technology, cat# 5425) for 1 h at room temperature. Polyclonal antibodies to cleaved caspase-3 (Asp175) (Cell Signalling Technology, cat# 9661, RRID: AB_2341188) and phospho-histone H3 (Ser10) (Cell Signalling Technology, cat# 9701, RRID: AB_331535) were applied at a 1:200 dilution in Signalstain Antibody Diluent (Cell Signalling Technology, cat# 8112) to the different sections respectively and incubated overnight at 4 °C. Dako visualization reagent (dextran polymer conjugated with horseradish peroxidase and goat anti-rabbit and mouse immunoglobulins) with 3,3′-diaminobenzidine tetrahydrochloride chromogen solution (Agilent Technologies, cat# K500711-2) was used as the detection system according to the manufacturer's protocol. Sections were also counterstained with hematoxylin.
Slides were scanned at 20 × magnification using the Vectra Polaris Automated Quantitative Pathology Imaging System (PerkinElmer), and regions of interests were identified by training the program with at least 10 training images from the dataset that were outlined manually. The total surface area of positive cleaved Caspase-3 staining and total counts of positive nuclear phospho-histone H3 (Ser10) were quantified using the inForm Tissue Finder version 2.4 and Phenochart version 1.0 software (PerkinElmer). Statistical significance was performed using Student's t-test.
2.23. Statistical analysis
Statistical analyses were performed using GraphPad Prism software. Details of replicates and statistical tests performed are all described in the respective figure legends.
2.24. Ethics statement
All animal work was carried out at the Biological Resource Centre (BRC), A*STAR under IACUC protocol number 151065. Details of the animal preclinical studies can be found in the ARRIVE author checklist associated with this manuscript.
2.25. Role of funders
The funders had no role in the study design, data collection, data analyses, interpretation, or writing of the report.
3. Results
3.1. HNSCC cells with wild-type EGFR undergo EMT upon gefitinib-induced resistance
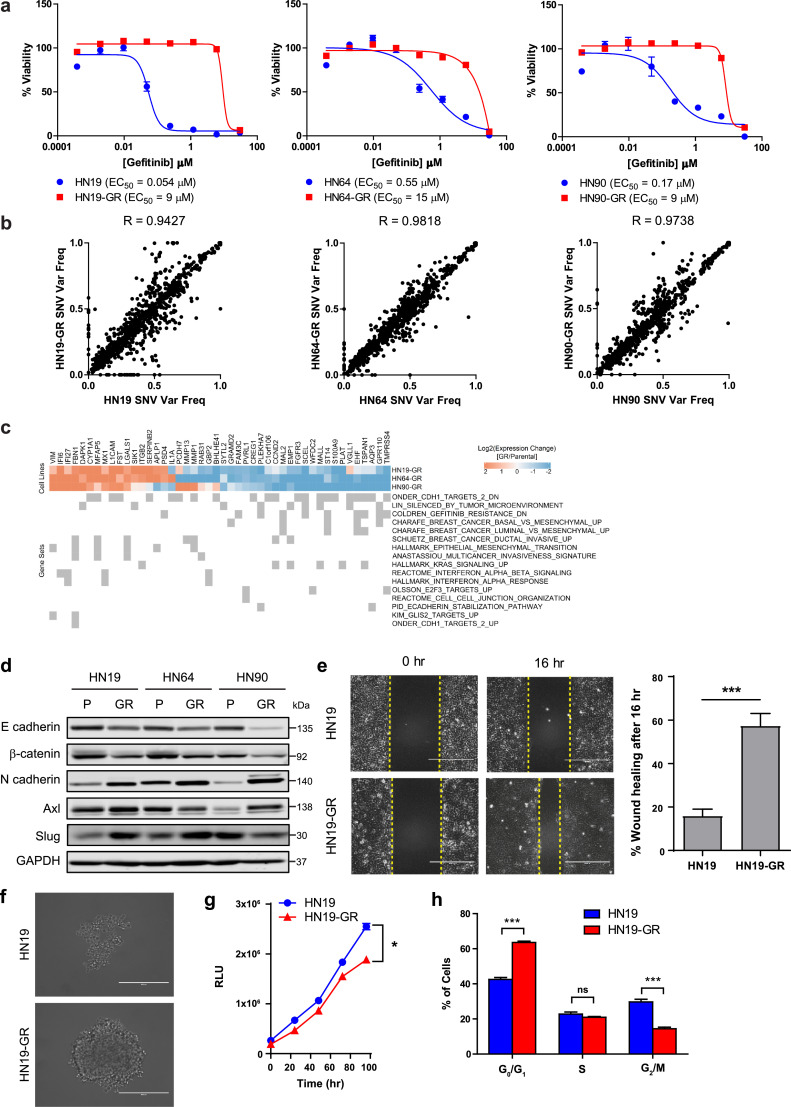
We identified three patient-derived HNSCC cell lines; NCC-HN19 (HN19), NCC-HN64 (HN64) and NCC-HN90 (HN90) which are highly sensitive to gefitinib in vitro. Through stepwise incremental exposure to gefitinib, we derived the corresponding gefitinib-resistant (GR) cell lines which are all more than 20-fold more resistant to gefitinib (Fig. 1a). Interestingly, while the parental patient-derived HNSCC cell lines were sensitive to gefitinib, our previous studies have shown that HN19 and HN64 parental cells are resistant to cetuximab [22].
Fig. 1.
Patient-derived HNSCC cells acquire resistance to gefitinib without EGFR T790M mutation. (a) Representative gefitinib dose response curves and EC50 values for NCC-HN19 (HN19, left panel), NCC-HN64 (HN64, middle panel), and NCC-HN90 (HN90, right panel) cell lines and their corresponding gefitinib resistant (GR) cell lines. Cells were treated with serial dilutions of gefitinib and cell viabilities were measured after 72 hr. Data represent n = 3 technical replicates and experiment was repeated at least twice with similar results. (b) SNV variant frequency distributions and Pearson correlations of SNVs detected through POLARIS Xplora Cancer panel for the HN19 (left panel), HN64 (middle panel), and HN90 (right panel) pairs of parental and GR cell lines. Data represents average of n = 2 biologically independent samples. (c) Selected differentially regulated genes in GR cells vs parental cells and the gene sets in which they are enriched. (d) Immunoblot analysis showing differential expression of EMT markers in HN19 and HN19-GR cells. (e) Scratch wound migration assay of HN19 and HN19-GR cells showing the relative wound healing at different time points. Scale bar represents 400 μm. Unpaired two-tailed Student's t test was carried out for percentage of wound healing after 16 h. *** (p < 0.001). (f) Spheroid formation of HN19 and HN19-GR cells after 4 days. Scale bar represents 400 μm. (g) Growth curve comparison between HN19 and HN19-GR cells. Paired Student's t-test was carried out between HN19 and HN19-GR. * (0.01 < p < 0.05). (h) Cell cycle profiles of HN19 and HN19-GR cells determined by FACS. Student's t-test was carried out between HN19 and HN19-GR at the different phases of the cell cycle. *** (p < 0.001). In all graphs, error bars are mean ± s.d.
