Abstract
Background
Trichomes play a key role in the development of plants and exist in a wide variety of species.
Results
In this paper, it was reviewed that the structure and morphology characteristics of trichomes, alongside the biological functions and classical regulatory mechanisms of trichome development in plants. The environment factors, hormones, transcription factor, non-coding RNA, etc., play important roles in regulating the initialization, branching, growth, and development of trichomes. In addition, it was further investigated the atypical regulation mechanism in a non-model plant, found that regulating the growth and development of tea (Camellia sinensis) trichome is mainly affected by hormones and the novel regulation factors.
Conclusions
This review further displayed the complex and differential regulatory networks in trichome initiation and development, provided a reference for basic and applied research on trichomes in plants.
Keywords: Trichome, Transcription factors, Environment, Hormones, Non-coding RNA
Background
Trichomes are defined as unicellular or multicellular appendages, which are an extension of the above-ground epidermal cells in plants [1]. These appendages play a key role in the development of plantsand occur in a wide variety of species [2]. Trichomes are a protective barrier against natural hazards, such as herbivores, ultraviolet (UV) irradiation, pathogen attacks, excessive transpiration, seed spread, and seed protection. The unicellular non-glandular trichomes of Arabidopsis serve as an excellent model to study the molecular mechanism of trichome development in plants [3–7]. Despite the small size of trichomes, they have a marked effect on plants and human health. Cotton seed trichomes are important raw materials in the textile industry [8–10], and tea trichomes are critical for tea breeding and tea quality as they are rich in nutrients [11]. The light-leaf and light-shell phenotypes of crop rice have missing trichomes, which are conducive to crop harvesting and subsequent processing; however, the leaf trichomes in Populus and Platanus are harmful to humans, easily causing respiratory tract infections, lung infections, fever, influenza, and in severe cases, potentially cause cancer [12].
Structure and morphology characteristics of trichomes
Trichomes are widely distributed on the surface of different organs/tissues in different plants, exhibiting various morphologies. Trichomes are generally divided into single-celled or multicellular, branched or unbranched, and glandular or non-glandular based on different characteristics and functions. Trichomes also have different shapes, such as head, star, hook and scale. Theobald and Barthlott further divided trichomes into three categories based on the distributions of leaves: large, small and glandular trichomes [13, 14]. Large trichomes are mainly distributed on the abaxial surface and margins and in the vascular bundles; small trichomes in the stomatal para cellular; and glandular trichomes are usually regularly distributed in all or part of the subepidermal tissue on the leaf surface [2]. The morphology of trichomes is associated with the spatial distribution of plant organs. The base of the tomato (Lycopersicon esculentum) stem is well-covered with long trichomes, whereas the upper parts of the stem has short and sparse trichomes [15]. Moreover, the density of trichomes varies in different organs. The density of trichomes on the leaf blade is significantly higher than that on the back of the blade [16]. Brewer found that different types of leaves had different densities of trichomes. In soybean, the adaxial surface of the leaf had a greater trichome density than the abaxial surface [17].
Biological functions of plant trichomes
Plants have evolved many defense mechanisms to protect against different abiotic and biotic stresses. The morphology and density of trichomes influence several aspects of plant physiology and ecology by mediating the interactions between the plant and its environment [18]. Trichomes, along with the stomata, cutin and wax on the epidermis, performed various protective functions through synthesizing, storing and secreting many important substances [19–21]. The cotton petal trichomes maintain the shape of the buds and ensure the production of seeds [22]. In addition, trichomes protect plants from herbivores, insects and pathogens by secreting repellents, alkaloids and toxic substances [21, 23–25]. Kim et al. (2011) examined the relationship between pepper trichomes and pepper mottle virus (PepMov) resistance and showed that the resistance to PepMoV-SNU1 strain is inherited by cross combinations among pepper cultivars CM334, Chilsungcho and ECW123R [15]. In rice, two TRICHOME BIREFRINGENCE (TBR)-like proteins play an essential role in the resistance to leaf spot disease [26].
Trichomes are also important in the response to abiotic stress [22]. The presence of trichomes increases the thickness of the epidermis, and the content of long-chain fatty acids is significantly higher than that in other epidermal cells, which is helpful to reduce evaporation and regulate temperature [20, 27]. In Brazil’s high-altitude rocky areas, the plants of Croton tiglium and Vriesea effectively absorb moisture and nutrients from the atmosphere through trichomes to improve water and fertilizer utilization [28, 29]. The high-density multi-branched Acanthophyllum squarrosum trichomes not only have high resistance to sand burial, but also reduce mechanical damage by wind and sand [30]. In addition, the trichomes of an aquatic plant, Salvinia molesta, play a hydrophobic role in maintaining normal respiration [14]. Trichomes also function in signal transmission. The cell wall of Arabidopsis trichomes gradually thins from the top to bottom, and this change makes the base of the trichomes extremely sensitive to external stimuli. The stimulation is transmitted to cells around the base of trichomes through changes in Ca 2+ content and pH, thereby regulating the synthesis of defensive substances [31, 32].
Regulatory mechanism of trichome development in plants
The development of plant trichomes is coordinated and regulated by a variety of factors, such as the environment, hormones, regulatory genes, and non-coding RNA. Among them, regulatory genes, including transcription factors and functional genes, play important roles in regulation of the initialization, growth and development of trichomes.
Regulatory mechanism of coding genes in trichome development
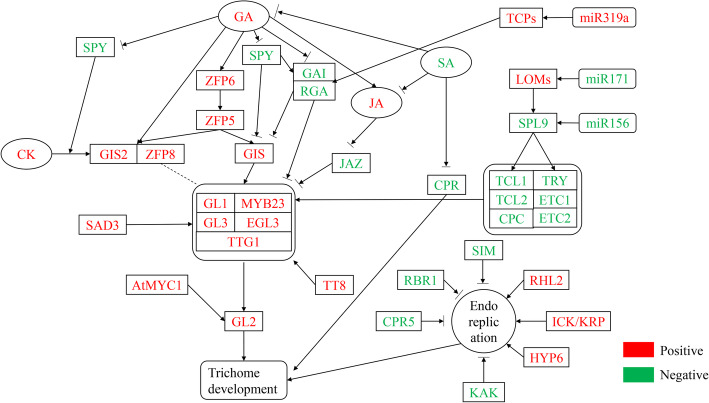
The development of plant trichomes are regulated by a complex molecular network, phytohormones, and environmental factors (Fig. 1). Previous studies have shown that many transcription factors are involved in regulating the initiation, growth and development of plant trichomes (Table 1). Positive regulatory factors include a WD40 family protein TTG1 (TRANSPARENT TESTA GLABRA1), four bHLH-like transcription factors GL3 (GLABRA3), EGL3 (ENHANCER OF GLABRA3), TT8 (TRANSPARENT TESTA), MYC-1, and three R2R3 MYB-related transcription factors (GL1, MYB23 and MYB5) [33, 41, 46, 47, 65–69]. These genes are functionally redundant and form an MYB-bHLH-TTG complex to bind to the promoter of GLABRA2 (GL2) [70]. TTG1 plays a vital role in regulating the development of trichomes. Bloomer et al. (2012) found that the regulatory roles of MYB23, GL1, GL2 and GL3 in trichome development require the participation of TTG1 [71]. Complete function mutations in GL1 and TTG1 result in a loss of trichome initiation. The gl3/egl3 double mutants also show a glabrous phenotype [72]. SAD2 (SENSITIVE TO ABA AND DROUGHT 2), an importin β-like protein, regulates trichome initiation through mediating GL3 function. In sad2 mutants, the transcript levels of MYB23, GL1, GL2 and TTG1 were increased with a decrease in the expression of GL3 and EGL3 [73].
Fig. 1.
Regulatory network during trichome development. Red one represents positive regulation factor, green one represents negative regulation factor. The arrow headed lines indicate the up-regulation, and the blunted lines indicate the downregulation or inhibition
Table 1.
