Abstract
The action of guanine nucleotide exchange factors (GEFs) on the ADP-ribosylation factor (ARF) family of small GTPases initiates intracellular transport pathways. This role requires ARF GEFs to be recruited from the cytosol to intracellular membrane compartments. An ARF GEF known as General receptor for 3-phosphoinositides 1 (Grp1) is recruited to the plasma membrane through its pleckstrin homology (PH) domain that recognizes phosphatidylinositol 3,4,5-trisphosphate (PIP3). Here, we find that the phosphorylation of Grp1 induces its PH domain to recognize instead phosphatidylinositol 4-phosphate (PI4P). This phosphorylation also releases an autoinhibitory mechanism that results in the coil–coil (CC) domain of Grp1 engaging two peripheral membrane proteins of the recycling endosome. Because the combination of these actions results in Grp1 being recruited preferentially to the recycling endosome rather than to the plasma membrane, our findings reveal the complexity of recruitment mechanisms that need to be coordinated in localizing an ARF GEF to an intracellular compartment to initiate a transport pathway. Our elucidation is also remarkable for having revealed that phosphoinositide recognition by a PH domain can be switched through its phosphorylation.
INTRODUCTION
The ADP-ribosylation factor (ARF) family of small GTPases recruits coat proteins from the cytosol to intracellular membrane compartments to initiate the formation of transport carriers. ARFs, in turn, are activated by guanine nucleotide exchange factors (GEFs). As such, ARF GEFs act as the ultimate initiators of intracellular transport pathways (Casanova, 2007; Donaldson and Jackson, 2011; Sztul et al., 2019). Recruitment from the cytosol to membrane represents a major mechanism of regulating ARF GEFs. Although recruitment mechanisms are being elucidated for the different ARF GEFs (Hurtado-Lorenzo et al., 2006; Cohen et al., 2007; Richardson et al., 2012; Hiester and Santy, 2013; Lowery et al., 2013; Tsai et al., 2013; Quilty et al., 2014; Karandur et al., 2017), the complexity of mechanisms that need to be coordinated in localizing an ARF GEF to a particular intracellular compartment with specificity remains an ongoing goal.
Several subclasses of ARF GEFs exist, with members of the cytohesin subclass being some of the best characterized (Casanova, 2007; Donaldson and Jackson, 2011; Sztul et al., 2019). They are the first ones to be identified (Chardin et al., 1996; Klarlund et al., 1997). A mechanistic understanding of how they act is being elucidated in detail (Cherfils et al., 1998; Goldberg, 1998; Ferguson et al., 2000; Lietzke et al., 2000). A cytohesin member known as General receptor for 3-phosphoinositides 1 (Grp1) has been shown to be recruited from the cytosol to membrane by recognizing phosphatidylinositol 3,4,5-triphosphate (PIP3; Klarlund et al., 1997), a phosphoinositide enriched at the plasma membrane (Di Paolo and De Camilli, 2006). This recognition is particularly well understood, as it has been elucidated in molecular detail through structural studies (Ferguson et al., 2000; Lietzke et al., 2000). Notably, however, we have found previously that Grp1 can also be recruited to the recycling endosome (Li et al., 2012). Studying the stimulation-dependent recycling of glucose transporter type 4 (glut4) in adipocytes, a process critical for glucose homeostasis (Foley et al., 2011; Kandror and Pilch, 2011; Leto and Saltiel, 2012), we found that insulin-induced signaling activates the protein kinase Akt, which phosphorylates Grp1 at threonine position 280 (T280), resulting in Grp1 being recruited to the recycling endosome to initiate glut4 recycling (Li et al., 2012).
The finding that phosphorylation of an ARF GEF can switch its localization to initiate a different transport pathway is remarkable. From the mechanistic perspective, such a finding also suggests an intriguing opportunity. By elucidating how this switch occurs, we may uncover the complexity of recruitment mechanisms that need to be coordinated in localizing an ARF GEF to an intracellular membrane compartment to initiate a transport pathway.
RESULTS
As a starting point, we noted that a previous study had suggested a mechanism for how phosphorylation of the T280 residue in Grp1 regulates its membrane recruitment. Studying a closely related cytohesin member, known as ARNO (ARF nucleotide site opener), the study found that the phosphorylation of the T276 residue in ARNO (which is equivalent to the T280 residue in Grp1) releases an autoinhibitory mechanism of membrane recruitment (Hiester and Santy, 2013). Specifically, the PH domain was found to bind the CC domain, with phosphorylation releasing this intramolecular interaction to allow the PH domain to engage the membrane (Hiester and Santy, 2013; also summarized in Supplemental Figure S1A). However, because only total membranes were examined in this study, it had remained unclear how the release of this autoinhibitory mechanism could explain the ability of the T280 phosphorylation in Grp1 to switch recruitment from the plasma membrane to the recycling endosome. Thus, we sought an approach that would achieve greater mechanistic resolution.
In recent years, liposomes generated with lipid composition that mimics those of organellar membranes have enabled detailed mechanistic dissection of how different transport factors act on intracellular membranes. Such an approach would be particularly attractive in the case of Grp1, as its recruitment involves a direct interaction with a lipid. Thus, to reconstitute the membrane recruitment of Grp1, we generated liposomes with major lipids of organellar membrane, having 48% phosphatidylcholine (PC), 20% phosphatidylethanolamine (PE), 4% phosphatidylserine (PS), 14% cholesterol, and 4% sphingomyelin, and also added 10% PIP3 as the relevant phosphoinositide. We first confirmed that the PH domain of Grp1 is recruited to these liposomes in a PIP3-dependent manner (Figure 1A). Moreover, as predicted by the previously proposed autoinhibitory mechanism (Hiester and Santy, 2013; see also Supplemental Figure S1A), we found that the full-length Grp1 is less efficiently recruited to these liposomes as compared with the PH domain alone (Figure 1B).
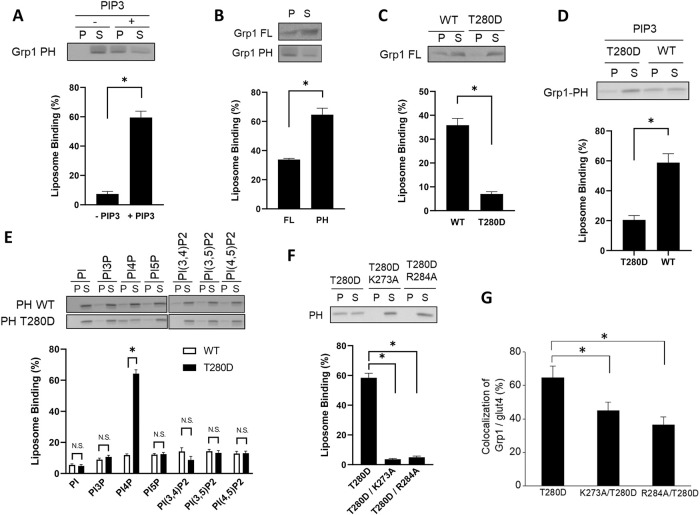
FIGURE 1:
T280 phosphorylation induces Grp1 to recognize a new phosphoinositide. Quantitative results are shown as mean with standard error: *, p < 0.05, NS p > 0.05, Student’s t test. The number of independent experiments performed is specified below. (A) PIP3-dependent recruitment of the Grp1 PH domain to liposomal membrane. The PH domain was incubated with liposomes containing PIP3, or not, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from four experiments is shown. (B) Full-length Grp1 shows reduced recruitment to PIP3-containing liposomes as compared with that of the PH domain. Constructs as indicated were incubated with PIP3-containing liposomes, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from three experiments is shown. (C) The T280D mutation further reduces the recruitment of full-length Grp1 to PIP3-containing liposomes. Grp1 constructs as indicated were incubated with PIP3-containing liposomes, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from four experiments is shown. (D) The T280D mutation also reduces the recruitment of the PH domain of Grp1 to PIP3-containing liposomes. PH domain constructs as indicated were incubated with PIP3-containing liposomes, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from four experiments is shown. (E) The T280D mutation induces the Grp1 PH domain to recognize PI4P. Liposomes were generated with major lipids of organellar membrane along with a particular phosphoinositide, PI3P, PI4P, PI5P, PI(3,4)P2, PI(3,5)P2, or PI(4,5)P2. The different liposomes were then incubated with PH domain constructs as indicated, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from three experiments is shown. (F) Mutating key residues in the phosphoinositide-binding pocket of the Grp1 PH domain prevents its recruitment to PI4P-containing liposomes. PH domain constructs as indicated were incubated with PI4P-containing liposomes, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from three experiments is shown. (G) Mutating key residues in the binding pocket of the PH domain reduces the localization of the Grp1-T280D at the recycling endosome. Adipocytes were stably transfected with constructs as indicated, and then the colocalization of different constructs with internal glut4 (marker for the recycling endosome in adipocytes) was quantified; n = 10 cells per experiment.
