Abstract
Iron‐sulfur clusters are required in a variety of biological processes. Biogenesis of iron‐sulfur clusters includes assembly of iron‐sulfur clusters on scaffold complexes and transfer of iron‐sulfur clusters to recipient apoproteins by iron‐sulfur carriers, such as nitrogen‐fixation‐subunit‐U (NFU)‐type proteins. Arabidopsis thaliana has three plastid‐targeted NFUs: NFU1, NFU2, and NFU3. We previously discovered that nfu2 −/− nfu3 −/− mutants are embryo lethal. The lack of viable nfu2 −/− nfu3 −/− mutants posed a serious challenge. To overcome this problem, we characterized nfu2‐1 −/− nfu3‐2+/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants. Simultaneous loss‐of‐function mutations in NFU2 and NFU3 have an additive effect on the declines of 4Fe‐4S‐containing PSI core subunits. Consequently, the sesquimutants had much lower PSI and PSII activities, much less chlorophyll, and much smaller plant sizes, than nfu2‐1 and nfu3‐2 single mutants. These observations are consistent with proposed roles of NFU3 and NFU2 in the biogenesis of chloroplastic 4Fe‐4S. By performing spectroscopic and in vitro reconstitution experiments, we found that NFU1 may act as a carrier for chloroplastic 4Fe‐4S and 3Fe‐4S clusters. In line with this hypothesis, loss‐of‐function mutations in NFU1 resulted in significant declines in 4Fe‐4S‐ and 3Fe‐4S‐containing chloroplastic proteins. The declines of PSI activity and 4Fe‐4S‐containing PSI core subunits in nfu1 mutants indicate that PSI is the main target of NFU1 action. The reductions in 4Fe‐4S‐containing PSI core proteins and PSI activity in nfu3‐2, nfu2‐1, and nfu1 single mutants suggest that all three plastid‐targeted NFU proteins contribute to the biogenesis of chloroplastic 4Fe‐4S clusters. Although different insertion sites of T‐DNA lines may cause variations in phenotypic results, mutation severity could be an indicator of the relative importance of the gene product. Our results are consistent with the hypothesis that NFU3 contributes more than NFU2 and NFU2 contributes more than NFU1 to the production of 4Fe‐4S‐containing PSI core subunits.
Keywords: Arabidopsis thaliana, chloroplast, iron‐sulfur clusters, photosynthesis, photosystem I
1. INTRODUCTION
Iron‐sulfur clusters are essential to all living organisms, because they participate in a variety of biological processes, such as oxidation‐reduction reactions, photosynthesis, respiration, and DNA metabolism (Kohbushi et al., 2009). Due to the varying redox potential of iron, iron‐sulfur clusters have the ability to transfer electrons (Beinert, 2000). In higher plants, iron‐sulfur clusters are best known for participating in photosynthetic electron transport in the chloroplast thylakoid membranes and respiratory electron transport in the inner mitochondrial membrane (Balk & Lobréaux, 2005; Balk & Pilon, 2011; Balk & Schaedler, 2014; Couturier et al., 2013; Johnson et al., 2005; Pilon et al., 2006). Common types of iron‐sulfur clusters found in chloroplasts include 2Fe‐2S, 3Fe‐4S, and 4Fe‐4S (Lu, 2018). Examples of chloroplastic iron‐sulfur proteins include 2Fe‐2S‐containing chloroplast ferredoxin (cFD; Hanke & Mulo, 2013; Hase et al., 2006); 3Fe‐4S‐containing ferredoxin‐glutamine oxoglutarate aminotransferase (FD‐GOGAT; Coschigano et al., 1998); and 4Fe‐4S‐containing Photosystem I core proteins PsaA, PsaB, and PsaC (Psa stands for Photosystem I; Amann et al., 2004; Golbeck, 2003; Lezhneva et al., 2004; Saenger et al., 2002; Schwenkert et al., 2010; Vassiliev et al., 1998).
The biogenesis of chloroplastic iron‐sulfur clusters is a two‐step process (Balk & Pilon, 2011; Couturier et al., 2013; Lill, 2009; Lu, 2018; Przybyla‐Toscano et al., 2018). The first step is the assembly of iron‐sulfur clusters on a scaffold complex by the sequential action of cysteine desulfurase and sulfur transferase (Léon et al., 2002, 2005; Singh et al., 2013; Turowski et al., 2012). The second step is the transfer of newly assembled iron‐sulfur clusters from the scaffold complex to recipient apoproteins by iron‐sulfur carriers, such as nitrogen‐fixation‐subunit‐U (NFU)‐type proteins (Léon et al., 2003). NFU‐type proteins exist ubiquitously in eukaryotes and nitrogen‐fixing prokaryotes (Léon et al., 2003; Lill, 2009). In the higher plant Arabidopsis thaliana, there are three nuclear‐encoded plastid‐targeted NFU proteins: NFU1, NFU2, and NFU3 (Léon et al., 2003; Lu, 2018; Przybyla‐Toscano et al., 2018; Yabe et al., 2004). Full‐length NFU1, NFU2, and NFU3 contain a plastid transit peptide, a redox‐active NFU domain with a conserved CXXC motif (C stands for cysteine, X stands for any amino acid), and a redox‐inactive NFU domain (Léon et al., 2003; Lu, 2018; Przybyla‐Toscano et al., 2018; Yabe et al., 2004). The chloroplast localization of NFU1, NFU2, and NFU3 had been demonstrated by fluorescent protein tagging and confocal microscopic analysis (Léon et al., 2003; Nath et al., 2017; Roland et al., 2020).
NFU2 was proposed to participate in the biogenesis of chloroplastic 4Fe‐4S and 2Fe‐2S clusters (Berger et al., 2020; Gao et al., 2013, 2018; Hu et al., 2017; Touraine et al., 2004, 2019; Yabe et al., 2004, 2008). The loss‐of‐function nfu2‐1 mutant had lower amounts of 4Fe‐4S‐containing PsaA, PsaB, and PsaC and 2Fe‐2S‐containing cFD (Touraine et al., 2004; Yabe et al., 2004). The reduction in the amounts of 4Fe‐4S‐containing PSI core proteins resulted in reduced PSI and PSII activities and a reduced plant size in the nfu2‐1 mutant (Touraine et al., 2004; Yabe et al., 2004). These observations suggest that 4Fe‐4S‐containing PSI core proteins and 2Fe‐2S‐containing chloroplastic proteins are the main targets of NFU2 action. In line with the hypothesized role of NFU2 in the biogenesis of chloroplastic 2Fe‐2S and 4Fe‐4S clusters (Touraine et al., 2004), the recombinant NFU2 protein was found to accommodate 2Fe‐2S and 4Fe‐4S clusters and is capable of transferring 2Fe‐2S and 4Fe‐4S clusters to recipient apoproteins, such as cFD (Berger et al., 2020; Gao et al., 2013, 2018; Touraine et al., 2019; Yabe et al., 2004). Furthermore, in vitro reconstitution experiments demonstrated that the recombinant NFU2 protein has an iron‐sulfur scaffold function (Gao et al., 2013).
NFU3 was proposed to be involved in the biogenesis of chloroplastic 4Fe‐4S and 3Fe‐4S clusters (Nath et al., 2016, 2017; Touraine et al., 2019). The loss‐of‐function nfu3‐1 and nfu3‐2 mutants had reduced amounts of 4Fe‐4S‐containing PSI core proteins PsaA, PsaB, and PsaC (Nath et al., 2016; Touraine et al., 2019). The substantial declines of PSI core subunits in the nfu3‐1 and nfu3‐2 mutants resulted in reduced PSI and PSII activities and a reduced plant size (Nath et al., 2016, 2017; Touraine et al., 2019). Consistent with the hypothesized role of NFU3, the recombinant NFU3 protein displayed features of 4Fe‐4S and 3Fe‐4S clusters (Nath et al., 2016). In addition, in vitro reconstitution experiments indicated that the recombinant NFU3 protein has an iron‐sulfur scaffold function (Nath et al., 2016).
In this work, we investigated the functional relationships among three plastid‐targeted NFU proteins in Arabidopsis. It was previously found that double homozygous nfu2 −/− nfu3 −/− mutants had an embryo‐lethal phenotype (Nath et al., 2016; Touraine et al., 2019). Due to the lack of viable nfu2 −/− nfu3 −/− mutants, we analyzed the phenotypes of the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1 +/‐ nfu3‐2 −/− sesquimutants in this study. Specifically, we investigated the plant size, the total chlorophyll content, PSI activity, and PSII activity in the sesquimutants and compared to those in the single mutants. Because NFU2 and NFU3 were both proposed to participate in the biogenesis of 4Fe‐4S clusters in the chloroplast, we determined the amounts of 4Fe‐4S‐containing PSI core subunits in the sesquimutants and compared to those in the single mutants. We found that concurrent loss‐of‐function mutations in the NFU2 and NFU3 genes had an additive effect on the declines of 4Fe‐4S‐containing PSI core subunits. Unlike NFU2 or NFU3, the function of NFU1 in the biogenesis of chloroplastic iron‐sulfur clusters had been understudied. Therefore, we investigated whether the recombinant NFU1 protein contains 4Fe‐4S and 3Fe‐3S clusters and possesses an iron‐sulfur scaffold function. This was accomplished via spectroscopic analysis and in vitro iron‐sulfur cluster reconstitution experiments of the recombinant NFU1 protein. We also studied whether loss‐of‐function mutations in the NFU1 gene result in reductions in the plant size, the total chlorophyll content, PSI activity, and PSII activity. To further investigate the function of NFU1, we determined the amounts of representative 4Fe‐4S‐ and 3Fe‐4S‐containing chloroplastic proteins in the nfu1 mutants. Results from these analyses suggest that NFU1 participates in the biogenesis of chloroplastic iron‐sulfur clusters (e.g., 4Fe‐4S and 3Fe‐4S). The significant declines of 4Fe‐4S‐containing PSI core subunits in the nfu3‐2, nfu2‐1, and nfu1 single mutants suggest that all three plastid‐targeted NFU proteins contribute to the biogenesis of 4Fe‐4S clusters in the chloroplast. The relative magnitudes of reductions in the abundances of 4Fe‐4S‐containing PSI core proteins in these single mutants allowed us to hypothesize the relative contributions of NFU3, NFU2, and NFU1 to the biogenesis of chloroplastic 4Fe‐4S clusters.