Important genes related to trichome development in plants
| Gene | Regulation | Functional description | References |
|---|---|---|---|
| GL1 |
Positive; Negative |
Regulate initiation of trichome, the mutant results in trichome defect, but overexpression leads to a reduction in trichome. | [33, 34] |
| TTG1 | Positive | Regulate the initiation of trichome, the mutant results in trichome defect. | [35, 36] |
| GL3 | Positive | Regulates the initiation of trichome, and the mutant results in fewer trichomes. | [37, 38] |
| GL2 | Positive | Regulates epidermal cell fate, the mutant results in fewer trichomes, and most of them are not branched. | [39] |
| EGL3 | Positive | Function redundant with GL3, and gl3/egl3 double mutants show no trichome. | [40] |
| MYB23 | Positive; Negative | myb23 single mutants exhibit reduced trichome branching but no obvious effect in trichome initiation and overexpression of the MYB23 gene causes ectopic trichome initiation. MYB23 has functional redundancy with GL1. | [41] |
| TRY | Negative | Trichome of mutant grows in clusters and branches increase. | [42] |
| CPC | Negative | The mutant exhibits reduced number of trichome and clustered trichome. | [43] |
| ETC1 | Negative | ETC1 overexpression causes a reduction in trichome formation, etc1 single mutant has no significant phenotype. Function of ETC1 is partially redundant with TRY and CPC. | [44] |
| ETC2 | Negative | ETC2 overexpression results in the suppression of trichomes, etc2 single mutant shows an increase in trichome number on leaves and petioles. ETC2 acts redundantly with TRY and CPC in trichome patterning. | [44] |
| TT8 | Positive | Particularly important for marginal trichome development, no trichomes could be detected at the margin of young developing tt8 leaves in the absence of exogenously added hormones. | [45] |
| MYC-1 | Positive | MYC1 is a positive regulator of GL2, the mutant with defect in trichome, and root hair pattern formation. | [46] |
| MYB5 | Positive | Constitutive expression of MYB5 resulted in the formation of smaller trichomes and ectopic trichomes and a reduction in total leaf trichome numbers and branching, the mutant displayed minimal changes in trichome morphology. | [47] |
| GIS | Positive | Regulates the differentiation of epidermal cells and inflorescences, and the trichome of the mutant is reduced. | [48] |
| RHL2 | Positive | Regulate trichome nucleus replication, regulate trichome morphology. | [49] |
| ICK/KRP | Positive | Regulate trichome nucleus replication. | [50] |
| HYP6 | Positive | The mutant has a small number of trichome branches and intranuclear replication (8C). | [51] |
| ZWI | Positive | The mutant has a small number of trichome branches. | [52] |
| SAD2 | Positive | Regulate the initiation of trichome, mutant sad2 presents a phenotype with reduced trichome. | [53] |
| SIM | Negative | Inhibits mitotic cycle, its mutants exhibit multicellular trichome, while other morphologies are normal. | [54] |
| KAK | Negative | The mutation of KAK resulted in excessively branched trichome and the nuclear DNA content reaches 64C. | [55] |
| SPY | Negative | SPY is a gibberellin signaling repressor and the SPY deletion mutant has increased trichome branching. | [4, 7] |
| CPR5 | Negative | The cpr5 mutant has a small number of trichome branches, the trichome volume decreases, and the intranuclear replication cycle stays at the end of the second cycle. | [56] |
| RBR1 | Negative | Through its interaction with members of the E2F family of transcription factors, regulates the balance between cell division and the endocycle, and the conditional inactivation of RBR1 results in trichomes with altered morphologies, which include more branches. | [57] |
| HDG11 | Negative | HDG11 plays negative regulatory roles in trichome branching and hdg11 mutants result in excess branching of trichomes. | [58] |
| GLH | Positive | GLH can induce the formation of papillae in the late stage of development, glh mutants showed defects in papillae formation and reduces in cellulose content. | [59] |
| TBR | Positive | TBR is required for secondary wall cellulose synthesis, the Arabidopsis tbr has severely reduced crystalline cellulose in trichomes and reduced trichome density, altered trichome shape and surface appearance, lack of trichome papillae and basal cells, altered stomata shape, and altered patterns of callose deposition. | [60] |
| NOK | Positive | The noeck (nok) mutant has trichomes with increased branching and a glassy transparent appearance but displays no increase in nuclear DNA content. | [61] |
| MIXTA | Positive | Overexpression of MIXTA led to the growth of a large number of trichomes on cotyledons, leaves, and stems in tobacco. | [62] |
| CotMYB | Positive | Overexpression of CotMYB does not change its phenotype in Arabidopsis but increases the number of tobacco trichomes. | [62] |
| GaMYB2 | Positive | Cotton fibrin GaMYB2 not only regulates the leaf and stem trichome development of the Arabidopsis gl1 mutant, but also induces the formation of seed trichome. | [63] |
| AnnGh3 | Positive | Overexpression of AnnGh3 isolated from cotton in Arabidopsis resulted in a significant increase in leaf trichome density and length. | [64] |
Negative regulators include CAPRICE (CPC), TRIPTYCHON (TRY), ENHANCER OF TRY AND CPC1 (ETC1), and ETC2, which show functional redundancy. Negative regulators that interact with GL3, EGL3, and TTG1 negatively regulate the development of trichomes [73]. In addition, ETC1 and ETC2 act as enhancers of TRY and CPC to promote the movement of TRY between cells and inhibit the initiation of trichome development by interacting with GL3 and TTG1. CPC and TRY have similar functions [42].
Presently, two models have been proposed for the molecular regulatory mechanism of trichome development that co-regulate the growth and development of trichomes in plants. The first is the activator-depletion model: the TTG1 has a dual role in trichome development, moves freely between cells and combines the GL3/EGL3 and GL1/MYB23 to form a GL1/MYB23-GL3/EGL3-TTG1 complex that positively regulates trichome development [74]. The second is the activator-inhibitor model: the MYB-bHLH-TTG complex activates the TRY/CPC and promotes TRY and CPC proteins to move into adjacent cells. This complex forms an inactive TRY/CPC-GL3/EGL3-TTG1 complex and negatively regulates trichome formation by replacing the positively regulated transcription factor GL1/MYB23 [75]. SAD2 maintains the stability of the GL1-GL3-TTG1 complex by regulating the accumulation of mRNAs of many trichome-related genes in plants, which in turn affects protein activity [53].
In addition, some regulatory factors affect trichome branching by modulating the nuclear replication of trichome cells. The formation of trichome branches occurs after four times of endoreplication of trichome nuclear DNA. Among them, the positive regulators affecting the DNA replication and branching processes are INTERACTORs of CDK/KIP-related protein (ICK/KRP), ROOT HAIRLESS 2 (RHL2), HYPOCO-TYL 6 (HYP6), ZWICHEL (ZWI), GLABROUS INFLORESCENCE STEMS (GIS), GL3 and TTG1.
Misexpression of the ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication, cell size, and induces cell death [50]. The zwi, rhl2 and hyp6 mutations reduce trichome branching [76]; AtFIP37 and GIS positively regulate plant trichome branching through an intranuclear replication cycle [77, 78]. Negative regulators of trichome branching include SIAMESE (SIM), KAKTUS (KAK), SPINDLY (SPY), CONSTITUTIVE PATHOGEN RESPONSE 5 (CPR5), RETINOBLASTOMA RELATED 1 (RBR), TRY, and HOMEODOMAIN GLABROUS 11 (HDG11). SIM, CPR5, and RBR all affect trichome branching by controlling nuclear replication [42, 56, 66]; KAK mainly regulates the expression pattern of GL3 and EGL3 causing reduced trichome branching [55]; SPY is a negative regulator controlled by gibberellin [79], and HDG11 can restore the phenotype of the gl2 mutant in the Arabidopsis trichome [58].
The development of trichome and plant organogenesis occurs synchronously. GLASSY HAIR (GLH), TBR, NOECK (NOK), and their protein complexes together induce the formation of trichomes. The mutant phenotypes of the three genes were all glassy trichomes [59–61]. The GLH can induce the formation of papillary hairs in the late development of trichomes, and the cellulose content of glh mutants was decreased in Arabidopsis [59]. In addition, the complex GL1-GL3-TTG1 affects the signal transmission between cells by activating negative regulatory genes of trichome development, such as CPC, TRY and ETC1, resulting in the inhibition of the activity of the GL1-GL3-TTG1 complex and promotion of the development of cells into non-trichome cells [50].
However, the regulatory mechanism of trichome development is different in various plants. Overexpression of Arabidopsis GL1 in tobacco (Nicotiana tabacum L) does not affect trichome development. Overexpression of the snapdragon MIXTA and CotMYB (a MIXTA homologous gene in cotton) genes increases the density of N. tabacum trichome, while transgenic Arabidopsis expressing the snapdragon MIXTA shows no change [68]. In addition, overexpression of the cotton TRY hindered the development of tobacco trichomes, while transgenic Arabidopsis has no change [64]. Arabidopsis trichome suppressor CPC has no effect on L. esculentum trichome traits [80]. Control of trichome initiation GL1 and trichome differentiation OCL4B (OUTER CELL LAYER 4) inhibition of maize [81] regulated the growth and development of the trichome only in Arabidopsis not in N. tabacum, P. hybrida L., and L esculentum [82]. Thus, all studies showed that there are different regulatory mechanisms during multicellular and unicellular trichome development. Nevertheless, there is a specific relationship between them. For example, the cotton trichome transcription factor GaMYB2 can restore the trichome phenotype of the gl1 mutant in Arabidopsis [63], and the protein AnnGh3, when isolated from cotton, promotes the trichome elongation of Arabidopsis [64].
