Abstract
When THP-1 cells are differentiated into adherent macro-phage-like cells, they respond to inflammatory stimuli by changing their phenotypes to an activation state and altering the expression of inflammation-related genes. Nitric oxide (NO) is a diatomic molecule implicating in various pathological conditions including tissue damage, ER stress, obesity, and cancer. The sustained inflammatory microenvironment leads to increased NO release through the activation of the inducible nitric oxide synthase (iNOS) gene in macrophages. Here, we provide a dataset on the optimized conditions for the THP-1 differentiation and the induction of NO/iNOS signaling under inflammatory stimulus. The human monocytic cells were differentiated into adherent macrophage-like phenotype by phorbol-12-myristate-13-acetate (PMA) stimulation under optimized conditions. In this study, NO/iNOS signaling capacity and the regulation of other pro-inflammatory genes including TNF-α, IL-1β, and COX-2 in the LPS-induced THP-1 were examined.
Keywords: Nitric Oxide, iNOS, Inflammation, THP-1 monocytic cells, PMA differentiation
Specifications Table
| Subject | Biochemistry or Cell Biology |
| Specific subject area | In vitro Mammalian Cell Culture |
| Type of data | Table Graph |
| How data were acquired | Cell Culture CO2 Incubator (CCL-170T-8, ESCO), Biological Safety Cabinets (Class II BSC, ESCO), Inverted Microscope (Euromex Oxion Serie), Microplate reader (Tecan Infinite® 200 PRO), Quantitative real-time PCR (Applied Biosystems™ 7500,Thermo Fisher Scientific) |
| Data format | Raw, Analyzed |
| Parameters for data collection | THP-1 monocytes were differentiated by PMA (10 ng/mL) stimulation and treated with 1 μg/mL of LPS for 24 h. RAW 264.7 murine macrophages were also activated by the same dose of LPS treatment. |
| Description of data collection | At the end of the appropriate incubation period, the cell culture media were saved for NO production assay. Besides, the cells were subjected to RNA isolation steps for quantitative PCR analyses. |
| Data source location | Institution: Molecular Biology Research Lab, Faculty of Art and Science, Canakkale Onsekiz Mart University City/Town/Region: Canakkale Country: Turkey |
| Data accessibility | The data are available with this article |
Value of the Data
-
•
THP-1 cells provide a practical in vitro model for the screening of compounds with immune-modulating activity, however, the data related to the NO/iNOS signaling capacity of this cell line under inflammatory conditions are limited in the literature.
-
•
Herein, we presented a compact dataset for the optimized conditions on the differentiation of THP-1 monocytes to LPS inducible adherent macrophage-like cells. Additionally, this dataset clarifies the capacity of differentiated THP-1 cells in terms of NO release and iNOS expression along with the other proinflammatory mediators upon inflammatory stimulus.
-
•
These data will guide other researchers who look for a practical cell model in the development of their experimental setup to identify the activities of candidate molecules on NO/iNOS signaling mechanism.
1. Data Description
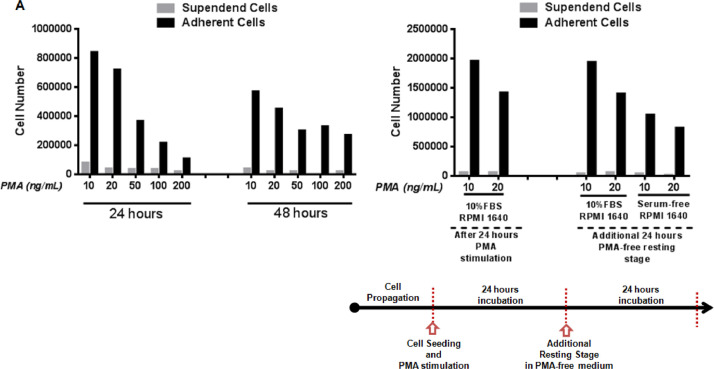
THP-1 cells are round suspension cells established in 1980 by Tsuchiya et al. from the peripheral blood of a 1-year-old human male with acute monocytic leukemia [1]. There is a variety of differentiation protocols for these monocytic cells into a macrophage-like phenotype by using different stimulation factors [2]. Here, we provide a dataset for validated protocol when THP-1 cells are differentiated into LPS inducible macrophages by using phorbol-12-myristate-13-acetate (PMA). The success of the differentiation procedure was validated with adherent cell numbers as compared to the suspended cells (Fig. 1, see S4-5 for raw data) and through examining the changes in the expression level of LPS-induced pro-inflammatory genes including TNF-α, IL-1β, and COX-2 (Fig. 2, see S1 for raw data) [2], [3], [4]. Accordingly, the suspended THP-1 cells were stimulated with different concentrations of PMA (10–200 ng/mL) for 24 and 48 h, separately. Increased PMA concentration resulted in a dose dependent decrease in the number of adherent cells for both incubation periods as shown in Fig. 1A. The number of adherent cells (∼8.4 × 105 cells) was highest in the 10 ng/mL PMA treatment group after 24 h incubation. However, in the same PMA concentration, the number of adherent cells (∼5.7 × 105 cells) was found to be less at the end of the two days incubation (Fig. 1A, see S4 for raw data). To assess the impact of serum supplementation in the culture medium during the resting step, the cells further maintained in the RPMI 1640 medium in the absence and presence of fetal bovine serum (FBS) for 24 h after PMA stimulation. The number of viable cells maintained in the serum-containing medium was almost identical to the viability of the PMA-stimulated cells (Fig. 1B, see S5 for raw data). However the lack of serum in the culture medium led to cell death during the resting stage.
