Abstract
Nuclear envelope proteins influence cell cytoarchitecure by poorly understood mechanisms. Here we show that small interfering RNA–mediated silencing of lamin A/C (LMNA) promotes contrasting stress fiber assembly and disassembly in individual cells and within cell populations. We show that LMNA-deficient cells have elevated myosin-II bipolar filament accumulations, irregular formation of actin comet tails and podosome-like adhesions, increased steady state nuclear localization of the mechanosensitive transcription factors MKL1 and YAP, and induced expression of some MKL1/serum response factor–regulated genes such as that encoding myosin-IIA (MYH9). Our studies utilizing live cell imaging and pharmacological inhibition of myosin-II support a mechanism of deregulated myosin-II self-organizing activity at the nexus of divergent actin cytoskeletal aberrations resulting from LMNA loss. In light of our results, we propose a model of how the nucleus, via linkage to the cytoplasmic actomyosin network, may act to control myosin-II contractile behavior through both mechanical and transcriptional feedback mechanisms.
INTRODUCTION
Mutations to LINC (linker of nucleoskeleton and cytoskeleton; Crisp et al., 2006) complex proteins or to proteins comprising the nuclear lamina, type A and B lamin intermediate filaments, give rise to pleiotropic human diseases collectively termed nuclear envelopathies (Burke and Stewart, 2006). The molecular and cellular pathogenesis of these diseases is not well understood but has been associated with perturbed gene regulation, altered cell mechanics, and impaired force transduction between the nucleus and the cytoskeleton (Broers et al., 2004; Lammerding et al., 2004; Stewart-Hutchinson et al., 2008). Mechanical signals originating with cytoskeletal filaments are transmitted to the nucleus via the LINC complex (Wallrath et al., 2016). The mechanotransduction LINC apparatus consists of outer nuclear membrane nesprins, which bind cytoskeletal filaments and also couple to inner nuclear membrane localized Sun-domain proteins, which in turn are tethered to the nuclear lamina through association with lamins. Cells deficient of lamins A and C, products of a single gene via alternative splicing, or cells with LINC complex impairment exhibit defects in cytoskeletal organization. However, there have been contrasting reports of cytoskeletal dysfunction following nuclear lamina disruption (via lamin A/C [LMNA] or LINC complex perturbations). LMNA-deficient cells, for instance, have variously been reported to have normal actin organization, increased fiber formation and increased contractile behavior, and alternatively, stress fiber loss with reduced contractility (Lammerding et al., 2004; Corne et al., 2017; van Loosdregt et al., 2017; Osmanagic-Myers et al., 2019). A rationale for this seemingly paradoxical collection of reports has not been identified.
Cytoarchitecture is directed by both F-actin arrangement and mechanical forces generated by the contractile activity of nonmuscle myosin-II (myo-II) molecular motors, which transport actin filaments relative to each other in an ATP-dependent cycle (Levayer and Lecuit, 2012). Individual myo-II motors are nonprocessive but form functional ensembles of variable numbers of bioriented units comprising single myo-II bipolar filaments. The activity of myo-II is regulated at multiple levels including phosphorylation by Rho-family effector kinases and through less characterized mechanisms such as via mechanical feedback from its own activity (Kasza and Zallen, 2011; Aguilar-Cuenca et al., 2014). Studies utilizing purified proteins in vitro, and of different cellular systems, have demonstrated the ability of myo-II to self-organize to diverse cytoskeletal patterns such as F-actin bundles, actomyosin asters, and networks with a dynamic steady state (Backouche et al., 2006; Levayer and Lecuit, 2012). Significantly, myo-II activity can drive both the assembly and disassembly of networks of coassembled F-actin and myosin.
Organization of the F-actin cytoskeleton significantly impacts nuclear function. For example, contingent upon F-actin assembly and mechanical signaling, key transcription coactivators such as YAP and MKL1 (also named MRTF-A and MAL) are either retained in the cytoplasm or shuttled to the nucleus, thereby modulating the expression of a broad array of genes important for various cell functions (Miralles et al., 2003; Olson and Nordheim, 2010; Aragona et al., 2013). Recently it was suggested that F-actin cytoskeletal perturbations resulting from LMNA deficiency may be the result of abrogated actin-dependent signaling that prevents proper translocation of MKL1 to the nucleus (Ho et al., 2013b). Loss of MKL1 coactivation of a master cytoskeletal transcriptional module mediated by serum response factor (SRF) and concomitant reduced expression of cytoskeletal genes, such as actin itself, was proposed as a mechanism for some cytoskeletal defects in LMNA-deficient cells. Nevertheless, alternative mechanisms must still be explored since, for instance, nuclear translocation of MKL1 is not impaired in all LMNA-deficient cells (Ho et al., 2013a). Furthermore, global reduced cytoskeletal gene expression appears insufficient to explain reported cases of enhanced contractile F-actin assembly in LMNA-deficient cells.
To begin to elucidate the manner by which nuclear envelope proteins may affect cytoskeletal organization, we have examined the effects of small interfering RNA (siRNA)-mediated depletion of LMNA in human cells. Remarkably, we find that depletion of LMNA, nesprins, or Sun proteins similarly produce localized and contrasting actomyosin cytoskeletal perturbations, even within single cells. We identify myosin-II assembly enhancement in LMNA-depleted cells. Our data link deregulated myosin-II and its self-organizing activity to both localized increased actomyosin assembly and disassembly associated with aberrant cytoskeletal phenotypes following LMNA depletion. We further demonstrate increased expression of myosin-2A in conjunction with elevated nuclear accumulation of both MKL1 and YAP, following LMNA depletion, in contrast to expectations based on some previous reports.
RESULTS
Depletion of nuclear envelope proteins induces divergent actomyosin structural anomalies
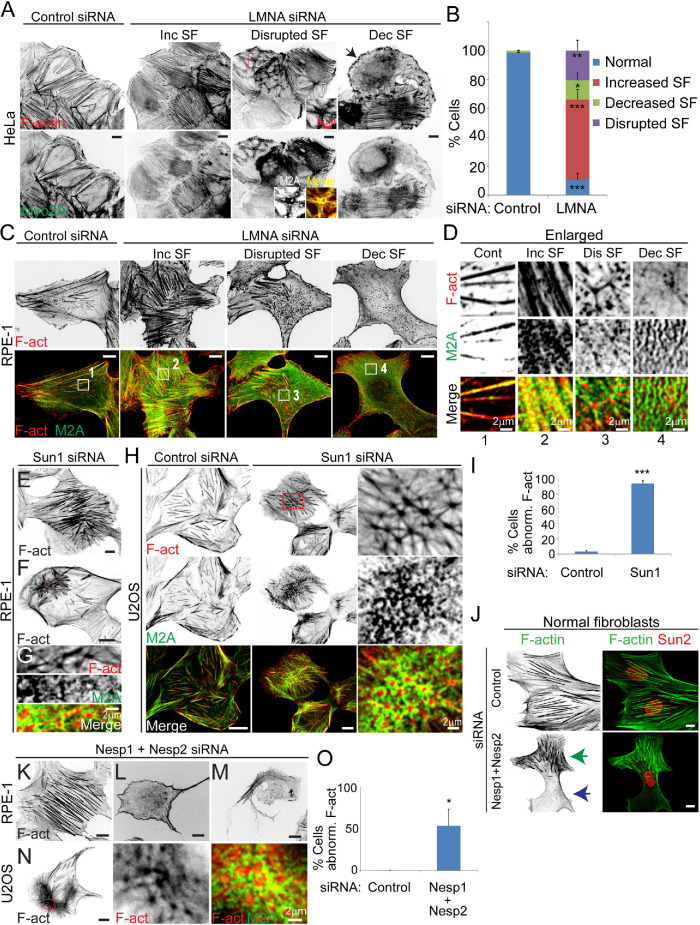
Using multiple siRNAs previously characterized to cause nuclear depletion of their respective targeted LINC complex proteins (Wiggan et al., 2017), we evaluated cytoskeletal organization following silencing of individual LINC complex proteins. Fascinatingly, silencing of LINC module components (hereafter referring to either outer nuclear membrane nesprin1 or nesprin2, inner nuclear membrane Sun1 or Sun2, and LMNA) in multiple human cell types including HeLa, U2OS, nontransformed RPE-1, and normal fibroblasts (GM-2149) resulted in similar divergent aberrant actomyosin cytoskeletal phenotypes (Figure 1). The majority of cells (∼55% for LMNA silencing) displayed increased numbers of thick basal stress fibers relative to controls. Additionally, we observed two other general phenotypic groups. One group showed much reduced levels or complete absence of basal stress fibers (particularly fibers beneath the nucleus), and the other had cells with a disrupted network of stress fibers; both groups were also characterized by the presence of actomyosin asters, F-actin condensates, or fragmented F-actin bundles (for examples see Figure 1, A, D, H, G, and N, enlargements). Remodeling of the actomyosin network occurred throughout the entire cell and was not restricted to basal stress fibers. Notably, however, actomyosin remodeling was regionally divergent such that individual cells simultaneously exhibited regions of increased F-actin bundling and those of stress fiber loss or fragmentation (e.g., Figure 1J, arrows).
FIGURE 1:
LINC complex disruption in multiple human cell types induces contrasting F-actin cytoskeletal anomalies. (A, B) Basal confocal immunofluorescence images of HeLa cells labeled for F-actin (phalloidin) and myosin-2A and quantification of F-actin phenotypes. Arrow depicts cell with decreased numbers of stress fibers (SF) and the presence of F-actin condensates. Insets show enlargements and merged images (bottom row) of boxed region depicting rings of myosin-II surrounding interconnected F-actin asters. Values are mean ± SD, control n = 1192, LMNA siRNA n = 473. (C–G) Confocal immunofluorescence images of RPE-1cells siRNA treated for depletion of LMNA (C, D) and Sun1 (E–G). Numbered boxed regions (C, bottom row) are shown enlarged (D). Cells with increased SF (E), disrupted SF network (F), relative to control (C) are illustrated. Boxed region (F) is enlarged in G. (H) Immunofluorescence images of U2OS cells. Boxed region is enlarged in last column and shows a disrupted network of actomyosin asters. (I) Quantification of total abnormal F-actin phenotypes in RPE-1 cells, as classified in B. Values are mean ± SD, control n = 180, Sun1 siRNA = 163. (J–N) Immunofluorescence images of normal human fibroblasts (J), RPE-1 (K–M), and U2OS (N). Increased SF formation (K), stress fiber loss (L, M), and disrupted stress fiber network (N) are shown. Green and blue arrows (J) depict regions of increased thick SF and stress fiber loss, respectively. Boxed region (N) is enlarged in subsequent columns. (O) Quantification of total abnormal F-actin phenotypes in RPE-1 cells, as classified in B. Values are mean ± SD, control n = 377, nesprin1 (Nesp1) + nesprin2 (Nesp2) siRNA = 256. Inc, increased; Dis, disrupted; Dec, decreased; F-act, F-actin; M2A, myosin-2A. * p < 0.05, ** p ≤ 0.01, *** p ≤ 0.001, ns, not significant, Welch’s t test. Bars, 10 μm except where noted.
