Abstract
Extracellular matrix (ECM) gives structure, support, and is the niche for several cells found in skeletal muscle. ECM is mainly produced by muscle connective tissue (CT) fibroblasts during development and regeneration. Stromal fibroadipogenic progenitors (FAPs) are CT fibroblasts-like mesenchymal progenitors (MPs) with important roles in regeneration and degeneration. Chronic damage restrains the normal regenerative behavior of muscle fibroblasts/FAPs. Thus, the isolation and study of these mesenchymal progenitors are of crucial importance for understanding their behavior and biology. We investigated whether adult muscle CT fibroblasts (hereafter referred to as adherent fibroblasts [aFbs]) cultured via pre-plating strategy belong to a heterogeneous population of FAPs. By combining microscopy, western blot analyses, flow cytometry, and FACS we determined that aFbs isolated from skeletal muscle largely overlap with FAPs. In addition, we used the PDGFRαEGFP mice in order to corroborate our results with EGFP+ FAPs. Moreover, our strategy allows the isolation of activated EGFP+ FAPs from the murine DMD model PDGFRαEGFP; mdx and PDGFRαEGFP denervated mice. Here we report that 1 h 30 min of pre-plating strategy allows the isolation and culture of a highly enriched population of aFbs. These cells are phenotypically and biochemically a FAPs-like population of adherent cells. In addition, aFbs respond in the same fashion as FAPs to Nilotinib, an inducer of FAPs apoptosis. Moreover, flow cytometry characterization of these aFbs suggests that 85% of them express the MP marker PDGFRα, and isolation of aFbs from the PDGFRαEGFP mice suggests that 75% of them show high EGFP expression. Furthermore, TGF-β1 induces aFbs proliferation, myofibroblast differentiation, and ECM production. We were also able to isolate activated aFbs from skeletal muscle of the DMD mice and from the PDGFRαEGFP mice 2-days after denervation. Our findings suggest that the in vitro pre-plating strategy allows the isolation and culture of a relatively pure aFbs population, which resembles FAPs in vitro.
Abbreviations: Adherent Fibroblasts, aFbs; Connective tissue, CT; Extracellular matrix, ECM; Fibroadipogenic progenitors, FAPs; Fluorescence-activated cell sorting, FACS; Mesenchymal Progenitors, MPs; Muscle stem cells, MuSCs; Platelet-derived growth factor receptor alpha, PDGFRα; Transforming growth factor type-beta, (TGF-β); Transcription factor, TF
Keywords: Skeletal muscle, PDGFRα, FAPs, Fibrosis, Mesenchymal progenitors, TGF-β signaling
Highlights
-
•
We are focused in the study of the mesenchymal progenitors in skeletal muscle.
-
•
We study whether muscle CT fibroblasts are similar to fibro/adipogenic progenitors.
-
•
Here we report a pre-plating strategy for the isolation and culture of an enriched population of FAPs-like adherent cells.
-
•
Adherent fibroblasts respond in the same fashion to TGF-β signaling and the tyrosine kinase inhibitor Nilotinib as FAPs.
-
•
In vitro pre-plating strategy allows the isolation and culture of CT fibroblasts, which resembles FAPs in vitro.
1. Introduction
Adult skeletal muscle is formed by multinucleated myofibers and their surrounding connective tissue (CT) that gives structure and joins the muscle fibers together. The CT is composed of stromal fibroblasts or mesenchymal progenitors (MPs) and extracellular matrix (ECM) [1]. The muscle milieu is highly complex in structure and different cell populations co-exist participating in tissue homeostasis and regeneration [2]. In order to understand the complex mechanisms controlling CT development, establishment, and muscle fibrosis it is important to consider the stromal MPs that regulate these processes. Muscle CT fibroblasts express the Wnt Tcf/Lef family member Tcf7l2 (Tcf4) transcription factor (TF) [3]. In mouse and chick embryos, CT fibroblasts are restricted to muscle CT and are organized along with the ECM component collagen type I [[3], [4], [5]]. In addition, the existence of human CT fibroblasts (Tcf7l2-expressing cells) was demonstrated and shown to expand after human muscle injury and stimulate human myogenesis [6].
A similar population of CT MPs called fibroadipogenic progenitors (FAPs), has been described to play an important role in mouse skeletal muscle regeneration, and mouse and human fibro-fatty degeneration [[7], [8], [9]]. Like CT fibroblasts, FAPs do not arise from the myogenic lineage [3,7,8]. These multipotent MPs can differentiate in vivo and in vitro to potentially become myofibroblasts, adipocytes, chondrogenic, and osteogenic cells [8,10,11]. CD140a (platelet-derived growth factor receptor α, PDGFRα) is widely used as MPs and FAPs marker, and CT fibroblasts also express this receptor [4,5,12]. In addition, the Odd-Skipped genes Osr1 and Osr2 are expressed in CT fibroblasts [13]. Osr1 is expressed in an embryonic MP population of stromal FAP-like cells found in limb muscles [12]. Additionally, Osr1 and Tcf7l2 (Tcf4) expression identifies distinct but overlapping muscle CT fibroblast populations [12]. Furthermore, we demonstrated that the number of MPs (Tcf7l2+/PDGFRα+) is increased in three different models of muscle fibrosis: mdx mice, barium chloride-induced chronic damage, and muscle denervation. These results suggest a prominent role for these cells in skeletal muscle fibrosis [4]. Moreover, Tcf7l2+/PDGFRα+ MPs are augmented in skeletal muscles of the symptomatic hSOD1G93A mice (amyotrophic lateral sclerosis (ALS) murine model), concomitantly with increased fibrosis and TGF-β levels [14]. Indeed, as these cells are necessary to fully resolve the damaged tissue during normal skeletal muscle regeneration is thought that their dysregulated activity in chronic fibrotic diseases is the main cause for fibro-fatty pathology and failed tissue repair [4,10,15]. Transforming growth factor type-beta (TGF-β) is a major cytokine regulating muscle fibrogenesis [16,17] and its actively regulated by ECM components [18,19]. Activated TGF-β signaling correlates with reduced tissue regeneration, angiogenesis and function, and increased fibrosis [[19], [20], [21]]. TGF-β signaling induces FAP and mesenchymal progenitor proliferation, ECM production and myofibroblastic differentiation [8,15,22]. Thus, understanding the role of TGF-β signaling in MPs and/or fibroblasts, the effector cells of fibrosis, is important for the development of new and effective anti-fibrotic approaches.
Human PDGFRα MPs have been found to be causative cells for fibro-fatty deposition in Duchenne Muscular Dystrophy (DMD) patients [9]. Because the starting volume of human tissue obtained from muscle biopsy is limited the authors used a pre-plating strategy. Thus, they enzymatically digested human muscle and then cultured the dissociated cells [9]. Pre-plating strategies have been extensively used to isolate, culture, and study CT fibroblasts, MPs and FAPs biology [5,9,23,24]. It is an affordable procedure for the isolation of highly pure MPs and for the rapid and selective growth of adherent stromal MPs. Different methods have been used to study MPs biology, which includes the use of in vivo approaches [25,26] and in vitro culture systems [27]. When in vivo approaches are used it is difficult to identify cell type functions and mechanisms. In vitro culture systems of MPs isolated from the tissue is an experimental approach with controlled in vitro conditions where different cytokines and inhibitors, among other elements, can be used in order to study their specific role or effects on these cells. The best example of this in vitro strategy is the monolayer study of myoblast precursors or myogenic progenitors [28]. In addition, pre-plating strategies for muscle MPs identification and isolation permits a rapid, high yield of cells. Moreover, flow cytometry or FACS are not required, which in turn avoids the potential effects of shear stresses on the desired cells, and finally, the relatively long time and cost involved in these procedures.