To investigate if the resistance to gefitinib is driven by genetic mutations, we examined the mutational profiles of all the cell lines using the POLARIS Xplora cancer panel. The Xplora cancer panel is an assay involving targeted DNA sequencing of a panel of 740 cancer associated genes (Table S1). Comparing the single nucleotide variations (SNVs) detected and their variant frequencies (Table S2), we observed that the GR cell lines have highly similar genetic profiles compared to their parental cells with Pearson correlation values of 0.9427 (HN19), 0.9818 (HN64), and 0.9738 (HN90) respectively (Fig. 1b). We also found that neither the EGFR T790M mutation, commonly associated with TKI resistance in NSCLC nor any other resistance alterations were present in all 3 pairs of parental and GR cell lines. Instead, EGFR R468K (R521K, rs2227983) polymorphism was observed at varying frequencies in the cell lines examined (Fig. S1a). Interestingly the R521K polymorphism has been linked to cetuximab resistance without influencing therapeutic vulnerability against TKIs, such as erlotinib [25]. Apart from EGFR, we also observed minimal alterations in the SNV variant frequencies of genes related to the RAS-RAF-MEK-ERK and PI3K-AKT-mTOR pathway in the 3 pairs of cell lines. While we observed a significant increase in MTOR R283T SNV (variant frequency 0 to 0.34) and a significant decrease in RPS6KA2 E156G SNV (variant frequency 0.43 to 0.08) in HN19-GR cells compared to the parental cells (Fig. S1a), not much is known about the clinical significance of these SNVs. The lack of significant variation between the SNV profiles of the parental and GR cells suggests that resistance in these cells may not be primarily driven by mutational changes.
We next investigated if differential sensitivities could be rationalised by gene expression changes (Table S3). Gene set enrichment analysis (GSEA) (Tables S4–S6) of the differentially expressed genes in the GR cells identified commonly enriched gene sets in all 3 pairs of cell lines (Fig. S1b). Pathways with positive normalized enrichment scores (NES > 1.5, FDR q value < 0.25) include those related to interferon signalling, epithelial to mesenchymal transition (EMT), and invasion (Fig. S1b). Genes associated with these pathways appear to be generally upregulated in the GR cells compared to parental cells (Fig. 1c). Conversely, gene sets with negative normalized enrichment scores (NES < −1.5, FDR q value < 0.25) include those that are associated with E-cadherin downregulation, and gefitinib-resistance (Fig. S1b). Genes enriched in these gene sets also tend to be downregulated in the GR cells compared to parental cells (Fig. 1c). Interestingly, differentially expressed genes in the GR cells were also enriched for genes downregulated by the tumour microenvironment (TME) (Fig. S1b). The TME has been shown to promote the dedifferentiation of cancer cells into cancer stem cells (CSCs) that have been shown to contribute to treatment resistance and metastasis [26]. Thus, based on gene expression analysis, the GR cells appear to adopt a more dedifferentiated and mesenchymal phenotype upon gaining resistance to gefitinib.
The differential gene expression observed was also validated by Western blot analysis showing decreased expression of epithelial/differentiation markers ZO1, E-cadherin, and β-catenin and increased expression of mesenchymal markers, including N-cadherin, Slug, and AXL in the GR cells (Fig. 1d). Immunofluorescence staining further validated the increased expression of N-cadherin in HN19-GR cells (Fig. S1c). Functionally, scratch wound migration assay demonstrated the increased migratory capacity of HN19-GR cells compared to HN19 cells (Fig. 1e). This EMT signature observed upon gaining resistance to gefitinib corroborated previous reports on gefitinib resistance in NSCLC [27], [28], [29] and HNSCC [30,31]. Moreover, HN19-GR cells form spheroids more readily compared to the parental HN19 cells (Fig. 1f). The HN19-GR cells in the presence of gefitinib also proliferated at a slower rate (Fig. 1g), and had lesser proportion of mitotic phase G2/M cells (Fig. 1h and Fig. S1d). These results further support the dedifferentiated, stem-like phenotype associated with treatment resistance.
3.2. Downregulation of EGFR contributes to resistance to gefitinib in HNSCC
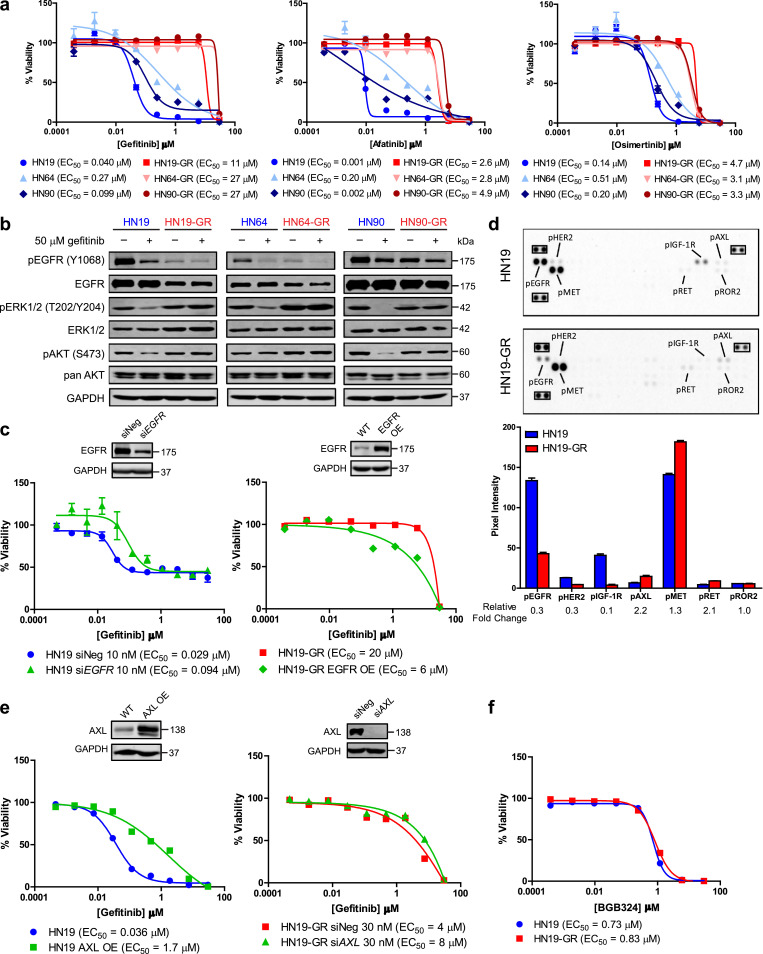
Given the lack of EGFR resistance mutations in GR cells, we tested if these can still be targeted by 2nd and 3rd generation TKIs. A comparison of the responses to afatinib (2nd generation), and osimertinib (3rd generation) revealed that the GR cells remained resistant to these drugs (Fig. 2a). Subsequent analysis of protein expression revealed that EGFR expression, and in particular EGFR phosphorylation, was downregulated in the GR cells (Fig. 2b). Despite the downregulation of EGFR in the GR cells, there was continued phosphorylation of ERK and AKT proteins in the GR cells (Fig. 2b). The levels of ERK and AKT phosphorylation in the GR cells were also comparable to that in the parental cells. Importantly, ERK and AKT phosphorylation was unaffected by gefitinib treatment, unlike in the parental cells (Fig. 2b). To investigate whether the loss of target engagement (i.e., EGFR) in the GR cells could account for their decreased sensitivity to not just gefitinib, but also to other TKIs, we either downregulated EGFR expression in parental HN19 cells via siRNA-mediated knockdown, or overexpressed EGFR in the HN19-GR cells. We observed that knockdown of EGFR in parental HN19 cells reduced its sensitivity to gefitinib, and overexpression of EGFR in HN19-GR cells reduced its resistance to gefitinib (Fig. 2c). Interestingly, further downregulation of EGFR expression in HN19-GR cells, through siRNA mediated knockdown of EGFR, could partially reduce ERK phosphorylation levels in these cells (Fig. S2a). However, the cells remained resistant to gefitinib, corroborating the notion that their survival was EGFR-independent (Fig. S2b). Altogether these results suggest that the GR cells may have adopted EGFR-independent pathway(s) to maintain ERK and AKT activation and are thus cross resistant to 2nd and 3rd generation EGFR TKIs.