Contrasting cytoskeletal phenotypes resulting from depletion of LINC module proteins could possibly result from different degrees of LINC protein silencing between cells. In disfavor of this hypothesis, immunofluorescence staining of residual LMNA levels in both HeLa and RPE-1 cells revealed low LMNA levels for cells displaying increased stress fibers comparable to those exhibiting stress fiber loss (Supplemental Figure S1, A and B). Further, quantification of basal F-actin accumulations revealed no significant correlations to residual cellular LMNA levels across cells. Curiously, heterogeneity in cytoskeletal defects within and between cells was a highlighted feature of fibroblasts from LMNA null mice in which all cells had the same level of genetic LMNA depletion (Broers et al., 2004). Together, these data suggest that differing cytoskeletal phenotypes are not coupled to varying residual levels of LINC proteins following siRNA-mediated depletion.
LMNA silencing induces increased contractile cell behavior without significant changes to p-MLC levels
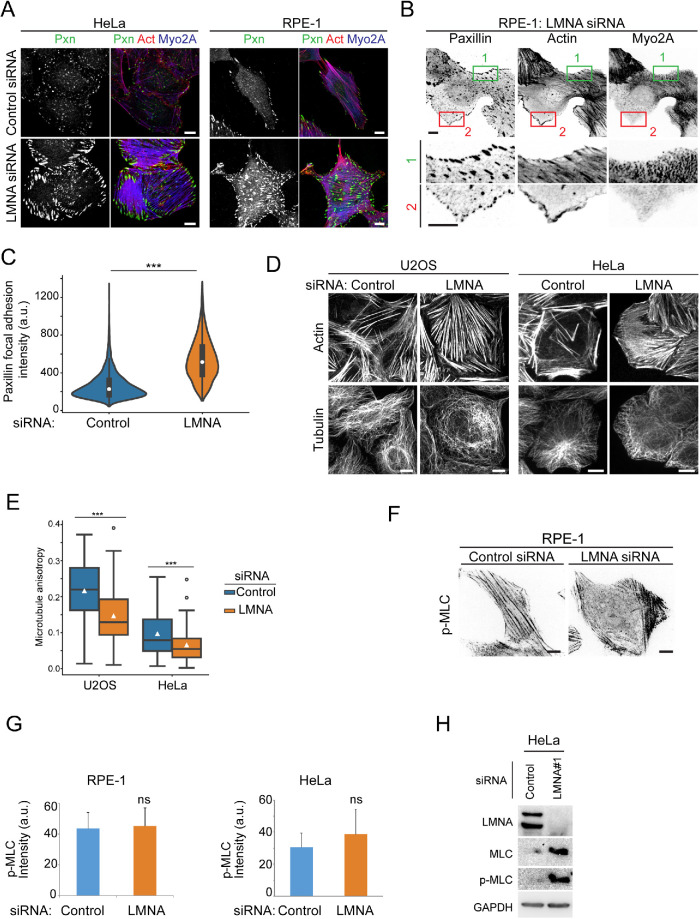
The predominance of cells with increased stress fiber formation following LMNA silencing suggested increased contractile myo-II activity in response to this treatment. Myosin contractility is essential for focal adhesion maturation, which is associated with enhanced recruitment of core focal adhesion proteins (Schwarz and Gardel, 2012). Examination of focal adhesions revealed overall significantly stronger accumulation (88% increase in RPE-1 cells) of the focal adhesion protein paxillin to adhesions in LMNA-depleted cells for both HeLa and RPE-1 cells (Figure 2, A–C). Interestingly, as observed for stress fibers, individual LMNA-depleted cells displayed heterogeneous localized regions of either focal adhesion enhancement or reduction, coincident with that of stress fiber organization. Figure 2B, for example, illustrates a region of increased actomyosin and paxillin accumulations (region 1) in contrast to a region of the same cell with stress fiber loss, which is associated with diminished paxillin accumulations (region 2). Heterogeneity in focal adhesion accumulations in LMNA-silenced cells was also manifested by the broader distribution of adhesion intensities for these cells relative to controls (Figure 2C). Given that myosin contractile activity can drive both actin network assembly and disassembly, the contrasting cytoskeletal rearrangements, evidenced by stress fibers and focal adhesions, are equally consistent with elevated contractile myo-II activity following LMNA disruption. We will return to discuss the seeming paradox of these concurrent opposing responses in more detail later.
FIGURE 2:
LMNA depletion elevates focal adhesion maturation without significantly altering global p-MLC levels. (A, B) Confocal immunofluoresence labeling for paxillin (pxn), F-actin (actin), and Myosin-2A (Myo2A). Boxed regions (B, top panels) are labeled correspondingly in magnified bottom panels. (C) Violin plot of focal adhesion paxillin intensity distributions for RPE-1 cells, n ≥ 30 cells/treatment. Bottom and top box horizontal lines of inner plot show 25th and 75th percentiles, respectively; white circle shows the median value. (D, E) Immunofluorescence confocal images and quantification of microtubule anisotropy. Box plots (E) show interquartile range, and white triangle is mean value. U2OS n ≥ 52, HeLa n ≥ 59. Immunofluorescence labeling (F), quantification of normalized cell p-MLC fluorescence intensity (G), and immunoblot (H) for active myosin-II (Ser-19 phosphorylated myosin light chain). Values are mean± SD, RPE-1 n ≥ 101, HeLa n ≥ 426. Bars, 10 μm except where noted. *** p < 0.001, ns, not significant, Welch’s t test.
Microtubule disassembly is known to stimulate formation of myo-II minifilaments, stress fibers, and focal adhesion (Liu et al., 1998; Krendel et al., 2002; Rafiq et al., 2019) so we assessed microtubule organization in different LMNA-depleted cell types. HeLa and U2OS LMNA-silenced cells displaying increased stressed fibers both maintained microtubules similar to controls (Figure 2D). Nevertheless, LMNA depletion induced microtubule reorganization, from a more ordered radial pattern in control cells to overlapping curved bundles manifested by increased microtubule fibrillar disorder (Figure 2E, reduced anisotropy, where 0 is isotropic and 1 is perfectly ordered; Boudaoud et al., 2014). These data show that LMNA deficiency contributes to altered microtubule patterning; however, the absence of gross microtubule loss in LMNA-depleted cells suggests that the latter feature was not a significant mechanistic factor for increased actomyosin assembly.
We next examined whether increased contractile behavior following LMNA depletion was associated with altered myo-II phospho-activation. Immunostaining for S19 phosphorylated myosin light chain (p-MLC, phospho-active myosin) revealed a trend toward increased p-MLC; however, p-MLC levels were not significantly different between controls and either RPE-1 or HeLa LMNA-depleted cells (Figure 2, F and G). Likewise, whereas immunoblots indicated increased (approximately fourfold) p-MLC levels for LMNA-depleted cells, this rise was in parallel to an approximately fivefold increase in total MLC levels, relative to controls (Figure 2H). Collectively, these data demonstrate that LMNA depletion stimulates increased cell contractile activity without apparent significant increases to steady state p-MLC levels.
LMNA deficiency induces aberrant membrane-associated actomyosin remodeling
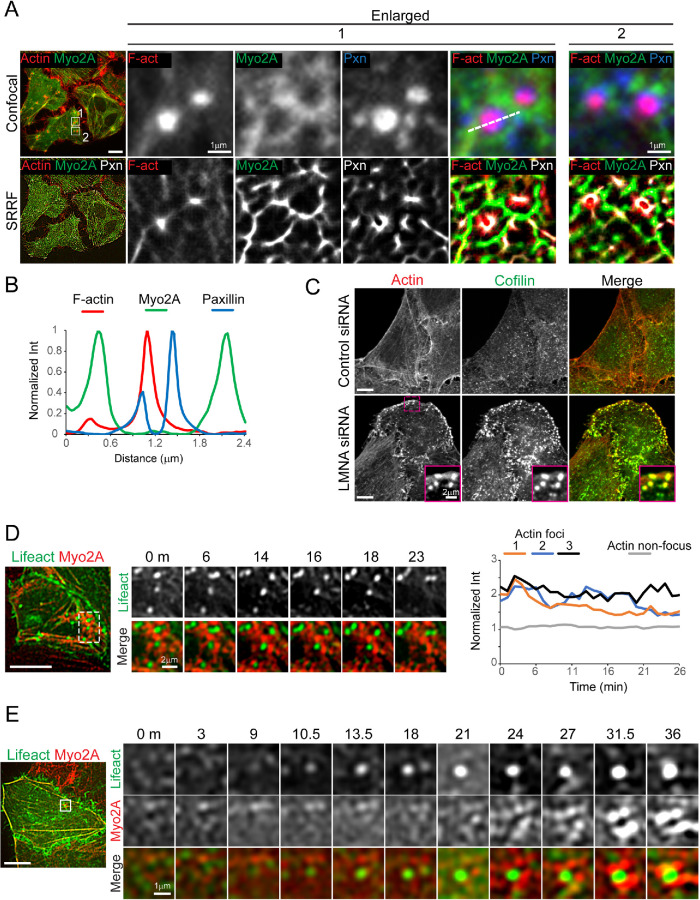
Myosin activity is important for dynamic remodeling of actin filaments in organization of various membrane structures (Koster et al., 2016; Vogel et al., 2017). If LMNA contributes to altered myo-II function, we hypothesized that LMNA deficiency may induce perturbations to membrane-associated actomyosin. In vitro, myosin self-organization can produce aster-like structures, where myo-II localizes to the center of the aster (Backouche et al., 2006; Soares e Silva et al., 2011); in contrast, aster-like structures at the basal plasma membrane in LMNA-depleted cells displayed a central core of actin surrounded by a ring of myo2A (see, for example, Figure 1, A, inset, and D, Dis SF). The latter organization is reminiscent of podosomes, plasma membrane–extracellular matrix adhesion structures, found in certain normal cell types and some transformed cells (Marchisio, 2012; Alonso et al., 2019). The actin core of podosomes is surrounded by outer rings of integrin-associated proteins, such as paxillin, and myo2A, respectively. Confocal images resolved below the diffraction limit using superresolution radial fluctuation (SRRF) computation (Gustafsson et al., 2016) revealed this organization among actin condensates in LMNA-depleted HeLa cells (Figure 3, A and B). Unlike in HeLa cells, podosome-like structures in RPE-1 cells did not assemble a ring of paxillin (Supplemental Figure S2), suggesting a difference between these two cell types in their ability to form full podosome-type adhesions. Other distinctive features of podosome-type adhesions, such as recruitment of the actin-binding protein cofilin to the actin core (Yamaguchi et al., 2005), were evident for podosome-like adhesions in HeLa cells (Figure 3C). Myosin contractile activity is essential for oscillatory intensity fluctuations of the actin core of podosomes (van den Dries et al., 2013), and this oscillatory behavior was displayed by podosome-like adhesions that formed in LMNA-deficient HeLa cells (Figure 3D; Supplemental Video S1). Live imaging suggested spontaneous formation of the actin core, which appeared to intensify and be stabilized by mutual assembly reinforcement with proximal encircling myo2A puncta (Figure 3E). These data highlight cell type–specific aberrant membrane-associated actomyosin remodeling in association with LMNA deficiency.