2. MATERIALS AND METHODS
2.1. Plant materials and growth conditions
Arabidopsis (Arabidopsis thaliana) T‐DNA insertion lines nfu1‐1, nfu1‐2, nfu2‐1, and nfu3‐2 used in this study were obtained from the Arabidopsis Biological Resource Center (stock numbers GABI_661F04, SALK_038073, SALK_039254, and GABI_791C01, respectively). All four single mutants are in the Columbia ecotype (Alonso et al., 2003; Kleinboelting et al., 2012). The genotypes of the nfu1‐1, nfu1‐2, nfu2‐1, and nfu3‐2 single mutants and the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were confirmed by PCR, using the Phire Plant Direct PCR kit (Thermo Scientific) and genotyping primers listed in Table S1. Plants were grown in a growth chamber on a 12‐hr‐light/12‐hr‐dark photoperiod. The light intensity was 150 μmol photons m−2 s−1, the temperature was 20°C, and the relative humidity was 50%.
2.2. Quantitative RT‐PCR
Quantitative RT‐PCR (qRT‐PCR) was carried out as described by Clark and Lu (2015). Total RNA was extracted from rosette leaves using the RNeasy plant mini kit (QIAGEN), digested with the RNase‐Free DNase I (QIAGEN), and reverse‐transcribed with random primers and Moloney murine leukemia virus reverse transcriptase (Promega). qPCR was performed on a StepOnePlus Real‐Time PCR System (Thermo Fisher) with Power SYBR Green PCR master mix (Thermo Fisher), and qRT‐PCR primers listed in Table S1. The transcript level of NFU1 was normalized by the transcript level of ACT2 (At3g18780).
2.3. Measurement of chlorophyll and carotenoid contents
Chlorophyll and carotenoid were extracted with 80% acetone in 2.5 mM HEPES‐KOH, pH7.5 and the amounts (mg) of chlorophyll and carotenoid per gram of fresh tissues were determined on a BioMate 3S spectrophotometer (Thermo Scientific) as described by Wellburn (1994).
2.4. Measurements of PSI activity
Measurements of PSI activity (i.e., P700 photooxidation) were performed on dark‐adapted detached leaves as described previously (Nath et al., 2016). The redox state of P700 was determined by monitoring the absorbance change at 830 nm (with an 875‐nm reference beam) on the Dual‐PAM‐100 measuring system (Heinz Walz GmbH). Far‐red light‐induced P700 photooxidation (ΔA 830 nm) is calculated as the absorbance change before and after a 39‐s illumination of saturating far‐red light (720 nm at the maximal light intensity corresponding to level 20 in the Dual‐PAM setting). After reaching a steady‐state level of P700 photooxidation by far‐red light, single‐turnover and multiple‐turnover flash pulses of white saturating light were applied.
2.5. Measurement of room temperature chlorophyll fluorescence
Chlorophyll fluorescence parameters were measured on dark‐adapted plants at room temperature with the MAXI Version of the IMAGING‐PAM M‐Series chlorophyll fluorescence system (Heinz Walz GmbH), as described by Lu (2011). The maximum photochemical efficiency of PSII (Fv/Fm) is calculated using the following equation: Fv/Fm = (F m – F o)/F m, where F v, F m, and F o are variable, maximal, and minimal fluorescence of dark‐adapted leaves, respectively.
2.6. Isolation of thylakoid membranes
Thylakoid membranes were isolated as described in Lu et al. (2011) with minor modifications. The entire aerial portion of plants (~2 g) was excised and ground into a fine powder in liquid nitrogen with a mortar and pestle. Freshly made grinding buffer (50 mM HEPES‐KOH, pH7.5, containing 330 mM sorbitol, 2 mM EDTA, 1 mM MgCl2, 5 mM ascorbate, 0.05% bovine serum albumin, 10 mM NaF, and 0.25 mg/ml of Pefabloc SC protease inhibitor) was added to the frozen powder (~25 ml/g tissues) and the sample was further homogenized by the repeated swirling of the pestle. The resulting homogenate was filtered through a layer of miracloth (EMD Millipore) and centrifuged at 2,500 g for 4 min at 4°C using a swing‐bucket rotor. The pellet was resuspended and centrifuged in resuspension buffer I (50 mM HEPES‐KOH, pH 7.5, containing 5 mM sorbitol, 10 mM NaF, and 0.25 mg/ml of Pefabloc SC). The resulting thylakoid pellet was resuspended and centrifuged in resuspension buffer II (50 mM HEPES‐KOH, pH 7.5, containing 100 mM sorbitol, 10 mM MgCl2, 10 mM NaF, and 0.25 mg/ml of Pefabloc SC. The final pellet was resuspended in a small volume of resuspension buffer II (~1 ml/2 g starting tissues). The chlorophyll in 20 µl of resuspended thylakoid membranes was extracted with 0.98 ml of 80% acetone in 2.5 mM HEPES‐KOH (pH 7.5) and the amount of chlorophyll was determined on a BioMate 3S spectrophotometer (Thermo Scientific) as described by Wellburn (1994). The remaining suspension was frozen in liquid nitrogen and stored at −80°C for further use.
2.7. Extraction of leaf total proteins
Leaf total proteins were extracted as described previously (Hackett et al., 2017). Mature rosette leaves (~50 mg) were excised, frozen in liquid nitrogen, and ground into fine powder with stainless steel beads and TissueLyser II (Qiagen). Freshly made plant protein extraction buffer (50 mM Tris‐HCl, pH 7.5, 150 mM NaCl, 1% Triton X‐100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA, 1 mM DTT, and 1% of plant protease inhibitor cocktail) was added to the frozen powder at 5 μl/mg tissues. The samples were further homogenized with TissueLyser II. The resulting homogenates were centrifuged at >10,000 g for 3 min at 4°C. The supernatants were transferred to new microfuge tubes and centrifuged again at >10,000 g for 3 min at 4°C to remove residual tissue debris. The protein concentrations were determined using the DC (Detergent Compatible) protein assay (Bio‐Rad) with 0 to 1.4 mg/ml of bovine serum albumin as standards.
2.8. SDS‐PAGE and immunoblot analysis
SDS‐PAGE and immunoblot analysis of thylakoid membrane proteins was carried out as described previously (Nath et al., 2016). Thylakoid membrane proteins from the wild type, nfu2 and nfu3 single mutants and sesquimutants were loaded on an equal fresh tissue weight basis, because the nfu2 and nfu3 single mutants and sesquimutants have lower chlorophyll contents than the wild type. Thylakoid membrane proteins from the wild‐type and the nfu1 single mutants were loaded on an equal chlorophyll basis, because the nfu1 single mutants have the same amount of chlorophyll as the wild type. Leaf total proteins from the wild‐type and the nfu1 single mutants were loaded on an equal total protein basis. Proteins were separated with SDS‐PAGE (15% polyacrylamide; 6 M urea), using a Mini PROTEAN® Tetra Cell vertical gel electrophoresis system (Bio‐Rad). After electrophoresis, the proteins were transferred to a polyvinylidene difluoride membrane (EMD Millipore) using the Trans‐Blot electrophoresis transfer cell (Bio‐Rad). The membrane was incubated in the blocking solution (5% nonfat dry milk, 0.1% Tween‐20 in 1X Tris Buffered Saline), then in a diluted primary antibody solution. All the primary antibodies were purchased from Agrisera. Immunodetection of proteins on the polyvinylidene difluoride membrane was performed using the SuperSignal West Pico rabbit IgG detecting kit (Thermo Fisher) and analyzed by the Gel Logic 1500 Imaging System (Kodak).
2.9. Expression and purification of the recombinant NFU1 protein
Expression and purification of the recombinant NFU1 protein in Escherichia coli were performed as described by Lu et al. (2006) with minor modifications. Total leaf RNA was extracted, digested with RNase‐free DNase I, and reverse‐transcribed with oligo(dT)15 primers and Moloney murine leukemia virus reverse transcriptase. Full‐length NFU1 cDNA (NFU1 1‐696 bp, corresponding to NFU11‐231 AA) and NFU1 cDNA lacking the plastid transit peptide (NFU1 208‐696 bp, corresponding to NFU170‐231 AA) were amplified using the mRNA‐cDNA hybrid, Phusion High‐Fidelity DNA Polymerase (New England Biolabs), forward primers NFU1_BamH1_ATG and NFU1_BamH1_noTP, and a reverse primer NFU1_Xho1_TAG (Table S1). The resulting PCR products were AT‐cloned into the pGEM‐T Easy Vector (Promega) and sequenced to confirm the absence of PCR errors. BamH1/Xho1‐digested NFU1 fragments were subcloned into the pET28a expression vector (Novagen) and were expressed in E. coli strain Rosetta 2 (DE3; Novagen). An overnight culture of Rosetta 2 (DE3) harboring the NFU1 208‐696 bp gene was diluted 1:20 and grown at 37°C for 1 hr. Expression of the recombinant NFU170‐231 AA protein was induced with 1 mM isopropyl β‐D‐thiogalactoside and cells were grown at 28°C overnight. The recombinant NFU170‐231 AA protein was affinity‐purified with nickel‐nitrilotriacetic acid (Ni‐NTA) agarose under native and aerobic conditions according to the QIAexpressionist protocol (QIAGEN).
2.10. Absorption spectra of the as‐purified, reduced, and reconstituted recombinant NFU1 protein
Absorption spectroscopy of the recombinant NFU170‐231 AA protein was carried out as described previously (Nakamaru‐Ogiso et al., 2002; Schwenkert et al., 2010; Yabe & Nakai, 2006). The absorption spectrum (300–700 nm) of the affinity‐purified recombinant NFU170‐231 AA protein was recorded on a BioMate 3S spectrophotometer (Thermo Scientific) before and after treating the protein with 10 mM sodium dithionite, a reducing agent capable of reducing iron‐sulfur clusters. In vitro reconstitution of iron‐sulfur clusters on the recombinant NFU170‐231 AA protein was performed in a Bactron anaerobic chamber as described by Yabe and Nakai (2006). The as‐purified recombinant NFU170‐231 AA protein was incubated in 100 μM ammonium ferrous sulfate and 100 μM sodium sulfide at 25°C for 2 hr in degassed buffer containing 50 mM Tris‐HCl (pH 7.5), 50 mM NaCl, and 5 mM dithiothreitol. This is followed by a desalting step using an Illustra NAP‐10 column (GE Healthcare Life Sciences) and the absorption spectrum of the reconstituted recombinant NFU170‐231 AA protein was recorded.