Fig. 2.
Decreased dependency of EGFR T790M negative HNSCCs to EGFR signalling pathway upon gaining gefitinib resistance. (a) Representative dose response curves and EC50 values for HN19, HN64, and HN90 pairs of parental and GR cell lines to gefitinib (left panel), afatinib (middle panel), and osimertinib (right panel). Cells were treated with serial dilutions of the respective compounds and cell viabilities were measured after 72 h. Data represent n = 3 technical replicates and experiment was repeated at least once with similar results. (b) Immunoblot analysis of EGFR signaling pathway activities of HN19 (left panel), HN64 (middle panel), and HN90 (right panel) and their corresponding GR cells in response to gefitinib treatment. Cells were treated with either gefitinib (+) or DMSO control (−) for 6 h. (c) Representative gefitinib dose response curves and EC50 values for HN19 (left panel) and HN19-GR (right panel) upon either EGFR knockdown (HN19) or EGFR overexpression (HN19-GR). Cells were transfected with the respective siRNAs for 48 h followed by treatment with serial dilutions of gefitinib for 72 h. Data represent n = 3 technical replicates. (d) Phospho-receptor tyrosine kinase array comparison between HN19 and HN19-GR cells. Top panel: annotated images of blots; reference spots are highlighted in black rectangles. Bottom panel: quantification of pixel intensities of spots with bars representing average pixel intensities and error bars representing standard deviation. Pixel intensity fold changes of HN19-GR over HN19 are labelled below the x-axis. (e) Representative dose response curves and EC50 values for HN19 (left panel) and HN19-GR (right panel) to gefitinib upon either AXL overexpression (HN19) or AXL knockdown (HN19-GR). Cells were transfected with the respective siRNAs for 48 h followed by treatment with serial dilutions of gefitinib for 72 h. Data represent n = 3 technical replicates. (f) Representative dose response curves and EC50 values for HN19 to BGB324 (R428). Cells were treated with serial dilutions of the respective compounds and cell viabilities were measured after 72 h. Data represent n = 3 technical replicates. In all graphs, error bars are mean ± s.d.
We next investigated if downregulation of EGFR in the GR cells could result in compensatory upregulation of other RTKs to maintain ERK and AKT activation. Using a phospho-RTK protein array, we compared the phospho-RTK profiles of HN19 and HN19-GR cells (Fig. 2d). Apart from phospho-EGFR downregulation, we also observed downregulation of phospho-HER2 and phospho-IGF-1R in the HN19-GR cells. On the other hand, phospho-AXL, phospho-MET, and phospho-RET were upregulated in the HN19-GR cells. Further validation of the results obtained from the phospho-RTK array revealed that protein expression and phosphorylation of other members of the ErbB family (i.e., HER2 and HER3) were downregulated in the GR cells (Fig. S2c); indicating that ERK phosphorylation in the GR cells was not maintained by compensatory upregulation of alternative ErbB pathways. MET amplification was also not observed in the GR cells (Fig. 2c), which was in contrast to a previous report by Engelman et al. [32]. Instead, we observed an upregulation of AXL phosphorylation in the HN19-GR cells which corresponds to the upregulation of AXL mentioned earlier (Fig. 1d and Fig. S2c). When we overexpressed AXL in the HN19 parental cells, we observed a decreased sensitivity to gefitinib (Fig. 2e). However, knockdown of AXL in the HN19-GR cells had no effect on their response to gefitinib (Fig. 2e). Furthermore, neither knockdown of AXL in HN19-GR cells (Fig. S2d) nor overexpression of AXL in HN19 cells (Fig. S2e) affected ERK phosphorylation in these cells, suggesting that the reactivation of ERK signalling in HN19-GR cells was likely not mediated by AXL upregulation. Moreover, both HN19 and HN19-GR were equally sensitive to a selective AXL inhibitor, BGB324 (R428) (Fig. 2f), suggesting that increased AXL expression in the HN19-GR cells was not the main driver of gefitinib resistance. Based on these results, we conclude that in HN19-GR cells, gefitinib resistance was not mediated through activation of alterative RTK pathways.
3.3. Gefitinib resistant cells are more sensitive to inhibitors of cell division
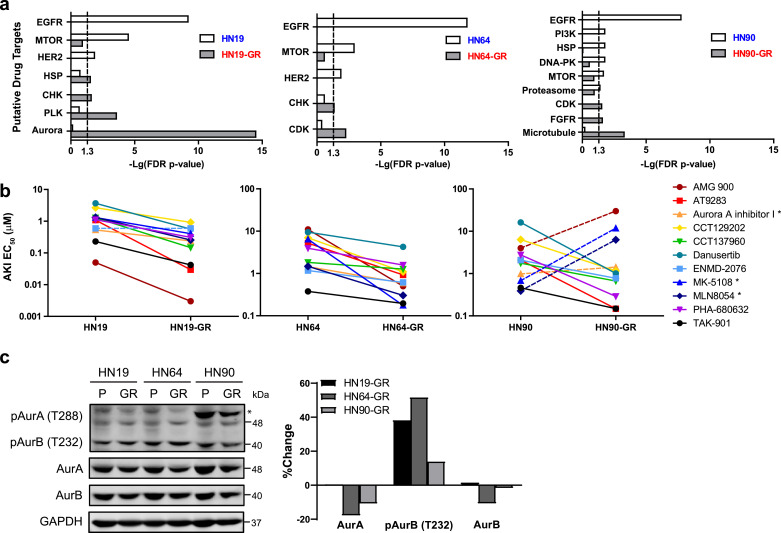
Since the GR cells can no longer be targeted by EGFR TKIs, we set out to identify alternative therapeutic vulnerabilities. To do this, we established a differential screening platform using gefitinib-sensitive and GR cells, with a panel of over 600 commercially available anti-cancer and kinase inhibitor compounds (Table S7). Enrichment of hit compounds against a particular protein target (Table S8) would enable us to identify alternative therapeutic options for these GR cells. Unsurprisingly, the results of the screen revealed that the parental cell lines were sensitive to inhibitors targeting EGFR, HER2, mTOR, and PI3K, all components of the EGFR pathway (Fig. 3a, and Fig. S3a). Interestingly, the GR cells were all sensitive to inhibitors targeting cell proliferation and cell cycle: Aurora kinase inhibitors (AKIs) for HN19-GR, cyclin-dependent kinase (CDK) inhibitors for HN64-GR, and microtubule inhibitors for HN90-GR (Fig. 3a, and Fig. S3a). Intriguingly, HN19-GR cells were highly enriched for a range of different aurora kinase inhibitors (AKIs) (FDR P value = 3.27 × 10−15). Secondary dose response EC50 studies validated these results, not only in HN19-GR cells but also HN64-GR cells, where the screen had identified sensitivity to CDK inhibitors (Fig. 3b). Interestingly, in the case of HN90-GR cells, the HN90 parental cells appeared to be more sensitive to Aurora kinase A specific inhibitors such as MK-5108 and MLN8054 while the HN90-GR cells were more sensitive to the pan Aurora kinase inhibitors (Fig. 3b).