Other transcription factors related to multicellular trichomes have also been studied. WO (WOLLY) affects the initial development of L. esculentum trichome [83]. Cucumber CsGL1 controls the development of trichome branch [84]. Moreover, transposable elements are involved in cotton fiber cell development [85]. m6A (methylations at position N6 of internal adenosines) residues are directly accommodated by its YTH reader. The ECT2 protein containing the YTH domain decodes the m6A signal during development and regulates trichome branching by controlling their ploidy levels [86, 87]. SlCycB2 overexpression results in L. esculentum plants with no non-glandular and glandular trichomes [88].
Regulation of non-coding genes
MicroRNAs (miRNAs) are endogenous non-coding small RNAs approximately 21 nucleotides in length with high complementarity sequences to their target mRNAs. These miRNAs control many aspects of cellular functions by modulating the expression level of their target genes at the post-transcriptional level [89, 90]. miRNAs regulate trichome development by modulating the expression of SQUAMOSA PROMOTER BINDING PROTEIN LIKE (SPL) [91]. miR156 is a potential graft-transmissible miRNA that modulates leaf morphology and produces long leaf trichomes [92]. In Arabidopsis, SPLs are negative regulators of trichome development in the inflorescence stem and floral organs. The Arabidopsis SPL gene family contains 17 members, 10 of which are targeted by miR156 [93]. The expression of miR156 and SPLs (SPL3/SPL9) are temporally regulated [94, 95]. The expression level of miR156 is the highest in young plants and declines as the plant ages, while the expression level of their targets, SPLs, are low in young plants and accumulates gradually during reproductive growth. The expression pattern was consistent with the gradual loss of trichomes on the stem and floral organs. Overexpression of a mimicry target of miR156 significantly decreased the density of stem trichomes. Similar phenotypes were found in transgenic plants overexpressing miR156-resistant forms of SPL3, SPL9 and SPL10. The SPL9, binding to the promoters of TCL1 and TRY, activates their expression and inhibits the formation of trichomes [91]. In addition, transgenic Arabidopsis constitutively overexpresses miR156 producing ectopic trichomes on the stem and floral organs. Transgenic alfalfa (Medicago sativa) overexpressing miR156 showed increased trichome density, delayed flowering time, and elevated biomass production owing to decrease the expression level of three target SPLs (SPL6, SPL12 and SPL13). We found that the miR171-targeted LOST MERISTEMS 1 (LOM1), LOM2 and LOM3 were also involved in regulating SPL activity. Reduced LOM abundance by overexpression of miR171 leads to decreased trichome density on the stems and floral organs. Conversely, constitutive expression of the miR171-resistant LOM (rLOM) promotes trichome production [96]. The recent study had revealed that the expression level of TCP, as a target of miR319a, was decreased in miR319a-Ox transgenic poplars and significantly increased the density of leaf trichomes. On the contrary, the use of short tandem target mimics (STTM) to repress miR319a, the transcription level of TCP was upregulated and decreased the trichome density (Table 2) [97].
Table 2.
MicroRNAs related to trichome development in plants
| MicroRNAs | Regulation | Functional description | References |
|---|---|---|---|
| miR156 | Negative | miR156-targeted SPL transcription factors, overexpression of a mimicry target of miR156 significantly increases the SPLs transcripts and decreases the density of stem trichomes. | [95] |
| miR171 | Negative | miR171-targeted LOM1, LOM2, and LOM3 regulating the SPL activity. Overexpression of miR171 led to decreased trichome density on stems and floral organs. | [96] |
| miR319a | Positive | miR319a-targeted TCPs, overexpression of miR319a decreased transcription levels of TCPs and significantly elevated leaf trichome density in transgenic poplar. | [97] |
Role of hormones in growth and development of trichomes
The differentiation of trichomes is closely related to its location, growth, development stage, and hormone level (Table 3). Studies have shown that gibberellin (GA) [101], cytokinin (CK) [102], salicylic acid (SA) [98], jasmonic acid (JA) [103], brassinolide (BR), and other hormones regulate the initiation, growth and development of plant trichomes. GA treatment increases trichome numbers on Arabidopsis leaves. GA enhances the expression levels of MYB23, GL1, GL3 and EGL3, while inhibiting the expression levels of TRY, ETC1 and ETC2 [74]. Analysis of Arabidopsis ga1-3, spy mutants and wild-type showed that an endogenous GA level and/or activity of the signal transduction pathway positively regulates the number and branching of trichomes [4, 97]. GA and CK control trichome formation primarily through C2H2 transcription factors, such as GIS1, GIS2, ZINC FINGER PROTEIN (ZFP) 5, ZFP6 and ZFP8, which act upstream of the GL3/EGL3-GL1-TTG1 transcriptional activator complex.
Table 3.
The phytohormones related to trichome development in plants
| Hormone | Functional description | References | |
|---|---|---|---|
| GA | Positive | GA enhances the expression levels of MYB23, GL1, GL3, and EGL3, but inhibits the expression levels of TRY, ETC1, and ETC2, and positively regulates the number and branching of trichomes. | [72] |
| CK | Positive | CKs increase trichome formation, regulate C2H2 transcription factors ZFP8 and GIS2 acting downstream of SPY and upstream of GL1. | [48] |
| SA | Negative | SA decreases both trichome density and number. | [98] |
| JA | Positive | JA degrades JAZ to release WD-repeat/bHLH/MYB complex and initiate trichome. | [99] |
| Ethylene | Positive | Ethylene affects trichome branching, Arabidopsis ethylene receptor mutant, etr2–3, has completely unbranched trichomes. | [99] |
| BR | Positive | - | [100] |
CKs promote trichome formation [45]. The GA response inhibitor SPY activates the CK signaling pathway in Arabidopsis [104]. GA and CK signaling pathways control trichome cell fate by integrating the Arabidopsis transcription factors GIS, ZFP8, GIS2 to regulate GL1 expression. The study revealed that ZFP8 and GIS2 are involved in the regulatory mechanism of CK on trichome production. GIS2 induced by CKs acts downstream of SPY but upstream of GL1.
SA reduces trichome formation [97]. The Arabidopsis cpr mutant that overproduces SA showed a low trichome density. SA reduced the positive effects of JA on trichome induction, suggesting negative crosstalk between the JA- and SA-dependent pathways. There is also evidence for interactions among GA, JA and SA pathways, and Arabidopsis treated with SA showed a 1/4 reduction in trichome density. In the induction of trichome formation, GA and JA act synergistically, but SA acts antagonistically to the effect of GA with respect to trichome density and numbers. Exogenous JA application significantly increased the density of trichomes in A. annua and L. esculentum [105]. The reduction of JA levels (through silencing of OPR3, a key enzyme in the biosynthesis of the precursor of JA) impairs glandular trichome development in L. esculentum [106]. The Jasmonate ZIM-domain (JAZ) proteins interact with bHLH transcription factors (GL3, EGL3 and TT8) and MYB transcription factors (MYB75 and GL1), which are degraded by JA to regulate the initiation and development of trichomes through releasing a WD-repeat/bHLH/MYB complex and activating downstream factors [99].
Ethylene increases trichome branching in cucumber [107]. Increasing ethylene synthesis enhances the length of trichome branches in cotton [108, 109]. The ethylene receptor ETR2 controls trichome branching by regulating the assembly of microtubules [110]. A recent study revealed that L. esculentum SlIAA15, acting as strong repressor of auxin-dependent transcription, is involved in regulating the initiation of trichomes [100]. Moreover, the application of BR in cotton increases the expression level of fiber-related genes in cotton ovules, while Brz (BR inhibitor) inhibits fiber-related gene expression [111].
Effects of environmental factors on trichome development
Trichomes help plants to deal with different environmental challenges. To better adapt to the environment, trichomes have evoluted to adapt to a complex and changing environment, such as salt, temperature, light, and other abiotic factors [112–114]. In a salt stress experiment, the trichome size and density of mint and marjoram increased significantly [115, 116]. The trichome density of Solanum habrochaites is affected by photoperiod and temperature. Under a long day of natural sunlight, trichome density increased; the density increased at temperatures of 20-25 °C and decreased at low temperatures [117]. Ning et al. (2016) selected two experimental sites (from wet to dry) along the Loess Plateau latitude gradient to observe changes in the trichome morphology of Caragana korshinskii [114]. Micro-phenomena through which trichomes grew denser and larger under reduced precipitation were observed using a scanning electron microscope. Evidence that trichomes have structural adaptations to low temperatures and enhance tolerance to freezing was obtained from a study on birch where frost rapidly increased the density of glandular trichomes [118]. Yan et al. (2012) found that exposure of Arabidopsis to UV-B could stimulate trichome formation [119]. Plants from higher latitudes tended to be shorter, have fewer branches, flower earlier, flower larger, develop fewer trichomes, and produce more highly lobed leaves than plants at lower latitudes [120]. The surface density of M. sativa increases after mechanical injury, drought stress or growing under water deficit conditions [121].