FIGURE 3:
LMNA silencing induces formation of podosome-type adhesions. (A) Confocal immunofluorescence (top panels) and SRRF images of LMNA siRNA–treated HeLa cells labeled for paxillin (pxn), F-actin (F-act), and myosin-2A (myo2A). Numbered boxed regions (left column) are shown enlarged, correspondingly. (B) Normalized line intensity profile plots, from line shown for region 1, top row, obtained from SRRF images. (C) Immunolabeled confocal images of fixed HeLa cells. Boxed region (LMNA siRNA) is shown magnified in insets. (D, E) Confocal video stills of HeLa Myo2-mCh cells, coexpressing GFP-Lifeact for F-actin labeling and siRNA treated for LMNA silencing. Boxed regions (left panels) are enlarged in time-series images (right panels). See also Supplemental Video S1. Graph (D) plots podosome-like structure actin core intensity with time. Scale bars, 10 μm.
Movie S1.
Time-lapse confocal microscopy of basal z-projection planes for HeLa cells expressing myosin-2A-mCherry and transiently transfected GFP-Lifeact (F-actin labeling). Images were captured at 1 min intervals, 48h post siRNA treatment and the video rate is 20 frames/s. Related to Figure 3.
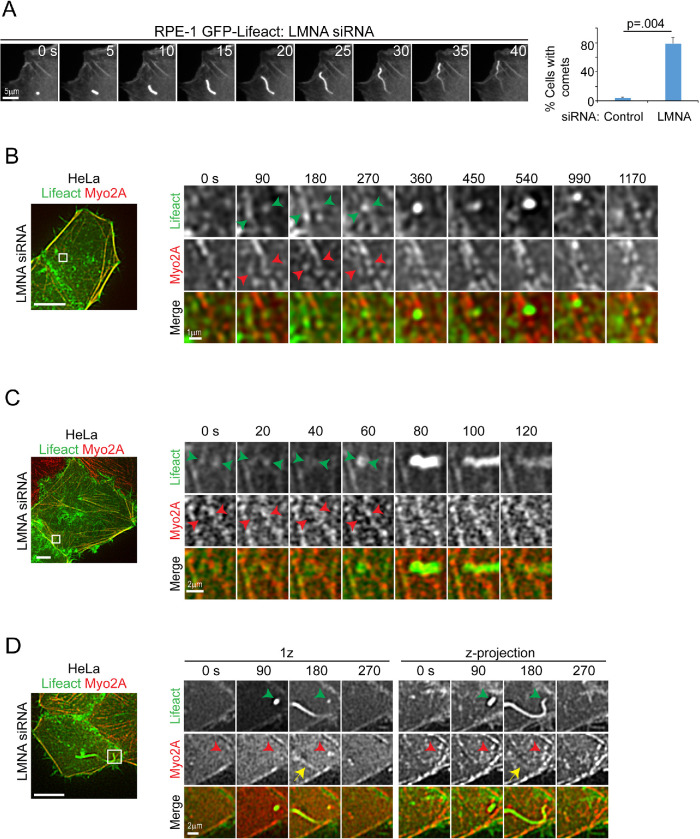
Live cell analyses of both HeLa and RPE-1 cells showed an increased presence of dynamic membrane-linked actin structures known as comet tails (Cameron et al., 2000) in LMNA-depleted cells relative to controls (Figure 4A; Supplemental Videos S2 and S3). In vitro models of actin comet formation do not show a necessity for myosin; however, some reports implicate association of myo-II with comet activity in cellular models of pathogen transmission (Rathman et al., 2000; Lum and Morona, 2014). We assessed whether myo-2A was associated with these actin assemblies in LMNA-silenced cells. As illustrated, Figure 4, B–D (arrowheads), we identified several instances in which translocation of actin-myo2A structures to a cluster was concurrent with the formation of an actin focus or comet. Figure 4D (yellow arrow; also see Supplemental Video S3) also shows that myo2A can incorporate to comet tails, further suggesting myo-II involvement in the formation of these structures in LMNA-depleted cells. Altogether, our analyses thus far are consistent with a hypothesis that deregulated actomyosin remodeling contributes to the major actin cytoskeletal defects, including those associated with cell membranes, obtained from LMNA deficiency.
FIGURE 4:
Localized actomyosin remodeling is associated with increased assembly of F-actin to comet tails in LMNA-depleted cells. (A) Confocal time-lapse series and quantification of F-actin comet formation in RPE-1 cells. Values are mean ± SD, n = 90/treatment. Welch’s t test. See also Supplemental Video S2. (B–D) Confocal video stills of HeLa Myo2-mCh cells, coexpressing GFP-Lifeact for F-actin labeling and siRNA treated for LMNA silencing. Left panels are basal cell z-projections, and boxed regions are enlarged in time series of selected z-planes (right panels). Arrowheads (B, C) depict movements that cluster actin-myosin structures at sites of heightened actin assembly. Arrowheads and yellow arrow (D) show myo2A accumulation (red arrowhead) at a site of comet tail formation (green arrowhead) and myo2A localization to the nascent comet tail (yellow arrow), respectively, for single and z-projected images. See also Supplemental Video S3. Scale bars, 10 μm except where noted.
Movie S2.
Time-lapse confocal microscopy of control and LMNA siRNA depleted RPE-1 cells depicting F-actin (GFP-Lifeact) comet formations (red circles). Images were captured at 5 sec intervals and the video rate is 10 frames/s. Related to Figure 4.
Movie S3.
Time-lapse confocal microscopy of basal z-projections for LMNA siRNA treated HeLa cells expressing myosin-2A-mCherry and transiently transfected GFP-Lifeact. Red arrow illustrates comet tail with myosin-2A labeling. Images were captured at 90 sec intervals and the video rate is 3 frames/s. Related to Figure 4.
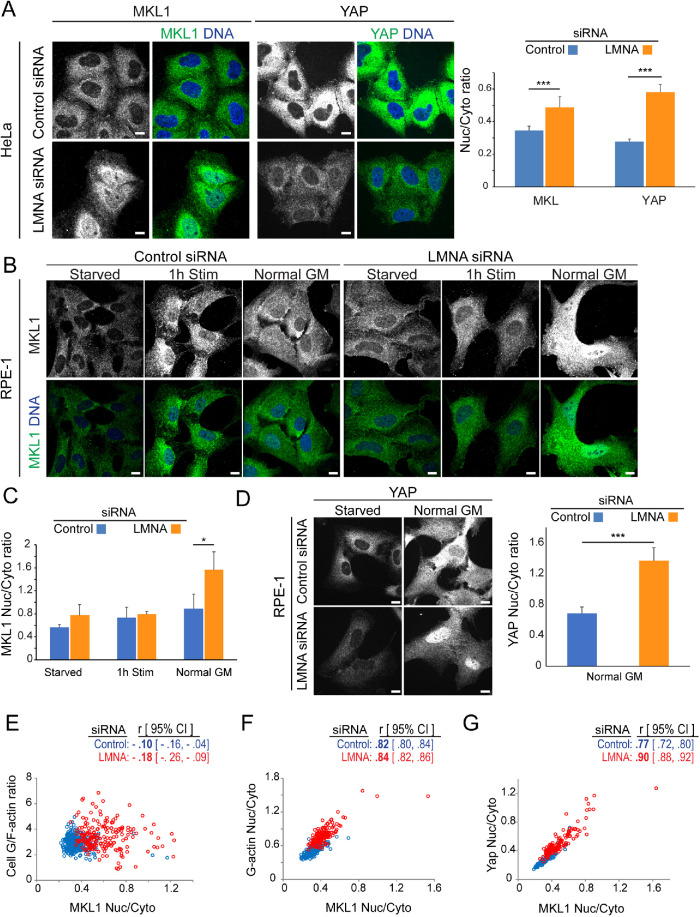
Increased nuclear accumulation for mechanosensitive transcription factors MKL1 and YAP in LMNA-depleted cells
Our results indicate that LMNA deficiency caused increased myo-II contractile behavior resulting, for example, in increased stress fibers and focal adhesion maturation. These data insinuated that LMNA deficiency may in fact stimulate mechanosensitive transcription pathways, such as for MKL1 and YAP. Furthermore, recent studies implicate mutual dependence of both MKL1 and YAP for regulation of their respective transcription targets (Foster et al., 2017). Examination of MKL1 and YAP cellular localization showed their increased nuclear localization following LMNA silencing for both HeLa (Figure 5A) and RPE-1 (Figure 5, B–D) cells grown in normal serum-containing medium. Our results for MKL1 in cells grown in normal growth medium differ from those previously reported for some LMNA-deficient cells stimulated from quiescence, where nuclear MKL1 accumulation was abrogated relative to controls (Ho et al., 2013b). However, at conditions of previous reports, that is, serum stimulation from quiescence, we did not detect any significant differences in MKL1 nuclear accumulation in LMNA-depleted cells relative to controls (Figure 5, B and C). These data imply that effects of LMNA deficiency on MKL1 nuclear translocation may be contextual. Separately, we uncovered strong correlations (Pearson’s r ≥ 0.8) between the nuclear/cytoplasmic ratio for MKL1 relative to those of both G-actin (Figure 5F) and YAP (Figure 5G), consistent with likely coordinate regulation of MKL1 localization with both factors. Interestingly, however, MKL1 nuclear/cytoplasmic distribution did not show a strong correlation to cellular G/F-actin ratios (Figure 5E), as might be expected based on current models of MKL1 activation. Altogether, these results advocate that LMNA depletion induced activation of both MKL1 and YAP nuclear translocation in the context of steady state cell growth conditions.
