Abstract
B cells are lymphocytes derived from hematopoietic stem cells and are a key component of the humoral arm of the adaptive immune system. They make attractive candidates for cell-based therapies because of their ease of isolation from peripheral blood, their ability to expand in vitro, and their longevity in vivo, Additionally, their normal biological function, to produce large amounts of protein in the form of antibodies, can be utilized to express very large amounts of recombinant protein in the form of recombinant antibody or enzyme for the treatment of enzymopathies and other diseases. Here, we provide detailed methods for isolating primary human B cells from PBMCs and activating/expanding isolated B cells in vitro. We then demonstrate the steps involved in using the CRISPR/Cas9 system for the site-specific KO of endogenous genes in B cells. This method allows for efficient KO of various genes which can be used to study the biological functions of a gene of interest. We then demonstrate the steps for using the CRISPR/Cas9 system together with recombinant adeno-associated viral (rAAV) vector for efficient site-specific integration and expression of a transgene expression cassette in B cells. Together, this protocol provides a step-by-step engineering platform that can be used in primary human B cells to study biological functions of genes in B cells as well for the development of B cell therapeutics.
Keywords: CRISPR/Cas9, genome engineering, recombinant AAV, gene editing, primary human B cells
SUMMARY:
Here, we provide a detailed step-by-step protocol for CRISPR/Cas9-based genome engineering of primary human B cells for gene knockout (KO) and knock-in (KI) to study biological functions of genes in B cells and the development of B cell therapeutics.
INTRODUCTION:
B cells are a subgroup of the lymphocytes linage derived from hematopoietic stem cells (HSCs). They perform a critical role in the adaptive humoral immune system by producing large amounts of antibodies in response to immune challenges1. B cells are also precursors of memory B cells and the terminally differentiated, long-lived plasma cells, thereby providing lasting humoral immunity2. Plasma cells, in particular, are unique among immune cells in their ability to produce large amount of antibody while surviving for years or decades3. Additionally, the ease of isolation from peripheral blood makes the B cell lineage an excellent candidate for novel cell-based therapies4.
Previously, random integration methods such as those using lentiviral vectors (LV) or a Sleeping Beauty (SB) transposon have been used to engineer B cells for transgene delivery and expression 5–8. However, the non-specific nature of these approaches makes it difficult to study the biological functions of a specific gene in the B cells, and also carries an inherent risk of insertional mutagenesis and variable transgene expression and/or silencing in the therapeutic setting.
The CRISPR/Cas9 system is a powerful genome engineering tool that allows researchers to precisely editing the genome of various cells in numerous species. Recently, two groups, including our own, have successfully developed methods for ex vivo expansion and targeted genome engineering of primary human B cells 9,10. Here, we demonstrate our updated and enhanced processes for isolating primary human B cells from peripheral blood mononuclear cells (PBMCs), expanding the isolated B cells in culture, engineering B cells with CRISPR/Cas9 for a gene KO, and engineering B cells using CRISPR/Cas9 together with recombinant AAV6 vector (rAAV6) for site-specific integration and expression of a transgene. These methods achieve up to a 44-fold expansion of the B cells in culture (Figure 1) with editing frequencies > 90% (Figure 2), while maintaining cell viability > 80% (Figure 2C) and site-specific transgene integrations rates of >60% (Figure 5A).
Figure 1. B cell expansion in vitro.