We then examined the effect of mutating the T280 residue in the full-length Grp1 to aspartate (T280D), as we had shown previously that this mutation mimics the effect of phosphorylation at the T280 residue (Li et al., 2012). Remarkably, whereas the previously proposed mechanism of autoinhibition predicts that membrane recruitment should be enhanced, because the T280 phosphorylation should release this inhibition (Hiester and Santy, 2013; also summarized in Supplemental Figure S1A), we found that the recruitment of the full-length Grp1 to the PIP3-containing liposomes is instead inhibited by the T280D mutation (Figure 1C). We also confirmed this finding by examining full-length Grp1 phosphorylated at the T280 residue. To achieve this phosphorylation, we performed the in vitro kinase assay, incubating Grp1 with Akt as previously described (Li et al., 2012). When phosphorylated Grp1 was incubated with PIP3-containing liposomes, we again observed inhibition of membrane recruitment (Supplemental Figure S1B). Thus, we next sought to sort out an explanation for this unexpected result.
Initially, we determined whether a similar inhibition would be observed when examining the recruitment of just the PH domain to PIP3-containing liposomes. Indeed, inhibition was also observed (Figure 1D). Thus, because the T280 residue is located near the phosphoinositide-binding pocket of the Grp1 PH domain (Supplemental Figure S1C), we next explored the intriguing possibility that the T280 phosphorylation could induce the PH domain to recognize a different phosphoinositide.
To screen among all physiologic forms of phosphoinositide, we generated liposomes that contain major lipids of organellar membrane, and also added a particular form of phosphoinositide individually. Thus, besides liposomes that contain PIP3, we also generated liposomes that contain PI3P (phosphatidylinositol 3-phosphate), PI4P (phosphatidylinositol 4-phosphate), PI5P (phosphatidylinositol 5-phosphate), PI(3,4)P2 (phosphatidylinositol 3,4-biphosphate), PI(3,5)P2 (phosphatidylinositol 3,5-biphosphate), or PI(4,5)P2 (phosphatidylinositol 4,5-biphosphate). We found that the T280D mutation induces the PH domain of Grp1 to have an appreciable recruitment to liposomes that contain PI4P (Figure 1E). We ruled out the possibility that the T280D mutation could be inducing the PH domain to bind PI4P outside of the canonical phosphoinositide-binding pocket. Key residues in this pocket have been defined previously through structural studies (Ferguson et al., 2000; Lietzke et al., 2000). We found that mutating these residues (summarized in Supplemental Figure S1C) prevents the T280D mutation from inducing the recruitment of the PH domain to PI4P-containing liposomes (Figure 1F). This result was also confirmed in cells, as mutating the key residues also reduced the localization of the full-length Grp1-T280D to the recycling endosome (Figure 1G and Supplemental Figure S1D).
We next sought to confirm that PI4P exists at the recycling endosome. Antibodies have been generated against different phosphoinositides. Using an antibody against PI4P, we found by confocal microscopy that PI4P in adipocytes shows significant colocalization with two markers of the recycling endosome (Figure 2A and Supplemental Figure S2A): Rab11 (Ullrich et al., 1996) and cellubrevin (Cbv; D’Souza-Schorey et al., 1998). Moreover, as we have shown previously that the internal pool of glut4 resides at the recycling endosome of adipocytes (Li et al., 2007), we found that PI4P also shows significant colocalization with internal glut4 in these cells (Figure 2A and Supplemental Figure S2A). In contrast, PIP3 exists at a minimal level at the recycling endosome, as assessed by its colocalization with Rab11, cellubrevin, and internal glut4 (Figure 2B and Supplemental Figure S2B). We also confirmed the presence of PI4P at the recycling endosome by another approach. PI4P biosensors have been developed based on protein domains that bind specifically to PI4P. Examining one such sensor, known as GFP-P4M-SidM (Hammond et al., 2014), we found that it also shows significant colocalization with internal glut4 in adipocytes (Supplemental Figure S2C).
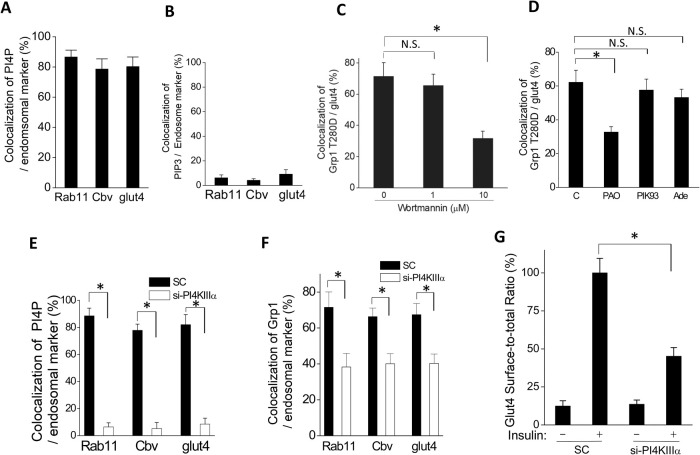
FIGURE 2:
Recruitment of Grp1 to the recycling endosome requires PI4KΙΙΙα. Quantitative results are shown as mean with standard error from three independent experiments: *, p < 0.05, NS p > 0.05, Student’s t test. (A) PI4P level at the recycling endosome was assessed through the colocalization of PI4P with different markers of the recycling endosome followed by quantitation; n = 10 cells per experiment. (B) PIP3 level at the recycling endosome was assessed through the colocalization of PIP3 with different markers of the recycling endosome followed by quantitation; n = 10 cells per experiment. (C) Wortmannin reduces the localization of Grp1-T280D at the recycling endosome. Adipocytes were treated with wortmannin at doses as indicated, and then the colocalization of Grp1-T280D with internal glut4 (marker for the recycling endosome in adipocytes) was quantified; n = 10 cells per experiment. (D) PAO, but not PIK93 or adenosine (Ade), significantly reduces the localization of Grp1-T280D at the recycling endosome. Adipocytes were treated with pharmacologic agents as indicated, and then the colocalization of Grp1-T280D with internal glut4 (marker for the recycling endosome in adipocytes) was quantified; n = 10 cells per experiment. (E) Knocking down PI4KΙΙΙα reduces the colocalization of PI4P with markers of the recycling endosome. Adipocytes were treated as indicated, and then the colocalization of PI4P with different markers of the recycling endosome (Rab11, cellubrevin, or internal glut4) was quantified; n = 10 cells per experiment. (F) Knocking down PI4KΙΙΙα reduces the colocalization of Grp1-T280D with markers of the recycling endosome. Adipocytes were treated as indicated, and then the colocalization of Grp1-T280D different markers of the recycling endosome (Rab11, cellubrevin, or internal glut4) was quantified; n = 10 cells per experiment. (G) Knocking down PI4KΙΙΙα reduces the ability of insulin to stimulate glut4 recycling. Adipocytes were treated as indicated (SC, scrambled siRNA), and then glut4 recycling was quantified; n = 10 cells per experiment.