Fig. 1.
PMA-stimulated THP-1 Differentiation. (A) Differentiation of monocytic cells into a macrophage-like phenotype by PMA stimulation. The number of adherent and suspended cells after 24 and 48 h of PMA treatment. (B) The effect of presence and absence of serum in the PMA-free culture medium during the resting stage on viable cell numbers. At first, cells were stimulated with PMA for 24 h (left side of Fig. 1B) and cells were maintained in serum-free or serum-containing medium for 24 h (right side of Fig. 1B).
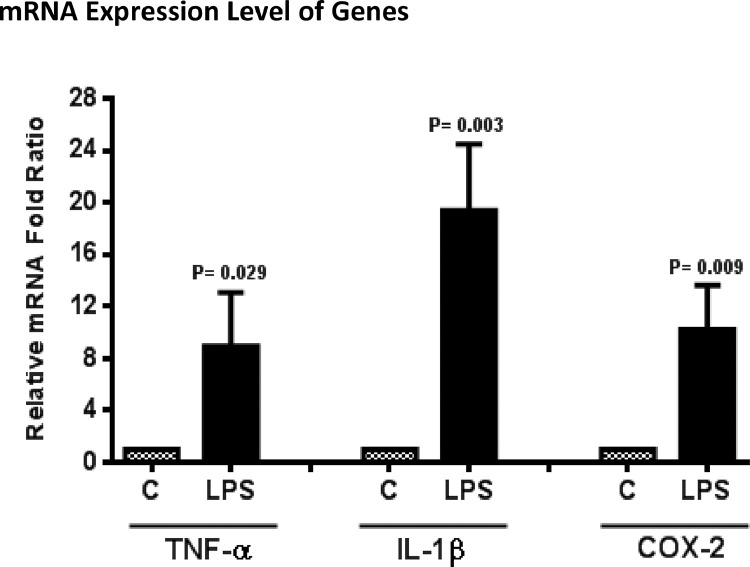
Fig. 2.
The relative mRNA fold ratio for pro-inflammatory genes in control and LPS-stimulated macrophage-like THP-1 cells.
The differentiated macrophage-like cells were stimulated with LPS (1 μg/mL) to induce the expression of pro-inflammatory genes. Quantitative real-time PCR analyses showed that the Ct values of TNF-α, IL-1β, and COX-2 were reduced by LPS treatment as compared to the control cells (Table 1, see S1 for raw data). Besides, β-actin was used as a housekeeping gene to normalize target gene expression and relative changes in the expression levels were evaluated by the comparative ΔΔCt method. According to the results, the mRNA expression levels of TNF-α, IL-1β, and COX-2 were up-regulated by almost 9, 19, and 10-fold, respectively, as compared to the LPS-free control group (Fig. 2, see S1 for raw data). mRNA Expression Level of Genes
Table 1.
The Ct values of pro-inflammatory genes in control and LPS-stimulated macrophage-like THP-1 cells.