Here, we describe the isolation and culture of adult muscle CT fibroblasts, and we discuss how this method could be useful to isolate aFbs, as these cells mostly resemble FAPs in monolayer culture. By using different methodologies and techniques, including flow cytometry and FACS, we studied whether aFbs recapitulate in vitro multiple behaviors described for FAPs. The PDGFRαEGFP reporter mouse strain [29], where PDGFRα-expressing cells (FAPs) are identified by EGFP expression, was useful to corroborate that a high proportion of the adherent cells are EGFP+ cells at passages 1 and 2. The aFbs in vitro population responds to Nilotinib in the same fashion as FAPs do, inducing their apoptosis. Western blot analyses of MPs markers (PDGFRα and αSMA) can differentiate this population of cells from muscle myogenic progenitors (Pax7+). In addition, aFbs stimulated with TGF-β1 responds by proliferating and differentiating into myofibroblasts along with increasing ECM production and deposition. This pre-plating strategy described here, also allowed us to isolate activated aFbs from muscles of the mdx mice and 2-day denervated muscles.
2. Methods
2.1. Mice and approval of the study
Mice protocols and experiments were conducted in strict accordance and with formal approval from the Animal Ethics Committee of the Pontificia Universidad Católica de Chile (ID protocol: 160512005). Mice were housed in standard cages under 12-h light-dark cycles and fed ad libitum with a standard chow diet. For wild type studies, 2- to 4-month-old C57Bl/10ScSc mice (hereafter referred to as wild type, WT) were used. In addition, 2 to 3-month-old Pdgfratm11(EGFP)Sor mice (JAX stock #007669 B6.129S4-Pdgfratm11(EGFP)Sor/J) were used [29]. PDGFRαEGFP mice were genotyped using the standard PCR protocol described by Jackson (https://www.jax.org/strain/007669). For aFbs isolation from mdx muscles, we crossed male C57Bl/10ScSn-mdx mice with hemizygous female B6.129S4-Pdgfratm11(EGFP)Sor/J mice. The mice used were the F1 males (2- to 3-month-old). All surgeries were performed after the mice had been anesthetized with 2.5% to 4% of isoflurane gas in pure oxygen. The mice were euthanized with cervical dislocation at the ages indicated in each figure, and the tissues were immediately processed for enzymatic digestion. In addition, muscles were processed either by direct freezing in liquid nitrogen for protein or in 2-methylbutane cooled with liquid nitrogen for histological analysis as described below.
2.2. Muscle denervation
Sciatic nerve denervation was performed unilaterally on 2- to 3-month-old PDGFRαeGFP mice. To constrain the sciatic nerve regeneration a section of 1 mm was removed [30]. Animals were euthanized 2-days after denervation, and the gastrocnemius, soleus, extensor digitorum longusm, and tibialis anterior muscles from the contralateral and denervated hind limbs were collected for enzymatic digestion and post-pre-plating strategy [4]. Contralateral muscles of the non-denervated limb were used as controls.
2.3. Reagents
Nilotinib (AMN-107) (CDS023093, Sigma-Aldrich, St. Louis, MO, USA) was reconstituted in DMSO (D2650, Sigma-Aldrich, USA), and cells were treated at final concentrations indicated in the corresponding figures. DMSO was used as a control. Cells were treated with recombinant hTGF-β1 (#580702, Biolegend, USA) in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% (v/v) fetal bovine serum and penicillin/streptomycin in a 5% CO2 atmosphere at the concentrations and times indicated in the corresponding figure legend.
2.4. Isolation of adult adherent muscle CT tissue fibroblasts (aFbs)
aFbs were isolated from limb muscles from 2- to 5-month-old mice, as previously described with some modifications [5,24]. Briefly, isolated muscles were washed once with PBS 1× and DMEM on ice. Tendons and adipose tissue were carefully discarded. Muscles were then cut in small pieces with scissors and smashed with blades (or scissors) on ice. Then, they were digested with collagenase type 1 (Worthington Biochemical, Lakewood, NJ) or Collagenase/Dispase (#11097113001, Sigma-Aldrich, St. Louis, MO, USA) in DMEM for 30–45 min in a thermo-regulated water bath with constant rotation at 37 °C. Of note, several collagenases were used showing similar results and yields. At the end of the incubation time, the muscle sample was vortexed, and DMEM 10% FBS (vol/vol) or FACS buffer (supplemented PBS containing 2 mM EDTA and 2% FBS) was added to stop the enzymatic digestion. The sample was then passed through 70-μm and 40-μm nylon mesh filters (Becton Dickinson, USA). After centrifugation at 1000g for 10 min, the supernatant was discarded, and the pellet resuspended in DMEM 10% FBS and then cultured for 1 h and 30 min in polystyrene 100 mm or 60 mm TC-treated Culture Dish (Corning, USA). Next, aFbs were maintained with DMEM 10% FBS and the supernatant discarded (other cells, i.e., myoblasts). aFbs were kept at 37 °C, 5% CO2, and 95% humidity. Isolated aFbs were used from passages 0 to 3. In general, passaging aFbs is associated with loss of multipotency. However, aFbs can be passaged 1–2 times at 70% confluence.
2.5. Flow cytometry/FACS
For flow cytometry and FACS of the aFbs, cells were washed two times with PBS 1× before the addition of extraction buffer (0.5% BSA, 1 mM EDTA, 1 mM EGTA, pH 7.4 (1 volume) + 0.05% Trypsin-EDTA (1 volume) (#15400054, Gibco Fisher Scientific, USA)). aFbs were incubated at 37 °C until they detached from the plate and then collected by pipetting, and centrifuged at 1000g for 8 min. After pellet resuspension in FACS buffer; cell preparations were passed through a 70-μm, and then 40-μm cell strainer (Becton Dickenson, USA). Resulting single cells were collected by centrifugation at 1000g for 5 min. Cell preparations were incubated with primary antibodies for 30 min at 4 °C in FACS buffer at ~1 × 107 cells per ml. We used the following monoclonal primary antibodies: anti-CD31 (clones MEC13.3, Becton Dickenson, and 390, Cedarlane Laboratories), anti-CD45 (clone 30-F11, Becton Dickenson), anti-Sca-1 (clone D7, eBiosciences) and anti-α7 integrin (clone R2F2) (AbLab). Typical antibody dilutions used were: anti-CD31, 1:400–500; anti-CD45, 1:400–500; anti-Sca-1, 1:2000–4000; anti-α7 integrin, 1:1400. For all antibodies, we performed fluorescence minus one control by staining with appropriate isotype antibodies. To assess viability, cells were stained with SYTOX™ Blue Dead Cell Stain, which detects dead cells, for flow cytometry following the provider's instructions (#S34857, Thermo Fisher Scientific, USA).
2.6. Crystal violet stain
For Fig. 1i and Fig. S1f, after Nilotinib treatment the cells were washed twice in cold PBS 1× and then fixed in 100% cold methanol (−20 °C) for 2 min. After methanol removal, Crystal Violet Staining Solution (0.5% w/v) (C3886, Sigma-Aldrich, St. Louis, MO, USA) was added and immediately washed with abundant distilled water. Stained cells were imaged in a Nikon Eclipse N600 microscope. We measured cell numbers in 5 randomly chosen fields from three independent experiments. Quantifications were done using ImageJ software with the cell counter plugin (version 1.46r, NIH, USA).