To complement the above findings, we next considered that PI4 kinase (PI4K) activity controls the PI4P level by converting PI to PI4P (Balla and Balla, 2006). As multiple isoforms of PI4K exist, we initially pursued a pharmacologic approach to determine whether PI4K activity is critical for Grp1 localizing to the recycling endosome. Wortmannin has been used in a dose-dependent manner to target different classes of PI kinases, with 1 µM inhibiting PI3K activity and 10 µM inhibiting PI4K activity (Balla and Balla, 2006). We found that the localization of Grp1-T280D at the recycling endosome is significantly reduced when cells are treated with 10 µM, but not 1 µM, of wortmannin (Figure 2C and Supplemental Figure S2D), suggesting that PI4K activity is needed for Grp1 recruitment to the recycling endosome.
As confirmation, we pursued further pharmacologic inhibition that targets subclasses of PI4K (Balla and Balla, 2006). We found that the Grp1-T280D localization to the recycling endosome was reduced, when cells were treated with phenylarsine oxide (PAO), but not with PIK93 or adenosine (Figure 2D and Supplemental Figure S2E). As this profile of pharmacologic inhibition suggested that the PI4KΙΙΙα isoform is being targeted (Balla and Balla, 2006), we sought further confirmation by treating cells with siRNA against PI4KΙΙΙα. We initially confirmed that this siRNA treatment is efficient in reducing the cellular level of PI4KΙΙΙα (Supplemental Figure S3A). We then found that the PI4P level at the recycling endosome is markedly reduced, as assessed by the colocalization of PI4P with Rab11 (Figure 2E and Supplemental Figure S3B), cellubrevin (Figure 2E and Supplemental Figure S3C), and internal glut4 (Figure 2E and Supplemental Figure S3D). Treated cells also showed a reduced level of the Grp1-T280D at the recycling endosome, as assessed by the colocalization of Grp1-T280D with Rab11 (Figure 2F and Supplemental Figure S3E), cellubrevin (Figure 2F and Supplemental Figure S3F), and internal glut4 (Figure 2F and Supplemental Figure S3G). Consistent with these findings, we found that the ability of insulin to induce glut4 recycling is also reduced in the treated cells (Figure 2G). Moreover, supporting the specificity of the siRNA targeting, we found that two other sequences against PI4KΙΙΙα also reduce Grp1-T280D localization to the recycling endosome (Supplemental Figure S3H). Thus, the collective results further support the importance of PI4P for the recruitment of Grp1 to the recycling endosome.
We then noted that, whereas siRNA against PI4KΙΙΙα markedly reduced the PI4P level at the recycling endosome, this treatment had a less dramatic effect on Grp1 localization to this compartment (compare Figure 2, E and F). Moreover, although key mutations in the lipid-binding pocket of the PH domain in Grp1 virtually abolished its recruitment to PI4P-containing liposomes, they had a lesser effect on the Grp1-T280D localization to the recycling endosome (compare Figure 1, F and G). Collectively, these observations suggested that additional mechanism(s), besides PI4P recognition by the PH domain, could be acting to promote the recruitment of Grp1 to the recycling endosome.
To pursue this possibility, we initially noted that ARF6 has been suggested to recruit cytohesin members to the plasma membrane (Cohen et al., 2007). However, we found that siRNA against ARF6 (Supplemental Figure S4A), using a sequence with previously documented specificity (Li et al., 2007), does not affect Grp1-T280D localization to the recycling endosome (Figure 3A and Supplemental Figure S4B). We next noted that cytohesin members have been found to interact with multiple peripheral membrane proteins, including CASP (Mansour et al., 2002), GRASP (Nevrivy et al., 2000), GRSP1 (Klarlund et al., 2001), and IPCEF1 (Venkateswarlu, 2003), but whether these interactions mediate cytohesin membrane recruitment have been unclear. Treating cells with siRNA, we found that targeting against GRASP and IPCEF1, but not against GRSP1 and CASP, results in appreciable reductions in Grp1-T280D localization to the recycling endosome (Figure 3A and Supplemental Figure S4C). The differential effects observed could not be attributed to differential efficiencies of siRNA treatment, as the mRNA levels of all four targeted proteins show a marked reduction (Figure 3B). Moreover, supporting the targeting specificity of the siRNA treatment, two other sequences for each target induced similar effects on Grp1-T280D localization to the recycling endosome (Supplemental Figure S5, A–D).
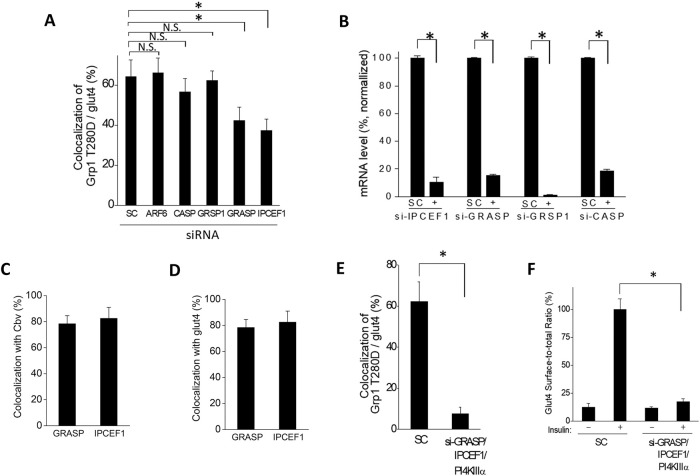
FIGURE 3:
Recruitment of Grp1 to the recycling endosome also requires GRASP and IPCEF1. Quantitative results are shown as mean with standard error from three independent experiments: *, p < 0.05, NS p > 0.05, Student’s t test. (A) The effect of knocking down candidate factors on the localization of Grp1-T280D at the recycling endosome. Adipocytes were treated with siRNA against different factors as indicated, and then the colocalization of Grp1-T280D with internal glut4 (marking the recycling endosome in adipocytes) was quantified; n = 10 cells per experiment. (B) Efficiency of knocking down GRASP, IPCEF1, CASP, and GRSP1. Cells were treated as indicated followed by quantitation of mRNA levels; n = 3 experiments. (C) Colocalization of GRASP or IPCEF1 with cellubrevin was assessed by confocal microscopy, followed by quantitation; n = 10 cells per experiment. (D) Colocalization of GRASP or IPCEF1 with internal glut4 was assessed by confocal microscopy, followed by quantitation; n = 10 cells per experiment. (E) The cumulative effect of knocking down GRASP, IPCEF1, and PI4KIIIα on the localization of Grp1-T280D at the recycling endosome. Adipocytes were treated with siRNA against factors as indicated, and then the colocalization of Grp1-T280D with internal glut4 (marking the recycling endosome in adipocytes) was quantified; n = 10 cells per experiment. (F) Knocking down the combination of GRASP, IPCEF1, and PI4KIIIα virtually eliminates the ability of insulin to stimulate glut4 recycling. Adipocytes were treated with siRNA against factors as indicated, and then glut4 recycling was quantified; n = 10 cells per experiment.