Research on trichomes in tea plants (C. sinensis)
The molecular mechanism of trichome formation has also been investigated in other model plants, such as C. sativus [122], L. esculentum [123], B. campestris [124] and B. napus [125]. The recent study had revealed that another regulatory mechanism might be involved in regulating trichome development in plants. Trichome (also referred to as ‘háo’ in tea) was generally regarded as one of the quality standards of tea due to secrete various secondary metabolites, such as flavonoids, tea polyphenols, and amino acids. The number of tea trichomes depends on degree of tenderness of tea leaves. The distribution of trichomes is especially thick on newly budded leaves and gradually decreases as the leaf develops [126]. Previous studies had revealed that over 20,000 differentially expressed genes (DEGs) were identified in FDDB (Fudingdabaicha, trichome-rich) and RCZ (Rongchunzao, trichome-less) [127]. However, the homologous sequences of Arabidopsis trichome-related genes were not significantly different between the two cultivars or was even undetected [127].
Two hypotheses were proposed to explain the above phenomenon. First, the low proportion of tea trichomes in collected leaves resulted in the relatively low transcripts of the trichome-related genes. Second, it is possible that a novel pathway has not been found about trichomes formation in tea plant. Previous study had revealed that the trichome-related genes, including four HB regulators (REVOLUTA, WOX1/WOX8, and ATHB-51), RAV1, SPLs and TCPs, were up-regulated in ‘RCZ’ tea cultivar. The expression level of ZFP4 gene was decreased in ‘RCZ’. These results were consistent with the previous studies that these genes played a key role in regulating trichome development in Arabidopsis [102, 128–133]. Phytohormones also play an important role in tea trichome development. Although the genes involved in each type of phytohormone signal were differentially expressed, those related to CKs and encoding the GA receptor GID1b were repressed in RCZ; however, DELLAs, which encode the major GA signaling repressors, were up-regulated. These results indicate that both GA and CK signals are suppressed in RCZ. Moreover, the functions of GA and CK could be influenced by SPINDLY, an O-linked N-acetylglucosamine transferase that suppresses GA signaling and promotes CK responses [48, 109, 132, 134]. The DEG dataset showed that the expression of SPINDLY was also down-regulated. Collectively, these results suggest that the hairlessness of RCZ might result in part from the repression of GA and CK signals. Trichomes contain cellulose for the establishment of thick cell walls, which is regulated by the trichome development phase [135]. In this study, we found that most TBR-like family (TBR and TBL) and cellulose synthase genes, which are important for cellulose synthesis, were repressed in RCZ, and this repression was markedly associated with the hairless phenotype of the RCZ cultivar.
Although tea trichomes have no significant effect on the content of leaf inclusions account for their small proportion in a leaf. But trichomes really exist some trichome-synthesized flavoring materials, including catechins, flavonols, amino acids, purine alkaloids and volatiles which are important ingredient in tea flavors. More importantly, some defensive metabolites and many anti-insect, anti-microbial genes highly accumulated and expressed in the trichomes, respectively, strongly indicated that trichomes may also play an important in protecting tea plants against biotic and abiotic stresses [11, 136].
Outlook
In recent years, although the great progress has been made in the molecular mechanism of trichome development in Arabidopsis, the research on the regulation mechanism of trichome development in non-model plants is still unclear. Further research is urgently needed to be explored the molecular mechanism of trichome development in non-model plants. With the development of next-generation sequencing technology, the combination of genomics and classical genetics provides a very powerful tool to identify candidate genes regulating trichome development. CRISPR/CAS9-mediated knockout and siRNA-mediated knockdown technologies was used to identify the function of candidate genes by comparing the difference between the mutant and control plants. Moreover, trichome, as a barrier for plants to resist different stresses, played an important role in coping with biotic and abiotic stresses. Through transcriptomics, proteomics combined with metabolomics were used to further clarify the complex signal conversion mechanism. The molecular mechanism of plants converts environmental stimuli into endogenous signals that in turn regulate the growth and development of trichome and affect the structure, density and function, which still unclear. On the other hand, an important feature of glandular trichome is that it specifically synthesizes a wide range of secondary metabolites. These secondary metabolites not only play an important role in response to biotic and abiotic stress, but also contribute to human production and life, having very important economic value. For example, artemisinin, a sesquiterpene lactone, is isolated from the glandular trichome of Artemisia annua and widely used for its high anti-malarial pharmacological effects [137]. The glandular trichome of C. sativa can synthesize and accumulate cannabi-noids, which are prenylated polyketides having important pharmacological effects in clinical studies [138]. Modern genetic engineering techniques are used to transform plant glandular trichome into ‘chemical factories’ to synthesize valuable compounds for humans also has great development prospects.
Conclusions
Trichomes have a marked effect on plants and human health, its growth and development are effected by environment factors, hormones, transcription factor, non-coding RNA, etc. Although several previous studies had summarized the factors that affected the formation and development of plant trichomes, their reviews lacked a comprehensive summary and only focused on transcription factors, hormones and environment in model plants. The aim of this review is not only to summarize the factors, such as environment factors, hormones, transcription factor, non-coding RNA, and functional genes, regulate the formation and development of plant trichomes in model plants, but also investigate the regulation mechanism of trichome development in non-model plants, such as tea. We are expected this review provided a reference for basic and applied research on trichomes in plants in the near future.
Acknowledgments
Not applicable.
Authors’ contributions
XJW, CS, PHM, GFT and LTL collected the manuscript data, XJW and CS wrote the manuscript, GFT, PHM and LTL approved the final manuscript, all authors have read and approved the final manuscript.
Funding
This study was financially supported by Guizhou Science and Technology Support Project (Qiankehe Support [2019]2257); Guizhou Science and Technology Plan Project (Qiankehe Platform Talents [2018] 5768-03); Guizhou Vegetable Varieties Creation and Industrialization (Qiankehe Platform Talents [2017] 5715); Guizhou High Level Innovative Talents Training, Hundred Level Talents Project (Qiankehe talent [2015] 4024); Guizhou Academy of Agricultural Sciences project (Guizhou Academy of Agricultural Sciences Germplasm Resources [2020] 10).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
There is no ethics approval and consent to participate in this manuscript.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xiaojing Wang and Chao Shen contributed equally to this work.
Contributor Information
Guofei Tan, Email: tagfei@foxmail.com.
Litang Lv, Email: ltlv@gzu.edu.cn.
References
- 1.Johnson HB. Plant pubescence: An ecological perspective. Bot Rev. 1975;41(3):233–258. doi: 10.1007/BF02860838. [DOI] [Google Scholar]
- 2.Xiao K, Mao XH, Lin YL, Xu HB, Zhu YS, Cai QH, et al. Trichome, a functional diversity phenotype in plant. Mol Biol. 2016;2017(1):1–6. [Google Scholar]
- 3.Lloyd A, Schena M, Walbot V, Davis R. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266(5184):436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- 4.Chien JC, Sussex IM. Differential regulation of trichome formation on the adaxial and abaxial leaf surfaces by gibberellins and photoperiod in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996;111(4):1321–1328. doi: 10.1104/pp.111.4.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larkin JC, Young N, Prigge M, Marks MD. The control of trichome spacing and number in Arabidopsis. Development. 1996;122(3):997–1005. doi: 10.1242/dev.122.3.997. [DOI] [PubMed] [Google Scholar]
- 6.Telfer A, Bollman KM, Poethig RS. Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development. 1997;124(3):645–654. doi: 10.1242/dev.124.3.645. [DOI] [PubMed] [Google Scholar]
- 7.Perazza D, Vachon G, Herzog M. Gibberellins promote trichome formation by up-regulating GLABROUS1 in Arabidopsis. Plant Physiol. 1998;117(2):375–383. doi: 10.1104/pp.117.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang G, Feng HJ, Sun JL, Du XM. Induction of cotton ovule culture fibre branching by co-expression of cotton BTL, cotton SIM, and Arabidopsis STI genes. J Exp Bot. 2013;64(14):4157–4168. doi: 10.1093/jxb/ert222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guan XY, Pang MX, Nah G, Shi X, Ye W, Stelly DM, et al. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun. 2014. 10.1038/ncomms4050. [DOI] [PubMed]
- 10.Li WQ, Wu JG, Weng SL, Zhang D, Zhang Y, Shi C. Characterization and fine mapping of the glabrous leaf and hull mutants (gl1) in rice (Oryza sativa L.) Plant Cell Rep. 2010;29(6):617–627. doi: 10.1007/s00299-010-0848-2. [DOI] [PubMed] [Google Scholar]
- 11.Li JL, Zeng LT, Liao YY, Tang J, Yang Z. Evaluation of the contribution of trichomes to metabolite compositions of tea (Camellia sinensis) leaves and their products. LWT Food Sci Technol. 2020.