Fig. 3.
High-throughput compound screen identifies alternative therapeutic sensitivity to inhibitors of cell proliferation in GR cells. (a) Differential target enrichment of hit compounds identified from compound screen for HN19 (left panel), HN64 (middle panel), and HN90 (right panel) parental and GR cells. Hit compounds were identified to have more than 50% toxicity at 1 μM after 72 hr. Target enrichment was carried out using Fisher's exact test and each graph shows the enriched targets with FDR p-value < 0.05 in either the parental or the GR cells. (b) Comparison of the EC50 values of different Aurora kinase inhibitors (AKIs) against HN19 (left panel), HN64 (middle panel), and HN90 (right panel) and their corresponding GR cells. Each line represents a unique AKI. Data represent n = 3 technical replicates. Unpaired one-tailed Student's t test was carried out for each pair of parental and GR cells. Dashed lines represent compounds for which the EC50 of the GR cells is not significantly (p < 0.05) less than that of the parental cells. (c) Immunoblot comparison and quantification of Aurora kinase expression and phosphorylation levels in HN19 parental, HN64 parental, HN90 parental, and their corresponding GR cell lines.
As the GR cells were more sensitive to AKIs, we first determined if this was due to differential expression of Aurora kinases in the parental and GR cells. Based on our RNA sequencing results, we found that expression of AURKA and AURKB genes were not significantly altered in the GR cells compared to the parental cells (Fig. S3b). Protein expression levels of Aurora kinase A (AurA) and Aurora kinase B (AurB) were also slightly downregulated in the GR cells compared to the parental cells (Fig. 3c). In contrast, we observed an upregulation of Aurora kinase B phosphorylation in the GR cells (Fig. 3c). The levels of phosphorylated Aurora kinase A in HN19-GR cells were also higher compared to that in HN19 parental cells (Fig. S3c). Gefitinib has been shown to induce G0/G1 cell cycle arrest in cancer cells [33]; it is thus likely that the GR cells upregulate their Aurora kinase phosphorylation to enable the cells to continue to proliferate. Consequently, this upregulation of Aurora kinase phosphorylation in the GR cells could then lead to their increased sensitivity to AKIs. Interestingly while the expression levels of Aurora kinase A were comparable across the 3 pairs of cell lines, the HN90 parental and HN90-GR cells had significantly higher levels of Aurora kinase A phosphorylation (Fig. 3c). Not only that, phospho-Aurora kinase A levels were also higher in the HN90 parental cell line compared to the HN90-GR cell line. This differential Aurora kinase A phosphorylation levels could thus account for the selective sensitivity of the HN90 parental cells to Aurora kinase A specific inhibitors. While the reason for the high phospho-Aurora kinase A levels in the HN90 cells is beyond the scope of this study, we speculate that a probable link could exist with the AURKA F31I (rs2273535) polymorphism as this SNV was detected only in the HN90 cells (Fig. S1a).
3.4. Aurora kinases are essential for cell survival in the EGFR-inhibited state
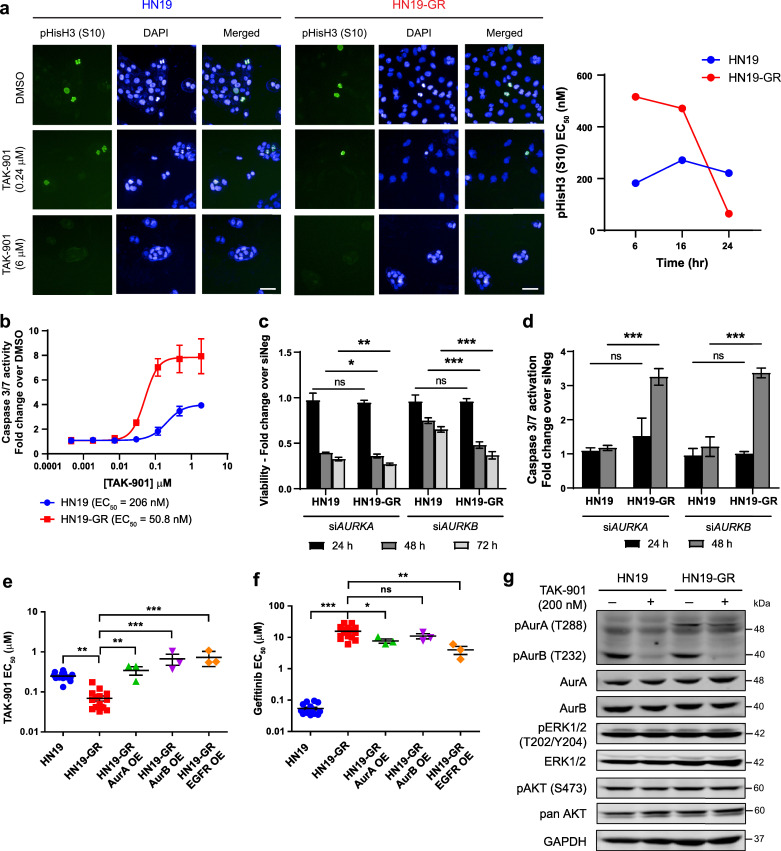
To further understand the differential sensitivity of the GR cells to AKIs, we focused on the HN19 pair of parental and GR cells. First, we investigated the effects of TAK-901, an investigational AKI [34], on the expression of phospho-histone H3 (S10) through immunofluorescence. Phospho-histone H3 is a mitotic marker as well as a target of Aurora kinases [35]. Treatment of both HN19 and HN19-GR cells with TAK-901 resulted in large, multinucleated cells that are characteristic of Aurora kinase B inhibition, as cells undergo mitosis without cytokinesis (Fig. 4a). Staining intensities for phospho-histone H3 also showed a dose dependent decrease with increasing concentrations of TAK-901 (Fig. S4a). Interestingly, the phospho-histone H3 EC50 values for HN19-GR cells showed a gradual decrease over 24 h while in comparison, the EC50 values for HN19 cells remained relatively constant during the same time period (Fig. 4a, and Fig. S4a). Apart from affecting histone H3 phosphorylation levels, we also observed a dose dependent increase in caspase 3/7 activity in both HN19 parental and HN19-GR cells upon treatment with TAK-901 (Fig. 4b). Notably, the EC50 for caspase 3/7 activation occurs at a lower concentration for HN19-GR cells compared to the parental cells, suggesting increased vulnerability of the HN19-GR cells towards AKIs.