We further noted that, whereas the overexpression of GRASP has been detected at the plasma membrane (Nevrivy et al., 2000), endogenous GRASP resides mostly at the recycling endosome (Venkataraman et al., 2012). Consistent with the latter finding, we found that endogenous GRASP and IPCEF1 reside mostly at the recycling endosome in adipocytes, as reflected by their colocalization with cellubrevin (Figure 3C and Supplemental Figure S4D) and internal glut4 (Figure 3D and Supplemental Figure S4E).
Importantly, when adipocytes were treated with the combination of siRNAs against PI4KΙΙΙα, GRASP and IPCEF1, the level of Grp1 at the recycling endosome was virtually abolished (Figure 3E and Supplemental Figure S4F). Correlated with this finding, we found that the ability of insulin to stimulate glut4 recycling was also virtually abolished (Figure 3F). Thus, these results suggested that, besides inducing the recognition of PI4P by the PH domain, the T280 phosphorylation also promotes Grp1 recruitment to the recycling endosome through GRASP and IPCEF1.
To elucidate how phosphorylation of this single residue in Grp1 could exert such multiple effects, we next sought to reconstitute Grp1 recruitment using purified proteins and defined liposomes, so that mechanistic details could be discerned. To incorporate GRASP and IPCEF1 onto liposomes, we pursued a previously established approach of localizing peripheral membrane proteins onto liposomal membrane, which involves tagging proteins with the 6x-His epitope and then incubating them with liposomes that contain a nickel-coupled lipid (Lee et al., 2005; Drin et al., 2008). We initially confirmed that such liposomes still support the PI4P-dependent recruitment of Grp1-T280D onto liposomal membrane (Figure 4A). When 6xHis-tagged GRASP was also added to these liposomes, we found that Grp1 recruitment is further enhanced (Figure 4A). The addition of 6xHis-tagged IPCEF1 to PI4P-containing liposomes also enhanced Grp1 recruitment (Figure 4A). Notably, when liposomes were endowed with all three recruitment factors (PI4P, GRASP, and IPCEF1), we found that soluble Grp1 becomes virtually all membrane-bound (Figure 4A).
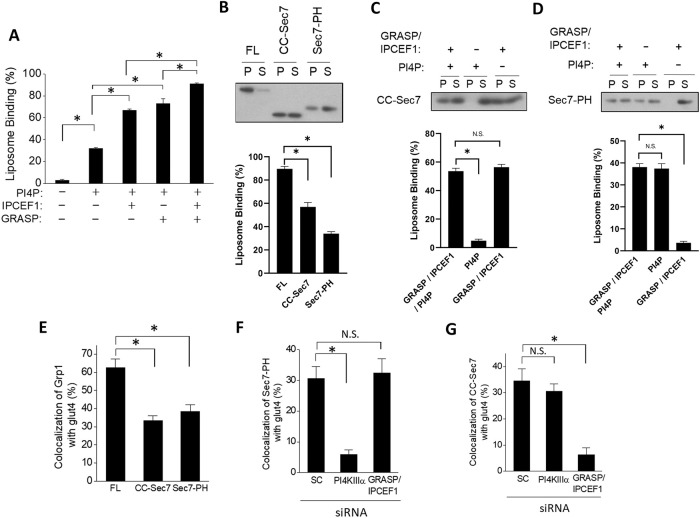
FIGURE 4:
Grp1 recruitment to the recycling endosome involves both lipid-based and protein-based mechanisms. Quantitative results are shown as mean with standard error: *, p < 0.05, NS p > 0.05, Student’s t test. The number of independent experiments performed is specified below. (A) Reconstituting the recruitment of Grp1 to liposomal membrane. Liposomes were endowed with different factors as indicated, and then incubated with full-length Grp1-T280D. The degree of recruitment was then quantified; n = 3 experiments. (B) Recruitment of different forms of Grp1 to liposomal membrane. Constructs as shown were incubated with fully endowed liposomes (containing PI4P, GRASP, and IPCEF1), followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S); FL (full-length Grp1-T280D), CC-Sec7 (Grp1 lacking the PH domain), Sec7-PH (Grp1-T280D lacking the CC domain). A representative result along with quantitation from three experiments is shown. (C) Recruitment of the CC-Sec7 construct to liposomal membrane requires GRASP/IPCEF1. The CC-Sec7 construct was incubated with liposomes that were endowed with different factors as indicated, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from three experiments is shown. (D) Recruitment of the Sec7-PH construct to liposomal membrane requires PI4P. The Sec7-PH construct was incubated with liposomes that were endowed with different factors as indicated, followed by centrifugation to detect distribution in the pellet (P) vs. supernatant (S). A representative result along with quantitation from three experiments is shown. (E) Recruitment of different forms of Grp1 to the recycling endosome. Constructs as shown were stably transfected into adipocytes, followed by confocal microscopy to assess the degree of localization to the recycling endosome; FL (full-length Grp1-T280D), CC-Sec7 (Grp1 lacking the PH domain), Sec7-PH (Grp1-T280D lacking the CC domain); n = 10 cells per experiment with three independent experiments performed. (F) Recruitment of the Sec7-PH construct to the recycling endosome requires PI4KIIIα. The Sec7-PH construct was stably transfected into adipocytes that were treated with siRNA as indicated, followed by confocal microscopy to assess the degree of localization to the recycling endosome; SC (scrambled siRNA); n = 10 cells per experiment with three independent experiments performed. (G) Recruitment of the CC-Sec7 construct to the recycling endosome requires GRASP/IPCEF1. The CC-Sec7 construct was stably transfected into adipocytes that were treated with siRNA as indicated, followed by confocal microscopy to assess the degree of localization to the recycling endosome; n = 10 cells per experiment with three independent experiments performed.
Having reconstituted all three mechanisms of Grp1 recruitment, we next sought to define in further detail how the T280 phosphorylation promotes these recruitment mechanisms. For this goal, we were led by a previous finding that GRASP interacts with the CC domain of Grp1 (Nevrivy et al., 2000). Thus, we explored whether the effects of T280 phosphorylation could be divided mechanistically into two parts: a lipid-based mechanism that involves the PH domain recognizing PI4P and a protein-based mechanism that involves the CC domain recognizing GRASP/IPCEF1.
