Abstract
Place a drop of pond water under the microscope, and you will likely find an ocean of extraordinary and diverse single-celled organisms called ciliates. This remarkable group of single-celled organisms wield microtubules, active systems, electrical signaling, and chemical sensors to build intricate geometrical structures and perform complex behaviors that can appear indistinguishable from those of macroscopic animals. Advances in computer vision and machine learning are making it possible to completely digitize and track the dynamics of complex ciliates and mine these data for the hidden structure, patterns, and motifs that are responsible for their behaviors. By deconstructing the diversity of ciliate behaviors in the natural world, themes for organizing and controlling matter at the microscale are beginning to take hold, suggesting new modular approaches for the design of autonomous molecular machines that emulate nature’s finest examples.
Cells are the most sophisticated molecular machines in existence, able to organize molecules and chemistry across multiple length and timescales to build extraordinary systems that move through, interact with, and respond to their environment like tiny robots. In contrast, our own ability to engineer dynamic and responsive systems at the microscale pales in comparison. Often it has been biology that has provided the critical insight and perspective needed to realize the best design practices and engineering approaches: enzymes revealed a path to build better catalysts (Knowles, 1991); spider silk inspired new approaches for synthetic textiles (Hinman et al., 2000); and the adaptive immune system provided diversification and selection strategies for generating potent binding molecules (Smith and Petrenko, 1997).
Here, I propose that the behavior of ciliates—a diverse group of unicellular protozoans whose bodies are covered with motile cilia and display a remarkable variety of morphologies, lifestyles, and animal-like functionalities—can provide a window into the systems biology that powers complex cellular dynamics and reveal strategies and blueprints for understanding and engineering complex autonomous microscopic machines. I review how the study of ciliate behavior evolved from early observational work to the critical recognition of specific patterns of behavior in Paramecium that could be targeted for genetic and molecular dissection. I point to advances in computer vision and machine learning that allow us to digitize the dynamic behavior of more complex and less tractable ciliates and identify the patterns of activity and motifs that drive their outputs. I propose that much of the diversity of ciliate behavior has been realized by a “modular toolkit” of structure, activity, and sensing, identifying new areas of focus for future investigation and suggesting design strategies that could ultimately be emulated in future engineering efforts to create a range of cell-like machines or devices.
Ciliate behavior: from “curious animalcules” to the genetics of specific behavioral motifs.
When Antonie van Leeuwenhoek first peered through his microscope and famously observed “curious animalcules” in the 1600s, it became clear that unicellular organisms like ciliates could build structures and organize their behavior to jump, contract, walk, forage, and even hunt on the same timescales that we do (van Leewenhoeck, 1677). As the field of microbiology developed over the next several hundred years, an ever increasing amount of observation and description of ciliates began to be catalogued: free-swimming cells ranging in length from fifty (Tetrahymena) to hundreds of microns (Paramecium) to even millimeter-sized “whales” that dwarf many multicellular animals (Spirostomum); sessile current-feeders shaped like flowers (Vorticella) or giant trumpets (Stentor); “cat-fish”-like cells that crawl or even “walk” along surfaces while scavenging food (Euplotes, Oxytricha); and even microscopic predators that use elaborate strategies to ensnare, capture, and prey on other ciliates as if in a microscopic Serengeti (Suctoria, Lacrymaria, Didinium; Woodruff, 1921; Noland and Finley, 1931; Rudzinska, 1973; Hara and Asai, 1980; Ishida and Shigenaka, 1988; Borror and Hill, 1995; Zoller et al., 2012; Slabodnick and Marshall, 2014; Ruehle et al., 2016; Coyle et al., 2019; Mathijssen et al., 2019). A high-resolution 4K video showing a sampling of our own field recordings of ciliates from the lakes of Wisconsin and their unique behaviors (along with hundreds of hours of additional related field video) has been included as a reference to illustrate the functional and structural diversity of ciliates and to pique the curiosity of the reader (https://youtu.be/v-KphVRo3dw).