3. RESULTS
3.1. Phenotypic characterization of the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants
As described previously, double homozygous nfu2 −/− nfu3 −/− mutants had an embryo‐lethal phenotype (Nath et al., 2016; Touraine et al., 2019). Due to the lack of viable nfu2 −/− nfu3 −/− mutants, it was challenging to investigate the functional relationship between NFU2 and NFU3. To overcome this problem, we isolated the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants. We crossed the homozygous nfu2‐1 mutant (Touraine et al., 2004; Yabe et al., 2004) with the homozygous nfu3‐2 mutant (Nath et al., 2016, 2017). The resulting F1 population was screened for double heterozygous nfu2 +/‐ nfu3 +/‐ mutants. Genotyping was performed by amplifying DNA from 2‐week‐old seedlings, using the Phire Plant Direct PCR kit (Thermo Scientific) and genotyping primers listed in Table S1. The self‐fertilized segregating F2 seeds were harvested from double heterozygous nfu2 +/‐ nfu3 +/‐ plants and were then sown on the soil. The resulting F2 seedlings were genotyped to screen for the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants. The genotype of each nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutant plant was carefully confirmed, prior to plant imaging, chlorophyll fluorescence imaging, or tissue harvesting. As shown in Figure 1, the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants had a pale green color and were much smaller than the nfu2‐1 and nfu3‐2 single mutants at the same age. These data demonstrate that the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants suffer further delays in plant growth and development than the nfu2‐1 and nfu3‐2 single mutants.
FIGURE 1.

Images of 4‐week‐old wild type, nfu2 and nfu3 single mutants and sesquimutants grown on a 12‐hr light/12‐hr dark photoperiod with an irradiance of 150 μmol photons m−2 s−1 during the light period. WT, wild type. All five plant images are on the same scale
3.2. The nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants displayed lower chlorophyll contents and PSI and PSII activities than the single mutants
To inspect whether the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants suffer additional decreases in chlorophyll, we determined total chlorophyll contents in 6‐week‐old plants (Figure 2). As shown in Figure 2a, the levels of total chlorophyll in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were ~47% lower than that in the wild type and ~18% lower than those in the nfu2‐1 and nfu3‐2 single mutants.
FIGURE 2.
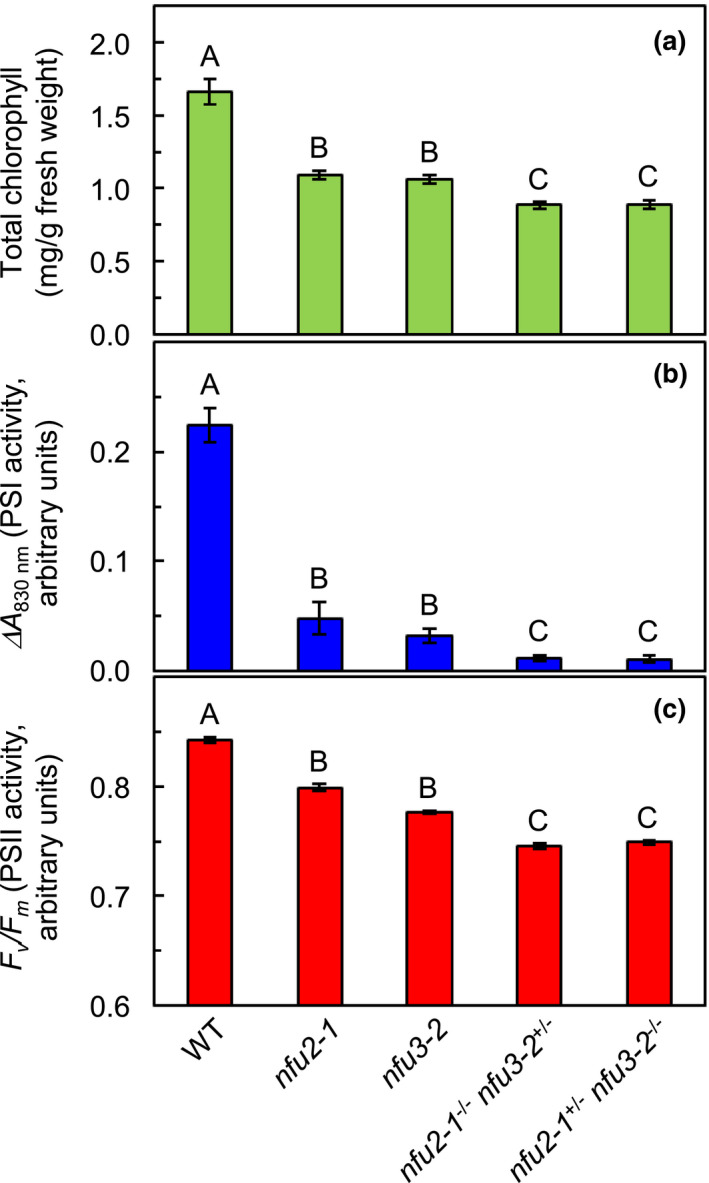
Total chlorophyll content, PSI activity, and PSII activity in the wild type, nfu2 and nfu3 single mutants and sesquimutants. (a) Total chlorophyll content. (b) PSI activity. (c) PSII activity. Values not connected by the same upper‐case letter are significantly different (Student's t test, p < .05). WT, wild type. Note that the Y‐axis in C starts from 0.6 to illustrate the differences between different genotypes. Six‐week‐old plants were used for chlorophyll, PSI activity, and PSII activity measurements in this figure
To investigate the molecular mechanism for the additional growth retardation in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants, we assessed PSI activity in 6‐week‐old plants. This was achieved by measuring far‐red light‐induced photooxidation of P700, where P700 is the PSI reaction‐center chlorophyll a molecule whose absorption spectrum peaks at 700 nm (Baker et al., 2007). As shown in Figure 2b, PSI activity in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants was 95% lower than that in the wild type, 77% lower than that in the nfu2‐1 single mutant, and 66% lower than that in the nfu3‐2 single mutant. This shows that the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants suffer further reductions in PSI activity than the nfu2‐1 and nfu3‐2 single mutants.
We also performed chlorophyll fluorescence measurements to determine the activity of PSII in 6‐week‐old plants. As shown in Figure 2c, the maximum photochemical efficiency of PSII (F v /F m) in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants was 11% lower than that in the wild type, 6.5% lower than that in the nfu2‐1 single mutant, and 4% lower than that in the nfu3‐2 single mutant. This shows that the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants suffer additional declines in PSII activity than the nfu2‐1 and nfu3‐2 single mutants.
3.3. The abundances of PSI core subunits PsaA, PsaB, and PsaC in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were lower than those in the single mutants
To investigate why the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants displayed additional decreases in the plant size, total chlorophyll content, PSI activity, and PSII activity, we performed SDS‐PAGE and immunoblot analysis on 4Fe‐4S‐containing PSI reaction‐center proteins PsaA, PsaB, and PsaC and other photosynthetic proteins, on an equal fresh tissue weight basis (Figure 3; Table 1). As shown in Figure 3a and Table 1, the average abundance of PSI core subunits PsaA, PsaB, and PsaC in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants was 96% lower than that in the wild type, 82% lower than that in the nfu2‐1 single mutant, and 67% lower than that in the nfu3‐2 single mutant. This shows that the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants suffer further reductions in the amounts of PSI core subunits than the nfu2‐1 and nfu3‐2 single mutants. This observation also suggests that the additional decrease of PSI activity in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants may result from the additional declines of PSI core subunits in the sesquimutants. Unlike PSI core proteins, the abundances of PSI‐associated light‐harvesting complex I proteins LHCA2 and LHCA4 in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were not substantially lower than those in the nfu2‐1 and nfu3‐2 single mutants (Figure 3a; Table 1). Similarly, the amounts of PSII core subunits D1, CP43, and CP47 and PSII‐associated light‐harvesting complex II proteins LHCB2 and LHCB4 in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were not substantially lower than those in the nfu2‐1 and nfu3‐2 single mutants (Figure 3b; Table 1). Therefore, the additional reductions of PSII activity in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants are likely a secondary effect of additional declines in PSI core subunits PsaA, PsaB, and PsaC in the sesquimutants.
FIGURE 3.
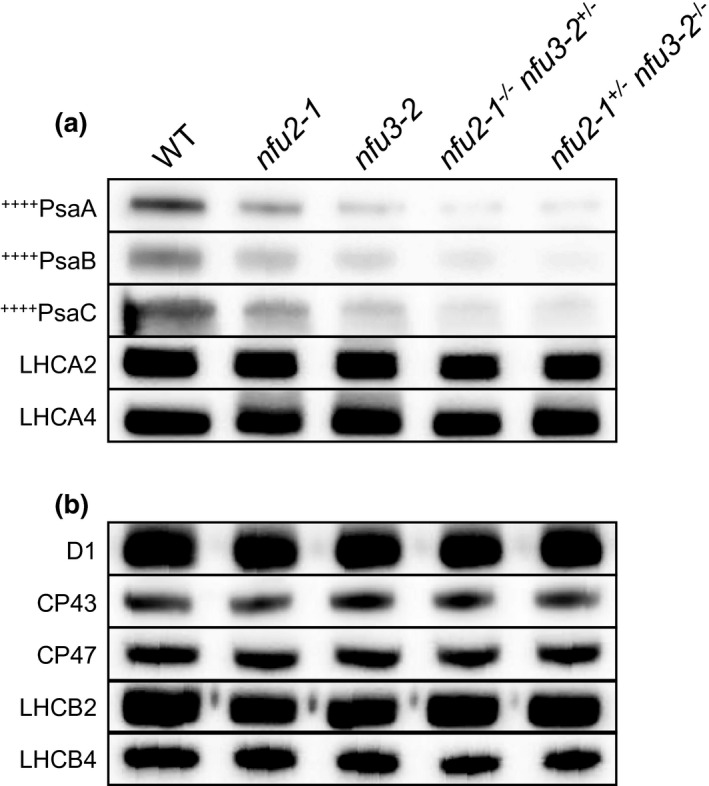
Immunoblots of representative PSI and PSII proteins in the wild type, nfu2 and nfu3 single mutants and sesquimutants. (a) Immunoblots of PSI reactive‐center proteins PsaA, PsaB, and PsaC, and PSI‐associated light‐harvesting complex I proteins LHCA2 and LHCA4. (b) Immunoblots of PSII reaction‐center proteins D1, CP43, and CP47 and PSII‐associated light‐harvesting complex II proteins LHCB2 and LHCB4. Thylakoid membrane proteins were used in (a‐b) and were loaded on an equal fresh tissue weight basis. Symbol ++++ indicates that the protein binds to 4Fe‐4S clusters. Five‐week‐old plants were used for SDS‐PAGE and immunoblot analysis in this figure
TABLE 1.