Fig. 4.
Aurora kinases are essential in the EGFR inhibited state. (a) Immunofluorescence staining for phospho-histone H3 (S10) in HN19 parental (left panel) and HN19-GR (middle panel) cells upon treatment with different concentrations of TAK-901 for 24 h. Scale bar represents 50 μm. Right panel: EC50 values of phospho-histone H3 (S10) staining upon treatment with TAK-901 at different time points for HN19 parental and HN19-GR cells respectively. (b) Representative caspase 3/7 activation curves upon TAK-901 treatment for HN19 parental and HN19-GR cells. Cells were treated with serial dilutions of TAK-901 for 24 h. Data represent n = 3 technical replicates. (c) and (d) Effects of AURKA and AURKB knockdown via siRNA on viability (c) and caspase 3/7 activity (d) in HN19 parental and HN19-GR cells. (e) and (f) Effects of Aurora kinase A, Aurora kinase B, and EGFR overexpression (OE) in HN19-GR cells on their EC50 values against TAK-901 (e) and gefitinib (f). Data represents values from at least 3 independent experiments. One-way ANOVA followed by Dunnett's multiple comparison test was carried out between cell lines. ns (p > 0.05), * (0.01 < p < 0.05), ** (0.001 < p < 0.01), *** (p < 0.001). Error bars represent mean ± SEM. (g) Immunoblot analysis of Aurora kinase phosphorylation and EGFR downstream signaling pathway activities in HN19 parental and HN19-GR cells in response to TAK-901 treatment. Cells were treated with either TAK-901 (+) or DMSO control (−) for 6 h.
Our results thus far indicated that the HN19-GR cells are more dependent on Aurora kinases for survival compared to the HN19 parental cells. To investigate this, we used siRNA to knockdown the AURKA and AURKB genes in HN19 parental and HN19-GR cells (Fig. S4b). Knockdown of AURKA rapidly decreased cell survival of HN19 and HN19-GR cells while knockdown of AURKB had a greater effect on HN19-GR cells (Fig. 4c). Correspondingly, we observed higher activation of caspase 3/7 in HN19-GR cells upon knockdown of AURKA and AURKB (Fig. 4d). These results support the increased dependency of HN19-GR cells to Aurora kinases for cell survival. Next, we wanted to investigate the link between increased Aurora kinase dependency and gefitinib resistance. Since knockdown of AURKA and AURKB genes were lethal to the cells, we overexpressed Aurora kinases A and B separately in the HN19 and HN19-GR cells (Fig. S4c) and investigated the effects on the cells’ response to TAK-901 and gefitinib treatment. Overexpression of either Aurora kinase A or B was sufficient to decrease the sensitivity of HN19-GR cells to TAK-901 (Fig. 4e). Phosphorylation levels of the Aurora kinases also increased upon overexpression of the respective proteins in the HN19-GR cells (Fig. S4c). However, overexpression of the same proteins in parental HN19 cells had no effect on their response to TAK-901 (Fig. S4d). The phosphorylation levels of Aurora kinases A and B were also unaffected by overexpression of the respective proteins in the HN19 parental cells (Fig. S4c). Interestingly, overexpression of EGFR in HN19-GR cells also decreased the sensitivity of these cells to TAK-901 (Fig. 4e). Our results thus suggest that in the absence of signalling through EGFR, the HN19-GR cells are dependent on Aurora kinases for survival. This was also supported by combination studies with gefitinib and TAK-901 in which greater synergistic effect was observed in HN19-GR cells compared to HN19 parental cells (Fig. S4e). In contrast, for HN19 parental cells which are dependent on EGFR signalling, overexpression of Aurora kinases does not confer additional survival benefit.
Having demonstrated that Aurora kinases are essential for survival in the EGFR inhibited state, we wanted to investigate if Aurora kinases could drive resistance to gefitinib in HN19-GR cells. Dose response studies with gefitinib on HN19-GR cells overexpressing either Aurora kinase A or B did not result in further gain in resistance to gefitinib (Fig. 4f). Overexpression of Aurora kinases A or B in HN19 parental cells also did not significantly change the cells’ sensitivity to gefitinib (Fig. S4d). Moreover, treatment of HN19 and HN19-GR cells with TAK-901 did not affect phosphorylation levels of ERK or AKT, even though Aurora kinase phosphorylation levels were reduced (Fig. 4g). Overexpression of Aurora kinases A or B also had no effects on ERK and AKT phosphorylation levels (Fig. S4c). These results showed that in the HN19-GR cells, ERK and AKT are not downstream targets of Aurora kinases. Similarly, the migration capability of HN19-GR cells was not affected by treatment with TAK-901 (Fig. S4f), indicating that the EMT phenotype observed upon gain in gefitinib resistance was not a consequence of increased Aurora kinase phosphorylation.