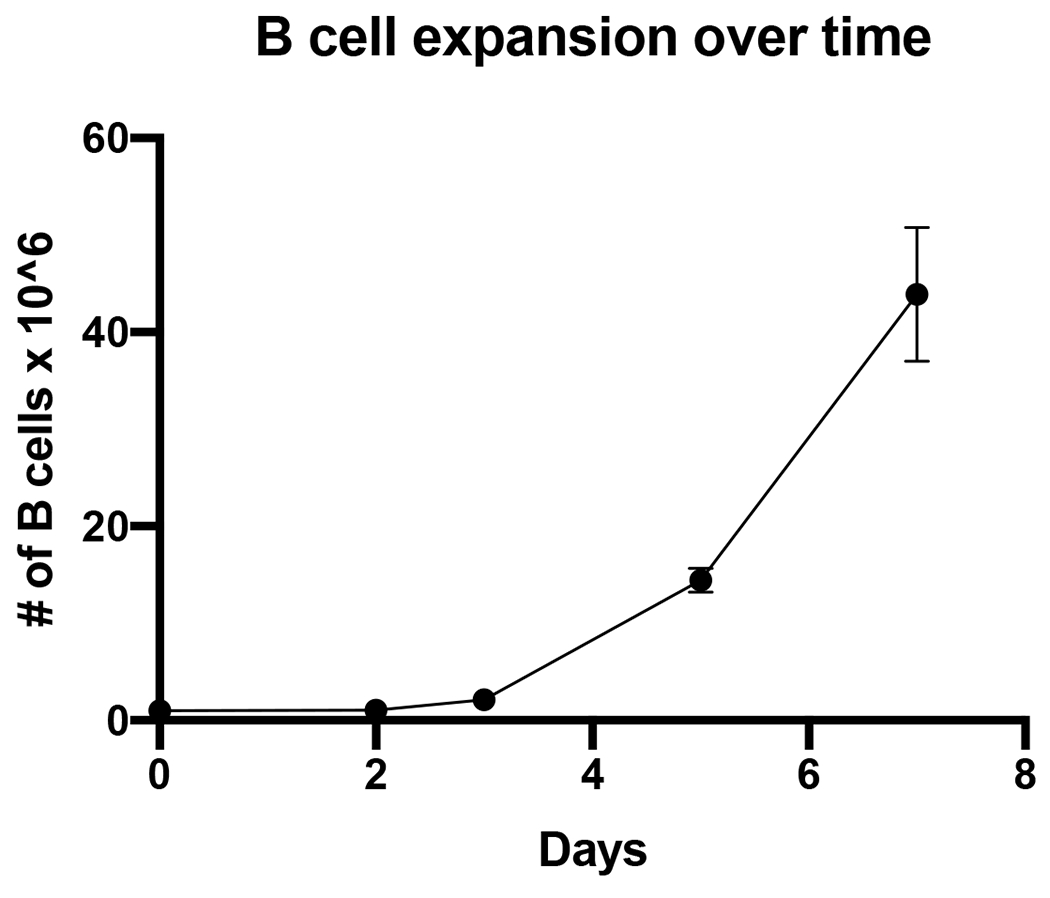
B cells were seeded at 1 x 106 cells on day 0 (zero) at a density of 5 x 105 per mL and expanded 44-fold in 7 days. (n=3 independent donors)
Figure 2. CRISPR/Cas9 mediated CD19 KO in B cells.
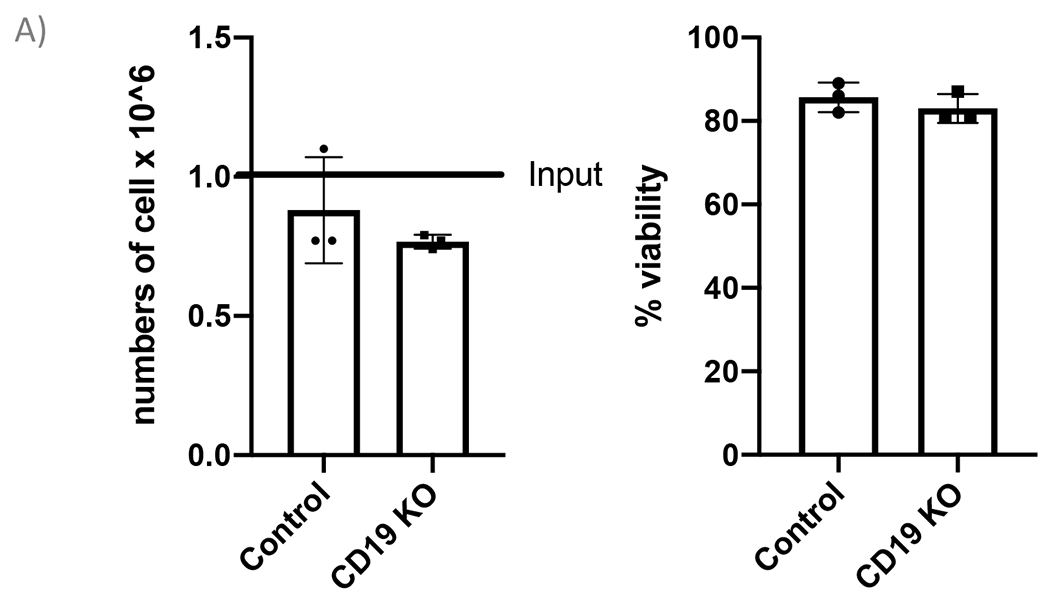
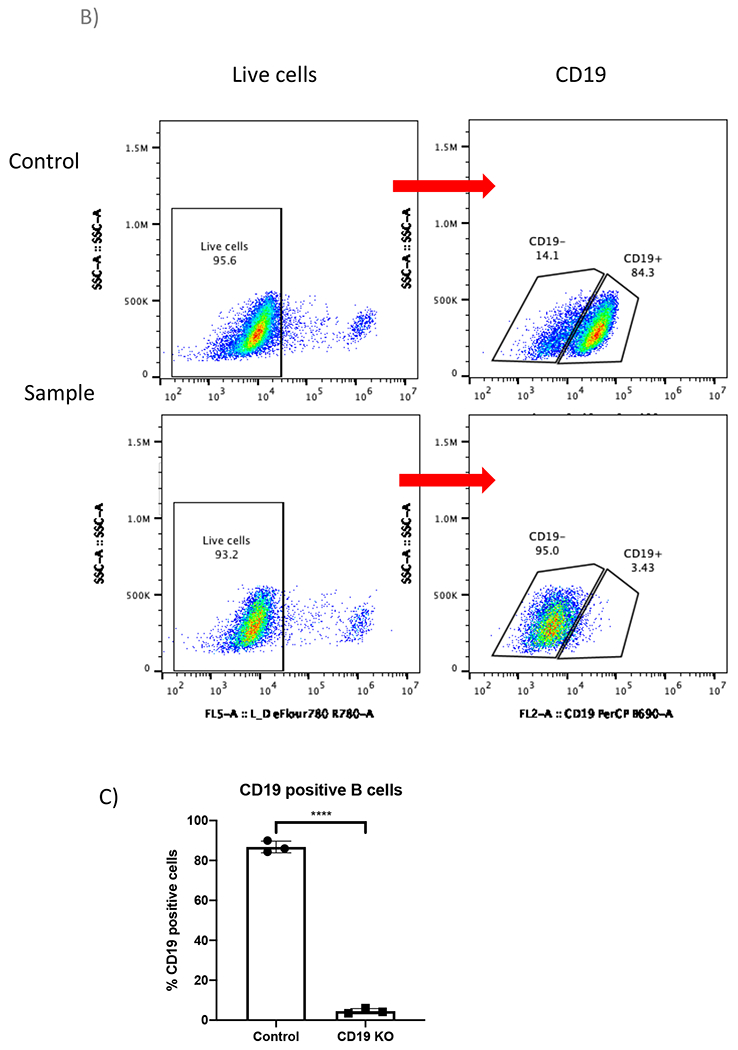
(A) Bar graph shows >70% cell recovery (left panel) and >80% viability (right panel) of cells post transfection were observed in both control and CD19 KO samples at 24-hour post electroporation. (B) Representative flow plots of CD19 gating of live cells shows 84.3% and 3.43% CD19 positive cells in the control sample and the CD19 KO sample, respectively. (C) Bar