- 12.Tang XL, You T, Wu W, Sun L, Li Z. Allergenic property and two kinds of common street trees in Nanjing and the control. J Jishou Univ (Natural Science Edition) 2014;35(3):69–73. [Google Scholar]
- 13.Theobald WL, Krahulik JL, Rollins RC. Trichome description and classification. Anatomy of the dicotyledons I. Oxford: Clarendon Press; 1980. pp. 40–53. [Google Scholar]
- 14.Barthlott W, Wiersch S, Colic ZC, Koch K. Classification of trichome types within species of the water fern Salvinia, and ontogeny of the egg-beater trichomes. Botany. 2009;87(9):830–836. doi: 10.1139/B09-048. [DOI] [Google Scholar]
- 15.Kim HJ, Han JH, Kim S, Lee HR, Shin J, Kim J, et al. Trichome density of main stem is tightly linked to PepMoV resistance in chili pepper (Capsicum annuum L.). Theor. Appl. Genet. 2011;122(6):1051–1058. doi: 10.1007/s00122-010-1510-7. [DOI] [PubMed] [Google Scholar]
- 16.Ascensão L, Marques N, Pais MS. Glandular trichomes on vegetative and reproductive organs of Leonotis leonurus (Lamiaceæ) Ann Bot. 1995;75(6):619–626. doi: 10.1006/anbo.1995.1067. [DOI] [Google Scholar]
- 17.Brewer CA, Smith WK. Influence of simulated dewfall on photosynthesis and yield in soybean isolines (Glycine max [L.] merr. cv Williams) with different trichome densities. Int J Plant Sci. 1994;155(4):460–466. doi: 10.1086/297183. [DOI] [Google Scholar]
- 18.Wagner GJ, Wang E, Shepherd RW. New approaches for studying and exploiting an old protuberance, the plant trichome. Ann Bot. 2004;93(1):3–11. doi: 10.1093/aob/mch011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDowell ET, Kapteyn J, Schmidt A, Li C, Kang J, Descour A, et al. Comparative functional genomic analysis of Solanum glandular trichome types. Plant Physiol. 2010;155(1):524–539. doi: 10.1104/pp.110.167114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegebarth D, Buschhaus C, Wu M, Bird DA, Jetter R. The composition of surface wax on trichomes of Arabidopsis thaliana differs from wax on other epidermal cells. The Plant J. 2016;88(5):762–774. doi: 10.1111/tpj.13294. [DOI] [PubMed] [Google Scholar]
- 21.Rakha M, Bouba N, Ramasamy S, Regnard J, Hanson P. Evaluation of wild tomato accessions (Solanum spp.) for resistance to two-spotted spider mite (Tetranychus urticae Koch) based on trichome type and acylsugar content. Genet. Resour. Crop Ev. 2017;64(5):1011–1022. doi: 10.1007/s10722-016-0421-0. [DOI] [Google Scholar]
- 22.Zhao Q, Chen XY. Development: a new function of plant trichomes. Nat Plants. 2016;2(7):16096. doi: 10.1038/nplants.2016.96. [DOI] [PubMed] [Google Scholar]
- 23.Kang JH, Liu G, Shi F, Jones AD, Beaudry RM, Howe GA. The tomato odorless-2 mutant is defective in trichome-based production of diverse specialized metabolites and broad-spectrum resistance to insect herbivores. Plant Physiol. 2010;154(1):262–272. doi: 10.1104/pp.110.160192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kang J, Shi F, Jones AD, Marks MD, Howe GA. Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot. 2009;61(4):1053–1064. doi: 10.1093/jxb/erp370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maes L, Van Nieuwerburgh F, Zhang Y, Reed DW, Pollier J, Casteele SV, et al. Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 2010;189(1):176–189. doi: 10.1111/j.1469-8137.2010.03466.x. [DOI] [PubMed] [Google Scholar]
- 26.Gao Y, He C, Zhang D, Liu X, Xu Z, Tian Y, et al. Two trichome birefringence-like proteins mediate xylan acetylation, which is essential for leaf blight resistance in rice. Plant Physiol. 2017;173(1):470–481. doi: 10.1104/pp.16.01618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Busta L, Hegebarth D, Kroc E, Jetter R. Changes in cuticular wax coverage and composition on developing Arabidopsis leaves are influenced by wax biosynthesis gene expression levels and trichome density. Planta. 2017;245(2):297–311. doi: 10.1007/s00425-016-2603-6. [DOI] [PubMed] [Google Scholar]
- 28.Vitarelli NC, Riina R, Cassino MF, Meira RM. Trichome-like emergences in croton of Brazilian highland rock outcrops: evidences for atmospheric water uptake. Perspect Plant Ecol. 2016;1:23–35. doi: 10.1016/j.ppees.2016.07.002. [DOI] [Google Scholar]
- 29.Vanhoutte B, Schenkels L, Ceusters J, De Proft MP. Water and nutrient uptake in vriesea cultivars: trichomes vs. roots. Environ Exp Bot. 2017;1:21–30. doi: 10.1016/j.envexpbot.2017.01.003. [DOI] [Google Scholar]
- 30.Chen G, Zhao J, Zhao X, Zhao P, Duan R, Nevo E, et al. A psammophyte Agriophyllum squarrosum (L.) Moq.: a potential food crop. Genet. Resour. Crop Ev. 2014;61(3):669–676. doi: 10.1007/s10722-014-0083-8. [DOI] [Google Scholar]
- 31.Liu H, Zhou LH, Jiao J, Liu S, Zhang ZM, Lu TJ, et al. Gradient mechanical properties facilitate Arabidopsis trichome as mechanosensor. Appl Mater Interfaces. 2016;8(15):9755–9761. doi: 10.1021/acsami.6b02253. [DOI] [PubMed] [Google Scholar]
- 32.Zhou LH, Liu SB, Wang P, Lu TJ, Xu F, Genin GM, et al. The Arabidopsis trichome is an active mechanosensory switch. Plant Cell Environ. 2017;40(5):611–621. doi: 10.1111/pce.12728. [DOI] [PubMed] [Google Scholar]
- 33.Oppenheimer DG, Herman PL, Sivakumaran S, Esch JJ, Marks MD. A MYB gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell. 1991;67(3):483–493. doi: 10.1016/0092-8674(91)90523-2. [DOI] [PubMed] [Google Scholar]
- 34.Dai X, Wang G, Yang DS, Tang Y, Broun P, Marks MD, et al. TrichOME: a comparative omics database for plant trichomes. Plant Physiol. 2009;152(1):44–54. doi: 10.1104/pp.109.145813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alahakoon UI, Taheri A, Nayidu NK, Epp DJ, Yu M, Parkin IA, et al. Hairy canola (Brasssica napus) re-visited: down-regulating TTG1 in an AtGL3-enhanced hairy leaf background improves growth, leaf trichome coverage, and metabolite gene expression diversity. BMC Plant Biol. 2016;16(1):e12. doi: 10.1186/s12870-015-0680-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ioannidi E, Rigas S, Tsitsekian D, Daras G, Alatzas A, Makris A, et al. Trichome patterning control involves TTG1 interaction with SPL transcription factors. Plant Mol Biol. 2016;92(6):675–687. doi: 10.1007/s11103-016-0538-8. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Shan X, Gao R, Yang S, Wang S, Gao X, et al. Two iiif clade-bHLHs from Freesia hybrida play divergent roles in flavonoid biosynthesis and trichome formation when ectopically expressed in Arabidopsis. Sci Rep. 2016;6(1):e30514. doi: 10.1038/srep30514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao C, Li D, Jin C, Duan S, Qi S, Liu K, et al. Genome-wide identification of GLABRA3 downstream genes for anthocyanin biosynthesis and trichome formation in Arabidopsis. Biochem Bioph Res Co. 2017;485(2):360–365. doi: 10.1016/j.bbrc.2017.02.074. [DOI] [PubMed] [Google Scholar]
- 39.Johnson CS, Kolevski B, Smyth DR. TRANSPARENT TESTA GLABRA2, a trichome and seed coat development gene of Arabidopsis, encodes a WRKY transcription factor. Plant Cell. 2002;14(6):1359–1375. doi: 10.1105/tpc.001404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai X, Zhou L, Zhang W, Cai L, Guo H, Tian H, et al. A single amino acid substitution in the R3 domain of GLABRA1 leads to inhibition of trichome formation in Arabidopsis without affecting its interaction with GLABRA3. Plant Cell Environ. 2016;39(4):897–907. doi: 10.1111/pce.12695. [DOI] [PubMed] [Google Scholar]
- 41.Tominagawada R, Nukumizu Y, Sato S, Kato T, Tabata S, Wada T. Functional divergence of MYB-related genes, WEREWOLF and AtMYB23 in Arabidopsis. Biosci Biotechnol Biochem. 2012;76(5):883–887. doi: 10.1271/bbb.110811. [DOI] [PubMed] [Google Scholar]
- 42.Esch JJ, Chen M, Sanders M. A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development. 2003;130(24):5885–5894. doi: 10.1242/dev.00812. [DOI] [PubMed] [Google Scholar]
- 43.Schellmann S, Schnittger A, Kirik V, Wada T, Okada K, Beermann A, et al. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002;21(19):5036–5046. doi: 10.1093/emboj/cdf524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kirik V, Simon M, Wester K, Schiefelbein J, Hulskamp M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol Biol. 2004;55(3):389–398. doi: 10.1007/s11103-004-0893-8. [DOI] [PubMed] [Google Scholar]