| Ct Values |
||
|---|---|---|
| Genes | Non-treated group | LPS-treated group |
| TNF-α | 30.39 ± 0.58 | 27.32 ± 0.92 |
| IL-1β | 24.84 ± 1.62 | 20.53 ± 1.75 |
| COX-2 | 29.54 ± 0.98 | 27.07 ± 0.36 |
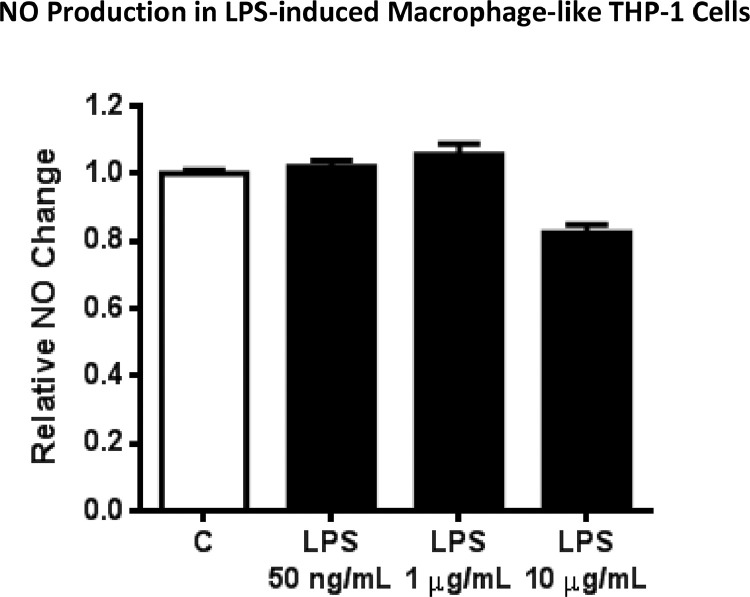
Nitric oxide (NO) is synthesized through the catalytic action of nitric oxide synthase (NOS) [5]. Mainly, three genes found in different chromosomes encode the NOS isoforms called endothelial NOS, neuronal NOS, and inducible NOS (iNOS) [6]. Apart from others, the expression of iNOS is triggered by various inflammatory stimuli such as bacterial lipopolysaccharides (LPS) and cytokines (IL-1β, TNF-α, IFN-γ, and endotoxins) [7,8]. Another main difference between isoforms is related to the quantity of released NO and their activity periods. While iNOS produces a large amount of NO in a prolonged time, other isoforms generate NO molecule in low concentrations with short-acting manner. Accordingly, sustained NO/iNOS signaling is found to be a risk factor for developing several inflammatory diseases and cancer [6]. Therefore, the inhibitory effects of potential anti-inflammatory compounds on NO productions have been extensively used as a screening tool in animal models and in vitro studies. In the literature, regarding the iNOS/NO signaling capacity of THP-1 cells upon inflammatory stimuli, the data are limited and inconclusive. In the study of Cam et al (2012) and Shin et al (2016), THP-1 cells were reported to increase the expression of iNOS and the ability for releasing NO upon 1 and 10 μg/mL LPS treatment, respectively [9,10]. However, in the study of Gross et al, they concluded that, in contrast to murine macrophages, human macrophages produce little detectable iNOS and NO in response to inflammatory stimuli due to epigenetic regulation around the NOS2 transcription start site [11]. In the present study, the Griess method was performed to quantify the NO concentration released by LPS-induced macrophage-like THP-1 and RAW 264.7 murine macrophage cells. The results showed that macrophage-like THP-1 released 0.30 ± 0.10 μM of NO in the culture medium and the level of NO was found almost identical (0.35 ± 0.01 μM) in the medium of LPS-treated cells (Table 2). Different concentrations (0.05, 1, and 10 μg/mL) of LPS treatment were tried in differentiated THP-1 cells, however, at even the highest LPS concentration the amount of NO release was not significantly different than that was found in the non-treated control group (Fig. 3, see S3 for raw data). Moreover, the expression of iNOS was not induced by LPS (1 μg/mL) in THP-1 cells. Unlike human macrophages, the iNOS expression level and NO production were triggered in RAW 264.7 murine macrophage cells. While non-treated cells released 0.74 ± 0.91 μM of NO, 1 μg/mL LPS stimulation increased the level to 37.68 ± 3.19 μM. Besides, the iNOS expression level was significantly induced in the LPS treatment group as compared to the control cells (Table 2, see S2 for raw data).
Table 2.
The released NO concentration and the Ct values of iNOS genes in non-treated and LPS (1 μg/mL) treated macrophages.
| Released NO Concentration (μM) |
Ct Values (iNOS) |
|||
|---|---|---|---|---|
| Group | THP-1 Cells | RAW 264.7 Cells | THP-1 Cells | RAW 264.7 Cells |
| Non-treated | 0.30 ± 0.10 | 0.74 ± 0.91 | 34.81 ± 0.25 | 28.74 ± 0.08 |
| LPS-treated | 0.35 ± 0.01 | 37.68 ± 3.19 | 35.96 ± 0.03 | 18.89 ± 1.40 |
Fig. 3.
Relative NO change in LPS-challenged human macrophage cells. C: Control.