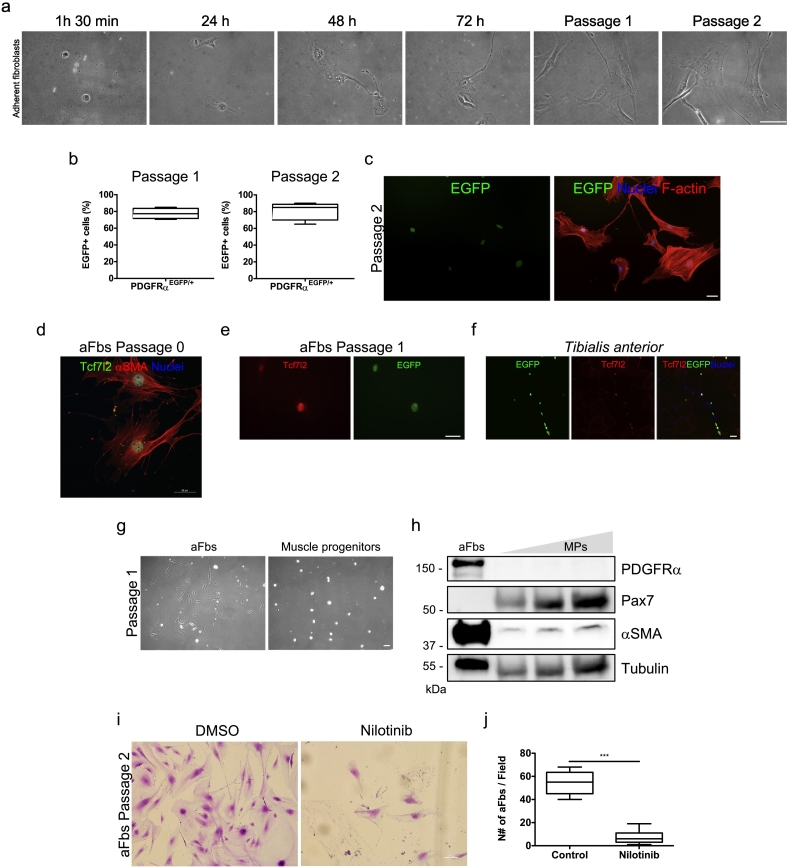
Fig. 1.
Adult adherent fibroblasts are likely fibro/adipogenic progenitors.
a. Representative bright-field images describing the time-course of adherent fibroblasts following the pre-plating strategy. Scale bar: 50 μm. b. PDGFRαEGFP reporter mice were used to quantify the % of adherent EGFP+ cells (FAPs) in the culture at passages 1 (left graph) and 2 (right graph). n = 3. The values correspond to the mean ± SEM. c. Representative immunofluorescence images showing adherent fibroblasts (EGFP+) following the pre-plating strategy at passage 2. F-actin (red) was used to label the cell structure. Hoechst (blue) was used to identify nuclei. Scale bar: 50 μm. d. aFbs expressing the Tcf7l2 (Tcf4) transcription factor (green) and αSMA (red) at passage 0. Hoechst (blue) was used to identify nuclei. Scale bar: 50 μm. e. aFbs expressing Tcf7l2 (red) and EGFP (green) at passage 1. Scale bar: 50 μm. f. Confocal immunofluorescence images showing EGFP+ FAPs and Tcf7l2-expressing cells in the tibialis anterior muscle. Hoechst (blue) was used to identify nuclei. Scale bar: 50 μm. g. Bright field images comparing aFbs with muscle progenitors (MPs) at passage 1. h. Western Blot analysis showing the relative protein levels of PDGFRα, Pax7, and αSMA in aFbs and muscle progenitors at passage 1. Tubulin was used as a loading control. i. Representative crystal violet staining images of aFbs survival after 72 h of Nilotinib treatment (5 μM) at passage 1. j. The graph shows the quantification of aFbs cell number per field after 72 h of Nilotinib treatment. n = 3; ***P < 0.001. The values correspond to the mean ± SEM.
2.7. Protein extraction and western blot analysis
Protein extracts from cells were obtained using RIPA 1× lysis buffer (#9806, Cell Signaling, MA, USA) plus protease/phosphatase inhibitors (#P8340/#P0044, Sigma-Aldrich, USA), and 1 mM phenylmethylsulfonyl fluoride (Sigma-Aldrich, USA). Then, the cells were sonicated for 10 s and centrifuged at 9000g. Proteins were quantified with the Micro BCA assay kit, following the manufacturer's instructions (Pierce, IL, USA). Extracts were subjected to SDS-PAGE electrophoresis in 9% polyacrylamide gels, transferred to PVDF membranes (Millipore, CA, USA), and probed with primary antibodies: goat anti-PDGFRα (1:1000) (#AF1062, R&D Systems, Minneapolis, MN, USA), mouse anti-αSMA (1:1000) (Sigma-Aldrich, St. Louis, MO, USA), rabbit anti-Integrin β1 (M-106) (1:1000) (sc-8978, Santa Cruz, USA), rabbit anti-fibronectin (1:10,000) (F3648, Sigma-Aldrich), mouse anti-Pax7-c (1:1000 from concentrate) (Developmental Studies Hybridoma Bank), mouse anti-GAPDH (1:5000) (#MAB374, Millipore, CA, USA), mouse anti-α-tubulin (1:5000) (#T5168, Sigma-Aldrich). Then, primary antibodies were detected with a secondary antibody (1:5000) conjugated to horseradish peroxidase: mouse anti-goat IgG, #31400; goat anti-rabbit IgG, #31460; and goat anti-mouse IgG, #31430 (Pierce, IL, USA). All immunoreactions were visualized by enhanced chemiluminescence Super- Signal West Dura (34075, Pierce, IL, USA) or Super Signal West Femto (34096, Pierce, IL, USA) by a ChemiDoc-It HR 410 imaging system (UVP, CA, USA).
2.8. Indirect immunofluorescence
For immunofluorescence analyses, the cells were seeded on 9.2 cm2 tissue culture dishes (TPP #93040). At the end of experiments, cells were washed three times with PBS 1×, fixed for 10 min in cold 4% paraformaldehyde, and washed with PBS again. Then, the cells were permeabilized with PBS, 0.1% Triton X-100 for 2 min, blocked for 30 min in blocking buffer (PBS, 0.1% Triton X-100, 1% BSA, 1% fish gelatin) and incubated with the primary antibody overnight: rabbit anti-Ki67 antibody (1:50) (#15580, Abcam), mouse anti-alpha-smooth muscle actin (αSMA) (1:200) (#A2547, Sigma-Aldrich), goat anti-PDGFRα (1:75) (#AF1062, R&D Systems, Minneapolis, MN, USA), goat anti-myogenin (F5D) (1:50) (#sc-12732, Santa Cruz), mouse anti- Pax7-c (1:50 from concentrate) (Developmental Studies Hybridoma Bank), rabbit anti-fibronectin (1:100; F3648; Sigma-Aldrich), rabbit anti-perilipin A/B (1:250) (#P1873, Sigma-Aldrich, USA), rabbit anti-Tcf7l2 (TCF4) (1:75) (#C48H11, Cell Signaling, USA). Next, the samples were washed with PBS and incubated for 1 h at room temperature with Alexa Fluor secondary antibodies (1:500) (Invitrogen, CA, USA). Next, Hoechst 33258 was added for 10 min for nuclei staining. Cells were washed with PBS, and DAKO fluorescent mounting medium (Dako North America Inc., CA, USA) was added. To stain F-actin Alexa Fluor 568 Phalloidin was added to the cells according to the provider's instructions (#A12380, Thermo- Fisher, MA, USA) for 10 min along with Hoechst. Confocal images were imaged on a Nikon Eclipse C2 si confocal spectral microscope using NIS-Elements AR software 4.00.00 (build 764) LO, 64 bits. The objective used was Plan Apo VC 60× Oil DIC N2 NA 1.4. Figs. 1c, e, i, 2c, 3d, Fig. S1a, c, e were imaged on a Nikon Eclipse E600 epifluorescence microscope.