graph shows significant reduction of CD19 in the CRISPR/Cas9 mediated CD19 KO group (p≤0.0001).
Figure 5. CRISPR/Cas9 and rAAV6 mediated site-specific integration of EGFP reporter cassette in B cells at day 12 post engineering.
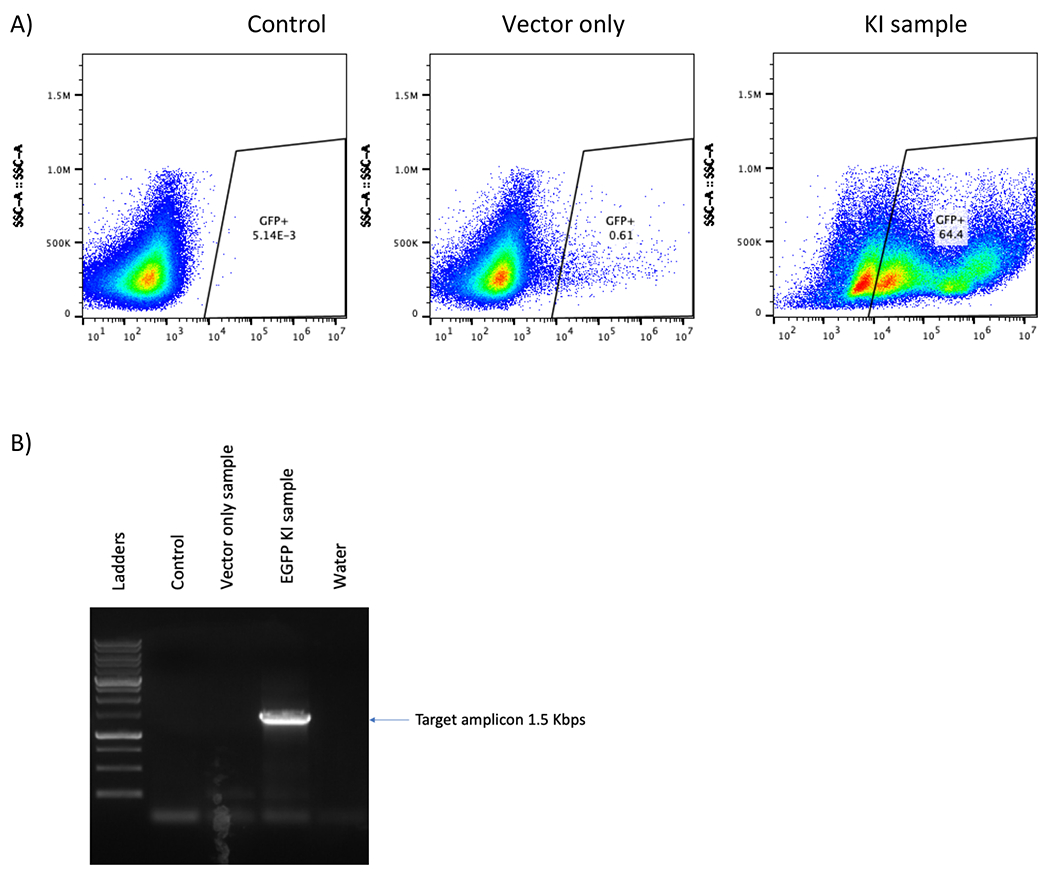
(A) Representative flow plot shows no EGFP-positive B cells in either the control or the “vector only” sample and 64.4% in EGFP B cell was observed in the KI sample. (B) Junction PCR of KI samples shows 1.5 Kbps band which is the predicted size of the amplicon while no band was found in the control or “vector only” samples. A water lane is used to ensure no contamination in the PCR process.