FIGURE 5:
LMNA silencing induces nuclear accumulation of mechanosensitive transcription factors. (A–D) Representative nuclear-level confocal immunofluorescence images and quantification of transcription factor nuclear-cytoplasmic ratios for HeLa cells (A) or RPE-1 cells (B–D) at 72 h post–siRNA treatments, as indicated. Cells (A and as specified) were cultured in normal growth medium (Normal GM) or serum stimulated (1 h stim) from serum starvation (starved) as indicated. Scale bars, 10 μm. Graph values are mean ± SD, n ≥ 115/treatment. (E–G) Representative per cell normalized fluorescence intensity correlation plots of HeLa cells treated as indicated. Values, r, are mean Pearson correlation coefficient and [95% confidence interval], n ≥ 283/treatment. Nuc, nuclear; Cyto, cytoplasmic. * p ≤ 0.05, *** p ≤ 0.001, Welch’s t test.
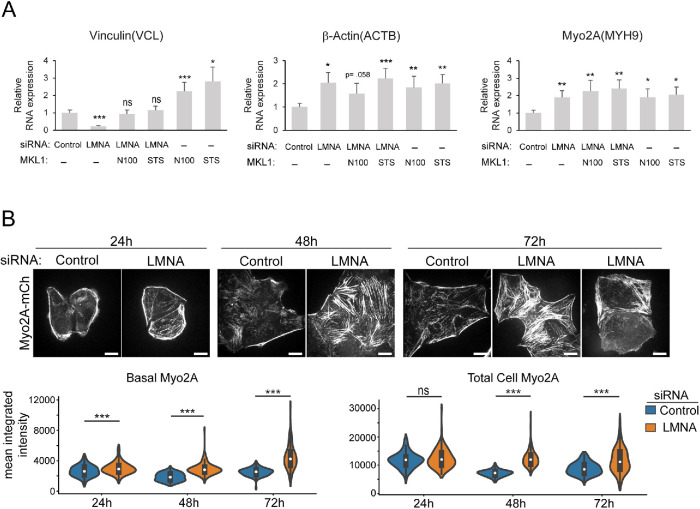
In light of observed increased nuclear MKL1 accumulation in LMNA-deficient cells, we examined whether known MKL1/SRF target genes were responsive to MKL1 regulation following LMNA depletion. Ectopic expression of two different constitutively active MKL1 mutant proteins (Flag-MKL1-N100, lacks an N-terminal actin-binding motif required for its G-actin–dependent cytoplasmic retention; and Flag-MKL1-STS/A, S449, S454, and T450 mutated to A,is refractory to ERK-regulated nuclear export; Muehlich et al., 2008) showed both nuclear and cytoplasmic localization (Supplemental Figure S3), as observed by others (Willer and Carroll, 2017). Significantly, active MKL1 mutants promoted expression of SRF target genes, for both control and LMNA-silenced cells, without mitigation of cytoskeletal anomalies resulting from LMNA silencing (Figure 6A; Supplemental Figure S3). For SRF targets assessed, LMNA silencing led to increased transcript levels for ACTB and MYH9; however, VCL levels were reduced fivefold (Figure 6A). These results suggest differential and complex regulation of MKL1/SRF target genes in response to LMNA deficiency.
FIGURE 6:
LMNA silencing alters MKL1/SRF target gene expression. (A) Quantitative PCR analysis of MKL1/SRF target gene expression, normalized to GAPDH, in HeLa cells treated as indicated and assessed at 72 h post–siRNA treatment. Values are mean ± SEM, n = 4. (B) Live cell fluorescence confocal images and fluorescence quantification of endogenous mCherry tagged myo2A (Myo2A-mCh) in HeLa cells at times post–siRNA treatment, as indicated. Basal myo2A corresponds to measurements from basal confocal planes and total cell to the entire z-volume. * p ≤ 0.05, **p ≤ 0.01, *** p ≤ 0.001, ns, not significant, Welch’s t test; n ≥ 96/treatment. Bars, 10 μm.
Congruent with increased myo2A transcript levels as measured at 72 h post–siRNA treatment (Figure 6A), myo2A protein levels increased progressively following LMNA silencing (Figure 6B). Myosin-2A fluorescence intensity measurements from cells expressing mCherry-tagged myo2A from the endogenous locus (HeLa Myo2A-mCh) provided a direct readout of myo2A protein levels throughout the period of LMNA silencing in a single population of live cells. By this procedure increased myo2A protein accumulation, both basally and cell-wide, was evident for LMNA-depleted cells by 24 h post–siRNA treatment, and this was maintained to 72 h posttreatment, relative to controls (Figure 6B). Our live cell studies also suggested progressive development of cytoskeletal anomalies in LMNA-depleted cells, with the presence of increased stress fibers becoming pronounced by 48 h post–siRNA treatment, whereas cells with central stress fiber loss or a disrupted network were more evident at 72 h (Figure 6B). Altogether, these data support activation of a contractile regulatory cascade entailing stimulation of mechanosensitive transcription factors with feedback to augmented myo2A expression.
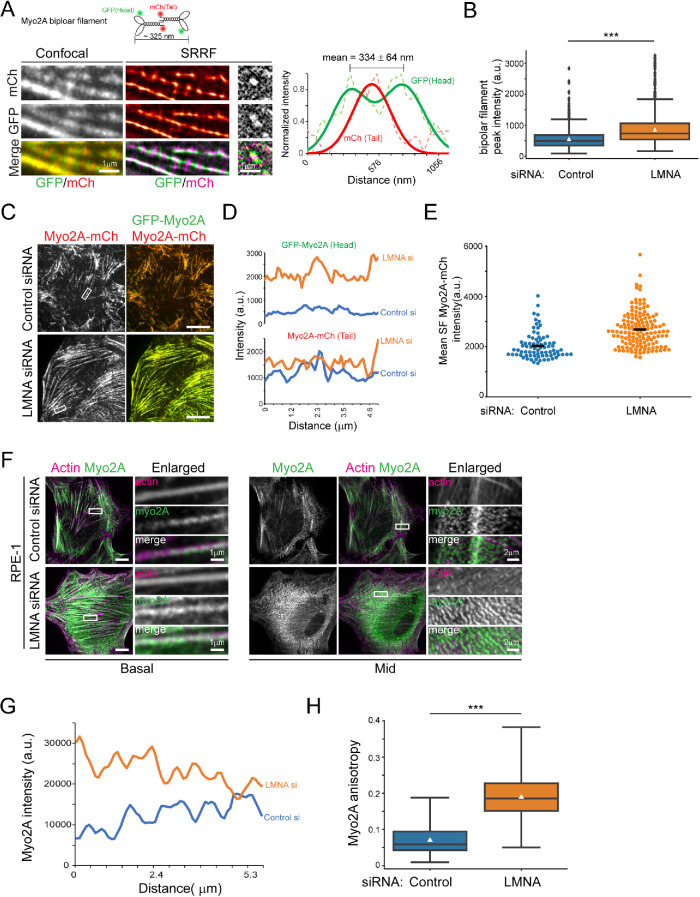
Increased myosin-II bipolar filament assembly in LMNA-depleted cells
Since our earlier results suggested that LMNA depletion may be associated with elevated myo-II contractile activity in the absence of significant increases to p-MLC levels, we interrogated other possible mechanisms linked to increased myo-II contractility. Computational models predict sharp sensitivities of myo-II force generation to bipolar filament ensemble size (Stam et al., 2015). Myosin-2A formed punctate structures in fluorescence images of both fixed cells labeled by antibodies to the C-terminal tail (e.g., Figure 1D) and live cells expressing C-terminal mCh-tagged myo2A. From SRRF superresolved live cell images where endogenous myo2A was labeled by both N-terminal green fluorescent protein (GFP) and C-terminal mCh, via CRISPR/Cas9 gene editing (Figure 7A), we determined that individual C-terminal myo2A labeled puncta match the profile of single bipolar filaments. As expected, C-terminal puncta were flanked on either side by spots corresponding to N-terminal head motifs. Accordingly, gaussian fits to intensity line scans of dual labeled myo2A puncta showed a mean bipolar filament length of 334 nm (n = 7; Figure 7A), in line with previous measurements made from electron microscopy ultrastructure studies (Niederman and Pollard, 1975). Measurements along stress fibers demonstrated more intense accumulation of myo2A to puncta for both HeLa and RPE-1 cells (Figure 7, C, D, F, and G). Myo2A-mCh peak intensities, a reflection of the quantity of myo2A motors in each bipolar filament or punctum, were on average 53% greater for LMNA-depleted HeLa cells (Figure 7B), indicating larger bipolar filament ensembles for this condition relative to control.