We generated two truncation constructs: one that lacks the PH domain (referred to hereon as the CC-Sec7 construct) and the other that lacks the CC domain (referred to hereon as the Sec7-PH construct; summarized in Supplemental Figure S6A). We compared the recruitment of these two truncation forms to that of the full-length form, with the T280D mutation incorporated into constructs that contain the PH domain (shown in Supplemental Figure S6A). When the different constructs were incubated with liposomes that possess all three recruitment factors (PI4P, GRASP, and IPCEF1), we found that both truncation forms showed reduced recruitment as compared with the full-length form (Figure 4B). Notably, we next found that liposomes possessing only PI4P could not recruit the CC-Sec7 construct (Figure 4C), while liposomes possessing only GRASP and IPCEF1 could not recruit the Sec7-PH construct (Figure 4D). Thus, these results confirmed that the effects of T280 phosphorylation could indeed be divided mechanistically into two parts, with one involving the PH domain engaging PI4P, and the other involving the CC domain engaging GRASP/IPCEF1.
We also pursued cell-based studies to confirm these findings. Parallel to the results from the reconstitution studies, we found that both truncation constructs showed reduced localization to the recycling endosome as compared with that of the full-length form (Figure 4E and Supplemental Figure S6B). We also found that siRNA against PI4KΙΙΙα reduces the localization of the Sec7-PH construct to the recycling endosome (Figure 4F and Supplemental Figure S6C), but does not affect the localization of the CC-Sec7 construct to this compartment (Figure 4G and Supplemental Figure S6D). Similarly, targeting against GRASP/IPCEF1 reduces the localization of the CC-Sec7 construct to the recycling endosome (Figure 4G and Supplemental Figure S6D), but does not affect the localization of the Sec7-PH construct to this compartment (Figure 4F and Supplemental Figure S6C). Thus, both in vitro and in vivo results suggested that the effect of the T280 phosphorylation on Grp1 recruitment could be divided mechanistically into two parts: a lipid-based mechanism that involves the PH domain targeting PI4P, and a protein-based mechanism that involves the CC domain targeting GRASP/IPCEF1.
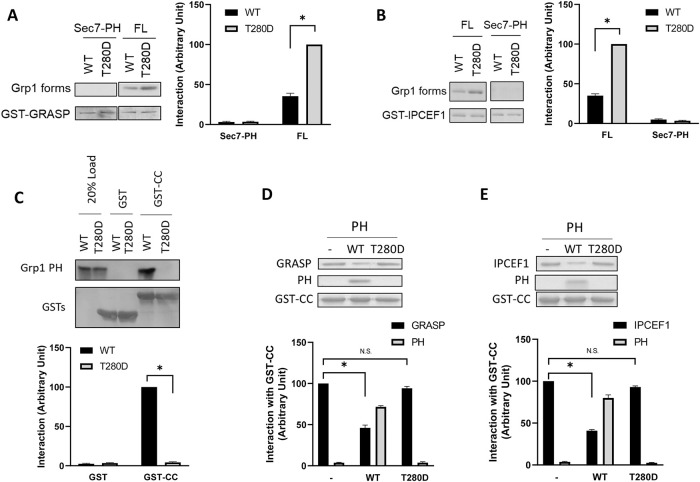
To characterize the protein-based mechanism in further detail, we next examined whether the CC domain interacts directly with GRASP/IPCEF1, and whether the T280 phosphorylation regulates such potential interactions. When GRASP is bound to beads followed by incubation with soluble Grp1 in a pull-down experiment, we confirmed that the T280D mutation enhances a direct interaction between full-length Grp1 and GRASP (Figure 5A). This enhancement requires the CC domain, as the deletion of this domain (resulting in the Sec7-PH construct) prevents Grp1 from interacting directly with GRASP (Figure 5A). Performing a similar experiment to examine IPCEF1, we found that the T280D mutation enhances a direct interaction between full-length Grp1 and IPCEF1 (Figure 5B). This enhancement also requires the CC domain, as the Sec7-PH construct shows no direct interaction with IPCEF1 (Figure 5B).
FIGURE 5:
T280 phosphorylation enhances the interaction between the CC domain of Grp1 and GRASP/IPCEF1. Quantitative results are shown as mean with standard error: *, p < 0.05, NS p > 0.05, Student’s t test. The number of independent experiments performed is specified below. (A) GRASP can interact directly with Grp1, which requires the CC domain in Grp1, and is modulated by the T280 phosphorylation. GRASP as a GST fusion protein was bound to beads and then incubated with various Grp1 constructs as indicated in pull-down experiments. A representative result along with quantitation from three experiments is shown. (B) IPCEF1 can interact directly with Grp1, which requires the CC domain in Grp1, and is modulated by the T280 phosphorylation. IPCEF1 as a GST fusion protein was bound to beads and then incubated with various Grp1 constructs as indicated in pull-down experiments. A representative result along with quantitation from three experiments is shown. (C) Reconstituting the intramolecular interaction between the CC domain and the PH domain in Grp1, and its release by the T280 phosphorylation. The CC domain as a GST fusion protein was bound to beads and then incubated with different forms of the PH domain as indicated in pull-down experiments. As negative control, a similar experiment was performed using GST alone. A representative result along with quantitation from three experiments is shown. (D) The direct interaction between the CC domain of Grp1 and GRASP is inhibited when the CC domain is preincubated with the wild-type, but not the T280D, form of the PH domain. The CC domain as a GST fusion protein was bound to beads and then incubated with GRASP in a pull-down experiment (left lane). The CC domain on beads was preincubated with the wild-type form of the PH domain, and then incubated with GRASP in a pull-down experiment (middle lane). The CC domain on beads was preincubated with the T280D form of the PH domain, and then incubated with GRASP in a pull-down experiment (right lane). A representative result along with quantitation from four experiments is shown. (E) The direct interaction between the CC domain of Grp1 and IPCEF1 is inhibited when the CC domain is preincubated with the wild-type, but not the T280D, form of the PH domain. The CC domain as a GST fusion protein was bound to beads and then incubated with IPCEF1 in a pull-down experiment (left lane). The CC domain on beads was preincubated with the wild-type form of the PH domain, and then incubated with IPCEF1 in a pull-down experiment (middle lane). The CC domain on beads was preincubated with the T280D form of the PH domain, and then incubated with IPCEF1 in a pull-down experiment (right lane). A representative result along with quantitation from four experiments is shown.
As the intramolecular interaction between the CC domain and the PH domain had only been known to inhibit the role of the PH domain in mediating the membrane recruitment of Grp1 (Hiester and Santy, 2013), we next examined whether this interaction could also affect the role of the CC domain in mediating membrane recruitment. Initially, we reconstituted the intramolecular interaction between the CC and PH domains by expressing them separately and then confirming that they could bind directly as purified components (Figure 5C). We also reconstituted the release of this interaction by the T280 phosphorylation, as introducing of the T280D mutation into the PH domain was sufficient in completely abrogating the interaction between the CC domain and the PH domain (Figure 5C). We next found that the direct interaction between the CC domain and GRASP is reduced when the CC domain is preincubated with the PH domain, and remarkably, this effect is abrogated when the T280D mutation is introduced into the PH domain (Figure 5D). Similarly, the direct interaction between the CC domain and IPCEF1 is reduced when the CC domain is preincubated with the PH domain, and this effect is also abrogated when the T280D mutation is introduced into the PH domain (Figure 5E). Thus, when taken altogether, the results revealed a previously unappreciated role for the intramolecular interaction between the CC domain and the PH domain. Rather than simply preventing the role of the PH domain in membrane recruitment, this interaction also prevents the role of the CC domain in membrane recruitment.