While historical writings provide an almost unlimited treasure trove of vivid observation, the complexity of the described behaviors seems at odds with the limitations of single cells. No matter how “cognitive” such a behavior might appear, there must be a concrete mechanistic basis for the observed behavior in the language of molecules and chemistry. A key insight came from Jennings, who recognized consistent patterns in the behavior in Paramecium (Jennings, 1906). He observed that when a cell encountered a physical obstruction it would perform an “avoidance reaction,” swimming backward briefly to reorient before resuming forward swimming in a new direction. Through repeated cycles of this behavior, the cell can resolve its frustration with the obstacles it encounters in its world and carry on with its other functions.
The recognition of a specific “behavioral motif” underlying Paramecium’s navigation provided a key starting point for more comprehensive mechanistic inquiry. Close inspection of the cilia during the avoidance reaction revealed that the direction and frequency of the ciliary beat-form changed during the reaction. Using electrophysiology, the transition between these ciliary states was shown to be regulated by the membrane potential across the cell’s surface (Kinosita et al.,1964), and a number of physical and chemical stimuli were found to trigger transient depolarization and an associated calcium influx that flips the ciliary orientation and swimming direction (Naitoh, 1966; Naitoh and Eckert, 1969). Kung and colleagues realized that these activities must be controlled by specific molecules, and in a pioneering set of papers used genetics to isolate mutants of Paramecium with specific defects in their avoidance responses, such as pawn mutants that fail to express voltage-gated calcium channels and thus can only swim forward (Kung and Naitoh, 1973; Chang et al., 1974; Kung et al., 1975; Lodh et al., 2016).
Once dissected and deconstructed, the seemingly inquisitive, probative, and responsive motility of Paramecium begins to look more like how a simple robot vacuum cleaner’s behavior might be realized. This is not said to disparage the evolutionary achievement of this cell! On the contrary, the fact that the types of simple mechanisms that we might use to program a macroscopic robot have been realized at the microscale using familiar molecular systems like ion channels, microtubules, and motor proteins makes the deconstruction of this and other ciliate behaviors a potentially powerful tool for understanding and engineering behavior at these length scales. However, the approach used on Paramecium avoidance has traditionally been difficult to extend to other ciliate behaviors, which can involve rapid and complex morphology changes, unfold over minutes or hours instead of seconds, and may occur in cells that are recalcitrant to lab culture and genetic dissection. This has made it difficult to identify the patterns and structure that underlie sophisticated ciliate behaviors and link them to molecular activity.
Computer vision and machine learning: quantifying ciliate behavior and mining it for patterns
Advances in machine vision and computational analysis techniques are helping the field overcome these challenges, allowing us to reverse engineer the inner-workings of ciliate behavior. Arbitrarily long high-frame-rate microscopy videos of ciliates recorded from low-density cultures or even field samples can now be digitized into a variety of compact quantitative descriptions of behavior. For example, the position of individual swimming cells can be followed indefinitely to generate a behavior track that can follow changes in speed and orientation throughout the cell’s lifetime. For more complex morphologically driven behaviors, the complete subcellular anatomy and posture of the cell can be quantified over time, revealing how different structures move and deform as the cell interacts with its environments.
The resulting tracks of behavior provide hundreds of thousands of temporally correlated observations of cell behavior that are well-suited to mine for patterns and structure using statistical methods, dimensionality reduction techniques, or application of machine learning. Similar methods have had success identifying patterns in the physical world (Deyle and Sugihara, 2011; Gilpin, 2020), the statistics and rhythmicity in the beats of individual cilia (Wan and Goldstein, 2014; Sartori et al., 2016), and the behavior of higher animals like Caenorhabditis elegans and Zebrafish (Stephens et al., 2008, 2011; Girdhar et al., 2015; Ahamed et al., 2019). By combining these techniques with recent advances for obtaining transcriptomic or genomic sequences from small numbers of protist cells (Kolisko et al., 2014), the field is poised for an explosion of understanding in how diverse ciliate behaviors are encoded.