Relative abundances of representative PSI and PSII proteins in the wild type, nfu2 and nfu3 single mutants and sesquimutants
| WT | nfu2−1 | nfu3−2 | nfu2−1 −/− nfu3−2 +/‐ | nfu2−1 +/‐ nfu3−2 −/− | |
|---|---|---|---|---|---|
| PsaA++++ | 1.000 ± 0.040A | 0.286 ± 0.018B | 0.113 ± 0.010C | 0.025 ± 0.006D | 0.023 ± 0.006D |
| PsaB++++ | 1.000 ± 0.041A | 0.224 ± 0.030B | 0.124 ± 0.012C | 0.036 ± 0.005D | 0.023 ± 0.006D |
| PsaC++++ | 1.000 ± 0.060A | 0.175 ± 0.032B | 0.133 ± 0.014BC | 0.067 ± 0.012C | 0.069 ± 0.021BC |
| LHCA2 | 1.000 ± 0.034A | 0.782 ± 0.023C | 0.878 ± 0.011B | 0.661 ± 0.010D | 0.728 ± 0.032CD |
| LHCA4 | 1.000 ± 0.120A | 0.643 ± 0.023B | 0.710 ± 0.062B | 0.581 ± 0.034B | 0.738 ± 0.016B |
| D1 | 1.000 ± 0.027A | 0.810 ± 0.033B | 0.873 ± 0.045B | 0.826 ± 0.016B | 0.832 ± 0.023B |
| CP43 | 1.000 ± 0.020A | 0.897 ± 0.027B | 1.013 ± 0.022A | 0.901 ± 0.011B | 0.885 ± 0.028B |
| CP47 | 1.000 ± 0.036A | 0.926 ± 0.074AB | 0.960 ± 0.030AB | 0.843 ± 0.033BC | 0.780 ± 0.051C |
| LHCB2 | 1.000 ± 0.062A | 0.651 ± 0.023B | 0.749 ± 0.032B | 0.899 ± 0.020A | 0.907 ± 0.019A |
| LHCB4 | 1.000 ± 0.032A | 0.818 ± 0.001B | 0.830 ± 0.020B | 0.650 ± 0.010C | 0.632 ± 0.045C |
The values (mean ± SE, n = 4) are given as ratios to the protein levels in the wild type (WT). Thylakoid membrane proteins for SDS‐PAGE and immunoblot analysis were loaded on an equal fresh tissue weight basis were used to determine the levels of proteins in this table. Symbols ++++ indicate 4Fe‐4S‐containing proteins. Values not connected by the same letter are significantly different (Student's t test, p < .05). Five‐week‐old plants were used for SDS‐PAGE and immunoblot analysis in this table.
Taken together, morphological, physiological, and biochemical characterizations of the nfu2 and nfu3 single mutants and sesquimutants demonstrate that simultaneous loss‐of‐function mutations in the NFU2 and NFU3 genes had additive effects on the abundances of 4Fe‐4S‐containing PSI‐core subunits, PSI activity, PSII activity, chlorophyll content, and plant size, and that NFU2 and NFU3 play major roles in the biogenesis of chloroplastic iron‐sulfur clusters.
3.4. The absorption spectrum of recombinant NFU1 showed features characteristic of 4Fe‐4S and 3Fe‐4S clusters
The NFU1 gene is predicted to encode a 231‐amino acid (AA) protein (Figure 4). As described previously, the coding region of NFU1 includes a plastid transit peptide (1‐69 AAs), a redox‐active NFU domain (90‐156 AAs) with the conserved CXXC motif, and a redox‐inactive NFU domain (168‐227 AAs) at the C‐terminus (Figure 4a). Fluorescent protein tagging and confocal microscopic analysis showed that NFU1 is targeted to the chloroplast (Léon et al., 2003; Roland et al., 2020). NFU1 was proposed to bind and deliver 4Fe‐4S clusters and possibly 3Fe‐4S clusters to recipient apoproteins (Roland et al., 2020). Therefore, we expressed the 6xHis‐tagged NFU1 protein in Escherichia coli strain Rosetta 2 (DE3) and purified the recombinant NFU1 protein with nickel‐charged agarose resin under native and aerobic conditions. The plastid transit peptide in NFU1 was removed to produce NFU170‐231 AA, to ensure protein solubility. The absorption spectrum of the as‐purified recombinant NFU170‐231 AA protein had a broad absorption peak around 410 nm (Figure 4b). This 410‐nm broad absorption peak is a feature of 4Fe‐4S and 3Fe‐4S clusters, not a feature of 2Fe‐2S clusters (Kennedy et al., 1984; Nakamaru‐Ogiso et al., 2002). Thus, the absorption spectrum of the recombinant NFU1 protein is consistent with the proposed role of NFU1 in the biogenesis of 4Fe‐4S and 3Fe‐4S clusters.
FIGURE 4.
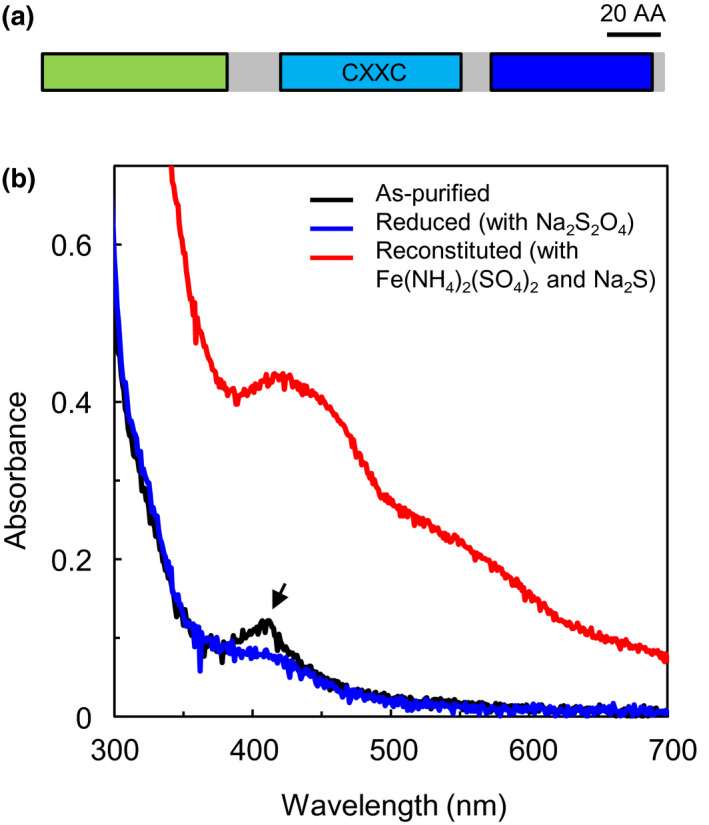
Spectroscopic and reconstitution assays of the recombinant NFU1 protein. (a) Domains in the full‐length NFU1 protein. The green box represents the plastid transit peptide; the cyan box represents the redox‐active NFU domain with the conserved CXXC motif; and the blue box represents the redox‐inactive NFU domain. AA, amino acids. (b) Absorption spectra of the as‐purified, reduced, and reconstituted recombinant NFU1 protein. Recombinant NFU170‐231 AA was purified aerobically, and an absorption spectrum was recorded (black line). The blue curve represents the absorption spectrum of recombinant NFU170‐231 AA after reduction with 10 mM sodium dithionite. The black arrow points to the absorption peak at 410 nm, a typical feature of 4Fe‐4S and 3Fe‐4S clusters, which disappears upon reduction by 10 mM sodium dithionite. The red curve represents the absorption spectrum of recombinant NFU170‐231 AA after reconstitution with ammonium ferrous sulfate and sodium sulfide
3.5. In vitro reconstitution indicated an iron‐sulfur scaffold function of NFU1
The 410‐nm broad absorption peak of the as‐purified NFU170‐231 AA protein disappeared after the addition of 1 mM reducing agent sodium dithionite (Figure 4b). This suggests that iron‐sulfur clusters bound to the as‐purified NFU170‐231 AA protein are redox‐sensitive (Nakamaru‐Ogiso et al., 2002). To test whether NFU1 has an iron‐sulfur scaffold function, we performed in vitro reconstitution of iron‐sulfur clusters on the recombinant NFU170‐231 AA protein, by applying the protein with an equimolar concentration of ferrous ion and sulfide. The 410‐nm broad absorption peak became more evident after the treatment (Figure 4b). The spectroscopic and in vitro reconstitution experiments demonstrated that NFU1 has an iron‐sulfur scaffold function, especially for 4Fe‐4S and 3Fe‐4S clusters.
3.6. Identification and phenotypic characterization of the T‐DNA insertion mutants nfu1‐1 and nfu1‐2
To study the function of NFU1, we isolated two homozygous T‐DNA insertion mutants of Arabidopsis: nfu1‐1 (GABI_661F04) and nfu1‐2 (SALK_038073; Figure 5). The nfu1‐1 and nfu1‐2 mutants carry the T‐DNA insertion in the first exon and the 50‐bp 5’‐untranslated region of At4g01940, respectively (Figure 5a). Quantitative reverse transcription (RT)‐PCR showed that the NFU1 transcript level is substantially decreased in the nfu1 mutants (Figure 5b). This confirms that nfu1‐1 and nfu1‐2 are loss‐of‐function mutants of the NFU1 gene. The nfu1‐1 mutant appeared to be phenotypically similar to the wild type (Figure 5c; Touraine et al., 2019). The nfu1‐2 mutant appeared to be smaller than the wild type (Figure 5c). It is unclear whether the subtle difference in the plant size among the two nfu1 mutants is due to allelic differences. In line with plant morphology, the aerial portion of the nfu1‐1 mutant had a similar fresh weight as the wild type, while the aerial portion of nfu1‐2 was significantly lighter than that of the wild type (Figure 5d).
FIGURE 5.