NO Production in LPS-induced Macrophage-like THP-1 Cells.
2. Experimental Design, Materials and Methods
2.1. Cell culture
THP-1, human monocytic cells were grown in RPMI-1640 medium containing 10% Fetal Bovine Serum (FBS), 1% Penicillin-Streptomycin. After reaching confluency, cells were seeded at 1 × 106 cells/mL density and stimulated with 10 ng/mL PMA for 24 h to differentiate them into macrophages. Then, they were maintained in PMA-free serum-containing RPMI 1640 medium for 24 h. Also, RAW 264.7 murine macrophages were maintained in culture flasks containing DMEM (Dulbecco's Modified Eagle's Serum) supplemented with 10% Fetal Bovine Serum (FBS), 1% Penicillin-Streptomycin at 37 °C under 5% CO2 in carbon dioxide incubator.
2.2. Determination of cell numbers
At the end of the PMA stimulation, the cell supernatants were collected, and adherent cells were scraped by using a sterile scraper. The number of adherent cells and suspended cells were counted by trypan blue staining under the inverted microscope.
2.3. RNA isolation and quantitative real-time PCR analyses
The manufacturer instructions of the PureLink™ RNA Mini Kit (12183018A, Invitrogen™) were applied with slight modifications. Briefly, THP-1 and Raw 264.7 cells were plated with 1 × 106 and 4 × 105 cells/mL densities, respectively. After appropriate cell culture treatments, cells were collected by TRIzol™ reagent (15596018, Invitrogen™). Once the sample was centrifuged, the aqueous phase was subjected to further steps for isolation. The purity and amount of RNA samples were evaluated by NanoQuant Plate™ (Tecan Infinite® 200 PRO, Switzerland) and samples having a ratio of OD 260/280 ≥ 2.0 and OD 260/230 ≥ 1.8 were considered to be good quality. cDNA synthesis was achieved by a series of reverse transcriptase reactions according to the manufacturer's instructions of the High-Capacity cDNA Reverse Transcription Kit (4368814, Applied Biosystems™). The effects of LPS treatment on target genes were investigated at the mRNA level by qPCR analysis. As mentioned previously [12], diluted cDNA samples were cycled on StepOnePlus® Real-Time PCR Systems (Applied Biosystems, Thermo Fisher Scientific) in the presence of specific TaqMan® probes (Life Technologies, Thermo Fisher Scientific) (Human IL-1β: Hs01555410_m1; Human TNF-α: Hs00174128_m1; Human COX-2: Hs00153133_m1; Human iNOS: Hs01075529_m1; Human β-Actin: Hs01060665_g1; Murine iNOS: Mm00440502-m1) and TaqMan™ Fast Advanced Master Mix (4444556, Applied Biosystems™) under recommended conditions. As endogenous control, the level of β-Actin was examined to normalize the gene expression levels. The results given as relative change were calculated by using the comparative ΔΔCt method.
2.4. NO production assay
An analytical method called Griess assay was carried out to investigate the effect on LPS-induced NO release as previously detailed [13]. RAW 264.7 macrophages were plated at a density of 5 × 104 cell/mL in 24- well plate in phenol red-free DMEM containing 10% FBS and then incubated with or without LPS (1 μg/mL) for 24 h. THP-1 cells were differentiated by PMA treatment (10 ng/mL) for 24 h and further incubated overnight for the resting stage. Once cells adhere, they were incubated in red phenol-free media for 24 h in the presence of LPS (1 µg/mL). Then, the amount of nitrite present in the supernatants was quantified with the Griess method [14]. Briefly, 100 μL of culture supernatants were transferred onto a 96-well plate and incubated with 100 μL of Griess reagent including 1% sulphanilamide and 0.1% naphthyl ethylenediamine in 5% phosphoric acid solution for 10 min at dark. For the assay, the reference curve was prepared by using nitrite standard (0 to 100 μM) from a stock of 0.1 M NaNO2. Then, absorbance was read at 540 nm by using a microplate reader (Tecan Infinite® 200 PRO, Switzerland).
2.5. Statistical analysis
Data were obtained through technical and biological replicated experiments. One-way ANOVA was performed for the expression level of three genes separately to test the differences between LPS-treated and control groups with JMP version 13.
CRediT Author Statement
Adem Ozleyen: Investigation, Writing - Original draft preparation, Visualization; Yakup Berkay Yilmaz: Investigation, Methodology; Tugba Boyunegmez Tumer: Conceptualization, Supervision, Writing - Review & Editing.
