Abstract
Cell-based approaches using tissue engineering and regenerative medicine to replace damaged renal tissue with 3D constructs is a promising emerging therapy for kidney disease. Besides living cells, a template provided by a scaffold based on biomaterials and bioactive factors is needed for successful kidney engineering. Nature's own template for a scaffolding system is the extracellular matrix (ECM). Research has focused on mapping the mature renal ECM; however, the developing fetal ECM matches more the active environment required in 3D renal constructs. Here, we characterized the differences between the human fetal and mature renal ECM using spectrometry-based proteomics of decellularized tissue. We identified 99 different renal ECM proteins of which the majority forms an overlapping core, but also includes proteins enriched in either the fetal or mature ECM. Relative protein quantification showed a significant dominance of EMILIN1 in the fetal ECM. We functionally tested the role of EMILIN1 in the ECM using a novel methodology that permits the reliable anchorage of native cell-secreted ECM to glass coverslips. Depletion of EMILIN1 from the ECM layer using siRNA mediated knock-down technologies does not affect renal epithelial cell growth, but does promote migration. Lack of EMILIN1 in the ECM layer reduces the adhesion strength of renal epithelial cells, shown by a decrease in focal adhesion points and associated stress fibers. We showed in this study the importance of a human renal fetal and mature ECM catalogue for identifying promising ECM components that have high implementation potential in scaffolds for 3D renal constructs.
Keywords: Extracellular matrix, Proteomics, Migration, Focal adhesion, EMILIN1
Highlights
-
•
Proteomics revealed the differences between the renal fetal and mature extracellular matrix.
-
•
EMILIN1 has a significant dominance in the fetal extracellular matrix.
-
•
EMILIN1 depletion from the extracellular matrix reduces the adhesion strength and promotes migration of renal epithelial cells.
Introduction
In search of a potential treatment for kidney disease, current investigations focus on cell-based approaches using tissue engineering and regenerative medicine (TERM) to replace damaged renal tissue with engineered 3D structures. One promising TERM-based approach is the use of organoids. Renal organoids derived from induced pluripotent stem cells that model the morphology and segmentation of human fetal nephrons have been developed [1,2]. Human primary renal cells can also grow into 3D kidney-like constructs with tubular structures. When implanted into mice, these constructs survived and maintained their renal phenotypes for up to 6 weeks [3]. However, these techniques cannot fully recapitulate the complex organization of the kidney and cannot be grown on a large scale yet.
Besides cells, another basic component for kidney engineering is a template provided by a scaffolding system based on biomaterials and bioactive factors that create a microenvironment which facilitates cell-specific behavior. Nature's own template for a scaffolding system is the extracellular matrix (ECM), which is a collection of molecules deposited by surrounding cells, making it a tissue-specific 3D structure in which cells are embedded. The ECM not only gives structural support, it also contains biochemical cues that influence many biological processes, including cell migration, adhesion, proliferation and differentiation [[4], [5], [6], [7]]. The matrices used to embed stem or primary renal cells are mostly collagen-based hydrogels or matrigel, which do not reflect the tissue-specific ECM of the kidney. Matrices containing renal-specific ECM cues could greatly advance TERM by promoting renal tissue formation. In order to create such a scaffolding system, the components of the human renal ECM first need to be fully characterized.
Proteomic techniques have been used to characterize the ECM composition in various tissues [8,9]. For the renal ECM, studies have focused on the human glomerular ECM and identified a highly connected network comprised of almost 200 different structural and regulatory ECM proteins [[10], [11], [12], [13]]. Despite previous work, ECM proteome research until now only focused on mapping the mature renal ECM. In the fetal microenvironment, the ECM balance is shifted towards an active environment [[14], [15], [16], [17]], which matches the need for tissue generation in 3D tissue constructs.
In this study, we characterized the differences between the developing fetal and the more static mature human renal ECM proteome. The resulting catalogue gives important insights into renal ECM structure and function and enables a detailed investigation of promising renal ECM molecules and their potential use for implementation in scaffolds.
Based on our proteome analysis, we identified Elastin Microfibril Interfacer 1 (EMILIN1) as an important component enriched in the fetal ECM. EMILIN1 is located in elastic fibers at the interface between elastin and the surrounding microfibrils [18]. The distinctive C-terminus gC1q structure of EMILIN1 interacts with integrins, thereby connecting cells to elastic fibers [19,20]. Many cell types depend on this gC1q-integrin interaction for cell adhesion, migration and proliferation [[21], [22], [23], [24]]. EMILIN1 is particularly abundant in the walls of blood and lymphatic vessels, where it is necessary for the formation of elastic fibers and anchoring filaments [25,26].
Despite studies implying that EMILIN1 is an important factor in maintaining cardiovascular health [[26], [27], [28], [29]], the role of EMILIN1 in the kidney is unknown. Here, we have shown for the first time that EMILIN1 is an important regulator of renal cell migration and adhesion. These findings validate the importance of our human renal fetal and mature ECM catalogue for identifying promising ECM components with high potential for implementation in scaffolds for 3D kidney constructs.
Results
Enrichment of extracellular matrix proteins
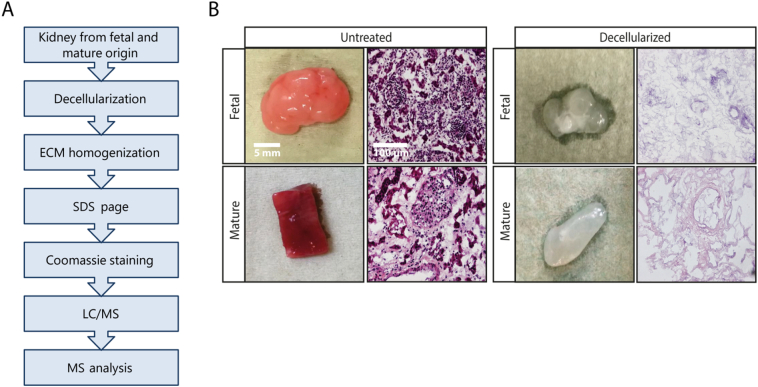
We analyzed healthy kidney samples from adult and fetal human donors. The ECM was enriched in these samples by decellularization (Fig. 1A). Cellular components were removed without disrupting the structure and morphology of the ECM (Fig. 1B). The obtained ECM extracts were separated by SDS-PAGE and liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to characterize the differences between the fetal and mature renal ECM proteome (Fig. 1a, Supplemental Fig. 1A).
Fig. 1.
Isolation of renal fetal and mature extracellular matrix proteins. (A) Workflow for the isolation of the extracellular matrix (ECM) from healthy human fetal and mature renal tissue. (B) Macroscopic appearance of representative human fetal and mature renal samples before and after decellularization (left panels). Scale bar represents 5 mm. Hematoxylin and eosin staining (200× magnification) of human fetal and mature renal tissue before and after decellularization demonstrates the successful removal of cellular components (right panels). Scale bar represents 100 μm.
Comparison of the renal proteome: fetal versus mature
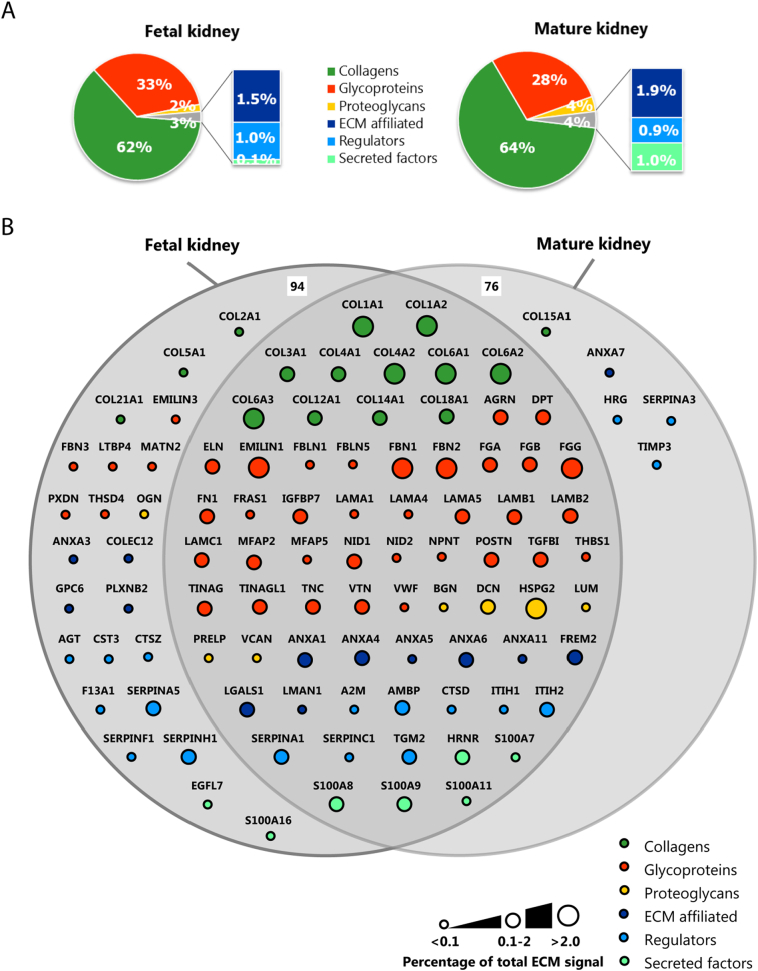
The obtained proteomic data were used to generate a catalogue of fetal and mature renal ECM proteins. Proteins that were identified in at least two replicates were included (Supplementary Fig. 2A). By crossreferencing with the Human Matrisome Project [8,9], we categorized our data proteins into core ECM proteins, including collagens, glycoproteins and proteoglycans, and ECM-associated proteins, including ECM-affiliated proteins, regulators and secreted factors. Relative protein quantification showed a dominance of collagens and glycoproteins in both the fetal and mature ECM: 62% of the fetal and 64% of the mature signal consisted of collagens, whereas 33% of the fetal and 28% of the mature selection consisted of glycoproteins (Fig. 2A).