PROTOCOL:
Leukophoresis samples from healthy donors were obtained from a local blood bank. All experiments described here were determined to be exempt research by the Institutional Review Board (IRB) and were approved by Institutional Biosafety Committee (IBC) at the University of Minnesota.
All experiments were done in compliance to the universal precaution for bloodborne pathogens, with sterile/aseptic technique and a proper biosafety level 2 (BSL2) equipment.
1. Prepare supplements for B-cell expansion medium.
1.1. Reconstitute CpG ODN2006 (ODN 7909) to a concentration of 1 mg/mL.
1.2. Reconstitute MEGA CD40L to a concentration of 100 μg/mL.
1.3. Reconstitute recombinant human IL-10 (rhIL-10) to a concentration of 50 μg/mL.
1.4. Reconstitute recombinant human IL-15 (rhIL-15) to a concentration of 10 μg/mL.
NOTE: Keep each supplement in small aliquots at −20°C to −80°C for up to 6 months.
2. Prepare basal medium
2.1. Combine Excellerate B cell medium with 5% (v/v) CTS Immune Cell SR and 1% (v/v) Penicillin and streptomycin.
2.2. Sterilize the basal medium using 0.22 micrometers filter adaptor into sterilized bottle.
2.3. Keep the basal medium at 4°C for up to 1 month.
3. Prepare B-cell expansion medium
3.1. Transfer required amount of the basal medium to culture the B cells at 5x105 cells/mL into a sterile container.
3.2. Supplement the basal medium with 1 μg/mL CpG, 100 ng/mL MEGA CD40L, 50 ng/mL rhIL-10 and 10 ng/mL rhIL-15.
3.3. Filter the B-cell expansion medium using 0.22 micrometer filter.
3.4. Equilibrate the B-cell expansion medium in the tissue culture incubator at 37°C, 5% CO2, with humidity for at least 30 minutes before use.
NOTE: Prepare fresh B-cell expansion medium to use for one day. Do not prepare the B-cell expansion medium to use for multiple days. This media recipe encourages proliferation of naïve B cells.
4. Human B cells purification and expansion
Add 99-100% Isopropanol to a “Mr. Frosty” following manufacturer’s instruction and keep at 4°C before starting step 4.1
4.1. Isolate PBMCs from Leukophoresis sample.
4.1.1. Transfer Leukophoresis sample (approximately 8-10 mL) to a sterile 50 mL conical tube.
4.1.2. Bring up the volume to 35 mL with sterile 1x PBS.
4.1.3. Layer 35 mL Leukophoresis sample on 15 mL Ficoll-Paque.
4.1.4. Centrifuge at 500 x g for 25 minutes without brake and collect the PBMCs from the interface and transfer to a new sterile 50 mL conical tube.
4.1.5. Bring up the PBMCs to 50 mL with 1x PBS
4.1.6. Centrifuge at 500 x g for 5 minutes without brake. Remove supernatant without disturbing the PBMC pellet. The pellet may appear red.
4.1.7. Add 7 mL ACK lysis buffer (see material list for catalog number), mix well and incubate at room temperature (RT) for 3 minutes.
4.1.8. Bring up the volume to 50 mL with 1x PBS.
4.1.9. Centrifuge at 400 x g for 5 minutes with low resistant brake. Remove supernatant without disturbing the pellet. The pellet should look pinkish or white.
NOTE: If you wish to continue to culture the freshly isolated B cells, prepare the B cell expansion medium before starting the B cell isolation (Step 4.2).