- 45.Maes L, Inze D, Goossens A. Functional specialization of the TRANSPARENT TESTA GLABRA1 network allows differential hormonal control of laminal and marginal trichome initiation in Arabidopsis rosette leaves. Plant Physiol. 2008;148(3):1453–1464. doi: 10.1104/pp.108.125385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao H, Wang X, Zhu D, Cui S, Li X, Cao Y, et al. A single amino acid substitution in IIIf subfamily of basic helix-loop-helix transcription factor AtMYC1 leads to trichome and root hair patterning defects by abolishing its interaction with partner proteins in Arabidopsis. J Biol Chem. 2012;287(17):14109–14121. doi: 10.1074/jbc.M111.280735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li SF, Milliken ON, Pham H, Seyit R, Napoli RS, Preston J, et al. The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell. 2009;21(1):72–89. doi: 10.1105/tpc.108.063503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gan Y, Liu C, Yu H, Broun P. Integration of cytokinin and gibberellin signalling by Arabidopsis transcription factors GIS, ZFP8 and GIS2 in the regulation of epidermal cell fate. Development. 2007;134(11):2073–2081. doi: 10.1242/dev.005017. [DOI] [PubMed] [Google Scholar]
- 49.Mittal A, Balasubramanian R, Cao J, Singh P, Subramanian S, Hicks GR, et al. TOPOISOMERASE 6B is involved in chromatin remodelling associated with control of carbon partitioning into secondary metabolites and cell walls, and epidermal morphogenesis in Arabidopsis. J Exp Bot. 2014;65(15):4217–4239. doi: 10.1093/jxb/eru198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schnittger A, Weinl C, Bouyer D, Schobinger U, Hulskamp M. Misexpression of the cyclin-dependent kinase inhibitor ICK1/KRP1 in single-celled Arabidopsis trichomes reduces endoreduplication and cell size and induces cell death. Plant Cell. 2003;15(2):303–315. doi: 10.1105/tpc.008342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugimotoshirasu K, Stacey N, Corsar J, Roberts K, Mccann MC. DNA topoisomerase VI is essential for endoreduplication in Arabidopsis. Curr Biol. 2002;12(20):1782–1786. doi: 10.1016/S0960-9822(02)01198-3. [DOI] [PubMed] [Google Scholar]
- 52.Mathur J, Chua NH. Microtubule stabilization leads to growth reorientation in Arabidopsis trichomes. Plant Cell. 2000;12(4):465–477. doi: 10.1105/tpc.12.4.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao Y, Gong XM, Cao WH, Zhao JF, Fu LQ, Wang XC, et al. SAD2 in Arabidopsis functions in trichome initiation through mediating GL3 function and regulating GL1, TTG1and GL2 expression. J Integr Plant Biol. 2008;50(7):906–917. doi: 10.1111/j.1744-7909.2008.00695.x. [DOI] [PubMed] [Google Scholar]
- 54.Churchman ML, Brown ML, Kato N, Kirik V, Hulskamp M, Inze D, et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. Plant Cell. 2006;18(11):3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perazza D, Herzog M, Hulskamp M, Brown SW, Dorne A, Bonneville J. Trichome cell growth in Arabidopsis thaliana can be derepressed by mutations in at least five genes. Genetics. 1999;152(1):461–476. doi: 10.1093/genetics/152.1.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kirik V, Bouyer D, Schobinger U, Bechtold N, Herzog M, Bonneville J, et al. CPR5 is involved in cell proliferation and cell death control and encodes a novel transmembrane protein. Curr Biol. 2001;134(4):1891–1895. doi: 10.1016/S0960-9822(01)00590-5. [DOI] [PubMed] [Google Scholar]
- 57.Morohashi K, Grotewold E. A systems approach reveals regulatory circuitry for Arabidopsis trichome initiation by the GL3 and GL1 selectors. PLoS Genet. 2009;5:2. doi: 10.1371/journal.pgen.1000396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Khosla A, Paper JM, Boehler AP, Bradley AM, Neumann TR, Schrick K. HD-zip proteins GL2 and HDG11 have redundant functions in Arabidopsis trichomes, and GL2 activates a positive feedback loop via MYB23. Plant Cell. 2014;26(5):2184–2200. doi: 10.1105/tpc.113.120360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suo BX, Seifert S, Kirik V. Arabidopsis GLASSY HAIR genes promote trichome papillae development. J Exp Bot. 2013;64(16):4981–4991. doi: 10.1093/jxb/ert287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bischoff V, Nita S, Neumetzler L, Schindelasch D, Urbain A, Eshed R, et al. TRICHOME BIREFRINGENCE and its homolog AT5G01360 encode plant-specific DUF231 proteins required for cellulose biosynthesis in Arabidopsis. Plant Physiol. 2010;153(2):590–602. doi: 10.1104/pp.110.153320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jakoby M, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, et al. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol. 2008;148(3):1583–1602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Payne T, Clement J, Arnold D, Lloyd AM. Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development. 1999;126(4):671–682. doi: 10.1242/dev.126.4.671. [DOI] [PubMed] [Google Scholar]
- 63.Shangguan X, Xu B, Yu Z, Wang L, Chen X. Promoter of a cotton fibre MYB gene functional in trichomes of Arabidopsis and glandular trichomes of tobacco. J Exp Bot. 2008;59(13):3533–3542. doi: 10.1093/jxb/ern204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li B, Li DD, Zhang J, Xia H, Wang XL, Li Y, et al. Cotton AnnGh3 encoding an annexin protein is preferentially expressed in fibers and promotes initiation and elongation of leaf trichomes in transgenic Arabidopsis. J Integr Plant Biol. 2013;55(10):902–916. doi: 10.1111/jipb.12063. [DOI] [PubMed] [Google Scholar]
- 65.Galway ME, Masucci JD, Lloyd AM, Walbot V, Davis RW, Schiefelbein J. The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev Biol. 1994;166(2):740–754. doi: 10.1006/dbio.1994.1352. [DOI] [PubMed] [Google Scholar]
- 66.Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, et al. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11(7):1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kirik V, Lee MM, Wester K, Herrmann U, Zheng Z, Oppenheimer DG, et al. Functional diversification of MYB23 and GL1 genes in trichome morphogenesis and initiation. Development. 2005;132(7):1477–1485. doi: 10.1242/dev.01708. [DOI] [PubMed] [Google Scholar]
- 68.Payne CT, Zhang F, Lloyd AM. GL3 encodes a bHLH protein that regulates trichome development in Arabidopsis through interaction with GL1and TTG1. Genetics. 2000;156(3):1349–1362. doi: 10.1093/genetics/156.3.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd AM. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130(20):4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 70.Wang SC, Chen JG. Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 2008;49(12):1792–1804. doi: 10.1093/pcp/pcn159. [DOI] [PubMed] [Google Scholar]
- 71.Bloomer RH, Juenger TE, Symonds VV. Natural variation in GL1and its effects on trichome density in Arabidopsis thaliana. Mol Ecol. 2012;1:3501–3515. doi: 10.1111/j.1365-294X.2012.05630.x. [DOI] [PubMed] [Google Scholar]
- 72.Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, et al. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell. 2014;21(14):1118–1133. doi: 10.1105/tpc.113.121731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szymanski DB, Lloyd AM, Marks MD. Progress in the molecular genetic analysis of trichome initiation and morphogenesis in Arabidopsis. Trends Plant Sci. 2000;5(5):214–219. doi: 10.1016/S1360-1385(00)01597-1. [DOI] [PubMed] [Google Scholar]
- 74.Meinhardt H, Gierer A. Applications of a theory of biological pattern formation based on lateral inhibition. J Cell Sci. 1974;15(2):321–346. doi: 10.1242/jcs.15.2.321. [DOI] [PubMed] [Google Scholar]
- 75.Yang CX, Ye ZB. Trichomes as models for studying plant cell differentiation. Cell Mol Life Sci. 2012;70(11):1937–1948. doi: 10.1007/s00018-012-1147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hartung F, Angelis KJ, Meister A, Schubert I, Melzer M, Puchta H. An archaebacterial topoisomerase homolog not present in other eukaryotes is indispensable for cell proliferation of plants. Curr Biol. 2002;12(20):1787–1791. doi: 10.1016/S0960-9822(02)01218-6. [DOI] [PubMed] [Google Scholar]