DISCUSSION
We have elucidated the complexity of recruitment mechanisms that need to be coordinated in localizing an ARF GEF to an intracellular compartment for its role in initiating a transport pathway. A previous study had proposed an autoinhibitory mechanism regulating the membrane recruitment of Grp1, which involves an intramolecular interaction between its PH and CC domains, with the T280 phosphorylation releasing this interaction to allow the PH domain to engage the membrane (Hiester and Santy, 2013). However, we find that the situation is more complex. Whereas only the PH domain is currently known to mediate membrane recruitment, we have uncovered that the CC domain is also involved. Specifically, we elucidate that the PH domain mediates a lipid-based mechanism of recruitment through phosphoinositide recognition, while the CC domain mediates a protein-based mechanism, which involves its interaction with two proteins at the recycling endosome, GRASP and IPCEF1. We also find that the intramolecular interaction between the PH domain and the CC domain inhibits this newly defined role of the CC domain, and thus uncovering an additional regulatory role for the autoinhibitory mechanism that had not been appreciated.
Even more remarkable, whereas the PH domain of Grp1 has been known to recognize PIP3, we find that the T280 phosphorylation induces this PH domain to recognize PI4P. Relevant to this finding, we note that there has been great interest in understanding phosphoinositide recognition by PH domains, as it represents a major mechanism by which many proteins are recruited from the cytosol to membrane (DiNitto and Lambright, 2006; Lemmon, 2008). Whereas the molecular details of how different PH domains recognize particular phosphoinositides are being achieved (DiNitto and Lambright, 2006; Lemmon, 2008), our results advance a novel understanding of this membrane recruitment by revealing that phosphoinositide recognition by the PH domain of Grp1 can be switched through its phosphorylation.
We further note that the crystal structures of the Grp1 and ARNO PH domains bound to the head groups of PIP3 and PIP2, respectively, have been solved. This allows us to perform molecular modeling in suggesting a possible explanation for the effect of T280 phosphorylation on the specificity of phosphoinositide binding (Figure 6). In the solved structure having the PIP3 head group, the 3-phosphate is located in the most electropositive region of the binding site, where it mediates critical polar interactions with arginine and lysine residues in a conserved basic motif characteristic of PH domains that bind phosphoinositides with high affinity (DiNitto and Lambright, 2006; Lemmon, 2008). Compensating for the lack of a 3-phosphate, the PIP2 head group binds in a rotated orientation in which the 4-phosphate mediates polar interactions with the conserved basic residues. Moreover, the side chain of T280 mediates an important polar interaction with the 1-phosphate of the PIP3 head group but does not contact the 1-phosphate of the PIP2 head group as a consequence of the rotated binding mode. Thus, a plausible molecular model for a complex that contains PI4P can be constructed based on the reasonable conjecture that PI4P binds in an orientation similar to PIP2, allowing the 4-phosphate to interact with the conserved basic residues. Whereas T280 phosphorylation is expected to create a major steric conflict with the 1-phosphate of PIP3, a minor if any steric clash would be predicted for PI4P. A more definitive assessment of this possibility will require a structure study in the future.
FIGURE 6:
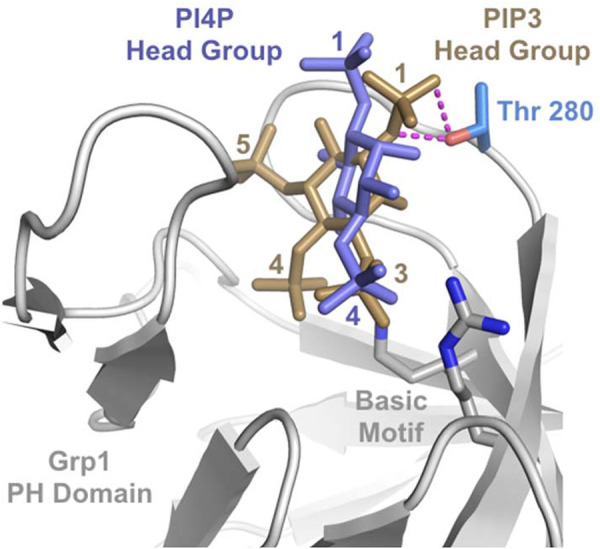
Structural model for the differential effect of T280 phosphorylation on PIP3 vs. PI4P binding to the Grp1 PH domain. The phosphoinositide-binding site in the crystal structure of the Grp1 PH domain bound to the PIP3 head group (PDB ID 1FGY) is compared with a hypothetical composite model for the PI4P head group. The PI4P head group was acquired from the crystal structure of the ARNO PH domain bound to the PIP2 head group (PDB ID 1U29) after alignment of the PH domains and deletion of the 5-phosphate. Magenta dashes represent hydrogen bonds between the T280 side chain hydroxyl group and the 1-phosphate of the PIP3 head group that are not observed for the PIP2 head group in the ARNO complex.
Besides ARF GEFs, multiple other classes of transport factors are also regulated by their recruitment from the cytosol to membranes (Sztul et al., 2019). Emerging from studies on all these factors has been a general appreciation that proper targeting to an intracellular compartment requires the coordination of multiple interactions, which has been termed coincidence detection (Sztul et al., 2019). Notably, besides providing targeting specificity, coincidence detection achieves another major purpose, allowing multiple weak interactions to synergize in forming an overall strong interaction. Along this line, we note that the effects of the T280 phosphorylation on individual Grp1 recruitment mechanisms (mediated by PI4P, GRASP, or IPCEF1) could be considered relatively modest. However, when the effects of the T280 phosphorylation are observed in the context of all three recruitment mechanisms, we have observed robust changes, as reflected by this phosphorylation converting Grp1 from its soluble form to becoming virtually all membrane-bound (Figure 4A).
MATERIALS AND METHODS
Reagents, proteins, and cells
Phospholipids and PIK93 were obtained from Avanti Polar Lipids (Alabaster, AL). Glutathione-sepharose 4B and PreScission protease were obtained from GE Healthcare (Pittsburgh, PA). Insulin, wortmannin, PAO, and adenosine were obtained from Sigma (St. Louis, MO). GST fusion proteins and 6xHis-tagged proteins were purified as described previously (Dai et al., 2004). Recombinant forms of Grp1 were generated by expressing them as GST fusion proteins in Escherichia coli, and then purified by binding to glutathione-sepharose resin, followed by PreScission protease cleavage according to the manufacturer’s instructions. 3T3-L1 fibroblast was obtained from ATCC (Rockville, MD). Their differentiation into adipocytes has been described previously (Li et al., 2007).
Antibodies
The following antibodies have been described previously (Li et al., 2007, 2012): mouse antibodies against Grp1 and Myc epitope (9E10), rabbit antisera against ARF6, cellubrevin, Grp1, and glut4, and anti-mouse and anti-rabbit secondary antibodies conjugated to HRP, Cy2, or Cy3. Newly acquired antibodies include mouse antibodies against PI4P (Z-P004) and PIP3 (Z-P345) from Echelon Biosciences (Salt Lake City, UT), mouse antibody against Rab11 (610656) from BD Transduction Laboratories (San Jose, CA), goat antibodies against GRASP (SC-55951) and IPCEF1 (SC-168195) from Santa Cruz (Dallas, TX), and anti-goat secondary antibodies conjugated to HRP (705-035-147) or Cy3 (705-165-147) from Jackson ImmunoResearch (West Grove, PA).