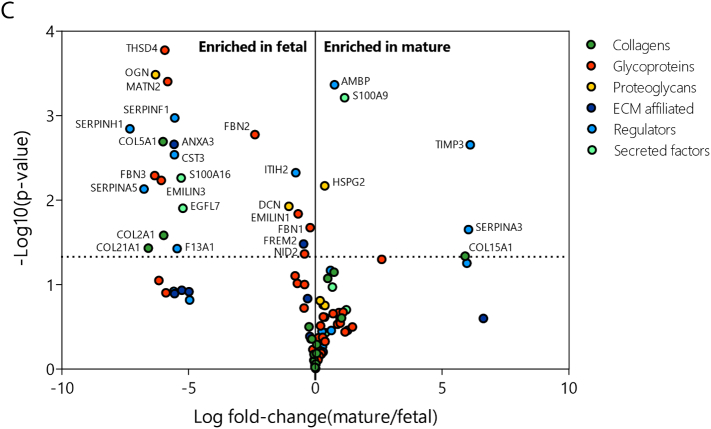
Fig. 2.
Defining the renal fetal and mature extracellular matrix proteome. Extracellular matrix (ECM) proteins identified by mass-spectrometry were classified as collagens, glycoproteins, proteoglycans, ECM affiliated, regulators or secreted factors and they were colored and arranged accordingly. (A) Pie charts summarizing the relative protein quantification for both the fetal (left) and mature (right) renal ECM proteome. (B) Euler-diagram visualizing the overlap and differences between the human fetal and mature renal ECM proteome. Each node represents a single protein and is labeled with the gene name. Node size is proportional to the abundance of the protein within the renal ECM proteome (<0.1%, between 0.1 and 2% or >2% of the total ECM signal in at least one dataset (fetal or mature)). (C) A volcano plot representation of the human fetal and mature ECM proteome showing the protein distribution between fetal and mature (log fold-change(mature/fetal), x-axis) and significance (P-value, y-axis) of all detected ECM proteins. Each circle represents a single protein. The horizontal dashed line indicates the threshold of statistical significance (P < 0.05).
We identified 94 fetal and 76 mature renal ECM proteins, from which the majority could be classified as core ECM proteins (60 and 51, respectively). The most abundant signal came from core ECM proteins as well, with many collagens comprising >2% of the total ECM signal (Fig. 2B, Supplemental Table 1). Four glycoproteins (EMILIN1, FGG, FBN1 and FBN2) and one proteoglycan (HSPG2) showed abundant signal, each comprising >2% of the total ECM signal (Fig. 2B, Supplemental Table 1). We identified 23 proteins that were significantly enriched in the fetal renal ECM compared to the mature ECM. Only 6 ECM proteins were significantly enriched in the mature renal ECM. Among the 23 proteins significantly enriched in the fetal selection were the abundant glycoproteins EMILIN1, fibrillin1 (FBN1) and fibrillin2 (FBN2) (Fig. 2C, Supplemental Table 1).
Lennon and colleagues published the composition of the human glomerular ECM [10]. By crossreferencing their dataset with the Human Matrisome Project [8,9] and subsequently with our dataset, we further classified our list in either glomerular or tubulointerstitial ECM proteins (Supplemental Fig. 3). By this means, we identified 55 glomerular ECM proteins, including 8 proteins detected exclusively in either the mature or fetal renal ECM. 29 glomerular ECM proteins were not detected by our MS analysis and were only found by Lennon et al. due to their extra glomeruli enrichment step prior to MS analysis [10]. Our remaining 54 ECM proteins could then be classified as mainly tubulointerstitial, including 19 proteins unique for the fetal renal ECM. One tubulointerstitial ECM protein was found to be unique for the mature renal ECM. These findings show that the fetal and mature renal ECM are complex and share a central overlap, but are also characterized by unique ECM proteins.
Validation of selected proteins within the renal extracellular matrix
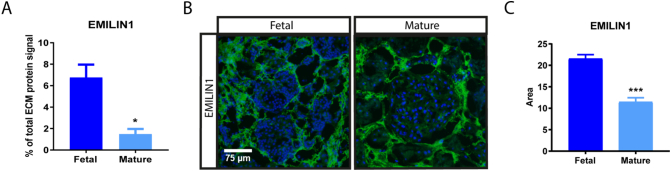
EMILIN1 was found to be one of the glycoproteins with the most abundant signal in the renal ECM and significantly enriched in the fetal selection, making it an interesting ECM protein for further investigation. EMILIN1 is a component of elastic fibers and previous studies have implied that its main function is to bind integrins, thereby connecting cells to the ECM [23,24]. In the fetal renal ECM, the EMILIN1 signal comprises 6.8% of the total ECM signal, compared to only 1.5% of the mature ECM (Fig. 3A, Supplemental Table 1). Protein verification was conducted using immunohistochemistry on non-decellularized renal tissue. Immunolocalization of EMILIN1 in mature and fetal human kidney showed localization mainly to the tubulointerstitial space (Fig. 3B). In concordance with the proteomic analysis, EMILIN1 was significantly enriched in the fetal kidney compared to the mature kidney (Fig. 3C).
Fig. 3.
Validation of selected ECM protein. (A) EMILIN1 signal identified with mass-spectrometry (MS) within fetal and mature renal ECM extracts. N = 3 MS analyses, each containing 3–5 kidney samples per group (fetal or mature). Shown is mean ± SEM; *P < 0.05. (B) Representative fluorescence immunohistochemistry images (200× magnification) demonstrate the localization and amount of EMILIN1 (green) in fetal and mature renal tissue. Scale bar represents 75 μm. (C) Quantification of normalized EMILIN1 area in both fetal and mature renal samples. N ≥ 25 fluorescent images derived from N = 3 samples. Shown is mean ± SEM; ***P < 0.001.
Another significantly abundant glycoprotein in the fetal renal ECM is FBN1, which is a major component of microfibrils that, together with elastins, form the elastic fibers [30]. The FBN1 signal comprises 11.1% of the fetal ECM signal, compared to 7.3% of the mature ECM (Supplemental Fig. 4A, Supplemental Table 1). Immunolocalization of FBN1 showed localization to both the glomerular and tubulointerstitial space, leading to no differences in fluorescent signal between the fetal and mature kidney (Supplemental Fig. 4B,C). mRNA levels on the other hand showed a significant increase of FBN1 in the fetal kidney compared to the mature kidney (Supplemental Fig. 4D). Elastic fiber components in general seemed to be increased in the fetal kidney (Supplemental Table 1). Verification by gene expression analysis indeed showed that MFAP2 was significantly increased in the fetal kidney compared to the adult kidney and the same trend was visible for ELN (Supplemental Fig. 4E, F). Based on these results, EMILIN1 likely plays a fundamental role within the fetal renal ECM.
Anchored cell-derived ECM can be modified by depleting specific ECM proteins
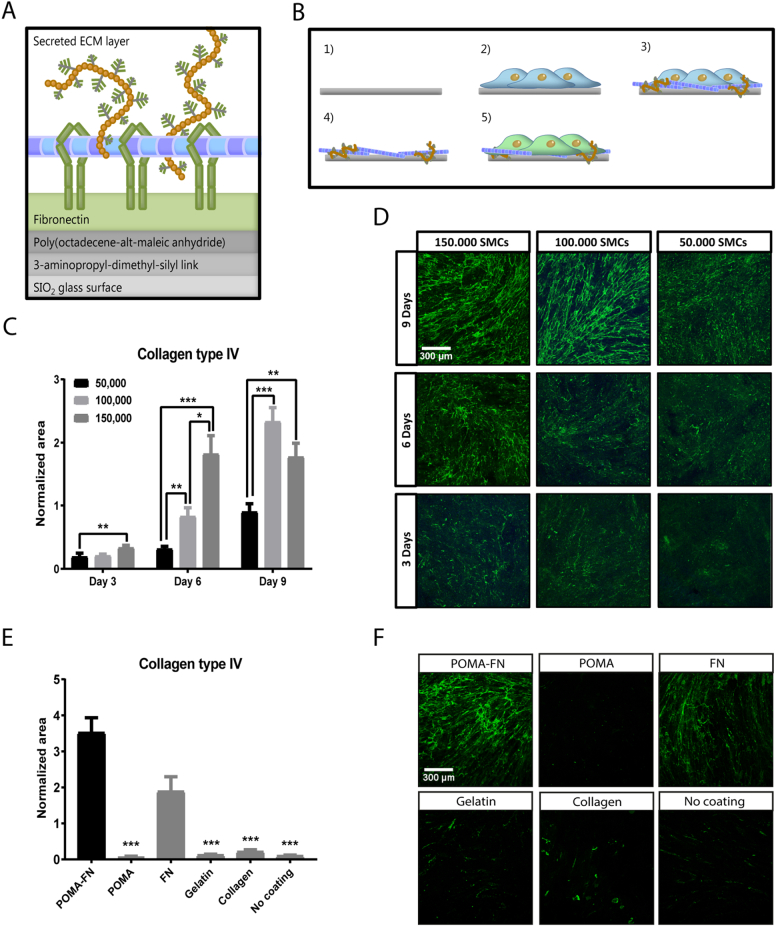
EMILIN1 was further investigated by using a method to reliably anchor native ECM through copolymer thin-film chemistry [[31], [32], [33]]. Poly(octadecene-alt-maleic anhydride) (POMA) was covalently coupled to aminosilanized glass coverslips [32]. Next, fibronectin (FN) was covalently attached via its lysine sidechain to the reactive anhydride moieties [33] (Fig. 4A). Immobilized FN allows the stable anchorage of the ECM via its binding domains to collagen, fibrin and heparin sulfate proteoglycans [31].
Fig. 4.
Reliable anchorage of native cell-secreted ECM to a glass surface. (A) Schematic overview of the chemical layers needed to reliably anchor cell-secreted ECM to a glass coverslip. Fibronectin (FN) is immobilized by a covalent linkage with poly(octadecene-alt-maleic anhydride (POMA) on an aminosilanized glass coverslip. Immobilized FN allows stable binding of cell-secreted ECM proteins. (B) Workflow for anchoring smooth muscle cell (SMC) ECM to glass coverslips: 1) FN is covalently immobilized on a POMA coverslip. 2) SMCs are grown to confluency. 3) During this culture time, the SMCs will deposit their own ECM. 4) The SMCs are removed by a decellularization process with NH4OH, leaving the secreted ECM attached to the POMA-FN coverslip. 5) Evaluation of renal cell function on captured ECM. (C) Quantification of collagen type IV area deposited by 50,000, 100,000 or 150,000 SMCs cultured for 3, 6 or 9 days on POMA-FN coverslips. Shown is mean ± SEM; **P < 0.01, ***P < 0.001. N ≥ 23 fluorescent Z-stacks derived from N = 3 samples. (D) Representative immunofluorescence Z-stacks (100× magnification) of anchored collagen type IV on POMA-FN coverslips, deposited by 50,000, 100,000 or 150,000 SMCs cultured for 3, 6 or 9 days. Scale bar represents 300 μm. (E) Quantification of collagen type IV area deposited by 150,000 SMCs cultured for 6 days on coverslips coated with POMA-FN, POMA, FN, gelatin or collagen or no coating. Shown is mean ± SEM; ***P < 0.001. N ≥ 19 fluorescent Z-stacks derived from N = 4 samples. (F) Representative immunofluorescence Z-stacks (100× magnification) of anchored collagen type IV deposited by 150,000 SMCs cultured for 6 days on coverslips coated with POMA-FN, POMA, FN, gelatin or collagen or no coating. Scale bar represents 300 μm.