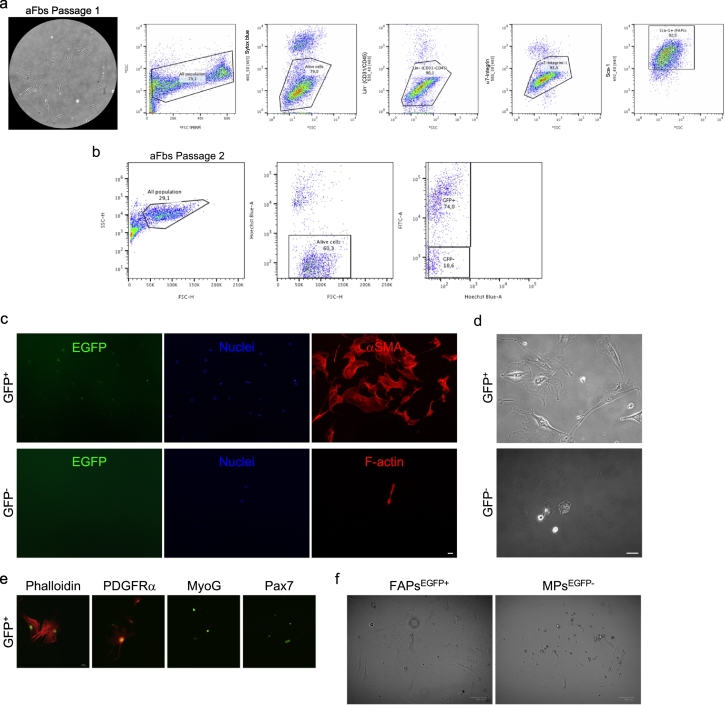
Fig. 2.
Prospective FACS analyses of adherent fibroblasts.
a. Sequential gating strategy used to distinguish FAPs from aFbs via pre-plating at passage 2. Common gating using FSC (linear scale) and SSC (logarithmic scale) to discard debris and identify the total cell population (all population). SSC vs Sytox blue was used to distinguish live cells. CD31/CD45/α7-integrin/Sca-1 vs SCC gating allows to discriminate between endothelial (CD31+), hematopoietic (CD45+), (α7-integrin+) muscle progenitors, and the Sca-1+ population (FAPs). Hence, aFbs are likely Sca-1+ FAPs (~85% of purity). b. PDGFRαEGFP reporter mice were used for FACS analyses of culture aFbs (EGFP+ cells) at passage 2. ~75% of the aFbs are EGFP+ FAPs c. Sorted EGFP+ cells from the pre-plating strategy express αSMA (red). eGFP− cells (lower panels) forming a single myocyte with 2-nuclei. d. Bright field images showing the cellular morphology of EGFP+ FAPs vs GFP− cells 5 days after FACS. e. Confocal images showing PDGFRα, MyoG, and Pax7 labeling in EGFP-sorted cells. f. Bright field images showing the cellular morphology of EGFP+ FAPs vs myogenic progenitor cells 3 days after FACS.
Fig. 3.
In vitro fibroadipogenic potential of adherent fibroblasts.
a. Quantification of FAPs (adherent EGFP+ cells) number per field. TGF-β1 (5 ng/mL) treatment for 24 h increases FAPs number. b. Evaluation of proliferation as the percentage (%) of Ki67-positive adherent EGFP+ cells per field in control or TGF-β1-treated adherent EGFP+ cells. The values correspond to the mean ± SEM (n = 3). *P < 0.05; Control vs TGF-β1; with two-tailed Student's t-test. c. qRT-PCR of Ctgf and αSMA (Acta2) mRNA levels in TGF-β1-treated aFbs. d. Immunofluorescent analyses of Fibronectin (red) in adherent EGFP+ cells under TGF-β1 treatment for 24 h. Fibronectin was used to identify the ECM of FAPs. Scale bar: 50 μm. e. Western Blot analysis showing the relative protein levels of β1-Integrin and fibronectin in control or TGF-β1-treated aFbs. Tubulin was used as a loading control. f. Representative Western Blot showing PDGFRα protein levels after serial passaging of aFbs (P0: Passage 0, P1: Passage 1, and P2: Passage 2). Tubulin was used as a loading control. g. Bright field images of aFbs following the pre-plating strategy 5 days after culture in growth media (GM) (left panel) or after 5 days in GM followed by 4 days in adipogenic media (Adip. M) (right panel). h. Immunofluorescence analyses of Perilipin (red) in EGFP+ cells after 7-day culture in GM. Hoechst (blue) was used to identify nuclei. Scale bar: 50 μm.
2.9. Immunofluorescence analyses of Ki67+ cells
The percentages of Ki67-positive cells were determined using ImageJ software (version 1.46r, NIH, USA) by analyzing particle function after image type transformation to 8-bit or by counting the positive cells individually per field using the cell counter plugin. Hoechst staining was used to determine total cell count. For each experimental condition, counts of 8 to 10 randomly chosen fields were averaged for three independent experiments.
2.10. RNA isolation, reverse transcription, and quantitative real-time polymerase chain reaction (RT-qPCR)
Total RNA from cultured cells was isolated using TRIzol (Invitrogen, CA, USA) according to the manufacturer's instructions. RNA integrity was corroborated as described before [31]. Two microgram RNA was reverse transcribed into cDNA using random primers and M-MLV reverse transcriptase (Invitrogen, CA, USA). RT-qPCR was performed in triplicate with the Eco Real-Time PCR System (Illumina, CA, USA), using primer sets for Ccn2/Ctgf, αSMA (Acta2), and the housekeeping gene 18S (used as a reference gene). The ΔΔCt method was used for quantification, and mRNA levels were expressed relative to the mean level of the control condition in each case. We analyzed and validated each RT-PCR expected gene product using a 2% agarose gel.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
|
Ccn2/Ctgf αSMA (Acta2) 18S |
CAGGCTGGAGAAGCAGAGTCGT TCCCTGGAGAGGAGCTACGA TGACGGAAGGGCACCACCAG |
CTGGTGCAGCCAGAAAGCTCAA CTTCTGCATCCTGTCAGCAA CACCACCACCCACGGAATCG |
2.11. Statistical analysis
Mean and SEM values, as well as the number of experiments performed, are indicated in each figure. The two-tailed Student t-test was performed when two conditions were compared. Differences were considered significant with a P value < 0.05. Data were collected in Microsoft Excel (Redmond, WA, USA), and statistical analyses were performed using Prism 5 software (GraphPad, CA, USA).
3. Results
3.1. Adult muscle adherent fibroblasts are likely fibroadipogenic progenitors in vitro
We isolated, through a pre-plating strategy and culture, aFbs from wild type skeletal muscle based on previous descriptions [24,32]. These cells adhere to the plastic plate after 1 h 30 min of culture and they do look like MPs with fibroblast morphology after 48 and 72 h of adhesion (Fig. 1a). Next, we took advantage of the PDGFRαEGFP mice as EGFP+ FAPs are found in limb skeletal muscles (Fig. S1a, b) [15]. Importantly, EGFP+ FAPs can be easily distinguished in our total tissue preparation after enzymatic digestion and before pre-plating because of EGFP expression (Fig. S1c). Hence, we cultured aFbs using the same pre-plating strategy described above and quantified the percentage of EGFP+ cells (hereafter referred to as FAPs) after passage 1 and 2 in the monolayer culture. Approximately, 80 to 85% of the aFbs were EGFP+ at passage 1 and 2, respectively (Fig. 1b, c). As previously suggested, a majority of these cells express the CT fibroblast marker Tcf7l2 (Tcf4) transcription factor [4,5,12], and αSMA [7] at passage 0 and 1 (Fig. 1d, e). Tissue-resident FAPs co-expressing EGFP and Tcf7l2 are present in the tibialis anterior muscle (Fig. 1f). Then, we simultaneously cultured aFbs and muscle progenitors via pre-plating and analyzed the protein levels of different markers for each cell type (Fig. 1g). Fig. 1h shows that aFbs expressed high levels of the mesenchymal progenitor markers PDGFRα and αSMA, while muscle progenitors express the paired box 7 protein (Pax7) at passage 1 [33]. Thus, our results suggest that the majority of the aFbs cultured via pre-plating are likely adult FAPs. In addition, when we analyzed the adherent cells from the PDGFRαEGFP reporter mice without applying the pre-plating strategy, a large proportion of small and rounded myoblast progenitors (EGFP−) can be observed along with adherent FAPs (EGFP+), which means that a mixed culture of different cell populations results (Fig. S1e). Finally, in order to corroborate that aFbs behave as FAPs in vitro, we treated them with the tyrosine kinase inhibitor Nilotinib. Nilotinib is known to induce FAPs apoptosis but does not have the same effect on myoblasts [15,31]. Thus, we asked whether Nilotinib could also be impairing aFbs survival in culture as it was described for FAPs [15,38]. Nilotinib treatment for 72 h significantly reduces the number of aFbs (Fig. 1i), suggesting that aFbs are likely to respond to Nilotinib as FAPs do. Nilotinib reduces aFbs number by 90% (Fig. 1j), probably by inducing aFbs growth arrest followed by its death (Fig. S1f) [15]. Overall, these data suggest that adult aFbs via pre-plating culture are FAPs-like cells in vitro.