3.5. Differential drug sensitivities in vitro could be recapitulated in vivo
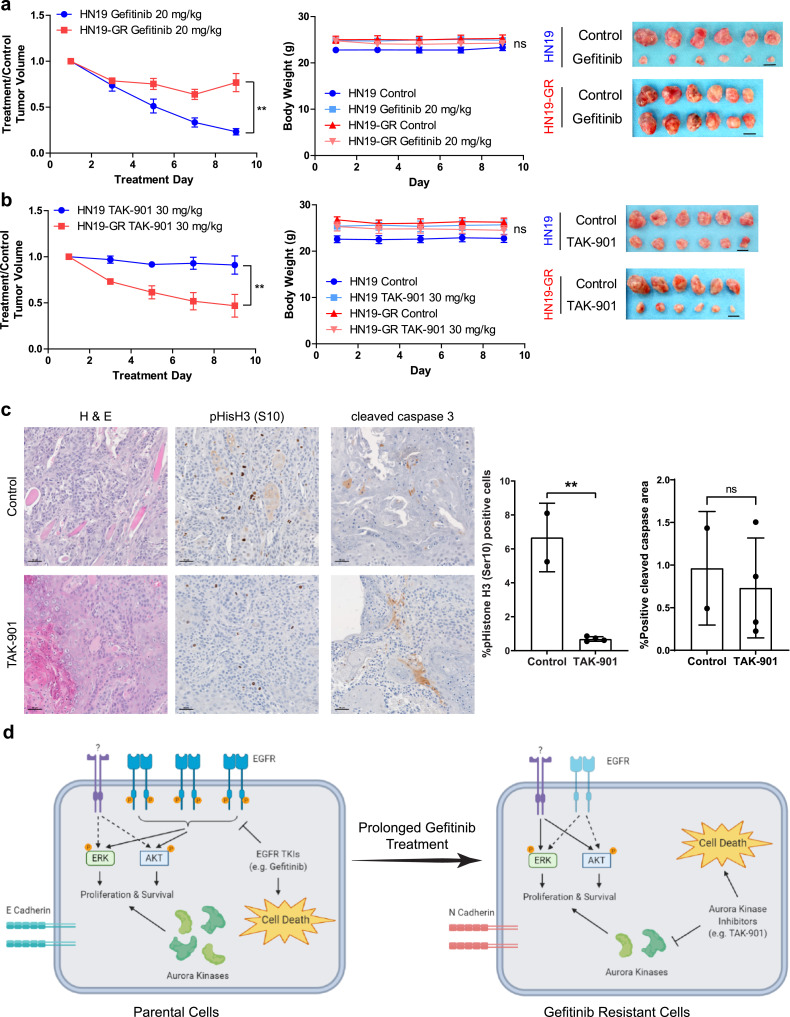
Finally, we demonstrate that the differential sensitivities of HN19 and HN19-GR cells in vitro were retained in vivo. When xenografts of HN19 and HN19-GR cells were treated with gefitinib, xenografts from HN19 cells displayed significantly better response than HN19-GR (Fig. 5a), which corresponds to their increased sensitivity to gefitinib in vitro. In contrast (and in support of the in vitro data above) xenografts from HN19-GR cells exhibited better response to TAK-901 compared to xenografts from HN19 cells (Fig. 5b). HN19-GR xenografts treated with TAK-901 also showed decreased phospho-histone H3 staining (Fig. 5c), demonstrating that the phenotypes observed in vivo is likely due to the effect of aurora kinase inhibition. While we did not observe increased cleaved caspase 3 in the TAK-901 treated HN19-GR xenografts (Fig. 5c), we believe this is likely due to the timing at which the xenografts were resected for immunohistochemistry.
Fig. 5.
Differential sensitivities of HN19 and HN19-GR cells in vitro can be recapitulated in vivo. (a) Differential sensitivities of HN19 and HN19-GR xenografts to gefitinib treatment (20 mg/kg). (b) Differential sensitivities of HN19 and HN19-GR xenografts to TAK-901 treatment (30 mg/kg). (a) and (b) left panel: normalised treatment over control tumour volumes of xenografts during treatment. (a) and (b) middle panel: change in mice body weights over time during treatment. (a) and (b) right panel: images of control and drug treated tumour sizes at the end of the treatment cycle. Data represents values from n = 6 mice per cell line per treatment condition. Two-way repeated measures ANOVA was carried out to determine the significance between the responses of the HN19 and HN19-GR xenografts to the respective drug treatments. Two-way repeated measures ANOVA was carried out to determine the significance between the body weights of mice during the course of the respective drug treatments. ns (p > 0.05), ** (0.001 < p < 0.05). Error bars represent mean ± SEM. Scale bar represents 1 cm. (c) Immunohistochemistry staining of HN19-GR tumours for phospho-histone H3 (S10) expression and cleaved caspase 3 activity upon treatment with TAK-901. Scale bar represents 50 μm. (d) Proposed model of gefitinib resistant in HNSCC cells with no EGFR T790M mutation. Artwork was generated using BioRender.
4. Discussion
In NSCLC, there have been a large number of studies that have examined the mechanisms of secondary resistance to 1st generation EGFR TKI therapy, for which the gain of EGFR-T790M mutation is the most common, accounting for around 50% of patients with acquired resistance [36]. There have been fewer studies that have unravelled mechanisms in the primary or innate resistance setting. In HNSCC all EGFR mutations, whether known for sensitizing or for promoting resistance are uncommon [18]. The failure of EGFR-TKI trials suggest an entirely different landscape/mechanism to escape EGFR targeting. Here, using 3 patient-derived HNSCC cell lines sensitive to gefitinib, we generated resistant (GR) lines and explored the non-genetic mechanisms of EGFR resistance (Fig. 5d).
First, we observed an upregulated EMT gene signature consistent with other previously reported studies and that was functionally reproducible [30,31]. Phenotypically the GR cells were also more migratory and more stem-like, corresponding to a more undifferentiated and slow proliferating cell state. Second, we observed a downregulation of the drug target EGFR, and decreased dependency on EGFR for downstream ERK and AKT signalling, resulting in resistance against 2nd and 3rd generation EGFR TKIs. Unlike in previous reports [32,37], we did not observe corresponding upregulation of HER2, HER3, or MET expression in the GR cells, suggesting that maintenance of ERK and AKT phosphorylation was not via activation of these alternative RTK signalling pathways. While upregulation of AXL expression was sufficient in conferring resistance to gefitinib in the HN19 parental cells, it was not a necessary driver of gefitinib resistance in the HN19-GR cells. This is in sharp contrast with a previous report in which AXL activation in a HNSCC cell line, HN5, mediated resistance to erlotinib [38]. This disparity in resistance mechanism highlights the intrinsic differences in HNSCC from patient to patient, and underscoring the need for personalised therapeutic strategies. Third, using high-throughput drug screening, we found that the GR cells are vulnerable to inhibitors of cellular division. In particular, HN19-GR and HN64-GR cells were significantly responsive to AKIs. Interestingly, HN90-GR cells were more responsive to Aurora kinase B inhibitors compared to Aurora kinase A inhibitors.
The involvement of Aurora kinases in resistance towards EGFR targeted therapy has previously been shown in the context of cetuximab resistance [39], [40], [41]. Reports by Hoellein et al. [39] and Boeckx et al. [40] both demonstrated that cetuximab resistance in HNSCC can be overcome by either single treatment with Aurora kinase inhibitors or in combination with cetuximab. Pickhard et al. [41] further went on to show that the response of HNSCC to cetuximab was associated with AURKA polymorphism. Interestingly the AURKA F31I polymorphism was found to be associated with cetuximab resistance. However, in our study, even though the AURKA F31I polymorphism was found in the HN90 parental cells, the cells were sensitive to gefitinib. Additionally, even though HN19 and HN64 were negative for the AURKA F31I polymorphism, they were resistant to cetuximab, indicating that cells can harness more than one mechanism to become resistant to therapy.