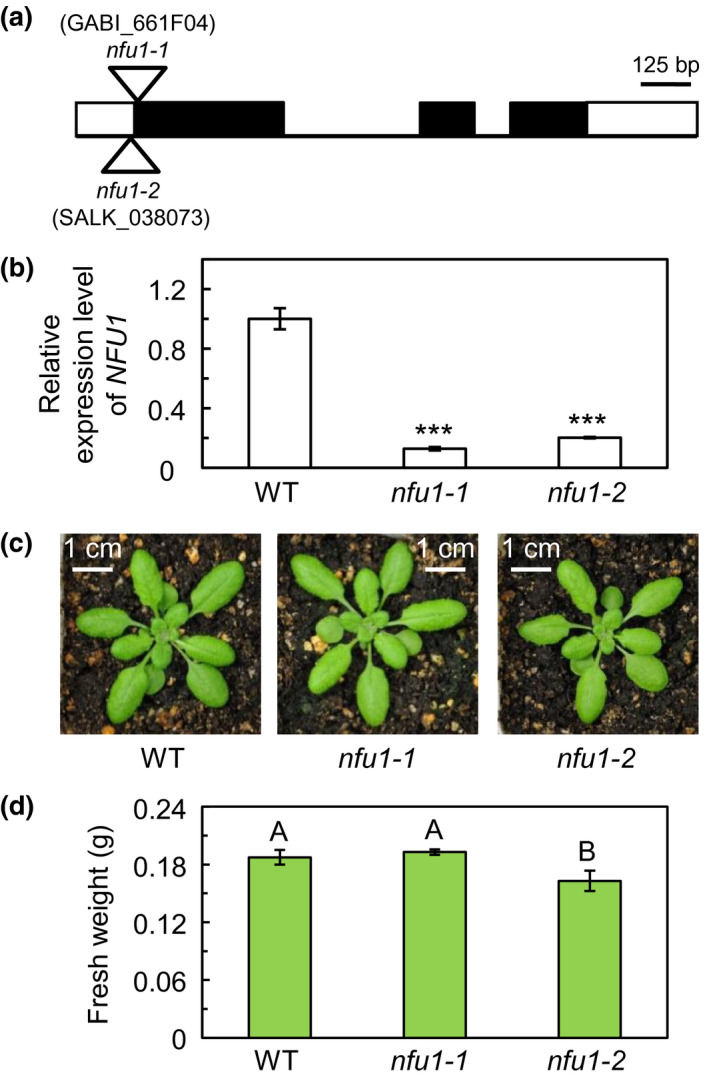
Identification and phenotypic characterization of the nfu1‐1 and nfu1‐2 mutants. (a) Structure of the NFU1 gene and locations of the nfu1‐1 and nfu1‐2 mutations. White boxes represent the untranslated regions; black boxes represent exons; and lines represent introns. The T‐DNA insertions in the nfu1‐1 and nfu1‐2 mutants are represented by triangles. bp, base pair. (b) Relative expression level of NFU1 determined by quantitative RT‐PCR in 4‐week‐old plants. The transcript level of NFU1 was normalized by the transcript level of ACT2 (At3g18780). The values (mean ± SE, n = 5) are given as ratios to the transcript level of NFU1 in the wild type. Asterisks indicate significant differences between the mutant and the wild type (WT; Student's t test; *, p < .05; **, p < .01; ***, p < .001). (c) Images of 4‐week‐old plants grown on a 12‐hr light/12‐hr dark photoperiod with an irradiance of 150 μmol photons m−2 s−1 during the light period. WT, wild type. (d) Fresh weight of the above‐ground portions of 4‐week‐old plants. Values are presented as mean ± SE (n = 7). Values not connected by the same upper‐case letter are significantly different (Student's t test, p < .05)
3.7. Pigment contents, PSI activity, and PSII activity in the nfu1‐1 and nfu1‐2 mutants
To further investigate the function of NFU1, we measured far‐red light‐induced photooxidation of P700, the PSI reaction‐center chlorophyll a molecule whose absorption peaks at 700 nm (Baker et al., 2007). This was achieved by monitoring the absorbance change in P700 at the wavelength of 830 nm, before and after a 39‐s saturating far‐red light illumination (Figure 6; Table 2). The far‐red light‐induced photooxidation of P700 in the nfu1‐1 and nfu1‐2 mutants was 10% and 22% lower than that in the wild type, respectively (Table 2), suggesting that loss‐of‐function mutations in the NFU1 gene cause decrease in PSI activity.
FIGURE 6.
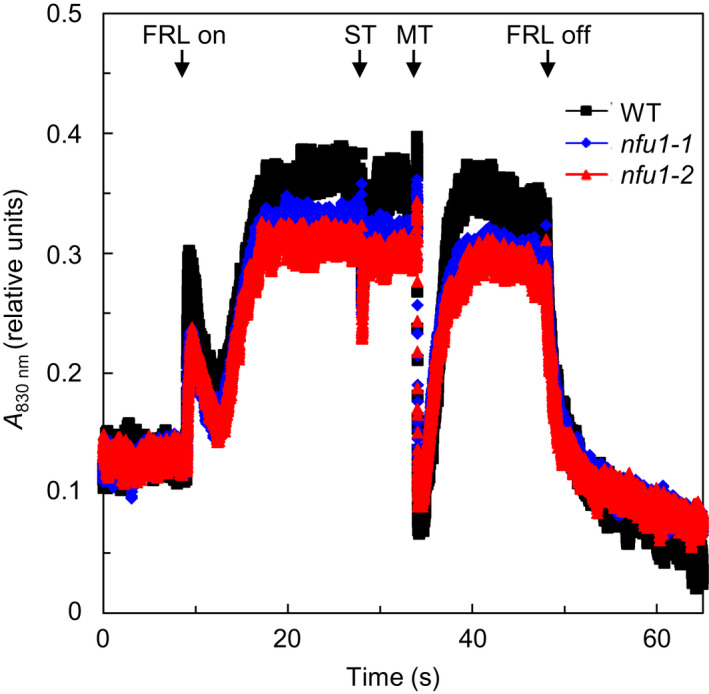
Kinetic measurements of PSI activity (P700 photooxidation) in the wild‐type and nfu1 mutants. Absorbance of P700 at 830 nm was used as a measurement of the PSI redox state. The measurement was performed on the Dual‐PAM‐100 measuring system (Heinz Waltz GmbH). Far‐red light‐induced P700 photooxidation (ΔA 830 nm) is calculated as the absorbance change before and after a 39‐s illumination of saturating far‐red light (FRL). After reaching a steady‐state level of P700 photooxidation by FRL, single‐turnover (ST) and multiple‐turnover (MT) flash pulses of white saturating light were applied. Representative traces are shown. Four‐week‐old plants were used for PSI activity measurements in this figure
TABLE 2.
Pigment contents and photosynthetic parameters in the wild‐type and nfu1 mutants
| WT | nfu1−1 | nfu1−2 | |
|---|---|---|---|
| ΔA 830 nm (PSI activity) | 0.216 ± 0.009A | 0.196 ± 0.012AB | 0.171 ± 0.007B |
| F v/F m (PSII activity) | 0.781 ± 0.002A | 0.782 ± 0.002A | 0.778 ± 0.003A |
| Total Chl (mg/g FW) | 1.267 ± 0.019A | 1.255 ± 0.019A | 1.211 ± 0.027A |
| Car (mg/g FW) | 0.302 ± 0.005A | 0.295 ± 0.006A | 0.287 ± 0.006A |
Measurements of P700 photooxidation were performed on dark‐adapted, detached leaves, with the Dual‐PAM‐100 measuring system (Heinz Waltz GmbH). Far‐red light‐induced P700 photooxidation (ΔA 830 nm) is calculated as the absorbance change before and after a 39‐s illumination of saturating far‐red light (720 nm). Measurements of chlorophyll fluorescence parameters were performed on dark‐adapted plants, with the IMAGING‐PAM M‐Series chlorophyll fluorescence system (Heinz Waltz GmbH). Chlorophyll and carotenoid were extracted and determined as described by Wellburn (1994). Data are presented as means ± SE (n = 6 for P700 photooxidation, n = 8 for chlorophyll fluorescence parameters, and n = 5 for pigment contents). Values not connected by the same letter are significantly different (Student's t test, p < .05). Four‐week‐old plants were used for PSI activity, PSII activity, and pigment measurements in this table.
Abbreviations: Chl, chlorophyll; Car, carotenoid; FW, fresh weight.
We also determined the maximum photochemical efficiency of PSII (F v /F m), the total chlorophyll content, and the carotenoid content in the nfu1 mutants and the wild type. Although F v /F m, the total chlorophyll content, and the carotenoid content in the nfu1 mutants were not statistically different from those in the wild type, they followed the same trend as PSI activity (Table 2).
3.8. The nfu1‐1 and nfu1‐2 mutants had reduced levels of 4Fe‐4S‐ and 3Fe‐4S‐containing chloroplastic proteins
To investigate the molecular mechanism for the reduced PSI activity in the nfu1 mutants, we extracted thylakoid membrane proteins and performed SDS‐PAGE and immunoblot analysis on several photosynthesis‐related proteins (Table 3; Figure S1). The level of PSI core protein PsaA in the nfu1‐1 and nfu1‐2 mutants was 7% and 13% lower than that in the wild type, respectively (Table 3). The average content of PSI core protein PsaC in the nfu1 mutants was ~ 25% lower than that in the wild type (Table 3). Although the amount of PSI core protein PsaB in the nfu1 mutants was not statistically different from that in the wild type, it followed the same trend as PsaA and PsaC (Table 3). The relative abundances of these three 4Fe‐4S‐containing PSI core subunits are consistent with the reduced PSI activity in the nfu1‐1 and nfu1‐2 mutants (Figure 6; Table 2). Unlike PSI core proteins, the amounts of PSII core subunits D1, CP43, and CP47 in the nfu1‐1 and nfu1‐2 mutants were not statistically different from those in the wild type (Table 3). This observation is in line with the lack of PSII‐related phenotype in the nfu1 mutants (Table 2). We also determined the relative abundances of two cytochrome b 6 f complex (Cyt b 6 f) proteins PetB and PetC (Photosynthetic electron transfer B and C). PetC is a Rieske iron‐sulfur protein, which contains a Rieske‐type 2Fe‐2S cluster (Hojka et al., 2014). The contents of PetB and PetC in the nfu1‐1 and nfu1‐2 mutants were statistically similar to those in the wild type (Table 3). Furthermore, we determined the relative abundances of cFD and FD‐GOGAT, which contain classic 2Fe‐2S and 3Fe‐4S, respectively (Coschigano et al., 1998; Hanke & Mulo, 2013). The level of 2Fe‐2S‐containing cFD in the nfu1 mutants did not appear to be statistically different from that in the wild type (Table 3). Interestingly, the abundance of 3Fe‐4S‐containing FD‐GOGAT in the nfu1‐1 and nfu1‐2 mutants was 10% and 5% lower than that in the wild type (Table 3).