4.2. B cell isolation from PBMCs using STEMCELL human primary B cell isolation kit.
4.2.1. Resuspend PBMCs with Isolation Buffer to a concentration of 5 x 107 cells/mL.
NOTE: If the total number of PBMCs is less than 5 x 107 cells, scale down the volume of isolation buffer to maintain 5 x 107 cells/mL. Minimum and maximum volumes of cell suspension are 0.25 mL and 8 mL, respectively.
4.2.2. Transfer up to 8 mL (5 x 107 cells/mL) to a sterile polypropylene round-bottom tube with cap.
4.2.3. Add 50 μL/mL of Cocktail Enhancer to the PBMCs.
4.2.4. Add 50 μL/mL of Isolation Cocktail to the PBMCs, cap the tube and invert 2-3 times to mix.
4.2.5. Incubate at RT for 5 minutes, while waiting, vortex RapidSpheres microbeads for at least 30 seconds.
4.2.6. Briefly vortex RapidSpheres again before the transfer 50 μL RapidSpheres per 1 mL of PBMCs, cap the tube and invert 2-3 times to mix.
4.2.7. Top up to 10 mL with Isolation buffer, and gently pipette up and down a 2-3 times.
4.2.8. Place the tube into the “Big Easy” Magnetic station and incubate at RT for 3 minutes.
4.2.9. Hold the magnetic and tube together, and in one motion, invert the magnet and tube together to pour cell suspension into the new tube. Discard the old tube.
4.2.10. Repeat step 4.2.8 (reduce incubation time to 2 minutes) and pour B cell suspension into a clean conical tube.
4.2.11. The enriched B cells are ready to use. If cells will be used immediately, continue to Human B cell expansion section. Check purity of Isolated B cells by flow cytometry (optional). If cells are to frozen before use continue to 4.2.12.
4.2.12. To freeze the B cells, centrifuge at 400 x g for 5 minutes and discard supernatant without disturbing the pellet.
4.2.13. Resuspend cells with Cryostor CS10 freezing medium at 107 cells/mL and aliquot 1 mL/cryovial.
4.2.14. Place the cryovial in a pre-chilled “Mr. Frosty” and store at −80°C overnight and transfer the frozen cryovial on the next day to a liquid nitrogen tank for up to 1 year.
4.3. Human B cells expansion
If using freshly isolated B cells, skip 4.3.1-4.3.5. Count the cells and transfer the required number of cells into a sterile conical tube and continue with step 4.3.6.
4.3.1. Prepare and pre-warm B cell expansion medium and FBS prior thawing the B cells.
4.3.2. Thaw B cells in a 37°C water bath. While waiting, transfer 2 mL of pre-warmed FBS into a sterile 15 mL conical tube.
4.3.3. Once B cells are completely thawed, add 1 mL pre-warmed FBS drop-wise into the sample. Incubate at RT for 1 minute.
4.3.4. Gently pipette to resuspend the sample and transfer the whole volume of sample, drop-wise, into a conical tube containing 2 mL pre-warmed FBS.
4.3.5. Bring up to 15 mL with sterile 1x PBS, cap, and invert the tube gently 2-3 times.
4.3.6. Centrifuge at 400 x g for 5 minutes.
4.3.7. Discard supernatant without disturbing the cell pellet, resuspend the cell pellet with 1 mL of pre-warmed B-cell expansion medium, and count cells.
4.3.8. Transfer cells into a flask containing pre-warmed B-cell expansion medium at cell concentration of 5x105 cells/mL.
4.3.9. Incubate the flask vertically in a tissue culture incubator at 37°C, 5% CO2, with humidity.
4.3.10. Refresh the expansion medium completely every 2 days by transferring the whole volume of the cells in to a sterile conical tube and repeat the step 4.3.6-4.3.9.
NOTE: T25 Flask can hold 10-20 mL of medium
T75 flask can hold up to 20-60 mL of medium
5. Primary human B cell engineering
B cells must be engineered at 48 ± 2 hours post expansion for the optimal results. Prepare B cell expansion medium, aliquot the medium to a tissue culture plate, and pre-warm in a tissue culture incubator prior transfection.
5.1. Prepare CRISPR/Cas9 reagent by mixing 1 μL (1 μg/μL) of chemically modified sgRNA and 1.5 μL (1 μg/μL) of chemically-modified Streptococcus pyogenes Cas 9 (S.p. Cas9) Nuclease mRNA for a total of 2.5 μL CRISPR/Cas9 reagent per 1 x 106 B cells.
For example: CD19 sgRNA is used for gene KO experiment (Figure 2 for result)
AAVS1 sgRNA is used for gene KI experiment (Figure 5 for result)
NOTE:
Design sgRNAs using online tool11
Design sgRNAs on an exon common to of all isoforms of the protein.
Order chemically modified sgRNA (e.g. Synthego, IDT).