We cultured smooth muscle cells (SMCs) to confluency on POMA-FN coverslips to capture their secreted ECM (Fig. 4B). SMCs were chosen as ECM production cells, since they expressed the highest amount of EMILIN1 and FBN1 compared to other cell types (Supplemental Fig. 5A). A culture period of 6 days and a cell density of 150,000 was found to be the ideal combination between ECM deposition and time (Fig. 4C,D). We verified the capacity of the POMA-FN coverslips to reliably anchor the SMC-secreted ECM by collagen type IV staining. When compared to a collagen or gelatin coating, solely a POMA layer, or no coating, the POMA-FN coverslips captured a significantly higher amount of homogeneously distributed ECM, with the rest exhibiting drastic delamination. With regards to a sole FN coating, almost double the amount of ECM was captured on POMA-FN coverslips (Fig. 4E–F). SMCs did not grow to confluency on solely POMA or uncoated coverslips after 6 days of culture, as indicated by a significant decrease in F-actin area (Supplemental Fig. 5B,D). However, no differences were found in collagen type IV production between SMCs grown on all coatings, as identified by intracellular staining (Supplemental Fig. 5C,D). Thus, although production level of collagen type IV by SMCs remained unaffected by the type of coating, the POMA-FN coverslips captured the most intact and highest amount of ECM.
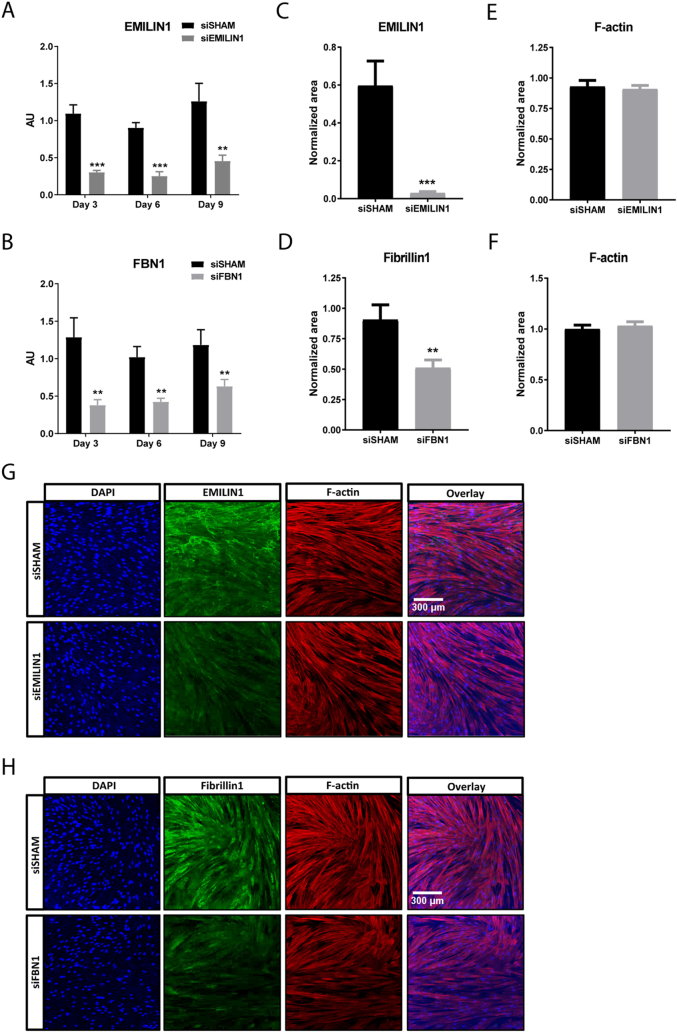
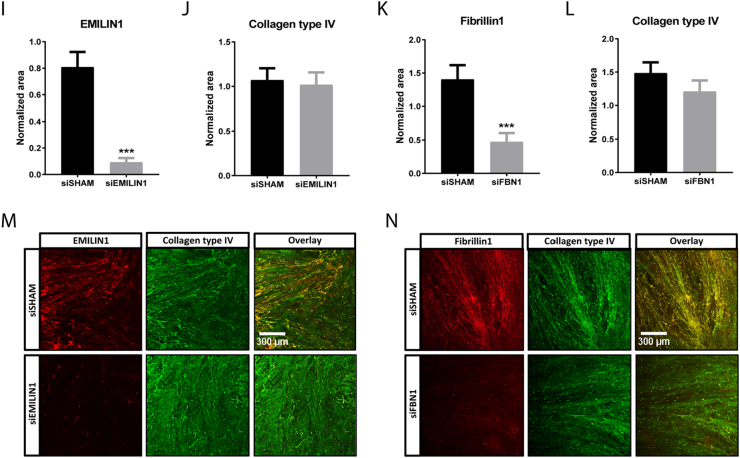
The next step was to deplete EMILIN1 from the anchored cell-secreted ECM. This was accomplished by using short interference RNA (siRNA) mediated silencing in the production cells. FBN1 was used as a second target to deplete from the ECM. QPCR analysis after 3, 6 and 9 days of culture showed efficient silencing of EMILIN1 or FBN1 expression in SMCs treated with a siRNA pool specific for EMILIN1 (siEMILIN1) or FBN1 (siFBN1), when compared to cells transfected with a pool of non-targeting siRNA sequences (siSHAM) (Fig. 5A,B). The mRNA levels of other EMILIN/multimerin and fibrillin family members were not affected by either siEMILIN1 or siFBN1 when compared to siSHAM, indicating that the siRNA-mediated silencing was specific for EMILIN1 and FBN1 (Supplemental Fig. 6A–H). Importantly, genes encoding for major ECM components, such as COL1A1, COL4A1, ELN and LAMA5 were not affected by EMILIN1 knockdown, implying that the absence of EMILIN1 did not greatly alter ECM composition (Supplemental Fig. 7A–D). Immunofluorescence confirmed a significant loss of EMILIN1 or FBN1 expression in siEMILIN1 or siFBN1 transfected SMCs when compared to siSHAM cells, with no variance in cellular F-actin area (Fig. 5C–H). Western blot analysis for EMILIN1 verified the loss of protein expression (Supplemental Fig. 8A,B). Additionally, their secreted ECM was significantly depleted of EMILIN1 or FBN1 and successfully anchored to the POMA-FN coverslips, with no difference in extracellular collagen type IV deposition (Fig. 5I–N).
Fig. 5.
EMILIN1 and FBN1 targeting siRNAs induce significant silencing intracellular and on secreted ECM level. Quantitative polymerase chain reaction (qPCR) of EMILIN1 (A) and FBN1 (B) in smooth muscle cells (SMCs) transfected with EMILIN1-targeting siRNA (siEMILIN1), FBN1-targeting siRNA (siFBN1) or non-targeting siRNA (siSHAM), cultured for 3, 6 or 9 days on POMA-FN coverslips. Shown are target gene/housekeeping gene (β-actin) ratios (AU). Control (non-transfected SMCs) values are set to 1 (not shown). Shown is mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001. N ≥ 5 qPCRs. Quantification of intracellular EMILIN1 (C), Fibrillin1 (D) or F-actin (E, F) area in confluent SMCs transfected with siSHAM, siEMILIN1 or siFBN1 cultured for 6 days on POMA-FN coverslips. Control (non-transfected SMCs) values are set to 1 (not shown). Shown is mean ± SEM; **P < 0.01, ***P < 0.001. N ≥ 29 fluorescent Z-stacks derived from N = 3 samples. Representative immunofluorescence Z-stacks (100× magnification) of SMCs transfected with siSHAM, siEMILIN1 (G) or siFBN1 (H) cultured for 6 days on POMA-FN coverslips and stained for F-actin (red), EMILIN1 or Fibrillin1 (green) and DAPI (blue). Scale bar represents 300 μm. Quantification of EMILIN1 (I), Fibrillin1 (J) or collagen type IV (K, L) area in the extracellular matrix (ECM) deposited by SMCs transfected with siSHAM, siEMILIN1 or siFBN1 cultured for 6 days on POMA-FN coverslips. Control (non-transfected SMCs) values are set to 1 (not shown). Shown is mean ± SEM; **P < 0.01, ***P < 0.001. N ≥ 29 fluorescent Z-stacks derived from N = 3 samples. Representative immunofluorescence Z-stacks (100× magnification) of the ECM deposited by SMCs transfected with siSHAM, siEMILIN1 (M) or siFBN1 (N) cultured for 6 days on POMA-FN coverslips and stained for collagen type IV (green) and EMILIN1 or Fibrillin1 (red). Scale bar represents 300 μm.