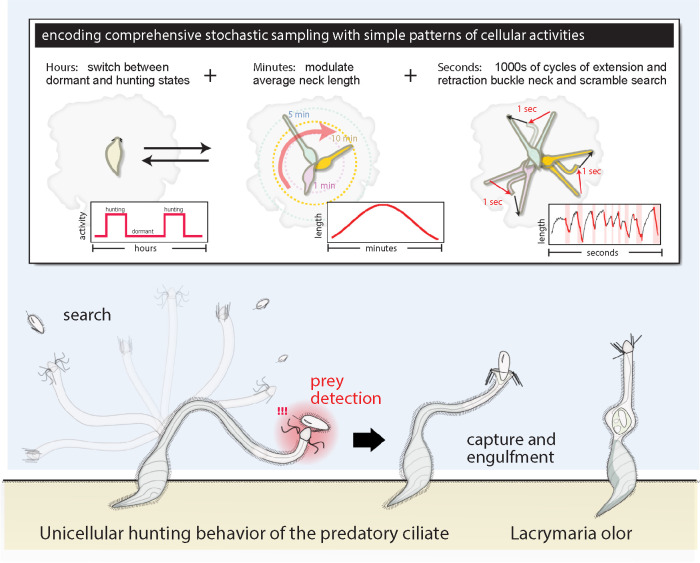
As a recent example, this approach was applied to analyze the hunting behavior of the highly distensible predatory ciliate Lacrymaria olor (Coyle et al., 2019). These fascinating cells attach their large bodies to debris in the environment and hunt for large prey by extending a slender neck structure hundreds of microns deep into the environment. The neck whips, bends, buckles, and darts about until the tip—a head-like structure that senses contact with prey—locates, strikes, and triggers engulfment of its prey targets (Figure 1 and Supplemental Video 1). The stunning morphological rearrangements and animal-like hunting behavior of these cells fascinated microbiologists for more than a century, but a quantitative understanding was lacking. Using a computer vision pipeline, the complete subcellular posture—body, head, and neck geometry—of actively hunting Lacrymaria was tracked and digitized over hours of real-time video, allowing a detailed statistical and computational analysis of the motion to be performed. Multiple distinguishable timescales of activity were found to contribute to the hunting behavior: extremely slow alternations between hunting and resting states over hours; slow timescale modulation of the neck length over minutes during active hunting; and rapid seconds timescale extension and contraction that buckle the neck and functioned to scramble the cell’s search trajectory (Figure 1).
FIGURE 1:
Lacrymaria hunting dynamics: encoding a complex cell behavior with patterns of different activities. Diagram of a Lacrymaria cell hunting for prey. Lacrymaria attaches a teardrop-shaped body to surfaces like debris and extends and whips its long slender neck throughout the local environment. When the tip of this structure hits and recognizes a suitable prey, it triggers the release of toxicysts and engulfment of the target (see also Supplemental Video 1). While the hunting appears animal-like, careful statistical analysis reveals this behavior is built up from multiple patterns of activity across different timescales that statistically encode comprehensive stochastic sampling of the environment to rapidly locate prey by chance alone.
Surprisingly, statistical analysis revealed that despite the animal-like appearance of its actions, Lacrymaria hunting was equivalent to rapid and stochastic sampling of the environment. Rather than carefully tracking and locating individual prey, the cell executes patterns of activity that collectively and probabilistically encode a comprehensive local search behavior that strikes at all nearby locations within a matter of minutes, maximizing chance encounter with nearby prey. Across different cells, changes to these patterns of activity appear to modulate the statistics, altering the duration, frequency, and search radius of the emergent hunting behavior. Thus, as with the Paramecium avoidance response, a careful analysis of behavior reveals hidden structure and simple motifs by which a more complex animal-like behavior emerges.
Paramecium and Lacrymaria behavior: similar mechanisms that produce different outcomes
Although Paramecium avoidance and Lacrymaria hunting appear quite different at the behavior level, there is an overlap in the mechanisms that drive these behaviors. In both cases, cycles of forward and reverse ciliary activity play a key role in driving the behavior. Given this, what contributes to the actual differences in the output behavior?