3.2. Prospective FACS analyses of adherent fibroblasts
FAPs can be distinguished in flow cytometry experiments based on the expression of CD34, stem cell antigen-1 (Sca1), PDGFRα, and on the lack of expression of the blood lineage (CD31−/CD45− (Lin−)) and the myoblast marker integrin-α7 (Intα7−) [2,7,8]. Thus, we performed flow cytometry analyses of aFbs at passage 2 (Fig. 2a). Fig. 2a shows that 85% of aFbs at passage 2 are FAPs-like cells being Lin−, and Intα7−, but expressing Sca-1. Next, we asked whether the PDGFRαEGFP mice could be useful for flow cytometry analyses of isolated and cultured EGFP+ aFbs. Fig. 2b shows flow cytometry identification of PDGFRα+ cells, based on EGFP expression, from aFbs population at passage 2. The analysis shows that ~75% of aFbs in culture expressed the fusion protein H2B-EGFP (Fig. 2b). Then, we decided to sort and culture the previously identified populations (EGFP+ vs EGFP−) and evaluate how they look in culture after sorting (Fig. 2c). We observed that almost all the EGFP+-sorted aFbs express αSMA and have fibroblast/myofibroblast features, such as prolonged cytoplasmic protrusions and large nuclei (some were binucleated) after 4 days in culture (Fig. 2c, d). However, none of the EGFP−-sorted cells have fibroblast/myofibroblast characteristics and they look like muscle progenitors/myocytes (Fig. 2c, d). EGFP+ sorted cells expressed PDGFRα, but not MyoG or Pax7 labeling was detected (Fig. 2e). The simultaneous FACS isolation of EGFP+ FAPs and EGFP− myoblast progenitors (Integrin-α7+), suggests that most of the EGFP− cells were myogenic-like cells (Fig. 2f). Altogether, our results suggest that aFbs can be easily isolated and cultured via pre-plating, representing an in vitro heterogenous population of previously characterized muscle FAPs.
3.3. In vitro fibroadipogenic potential of adherent fibroblasts
It has been demonstrated that FAPs respond to TGF-β1, proliferating and differentiating into a myofibroblast phenotype in vitro and in vivo [8,15]. Hence, we decided to evaluate whether aFbs behave similarly to FAPs in response to TGF-β1. Thus, we isolated aFbs via pre-plating technique from the PDGFRαEGFP mice and treated them with TGF-β1 for 24 h. Next, we evaluated the number of EGFP+ cells, their proliferation rate (Ki67+), and the expression of the matricellular protein CCN2/CTGF, αSMA (myofibroblast marker), and the ECM-related proteins β1-Integrin and fibronectin. Of note, all of these are TGF-β-target genes. TGF-β1 induces adherent fibroblasts (EGFP+) expansion and proliferation, as evaluated by their total numbers and Ki67 labeling (Fig. 3a, b). TGF-β1 stimulation also increases aFbs CCN2/CTGF expression and influences their differentiation towards a myofibroblast phenotype as evaluated by αSMA, β1-Integrin, and fibronectin expression after 24 h (Fig. 3c, d, e). Increased fibronectin immunofluorescence signal also demonstrates that TGF-β1 enhances ECM deposition by aFbs (Fig. 3d). Hence, TGF-β1 promotes aFbs proliferation, myofibroblast differentiation, and ECM deposition. Next, to discriminate the effects of serial passaging in aFbs/FAPs fate, we determined the protein levels of PDGFRα as an indicator of aFbs/FAPs loss of multipotency. PDGFRα protein levels diminished after serial passaging (Fig. 3f). Thus, we decided to use aFbs no further than passage 2, because they began to become activated, senescent, and more differentiated towards a myofibroblast phenotype after sequential passaging (Fig. S1d). Finally, we asked whether aFbs are able to differentiate into adipocytes in culture. After reaching confluence and adding adipogenic media for 4 days to the aFbs population, we observed some lipid-rich adipocytes emerging in the monolayer culture at passage 0 (Fig. 3f). In addition, both myofibroblasts (αSMA+) and adipocytes (perilipin+) were observed 10 days after culturing aFbs (Fig. 3g). Thus, aFbs retain their fibroadipogenic potential at passage 0 but loss their multipotency at later passages.
3.4. Activated adherent fibroblasts are isolated from the mdx; PDGFRαEGFP and PDGFRαEGFP denervated mice
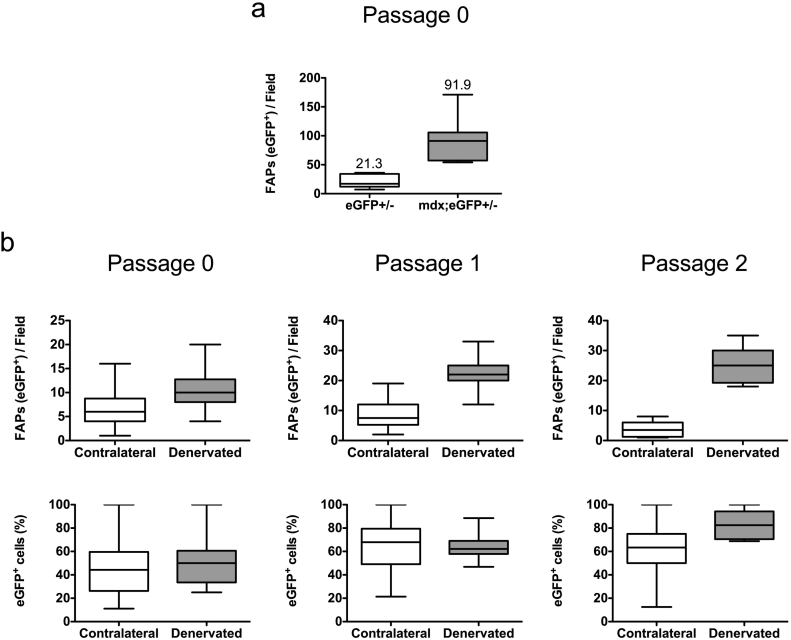
It is known that wild type and mdx muscle FAPs behave differently and are in distinct functional states [[34], [35], [36]]. Therefore, using the same strategy described above we isolated and cultured mdx;PDGFRαEGFP aFbs and compared their relative numbers to their PDGFRαEGFP littermates aFbs. Fig. 4a shows that after 4 days in culture (passage 0) the yield of aFbs isolated from the mdx;PDGFRαEGFP is 4- to 5-times more than the yield from PDGFRαEGFP aFbs. Hence, the pre-plating strategy presented here allows us to corroborate that mdx aFbs (EGFP+) behave differently in culture since there are more cells by day 4 post-culture when compared to their non-mdx littermates. We previously described that FAPs are activated as early as two days after denervation [4]. Thus, the isolation of aFbs from day 2 denervated limb from PDGFRαEGFP mice could be informative for evaluating whether these cells also behave differently in vitro. Initially, we could not see a significant difference in aFbs (EGFP+) at passage 0 (Fig. 4b). However, differences in aFbs numbers and proportion (%) are observed after passage 1 where more EGFP+ cells are found in the 2-day denervation culture, being the difference more marked at passage 2 (Fig. 4b). Therefore, denervated aFbs (FAPs) have a different in vitro behavior, when compared with aFbs isolated from to the contralateral limb; being this difference more pronounced after passaging.