Plasmids and transfections
Myc-tagged mouse Grp1 (wild-type and T280D) in pENTR and pCDNA3.1 that are siRNA resistant have been described (Li et al., 2012). A PI4P biosensor, known as GFP-P4M-SidM, has also been described (Hammond et al., 2014), and was obtained from Addgene (plasmid #51469). 6xHis-tagged GRASP and IPCEF1 were generated by subcloning the coding sequence of GRASP and IPCEF1 into pET-15b vector (EMD Millipore, Billerica, MA). To generate GST fusions of Grp1, GRASP, and IPCEF1, the coding sequences of proteins were amplified by PCR, and then subcloned into pGEX-6P-1 vector (GE Healthcare, Pittsburgh, PA). Different forms of Myc-tagged Grp1 were subcloned and inserted into pENTR (Invitrogen, Carlsbad, CA). Point mutants were then generated by a QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Myc-Grp1 constructs in pLenti6.2 were further generated by recombination according to the Gateway protocol provided (Invitrogen). Lentiviral particles expressing Myc-Grp1 constructs were generated using a ViraPower Lentiviral Expression System (Invitrogen). 3T3-L1 fibroblast cell lines that stably express different forms of Myc-Grp1 were generated by lentiviral transduction followed by selection in 10 μg/ml blasticidin (Invitrogen).
RNA silencing
The siRNA smartpools for mouse PI4KIIIα, GRASP, IPCEF1, GRSP1, and CASP were obtained from Dharmacon (Chicago, IL). Individual oligonucleotides in smartpools that are documented in this study to have efficient knockdown are CCGAATGTTCAATGAGCAT, AGAAAGCACAGCTCGGAAA, and GATATGACAAGATGGGCTA for PI4KIIIα; ACGCAGCACTGGAGGACTA, GGGAGATTGTCGATATCAT, and GGAGAATACAGGTCACTTA for GRASP; CAGAAGGTGGTTTGTTGAA, TCAATCACCCACAGATCAA, and ACTCAAGCATATTTGCCAA for IPCEF1; GCACGGAGATCCTTGACGA, GGAGAGCAATTTCCTGATA, and GCACTTCGCTGTCAGGTTC for GRSP1; and GAAGACAACCGAAGGATTC, GGGAAGAGATGTGCGTTTA, and GAGACGAGATTCTGCGGAA for CASP. The first sequence against each target is used for all subsequent studies. A scrambled sequence was used as control, and was obtained from Ambion (Austin, TX). Transfection of siRNAs into 3T3-L1 adipocytes was achieved by using a Deliver-X Plus delivery kit (Affymetrix, Santa Clara, CA) according to the manufacturer’s protocol. Cells were transfected for 48 h before examination.
The efficiency of siRNA treatment was verified by either Western blotting or quantitative polymerase chain reaction (qPCR). For qPCR, total mRNAs were extracted from cells by using a Purelink RNA mini kit (Invitrogen), and then total cDNAs were synthesized by using a QuantiTect Reverse Transcription kit (QIAGEN, Germantown, MD). Quantitation of mRNA level was performed by using a Brilliant III Ultra-Fast SYBR Green QPCR Master Mix kit (Agilent Technologies, San Diego, CA) on a Stratagene MX3000P qPCR machine. The primer pairs for qPCR are GCA GCA CTG GAG GAC TAT C and TCT CCA AAG TTA GAA CCT TCC G for GRASP; CGT GTG CTT CCT GGG AAC and TCT CTG CAT TGC CTT CTG TG for IPCEF1; TCG GCA AAG ATG ACA GAA GG and AAG TAC TCC TTC TCT TTC AGG TTG for GRSP1; and TCT TTG CCC ACA ATG TCT CTG and TGA ATC CTT CGG TTG TCT TCC for CASP. The mRNA levels were normalized to respective scrambled control.
In vivo assays
The glut4 recycling assay has been previously described (Li et al., 2007). Colocalization studies were performed using a Nikon C1 plus confocal system and have also been described previously (Li et al., 2007, 2012). We examined noninsulin stimulated adipocytes for colocalization studies, which allows internal glut4 to be used as an additional marker of the recycling endosome, as previously described (Li et al., 2007, 2012). Quantitation was performed using Adobe Photoshop CS6 and NIH image analysis software packages and Image J (v. 1.50e) with a colocalization plug-in. For each condition, 10 cells were examined for quantitation.
In vitro assays
Pull-down assays using GST fusion proteins have also been described (Dai et al., 2004). In these experiments, the GST fusion protein is detected with Coomassie blue staining of gels while the soluble proteins bound to the GST fusion are detected with antibodies through immunoblotting, which are anti-Grp1 antibody to detect different Grp1 forms and anti-6xHis antibody to detect 6xHis-tagged GRASP and IPCEF1. The liposome recruitment assay was performed essentially as previously described (Yang et al., 2008). Briefly, lipids were mixed in molar ratios to mimic the composition of organellar membrane (PC 58%, PE 20%, cholesterol 14%, PS 4%, and sphingomyelin 4%). Unilamellar liposomes were then generated in a buffer containing 20 mM HEPES, pH 7.4, and 170 mM sucrose, and resuspended in a buffer containing 20 mM HEPES, pH 7.4, and 100 mM KCl. For recruitment studies involving specific phosphoinositide, liposomes were generated with 10% of a particular phosphoinositide, with the level of PC reduced proportionally. For studies involving a nickel-conjugated lipid, liposomes also contained 10% of 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (nickel salt).
Supplementary Material
Acknowledgments
We thank Andrew Malaby for pilot experiments that led to this study, and Jia-Shu Yang, Seung-Yeol Park, and Jia-Wei Hsu for advice and comments. This work is supported by grants from the National Institutes of Health to V.W.H. (Grant no. GM-115683) and D.G.L. (Grant no. GM-056324), and from the American Heart Association to J.L. (Grant no. 13SDG13920005).
Abbreviations used:
- ARF
ADP-ribosylation factor
- ARNO
ARF nucleotide site opener
- Cbv
cellubrevin
- CC
coil-coil domain
- GEF
guanine nucleotide exchange factor
- glut4
glucose transporter type 4
- Grp1
general receptor for 3-phosphoinositides 1
- PAO
phenylarsine oxide
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PH
pleckstrin homology domain
- PI(3,4)P2
phosphatidylinositol 3,4-biphosphate
- PI(3,5)P2
phosphatidylinositol 3,5-biphosphate
- PI(4,5)P2
phosphatidylinositol 4,5-biphosphate
- PI3P
phosphatidylinositol 3-phosphate
- PI4K
PI4 kinase
- PI4P
phosphatidylinositol 4-phosphate
- PI5P
phosphatidylinositol 5-phosphate
- PIP3
phosphatidylinositol 3,4,5-trisphosphate
- PS
phosphatidylserine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-03-0173) on October 7, 2020.