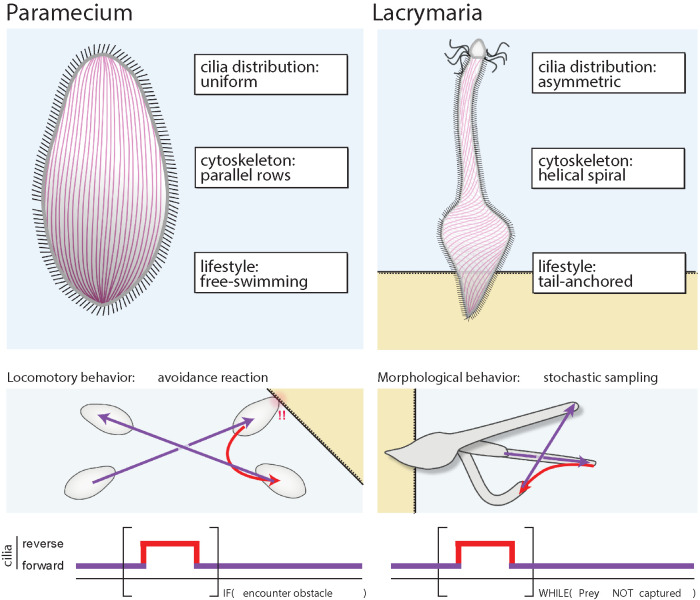
First, there are differences in the structure of the cell on which these activities operate and the associated geometrical and physical constraints. Paramecium is free-swimming with a cytoskeletal geometry that has little helical character and uniformly distributed cilia (Figure 2). In contrast, Lacrymaria anchors itself to surfaces and possesses a cytoskeletal geometry in which its microtubules are arranged in a helical spiral that resembles a slinky, with a powerful crown of cilia at the anterior head of the cell (Figure 2; Yanase et al., 2018; Coyle et al., 2019). Thus, while cycles of forward and backward ciliary activity drive Paramecium forward and backward, something different happens in Lacrymaria. The anchoring creates resistance to translation that prevents the cell from moving forward, and its helical cytoskeleton—much like a spring—begins to unwind in response to the applied force, allowing the neck to extend. Thus, the same activity that drives translation in Paramecium can instead produce morphological rearrangement in Lacrymaria.
FIGURE 2:
Paramecium and Lacrymaria use different combinations of structure, activity, and sensing to program unique behaviors from a similar toolbox. Diagram showing the different uses of cortical microtubule geometry (purple lines), ciliary distribution, and ciliary activity in Paramecium and Lacrymaria to encode distinct behaviors from a common toolbox of molecular components and activities. Paramecium is a free-swimming ciliate that arranges its cytoskeleton in parallel rows and distributes it cilia uniformly along these rows. Lacrymaria is a tail-anchored cell that arranges its cytoskeleton in a helical spiral with small cilia along these rows and a dense crown of longer cilia at the anterior end. Both cells use cycles of forward and reverse ciliary activity to exploit these differences and program distinct behaviors: stimulus-triggered reversal to encode a locomotive “avoidance reaction” in Paramecium; and continuous cycling in Lacrymaria to encode morphology-driven stochastic sampling to locate and strike prey.
Second, there are differences in the patterns of activity that are applied by the cell. Changes in the ciliary activity of Paramecium are a reaction to the environment. In contrast, when Lacrymaria is hunting, these cycles occur spontaneously and nearly constantly, performing thousands of cycles over several minutes to drive the stochastic environmental sampling needed to locate prey. Thus, the ground-state pattern of activity in Paramecium is to swim forward until a disruption triggers reversal, while in Lacrymaria the ground-state pattern is to constantly switch between forward and reverse modes. In this way, the same fundamental activities are deployed in different “programs” by the two cells (Figure 2).