Fig. 4.
In vitro behavior of adherent fibroblasts isolated from mdx;PDGFRαEGFP and sciatic-denervated PDGFRαEGFP mice.
a. Quantification of two representative experiments, evaluating the number of EGFP+ cells (FAPs) from EGFP and mdx;EGFP mice after the pre-plating strategy following 4 days in culture in growth media. b. Quantification of two representative experiments, evaluating the number of EGFP+ cells from PDGFRαEGFP denervated mice after the pre-plating strategy following sequential passaging in growth media. Cells were counted 24 h after passaging and their number expressed per field or as a percentage (%). Contralateral limb muscles were used as controls. The values correspond to the mean ± SEM.
4. Discussion
After the exclusion of endothelial and hematopoietic cells, FAPs can be identified and purified from skeletal muscle based on the expression of cell surface markers, such as: Sca-1 (not present in humans), PDGFRα, and CD34. However, the convergent population of cells such as Osr1+ CT fibroblasts (also referred as FAPs) or Tcf7l2-(Tcf4)+ CT fibroblasts appears to make up a population of stromal MPs with multiple fate potentials [12]. Furthermore, the overlapping but not an identical expression of PDGFRα, Tcf7l2 (Tcf4), and Osr1 in embryonic FAPs make difficult a unique definition of these cells [12]. In addition, adult FAPs express Osr1 at low levels and frequency, but its expression is activated upon acute muscle injury [37]. Here, we support the idea that most adult muscle CT fibroblasts express Tcf7l2 and PDGFRα, and therefore, muscle CT fibroblasts and FAPs are likely the same populations. However, the possibility that FAPs, stromal mesenchymal progenitors, and CT fibroblast are a heterogeneous mixture of multipotent progenitors with molecular heterogeneity and also with overlapping but distinct roles during regeneration and pathology undoubtedly exist [35]. However, this hypothesis still needs to be further validated and studied in vivo as well as in vitro. Moreover, further lineage-tracing studies are also needed to clarify whether adult FAPs, stromal mesenchymal progenitors, and CT fibroblasts are the same population of cells expressing an overlapping array of markers.
Here, we showed that adult wild type adherent fibroblasts, isolated via pre-plating, are likely a heterogeneous population of mesenchymal adherent FAPs. The use of the PDGFRαEGFP reporter mice suggests that CT aFbs are likely an overlapping population of previously uncharacterized in vitro EGFP+ FAPs. Thus, a high proportion of adherent cells retain EGFP expression after serial passaging as observed in passage 1 and 2, which suggests that EGFP is not lost after culture. Moreover, we suggested that serial passaging reduces PDGFRα expression and negatively affects aFbs/FAPs multipotency. In addition, Nilotinib treatment induces aFbs death. It has been reported that Nilotinib also promotes FAPs apoptosis [15,38]. Furthermore, adult aFbs respond to TGF-β1 as wild type FAPs do [8,15]. We demonstrated that TGF-β1 enhances aFbs proliferation, myofibroblast differentiation, and ECM expression. Altogether, these results suggest that aFbs respond to TGF-β signaling in the same fashion as FAPs in vitro. Thus, our results suggest that the population of adherent fibroblasts isolated is likely to be FAPs.
On the other hand, breeding the PDGFRαEGFP colony with the mdx mice results in the easy identification of EGFP+ cells in the mdx background. Moreover, the fact that after passaging denervated EGFP+ aFbs behave differently from their contralateral counterpart suggests that 2-day denervation induces early activation of resident EGFP+ cells that can be seen in vitro. Whether early denervation promotes more in vitro survival of aFbs or better attachment to the plastic plate was not fully addressed here and needs to be further investigated. Although, our results suggest that denervation improves aFbs survival. The same argument applies to the isolated mdx aFbs. In a recent work, signaling pathway analyses based on RNA-seq from denervated-FAPs at day 15 revealed different signatures compared to cardiotoxin induced FAPs [39]. Interestingly, in C2C12 myotube co-culture experiments, denervated-FAPs but not cardiotoxin-FAPs cause C2C12 muscle atrophy. Thus, we propose that further studies are needed in order to understand the early (2 days or before) activation of FAPs following muscle denervation.
Having a large number of cells is crucial for western blot, flow cytometry, and biochemical analyses. The advantage of serial passaging of adherent cells is that it provides enough cells to perform several experiments. However, further studies need to be done in order to corroborate whether FAPs retains their multipotent capabilities after passaging. It is possible that sequential passaging alters the multipotent properties of stromal mesenchymal fibroblasts as it has been reported for human skin fibroblasts, and also seen in our hands [40]. Another advantage of pre-plating aFbs is the fact that less time is involved in the procedure when compared to FAPs flow cytometry and FACS. Having less time between the enzymatic digestion and culture may be critical to keep the normal biological functions of the desired cells. In addition, adhesion by pre-plating is a relatively low-cost procedure when compared to common FAPs isolation, which requires the use of a different set of antibodies, dyes, cytometers, sorters, and people involved in the equipment management.
Whereas muscle myogenic progenitors are essential mediators for damaged muscle replacement, regeneration also relies on the role of non-myogenic progenitors that reside in the muscle stroma. The aberrant regulation of the stromal compartment, CT fibroblasts and/or FAPs, underlies the degenerative progression of myopathies and aging [41]. Thus, under chronic damage, these fibrotic cells become activated producing exacerbated amounts of ECM, which in turn impairs muscle function [20]. However, their normal behavior during muscle regeneration is of importance as these progenitors are also key mediators of this process. Due to the lack of in vitro characterization, genetic labeling, single markers, and agreement in the definition of nomenclature fibroblasts still remain as a population of elusive mesenchymal progenitors with potential for further in vivo and in vitro studies [42,43]. We hypothesized that FAPs can actively regulate the expression of their markers, therefore, making their identification dynamic and not fully precise. Hence, understanding how the cellular milieu maintains muscle homeostasis and function, as well as how it participates in disease progression is of great importance to developing new ideas, methodologies, and knowledge to comprehend skeletal muscle biology.
The following is the supplementary data related to this article.
Adult muscle adherent fibroblasts are likely fibro/adipogenic progenitors.
a. Fluorescent images of tissue sections from tibialis anterior (left panel) and gastrocnemius (right panel) muscle from a 4-month-old PDGFRαEGFP mouse. b. Representative western blot, showing the relative expression of PDGFRα, Tcf7l2, Fibronectin, and β-Catenin. GAPDH was used as the loading control. n = 3. c. Fluorescent images of the enzymatic tissue preparation after cell dissociation. aFbs/FAPs cells can be easily distinguished in an epifluorescence microscope by EGFP expression. d. Bright field images of aFbs from passage 3 at 60–70% confluency. At passage 3 most of the resultant adherent cells are likely fibroblasts with an activated myofibroblast phenotype. Almost no myoblast-like cells are seen at passage 3. e. Representative images of cultured cells after 7 days without pre-plating them for 1 h and 30 min. Images show a mix of EGFP cells (aFbs/FAPs) along with small and rounded myoblast-like cells (EGFP-negative). Dashed lines show the myoblast-like cells. Nuclei (blue) were stained with Hoechst. F-actin staining (red) was used to visualize adherent cells. e. Representative image showing aberrant morphology of aFbs after Nilotinib treatment for 72 h at passage 2. Ghost/death cells can be identified along with cell vacuolizations in Nilotinib-treated aFbs.
Acknowledgments
Acknowledgments
We are grateful to the Unidad de Microscopía Avanzada (UMA) of Pontificia Universidad Católica de Chile for its support in image acquisition. The authors acknowledge the services provided by UC CINBIOT Animal Facility funded by PIA CONICYT* ECM-07 Program for Associative Research, of the Chilean National Council for Science and Technology. We thank the animal unit staff member Ms. Micaela Ricca of the Pontificia Universidad Católica de Chile. We acknowledge Camilo Riquelme-Guzmán for proofreading the article. For administrative assistance, we thank Ms. Vanessa Morales. We also acknowledge Eduardo Ramirez and Darling Vera for their technical support.