TABLE 3.
Relative abundances of representative iron‐sulfur cluster‐containing proteins and other photosynthetic proteins in the wild‐type and nfu1 mutants
| WT | nfu1−1 | nfu1−2 | |
|---|---|---|---|
| PsaA++++ | 1.00 ± 0.03A | 0.93 ± 0.02AB | 0.87 ± 0.02B |
| PsaB++++ | 1.00 ± 0.05A | 0.87 ± 0.05A | 0.92 ± 0.03A |
| PsaC++++ | 1.00 ± 0.10A | 0.74 ± 0.06B | 0.75 ± 0.04B |
| D1 | 1.00 ± 0.05A | 1.12 ± 0.03A | 1.08 ± 0.04A |
| CP43 | 1.00 ± 0.06A | 1.06 ± 0.04A | 1.10 ± 0.03A |
| CP47 | 1.00 ± 0.04A | 1.04 ± 0.00A | 1.04 ± 0.02A |
| PetB | 1.00 ± 0.03A | 0.94 ± 0.02A | 0.91 ± 0.02A |
| PetC†† | 1.00 ± 0.10A | 1.08 ± 0.04A | 1.05 ± 0.03A |
| cFD++ | 1.00 ± 0.03A | 0.93 ± 0.02A | 0.98 ± 0.00A |
| FD‐GOGAT+++ | 1.00 ± 0.02A | 0.90 ± 0.03B | 0.95 ± 0.02AB |
Proteins were immunodetected as in Figure S1. The values (mean ± SE, n = 4) are given as ratios to the protein levels in the wild type (WT). Leaf total proteins loaded on an equal total protein basis were used to determine the abundances of cFD and FD‐GOGAT. Because the nfu1 mutants did not display changes in chlorophyll contents, thylakoid membrane proteins loaded on an equal chlorophyll basis were used to determine the levels of other proteins in this table. Symbols ++, ††, +++, and ++++ indicate that the protein binds classic 2Fe‐2S, Rieske‐type 2Fe‐2S, 3Fe‐4S, and 4Fe‐4S, respectively. Values not connected by the same letter are significantly different (Student's t test, p < .05). Four‐week‐old plants were used for SDS‐PAGE and immunoblot analysis in this table.
Taken together, spectroscopic and reconstitution analyses of the recombinant NFU1 protein plus morphological, physiological, and biochemical characterizations of the nfu1 mutants collectively demonstrate that NFU1 also participates in the biogenesis of chloroplastic iron‐sulfur clusters (e.g., 4Fe‐4S and 3Fe‐4S).
4. DISCUSSION
The Arabidopsis chloroplast contains three nuclear‐encoded NFU proteins: NFU1, NFU2, and NFU3 (Léon et al., 2003; Lu, 2018; Yabe et al., 2004). As discussed below, we showed that concurrent loss‐of‐function mutations in the NFU2 and NFU3 genes had an additive effect on the declines of 4Fe‐4S‐containing PSI core proteins. Consequently, the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1 +/‐ nfu3‐2 −/− sesquimutants had much lower PSI and PSII activities, much less chlorophyll, and a much smaller plant size than the nfu2‐1 and nfu3‐2 single mutants. Spectroscopic and reconstitution experiments of the recombinant NFU1 protein, as well as physiological and biochemical characterizations of the loss‐of‐function nfu1 mutants, suggest that NFU1 may act as a carrier for chloroplastic 4Fe‐4S and 3Fe‐4S clusters. Furthermore, we discussed the relative contributions of NFU3, NFU2, and NFU1 to the biogenesis of chloroplastic 4Fe‐4S clusters, according to the magnitudes of reductions of 4Fe‐4S‐containing PSI core proteins in the corresponding single mutants.
4.1. Simultaneous loss‐of‐function mutations in NFU2 and NFU3 have additive effects on 4Fe‐4S‐containing PSI core subunits, PSI activity, PSII activity, and mutant phenotypes
As shown in Figure 1, the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were much smaller than the nfu2‐1 and nfu3‐2 single mutants at the same age. Consistent with the smaller plant size and paler leaf color, the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants had significantly less chlorophyll than the nfu2‐1 and nfu3‐2 single mutants (Figure 2a). We also found that PSI and PSII activities in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were significantly lower than those in the nfu2‐1 and nfu3‐2 single mutants (Figure 2b,c). In line with the substantial reductions of PSI activity in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants, the abundances of 4Fe‐4S‐containing PSI core subunits PsaA, PsaB, and PsaC in these sesquimutants were substantially lower than those in the nfu2‐1 and nfu3‐2 single mutants (Figure 3a; Table 1). These observations collectively demonstrate that concurrent loss‐of‐function mutations in NFU2 and NFU3 have additive effects on the abundances of 4Fe‐4S‐containing PSI core subunits, PSI activity, PSII activity, and mutant phenotypes. This is probably because both NFU2 and NFU3 play major roles in the maturation of 4Fe‐4S‐containing PSI core subunits.
Interestingly, the magnitude of reduction in PSI activity in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants was much more evident than the magnitude of reduction in PSII activity (Figure 2b,c). This suggests that the decline of PSII activity in these sesquimutants is likely the result of the substantial reduction in PSI activity. Consistent with this hypothesis, the abundances of PSII core subunits D1 and CP43 in the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants were statistically similar to those in the single mutants (Figure 3b; Table 1). This is in sharp contrast to the substantial reductions of 4Fe‐4S‐containing PSI core subunits PsaA, PsaB, and PsaC in the sesquimutants (Figure 3a; Table 1). Such differential reductions in PSI and PSII activities and core protein abundances had also been observed in the nfu2 and nfu3 single mutants (Nath et al., 2016; Touraine et al., 2004). It is established that defects in PSI or intersystem electron transport may result in secondary reductions in PSII activity (Amann et al., 2004; Lennartz et al., 2001; Walters et al., 2004).
4.2. NFU1 contributes to the biogenesis of 4Fe‐4S and 3Fe‐4S clusters in the chloroplast
It was proposed that NFU1 may bind and deliver iron‐sulfur clusters (i.e., 4Fe‐4S and 3Fe‐4S) to recipient apoproteins (Roland et al., 2020). Using spectroscopic analysis, we found that the affinity‐purified recombinant NFU1 protein had a broad absorption peak at ~410 nm (Figure 4b), a feature of 4Fe‐4S and 3Fe‐4S clusters (Kennedy et al., 1984; Nakamaru‐Ogiso et al., 2002). Such feature had also been observed in NFU3, a known carrier for 4Fe‐4S and 3Fe‐4S clusters in the chloroplast (Nath et al., 2016, 2017). As shown in Figure 4b, the 410‐nm broad absorption peak of the as‐purified NFU1 protein disappeared after the addition of the reducing agent sodium dithionite, which suggests that the iron‐sulfur clusters bound to NFU1 are redox‐labile (Nakamaru‐Ogiso et al., 2002). Furthermore, in vitro reconstitution experiments showed that NFU1 has an iron‐sulfur scaffold function. These experiments led to the hypothesis that NFU1 may act as a carrier in the biogenesis of 4Fe‐4S and 3Fe‐4S clusters in the chloroplast. Consistent with this hypothesis, loss‐of‐function mutations in the NFU1 gene resulted in reductions in the levels of 4Fe‐4S‐containing PSI core subunits (e.g., PsaA and PsaC) and 3Fe‐4S‐containing FD‐GOGAT (Table 3; Figure S1). The nfu1‐1 and nfu1‐2 mutants displayed an average of 13% reduction in these 4Fe‐4S‐ and 3Fe‐4S‐containing chloroplastic proteins.
4.3. PSI is a target of NFU1 action
The nfu1‐1 and nfu1‐2 mutants showed an average of 15% reduction in PSI core subunits such as PsaA and PsaC (Table 3). In line with the declines in PSI core subunits, the nfu1‐1 and nfu1‐2 mutants displayed a ~16% reduction in PSI activity (Figure 6; Table 2). These data suggest that NFU1 contributes to the biogenesis of 4Fe‐4S clusters present in PSI core subunits and that PSI is a target of NFU1 action. Such reductions in PSI core subunit abundances and PSI activity did not result in noticeable declines in PSII activity or total chlorophyll content (Table 2), presumably due to the relatively modest magnitude of the reduction. Although immunoblot analysis and PSI activity measurements of the nfu1 mutants (Figure 6; Tables 2 and 3) suggest that NFU1 participates in the maturation of 4Fe‐4S‐containing PSI core subunits, we should not exclude the possibility of NFU1 being involved in the maturation of other 4Fe‐4S‐containing proteins in the chloroplast. Bimolecular fluorescence complementation assays showed that NFU1 may interact with other chloroplastic 4Fe‐4S‐containing proteins (Berger et al., 2020).
4.4. Relative contributions of NFU1, NFU2, and NFU3 in the biogenesis of chloroplastic 4Fe‐4S clusters
The nfu1‐1 and nfu1‐2 mutants displayed an average of 15% reduction in the abundances of 4Fe‐4S‐containing PSI core subunits (Table 3). In this study, the nfu2‐1 mutant displayed an average of 77% reduction in the abundances of such PSI core subunits, and the nfu3‐2 mutant displayed an average of 88% reduction (Table 1). Similar trends in the abundances of PSI core subunits had been previously observed in the nfu2‐1 and nfu3 single mutants (Nath et al., 2016; Touraine et al., 2004). These results suggest that all three plastid‐targeted NFU proteins contribute to the biogenesis of 4Fe‐4S clusters in the chloroplast. Although different locations of T‐DNA insertions in a gene may result in phenotypic variations, mutation severity is generally a good indicator of the relative importance of the gene product. The relative magnitudes of reductions in 4Fe‐4S‐containing PSI core proteins in the loss‐of‐function mutants are consistent with the hypothesis that NFU3 contributes more than NFU2 and NFU2 contributes more than NFU1. In line with the presumed contributions of the three plastid‐targeted NFU proteins, the nfu3‐2, nfu2‐1, and nfu1 single mutants displayed 86%, 78%, and 16% reduction in PSI activity, respectively (Figure 2b; Table 2). The relative contributions of three plastid‐targeted NFU proteins in the biogenesis of chloroplastic 4Fe‐4S clusters could reflect their protein expression levels in photosynthetic tissues. According to immunoblot analysis, the three plastid‐targeted NFU proteins are differentially expressed in different tissues (Touraine et al., 2019; Yabe et al., 2004). For example, the NFU2 protein is expressed in true leaves, cauline leaves, flower stalks, flowers, green siliques, and roots (Touraine et al., 2019; Yabe et al., 2004). The unique expression of NFU2 in the roots led to the discovery that NFU2 participates in the maturation of 2Fe‐2S‐containing dihydroxyacid dehydratase and is required for the synthesis of branched‐chain amino acids in Arabidopsis roots (Touraine et al., 2019). Unlike NFU2 or NFU3, the NFU1 protein appeared to be expressed at a very low level in leaves (Yabe et al., 2004).