REFERENCES
- Balla A, Balla T (2006). Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol , 351–361. [DOI] [PubMed] [Google Scholar]
- Casanova JE (2007). Regulation of Arf activation: the Sec7 family of guanine nucleotide exchange factors. Traffic , 1476–1485. [DOI] [PubMed] [Google Scholar]
- Chardin P, Paris S, Antonny B, Robineau S, Beraud-Dufour S, Jackson CL, Chabre M (1996). A human exchange factor for ARF contains Sec7- and pleckstrin-homology domains. Nature , 481–484. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Menetrey J, Mathieu M, Le Bras G, Robineau S, Beraud-Dufour S, Antonny B, Chardin P (1998). Structure of the Sec7 domain of the Arf exchange factor ARNO. Nature , 101–105. [DOI] [PubMed] [Google Scholar]
- Cohen LA, Honda A, Varnai P, Brown FD, Balla T, Donaldson JG (2007). Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol Biol Cell , 2244–2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Li J, Bos E, Porcionatto M, Premont RT, Bourgoin S, Peters PJ, Hsu VW (2004). ACAP1 promotes endocytic recycling by recognizing recycling sorting signals. Dev Cell , 771–776. [DOI] [PubMed] [Google Scholar]
- DiNitto JP, Lambright DG (2006). Membrane and juxtamembrane targeting by PH and PTB domains. Biochim Biophys Acta , 850–867. [DOI] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006). Phosphoinositides in cell regulation and membrane dynamics. Nature , 651–657. [DOI] [PubMed] [Google Scholar]
- Donaldson JG, Jackson CL (2011). ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat Rev Mol Cell Biol , 362–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drin G, Morello V, Casella JF, Gounon P, Antonny B (2008). Asymmetric tethering of flat and curved lipid membranes by a golgin. Science , 670–673. [DOI] [PubMed] [Google Scholar]
- D’Souza-Schorey C, van Donselaar E, Hsu VW, Yang C, Stahl PD, Peters PJ (1998). ARF6 targets recycling vesicles to the plasma membrane: insights from an ultrastructural investigation. J Cell Biol , 603–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson KM, Kavran JM, Sankaran VG, Fournier E, Isakoff SJ, Skolnik EY, Lemmon MA (2000). Structural basis for discrimination of 3-phosphoinositides by pleckstrin homology domains. Mol Cell , 373–384. [DOI] [PubMed] [Google Scholar]
- Foley K, Boguslavsky S, Klip A (2011). Endocytosis, recycling, and regulated exocytosis of glucose transporter 4. Biochemistry , 3048–3061. [DOI] [PubMed] [Google Scholar]
- Goldberg J (1998). Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell , 237–248. [DOI] [PubMed] [Google Scholar]
- Hammond GR, Machner MP, Balla T (2014). A novel probe for phosphatidylinositol 4-phosphate reveals multiple pools beyond the Golgi. J Cell Biol , 113–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiester KG, Santy LC (2013). The cytohesin coiled-coil domain interacts with threonine 276 to control membrane association. PLoS One , e82084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado-Lorenzo A, Skinner M, El Annan J, Futai M, Sun-Wada GH, Bourgoin S, Casanova J, Wildeman A, Bechoua S, Ausiello DA, et al. (2006). V-ATPase interacts with ARNO and Arf6 in early endosomes and regulates the protein degradative pathway. Nat Cell Biol , 124–136. [DOI] [PubMed] [Google Scholar]
- Kandror KV, Pilch PF (2011). The sugar is sIRVed: sorting Glut4 and its fellow travelers. Traffic , 665–671. [DOI] [PubMed] [Google Scholar]
- Karandur D, Nawrotek A, Kuriyan J, Cherfils J (2017). Multiple interactions between an Arf/GEF complex and charged lipids determine activation kinetics on the membrane. Proc Natl Acad Sci USA , 11416–11421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klarlund JK, Guilherme A, Holik JJ, Virbasius JV, Chawla A, Czech MP (1997). Signaling by phosphoinositide-3,4,5-trisphosphate through proteins containing pleckstrin and Sec7 homology domains. Science , 1927–1930. [DOI] [PubMed] [Google Scholar]
- Klarlund JK, Holik J, Chawla A, Park JG, Buxton J, Czech MP (2001). Signaling complexes of the FERM domain-containing protein GRSP1 bound to ARF exchange factor GRP1. J Biol Chem , 40065–40070. [DOI] [PubMed] [Google Scholar]
- Lee MC, Orci L, Hamamoto S, Futai E, Ravazzola M, Schekman R (2005). Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell , 605–617. [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2008). Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol , 99–111. [DOI] [PubMed] [Google Scholar]
- Leto D, Saltiel AR (2012). Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol , 383–396. [DOI] [PubMed] [Google Scholar]
- Li J, Malaby AW, Famulok M, Sabe H, Lambright DG, Hsu VW (2012). Grp1 plays a key role in linking insulin signaling to glut4 recycling. Dev Cell , 1286–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Peters PJ, Bai M, Dai J, Bos E, Kirchhausen T, Kandror KV, Hsu VW (2007). An ACAP1-containing clathrin coat complex for endocytic recycling. J Cell Biol , 453–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lietzke SE, Bose S, Cronin T, Klarlund J, Chawla A, Czech MP, Lambright DG (2000). Structural basis of 3-phosphoinositide recognition by pleckstrin homology domains. Mol Cell , 385–394. [DOI] [PubMed] [Google Scholar]
- Lowery J, Szul T, Styers M, Holloway Z, Oorschot V, Klumperman J, Sztul E (2013). The Sec7 guanine nucleotide exchange factor GBF1 regulates membrane recruitment of BIG1 and BIG2 guanine nucleotide exchange factors to the trans-Golgi network (TGN). J Biol Chem , 11532–11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour M, Lee SY, Pohajdak B (2002). The N-terminal coiled coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with the scaffolding protein CASP. J Biol Chem , 32302–32309. [DOI] [PubMed] [Google Scholar]
- Nevrivy DJ, Peterson VJ, Avram D, Ishmael JE, Hansen SG, Dowell P, Hruby DE, Dawson MI, Leid M (2000). Interaction of GRASP, a protein encoded by a novel retinoic acid-induced gene, with members of the cytohesin family of guanine nucleotide exchange factors. J Biol Chem , 16827–16836. [DOI] [PubMed] [Google Scholar]
- Quilty D, Gray F, Summerfeldt N, Cassel D, Melancon P (2014). Arf activation at the Golgi is modulated by feed-forward stimulation of the exchange factor GBF1. J Cell Sci , 354–364. [DOI] [PubMed] [Google Scholar]
- Richardson BC, McDonold CM, Fromme JC (2012). The Sec7 Arf-GEF is recruited to the trans-Golgi network by positive feedback. Dev Cell , 799–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztul E, Chen PW, Casanova JE, Cherfils J, Dacks JB, Lambright DG, Lee FS, Randazzo PA, Santy LC, Schurmann A, et al. (2019). ARF GTPases and their GEFs and GAPs: concepts and challenges. Mol Biol Cell , 1249–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PC, Hsu JW, Liu YW, Chen KY, Lee FJ (2013). Arl1p regulates spatial membrane organization at the trans-Golgi network through interaction with Arf-GEF Gea2p and flippase Drs2p. Proc Natl Acad Sci USA , E668–E677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol , 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkataraman A, Nevrivy DJ, Filtz TM, Leid M (2012). Grp1-associated scaffold protein (GRASP) is a regulator of the ADP ribosylation factor 6 (Arf6)-dependent membrane trafficking pathway. Cell Biol Int , 1115–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkateswarlu K (2003). Interaction protein for cytohesin exchange factors 1 (IPCEF1) binds cytohesin 2 and modifies its activity. J Biol Chem , 43460–43469. [DOI] [PubMed] [Google Scholar]
- Yang JS, Gad H, Lee SY, Mironov A, Zhang L, Beznoussenko GV, Valente C, Turacchio G, Bonsra AN, Du G, et al. (2008). A role for phosphatidic acid in COPI vesicle fission yields insights into Golgi maintenance. Nat Cell Biol , 1146–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.