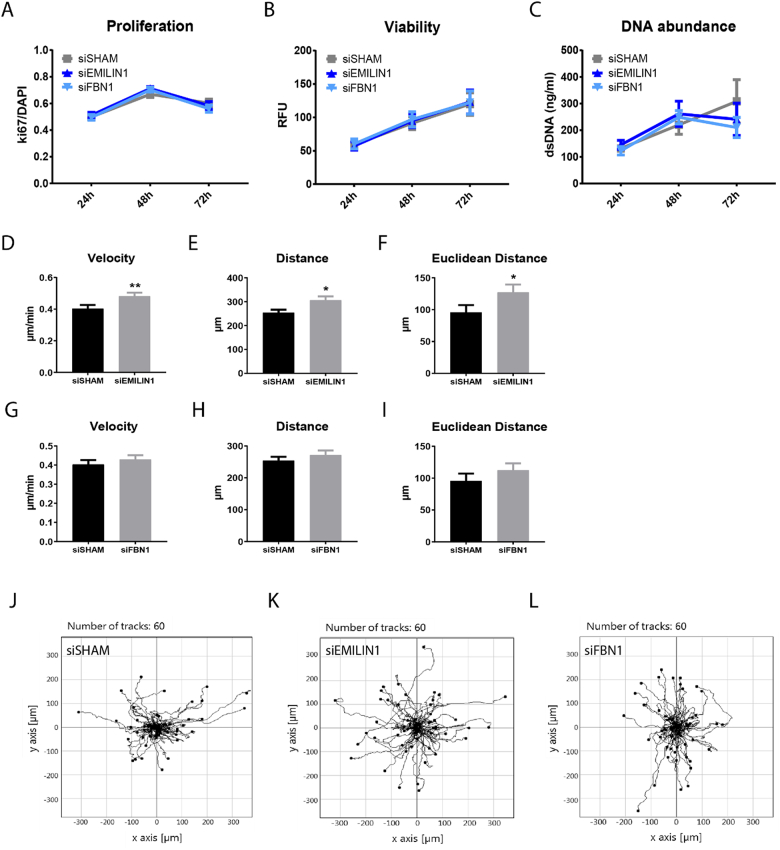
Depletion of EMILIN1 from the ECM promotes renal epithelial cell migration, but does not affect cell growth
Previous studies have shown that EMILIN1 is important for migration and growth of various cell types through the interaction of its gC1q domain with integrins [[21], [22], [23], [24]]. Therefore, we studied the migration and growth of renal epithelial cells cultured on an ECM layer depleted of EMILIN1. Depletion of either EMILIN1 or FBN1 from the ECM did not affect proliferation, viability and DNA abundance of renal epithelial cells, when compared to cells cultured on siSHAM ECM (Fig. 6A–C). Next, the migration capacity was assessed using live-cell tracking. A significant increase in velocity and distance covered by renal epithelial cells cultured on siEMILIN1 ECM was observed when compared to cells cultured on siSHAM ECM (Fig. 6D–F, J–K). This positive effect was absent in renal epithelial cells cultured on siFBN1 ECM (Fig. 6G–J, L), hinting towards a link between enhanced renal cell migration and disruption of gC1q-intergrin interaction.
Fig. 6.
Effect of EMILIN1 or Fibrillin1 depletion from the extracellular matrix on renal epithelial cell growth and migration. (A) ki67/DAPI ratio as measure for cell proliferation at 24, 48 or 72 h post seeding in HRPTECs cultured on siSHAM, siEMILIN1 and siFBN1 ECM. Shown is mean ± SEM. N ≥ 26 fluorescent images derived from N = 3 samples. (B) Viability measurements using the PrestoBlue assay after 24, 48 or 72 h post-seeding in HRPTECs on siSHAM, siEMILIN1 and siFBN1 ECM. Shown is mean ± SEM. N = 6 assays. (C) DNA abundance measurements using the PicoGreen assay after 24, 48 or 72 h post-seeding in HRPTECs on siSHAM, siEMILIN1 and siFBN1 ECM. Shown is mean ± SEM. N = 5 assays. Quantified live-cell migration assay results showing velocity and covered and Euclidean distance of HK2 cells on either siEMILIN1 (D-F) and siFBN1 (G-I) ECM compared to cells on siSHAM ECM. The migration of HK2 cells on the ECM layer was tracked overnight with a confocal microscope. Shown is mean ± SEM; *P < 0.05, **P < 0.01. N = 60 individual cells tracked derived from N = 4 assays. Migration plots showing multiple tracks of individual HK2 cells on siSHAM (J), siEMILIN1 (K) and siFBN1 ECM (L).
EMILIN1 is important for binding of renal epithelial cells to the ECM and for focal adhesion assembly
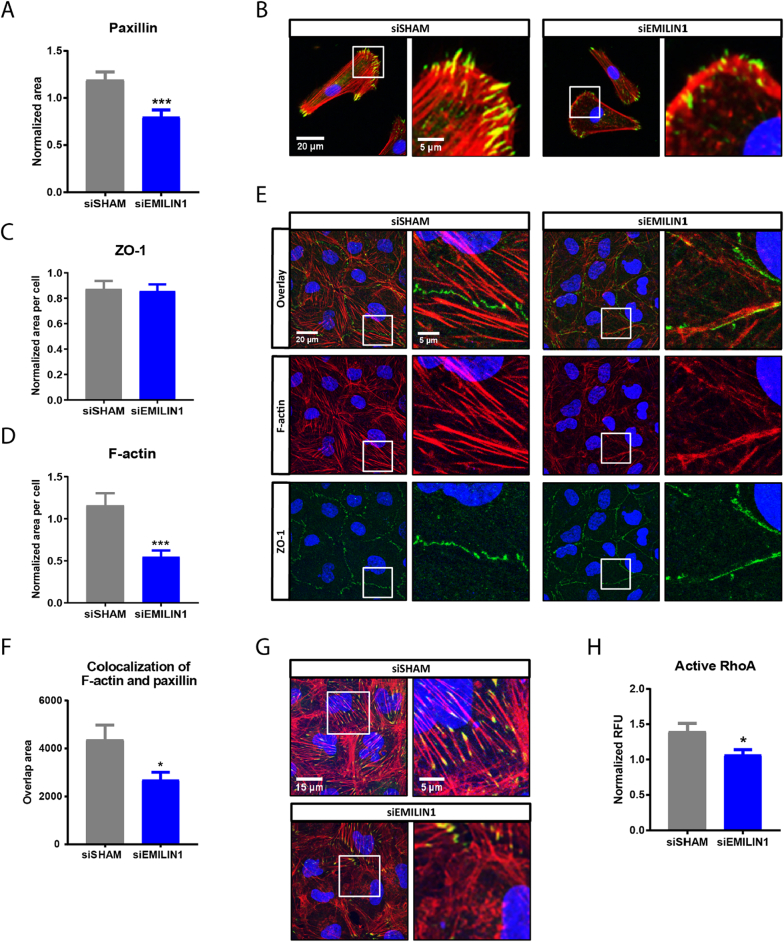
Integrins form part of large dynamic protein complexes that connect the cell cytoskeleton to the ECM, called focal adhesion complexes. We assessed whether EMILIN1 in the ECM is required for assembly of focal adhesion complexes during the initial binding of renal epithelial cells to the ECM. Depletion of EMILIN1 from the ECM caused defects in the initial assembly of the focal adhesion-structural protein paxillin. Renal cells cultured for only 2 h on siEMILIN1 ECM exhibited a significant decrease of paxillin area when compared to cells cultured on siSHAM ECM, indicating a reduced spreading ability (Fig. 7A–B). Cells cultured on siFBN1 ECM for 2 h did not show a decrease in paxillin area (Supplemental Fig. 9A–B), verifying that the putative effect is specific for EMILIN1 depletion. Next, we assessed the capacity of renal epithelial cells to form a tight monolayer when cultured on siEMILIN1 ECM. The tight junction protein ZO-1 was used to measure barrier formation. Intracellular staining revealed no changes in ZO-1 protein expression in renal cell cultured for 48 h on siEMILIN1 ECM compared to siSHAM ECM, indicating that these cells are capable of creating a connected monolayer (Fig. 7C). However, silencing of EMILIN1 in the ECM appeared to alter the junctional pattern of ZO-1 into a linear instead of zigzagged configuration (Fig. 7E). A significant decrease in F-actin area was also observed in cells grown on siEMILIN1 ECM compared to siSHAM ECM (Fig. 7D). Immunofluorescence visualization of F-actin revealed that formation of stress fibers was affected (Fig. 7E). The assembly of focal adhesion complexes in confluent renal epithelial monolayers was impaired as well. Intracellular staining revealed that EMILIN1-depleted ECM significantly inhibited formation of paxillin points and stress fibers in confluent renal epithelial monolayers, since less paxillin colocalized with F-actin (Fig. 7F–G). Cells cultured to confluency on FBN1-depleted ECM did not exhibit a significant reduction in colocalization of paxillin with F-actin (Supplemental Fig. 9C–D), further verifying that the impairment of focal adhesion assembly is specific for EMILIN1 depletion. Actin cytoskeleton remodeling upon integrin mediated focal adhesion formation is regulated by RhoA signaling. Thus, we assessed the RhoA activity in renal epithelial cells grown on siEMILIN1 ECM for 48 h when compared to cells grown on siSHAM ECM. In concordance with our previous results, EMILIN1 silencing in the ECM significantly reduced RhoA activation in renal cells (Fig. 7H). All together, these findings indicate that EMILIN1 in the ECM is important for assembly of focal adhesion complexes and subsequent actin-cytoskeleton adaptation in renal epithelial cells.
Fig. 7.
Depletion of EMILIN1 from the ECM layer reduces paxillin area and stress fibers in renal epithelial cells. (A) Quantified results showing the paxillin area in HRPTECs on siSHAM or siEMILIN1 ECM during initial binding to the ECM (cultured for 2 h). Shown is mean ± SEM; ***P < 0.001. N = 30 fluorescent images derived from N = 3 samples. (B) Representative immunofluorescence images (400× magnification) of HRPTECs after 2 h of binding on siSHAM or siEMILIN1 ECM and stained for paxillin (green), F-actin (red) and DAPI (blue). Scale bar represents 20 μm (overview image, left) and 5 μm (zoomed-in image, right). Quantified results showing the ZO-1 (C) and F-actin area (D) in HK2 cells cultured to confluency on siSHAM or siEMILIN1 ECM. Shown is mean ± SEM; ***P < 0.001. N = 20 fluorescent Z-stacks derived from N = 4 samples. (E) Representative immunofluorescence Z-stacks (630× magnification) of HK2 cells cultured to confluency (cultured for 48 h) on siSHAM or siEMILIN1 ECM and stained for ZO-1 (green), F-actin (red) and DAPI (blue). Scale bar represents 20 μm (overview image, left) and 5 μm (zoomed-in image, right). (F) Colocalization of paxillin and F-actin in HK2 cells cultured to confluency on siSHAM or siEMILIN1 ECM. Shown is mean ± SEM; *P < 0.05. N ≥ 25 fluorescent Z-stacks, from N ≥ 5 samples. (G) Representative immunofluorescence Z-stacks (630× magnification) of HK2 cells cultured to confluency on siSHAM or siEMILIN1 ECM and stained for paxillin (green), F-actin (red) and DAPI (blue). Colocalization is displayed as yellow. Scale bar represents 15 μm (overview image, left) and 5 μm (zoomed-in image, right). (H) RhoA activation levels in HK2 cells cultured for 48 h on siSHAM or siEMILIN1 ECM. Shown is mean ± SEM; *P < 0.05. N = 5 assays.