FIGURE 7:
Increased myosin-II bipolar filament assembly in LMNA-depleted cells. (A) Confocal and SRRF images of myosin-II bipolar filament organization in HeLa cells expressing dual tagged (as illustrated in cartoon) endogenous myo2A. Graph shows line intensity profile (marked by white line in right column) of an isolated bipolar filament. Plots (dashed lines) and gaussian fits (solid lines) are shown. (B) Plots of bipolar filament peak intensities as obtained from mCh (myo2A tail) images, n ≥ 968/treatment. (C,D) Basal confocal fluorescence images and line intensity profile plots of stress fibers shown in boxed region (C) for dual labeled myo2A in HeLa cells. (E) Quantification of myo2A intensity along stress fibers assembled following blebbistatin treatment and washout, n ≥ 78/treatment. Horizontal lines are mean values. (F) Basal and mid–cell level confocal immunofluorescence images of RPE-1 cells. Boxed regions are shown in enlargements. (G) Myo2A line intensity plots along stress fibers shown in basal boxed regions (F). (H) Quantification of myo2A filament anisotropy, n ≥ 83/treatment. White triangles are mean values. Scale bars, 10 μm or as indicated. *** p ≤ 0.001, Welch’s t test.
In addition to the increase of myo2A to basal stress fibers, for RPE-1 LMNA-depleted cells there was also pronounced accumulation and ordering of myo2A to elongated arrays of bipolar filaments in features previously characterized as myosin “ribbons” or “stacks” (Figure 7F; Verkhovsky et al., 1995; Fenix et al., 2016). Measurements of the fibrillar ordering of myo2A affirmed augmented anisotropy for LMNA-silenced RPE-1 cells relative to controls (Figure 7H). In contrast to myo2A bipolar filaments in stress fibers, myo2A stacks concentrated at mid to apical regions of cells and did not appear to be associated with contractile actin structures; instead, these stacks were arranged as orthogonal arrays to sheets of thin of F-actin bundles (Figure 7F). Our data here establish that LMNA deficiency results in enhanced myo-II bipolar filament assembly and suggest the involvement of cell type–specific variables in accompanying filament stack expansion.
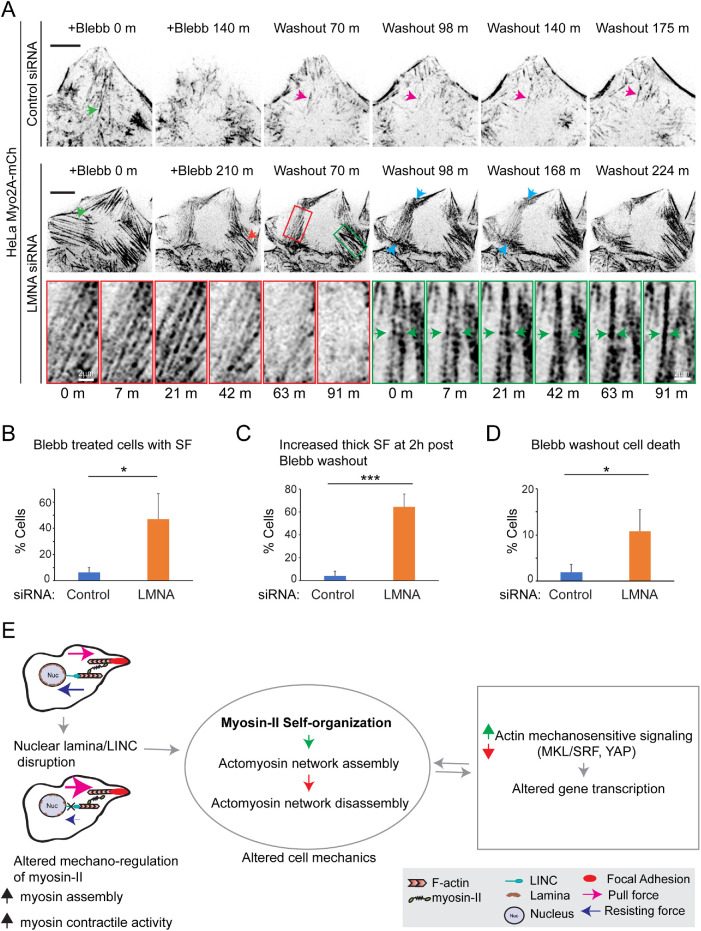
Stress fiber disorder in LMNA-deficient cells requires myosin-II contractile activity
To further define the requirement for myosin-II in the induction of contractile structures such as stress fibers in LMNA-depleted cells, we evaluated whether myo-II inhibition altered assembly of such features. HeLa Myo2A-mCh cells were siRNA cotreated with blebbistatin (blebb) (Straight et al., 2003), an inhibitor of myo-II that blocks motor ATPase activity and maintains myosin in a weak actin-binding state, followed by live cell image analysis. Blebb treatment resulted in robust stress fiber disassembly for both LMNA-silenced and control cells (Figure 8A; Supplemental Figure S4A; Supplemental Video S4). Strikingly, however, whereas only 6% of control cells had any remaining myo2A-labeled stress fibers at 2 h post–blebb treatment, 47% of LMNA-silenced cells displayed the persistence of some stress fiber labeling at this time point (Figure 8, A and B; Supplemental Figure S4A, red arrows). Thus, while myo-II activity is required for the presence of increased stress fibers in LMNA-depleted cells, these data also show that LMNA deficiency results in stronger myo-II-actin interaction that is more resistant to blebb inhibition. Consistently, within 2 h following washout of blebb, a significantly greater percentage of LMNA-silenced cells had developed increased numbers of thick stress fibers relative to control cells (Figure 8, A and C; Supplemental Figure S4A), and these stress fibers accumulated more myo2A (32% increase, p < 0.0001, Welch’s ttest; Figure 7E) in comparison to those formed in control cells. Of note, following blebb washout, as cells reassembled stress fibers, LMNA-depleted cells were more prone to contract and undergo apoptosis relative to control cells (Figure 8D; Supplemental Figure S4B). The latter result suggested greater sensitivity to mechanical stress for these LMNA-deficient cells.
FIGURE 8:
Myosin-II contractile activity instigates localized dynamic actomyosin remodeling in LMNA-deficient cells. (A) Fluorescence time-lapse series of blebbistatin treatment (+Blebb) and drug washout. Green arrowheads depict stress fibers (SF), which disassembe during bleb treatment. Red arrowhead shows SF, which persists at 2 h post–blebb treatment. Magenta arrowhead illustrates dynamics of a new, post–drug washout SF for a control cell. Red and green boxed regions (LMNA siRNA, top panels) are magnified in bottom panels and highlight localized actomyosin remodeling to SF disassembly and SF bundling, respectively, at ∼1 h post–blebb washout. Blue arrowheads depict regions of apparent focal myosin contractile activity. Green arrowheads depict merger of two adjacent SFs. See also Supplemental Video S4. (B–D) Quantification of cells with SFs at 2 h post–blebb treatment (B), quantification of increased thick SF assembly at 2 h post–blebb washout (C), and quantification of cell death occurring within 3 h post–blebb washout (D). Scale bars, 10 μm. Values are mean ± SD, n ≥ 242/treatment. * p ≤ 0.05, *** p ≤ 0.001; Welch’s t test. (E) Proposed mechanistic model for actomyosin organization dysfunction in response to disruption of the nuclear LINC module.
Movie S4.
Time-lapse confocal microscopy of basal z-projections for control or LMNA siRNA treated HeLa cells expressing myosin-2A-mCherry. Cells were treated with 100 M blebbistatin and washed as indicated. Images were captured at 7 min intervals and the video rate is 20 frames/s. Related to Figure 8.
Finally, we assessed signs of myo-II self-organization to different patterns from live cell time series images, following blebb washout. Myo2A structures were dynamic for both control and LMNA-depleted cells following blebb removal. For example, Figure 8A (control siRNA, magenta arrow) illustrates fluctuations in the organization of a newly formed stress fiber in a control cell and that this stress fiber persisted in some form for more than 1 h. For LMNA-depleted cells, we also detected dynamic myo2A behaviors, which apparently contributed to simultaneous localized stress fiber bundling (Figure 8A, green boxed region) and stress fiber disassembly (red boxed region) within a single cell. Stress fiber bundling looked to entail the translocation of adjacent stress fibers and their merger into a single thicker bundle (Figure 8A, green arrows), concomitant with the merger of myo2A puncta to more intense filaments. Localized stress fiber disassembly in this instance seemed to involve opposing contractile forces focused at either end of the stress fiber bundles (Figure 8A, blue arrows). Both forms of myo-II patterning behaviors occurred over a period of tens of minutes, suggesting a relatively nonrapid timescale for myo-II self-organization in this cellular context. Combined, our live cell analyses support a pivotal role for deregulated myo-II motor activity and consequent exacerbated myo-II self-organizing behaviors in the etiology of actin cytoskeletal disorganization resulting from LMNA deficiency.
DISCUSSION
The opposing actin cytoskeletal alterations we observe in single populations of cells, following LINC module perturbations, have been reported independently as distinct aberrations by previous seemingly conflicting reports. In our experimental system, a simple explanation may have been that different residual levels of LMNA depletion induced different cytoskeletal anomalies. However, the totality of our data, including Supplemental Figure S1, is unsupportive of this explanation. Furthermore, this principle cannot account for similar heterogeneous cytoskeletal abnormalities reported within individuals and across a population of germline LMNA knockout cells (Broers et al., 2004). Blending our data with those from other studies, we offer a rationale for the superficial paradox. We conclude that divergent actomyosin phenotypes are the result of a common process, myosin-II self-organization. Differing actomyosin arrangements within and across cells represent varying phases of a dynamic self-organizing contractile process that can transition between localized states of actomyosin assembly and disassembly. Our showing of the increased presence of stress fibers, focal adhesions, and actomyosin aster-like structures following LMNA depletion in various cell types are all consistent with elevated myo-II contractile activity. Less appreciated, but anticipated based on numerous prior studies, actomyosin disassembly is also consistent with increased myo-II contractile activity (Haviv et al., 2008; Wilson et al., 2010; Blanchoin et al., 2014). Thus, both stress fiber assembly and loss are possible, even within a single cell, in response to increased myosin contractility (e.g., see Figure 8A). Expression of different steady state localized patterns driven by myosin self-organization is expected to be highly influenced by mechanical context, based on in vitro studies that imply important roles for factors such as localized availability of actin cross-linking proteins and network architecture (Backouche et al., 2006; Reymann et al., 2012; Koenderink and Paluch, 2018). These parameters, in addition to the magnitude of contractile forces experienced by local cytoskeletal structures, are likely to dictate whether persistent actomyosin assembly versus dissolution is realized over different length- and timescales.