Thirdly, there are differences in how sensing is incorporated into the regulation of these activity patterns. For Paramecium, the avoidance response acts as a generic behavior module that can be triggered by activation of different chemical, mechanical, or thermal sensors. This makes it a flexible system to resolve the range of conflicts it might encounter. For Lacrymaria’s predatory behavior, the cell must balance moving its neck through complicated and sometimes dangerous environments, while simultaneously being ready at a moment’s notice to capture detected prey. Like Paramecium, if Lacrymaria strikes an object in its environment that is not edible, it triggers ciliary reversal and instantaneous contraction of the neck—essentially an avoidance reaction. However, if the head strikes prey, the cell responds with toxicyst release, amplified ciliary activity, and prey engulfment. The identity of the prey-specific sensors and the mechanisms by which they trigger this distinct response are currently unknown, but the timescale suggests action through the excitable plasma membrane, not unlike the avoidance reaction. Thus, different sensors can signal to and regulate a cell’s ground-state pattern of ciliary activity and can facilitate specific organismal needs: coherent, guided motility in Paramecium; and random sampling and prey detection for hunting Lacrymaria (Figure 2).
Structure, activity, and sensing: a modular toolkit for ciliate behavior and beyond
These parallels suggest that the diversity of ciliate behaviors seen in the natural world may have been realized through different application of a “modular toolkit” available to them during evolution: structure, activity, and sensing. This echoes how different biochemical activities and targeting domains have been harnessed to diversify cell signaling outputs (Bhattacharyya et al., 2006; Coyle and Lim, 2016), or how the Hox genes have been used to diversify body plans in animals (Dassow and Munro, 1999).
The diverse geometries ciliates build lay out the overall cell structure and the locations of the active components that support different behaviors. It remains an open question in ciliate biology how these fantastic microtubule structures and geometries are created and maintained by the cell, but emerging model systems for cell geometry like Stentor show promise to provide answers (Slabodnick et al., 2014, 2017). These structures are acted upon by patterns of ciliary and contractile activity to drive different ground-state behaviors of the cell. Hans Machemer previously recognized the power of forward/reverse cycles to encode the diversity of different swimming trajectories seen in protists and bacteria (Machemer, 2001). This idea can be extended to easily deformed structures like Lacrymaria’s helical cytoskeleton to produce the morphological rearrangements critical for execution of certain functional behaviors, or to other locomotory modes like the walking gait of Euplotes and other hypotrichs. How specific timescales and patterns of activity are generated and maintained by the cell is an exciting systems-level question that demands further inquiry. Finally, these ground-state patterns of activity are modulated by different sensory inputs to the cell. Sense data can induce electrical signals across the membrane to trigger rapid ciliary responses or ultrafast contractions (Naitoh and Eckert, 1969; Mathijssen et al., 2019); or they can act through cyclic nucleotide production that modulates ciliary amplitudes and orientation (Pech, 1995). While some sensors are already known in Paramecium and Tetrahymena, the multitiered systems-level decision making observed in predatory ciliates like Lacrymaria (Coyle et al., 2019) or the hierarchical avoidance responses seen in some Stentor species (Dexter et al., 2019) has the potential to reveal how fast timescale logic/decision trees are realized by molecular circuitry in single cells.
The integration of structure, activity, and sensing that underlies the diversity of ciliate behaviors resembles the toolbox of core functions by which simple macroscopic autonomous robotic systems are engineered but that have not yet been implemented effectively at the microscale. Looking ahead to the challenge of developing autonomous microscopic machines, our own designs would likely benefit by emulating the strategies of ciliates. For example, we should find ways to incorporate structure and geometry into our designs, such that the machines we build bend, stretch, and rearrange in ways that synergize with their intended functions. We should also explore ways to drive the behavior of these structures statistically through patterns or ensembles of activities rather than deterministically. Using these design strategies, sense data would modulate behavior not through centralized information processing but by modulating the distribution of activities and patterns that organize the emergent statistics of the behavior instead.
Such approaches can seem daunting because they run contrary to the deterministic approaches to building machines that are far more intuitive. However, these ideas are simply an extension of Kirschner and Mitchison’s famous realization that microtubules harness dynamic instability to stochastically locate their target binding sites inside the cell (Mitchison and Kirschner, 1984). Ultimately, we must embrace the challenges of the microscopic world and adapt to it. Fortunately, the ponds and oceans are full of extraordinary ciliates that can provide us the examples we need to help inspire our designs, and the technology of the 21st century has given us a powerful collection of new tools to finally understand and deconstruct them.