Discussion
This study presents to date the first catalogue that compares the human fetal and mature renal ECM. A total of 99 different ECM proteins was detected by proteomic analysis of which the majority forms an overlapping core, but also includes many renal ECM proteins that are enriched in either the fetal or mature condition.
Using immunohistochemistry, we confirmed the identification of EMILIN1 and FBN1 as key proteins in renal ECM. EMILIN1 mainly localizes to the tubulointerstitial space, but is also classified as a structural glomerular protein by Lennon et al [10]. However, they solely focused on identifying glomerular proteins and did not analyze the remainder from their glomerular enrichment strategy. Immunolocalization of FBN1 showed its presence in both glomeruli and tubulointerstitial space, which is in concordance with Lennon et al., who classified it as a glomerular basement membrane protein.
Our proteome comparison between the fetal and mature renal ECM has the potential to mark novel molecular players by which the renal ECM can influence cellular behavior. Percentile ranking by LFQ intensity proved effective at finding proteins enriched in the fetal ECM. One striking difference between the fetal and mature renal ECM proteome is the enrichment of elastic fiber components in the fetal ECM, including fibrillin1, MFAP2 and elastin. Particularly the glycoprotein EMILIN1 was abundant in the fetal ECM. Many other ECM proteins differ significantly between fetal and mature tissue, however most of them are present in low quantities in the renal ECM (<2%) and are therefore less likely to play a fundamental role (for example EMILIN3).
The importance of EMILIN1 has been previously studied using EMILIN−/− mice, which display several vascular defects, including impaired lymph drainage, increased leakage and elevated blood pressure associated with narrower vessels [[26], [27], [28], [29]]. The latter is due to an increased presence of transforming growth factor β (TGF-β), since EMILIN1 prevents TGF-β processing [[27], [28], [29]]. In aortic valves, increased TGF-β signaling due to EMILIN1 deficiency causes elastic fiber fragmentation and subsequent aortic valve disease [34]. Both these mutant mice and aged mice exhibit ECM stiffness, fibrosis, increased collagen expression and cell adhesion and fibronectin alterations [35]. This suggests that loss of EMILIN1 and associated phenotype is linked to a mature state, which is in concordance with our proteomic data. Despite several studies described the function of EMILIN1, a possible renal function remains unclear.
Lack of EMILIN1 is associated with altered vascular and lymphatic cell anchorage, patterning and morphology [25,26]. Previous studies have reported that the gC1q domain of EMILIN1 is responsible for promoting cell adhesion [23,24,36]. Our study showed that renal epithelial cells exhibit a weaker adhesion pattern on EMILIN1-depleted ECM, indicated by fewer focal adhesion points. Focal adhesions provide linkage points between the cell cytoskeleton and the ECM, playing a central role in adhesion [37]. Renal epithelial cells grown on EMILIN1-depleted ECM exhibit a low number of actin stress fibers and less paxillin, a focal adhesion adaptor protein that localizes to large focal contacts at the tips of these fibers. More evidence for the role of EMILIN1 in renal cell adhesion comes from the activation of signaling molecule RhoA, that is recruited and activated upon integrin-ECM binding and leads to maturation of focal adhesions [37]. Less RhoA was activated in renal epithelial cells grown on EMILIN1-depleted ECM, indicating reduced stabilization of focal structures. Furthermore, the junctional pattern of ZO-1 in these renal cells appeared linear instead of zigzagged. High bioavailability of ZO-1 at tight junctions induces zigzag patterning and increases the overall permeability of epithelia [38]. Actin filaments terminate at tight junctions, thereby linking them with focal adhesions [39]. This suggests that defects in focal adhesion assembly by EMILIN1 depletion in the ECM can result in F-actin stress fiber dysregulation and subsequent reduction of functional tight junctions, affecting renal epithelial barrier function. Indeed, previous studies showed that ZO-1 interaction with F-actin at tight junctions is important for epithelial polarization and formation of single lumens [40]. Altogether, our study indicates that EMILIN1 could be a promising candidate for implementation in renal scaffolds for the enhancement of cell attachment and barrier formation.
Our findings are not in line with previous studies, in which actin organization in cells attached to EMILIN1 was described to be organized along the cell periphery without any apparent focal contacts [24,36]. More in depth experiments have shown that in these cells, the gC1q domain of EMILIN1 promotes cell adhesion by interacting with α4/α9/β1 integrins [23,24,36]. The reported synovial sarcoma cells and T lymphocytes use α4β1 integrins for gC1q-mediated cell attachment, whereas lymphatic endothelial cells (LECs) and microvascular ECs attachment to EMILIN1 is mediated by α9β1 integrins [23,24]. These reports suggest that different cell types express a specific selection of integrins to react to environmental stimuli.
β1 is the most widely expressed renal integrin subunit and is responsible for interactions with the cell cytoskeleton via binding with several focal adhesion complex proteins [41,42]. Different α subunits have been demonstrated to be expressed by renal tissue, which all can form heterodimers with β1: α1, α2, α3, α5, α6, α8 and αv [41,42]. The renal cells used in our migration and confluent adhesion assays showed high expression of the integrin subunits α3, α5, αv, and β1, modest levels of α2, α4, α6 and non-detectable expression of α9 (data not shown). EMILIN1 may exerts its effect in renal cells through another αβ1 integrin heterodimer than the α4/α9/β1-gC1q interaction described, which might explain the distinct effect we found of EMILIN1 on renal epithelial cell adhesion strength.
The dynamic assembly and disassembly of focal adhesions play a central role in cell migration and proliferation. EMILIN1 also plays an important role in these processes: fibroblasts, keratinocytes and LECs obtained from EMILIN1−/− mice retain a high proliferation rate compared to wildtype cells [22,23]. This reduction in proliferation is associated with the gC1q–α9 integrin interaction, which also stimulates LEC migration [23]. Furthermore, the gC1q–α4 integrin interaction promotes haptotactic directional migration of trophoblasts [36], indicating that EMILIN1 can promote cell proliferation and migration by interacting with α4/α9/β1 integrins.
In contrast, the present study shows that EMILIN1-deficient ECM does not influence proliferation of renal epithelial cells and absence of EMILIN1 stimulates renal epithelial cell migration. Hence, the effect of EMILIN1 seems to be cell type specific. Indeed, Colombatti and colleagues indicated that the effect of EMILIN1 on cell adhesion and migration can be either enhancing or reducing depending on the cell type investigated [43], which might be linked to different integrin expression profiles.
Strong adhesion through focal adhesion formation stabilized by stress fibers is considered to be unfavorable for cell detachment needed for migration [44]. In 1979, Couchman and Rees already made the observation that fibroblasts displayed little to no focal adhesions during the initial period of rapid migration. Focal adhesions and associated ventral stress fibers only developed when migration slowed down [45]. This observation implies that focal adhesion disassembly facilitates rapid migration. Indeed, we observed that renal epithelial cells migrate faster and cover more distance when less EMILIN1 is present in the ECM. Therefore, this rapid migration pattern is most likely stimulated by the reduction of focal adhesions and stress fibers seen in these renal cells as well.
The motility phenotype of cells also depends on which integrins are available on the cell membrane. For example, previous research has observed that the α4/α9/β1-gC1q interaction encourages a cellular phenotype that lacks stress fibers and promotes lamellipodia formation, thereby facilitating cell spreading [23,24,36]. The α4/α9 subfamily is a specific subset of α subunits based on evolutionary relationships, that share a high sequence similarity and ligand specificities [46]. In contrast, the present study showed that lack of EMILIN1 interaction stimulates cell migration. Renal cells express other α subunits in high quantities [41,42]; suggesting that they likely express subunits that stabilize focal adhesions. For example, the renal cells used in our migration assay highly express integrin subunits α5 and β1, which as a heterodimer is known to mediate stabilization of mature focal adhesions [47].
Apparent differences between the renal epithelial cell behavior seen in response to EMILIN1 in this study and the results of earlier studies may also be due to differences in study design. We cultured our renal cells on a complex ECM network depleted from a protein of interest from the beginning, rather than culturing on a coating of one specific ECM protein. It is inevitable that this rich ECM background from the start will have an influence on focal adhesion composition, mostly the integrin expression profile, and subsequent cell behavior. This environment mimics more the in vivo ECM complexity, even though it lacks a 3D configuration. Nevertheless, this 2D ECM model is a valuable resource for additional investigation and thereby aids in the search for promising ECM components to implement in 3D culture.
In conclusion, based on our proteome analysis we provide evidence for EMILIN1 as a promising candidate for implementation in renal scaffolds as a bioactive factor. Here, it would most likely stimulate an adhesive phenotype of the embedded renal cells. Matrices containing organ specific ECM cues to direct cell adhesion may ultimately allow the generation of whole 3D kidney constructs for implantation.
Experimental procedures
Antibodies
Polyclonal antibodies used were against EMILIN1 (HPA002822; Sigma-Aldrich), fibrillin1 (HPA021057; Sigma-Aldrich) and collagen type IV (AB769; Merck Millipore). Monoclonal antibodies used were against ki-67 (RM-9106-R7, ThermoFisher), paxillin (ab32084, Abcam), ZO-1 (610967; BD Biosciences) and β-actin (ab8226, Abcam). Rhodamine phalloidin conjugate was used for staining F-actin (R415; ThermoFischer). Secondary antibodies used for immunohistochemistry were goat-anti-rabbit Alexa Fluor 488 (A11034; ThermoFisher), donkey-anti-rabbit Alexa Fluor 568 (A10042; ThermoFisher), donkey-anti-goat Alexa Fluor 488 (A11055; ThermoFisher) and goat-anti-mouse Alexa Fluor 488 (A11029; ThermoFisher). Secondary antibodies used for Western blot were goat-anti-rabbit IRDye 800CW (926-32211; Li-cor Biosciences) and goat-anti-mouse IRDye 680RD (926-68070; Li-cor Biosciences).
Human samples
Normal human renal tissue from healthy adult donors was obtained from the Erasmus MC Tissue Bank (n = 13; gender: 8 male, 5 female; age [mean ± SD]: 55 ± 24 years). Normal human kidneys from healthy fetal donors were received from the department of Molecular Cell Biology from the Leiden UMC (n = 13; age range: 18–24 weeks of pregnancy).