By what mechanism does loss of LINC module proteins cause increased myo-II contractile behaviors? We did not observe LMNA depletion to be associated with significant global increases to p-MLC levels in either HeLa or RPE-1 cells, although localized gain of p-MLC may be pertinent. Myosin-II activation has been best characterized in response to phosphorylation; however, increased myosin contractile activity in the absence of greater MLC (S19) phosphorylation is not unprecedented (Giuliano et al., 1992; Obara et al., 1995). Indeed, our results parallel those of a previous report that also suggested greater cell contractile activity, but without increased p-MLC levels, for cells depleted of the LINC module protein nesprin1 (Chancellor et al., 2010). We propose that altered myo-II contractile activity in cells deficient of LINC module proteins is mediated by perturbation of a feedback regulatory cascade involving both mechanical and transcriptional control of myo-II, as illustrated (Figure 8E).
Mechanical control of myosin function, likely through the effects of load in modifying myosin-actin catch bond behavior, is well supported by computational models and studies from cellular systems (Kasza and Zallen, 2011; Levayer and Lecuit, 2012; Greenberg et al., 2016). For instance, applied forces have been shown to control myo-II assembly and activity during fly development (Fernandez-Gonzalez et al., 2009). Our previous work and those of others suggest that the cytoplasmic actomyosin network generates tensile forces on the nucleus (Arsenovic et al., 2016; Wiggan et al., 2017). We suggest that obligate resisting forces provided by the nucleus, via LINC module mechanocoupling, provide important mechanical feedback signaling cues to help control the contractile activity of myo-II across the cytoplasmic actomyosin network. LINC module disruption is proposed to impair physical mechanocoupling and feedback that likely entails the activity of the nuclear envelope protein emerin, which contributes to tuning of nuclear mechanical resistance (Guilluy et al., 2014). Further studies utilizing more rapid and acute procedures for nuclear envelope disruption are required to examine these initial myo-II activation events. Nevertheless, a model for the nucleus playing a general role in mechanoregulation of the cytoplasmic cytoskeletal network is supported by recent data (Ambrosini et al., 2019). Distinct roles for individual Sun and nesprin LINC complex proteins, nonactomyosin cytoskeletal elements, and chromatin remodeling are feasible (Hamouda et al., 2020).
We further propose that increased contractile activity and actomyosin assembly, pursuant to mechanoactivation of myo-II, instigates nuclear translocation of mechanosensitive transcription factors such as MKL1 and YAP, as we observe for LMNA-deficient cells in this study. Our results for YAP are similar to those shown for human myoblasts harboring mutant LMNA (Bertrand et al., 2014). Our finding that myo2A and β-actin transcript levels increased following LMNA silencing is consistent with activation of MKL1/SRF signaling, as both genes are transcriptional targets of MKL1/SRF (Esnault et al., 2014; Foster et al., 2017). Likewise, transcripts for both β-actin and myo2A were recently shown to be up-regulated in cardiomyocytes from a human laminopathic model (Bertero et al., 2019), and in yet another laminopathic model, increased β-actin transcript levels coupled to aberrant MKL1-SRF activation have been observed (Osmanagic-Myers et al., 2019). Altogether, this body of work strongly supports deregulated MKL1/SRF signaling as a commonality across models of LMNA deficiency. Our results (Figure 5A) and those of others (Willer and Carroll, 2017) nonetheless suggest complex and differential regulation of MKL1/SRF gene targets in LMNA-deficient cells, as some genes such as VCL are repressed at steady state and not activated. Repression of some SRF targets like VCL in the face of increased MKL signaling may involve other gene-specific regulatory factors. In preliminary studies we have found that LMNA-depleted cells show increased nuclear targeting of the transcription factor YY1, which also shuttles between the nucleus and cytoplasm dependent on actin organization, analogous to MKL1 and YAP (Ellis et al., 2002). YY1 acts predominantly as a transcriptional repressor and is known to antagonize expression of some SRF-regulated genes via overlapping gene promoter binding elements (Chen and Schwartz, 1997). Interestingly, the proximal promoter of VCL contains both SRF and YY1 binding sites, suggesting this hypothesis as a plausible mechanism.
The aforementioned evidence suggesting increased MKL1 activation in LMNA-deficient cells is not necessarily in conflict with an initial report by Ho et al. (2013b), which advocated abrogated MKL1 signaling. Combined results from other reports suggest that impairment of mechanosensitive transcription factor signaling in LINC module–disrupted cells may be contextual to factors such as cell and extracellular matrix mechanical properties (Bertrand et al., 2014; Willer and Carroll, 2017). For example, impaired MKL1 and YAP nuclear translocation may be anticipated in mechanical contexts that favor steady states of self-organized actomyosin network disassembly.
In LMNA-disrupted cells, induction of mechanosensitive transcription programs, such as for MKL1 and YAP, ostensibly generates a critical positive feedback loop contributing to the increased myo2A expression and assembly we uncovered. Interestingly, enucleated cell cytoplasts show impaired responses to mechanical cues and are less contractile than intact cells (Graham et al., 2018). These data support the notion that the presence of the nucleus may be relevant not only for direct mechanical feedback to actomyosin but also for gene transcription toward sustained contractile actomyosin reinforcement.
In accord with our model that deregulated actomyosin assembly is a key feature of LMNA deficiency, we showed that loss of LMNA resulted in ectopic myo2A accumulation to dynamic cell membrane structures, in the form of podosome-type adhesions and actin comet tails. Previous studies have postulated possible roles for myo-II in both podosome assembly and disassembly (Bhuwania et al., 2012); our live cell image analyses implicate roles for myo-II contractile activity in promoting F-actin polymerization at both types of membrane structures. Increased membrane-associated F-actin assembly may occur through activation of lipid-bound actin assembly factors brought together by myosin-induced clustering, as previously characterized (Koster et al., 2016; Vogel et al., 2017). Formation of podosome-like adhesions following LMNA loss in HeLa cells and not RPE-1 cells suggests that deregulated myosin activity may contribute to different tissue-specific phenotypes observed across the pleotropic traits observed for laminopathies. Intriguingly, in human pathologies, myosin-II plays an outsized role in one of the predominant tissues affected by LMNA deficiency, that being muscle. Further, we speculate that deviant membrane-associated actomyosin remodeling provokes, for example, altered cellular features like podosome formation in normally competent cell types, such as osteoclasts. Modulation of podosomes, sites of extracelluar matrix degradation, may contribute to tissue specific laminopathic defects such as osteolysis.
In conclusion, our work establishes deregulation of myosin-II as a key mechanistic factor in cytoskeletal anomalies arising from disruption of nuclear LMNA. Myosin-II and its regulatory cascade appear to be attractive candidates for further therapeutic investigations toward treatment of nuclear envelopathies.
MATERIALS AND METHODS
Cells
RPE-1, HeLa (Kyoto strain), U2OS, and GM-2149 (normal fibroblasts) cells were grown in DMEM supplemented with 10% fetal bovine serum (Atlas Biologicals).
Transfections
Cells were transfected with 40–50 nM of siRNAs at the time of plating and again 24 h later with either Lipofectamine RNAiMax or Lipofectamine 2000 (Invitrogen) using manufacturer’s protocols.
siRNA
siRNA oligonucleotides targeted to human Sun1, Sun2, nesprin1, nesprin2, lamin A/C, and a control siRNA targeted to luciferase (GL2), as described (Wiggan et al., 2017), were obtained from Qiagen. siRNAs for individual targets produced similar phenotypes and were used interchangeably between experiments.
Antibodies
Antibodies and sources were Sun1, Sun2, nesprin1, nesprin2, myosin-2A, Flag-M2, and α-tubulin (Sigma); p-myosin light chain (Ser-19 and Thr-18/Ser-19), (Cell Signaling); p-MLC (immunoblots, gift of R. Wysolmerski, West Virginia University); lamin A/C (sc-7292), MKL1 (c-19), MKL1 (H-140), YAP (sc-101199), and MLC (sc-4814) (Santa Cruz); GAPDH (Millipore); and ADF/cofilin (1439; Shaw et al., 2004).
Gene expression analysis
Total mRNA was isolated using the RNeasy Mini Kit (Qiagen) and cDNA prepared using the iScript cDNA synthesis kit (Bio-Rad) following the manufacturer’s procedures. Quantitative PCR (qPCR) was performed in duplicate or triplicate using the iQ SYBR green reagent with a Bio-Rad CFX 96 thermocycler. Target gene expression was normalized to GAPDH and measured using the ∆∆Ct method. Primers used for qPCR are listed in Supplemental Table S1.
Gene editing
Single-guide RNAs (sgRNAs) targeting N- and C-terminal coding exons of MYH9 were designed using CCTop (Stemmer et al., 2015). Guide RNAs were cloned into the plasmid pX330-U6-Chimeric_BB-CBh-hSpCas9 (Addgene plasmid 42230; Cong et al., 2013) using procedures as described (Bauer et al., 2015). Homology-directed repair donor plasmids were generated using a combination of gene synthesized DNA sequences for ∼225-base-pair homology arms and either mAID-mCherry2 derived from plasmid pMK292 (Addgene plasmid 72830) for C-terminal tagging or sfGFP for N-terminal tagging, as described (Natsume et al., 2016). Cells were transfected with plasmids for sgRNA and donor repair using Lipofectamine 2000. Following neomycin selection, mCherry-positive cells were confirmed for proper recombination by genomic DNA PCR genotyping, as described (Bauer et al., 2015), and by visual evaluation of expected mCherry-protein cellular distribution through fluorescence microscopy. Confirmed mCherry-expressing cells were subjected to a second round of CRISPR editing with plasmids for N-terminal sfGFP tagging. Dual GFP/mCherry-positive cells were isolated by fluorescence-activated cell sorting and characterized as described earlier. Sequences for sgRNAs, homology arms, and PCR primers for genotyping are listed in Supplemental Tables S2 and S3.