Funding
FONDECYT 1150106, 1190144 grants and CONICYT AF- B170005 grant to E.B., CIHR FDN-159908 grant to F.M.R., and Beca de Doctorado Nacional Folio 21140378 from CONICYT to O.C. sup- ported this work. The funding agencies had no role in the design of the study, data collection, and analysis, the decision to publish, or preparation of the manuscript.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors' individual contributions
OC conceived the concepts, designed the study, performed all the experiments, formal analysis, drafted the initial manuscript, and analyzed the data. EB helped with conceptualization and design of the study, reviewed and edited the manuscript and provided funding. FMR reviewed and edited the manuscript and provided funding. All the authors read and approved the final manuscript.
Ethics approval and consent to participate.
Not applicable.
Consent for publication.
Not applicable.
Competing interests
The authors declare that they have no competing financial interests.
Contributor Information
Osvaldo Contreras, Email: oicontre@uc.cl.
Fabio M. Rossi, Email: fabio@brc.ubc.ca.
Enrique Brandan, Email: ebrandan@bio.puc.cl.
References
- 1.Nassari S., Duprez D., Fournier-Thibault C. Non-myogenic contribution to muscle development and homeostasis: the role of connective tissues. Frontiers in Cell and Development Biology. 2017;5:22. doi: 10.3389/fcell.2017.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wosczyna M.N., Rando T.A. A muscle stem cell support group: coordinated cellular responses in muscle regeneration. Developmental Cell. 2018;46(2):135–143. doi: 10.1016/j.devcel.2018.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kardon G., Harfe B.D., Tabin C.J. A Tcf4-positive mesodermal population provides a prepattern for vertebrate limb muscle patterning. Developmental Cell. 2003;5(6):937–944. doi: 10.1016/s1534-5807(03)00360-5. [DOI] [PubMed] [Google Scholar]
- 4.Contreras O., Rebolledo D.L., Oyarzun J.E., Olguin H.C., Brandan E. Connective tissue cells expressing fibro/adipogenic progenitor markers increase under chronic damage: relevance in fibroblast-myofibroblast differentiation and skeletal muscle fibrosis. Cell and Tissue Research. 2016;364(3):647–660. doi: 10.1007/s00441-015-2343-0. [DOI] [PubMed] [Google Scholar]
- 5.Mathew S.J., Hansen J.M., Merrell A.J., Murphy M.M., Lawson J.A., Hutcheson D.A., Hansen M.S., Angus-Hill M., Kardon G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development. 2011;138(2):371–384. doi: 10.1242/dev.057463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mackey A.L., Magnan M., Chazaud B., Kjaer M. Human skeletal muscle fibroblasts stimulate in vitro myogenesis and in vivo muscle regeneration. The Journal of Physiology. 2017;595(15):5115–5127. doi: 10.1113/JP273997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joe A.W., Yi L., Natarajan A., Le Grand F., So L., Wang J., Rudnicki M.A., Rossi F.M. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature Cell Biology. 2010;12(2):153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uezumi A., Ito T., Morikawa D., Shimizu N., Yoneda T., Segawa M., Yamaguchi M., Ogawa R., Matev M.M., Miyagoe-Suzuki Y., Takeda S., Tsujikawa K., Tsuchida K., Yamamoto H., Fukada S. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. Journal of Cell Science. 2011;124:3654–3664. doi: 10.1242/jcs.086629. Pt 21. [DOI] [PubMed] [Google Scholar]
- 9.Uezumi A., Ikemoto-Uezumi M., Tsuchida K. Roles of nonmyogenic mesenchymal progenitors in pathogenesis and regeneration of skeletal muscle. Fontiers in Physiology. 2014;5:68. doi: 10.3389/fphys.2014.00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uezumi A., Fukada S., Yamamoto N., Takeda S., Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature Cell Biology. 2010;12(2):143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 11.Wosczyna M.N., Biswas A.A., Cogswell C.A., Goldhamer D.J. Multipotent progenitors resident in the skeletal muscle interstitium exhibit robust BMP-dependent osteogenic activity and mediate heterotopic ossification. Journal of Bone and Mineral Research. 2012;27(5):1004–1017. doi: 10.1002/jbmr.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vallecillo-Garcia P., Orgeur M., Hofe-Schneider S. Vom, Stumm J., Kappert V., Ibrahim D.M., Borno S.T., Hayashi S., Relaix F., Hildebrandt K., Sengle G., Koch M., Timmermann B., Marazzi G., Sassoon D.A., Duprez D., Stricker S. Odd skipped-related 1 identifies a population of embryonic fibro-adipogenic progenitors regulating myogenesis during limb development. Nature Communications. 2017;8(1):1218. doi: 10.1038/s41467-017-01120-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stricker S., Mathia S., Haupt J., Seemann P., Meier J., Mundlos S. Odd-skipped related genes regulate differentiation of embryonic limb mesenchyme and bone marrow mesenchymal stromal cells. Stem Cells and Development. 2012;21(4):623–633. doi: 10.1089/scd.2011.0154. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez D., Contreras O., Rebolledo D.L., Espinoza J.P., van Zundert B., Brandan E. ALS skeletal muscle shows enhanced TGF-beta signaling, fibrosis and induction of fibro/adipogenic progenitor markers. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lemos D.R., Babaeijandaghi F., Low M., Chang C.K., Lee S.T., Fiore D., Zhang R.H., Natarajan A., Nedospasov S.A., Rossi F.M. Nilotinib reduces muscle fibrosis in chronic muscle injury by promoting TNF-mediated apoptosis of fibro/adipogenic progenitors. Nature Medicine. 2015;21(7):786–794. doi: 10.1038/nm.3869. [DOI] [PubMed] [Google Scholar]
- 16.Ceco E., McNally E.M. Modifying muscular dystrophy through transforming growth factor-beta. The FEBS Journal. 2013;280(17):4198–4209. doi: 10.1111/febs.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mann C.J., Perdiguero E., Kharraz Y., Aguilar S., Pessina P., Serrano A.L., Munoz-Canoves P. Aberrant repair and fibrosis development in skeletal muscle. Skeletal Muscle. 2011;1(1):21. doi: 10.1186/2044-5040-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Droguett R., Cabello-Verrugio C., Riquelme C., Brandan E. Extracellular proteoglycans modify TGF-beta bio-availability attenuating its signaling during skeletal muscle differentiation. Matrix Biology. 2006;25(6):332–341. doi: 10.1016/j.matbio.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 19.Gyorfi A.H., Matei A.E., Distler J.H.W. Targeting TGF-beta signaling for the treatment of fibrosis. Matrix Biology. 2018;68-69:8–27. doi: 10.1016/j.matbio.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 20.Pessina P., Kharraz Y., Jardi M., Fukada S., Serrano A.L., Perdiguero E., Munoz-Canoves P. Fibrogenic cell plasticity blunts tissue regeneration and aggravates muscular dystrophy. Stem Cell Reports. 2015;4(6):1046–1060. doi: 10.1016/j.stemcr.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal B., Serrano A.L., Tjwa M., Suelves M., Ardite E., De Mori R., Baeza-Raja B., Martinez de Lagran M., Lafuste P., Ruiz-Bonilla V., Jardi M., Gherardi R., Christov C., Dierssen M., Carmeliet P., Degen J.L., Dewerchin M., Munoz-Canoves P. Fibrinogen drives dystrophic muscle fibrosis via a TGFbeta/alternative macrophage activation pathway. Genes & Development. 2008;22(13):1747–1752. doi: 10.1101/gad.465908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinz B., Phan S.H., Thannickal V.J., Galli A., Bochaton-Piallat M.L., Gabbiani G. The myofibroblast: one function, multiple origins. Am J Pathol. 2007;170(6):1807–1816. doi: 10.2353/ajpath.2007.070112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goetsch K.P., Snyman C., Myburgh K.H., Niesler C.U. Simultaneous isolation of enriched myoblasts and fibroblasts for migration analysis within a novel co-culture assay. Biotechniques. 2015;58(1):25–32. doi: 10.2144/000114246. [DOI] [PubMed] [Google Scholar]