Cell culture
Primary aortic smooth muscle cells (aSMCs) were purchased from Lonza (CC-2571) and maintained in Smooth Muscle Growth Medium (CC-3182; Lonza). Primary Human Renal Proximal Tubular Epithelial Cells (HRPTECs) were purchased from ScienCell (#4100) and maintained in Epithelial Cell Medium (#4101; ScienCell). Immortalized Human Kidney [48] (HK2) cells were maintained in RPMI-1640 medium with glutamine (61,870,010; ThermoFisher), supplemented with 10% (vol/vol) FCS (Biowest) and 100 UmL−1 PS (Gibco). All cells were cultured at 37 °C in a humidified atmosphere containing 5% CO2. Primary cell lines were used between passages 3 and 8.
Tissue decellularization and ECM homogenization
Tissue decellularization was used to reduce the complexity of samples by removing cellular components and thereby enriching for ECM proteins. Renal tissue was cut into 4 pieces of uniform thickness and one piece was fixed and saved for hematoxylin and eosin (H&E) staining to asses tissue morphology. The other pieces were decellularized overnight in 1% SDS under constant rotation. Next, a shorter decellularization step was performed for 1 h with 1% Triton X-100 under constant rotation. Tissue was washed thoroughly in PBS for 2 h under constant rotation to remove as much residual detergent as possible before proceeding with tissue lysis. One decellularized piece was fixed and saved for H&E staining to validate the removal of all cellular content and assess preservation of the ECM architecture. Tissue was homogenized in lysis buffer (10 mM Tris [pH 7.4], 100 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, 1% Triton X-100, 1 mM EGTA, 1 mM EDTA, 10% glycerol, 1 mM NaF, 1 mM Na orthovanadate, supplemented with a protein inhibitor cocktail (cOmplete, Mini Protease Inhibitor Tablets)) using an Ultra-Turrax (IKA) and lysed for an hour at 4 °C under rotation. After homogenizing a second time, tissue lysates were centrifuged [10 min, 1000 ×g, 4 °C] and supernatant was saved at −80 °C until further processing.
MS data acquisition and analyses
ECM extracts were separated by SDS-PAGE. Three gels were loaded with a fetal and mature ECM extract of the same protein concentration, each containing 3–5 pooled kidney samples that were randomly selected (mixed age and gender). The separated samples were prepared for LC-MS/MS by the Proteomics Centre of the Erasmus MC as follows. 1D SDS-PAGE gel was visualized with a Coomassie staining and lanes were cut into 2-mm slices using an automatic gel slicer and subjected to in-gel reduction with dithiothreitol, alkylation with iodoacetamide and digestion with trypsin (Promega, sequencing grade), essentially as described previously [49]. Supernatants were stored in glass vials at −20 °C until LC-MS. Nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS) was performed on an 1100 series capillary LC system (Agilent Technologies) coupled to an LTQ-Orbitrap XL mass spectrometer (Thermo) operating in positive mode and equipped with a nanospray source, essentially as previously described [50]. Peptide mixtures were trapped on a ReproSil C18 reversed phase column (Dr Maisch GmbH; column dimensions 1.5 cm × 100 μm, packed in-house) at a flow rate of 8 μl/min. Peptide separation was performed on ReproSil C18 reversed phase column (Dr Maisch GmbH; column dimensions 15 cm × 50 μm, packed in-house) using a linear gradient from 0 to 80% B (A = 0.1% formic acid; B = 80% (v/v) acetonitrile, 0.1% formic acid) in 70 min and at a constant flow rate of 200 nl/min using a splitter. The column eluent was directly sprayed into the ESI source of the mass spectrometer. Mass spectra were acquired in continuum mode; fragmentation of the peptides was performed in data-dependent mode.
Raw data files were analyzed using the MaxQuant software as described by Cox et al [51]. Proteins that were identified in at least two of the three fetal or mature replicates were used for further analysis. By crossreferencing with the Human Matrisome Project [8,9], we categorized the obtained proteins into ECM core and associated proteins. The label free quantification (LFQ) module was used to quantify the relative abundances of fetal and mature renal ECM proteins.
POMA-fibronectin coverslips preparation
Circular glass coverslips were cleaned to remove organic residues with a quick rinse in acetone and sonication in 50% methanol for 20 min and in chloroform for another 20 min. Coverslips were oxidized with piranha solution [70% sulphuric acid (99% w/v) and 30% hydrogen peroxide (35% w/v)] for 20 min and sonicated in chloroform for 10 min. To ensure complete removal of the piranha solution, coverslips were washed by sonication with ultrapure water for 5 min and with chloroform for another 5 min in an alternating fashion and repeated for at least 5 times [32]. Freshly oxidized coverslips were aminosilanized with a 2% (3-Aminopropyl)triethoxysilane (APTES, Sigma-Aldrich) solution in 95% ethanol for 10 min. Excess APTES was removed by 4 washings with 95% ethanol for 5 min. The APTES layer was cured at 120 °C for 30 min [31,33]. In order to covalently bind fibronectin (FN), the coverslips were coated with a thin layer of 0.16% (w/v) poly(octadecene-alt-maleic anhydride (POMA; Sigma-Aldrich) solution in tetrahydrofuran (Sigma-Aldrich) using a spin-coater (Brewer Science) [1500 rpm s−1, 30 s, 4000 rpm]. The polymer coating was cured at 120 °C for 2 h [31,33]. The POMA-coated coverslips were stored in the dark for up to 3 months. Heat activation of the anhydride moieties needed to be repeated if coverslips were stored for longer than 2 weeks. Prior to use in cell culture, the POMA-coated coverslips were sterilized under UV-light for 30 min. Lastly, the coverslips were coated with sterile 50 μg/mL fibronectin (FN; Roche) in PBS for 1 h at 37 °C to allow stable anchorage of cell secreted ECM. POMA-FN coverslips were rinsed twice with sterile PBS to remove excess FN and to achieve a homogeneous coating before seeding with cells [33].
Anchored cell-secreted ECM layer preparation
To create cell derived ECM layers anchored to POMA-FN coverslips, SMCs were seeded at 40,000 cells per cm2. The cultures were decellularized at day 6 using warm 20 mM NH4OH by gentle agitation for 15 min, which disrupts lipid interactions but preserves protein interactions. The resulting ECM layers were treated with 10% DNase I (Qiagen) for 10 min to remove DNA traces. Cell remnants were removed by washing thrice with ultrapure water and twice with PBS. The decellularized ECM layer was immediately re-seeded with renal cells or fixated for immunofluorescence analysis. The presence of nucleic acids was assessed with DAPI staining. After the decellularization procedure, cellular components such as DNA remnants were barely detected.
Immunofluorescence
Sucrose cryoprotected renal tissue was embedded in an OCT compound (TissueTek) and stored at −80 °C. Frozen samples were cut into 7-μm-thick sections, fixated with acetone and stained with H&E or used for immunofluorescence stainings. Cells or ECM attached to coverslips were washed with PBS and fixed with 4% paraformaldehyde for 30 min. Blocking of tissue, cells or ECM occurred with 1% BSA/PBS for 30 min before incubation with primary and secondary antibodies for 1 h at room temperature. Cells and tissue were also permeabilized with 0.5% Triton X-100/PBS for 30 min before antibody incubation. DAPI was used as a nuclear counterstaining. Images of fluorescent-labeled markers were obtained with a Leica TCS SP8 X microscope. For quantification of the total area, 3D images were obtained by scanning multiple XY planes in the Z direction. Serial pictures along the Z-axis were combined to create a stacked XY image that was further analyzed using ImageJ 1.47v. Single XY pictures were taken with the Olympos BX51 upright microscope.
Quantitative PCR
Total RNA was isolated from SMCs using the ISOLATE II RNA mini kit (Bioline) according to manufacturer's instructions and from human renal tissue using TRIzol. Briefly, tissue was homogenized in 1 mL TRIzol using an Ultra-Turrax (IKA) on ice. 200 μl chloroform was added per 1 mL TRIzol, mixed and incubated for 3 min at room temperature. The aqueous phase was separated from the phenol by centrifugation [15 min, 12,000 ×g, 4 °C] and transferred to a new tube. RNA was precipitated by mixing the aqueous phase with 600 μl isopropanol and 1 μl of glycogen (20 mg/mL). After incubation for 10 min at room temperature, the precipitate was pelleted by centrifugation [10 min, 12,000 ×g, 4 °C]. The pellet was washed with 600 μl ice-cold 75% ethanol and pelleted again [5 min, 7500 ×g, 4 °C]. The pellet was dried at room temperature and the RNA was precipitated over night at −20 °C to increase yield by mixing the pellet with 200 μl Nuclease-free water (Qiagen), 20 μl 3 M Sodium acetate and 600 μl 75% ice-cold ethanol. The precipitate was pelleted by centrifugation [30 min, 15,000 ×g, 4 °C]. The pellet was washed with 200 μl ice-cold 75% ethanol and pelleted again [5 min, 15,000 ×g, 4 °C]. The RNA pellet was dried at room temperature and dissolved in 50 μl Nuclease-free water (Qiagen). RNA concentrations were assessed with a nanodrop spectrophotometer. Total RNA was reverse-transcribed using the SensiFAST cDNA synthesis kit (Bioline) according to manufacturer's instructions. Gene expression was determined using quantitative real-time PCR (qPCR) by loading samples in duplicate on a CFX96 Real-Time PCR Detection System (Biorad). The primer sequences used are listed in Supplemental Table 2. Data was normalized for the expression of housekeeping gene β-actin.
Short interference RNA
EMILIN1 or FBN1 knockdown in SMCs was achieved by using a mix of 4 complementary siRNA sequences directed against the mRNA of either EMILIN1 or FBN1 (ThermoScientific). As a negative control, cells were either untreated or transfected with a mix of 4 non-targeting siRNA sequences (ThermoScientific). The siRNA sequences used are listed in Supplemental Table 3.