Cell culture, immunofluorescence staining, and microscopy
Fluorescence images were acquired with an Olympus IX81 spinning disk confocal (CSU22 head) microscope with either 100x/1.40 NA, 60x/1.42 NA, or 40x/1.35 NA objectives. Images were acquired with a Photometrics Cascade II CCD (charge-coupled device) camera using SlideBook (Intelligent Imaging Innovations). Some live cell images were captured in total internal reflection fluorescence mode on a Nikon Eclipse TiE inverted microscope with 100x/1.40 NA to an Andor iXon Ultra 888 EMCCD or Clara camera.
For live cell imaging, cells plated on glass bottom 35 mm dishes were housed in a stage incubator at 37°C with CO2 in regular growth medium. Cells were cotransfected with siRNAs and/or plasmids encoding either GFP-Lifeact (Riedl et al., 2008), pCMV-Flag-MKL1-N100 (derived from Addgene plasmid 19848), or FLAG-MKL1 or S449A/T450A/S454A (Addgene plasmid 19845, Muehlich et al., 2008). Phenotypic analyses of siRNA-treated cells were at 48–72 h post–initial siRNA treatment. In serum stimulation experiments cells were starved for 24 h in medium containing 0.3% serum and stimulated by replacing with medium containing 15% serum. Cells were treated with 100 μM blebbistatin (Sigma) or with dimethyl sulfoxide (DMSO) for controls in experiments assessing myosin-II inhibition. For fixed-cell microscopy, cells grown on glass coverslips were fixed in 4% formaldehyde in CBS buffer (10 mM 4-Morpholineethanesulfonic acid, pH 6.1, 138 mM KCl, 3 mM MgCl2, 2 mM ethyleneglycol-bis(β-aminoethyl)-N,N,N′,N′-tetraacetic acid, 0.32 M sucrose) for 20 min at room temperature. For cytoskeletal staining 0.4% Triton X-100 was included in the fix buffer. Fluorescently labeled phalloidin (F-actin labeling), Alexa-594–conjugated DNaseI (G-actin labeling), and secondary antibodies were from Invitrogen. DNA was labeled by 4′,6-diamidino-2-phenylindole (DAPI). Confocal z-stacks were acquired for analyses of both live and fixed cells.
Image and data analysis
Quantification of fluorescence intensity and image data analyses were performed using CellProfiler (Carpenter et al., 2006), ImageJ, Python, and open-source Python packages including Pyqtgraph and Pandas. Focal adhesions were segmented and quantified by first applying a difference of gaussian filter to basal confocal fluorescent images of paxillin labeling. Adhesions from all cells in each captured field were then segmented by the triangle thresholding method. Intensity measurements were made from background-subtracted images on adhesions of size ≥0.5 μm2. For nuclear/cytoplasmic measurements, projection images from three nuclear midpoint planes based on DAPI-labeled confocal z-stacks captured at 0.2–0.3 μm steps were utilized. Intensity values for each experiment were generally normalized to range between 0.01 and 1. Maximum-intensity projection images of the entire z-stack were utilized for whole cell measurements. G/F-actin ratios were measured from cells dual labeled with phalloidin and DNaseI. Images were processed by a CellProfiler pipeline, as described (Wiggan et al., 2017), for segmentation and data extraction. Myosin-II filament intensities were measured from line profile peak intensities of fluorescent images of C-terminal mCherry-labeled Myo2A puncta along stress fibers. Microtubule and myosin filament anisotropy was measured using the ImageJ plugin, FibrilTool, from mid–cell level confocal z-stack planes of randomly selected ∼6 μm2 cell regions of myo2A or tubulin immunostained images, as detailed (Boudaoud et al., 2014). Live cell fluorescence images of cells expressing Myo2A-mCh and GFP-Lifeact were enhanced by a Laplacian of Gaussian filter. SRRF images were generated using the default settings of the ImageJ plugin, NanoJ-SRRF (Gustafsson et al., 2016), from 100 confocal frames captured at 10–250 ms exposures.
F-actin abnormality, based on comparison to normal control cells, was assessed as follows: cells with evident increased numbers of basal stress fibers (SF) that were maintained over the central region of the cells were classified as “increased SF”; cells with much reduced levels or complete absence of basal stress fibers were classified as “decreased SF”; and cells with a mixture of SF organizations often characterized by a disrupted network of stress fibers, the presence of actomyosin asters, F-actin condensates, fragmented F-actin bundles, and F-actin bundles with frayed ends were classified as “disrupted SF.” Mean Pearson’s correlation coefficients and 95% confidence intervals were calculated following Fisher’s Z-transformation. Statistical analyses were conducted using either Python or R statistical packages. Data were tabulated from three or more experiments with the exception of Figures 2E and 5C (starved and 1 h serum stimulation), which were from two experiments. Where appropriate either a two-sided Welch’s t test or one-way analysis of variance followed by either Tukey–Kramer or Dunnett’s post hoc (for comparisons to control) were utilized.
Supplementary Material
Acknowledgments
We thank Jennifer Skinner for assistance with experiments and R. Wysolmerski for sharing antibodies. The work was supported in part by a Colorado State University Core Infrastructure Grant for Microscopy, a Boettcher Foundation Webb-Waring Biomedical Research Program award (T.J.S.), a grant from the Pew Biomedical Scholars Program (J.G.D.), and National Institutes of Health grants NS064217 (O.W.), GM130365 (J.G.D.), R35GM119728 (T.J.S.), and R01AG049668, 1S10OD025127 and AG044812 (J.R.B.).
Abbreviations used:
- blebb
blebbistatin
- LINC
linker of nucleoskeleton and cytoskeleton
- mCh
m-Cherry fluorescent protein
- myo-II
non muscle myosin-II
- myo2A
myosin-2A
- p-MLC
phosphorylated-myosin light chain
- SF
stress fiber
- SRF
serum response factor
- SRRF
super-resolution radial fluctuations.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-01-0017-T) on August 20, 2020.
REFERENCES
- Aguilar-Cuenca R, Juanes-Garcia A, Vicente-Manzanares M (2014). Myosin II in mechanotransduction: master and commander of cell migration, morphogenesis, and cancer. Cell Mol Life Sci , 479–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso F, Spuul P, Daubon T, Kramer I, Genot E (2019). Variations on the theme of podosomes: a matter of context. Biochim Biophys Acta Mol Cell Res , 545–553. [DOI] [PubMed] [Google Scholar]
- Ambrosini A, Rayer M, Monier B, Suzanne M (2019). Mechanical function of the nucleus in force generation during epithelial morphogenesis. Dev Cell , 197–211.e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aragona M, Panciera T, Manfrin A, Giulitti S, Michielin F, Elvassore N, Dupont S, Piccolo S (2013). A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell , 1047–1059. [DOI] [PubMed] [Google Scholar]
- Arsenovic PT, Ramachandran I, Bathula K, Zhu R, Narang JD, Noll NA, Lemmon CA, Gundersen GG, Conway DE (2016). Nesprin-2G, a component of the nuclear LINC complex, is subject to myosin-dependent tension. Biophys J , 34–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Backouche F, Haviv L, Groswasser D, Bernheim-Groswasser A (2006). Active gels: dynamics of patterning and self-organization. Phys Biol , 264–273. [DOI] [PubMed] [Google Scholar]
- Bauer DE, Canver MC, Orkin SH (2015). Generation of genomic deletions in mammalian cell lines via CRISPR/Cas9. J Vis Exp 2015, e52118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertero A, Fields PA, Smith AST, Leonard A, Beussman K, Sniadecki NJ, Kim DH, Tse HF, Pabon L, Shendure J, et al. (2019). Chromatin compartment dynamics in a haploinsufficient model of cardiac laminopathy. J Cell Biol , 2919–2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertrand AT, Ziaei S, Ehret C, Duchemin H, Mamchaoui K, Bigot A, Mayer M, Quijano-Roy S, Desguerre I, Laine J, et al. (2014). Cellular microenvironments reveal defective mechanosensing responses and elevated YAP signaling in LMNA-mutated muscle precursors. J Cell Sci , 2873–2884. [DOI] [PubMed] [Google Scholar]
- Bhuwania R, Cornfine S, Fang Z, Krüger M, Luna EJ, Linder S (2012). Supervillin couples myosin-dependent contractility to podosomes and enables their turnover. J Cell Sci , 2300–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J (2014). Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev , 235–263. [DOI] [PubMed] [Google Scholar]
- Boudaoud A, Burian A, Borowska-Wykret D, Uyttewaal M, Wrzalik R, Kwiatkowska D, Hamant O (2014). FibrilTool, an ImageJ plug-in to quantify fibrillar structures in raw microscopy images. Nat Protoc , 457–463. [DOI] [PubMed] [Google Scholar]
- Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, Oomens CW, Baaijens FP, Ramaekers FC (2004). Decreased mechanical stiffness in LMNA-/- cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum Mol Genet , 2567–2580. [DOI] [PubMed] [Google Scholar]
- Burke B, Stewart CL (2006). The laminopathies: the functional architecture of the nucleus and its contribution to disease. Annu Rev Genomics Hum Genet , 369–405. [DOI] [PubMed] [Google Scholar]