- 24.Riquelme-Guzman C., Contreras O., Brandan E. Expression of CTGF/CCN2 in response to LPA is stimulated by fibrotic extracellular matrix via the integrin/FAK axis. American Journal of Physiology. Cell Physiology. 2018;314(4):C415–C427. doi: 10.1152/ajpcell.00013.2017. [DOI] [PubMed] [Google Scholar]
- 25.Scott R.W., Underhill T.M. Methods and strategies for lineage tracing of mesenchymal progenitor cells. Methods in Molecular Biology. 2016;1416:171–203. doi: 10.1007/978-1-4939-3584-0_10. [DOI] [PubMed] [Google Scholar]
- 26.Swonger J.M., Liu J.S., Ivey M.J., Tallquist M.D. Genetic tools for identifying and manipulating fibroblasts in the mouse. Differentiation. 2016;92(3):66–83. doi: 10.1016/j.diff.2016.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ciuffreda M.C., Malpasso G., Musaro P., Turco V., Gnecchi M. Protocols for in vitro differentiation of human mesenchymal stem cells into osteogenic, chondrogenic and adipogenic lineages. Methods in Molecular Biology. 2016;1416:149–158. doi: 10.1007/978-1-4939-3584-0_8. [DOI] [PubMed] [Google Scholar]
- 28.Chal J., Pourquie O. Making muscle: skeletal myogenesis in vivo and in vitro. Development. 2017;144(12):2104–2122. doi: 10.1242/dev.151035. [DOI] [PubMed] [Google Scholar]
- 29.Hamilton T.G., Klinghoffer R.A., Corrin P.D., Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Molecular and Cellular Biology. 2003;23(11):4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rebolledo D.L., González D., Faundez-Contreras J., Contreras O., Vio C.P., Murphy-Ullrich J.E., Lipson K.E., Brandan E. Denervation-induced skeletal muscle fibrosis is mediated by CTGF/CCN2 independently of TGF-β. Matrix Biology. 2019 doi: 10.1016/J.MATBIO.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 31.Contreras O., Villarreal M., Brandan E. Nilotinib impairs skeletal myogenesis by increasing myoblast proliferation. Skeletal Muscle. 2018;8(1):5. doi: 10.1186/s13395-018-0150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy M.M., Lawson J.A., Mathew S.J., Hutcheson D.A., Kardon G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development. 2011;138(17):3625–3637. doi: 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olguin H.C., Olwin B.B. Pax-7 up-regulation inhibits myogenesis and cell cycle progression in satellite cells: a potential mechanism for self-renewal. Developmental Biology. 2004;275(2):375–388. doi: 10.1016/j.ydbio.2004.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mozzetta C., Consalvi S., Saccone V., Tierney M., Diamantini A., Mitchell K.J., Marazzi G., Borsellino G., Battistini L., Sassoon D., Sacco A., Puri P.L. Fibroadipogenic progenitors mediate the ability of HDAC inhibitors to promote regeneration in dystrophic muscles of young, but not old Mdx mice. EMBO Molecular Medicine. 2013;5(4):626–639. doi: 10.1002/emmm.201202096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malecova B., Gatto S., Etxaniz U., Passafaro M., Cortez A., Nicoletti C., Giordani L., Torcinaro A., De Bardi M., Bicciato S., De Santa F., Madaro L., Puri P.L. Dynamics of cellular states of fibro-adipogenic progenitors during myogenesis and muscular dystrophy. Nature Communications. 2018;9(1):3670. doi: 10.1038/s41467-018-06068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marinkovic M., Sacco F., Spada F., Petrilli L.L., Fuoco C., Micarelli E., Pavlidou T., Castagnoli L., Mann M., Gargioli C., Cesareni G. 2017. Skeletal Muscle Fibro-adipogenic Progenitors of Dystrophic Mice are Insensitive to NOTCH-dependent Regulation of Adipogenesis bioRxiv. [Google Scholar]
- 37.Stumm J., Vallecillo-Garcia P., Vom Hofe-Schneider S., Ollitrault D., Schrewe H., Economides A.N., Marazzi G., Sassoon D.A., Stricker S. Odd skipped-related 1 (Osr1) identifies muscle-interstitial fibro-adipogenic progenitors (FAPs) activated by acute injury. Stem Cell Research. 2018;32:8–16. doi: 10.1016/j.scr.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 38.Fiore D., Judson R.N., Low M., Lee S., Zhang E., Hopkins C., Xu P., Lenzi A., Rossi F.M., Lemos D.R. Pharmacological blockage of fibro/adipogenic progenitor expansion and suppression of regenerative fibrogenesis is associated with impaired skeletal muscle regeneration. Stem Cell Research. 2016;17(1):161–169. doi: 10.1016/j.scr.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 39.Madaro L., Passafaro M., Sala D., Etxaniz U., Lugarini F., Proietti D., Alfonsi M.V., Nicoletti C., Gatto S., De Bardi M., Rojas-Garcia R., Giordani L., Marinelli S., Pagliarini V., Sette C., Sacco A., Puri P.L. Denervation-activated STAT3-IL-6 signalling in fibro-adipogenic progenitors promotes myofibres atrophy and fibrosis. Nature Cell Biology. 2018;20(8):917–927. doi: 10.1038/s41556-018-0151-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klingberg F., Chow M.L., Koehler A., Boo S., Buscemi L., Quinn T.M., Costell M., Alman B.A., Genot E., Hinz B. Prestress in the extracellular matrix sensitizes latent TGF-beta1 for activation. The Journal of Cell Biology. 2014;207(2):283–297. doi: 10.1083/jcb.201402006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith L.R., Barton E.R. Regulation of fibrosis in muscular dystrophy. Matrix Biology. 2018;68–69:602–615. doi: 10.1016/j.matbio.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chapman M.A., Meza R., Lieber R.L. Skeletal muscle fibroblasts in health and disease. Differentiation. 2016;92(3):108–115. doi: 10.1016/j.diff.2016.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karamanos N.K., Theocharis A.D., Neill T., Iozzo R.V. Matrix modeling and remodeling: a biological interplay regulating tissue homeostasis and diseases. Matrix Biology. 2019;75–76:1–11. doi: 10.1016/j.matbio.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Adult muscle adherent fibroblasts are likely fibro/adipogenic progenitors.
a. Fluorescent images of tissue sections from tibialis anterior (left panel) and gastrocnemius (right panel) muscle from a 4-month-old PDGFRαEGFP mouse. b. Representative western blot, showing the relative expression of PDGFRα, Tcf7l2, Fibronectin, and β-Catenin. GAPDH was used as the loading control. n = 3. c. Fluorescent images of the enzymatic tissue preparation after cell dissociation. aFbs/FAPs cells can be easily distinguished in an epifluorescence microscope by EGFP expression. d. Bright field images of aFbs from passage 3 at 60–70% confluency. At passage 3 most of the resultant adherent cells are likely fibroblasts with an activated myofibroblast phenotype. Almost no myoblast-like cells are seen at passage 3. e. Representative images of cultured cells after 7 days without pre-plating them for 1 h and 30 min. Images show a mix of EGFP cells (aFbs/FAPs) along with small and rounded myoblast-like cells (EGFP-negative). Dashed lines show the myoblast-like cells. Nuclei (blue) were stained with Hoechst. F-actin staining (red) was used to visualize adherent cells. e. Representative image showing aberrant morphology of aFbs after Nilotinib treatment for 72 h at passage 2. Ghost/death cells can be identified along with cell vacuolizations in Nilotinib-treated aFbs.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.