- 77.An L, Zhou Z, Su S, Yan A, Gan Y. GLABROUS INFLORESCENCE STEMS (GIS) is required for trichome branching through gibberellic acid signaling in Arabidopsis. Plant Cell Physiol. 2011;53(2):457–469. doi: 10.1093/pcp/pcr192. [DOI] [PubMed] [Google Scholar]
- 78.Vespa L, Vachon G, Berger F, Perazza D, Faure J, et al. The immunophilin-interacting protein AtFIP37 from Arabidopsis is essential for plant development and is involved in trichome endoreduplication. Plant Physiol. 2004;134(4):1283–1292. doi: 10.1104/pp.103.028050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jacobsen SE, Binkowski KA, Olszewski NE. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. P Natl Acid Sci USA. 1996;93(17):9292–9296. doi: 10.1073/pnas.93.17.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tominagawada R, Nukumizu Y, Sato S, Wada T. Control of plant trichome and root-hair development by a tomato (Solanum lycopersicum) R3 MYB transcription factor. PLoS One. 2013;8:1. doi: 10.1371/journal.pone.0054019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vernoud V, Laigle G, Rozier F, Meeley RB, Perez P, Rogowsky PM. The HD-ZIP IV transcription factor OCL4 is necessary for trichome patterning and anther development in maize. Plant J. 2009;59(6):883–894. doi: 10.1111/j.1365-313X.2009.03916.x. [DOI] [PubMed] [Google Scholar]
- 82.Lloyd AM, Walbot V, Davis RW. Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science. 1992;258(5089):1773–1775. doi: 10.1126/science.1465611. [DOI] [PubMed] [Google Scholar]
- 83.Spyropoulou EA, Haring MA, Schuurink RC. RNA sequencing on Solanum lycopersicum trichomes identifies transcription factors that activate terpene synthase promoters. BMC Genomics. 2014;15(1):402. doi: 10.1186/1471-2164-15-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Q, Cao C, Zhang C, Zheng S, Wang Z, Wang L, et al. The identification of Cucumis sativus Glabrous 1 (CsGL1) required for the formation of trichomes uncovers a novel function for the homeodomain-leucine zipper I gene. J Exp Bot. 2015;66(9):2515–2526. doi: 10.1093/jxb/erv046. [DOI] [PubMed] [Google Scholar]
- 85.Wang K, Huang G, Zhu YX. Transposable elements play an important role during cotton genome evolution and fiber cell development. Sci China Life Sci. 2015;59(2):112–121. doi: 10.1007/s11427-015-4928-y. [DOI] [PubMed] [Google Scholar]
- 86.Scutenaire J, Deragon J, Jean V, Benhamed M, Raynaud C, Favory J, et al. The YTH domain protein ECT2 is an m6a reader required for normal trichome branching in Arabidopsis. Plant Cell. 2018;30(5):986–1005. doi: 10.1105/tpc.17.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wei L, Song P, Wang Y, Lu Z, Tang Q, Yu Q, et al. The m6A reader ECT2 controls trichome morphology by affecting mRNA stability in Arabidopsis. Plant Cell. 2018;30(5):968–985. doi: 10.1105/tpc.17.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao S, Gao Y, Xiong C, Yu G, Chang J, Yang Q, et al. The tomato B-type cyclin gene, SlCycB2, plays key roles in reproductive organ development, trichome initiation, terpenoids biosynthesis and Prodenia litura defense. Plant Sci. 2017;2017:103–114. doi: 10.1016/j.plantsci.2017.05.006. [DOI] [PubMed] [Google Scholar]
- 89.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136(4):669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 90.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79(1):351–379. doi: 10.1146/annurev-biochem-060308-103103. [DOI] [PubMed] [Google Scholar]
- 91.Yu N, Cai W, Wang S, Shan C, Wang L, Chen X. Temporal control of trichome distribution by microRNA156-targeted SPL genes in Arabidopsis thaliana. Plant Cell. 2010;22(7):2322–2335. doi: 10.1105/tpc.109.072579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bhogale S, Mahajan A, Natarajan B, Rajabhoj M, Thulasiram HV, Banerjee A. MicroRNA156: a potential graft-transmissible microRNA that modulates plant architecture and tuberization in Solanum tuberosum ssp. andigena. Plant Physiol. 2014;164(2):1011–1027. doi: 10.1104/pp.113.230714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP. Prediction of plant microRNA targets. Cell. 2002;110(4):513–520. doi: 10.1016/S0092-8674(02)00863-2. [DOI] [PubMed] [Google Scholar]
- 94.Wu G. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development. 2006;133(18):3539–3547. doi: 10.1242/dev.02521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Aung B, Gruber MY, Amyot L, Omari KW, Bertrand A, Hannoufa A. MicroRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol J. 2015;13(6):779–790. doi: 10.1111/pbi.12308. [DOI] [PubMed] [Google Scholar]
- 96.Xue X, Zhao B, Chao L, Chen D, Cui W, Mao Y, et al. Interaction between two timing microRNAs controls trichome distribution in Arabidopsis. PLoS Genet. 2014;10:4. doi: 10.1371/journal.pgen.1004266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan D, Ran LY, Hu J, Ye X, Xu D, Li J, et al. miR319a/TCP module and DELLA protein regulate trichome initiation synergistically and improve insect defenses in Populus tomentosa. New Phytol. 2020;227(3):867–883. doi: 10.1111/nph.16585. [DOI] [PubMed] [Google Scholar]
- 98.Traw MB, Bergelson J. Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 2003;133(3):1367–1375. doi: 10.1104/pp.103.027086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, et al. The Jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011;23(5):1795–1814. doi: 10.1105/tpc.111.083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Deng W, Yang Y, Ren Z, Audrandelalande C, Mila I, Wang X, et al. The tomato SlIAA15 is involved in trichome formation and axillary shoot development. New Phytol. 2012;194(2):379–390. doi: 10.1111/j.1469-8137.2012.04053.x. [DOI] [PubMed] [Google Scholar]
- 101.Inthima P, Nakano M, Otani M, Niki T, Nishijima T, Koshioka M, et al. Overexpression of the gibberellin 20-oxidase gene fromTorenia fournieriresulted in modified trichome formation and terpenoid metabolities of Artemisia annua L. Plant Cell Tiss Org. 2017;129(2):223–236. doi: 10.1007/s11240-017-1171-1. [DOI] [Google Scholar]
- 102.Zhou Z, Sun L, Zhao Y, An L, Yan A, Meng X, et al. Zinc finger protein 6 (ZFP6) regulates trichome initiation by integrating gibberellin and cytokinin signaling in Arabidopsis thaliana. Plant Physiol. 2013;198(3):699–708. doi: 10.1111/nph.12211. [DOI] [PubMed] [Google Scholar]
- 103.Xia X, Hu Q, Li W, Chen Y, Han L, Tao M, et al. Cotton (Gossypium hirsutum) JAZ3 and SLR1 function in jasmonate and gibber-ellin mediated epidermal cell differentiation and elongation. Plant Cell Tiss Org. 2018;133(2):249–262. doi: 10.1007/s11240-018-1378-9. [DOI] [Google Scholar]
- 104.Greenboim-Wainberg Y, Maymon I, Borochov R, et al. Cross talk between gibberellin and cytokinin: the Arabidopsis GA response inhibitor SPINDLY plays a positive role in cytokinin signaling. Plant Cell. 2005;17(1):92–102. doi: 10.1105/tpc.104.028472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yan T, Chen M, Shen Q, Li L, Fu X, Pan Q, et al. HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Arte-misia annua. New Phytol. 2017;213(3):1145–1155. doi: 10.1111/nph.14205. [DOI] [PubMed] [Google Scholar]