During the course of our study, several reports have been published that correlate alterations in cell cycle regulators and resistance to EGFR TKI therapy [42,43]. We have also recently observed that high expression of Aurora kinase A in HNSCC patients correlated with worse prognosis [44]. Notably a study reported by Shah et al. [43] described the synergistic effects of Aurora kinase A inhibitors and 3rd generation EGFR kinase inhibitors in targeting resistant cells in lung cancer. In their study, they observed that upon chronic EGFR inhibition, activation of Aurora kinase A (i.e., increased phosphorylation) occurred which conferred resistance to EGFR TKIs. While the resistant cells had similar protein expression levels of Aurora kinases A and B compared to parental cells, Aurora kinase A phosphorylation levels in the resistant cells were increased due to increased protein expression of the Aurora kinase A activator TPX2. In our study, both Aurora kinase A and B phosphorylation levels were increased in HN19-GR cells (Fig. 3c, and Fig. S3c). However, we did not observe increased TPX2 gene expression in all 3 pairs of cell lines (Table S3). Overexpression of Aurora kinase A or B in parental HN19 cells was also not sufficient to confer resistance to gefitinib (Fig. S4d). Importantly, our studies suggest that in HN19-GR cells, increased sensitivity to Aurora kinase inhibitors occur in the EGFR-inhibited state. Nonetheless, Aurora kinases represent previously unsuspected and useful therapeutic targets in these GR cells. Extending these observations to the other models described in this study, this convergence to limiting cell proliferation and cell cycle pathways may offer a novel therapeutic vulnerability that can be targeted in the clinical setting.
Declaration of Interests
DSWT received honoraria from Bristol-Myers Squibb, Takeda Pharmaceuticals, Novartis, Roche, and Pfizer; and has consulting or advisory role in Novartis, Merck, Loxo Oncology, AstraZeneca, Roche, and Pfizer. DSWT also received research funding from Novartis (Inst), GlaxoSmithKline (Inst), and AstraZeneca (Inst), outside this submitted work.
Acknowledgments
Acknowledgements
This work was supported by grants from the Agency for Science, Technology, and Research (A*STAR), and the National Medical Research Council (NMRC). RD was supported in part by NIH/NCI #1R01CA155125-01, and by Genome Institute of Singapore (GIS, A*STAR) core funds (BMRC/A*STAR). NGI is supported by NMRC Clinician-Scientist Awards (NMRC/CSA-INV/0011/2016 and MOH-000325-00). RD and NGI were also supported by NMRC/CIRG/1434/2015.
We would like to thank the Centre for High Throughput Phenomics (CHiP-GIS) for their support with the high-throughput compound screens. Many thanks to A*STAR POLARIS for mutational profiling of the cell lines. We would also like to thank the Biological Resource Centre (BRC) at A*STAR for providing the facilities to support the in vivo studies. Lastly, we would like to thank the Advanced Molecular Pathology Laboratory (AMPL) for their assistance with our histology requests.
Data sharing
The RNA sequencing datasets generated during the course of this study have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE157374. All other data are available upon request to the corresponding authors.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103220.
Contributor Information
N Gopalakrishna Iyer, Email: gopaliyer@singhealth.com.sg.
Ramanuj DasGupta, Email: dasguptar@gis.a-star.edu.sg.
Appendix. Supplementary materials
References
- 1.Chow LQM. Head and neck cancer. N Eng J Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 2.Kundu SK, Nestor M. Targeted therapy in head and neck cancer. Tumor Biol. 2012;33:707–721. doi: 10.1007/s13277-012-0350-2. [DOI] [PubMed] [Google Scholar]
- 3.Yarden Y, Sliwkowski MX. Untangling the ErbB signalling network. Nat Rev Mol Cell Biol. 2001;2:127–137. doi: 10.1038/35052073. [DOI] [PubMed] [Google Scholar]
- 4.Grandis JR, Tweardy DJ. Elevated levels of transforming growth factor α and epidermal growth factor receptor messenger RNA are early markers of carcinogenesis in head and neck cancer. Cancer Res. 1993;53:3579–3584. [PubMed] [Google Scholar]
- 5.Dassonville O, Formento JL, Francoual M, Ramaioli A, Santini J, Schneider M. Expression of epidermal growth factor receptor and survival in upper aerodigestive tract cancer. J Clin Oncol. 1993;11:1873–1878. doi: 10.1200/JCO.1993.11.10.1873. [DOI] [PubMed] [Google Scholar]
- 6.Keren S, Shoude Z, Lu Z, Beibei Y. Role of EGFR as a prognostic factor for survival in head and neck cancer: a meta-analysis. Tumor Biol. 2014;35:2285–2295. doi: 10.1007/s13277-013-1303-0. [DOI] [PubMed] [Google Scholar]
- 7.Ang KK, Berkey BA, Tu X, Zhang HZ, Katz R, Hammond EH. Impact of epidermal growth factor receptor expression on survival and pattern of relapse in patients with advanced head and neck carcinoma. Cancer Res. 2002;62:7350–7356. [PubMed] [Google Scholar]
- 8.Chung CH, Ely K, McGavran L, Varella-Garcia M, Parker J, Parker N. Increased epidermal growth factor receptor gene copy number is associated with poor prognosis in head and neck squamous cell carcinomas. J Clin Oncol. 2006;24:4170–4176. doi: 10.1200/JCO.2006.07.2587. [DOI] [PubMed] [Google Scholar]
- 9.Taberna M, Oliva M, Mesía R. Cetuximab-containing combinations in locally advanced and recurrent or metastatic head and neck squamous cell carcinoma. Front Oncol. 2019;9:383. doi: 10.3389/fonc.2019.00383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen EEW, Kane MA, List MA, Brockstein BE, Mehrotra B, Huo D. Phase II trial of gefitinib 250 mg daily in patients with recurrent and/or metastatic squamous cell carcinoma of the head and neck. Clin Cancer Res. 2005;11:8418–8424. doi: 10.1158/1078-0432.CCR-05-1247. [DOI] [PubMed] [Google Scholar]
- 11.Perez CA, Song H, Raez LE, Agulnik M, Grushko TA, Dekker A. Phase II study of gefitinib adaptive dose escalation to skin toxicity in recurrent or metastatic squamous cell carcinoma of the head and neck. Oral Oncol. 2012;48:887–892. doi: 10.1016/j.oraloncology.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Thariat J, Bensadoun RJ, Etienne-Grimaldi MC, Grall D, Penault-Llorca F, Dassonville O. Contrasted outcomes to gefitinib on tumoral IGF1R expression in head and neck cancer patients receiving postoperative chemoradiation (GORTEC trial 2004-02) Clin Cancer Res. 2012;18:5123–5133. doi: 10.1158/1078-0432.CCR-12-1518. [DOI] [PubMed] [Google Scholar]
- 13.Tan EH, Goh C, Lim WT, Soo KC, Khoo ML, Tan T. Gefitinib, cisplatin, and concurrent radiotherapy for locally advanced head and neck cancer: EGFR FISH, protein expression, and mutational status are not predictive biomarkers. Ann Oncol. 2012;23:1010–1016. doi: 10.1093/annonc/mdr327. [DOI] [PubMed] [Google Scholar]