5.2. Mix gently and set aside at RT
5.3. Turn on Amaxa 4D nucleofector (a.k.a. Lonza 4D electroporator) and prepare transfection reagents following Table 1.
Table 1.
gRNAs sequences
| Name | gRNA sequence |
|---|---|
| CD19 | 5′-GCTGTGCTGCAGTGCCTCAA-3′ |
| AAVS1 | 5′-GTCACCAATCCTGTCCCTAG-3′ |
NOTE:
This is a good step for pause if needed by putting all reagents on ice. Remove all reagents from ice when ready to resume the experiment.
If using S.p. Cas9 protein (IDT), mix 1 ug of sgRNA to 5 ug of Cas9 protein, while avoiding bubbles, and incubate the mixture at RT for at least 20 minutes before use.
5.4. Count and transfer the required number B cells into a sterile 15 mL conical tube.
5.5. Bring up the volume to 15 mL with sterile 1x PBS and centrifuge at 400 x g for 5 minutes. While waiting, prepare P3 primary cell transfection reagent (see table 2.) and set aside at RT.
Table 2.
Prepare Nucleofection reagent mix
| Reagents | Volume per 1 reaction |
|---|---|
| P3 Primary Cell solution | 16.4 μL |
| Supplement 1 | 3.6 μL |
| Total | 20 μL |
NOTE: One reaction = 1x106 cells. Scale up the reagents to meet the number of reactions.
5.6. Discard supernatant without disturbing the cell pellet.
5.7. Resuspend the cell pellet with 1 mL sterile 1x PBS and transfer to 1.5 mL microcentrifuge tube. Centrifuge at 400 x g for 5 minutes.
5.8. Discard the supernatant completely without disturbing the cell pellet.
5.9. Resuspend the cell pellet with P3 primary cell transfection reagent to a concentration of 1x106 cells/20 μL and transfer into a PCR tube.
5.10. Add 0.5 μg of chemically-modified GFP mRNA (as a transfection reporter). This step is optional. Modify experiment to match your needs.
5.11. Transfer 2.5 μL CRISPR/Cas9 reagent into the 20 μL cell suspension.
5.12. Pipette up and down once to mix the RNPs and the cell suspension and transfer the entire volume into a Lonza 4D cuvette. Cap and tap the cuvette on the bench gently to ensure the liquid covers the bottom of the cuvette.
5.13. Use EO117 protocol for transfection.
NOTE: The Lonza 4D electroporator can be used outside tissue culture hood. Capping the cuvette ensure sterility.
5.14. Incubate the cuvette at RT for 15 minutes.
5.15. Transfer 80 μL of pre-warmed medium from the tissue culture plate into the transfection reaction in the cuvette. Place the cuvette in the tissue culture incubator for 30 min.
5.16. Transfer the whole volume of the sample from the cuvette to an appropriate well of a 48-well tissue culture plate containing 1 mL B cell expansion medium. The final concentration of the cells should be 5 x 105 cells/mL.
5.17. If performing gene KI experiment transfer rAAV6 vector at 500,000 MOI into the appropriate well. See an example rAAV6 vector construct on Figure 4A.
Figure 4. rAAV6 AAVS1 MND-GFP vector construct.

Expression cassette contains a strong synthetic promoter (MND) sequence immediately followed by EGFP coding sequence and poly adenylation (Poly A) sequence. AAVS1 homology arms flank upstream of the MND promoter and downstream of the poly A sequences. EGFP will be expressed under the regulation of the MND promoter. Sequence lengths are indicated above each component of the construct.
NOTE: rAAV6 must contain homology arms up- and downstream of the CRISPR/Cas9 target double-strand break (DSB) site for homology directed repair.
5.18. Place the plate in a tissue culture incubator at 37°C and 5% CO2 with humidity.
5.19. Refresh B cell expansion medium every 2 days by transferring the whole volume of the cells into a clean 15 mL conical tube, centrifuge at 400 x g for 5 minutes, discard supernatant without disturbing the pellet, resuspend cells with 100 uL fresh B cell expansion medium and transfer to a tissue culture plate. Bring up the medium volume to have cell concentration at 5 x 105 cells/mL.
NOTE: 48-well plate can hold media up to 1 mL/well.
24-well plate can hold media up to 2 mL/well.
12-well plate can hold up media to 4 mL/well.
6-well plate can hold media up to 8 mL/well.