Western blot
SMCs received a PBS wash twice and were harvested in lysis buffer (50 mM Tris [pH 8.0], 1% NP-40, 150 mM NaCl, 0.1% SDS, 0.5% Na deoxycholate, supplemented with a protein inhibitor cocktail (cOmplete, Mini Protease Inhibitor Tablets)). The lysates were incubated on ice for 20 min, centrifuged [12 min, 13,000 ×g 4 °C] and the supernatant was stored at −80 °C until further processing. Total protein concentration was determined by using the Pierce® BCA Protein Assay Kit (Thermo Scientific). Lysates were denaturated in Laemmli buffer (60 mM Tris [pH 6.8], 2% SDS, 10% glycerol, 5% β-mercaptoethanol, 0.01% bromophenol blue) at 90 °C for 5 min. Equal amounts of sample were separated by electrophoresis on a 10% SDS-PAGE gel and transferred onto a nitrocellulose membrane (Pierce) at 4 °C overnight. Membranes were blocked and probed with primary antibodies. Protein bands were visualized with Li-Cor secondary antibodies and detection system (Westburg) according to manufacturer's instructions.
Live-cell tracking
POMA-FN coverslips were placed into a 6-wells plate containing a No. 0 coverslip glass bottom (P06G-0-20-F, MatTek) to allow live-cell tracking. SMCs were transfected with siRNA after 24 h and coverslips were decellularized 6 days post-transfection. Before reseeding with 3700 HK2 cells per cm2, HK2 cells were visualized for fluorescent tracking by incubating the cells for 15 min at 37 °C in basal RMPI medium containing 4 μM CellTracker Blue CMAC dye (ThermoFisher). 24 h post-seeding, single cell migration was tracked overnight at 37 °C in a humidified atmosphere containing 5% CO2 using a Leica SP8X confocal microscope. Per condition, three positions were selected and imaged every 15 min for 12 h. Obtained videos were analyzed with ImageJ (v1.47) Manual Tracking and Chemotaxis Tool.
In vitro assays
Prior to all in vitro assays, SMCs were seeded on POMA-FN coverslips and transfected with siRNA after 24 h. The coverslips were decellularized 6 days post-transfection and reseeded with renal cells.
Cell adhesion assay
Captured ECM was reseeded with 6500 HRPTECs per cm2 for testing initial binding to the ECM or 25,000 HK2 per cm2 for measuring cell-ECM adhesion of a confluent monolayer. For testing initial binding, HRPTECs were left to adhere for 2 h. HK2 were cultured to confluence (48 h) on the captured ECM. Fixated cells were stained for either paxillin, a focal adhesion complex protein, or for ZO-1, a tight junction protein, and for F-actin.
PrestoBlue assay
Captured ECM was reseeded with 6500 HRPTECs per cm2. Cell viability was measured 24, 48 and 72 h post-seeding using PrestoBlue Cell Viability Reagent (ThermoScientific) according to manufacturer's protocol.
Ki67 nuclear staining
Captured ECM was reseeded with 6500 HRPTECs per cm2. Cell proliferation was visualized 24, 48 and 72 h post-seeding by immunofluorescence labelling with the proliferation marker ki67. The images were analyzed with ImageJ (v 1.47) to obtain the percentage of proliferating cells by dividing the amount of ki67+ cells by the DAPI+ count.
PicoGreen assay
Captured ECM was reseeded with 6500 HRPTECs per cm2. To quantify the amount of double stranded DNA, a Quant-iT™ PicoGreen™ dsDNA Assay (ThermoFisher) was performed 24, 48 and 72 h post-seeding according to manufacturer's protocol.
RhoA activation assay
SMCs were seeded on POMA-FN coverslips and transfected with siRNA after 24 h. The coverslips were decellularized 6 days post-transfection and reseeded with 25,000 HK2 per cm2. The activation of the small G-protein RhoA was determined 48 h post-seeding using the G-LISA RhoA Activation Assay Biochem Kit (Cytoskeleton, BK124) according to manufacturer's instructions. HK2 cells were serum starved overnight (in RPMI containing 0.2% FCS) and stimulated with full RPMI medium (containing the standard 20% FCS) for 1 min before starting the RhoA activation assay to modulate GTP-RhoA levels.
Statistical analyses
Graphpad Prism (version 7.02) was used to perform the statistical analyses. To test if values came from a Gaussian distribution, either the D'Agostino-Pearson omnibus or Shapiro-Wilk normality test was used. The ordinary one-way ANOVA or unpaired t-test was used if the values were normally distributed. In case the values did not pass the normality test, either Kruskal-Wallis or the Mann-Whitney test was used as non-parametric tests. P-values <0.05 were considered significant. All measurements are shown as mean ± SEM.
CRediT authorship contribution statement
Laura Louzao-Martinez:Conceptualization, Methodology, Validation, Investigation, Formal analysis, Writing - original draft, Visualization, Project administration.Christian G.M. van Dijk:Conceptualization, Methodology, Validation, Investigation, Writing - review & editing.Yan Juan Xu:Investigation.Amber Korn:Investigation.Nicolaas J. Bekker:Investigation.Romi Brouwhuis:Investigation.Maria Novella Nicesse:Investigation.Jeroen A.A. Demmers:Methodology, Investigation, Resources, Writing - review & editing.Marie-José T.H. Goumans:Resources, Writing - review & editing.Rosalinde Masereeuw:Writing - review & editing.Dirk J. Duncker:Writing - review & editing, Funding acquisition.Marianne C. Verhaar:Writing - review & editing, Supervision, Funding acquisition.Caroline Cheng:Conceptualization, Validation, Resources, Supervision, Writing - review & editing, Project administration, Funding acquisition.
Declaration of Competing Interest
None.
Acknowledgements
We would like to thank the Proteomics Center of the Erasmus University Medical Center for excellent technical support.
This work was supported by the Ministry of Education, Culture and Science (Gravitation Program 024.003.013), the Netherlands Foundation for Cardiovascular Excellence [to C.C.], NWO VIDI grant [no. 91714302 to C.C.], the RM fellowship grant of the UMC Utrecht [to C.C.] and the Netherlands Cardiovascular Research Initiative: An initiative with support of the Dutch Heart Foundation [CVON2014-11 RECONNECT to C.C., M.V. and D.D.].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2019.100011.
Appendix A. Supplementary data
Supplementary material
References
- 1.Takasato M., Er P.X., Becroft M., Vanslambrouck J.M., Stanley E.G., Elefanty A.G., Little M.H. Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nature Cell Biology. 2014;16(1):118–126. doi: 10.1038/ncb2894. [DOI] [PubMed] [Google Scholar]
- 2.Takasato M., Er P.X., Chiu H.S., Maier B., Baillie G.J., Ferguson C., Parton R.G., Wolvetang E.J., Roost M.S., de Sousa Lopes S.M. Chuva, Little M.H. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature. 2015;526(7574):564–568. doi: 10.1038/nature15695. [DOI] [PubMed] [Google Scholar]
- 3.Guimaraes-Souza N.K., Yamaleyeva L.M., AbouShwareb T., Atala A., Yoo J.J. In vitro reconstitution of human kidney structures for renal cell therapy. Nephrology, Dialysis, Transplantation. 2012;27(8):3082–3090. doi: 10.1093/ndt/gfr785. [DOI] [PubMed] [Google Scholar]
- 4.Flaim C.J., Chien S., Bhatia S.N. An extracellular matrix microarray for probing cellular differentiation. Nature Methods. 2005;2(2):119–125. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 5.Plotnikov S.V., Pasapera A.M., Sabass B., Waterman C.M. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151(7):1513–1527. doi: 10.1016/j.cell.2012.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vorotnikova E., McIntosh D., Dewilde A., Zhang J., Reing J.E., Zhang L., Cordero K., Bedelbaeva K., Gourevitch D., Heber-Katz E., Badylak S.F., Braunhut S.J. Extracellular matrix-derived products modulate endothelial and progenitor cell migration and proliferation in vitro and stimulate regenerative healing in vivo. Matrix Biology. 2010;29(8):690–700. doi: 10.1016/j.matbio.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 7.van Genderen A.M., Jansen J., Cheng C., Vermonden T., Masereeuw R. Renal tubular- and vascular basement membranes and their mimicry in engineering vascularized kidney tubules. Advanced Healthcare Materials. 2018;7(19) doi: 10.1002/adhm.201800529. [DOI] [PubMed] [Google Scholar]
- 8.Naba A., Clauser K.R., Ding H., Whittaker C.A., Carr S.A., Hynes R.O. The extracellular matrix: tools and insights for the "omics" era. Matrix Biology. 2016;49:10–24. doi: 10.1016/j.matbio.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., Hynes R.O. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Molecular & Cellular Proteomics. 2012;11(4) doi: 10.1074/mcp.M111.014647. (M111.014647) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lennon R., Byron A., Humphries J.D., Randles M.J., Carisey A., Murphy S., Knight D., Brenchley P.E., Zent R., Humphries M.J. Global analysis reveals the complexity of the human glomerular extracellular matrix. Journal of the American Society of Nephrology: JASN. 2014;25(5):939–951. doi: 10.1681/ASN.2013030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshida Y., Miyazaki K., Kamiie J., Sato M., Okuizumi S., Kenmochi A., Kamijo K., Nabetani T., Tsugita A., Xu B., Zhang Y., Yaoita E., Osawa T., Yamamoto T. Two-dimensional electrophoretic profiling of normal human kidney glomerulus proteome and construction of an extensible markup language (XML)-based database. Proteomics. 2005;5(4):1083–1096. doi: 10.1002/pmic.200401075. [DOI] [PubMed] [Google Scholar]