- 14.Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Eng J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 15.Paez JG, Janne PA, Tracy S, Greulich H, Gabriel S, Herman P. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 16.Martin D, Abba MC, Molinolo AA, Vitale-Cross L, Wang Z, Zaida M. The head and neck cancer cell oncogenome: a platform for the development of precision molecular therapies. Oncotarget. 2014;5:8906–8923. doi: 10.18632/oncotarget.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence MS, Sougnez C, Lichtenstein L, Cibulskis K, Lander E, Gabriel SB. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perisanidis C. Prevalence of EGFR tyrosine Kinase domain mutations in head and neck squamous cell carcinoma: cohort study and systematic review. In Vivo. 2017;31:23–34. doi: 10.21873/invivo.11020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai AZ, Abella JV, Park M. Crosstalk in met receptor oncogenesis. Trends Cell Biol. 2009;19:542–551. doi: 10.1016/j.tcb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 20.Veeken J, Oliveira S, Schiffelers R, Storm G, Henegouwen PMP, Roovers R. Crosstalk between epidermal growth factor receptor- and insulin-like growth factor-1 receptor signaling: implications for cancer therapy. Curr Cancer Drug Targets. 2009;9:748–760. doi: 10.2174/156800909789271495. [DOI] [PubMed] [Google Scholar]
- 21.Leong HS, Chong FT, Sew PH, Lau DP, Wong BH, Teh B-T. Targeting cancer stem cell plasticity through modulation of epidermal growth factor and insulin-like growth factor receptor signaling in head and neck squamous cell cancer. Stem Cells Transl Med. 2014;3:1055–1065. doi: 10.5966/sctm.2013-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan DSW, Chong FT, Leong HS, Toh SY, Lau DP, Kwang XL. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat Med. 2017;23:1167–1175. doi: 10.1038/nm.4401. [DOI] [PubMed] [Google Scholar]
- 23.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ianevski A, Giri AK, Aittokallio T. SynergyFinder 2.0: visual analytics of multi-drug combination synergies. Nucleic Acids Res. 2020;48:W488–W493. doi: 10.1093/nar/gkaa216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braig F, Kriegs M, Voigtlaender M, Habel B, Grob T, Biskup K. Cetuximab resistance in head and neck cancer is mediated by EGFR-K521 polymorphism. Cancer Res. 2017;77:1188–1199. doi: 10.1158/0008-5472.CAN-16-0754. [DOI] [PubMed] [Google Scholar]
- 26.Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234:8381–8395. doi: 10.1002/jcp.27740. [DOI] [PubMed] [Google Scholar]
- 27.Cao X, Lai S, Hu F, Li G, Wang G, Luo X. miR-19a contributes to gefitinib resistance and epithelial mesenchymal transition in non-small cell lung cancer cells by targeting c-Met. Sci Rep. 2017;7:2939. doi: 10.1038/s41598-017-01153-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jakobsen KR, Demuth C, Madsen AT, Hussmann D, Vad-Nielsen J, Nielsen AL. MET amplification and epithelial-to-mesenchymal transition exist as parallel resistance mechanisms in erlotinib-resistant, EGFR-mutated, NSCLC HCC827 cells. Oncogenesis. 2017;6:e307. doi: 10.1038/oncsis.2017.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Fleur L, Johansson AC, Roberg K. A CD44high/EGFRlow subpopulation within head and neck cancer cell lines shows an epithelial-mesenchymal transition phenotype and resistance to treatment. PLoS One. 2012;7:e44071. doi: 10.1371/journal.pone.0044071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haddad Y, Choi W, McConkey DJ. Delta-crystallin enhancer binding factor 1 controls the epithelial to mesenchymal transition phenotype and resistance to the epidermal growth factor receptor inhibitor erlotinib in human head and neck squamous cell carcinoma lines. Clin Cancer Res. 2009;15:532–542. doi: 10.1158/1078-0432.CCR-08-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Joon OP. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–1043. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 33.Sgambato A, Camerini A, Faraglia B, Ardito R, Bianchino G, Spada D. Targeted inhibition of the epidermal growth factor receptor-tyrosine kinase by ZD1839 ('Iressa’) induces cell-cycle arrest and inhibits proliferation in prostate cancer cells. J Cell Physiol. 2004;201:97–105. doi: 10.1002/jcp.20045. [DOI] [PubMed] [Google Scholar]
- 34.Farrell P, Shi L, Matuszkiewicz J, Balakrishna D, Hoshino T, Zhang L. Biological characterization of TAK-901, an investigational, novel, multitargeted aurora B kinase inhibitor. Mol Cancer Ther. 2013;12:460–470. doi: 10.1158/1535-7163.MCT-12-0657. [DOI] [PubMed] [Google Scholar]
- 35.Crosio C, Fimia GM, Loury R, Kimura M, Okano Y, Zhou H. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian aurora kinases. Mol Cell Biol. 2002;22:874–885. doi: 10.1128/MCB.22.3.874-885.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chong CR, Jänne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19:1389–1400. doi: 10.1038/nm.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Yu S, Zhao W, Qin S, Chu Q, Wu K. EGFR-TKIs resistance via EGFR-independent signaling pathways. Mol Cancer. 2018;17:53. doi: 10.1186/s12943-018-0793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giles KM, Kalinowski FC, Candy PA, Epis MR, Zhang PM, Redfern AD. Axl mediates acquired resistance of head and neck cancer cells to the epidermal growth factor receptor inhibitor erlotinib. Mol Cancer Ther. 2013;12:2541–2558. doi: 10.1158/1535-7163.MCT-13-0170. [DOI] [PubMed] [Google Scholar]
- 39.Hoellein A, Pickhard A, von Keitz F, Schoeffmann S, Piontek G, Rudelius M. Aurora kinase inhibition overcomes cetuximab resistance in squamous cell cancer of the head and neck. Oncotarget. 2011;2:599–609. doi: 10.18632/oncotarget.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boeckx C, op de Beeck K, Wouters A, Deschoolmeester V, Limame R, Zwaenepoel K. Overcoming cetuximab resistance in HNSCC: the role of AURKB and DUSP proteins. Cancer Lett. 2014;354:365–377. doi: 10.1016/j.canlet.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 41.Pickhard A, Siegl M, Baumann A, Huhn M, Wirth M, Reiter R. The response of head and neck squamous cell carcinoma to cetuximab treatment depends on Aurora kinase A polymorphism. Oncotarget. 2014;5:5428–5438. doi: 10.18632/oncotarget.2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou J, Wu Z, Wong G, Pectasides E, Nagaraja A, Stachler M. CDK4/6 or MAPK blockade enhances efficacy of EGFR inhibition in oesophageal squamous cell carcinoma. Nat Commun. 2017;8:13897. doi: 10.1038/ncomms13897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25:111–118. doi: 10.1038/s41591-018-0264-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lek SM, Li K, Tan QX, Shannon NB, Ng WH, Hendrikson J. Pairing a prognostic target with potential therapeutic strategy for head and neck cancer. Oral Oncol. 2020;111 doi: 10.1016/j.oraloncology.2020.105035. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.