Ethics Statement
The authors declare that the paper provides the original materials which were found out through their own original work. All authors have been actively involved in the study and credited for their meaningful contributions. The policies of Elsevier outlined in the Guide for Authors and in the Ethics Statement were considered during the preparation of the paper.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Acknowledgments
This study was partially supported by a research grant from Çanakkale Onsekiz Mart University (Scientific Research Projects, Project ID: FYL-2019-3123 and FYL-2019-3054).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2021.106786.
Contributor Information
Adem Ozleyen, Email: ademozleyen@gmail.com.
Tugba Boyunegmez Tumer, Email: tumertb@comu.edu.tr.
Appendix. Supplementary materials
References
- 1.Tsuchiya S. Establishment and characterization of a human acute monocytic leukemia cell line (THP-1) Int. J. Cancer. 1980;26(2):171–176. doi: 10.1002/ijc.2910260208. [DOI] [PubMed] [Google Scholar]
- 2.Chanput W., Mes J.J., Wichers H.J. THP-1 cell line: An in vitro cell model for immune modulation approach. Int. Immunopharmacol. 2014;23(1):37–45. doi: 10.1016/j.intimp.2014.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Park E. Optimized THP-1 differentiation is required for the detection of responses to weak stimuli. Inflamm. Res. 2007;56(1):45–50. doi: 10.1007/s00011-007-6115-5. [DOI] [PubMed] [Google Scholar]
- 4.Chanput W. Characterization of polarized THP-1 macrophages and polarizing ability of LPS and food compounds. Food Funct. 2013;4(2):266–276. doi: 10.1039/C2FO30156C. [DOI] [PubMed] [Google Scholar]
- 5.Janakiram N.B., Rao C.V. iNOS-selective inhibitors for cancer prevention: promise and progress. Futur. Med. Chem. 2012;4(17):2193–2204. doi: 10.4155/fmc.12.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mollace V. Modulation of prostaglandin biosynthesis by nitric oxide and nitric oxide donors. Pharmacol. Rev. 2005;57(2):217–252. doi: 10.1124/pr.57.2.1. [DOI] [PubMed] [Google Scholar]
- 7.Bonaiuto C. Activation of nuclear factor-κB by β-amyloid peptides and interferon-γ in murine microglia. J. Neuroimmunol. 1997;77(1):51–56. doi: 10.1016/S0165-5728(97)00054-4. [DOI] [PubMed] [Google Scholar]
- 8.Hartlage-Rübsamen M., Lemke R., Schliebs R. Interleukin-1β, inducible nitric oxide synthase, and nuclear factor-κB are induced in morphologically distinct microglia after rat hippocampal lipopolysaccharide/interferon-γ injection. J. Neurosci. Res. 1999;57(3):388–398. . doi:10.1002/(SICI)1097-4547(19990801)57:3<388::AID-JNR11>3.0.CO;2-2. [PubMed] [Google Scholar]
- 9.Cam, A. and E.G. de Mejia, RGD-peptide lunasin inhibits Akt-mediated NF-κB activation in human macrophages through interaction with the αVβ3 integrin. Molecular nutrition & food research, 56(10) (2012) 1569–1581. doi: 10.1002/mnfr.201200301. [DOI] [PubMed]
- 10.Shin J.-S. Berberine decreased inducible nitric oxide synthase mRNA stability through negative regulation of human antigen R in lipopolysaccharide-induced macrophages. J. Pharmacol. Exp. Ther. 2016;358(1):3–13. doi: 10.1124/jpet.115.231043. [DOI] [PubMed] [Google Scholar]
- 11.Gross T.J. Epigenetic silencing of the human NOS2 gene: rethinking the role of nitric oxide in human macrophage inflammatory responses. J. Immunol. 2014;192(5):2326–2338. doi: 10.4049/jimmunol.1301758. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tumer T.B. GR24, a synthetic analog of Strigolactones, alleviates inflammation and promotes Nrf2 cytoprotective response: In vitro and in silico evidences. Comput. Biol. Chem. 2018;76:179–190. doi: 10.1016/j.compbiolchem.2018.07.014. [DOI] [PubMed] [Google Scholar]
- 13.Kapche D.W.F.G. Aryl benzofuran derivatives from the stem bark of Calpocalyx dinklagei attenuate inflammation. Phytochemistry. 2017;141:70–79. doi: 10.1016/j.phytochem.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Griess P. Bemerkungen zu der Abhandlung der HH. Weselsky und Benedikt „Ueber einige Azoverbindungen. Ber. Dtsch. Chem. Ges. 1879;12(1):426–428. doi: 10.1002/cber.187901201117. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.