- Cameron LA, Giardini PA, Soo FS, Theriot JA (2000). Secrets of actin-based motility revealed by a bacterial pathogen. Nat Rev Mol Cell Biol , 110–119. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol , R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chancellor TJ, Lee J, Thodeti CK, Lele T (2010). Actomyosin tension exerted on the nucleus through nesprin-1 connections influences endothelial cell adhesion, migration, and cyclic strain-induced reorientation. Biophys J , 115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ (1997). Competition between negative acting YY1 versus positive acting serum response factor and tinman homologue Nkx-2.5 regulates cardiac alpha-actin promoter activity. Mol Endocrinol , 812–822. [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013). Multiplex genome engineering using CRISPR/Cas systems. Science , 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corne TDJ, Sieprath T, Vandenbussche J, Mohammed D, Te Lindert M, Gevaert K, Gabriele S, Wolf K, De Vos WH (2017). Deregulation of focal adhesion formation and cytoskeletal tension due to loss of A-type lamins. Cell Adh Migr , 447–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D (2006). Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol , 41–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis PD, Martin KM, Rickman C, Metcalfe JC, Kemp PR (2002). Increased actin polymerization reduces the inhibition of serum response factor activity by Yin Yang 1. Biochem J , 547–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Stewart A, Gualdrini F, East P, Horswell S, Matthews N, Treisman R (2014). Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev , 943–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenix AM, Taneja N, Buttler CA, Lewis J, Van Engelenburg SB, Ohi R, Burnette DT (2016). Expansion and concatenation of non-muscle myosin IIA filaments drive cellular contractile system formation during interphase and mitosis. Mol Biol Cell , 1465–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez R, Simoes Sde M, Roper JC, Eaton S, Zallen JA (2009). Myosin II dynamics are regulated by tension in intercalating cells. Dev Cell , 736–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster CT, Gualdrini F, Treisman R (2017). Mutual dependence of the MRTF-SRF and YAP-TEAD pathways in cancer-associated fibroblasts is indirect and mediated by cytoskeletal dynamics. Genes Dev , 2361–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano KA, Kolega J, DeBiasio RL, Taylor DL (1992). Myosin II phosphorylation and the dynamics of stress fibers in serum-deprived and stimulated fibroblasts. Mol Biol Cell , 1037–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DM, Andersen T, Sharek L, Uzer G, Rothenberg K, Hoffman BD, Rubin J, Balland M, Bear JE, Burridge K (2018). Enucleated cells reveal differential roles of the nucleus in cell migration, polarity, and mechanotransduction. J Cell Biol , 895–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, Arpag G, Tuzel E, Ostap EM (2016). A perspective on the role of myosins as mechanosensors. Biophys J , 2568–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilluy C, Osborne LD, Van Landeghem L, Sharek L, Superfine R, Garcia-Mata R, Burridge K (2014). Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol , 376–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson N, Culley S, Ashdown G, Owen DM, Pereira PM, Henriques R (2016). Fast live-cell conventional fluorophore nanoscopy with ImageJ through super-resolution radial fluctuations. Nat Commun , 12471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda MS, Labouesse C, Chalut KJ (2020). Nuclear mechanotransduction in stem cells. Curr Opin Cell Biol , 97–104. [DOI] [PubMed] [Google Scholar]
- Haviv L, Gillo D, Backouche F, Bernheim-Groswasser A (2008). A cytoskeletal demolition worker: myosin II acts as an actin depolymerization agent. J Mol Biol , 325–330. [DOI] [PubMed] [Google Scholar]
- Ho CY, Jaalouk DE, Lammerding J (2013a). Novel insights into the disease etiology of laminopathies. Rare Dis , e27002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Jaalouk DE, Vartiainen MK, Lammerding J (2013b). Lamin A/C and emerin regulate MKL1-SRF activity by modulating actin dynamics. Nature , 507–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasza KE, Zallen JA (2011). Dynamics and regulation of contractile actin-myosin networks in morphogenesis. Curr Opin Cell Biol , 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenderink GH, Paluch EK (2018). Architecture shapes contractility in actomyosin networks. Curr Opin Cell Biol , 79–85. [DOI] [PubMed] [Google Scholar]
- Koster DV, Husain K, Iljazi E, Bhat A, Bieling P, Mullins RD, Rao M, Mayor S (2016). Actomyosin dynamics drive local membrane component organization in an in vitro active composite layer. Proc Natl Acad Sci USA , E1645–E1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krendel M, Zenke FT, Bokoch GM (2002). Nucleotide exchange factor GEF-H1 mediates cross-talk between microtubules and the actin cytoskeleton. Nat Cell Biol , 294–301. [DOI] [PubMed] [Google Scholar]
- Lammerding J, Schulze PC, Takahashi T, Kozlov S, Sullivan T, Kamm RD, Stewart CL, Lee RT (2004). Lamin A/C deficiency causes defective nuclear mechanics and mechanotransduction. J Clin Invest , 370–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levayer R, Lecuit T (2012). Biomechanical regulation of contractility: spatial control and dynamics. Trends Cell Biol , 61–81. [DOI] [PubMed] [Google Scholar]
- Liu BP, Chrzanowska-Wodnicka M, Burridge K (1998). Microtubule depolymerization induces stress fibers, focal adhesions, and DNA synthesis via the GTP-binding protein Rho. Cell Adhes Commun , 249–255. [DOI] [PubMed] [Google Scholar]
- Lum M, Morona R (2014). Myosin IIA is essential for Shigella flexneri cell-to-cell spread. Pathog Dis , 174–187. [DOI] [PubMed] [Google Scholar]
- Marchisio PC (2012). Fortuitous birth, convivial baptism and early youth of podosomes. Eur J Cell Biol , 820–823. [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R (2003). Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell , 329–342. [DOI] [PubMed] [Google Scholar]
- Muehlich S, Wang R, Lee SM, Lewis TC, Dai C, Prywes R (2008). Serum-induced phosphorylation of the serum response factor coactivator MKL1 by the extracellular signal-regulated kinase 1/2 pathway inhibits its nuclear localization. Mol Cell Biol , 6302–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, Kanemaki MT (2016). Rapid protein depletion in human cells by auxin-inducible degron tagging with short homology donors. Cell Rep , 210–218. [DOI] [PubMed] [Google Scholar]
- Niederman R, Pollard TD (1975). Human platelet myosin. II. In vitro assembly and structure of myosin filaments. J Cell Biol , 72–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obara K, Nikcevic G, Pestic L, Nowak G, Lorimer DD, Guerriero V Jr, Elson EL, Paul RJ, de Lanerolle P (1995). Fibroblast contractility without an increase in basal myosin light chain phosphorylation in wild type cells and cells expressing the catalytic domain of myosin light chain kinase. J Biol Chem , 18734–18737. [DOI] [PubMed] [Google Scholar]
- Olson EN, Nordheim A (2010). Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol , 353–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S, Kiss A, Manakanatas C, Hamza O, Sedlmayer F, Szabo PL, Fischer I, Fichtinger P, Podesser BK, Eriksson M, Foisner R (2019). Endothelial progerin expression causes cardiovascular pathology through an impaired mechanoresponse. J Clin Invest , 531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiq NBM, Nishimura Y, Plotnikov SV, Thiagarajan V, Zhang Z, Shi S, Natarajan M, Viasnoff V, Kanchanawong P, Jones GE, Bershadsky AD (2019). A mechano-signalling network linking microtubules, myosin IIA filaments and integrin-based adhesions. Nat Mater , 638–649. [DOI] [PubMed] [Google Scholar]
- Rathman M, de Lanerolle P, Ohayon H, Gounon P, Sansonetti P (2000). Myosin light chain kinase plays an essential role in S. flexneri dissemination. J Cell Sci (Pt 19), 3375–3386. [DOI] [PubMed] [Google Scholar]
- Reymann AC, Boujemaa-Paterski R, Martiel JL, Guerin C, Cao W, Chin HF, De La Cruz EM, Thery M, Blanchoin L (2012). Actin network architecture can determine myosin motor activity. Science , 1310–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods , 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz US, Gardel ML (2012). United we stand: integrating the actin cytoskeleton and cell-matrix adhesions in cellular mechanotransduction. J Cell Sci , 3051–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw AE, Minamide LS, Bill CL, Funk JD, Maiti S, Bamburg JR (2004). Cross-reactivity of antibodies to actin-depolymerizing factor/cofilin family proteins and identification of the major epitope recognized by a mammalian actin-depolymerizing factor/cofilin antibody. Electrophoresis , 2611–2620. [DOI] [PubMed] [Google Scholar]
- Soares e Silva M, Depken M, Stuhrmann B, Korsten M, MacKintosh FC, Koenderink GH (2011). Active multistage coarsening of actin networks driven by myosin motors. Proc Natl Acad Sci USA , 9408–9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam S, Alberts J, Gardel ML, Munro E (2015). Isoformsconfer characteristic force generation and mechanosensation by myosin II filaments. Biophys J , 1997–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, Mateo JL (2015). CCTop: an intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One , e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart-Hutchinson PJ, Hale CM, Wirtz D, Hodzic D (2008). Structural requirements for the assembly of LINC complexes and their function in cellular mechanical stiffness. Exp Cell Res , 1892–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ (2003). Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science , 1743–1747. [DOI] [PubMed] [Google Scholar]
- van den Dries K, Meddens MB, de Keijzer S, Shekhar S, Subramaniam V, Figdor CG, Cambi A (2013). Interplay between myosin IIA-mediated contractility and actin network integrity orchestrates podosome composition and oscillations. Nat Commun , 1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loosdregt I, Kamps MAF, Oomens CWJ, Loerakker S, Broers JLV, Bouten CVC (2017). Lmna knockout mouse embryonic fibroblasts are less contractile than their wild-type counterparts. Integr Biol , 709–721. [DOI] [PubMed] [Google Scholar]
- Verkhovsky AB, Svitkina TM, Borisy GG (1995). Myosin II filament assemblies in the active lamella of fibroblasts: their morphogenesis and role in the formation of actin filament bundles. J Cell Biol , 989–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SK, Greiss F, Khmelinskaia A, Schwille P (2017). Control of lipid domain organization by a biomimetic contractile actomyosin cortex. eLife . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallrath LL, Bohnekamp J, Magin TM (2016). Cross talk between the cytoplasm and nucleus during development and disease. Curr Opin Genet Dev , 129–136. [DOI] [PubMed] [Google Scholar]
- Wiggan O, Schroder B, Krapf D, Bamburg JR, DeLuca JG (2017). Cofilin regulates nuclear architecture through a myosin-II dependent mechanotransduction module. Sci Rep , 40953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer MK, Carroll CW (2017). Substrate stiffness-dependent regulation of the SRF-Mkl1 co-activator complex requires the inner nuclear membrane protein emerin. J Cell Sci , 2111–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Tsuchida MA, Allen GM, Barnhart EL, Applegate KT, Yam PT, Ji L, Keren K, Danuser G, Theriot JA (2010). Myosin II contributes to cell-scale actin network treadmilling through network disassembly. Nature , 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Lorenz M, Kempiak S, Sarmiento C, Coniglio S, Symons M, Segall J, Eddy R, Miki H, Takenawa T, Condeelis J (2005). Molecular mechanisms of invadopodium formation: the role of the N-WASP-Arp2/3 complex pathway and cofilin. J Cell Biol , 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.