Footnotes
REFERENCES
- Ahamed T, Costa AC, Stephens GJ (2019). Capturing the continuous complexity of behavior in C. elegans, Animal Behavior and Cognition, https://www.biorxiv.org/content/10.1101/827535v1. [Google Scholar]
- Bhattacharyya RP, Reményi A, Yeh BJ, Lim WA (2006). Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu Rev Biochem , 655–680. [DOI] [PubMed] [Google Scholar]
- Borror AC, Hill BF (1995). The order euplotida (ciliophora): taxonomy, with division of euplotes into several genera. J Eukaryot Microbiol , 457–466. [Google Scholar]
- Chang S-Y, Houten JV, Robles LJ, Lui SS, Kung C (1974). An extensive behavioural and genetic analysis of the Pawn mutants in Paramecium aurelia. Genet Res , 165–173. [DOI] [PubMed] [Google Scholar]
- Coyle SM, Flaum EM, Li H, Krishnamurthy D, Prakash M (2019). Coupled active systems encode an emergent hunting behavior in the unicellular predator Lacrymaria olor. Curr Biol , 3838–3850.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle SM, Lim WA (2016). Mapping the functional versatility and fragility of Ras GTPase signaling circuits through in vitro network reconstitution. ELife , e12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassow G, Munro E (1999). Modularity in animal development and evolution: elements of a conceptual framework for EvoDevo. J Exp Zool , 307–325. [PubMed] [Google Scholar]
- Dexter JP, Prabakaran S, Gunawardena J (2019). A complex hierarchy of avoidance behaviors in a single-cell eukaryote. Curr Biol , 4323–4329.e2. [DOI] [PubMed] [Google Scholar]
- Deyle ER, Sugihara G (2011). Generalized theorems for nonlinear state space reconstruction. PLoS One , e18295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilpin W (2020). Deep reconstruction of strange attractors from time series. ArXiv200205909 Nlin Physicsphysics Q-Bio Stat, https://arxiv.org/abs/2002.05909.
- Girdhar K, Gruebele M, Chemla YR (2015). The behavioral space of Zebrafish locomotion and its neural network analog. PLoS One , e0128668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R, Asai H (1980). Electrophysiological responses of Didinium nasutum to Paramecium capture and mechanical stimulation. Nature , 869–870.6253817 [Google Scholar]
- Hinman MB, Jones JA, Lewis RV (2000). Synthetic spider silk: a modular fiber. Trends Biotechnol , 374–379. [DOI] [PubMed] [Google Scholar]
- Ishida H, Shigenaka Y (1988). Cell model contraction in the ciliate spirostomum. Cell Motil , 278–282. [Google Scholar]
- Jennings HS (1906). Behavior of the Lower Organisms, New York: Columbia University Press. [Google Scholar]
- Kinosita H, Dryl S, Naitoh Y (1964). Changes in the membrane potential and the response to stimuli in Paramecium. J Fac Sci Univ Tokyo Sect IV , 291. [Google Scholar]
- Knowles JR (1991). Enzyme catalysis: not different, just better. Nature , 121–124. [DOI] [PubMed] [Google Scholar]
- Kolisko M, Boscaro V, Burki F, Lynn DH, Keeling PJ (2014). Single-cell transcriptomics for microbial eukaryotes. Curr Biol , R1081–R1082. [DOI] [PubMed] [Google Scholar]
- Kung C, Chang S-Y, Satow Y, van Houten J, Hansma H (1975). Genetic dissection of behavior in Paramecium. Science , 898–904. [PubMed] [Google Scholar]
- Kung C, Naitoh Y (1973). Calcium-induced ciliary reversal in the extracted models of “Pawn,” a behavioral mutant of Paramecium. Science , 195–196. [DOI] [PubMed] [Google Scholar]
- Lodh S, Yano J, Valentine MS, Houten JLV (2016). Voltage-gated calcium channels of Paramecium cilia. J Exp Biol , 3028–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machemer H (2001). The swimming cell and its world: structures and mechanisms of orientation in protists. Eur J Protistol , 3–14. [Google Scholar]