5.20. Count cells and record viability at day 1 post engineering.
5.21. Expand the engineered cells for at least 5 days before downstream analysis.
REPRESENTATIVE RESULTS:
B cells proliferated rapidly in culture and reached 44-fold expansion in 7 days (Figure 1). In the KO experiment, B cells were collected on day 5 post transfection for flow cytometry analysis and the genomic DNA of the same samples were extracted for insertion/deletion (indel) analysis. Trypan blue staining showed more than 80% viable cells in both the control and the CD19 KO samples at 24 hours post electroporation (Figure 2A). Representative flow plots of the CD19 KO sample and control showed 95% and 11% CD19 negative cells, respectively (Figure 2A). There is a significant reduction of CD19 in the CD19 KO samples compared with control samples at day 5 post engineering (Figure 2B; p≤0.0001). Chromatograms of genomic sequencing (see primer sequences in table 3) showed double peaks in the CD19 KO B cells (Figure 3A), while single peaks were observed in the control, as expected (Figure 3B). Indel analysis of the chromatograph (ice.synthego.com) of the KO samples showed high % indels (>90%) at CD19 locus that matched well with the % CD19 protein loss (Figure 3C; p≥0.05). 64% of EGFP-positive cells were observed in the sample that received AAVS1 CRISPR/Cas9 system together with rAAV6, while No EGFP-positive cell was found in either control or the sample received vector only (without AAVS1 CRISPR/Cas9) at day 12 post engineering (Figure 5A). A junction PCR amplification (see primer sequences in table 3) showed 1.5 Kbps amplicons only in the KI sample (Figure 5B), while no PCR products in either the control or vector only.
Table 3.
Primers used
| Description | Sequence | Use for |
|---|---|---|
| CD19 forward primers | 5′-AAATTCAGGAAAGGGTTGGAAG-3′ | Amplify the CD19 locus for Indel analysis |
| CD19 Reverse primer | 5′-GCGGACCTCTTCTGTCCATG-3′ | Amplify the CD19 locus for Indel analysis |
| Junction PCR forward primer | 5′-GGACGAGCTGTACAAGTAACG-3′ | Junction PCR |
| Junction PCR Reverse primer | 5′-GAGACAGTGACCAACCATCC-3 | Junction PCR |
Figure 3. CD19 protein loss vs indel formation.
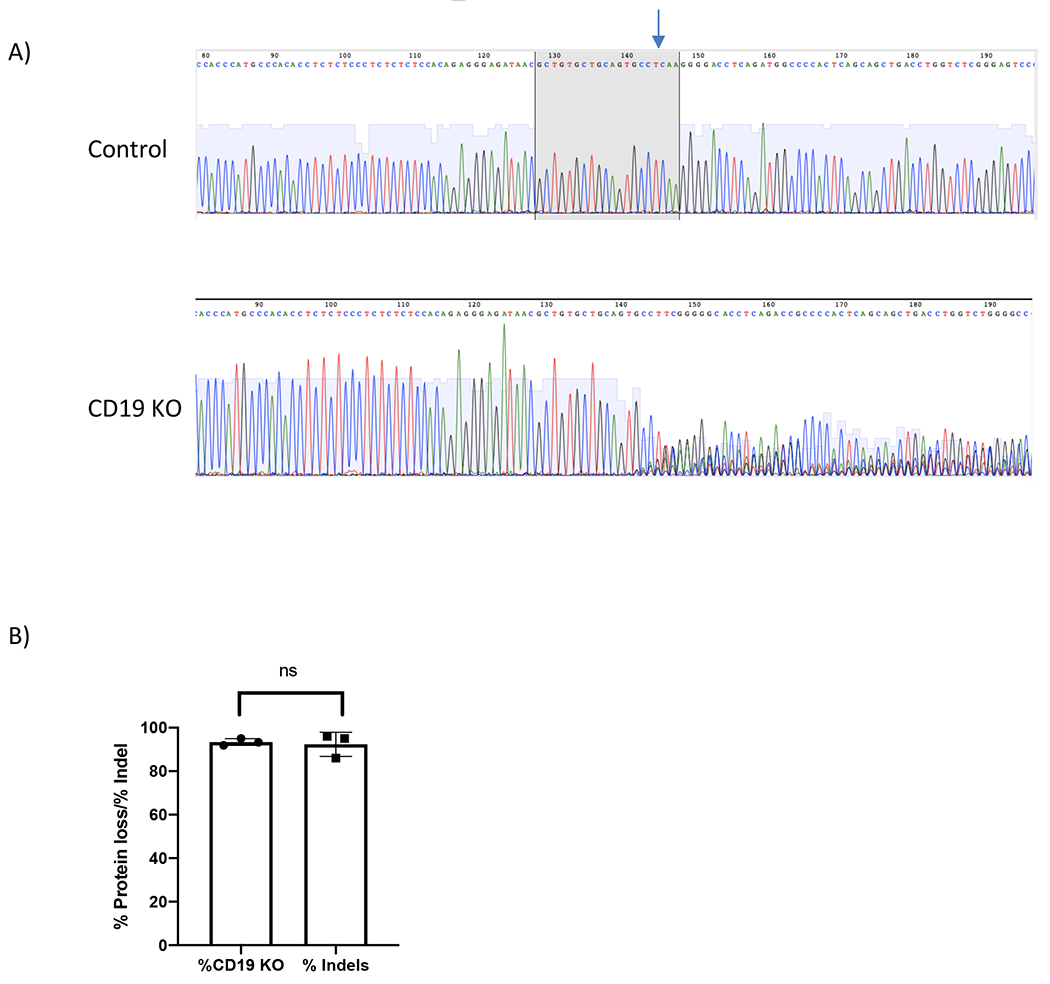
(A) Chromatograms depict sequencing peaks of control and CD19 KO sample. Gray box on the control peaks highlights the target sequence of CD19 gRNA with the predicted cut site indicated (blue arrow). CD19 KO showed “double peaks” sequencing around the predicted cut site. (B) Bar graph showing consistent results between % CD19 protein loss and % indels formation at CD19 locus (p≥0.05).