- 106.Bosch M, Wright LP, Gershenzon J, Wasternack C, Hause B, Schaller A, et al. Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiol. 2014;166(1):396–410. doi: 10.1104/pp.114.237388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kazama H, Dan H, Imaseki H, Wasteneys GO. Transient exposure to ethylene stimulates cell division and alters the fate and polarity of hypocotyl epidermal cells. Plant Physiol. 2004;134(4):1614–1623. doi: 10.1104/pp.103.031088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shi Y, Zhu S, Mao X, Feng J, Qin Y, Zhang L, et al. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18(3):651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Qin Y, Hu C, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu Y. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell. 2007;19(11):3692–3704. doi: 10.1105/tpc.107.054437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Plett JM, Mathur J, Regan S. Ethylene receptor ETR2 controls trichome branching by regulating microtubule assembly in Arabidopsis thaliana. J Exp Bot. 2009;60(13):3923–3933. doi: 10.1093/jxb/erp228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu R, Zhang J, Liu D, Wei Y, Wang Y, Li X. Characterization of bHLH/HLH genes that are involved in brassinosteroid (BR) signaling in fiber development of cotton (Gossypium hirsutum) BMC Plant Biol. 2018;18(1):1–13. doi: 10.1186/s12870-017-1213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hendrick MF, Finseth FR, Mathiasson ME, Palmer KA, Broder EM, Breigenzer P, et al. The genetics of extreme microgeographic adaptation: an integrated approach identifies a major gene underlying leaf trichome divergence in Yellowstone Mimulus guttatus. Mol Ecol. 2016;25(22):5647–5662. doi: 10.1111/mec.13753. [DOI] [PubMed] [Google Scholar]
- 113.Mazie AR, Baum DA. Clade-specific positive selection on a developmental gene: BRANCHLESS TRICHOME and the evolution of stellate trichomes in Physaria (Brassicaceae) Mol Phyogenew Evol. 2016;1:31–40. doi: 10.1016/j.ympev.2016.03.027. [DOI] [PubMed] [Google Scholar]
- 114.Ning P, Wang J, Zhou Y, Gao L, Wang J, Gong C. Adaptional evolution of trichome in Caragana korshinskii to natural drought stress on the loess plateau, China. Nat Ecol Evol. 2016;6(11):3786–3795. doi: 10.1002/ece3.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karraybouraoui N, Rabhi M, Neffati M, Baldan B, Ranieri A, Marzouk B, et al. Salt effect on yield and composition of shoot essential oil and trichome morphology and density on leaves of Mentha pulegium. Ind Crop Prod. 2009;30(3):338–343. doi: 10.1016/j.indcrop.2009.06.003. [DOI] [Google Scholar]
- 116.Olfa B, Imen T, Mohamed C. Essential oil and trichome density from Origanum majorana L. shoots affected by leaf age and salinity = Óleo essencial e densidade do tricoma dos rebentos da Origanum majorana L. afetados pela idade da folhagem e pela salinidade. Biosci J. 2016;32:1. doi: 10.14393/BJ-v32n1a2016-30323. [DOI] [Google Scholar]
- 117.Tian D, Peiffer M, De Moraes CM, Felton GW. Roles of ethylene and jasmonic acid in systemic induced defense in tomato (Solanum lycopersicum) against Helicoverpa zea. Planta. 2014;239(3):577–589. doi: 10.1007/s00425-013-1997-7. [DOI] [PubMed] [Google Scholar]
- 118.Prozherina N, Freiwald V, Rousi M, Oksanen E. Effect of spring-time frost and elevated ozone on early growth, foliar injuries and leaf structure of birch (Betula pendula Roth) genotypes. New Phytol. 2003;159(3):623–636. doi: 10.1046/j.1469-8137.2003.00828.x. [DOI] [PubMed] [Google Scholar]
- 119.Yan A, Pan J, An L, Gan Y, Feng H. The responses of trichome mutants to enhanced ultraviolet-B radiation in Arabidopsis thaliana. J Photochem Photobiol B. 2012:29–35. [DOI] [PubMed]
- 120.Kooyers NJ, Greenlee AB, Colicchio JM, Oh M, Blackman BK. Replicate altitudinal clines reveal that evolutionary flexibility underlies adaptation to drought stress in annual Mimulus guttatus. New Phytol. 2015;206(1):152–165. doi: 10.1111/nph.13153. [DOI] [PubMed] [Google Scholar]
- 121.Scoville AG, Barnett LL, Bodbylroels SA, Kelly J, Hileman LC. Differential regulation of a MYB transcription factor is correlated with transgenerational epigenetic inheritance of trichome density in Mimulus guttatus. New Phytol. 2011;191(1):251–263. doi: 10.1111/j.1469-8137.2011.03656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Cui J, Miao H, Ding L, Wehner TC, Liu P, Wang Y, et al. A new glabrous gene (csgl3) identified in trichome development in cucumber (Cucumis sativus L.) PLoS One. 2016;11:2. doi: 10.1371/journal.pone.0148422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yang C, Li H, Zhang J, Luo Z, Gong P, Zhang C, et al. A regulatory gene induces trichome formation and embryo lethality in tomato. P Natl A SCI. 2011;108(29):11836–11841. doi: 10.1073/pnas.1100532108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang J, Lu Y, Yuan Y, Zhang X, Geng J, Chen Y, et al. Map-based cloning and characterization of a gene controlling hairiness and seed coat color traits in Brassica rapa. Plant Mol Biol. 2009;69(5):553–563. doi: 10.1007/s11103-008-9437-y. [DOI] [PubMed] [Google Scholar]
- 125.Gruber M, Alahakoon U, Taheri A, Nagubushana N, Zhou R, Aung B, et al. The biochemical composition and transcriptome of cotyledons from Brassica napus lines expressing the AtGL3 transcription factor and exhibiting reduced flea beetle feeding. BMC Plant Biol. 2018;18(1):1–19. doi: 10.1186/s12870-018-1277-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sun B, Zhu Z, Liu R, Wang L, Dai F, Cao F, et al. TRANSPARENT TESTA GLABRA1 (TTG1) regulates leaf trichome density in tea Camellia sinensis. Nord J Bot. 2020;38:1. [Google Scholar]
- 127.Yue C, Cao H, Chen D, Lin H, Wang Z, Hu J, et al. Comparative transcriptome study of hairy and hairless tea plant (Camellia sinensis) shoots. Plant Physiol. 2018:41–52. [DOI] [PubMed]
- 128.Zhou Z, An L, Sun L, Zhu S, Xi W, Broun P, et al. Zinc finger protein5 is required for the control of trichome initiation by acting upstream of zinc finger protein8 in Arabidopsis. Plant Physiol. 2011;157(2):673–682. doi: 10.1104/pp.111.180281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yan A, Wu M, Zhao Y, Zhang A, Liu B, Schiefelbein J, et al. Involvement of C2H2 zinc finger proteins in the regulation of epidermal cell fate determination in Arabidopsis. J Integr Plant Biol. 2014;56(12):1112–1117. doi: 10.1111/jipb.12221. [DOI] [PubMed] [Google Scholar]
- 130.Matías-Hernández L, Aguilar-Jaramillo AE, et al. TEMPRANILLO reveals the mesophyll as cucial for epidermal trichome formation. Plant Physiol. 2016;170(3):1624–1639. doi: 10.1104/pp.15.01309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang M, Zhao P, Cheng H, Han L, Wu X, Gao P, et al. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 2013;162(3):1669–1680. doi: 10.1104/pp.113.215673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Steiner E, Efroni I, Gopalraj M, Saathoff K, Tseng TS, Kieffer M, et al. The Arabidopsis O-linked N-Acetylglucosamine transferase SPINDLY interacts with class I TCPs to facilitate cytokinin responses in leaves and flowers. Plant Cell. 2012;24(1):96–108. doi: 10.1105/tpc.111.093518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Vadde BV, Challa KR, Nath U. The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. 2018;93(2):259–269. doi: 10.1111/tpj.13772. [DOI] [PubMed] [Google Scholar]
- 134.Shimada A, Ueguchi-Tanaka M, Sakamoto T, et al. The rice SPINDLY gene functions as a negative regulator of gibberellin signaling by controlling the suppressive function of the DELLA protein, SLR1, and modulating brassinosteroid synthesis. Plant J. 2006;48(3):390–402. doi: 10.1111/j.1365-313X.2006.02875.x. [DOI] [PubMed] [Google Scholar]
- 135.Marks MD, Betancur L, Gilding EK, Chen F, Bauer S, Wenger JP, et al. A new method for isolating large quantities of Arabidopsis trichomes for transcriptome, cell wall and other types of analyses. Plant J. 2008;56(3):483–492. doi: 10.1111/j.1365-313X.2008.03611.x. [DOI] [PubMed] [Google Scholar]
- 136.Li P, Xu Y, Zhang Y, Fu J, Yu S, Guo H, et al. Metabolite profiling and transcriptome analysis revealed the chemical contributions of tea trichomes to tea flavors and tea plant defenses. J Agric Food Chem. 2020. 10.1021/acs.jafc.0c04075. [DOI] [PubMed]
- 137.Brown GD. The biosynthesis of Artemisinin (Qinghaosu) and the Phytochemistry of Artemisia annua L. (Qinghao) Molecules. 2010;15(11):7603–7698. doi: 10.3390/molecules15117603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sirikantaramas S, Morimoto S, Shoyama Y, Ishikawa Y, Wada Y, Shoyama Y, et al. The gene controlling marijuana Psychoactivity. J Biol Chem. 2004;279(38):39767–39774. doi: 10.1074/jbc.M403693200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.