- Mathijssen A, Culver J, Bhamla MS, Prakash M (2019). Collective intercellular communication through ultra-fast hydrodynamic trigger waves. Nature , 560–564. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M (1984). Dynamic instability of microtubule growth. Nature , 237–242. [DOI] [PubMed] [Google Scholar]
- Naitoh Y (1966). Reversal response elicited in nonbeating cilia of Paramecium by membrane depolarization. Science , 660–662. [DOI] [PubMed] [Google Scholar]
- Naitoh Y, Eckert R (1969). Ionic mechanisms controlling behavioral responses of Paramecium to mechanical stimulation. Science , 963–965. [DOI] [PubMed] [Google Scholar]
- Noland LE, Finley HE (1931). Studies on the taxonomy of the genus vorticella. Trans Am Microsc Soc , 81–123. [Google Scholar]
- Pech LL (1995). Regulation of ciliary motility in Paramecium by cAMP and cGMP. Comp Biochem Physiol A Physiol , 31–37. [Google Scholar]
- Rudzinska MA (1973). Do suctoria really feed by suction? BioScience , 87–94. [Google Scholar]
- Ruehle MD, Orias E, Pearson CG (2016). Tetrahymena as a unicellular model eukaryote: genetic and genomic tools. Genetics , 649–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori P, Geyer VF, Scholich A, Jülicher F, Howard J (2016). Dynamic curvature regulation accounts for the symmetric and asymmetric beats of Chlamydomonas flagella. ELife , e13258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM, Ruby GJ, Reiff SB, Swart EC, Gosai S, Prabakaran S, Witkowska E, Larue GE, Fisher S, Freeman RM, et al. (2017). The macronuclear genome of Stentor coeruleus reveals tiny introns in a giant cell. Curr Biol , 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM, Marshall WF (2014). Stentor coeruleus. Curr Biol , R783–R784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabodnick MM, Ruby JG, Dunn JG, Feldman JL, DeRisi JL, Marshall WF (2014). The kinase regulator Mob1 acts as a patterning protein for stentor morphogenesis. PLoS Biol , e1001861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP, Petrenko VA (1997). Phage display. Chem Rev , 391–410. [DOI] [PubMed] [Google Scholar]
- Stephens GJ, Johnson-Kerner B, Bialek W, Ryu WS (2008). Dimensionality and dynamics in the behavior of C. elegans. PLoS Comput Biol , e1000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens GJ, Mesquita MB, Ryu WS, Bialek W (2011). Emergence of long timescales and stereotyped behaviors in Caenorhabditis elegans. Proc Natl Acad Sci USA , 7286–7289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leewenhoeck A (1677). Observations, communicated to the publisher by Mr. Antony van Leewenhoeck, in a Dutch Letter of the 9th of Oct. 1676. Here, in English’d: concerning little animals by him observed in rain-well-sea and snow water; as also in water wherein pepper had lain infused. Philos Trans 1665–1678 , 821–831. [Google Scholar]
- Wan KY, Goldstein RE (2014). Rhythmicity, recurrence, and recovery of flagellar beating. Phys Rev Lett , 238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodruff LL (1921). The structure, life history, and intrageneric relationships of Paramecium calkinsi, sp. nov. Biol Bull , 171–180. [Google Scholar]
- Yanase R, Nishigami Y, Ichikawa M, Yoshihisa T, Sonobe S (2018). The neck deformation of Lacrymaria olor depending upon cell states. J Protistol , 1–6. [Google Scholar]
- Zoller SD, Hammersmith RL, Swart EC, Higgins BP, Doak TG, Herrick G, Landweber LF (2012). Characterization and taxonomic validity of the ciliate Oxytricha trifallax (class Spirotrichea) based on multiple gene sequences: limitations in identifying genera solely by morphology. Protist , 643–657. [DOI] [PMC free article] [PubMed] [Google Scholar]