5. CONCLUSIONS
As discussed above, concurrent loss‐of‐function mutations in the NFU2 and NFU3 genes have additive effects on the declines of 4Fe‐4S‐containing PSI core subunits. Thus, the nfu2‐1 −/− nfu3‐2 +/‐ and nfu2‐1+/‐ nfu3‐2 −/− sesquimutants had much lower PSI and PSII activities, much less chlorophyll, and a much smaller plant size than the nfu2‐1 and nfu3‐2 single mutants. These observations also suggest that NFU2 and NFU3 play major roles in the biogenesis of chloroplastic iron‐sulfur clusters. Spectroscopic and in vitro reconstitution experiments of the recombinant NFU1 protein led to the hypothesis that NFU1 may act as a carrier in the biogenesis of 4Fe‐4S and 3Fe‐4S clusters in the chloroplast. Consistent with this hypothesis, the loss‐of‐function nfu1 mutants displayed significant reductions in the abundances of 4Fe‐4S‐containing PSI core proteins and 3Fe‐4S‐containing FD‐GOGAT. The significant declines of PSI activity and 4Fe‐4S‐containing PSI core subunits in the nfu1 mutants suggest that PSI is a main target of NFU1. Results in this and previous studies showed that all three plastid‐targeted NFU proteins contribute to the biogenesis of 4Fe‐4S clusters in the Arabidopsis chloroplast. The relative magnitudes of reductions in 4Fe‐4S‐containing PSI core proteins in the loss‐of‐function mutants are in line with the hypothesis that NFU3 contributes more than NFU2 and NFU2 contributes more than NFU1. The relative contributions of NFU3, NFU2, and NFU1 to chloroplastic 4Fe‐4S biogenesis are consistent with the plant size, chlorophyll content, PSI activity, and PSI activity of the corresponding loss‐of‐function mutants.
ACCESSION NUMBERS
Sequence data of related genes/proteins can be found in the GenBank/EMBL databases under the following accession numbers: N FU1, At4g01940; NFU2, At5g49940; NFU3, At4g25910; and ACT2, At3g18780.
CONFLICTS OF INTERESTS
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
M.B.S. and Jun Zhao performed most of the experiments and analyzed most of the data; Jessica Zhang performed some experiments; F.Y. edited the article; Y.L. conceived the project, conducted some specific experiments, analyzed the corresponding data, and wrote and edited the article.
Supporting information
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Krishna Nath, James P. O'Donnell, Audrey M. Searing, and Michael J. Voyt (Western Michigan University [WMU]) for technical assistance, Christopher D. Jackson (WMU) for growth chamber management, and Jian Yao (WMU) for comments on experimental design.
Satyanarayan MB, Zhao J, Zhang J, Yu F, Lu Y. Functional relationships of three NFU proteins in the biogenesis of chloroplastic iron‐sulfur clusters. Plant Direct. 2021;5:e00303 10.1002/pld3.303
Manasa B. Satyanarayan, and Jun Zhao contributed equally to this work.
Funding information
This work was supported by the U.S. National Science Foundation under Grant Number MCB‐1244008 (to Y. L.).
DATA AVAILABILITY STATEMENT
All relevant data can be found within the manuscript and its supporting materials.
REFERENCES
- Alonso, J. M. , Stepanova, A. N. , Leisse, T. J. , Kim, C. J. , Chen, H. M. , Shinn, P. , Stevenson, D. K. , Zimmerman, J. , Barajas, P. , Cheuk, R. , Gadrinab, C. , Heller, C. , Jeske, A. , Koesema, E. , Meyers, C. C. , Parker, H. , Prednis, L. , Ansari, Y. , Choy, N. , … Ecker, J. R. (2003). Genome‐wide insertional mutagenesis of Arabidopsis thaliana . Science, 301, 653–657. 10.1126/science.1086391 [DOI] [PubMed] [Google Scholar]
- Amann, K. , Lezhneva, L. , Wanner, G. , Herrmann, R. G. , & Meurer, J. (2004). ACCUMULATION OF PHOTOSYSTEM ONE1, a member of a novel gene family, is required for accumulation of [4Fe‐4S] cluster‐containing chloroplast complexes and antenna proteins. The Plant Cell, 16, 3084–3097. 10.1105/tpc.104.024935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, N. R. , Harbinson, J. , & Kramer, D. M. (2007). Determining the limitations and regulation of photosynthetic energy transduction in leaves. Plant, Cell and Environment, 30, 1107–1125. 10.1111/j.1365-3040.2007.01680.x [DOI] [PubMed] [Google Scholar]
- Balk, J. , & Lobréaux, S. (2005). Biogenesis of iron‐sulfur proteins in plants. Trends in Plant Science, 10, 324–331. 10.1016/j.tplants.2005.05.002 [DOI] [PubMed] [Google Scholar]
- Balk, J. , & Pilon, M. (2011). Ancient and essential: The assembly of iron‐sulfur clusters in plants. Trends in Plant Science, 16, 218–226. 10.1016/j.tplants.2010.12.006 [DOI] [PubMed] [Google Scholar]
- Balk, J. , & Schaedler, T. A. (2014). Iron cofactor assembly in plants. Annual Review of Plant Biology, 65, 125–153. 10.1146/annurev-arplant-050213-035759 [DOI] [PubMed] [Google Scholar]
- Beinert, H. (2000). Iron‐sulfur proteins: Ancient structures, still full of surprises. JBIC Journal of Biological Inorganic Chemistry, 5, 2–15. 10.1007/s007750050002 [DOI] [PubMed] [Google Scholar]
- Berger, N. , Vignols, F. , Przybyla‐Toscano, J. , Roland, M. , Rofidal, V. , Touraine, B. , Zienkiewicz, K. , Couturier, J. , Feussner, I. , Santoni, V. , Rouhier, N. , Gaymard, F. , & Dubos, C. (2020). Identification of client iron–sulfur proteins of the chloroplastic NFU2 transfer protein in Arabidopsis thaliana . Journal of Experimental Botany, 71, 4171–4187. 10.1093/jxb/eraa166 [DOI] [PubMed] [Google Scholar]
- Clark, T. J. , & Lu, Y. (2015). Analysis of loss‐of‐function mutants in aspartate kinase and homoserine dehydrogenase genes points to complexity in the regulation of aspartate‐derived amino acid contents. Plant Physiology, 168, 1512–1526. 10.1104/pp.15.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coschigano, K. T. , Melo‐Oliveira, R. , Lim, J. , & Coruzzi, G. M. (1998). Arabidopsis gls mutants and distinct Fd‐GOGAT genes. Implications for photorespiration and primary nitrogen assimilation. The Plant Cell, 10, 741–752. 10.1105/tpc.10.5.741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier, J. , Touraine, B. , Briat, J. F. , Gaymard, F. , & Rouhier, N. (2013). The iron‐sulfur cluster assembly machineries in plants: Current knowledge and open questions. Frontiers in Plant Science, 4, 259 10.3389/fpls.2013.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Azam, T. , Randeniya, S. , Couturier, J. , Rouhier, N. , & Johnson, M. K. (2018). Function and maturation of the Fe‐S center in dihydroxyacid dehydratase from Arabidopsis. Journal of Biological Chemistry, 293, 4422–4433. 10.1074/jbc.RA117.001592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, H. , Subramanian, S. , Couturier, J. , Naik, S. G. , Kim, S. K. , Leustek, T. , Knaff, D. B. , Wu, H. C. , Vignols, F. , Huynh, B. H. , Rouhier, N. , & Johnson, M. K. (2013). Arabidopsis thaliana Nfu2 accommodates [2Fe‐2S] or [4Fe‐4S] clusters and is competent for in vitro maturation of chloroplast [2Fe‐2S] and [4Fe‐4S] cluster‐containing proteins. Biochemistry, 52, 6633–6645. 10.1021/bi4007622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golbeck, J. H. (2003). The binding of cofactors to Photosystem I analyzed by spectroscopic and mutagenic methods. Annual Review of Biophysics and Biomolecular Structure, 32, 237–256. 10.1146/annurev.biophys.32.110601.142356 [DOI] [PubMed] [Google Scholar]
- Hackett, J. B. , Shi, X. , Kobylarz, A. T. , Lucas, M. K. , Wessendorf, R. L. , Hines, K. M. , Bentolila, S. , Hanson, M. R. , & Lu, Y. (2017). An Organelle RNA Recognition Motif protein is required for photosystem II subunit psbF transcript editing. Plant Physiology, 173, 2278–2293. 10.1104/pp.16.01623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke, G. U. Y. , & Mulo, P. (2013). Plant type ferredoxins and ferredoxin‐dependent metabolism. Plant, Cell and Environment, 36, 1071–1084. 10.1111/pce.12046 [DOI] [PubMed] [Google Scholar]
- Hase, T. , Schürmann, P. , & Knaff, D. B. (2006). The interaction of ferredoxin with ferredoxin‐dependent enzymes In Golbeck J. H. (Ed.), Photosystem I: The Light‐Driven Plastocyanin:Ferredoxin Oxidoreductase (pp. 477–498). Springer Netherlands; 10.1007/978-1-4020-4256-0_28 [DOI] [Google Scholar]
- Hojka, M. , Thiele, W. , Toth, S. Z. , Lein, W. , Bock, R. , & Schottler, M. A. (2014). Inducible repression of nuclear‐encoded subunits of the cytochrome b6f complex in tobacco reveals an extraordinarily long lifetime of the complex. Plant Physiology, 165, 1632–1646. 10.1104/pp.114.243741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X. , Kato, Y. , Sumida, A. , Tanaka, A. , & Tanaka, R. (2017). The SUFBC2D complex is required for the biogenesis of all major classes of plastid Fe‐S proteins. The Plant Journal, 90, 235–248. 10.1111/tpj.13483 [DOI] [PubMed] [Google Scholar]
- Johnson, D. C. , Dean, D. R. , Smith, A. D. , & Johnson, M. K. (2005). Structure, function, and formation of biological iron‐sulfur clusters. Annual Review of Biochemistry, 74, 247–281. 10.1146/annurev.biochem.74.082803.133518 [DOI] [PubMed] [Google Scholar]