DISCUSSION:
Precise genome engineering in primary human B cells has been challenging until recently9–10. We previously published protocols using CRISPR/Cas9 to engineer primary human B cells9. Here, we outline improved protocols for B cell isolation, expansion, and engineering to allow for efficient KO of a gene of interest or for knocking-in a transgene. Here we demonstrate that high CD19 KO efficiency can be achieved by our improved protocol. We also demonstrate that CRISPR/Cas9 together with rAAV6 can be used to efficiently mediated targeted knocked-in and expression of EGFP at the AAVS1 locus.
This improved methods of efficiently knocking out any gene of interest can be used to study biological functions of the genes in B cells or to improve the efficacy of a therapeutic product. Similarly, the ability to KI a transgene can also be used for therapeutic purposes. This engineering method can be scaled up for larger numbers of B cells using larger commercially available cuvettes (data not shown).
In summary, we demonstrate comprehensive step-by-step processes from B cells isolation, expansion, and engineering that resulted in high gene modification efficiencies in B cells. This engineering method can be used to study B cells or use B cells as a novel cell-based therapies.
Table 4.
Flow cytometry antibody and viability dye
| Description | Fluorophore | Clone | Vender |
|---|---|---|---|
| Anti- human CD19 | PerCP | HIB19 | Biolegend |
| Viability Dye | eFlour 780 | - | eBiosciences |
ACKNOWLEDGMENTS:
This work was funded by the Children’s Cancer Research Fund (CCRF) and NIH R01 AI146009 to B.S.M.
Footnotes
DISCLOSURES:
A patent has been filed on the methods of making and using genome- edited B cells with M.J.J, K.L., and B.S.M. as inventors. B.S.M is a consultant for and owns stock in Immusoft Inc. Immusoft Inc has sponsored research in the lab of B.S.M.
REFERENCES:
- 1.Lebien TW, Thomas * & Tedder F B lymphocytes: how they develop and function. ashpublications.org (2008). doi: 10.1182/blood [DOI] [PMC free article] [PubMed]
- 2.Nutt SL, Hodgkin PD, Tarlinton DM & Corcoran LM The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 15, 160–171 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Slifka MK, Antia R, Whitmire JK & Ahmed R Humoral immunity due to long-lived plasma cells. Immunity 8, 363–72 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Spriggs MK et al. Recombinant human CD40 ligand stimulates B cell proliferation and immunoglobulin E secretion. J. Exp. Med. 176, 1543–1550 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fusil F et al. A Lentiviral Vector Allowing Physiologically Regulated Membrane-anchored and Secreted Antibody Expression Depending on B-cell Maturation Status. Mol. Ther. 23, 1734–1747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo XM et al. Engineering human hematopoietic stem/progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Blood 113, 1422–1431 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Mock U, Thiele R, Uhde A, Fehse B & Horn S Efficient Lentiviral Transduction and Transgene Expression in Primary Human B Cells. Hum. Gene Ther. Methods 23, 408–415 (2012). [DOI] [PubMed] [Google Scholar]
- 8.Heltemes-Harris LM et al. Sleeping Beauty transposon screen identifies signaling modules that cooperate with STAT5 activation to induce B-cell acute lymphoblastic leukemia. Oncogene 35, 3454–3464 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson MJ, Laoharawee K, Lahr WS, Webber BR & Moriarity BS Engineering of Primary Human B cells with CRISPR/Cas9 Targeted Nuclease. Sci. Rep 8, 12144 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hung KL et al. Engineering Protein-Secreting Plasma Cells by Homology-Directed Repair in Primary Human B Cells. Mol. Ther. 26, 456–467 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y, Xu J, Cheng M, Liao X & Peng S Review of CRISPR/Cas9 sgRNA Design Tools. Interdisciplinary Sciences: Computational Life Sciences 10, 455–465 (2018). [DOI] [PubMed] [Google Scholar]