- Kennedy, M. C. , Kent, T. A. , Emptage, M. , Merkle, H. , Beinert, H. , & Munck, E. (1984). Evidence for the formation of a linear [3Fe‐4S] cluster in partially unfolded aconitase. Journal of Biological Chemistry, 259, 14463–14471. 10.1016/S0021-9258(17)42622-6 [DOI] [PubMed] [Google Scholar]
- Kleinboelting, N. , Huep, G. , Kloetgen, A. , Viehoever, P. , & Weisshaar, B. (2012). GABI‐Kat SimpleSearch: New features of the Arabidopsis thaliana T‐DNA mutant database. Nucleic Acids Research, 40, D1211–1215. 10.1093/nar/gkr1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohbushi, H. , Nakai, Y. , Kikuchi, S. , Yabe, T. , Hori, H. , & Nakai, M. (2009). Arabidopsis cytosolic Nbp35 homodimer can assemble both [2Fe–2S] and [4Fe–4S] clusters in two distinct domains. Biochemical and Biophysical Research Communications, 378, 810–815. 10.1016/j.bbrc.2008.11.138 [DOI] [PubMed] [Google Scholar]
- Lennartz, K. , Plucken, H. , Seidler, A. , Westhoff, P. , Bechtold, N. , & Meierhoff, K. (2001). HCF164 encodes a thioredoxin‐like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. The Plant Cell, 13, 2539–2551. 10.1105/tpc.010245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon, S. , Touraine, B. , Briat, J. F. , & Lobréaux, S. (2002). The AtNFS2 gene from Arabidopsis thaliana encodes a NifS‐like plastidial cysteine desulphurase. The Biochemical Journal, 366, 557–564. 10.1042/bj20020322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Léon, S. , Touraine, B. , Briat, J.‐F. , & Lobréaux, S. (2005). Mitochondrial localization of Arabidopsis thaliana Isu Fe–S scaffold proteins. FEBS Letters, 579, 1930–1934. 10.1016/j.febslet.2005.02.038 [DOI] [PubMed] [Google Scholar]
- Léon, S. , Touraine, B. , Ribot, C. , Briat, J. F. , & Lobréaux, S. (2003). Iron‐sulphur cluster assembly in plants: Distinct NFU proteins in mitochondria and plastids from Arabidopsis thaliana . The Biochemical Journal, 371, 823–830. 10.1042/bj20021946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva, L. , Amann, K. , & Meurer, J. (2004). The universally conserved HCF101 protein is involved in assembly of [4Fe‐4S]‐cluster‐containing complexes in Arabidopsis thaliana chloroplasts. The Plant Journal, 37, 174–185. 10.1046/j.1365-313X.2003.01952.x [DOI] [PubMed] [Google Scholar]
- Lill, R. (2009). Function and biogenesis of iron‐sulphur proteins. Nature, 460, 831–838. 10.1038/nature08301 [DOI] [PubMed] [Google Scholar]
- Lu, Y. (2011). The occurrence of a thylakoid‐localized small zinc finger protein in land plants. Plant Signal Behav, 6, 1181–1185. 10.4161/psb.6.12.18022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. (2018). Assembly and transfer of iron–sulfur clusters in the plastid. Front Plant Sci, 9, 336 10.3389/fpls.2018.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. , Hall, D. A. , & Last, R. L. (2011). A small zinc finger thylakoid protein plays a role in maintenance of Photosystem II in Arabidopsis thaliana . The Plant Cell, 23, 1861–1875. 10.1105/tpc.111.085456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. , Steichen, J. M. , Yao, J. , & Sharkey, T. D. (2006). The role of cytosolic a‐glucan phosphorylase in maltose metabolism and the comparison of amylomaltase in Arabidopsis and Escherichia coli . Plant Physiology, 142, 878–889. 10.1104/pp.106.086850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamaru‐Ogiso, E. , Yano, T. , Ohnishi, T. , & Yagi, T. (2002). Characterization of the iron‐sulfur cluster coordinated by a cysteine cluster motif (CXXCXXXCX27C) in the Nqo3 subunit in the proton‐translocating NADH‐quinone oxidoreductase (NDH‐1) of Thermus thermophilus HB‐8. Journal of Biological Chemistry, 277, 1680–1688. 10.1074/jbc.M108796200 [DOI] [PubMed] [Google Scholar]
- Nath, K. , O'Donnell, J. P. , & Lu, Y. (2017). Chloroplastic iron‐sulfur scaffold protein NFU3 is essential to overall plant fitness. Plant Signal Behav, 12, e1282023 10.1080/15592324.2017.1282023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath, K. , Wessendorf, R. L. , & Lu, Y. (2016). A nitrogen‐fixing subunit essential for accumulating 4Fe‐4S‐containing Photosystem I core proteins. Plant Physiology, 172, 2459–2470. 10.1104/pp.16.01564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon, M. , Abdel‐Ghany, S. E. , Van Hoewyk, D. , Ye, H. , & Pilon‐Smits, E. A. (2006). Biogenesis of iron‐sulfur cluster proteins in plastids. Genetic Engineering, 27, 101–117. 10.1007/0-387-25856-6_7 [DOI] [PubMed] [Google Scholar]
- Przybyla‐Toscano, J. , Roland, M. , Gaymard, F. , Couturier, J. , & Rouhier, N. (2018). Roles and maturation of iron‐sulfur proteins in plastids. Journal of Biological Inorganic Chemistry, 23, 545–566. 10.1007/s00775-018-1532-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland, M. , Przybyla‐Toscano, J. , Vignols, F. , Berger, N. , Azam, T. , Christ, L. , Santoni, V. , Wu, H.‐C. , Dhalleine, T. , Johnson, M. K. , Dubos, C. , Couturier, J. , & Rouhier, N. (2020). The plastidial Arabidopsis thaliana NFU1 protein binds and delivers [4Fe‐4S] clusters to specific client proteins. Journal of Biological Chemistry, 295, 1727–1742. 10.1074/jbc.RA119.011034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenger, W. , Jordan, P. , & Krauss, N. (2002). The assembly of protein subunits and cofactors in Photosystem I. Current Opinion in Structural Biology, 12, 244–254. 10.1016/S0959-440X(02)00317-2 [DOI] [PubMed] [Google Scholar]
- Schwenkert, S. , Netz, D. J. , Frazzon, J. , Pierik, A. J. , Bill, E. , Gross, J. , Lill, R. , & Meurer, J. (2010). Chloroplast HCF101 is a scaffold protein for [4Fe‐4S] cluster assembly. The Biochemical Journal, 425, 207–214. 10.1042/BJ20091290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, H. , Dai, Y. , Outten, F. W. , & Busenlehner, L. S. (2013). Escherichia coli SufE sulfur transfer protein modulates the SufS cysteine desulfurase through allosteric conformational dynamics. Journal of Biological Chemistry, 288, 36189–36200. 10.1074/jbc.M113.525709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touraine, B. , Boutin, J. P. , Marion‐Poll, A. , Briat, J. F. , Peltier, G. , & Lobréaux, S. (2004). Nfu2: A scaffold protein required for [4Fe‐4S] and ferredoxin iron‐sulphur cluster assembly in Arabidopsis chloroplasts. The Plant Journal, 40, 101–111. 10.1111/j.1365-313X.2004.02189.x [DOI] [PubMed] [Google Scholar]
- Touraine, B. , Vignols, F. , Przybyla‐Toscano, J. , Ischebeck, T. , Dhalleine, T. , Wu, H.‐C. , Magno, C. , Berger, N. , Couturier, J. , Dubos, C. , Feussner, I. , Caffarri, S. , Havaux, M. , Rouhier, N. , & Gaymard, F. (2019). Iron–sulfur protein NFU2 is required for branched‐chain amino acid synthesis in Arabidopsis roots. Journal of Experimental Botany, 70, 1875–1889. 10.1093/jxb/erz050 [DOI] [PubMed] [Google Scholar]
- Turowski, V. R. , Busi, M. V. , & Gomez‐Casati, D. F. (2012). Structural and functional studies of the mitochondrial cysteine desulfurase from Arabidopsis thaliana . Molecular Plant, 5, 1001–1010. 10.1093/mp/sss037 [DOI] [PubMed] [Google Scholar]
- Vassiliev, I. R. , Jung, Y.‐S. , Yang, F. , & Golbeck, J. H. (1998). PsaC subunit of Photosystem I is oriented with iron‐sulfur cluster FB as the immediate electron donor to ferredoxin and flavodoxin. Biophysical Journal, 74, 2029–2035. 10.1016/S0006-3495(98)77909-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, R. G. , Ibrahim, D. G. , Horton, P. , & Kruger, N. J. (2004). A mutant of Arabidopsis lacking the triose‐phosphate/phosphate translocator reveals metabolic regulation of starch breakdown in the light. Plant Physiology, 135, 891–906. 10.1104/pp.104.040469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellburn, A. R. (1994). The spectral determination of chlorophyll a and chlorophyll b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. Journal of Plant Physiology, 144, 307–313. [Google Scholar]
- Yabe, T. , Morimoto, K. , Kikuchi, S. , Nishio, K. , Terashima, I. , & Nakai, M. (2004). The Arabidopsis chloroplastic NifU‐like protein CnfU, which can act as an iron‐sulfur cluster scaffold protein, is required for biogenesis of ferredoxin and Photosystem I. The Plant Cell, 16, 993–1007. 10.1105/tpc.020511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe, T. , & Nakai, M. (2006). Arabidopsis AtIscA‐I is affected by deficiency of Fe‐S cluster biosynthetic scaffold AtCnfU‐V. Biochemical and Biophysical Research Communications, 340, 1047–1052. 10.1016/j.bbrc.2005.12.104 [DOI] [PubMed] [Google Scholar]
- Yabe, T. , Yamashita, E. , Kikuchi, A. , Morimoto, K. , Nakagawa, A. , Tsukihara, T. , & Nakai, M. (2008). Structural analysis of Arabidopsis CnfU protein: An iron‐sulfur cluster biosynthetic scaffold in chloroplasts. Journal of Molecular Biology, 381, 160–173. 10.1016/j.jmb.2008.05.072 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Data Availability Statement
All relevant data can be found within the manuscript and its supporting materials.