- 12.Cui Z., Yoshida Y., Xu B., Zhang Y., Nameta M., Magdeldin S., Makiguchi T., Ikoma T., Fujinaka H., Yaoita E., Yamamoto T. Profiling and annotation of human kidney glomerulus proteome. Proteome Science. 2013;11(1):13. doi: 10.1186/1477-5956-11-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byron A., Randles M.J., Humphries J.D., Mironov A., Hamidi H., Harris S., Mathieson P.W., Saleem M.A., Satchell S.C., Zent R., Humphries M.J., Lennon R. Glomerular cell cross-talk influences composition and assembly of extracellular matrix. Journal of the American Society of Nephrology: JASN. 2014;25(5):953–966. doi: 10.1681/ASN.2013070795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Virtanen I., Laitinen L., Korhonen M. Differential expression of laminin polypeptides in developing and adult human kidney. The Journal of Histochemistry and Cytochemistry: Official Journal of the Histochemistry Society. 1995;43(6):621–628. doi: 10.1177/43.6.7769233. [DOI] [PubMed] [Google Scholar]
- 15.Guinobert I., Viltard M., Piquemal D., Elalouf J.M., Marti J., Lelievre-Pegorier M. Identification of differentially expressed genes between fetal and adult mouse kidney: candidate gene in kidney development. Nephron. Physiology. 2006;102(3–4):p81–p91. doi: 10.1159/000090054. [DOI] [PubMed] [Google Scholar]
- 16.Upadhyay K.K., Silverstein D.M. Renal development: a complex process dependent on inductive interaction. Current Pediatric Reviews. 2014;10(2):107–114. doi: 10.2174/157339631002140513101950. [DOI] [PubMed] [Google Scholar]
- 17.Wang P., Chen Y., Yong J., Cui Y., Wang R., Wen L., Qiao J., Tang F. Dissecting the global dynamic molecular profiles of human fetal kidney development by single-cell RNA sequencing. Cell Reports. 2018;24(13):3554–3567.e3. doi: 10.1016/j.celrep.2018.08.056. [DOI] [PubMed] [Google Scholar]
- 18.Bressan G.M., Daga-Gordini D., Colombatti A., Castellani I., Marigo V., Volpin D. Emilin, a component of elastic fibers preferentially located at the elastin-microfibrils interface. The Journal of Cell Biology. 1993;121(1):201–212. doi: 10.1083/jcb.121.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Colombatti A., Spessotto P., Doliana R., Mongiat M., Bressan G.M., Esposito G. The EMILIN/multimerin family. Frontiers in Immunology. 2011;2:93. doi: 10.3389/fimmu.2011.00093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Verdone G., Corazza A., Colebrooke S.A., Cicero D., Eliseo T., Boyd J., Doliana R., Fogolari F., Viglino P., Colombatti A., Campbell I.D., Esposito G. NMR-based homology model for the solution structure of the C-terminal globular domain of EMILIN1. Journal of Biomolecular NMR. 2009;43(2):79–96. doi: 10.1007/s10858-008-9290-y. [DOI] [PubMed] [Google Scholar]
- 21.Verdone G., Doliana R., Corazza A., Colebrooke S.A., Spessotto P., Bot S., Bucciotti F., Capuano A., Silvestri A., Viglino P., Campbell I.D., Colombatti A., Esposito G. The solution structure of EMILIN1 globular C1q domain reveals a disordered insertion necessary for interaction with the alpha4beta1 integrin. The Journal of Biological Chemistry. 2008;283(27):18947–18956. doi: 10.1074/jbc.M801085200. [DOI] [PubMed] [Google Scholar]
- 22.Danussi C., Petrucco A., Wassermann B., Pivetta E., Modica T.M., Del Bel Belluz L., Colombatti A., Spessotto P. EMILIN1-alpha4/alpha9 integrin interaction inhibits dermal fibroblast and keratinocyte proliferation. The Journal of Cell Biology. 2011;195(1):131–145. doi: 10.1083/jcb.201008013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danussi C., Del Bel Belluz L., Pivetta E., Modica T.M., Muro A., Wassermann B., Doliana R., Sabatelli P., Colombatti A., Spessotto P. EMILIN1/alpha9beta1 integrin interaction is crucial in lymphatic valve formation and maintenance. Molecular and Cellular Biology. 2013;33(22):4381–4394. doi: 10.1128/MCB.00872-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spessotto P., Cervi M., Mucignat M.T., Mungiguerra G., Sartoretto I., Doliana R., Colombatti A. beta 1 Integrin-dependent cell adhesion to EMILIN-1 is mediated by the gC1q domain. Journal of Biological Chemistry. 2003;278(8):6160–6167. doi: 10.1074/jbc.M208322200. [DOI] [PubMed] [Google Scholar]
- 25.Zanetti M., Braghetta P., Sabatelli P., Mura I., Doliana R., Colombatti A., Volpin D., Bonaldo P., Bressan G.M. EMILIN-1 deficiency induces elastogenesis and vascular cell defects. Molecular and Cellular Biology. 2004;24(2):638–650. doi: 10.1128/MCB.24.2.638-650.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Danussi C., Spessotto P., Petrucco A., Wassermann B., Sabatelli P., Montesi M., Doliana R., Bressan G.M., Colombatti A. Emilin1 deficiency causes structural and functional defects of lymphatic vasculature. Molecular and Cellular Biology. 2008;28(12):4026–4039. doi: 10.1128/MCB.02062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raman M., Cobb M.H. TGF-beta regulation by Emilin1: new links in the etiology of hypertension. Cell. 2006;124(5):893–895. doi: 10.1016/j.cell.2006.02.031. [DOI] [PubMed] [Google Scholar]
- 28.Zacchigna L., Vecchione C., Notte A., Cordenonsi M., Dupont S., Maretto S., Cifelli G., Ferrari A., Maffei A., Fabbro C., Braghetta P., Marino G., Selvetella G., Aretini A., Colonnese C., Bettarini U., Russo G., Soligo S., Adorno M., Bonaldo P., Volpin D., Piccolo S., Lembo G., Bressan G.M. Emilin1 links TGF-beta maturation to blood pressure homeostasis. Cell. 2006;124(5):929–942. doi: 10.1016/j.cell.2005.12.035. [DOI] [PubMed] [Google Scholar]
- 29.Litteri G., Carnevale D., D'Urso A., Cifelli G., Braghetta P., Damato A., Bizzotto D., Landolfi A., Ros F.D., Sabatelli P., Facchinello N., Maffei A., Volpin D., Colombatti A., Bressan G.M., Lembo G. Vascular smooth muscle Emilin-1 is a regulator of arteriolar myogenic response and blood pressure. Arteriosclerosis, Thrombosis, and Vascular Biology. 2012;32(9):2178–2184. doi: 10.1161/ATVBAHA.112.254664. [DOI] [PubMed] [Google Scholar]
- 30.Kielty C.M., Sherratt M.J., Shuttleworth C.A. Elastic fibres. Journal of Cell Science. 2002;115(Pt 14):2817–2828. doi: 10.1242/jcs.115.14.2817. [DOI] [PubMed] [Google Scholar]
- 31.Prewitz M.C., Seib F.P., von Bonin M., Friedrichs J., Stissel A., Niehage C., Muller K., Anastassiadis K., Waskow C., Hoflack B., Bornhauser M., Werner C. Tightly anchored tissue-mimetic matrices as instructive stem cell microenvironments. Nature Methods. 2013;10(8):788–794. doi: 10.1038/nmeth.2523. [DOI] [PubMed] [Google Scholar]
- 32.Labit H., Goldar A., Guilbaud G., Douarche C., Hyrien O., Marheineke K. A simple and optimized method of producing silanized surfaces for FISH and replication mapping on combed DNA fibers. BioTechniques. 2008;45(6) doi: 10.2144/000113002. (649-52, 654, 656-8) [DOI] [PubMed] [Google Scholar]
- 33.Pompe T., Salchert K., Alberti K., Zandstra P., Werner C. Immobilization of growth factors on solid supports for the modulation of stem cell fate. Nature Protocols. 2010;5(6):1042–1050. doi: 10.1038/nprot.2010.70. [DOI] [PubMed] [Google Scholar]
- 34.Munjal C., Opoka A.M., Osinska H., James J.F., Bressan G.M., Hinton R.B. TGF-beta mediates early angiogenesis and latent fibrosis in an Emilin1-deficient mouse model of aortic valve disease. Disease Models & Mechanisms. 2014;7(8):987–996. doi: 10.1242/dmm.015255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angel P.M., Narmoneva D.A., Sewell-Loftin M.K., Munjal C., Dupuis L., Landis B.J., Jegga A., Kern C.B., Merryman W.D., Baldwin H.S., Bressan G.M., Hinton R.B. Proteomic alterations associated with biomechanical dysfunction are early processes in the Emilin1 deficient mouse model of aortic valve disease. Annals of Biomedical Engineering. 2017;45(11):2548–2562. doi: 10.1007/s10439-017-1899-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spessotto P., Bulla R., Danussi C., Radillo O., Cervi M., Monami G., Bossi F., Tedesco F., Doliana R., Colombatti A. EMILIN1 represents a major stromal element determining human trophoblast invasion of the uterine wall. Journal of Cell Science. 2006;119(Pt 21):4574–4584. doi: 10.1242/jcs.03232. [DOI] [PubMed] [Google Scholar]
- 37.Parsons J.T., Horwitz A.R., Schwartz M.A. Cell adhesion: integrating cytoskeletal dynamics and cellular tension. Nature Reviews. Molecular Cell Biology. 2010;11(9):633–643. doi: 10.1038/nrm2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokuda S., Higashi T., Furuse M. ZO-1 knockout by TALEN-mediated gene targeting in MDCK cells: involvement of ZO-1 in the regulation of cytoskeleton and cell shape. PLoS One. 2014;9(8) doi: 10.1371/journal.pone.0104994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itoh M., Nagafuchi A., Moroi S., Tsukita S. Involvement of ZO-1 in cadherin-based cell adhesion through its direct binding to alpha catenin and actin filaments. The Journal of Cell Biology. 1997;138(1):181–192. doi: 10.1083/jcb.138.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Odenwald M.A., Choi W., Buckley A., Shashikanth N., Joseph N.E., Wang Y., Warren M.H., Buschmann M.M., Pavlyuk R., Hildebrand J., Margolis B., Fanning A.S., Turner J.R. ZO-1 interactions with F-actin and occludin direct epithelial polarization and single lumen specification in 3D culture. Journal of Cell Science. 2017;130(1):243–259. doi: 10.1242/jcs.188185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreidberg J.A., Symons J.M. Integrins in kidney development, function, and disease. American Journal of Physiology. Renal Physiology. 2000;279(2):F233–F242. doi: 10.1152/ajprenal.2000.279.2.F233. [DOI] [PubMed] [Google Scholar]
- 42.Mathew S., Chen X., Pozzi A., Zent R. Integrins in renal development. Pediatric Nephrology. 2012;27(6):891–900. doi: 10.1007/s00467-011-1890-1. [DOI] [PubMed] [Google Scholar]
- 43.Colombatti A., Doliana R., Bot S., Canton A., Mongiat M., Mungiguerra G., Paron-Cilli S., Spessotto P. The EMILIN protein family. Matrix Biology. 2000;19(4):289–301. doi: 10.1016/s0945-053x(00)00074-3. [DOI] [PubMed] [Google Scholar]
- 44.Palecek S.P., Loftus J.C., Ginsberg M.H., Lauffenburger D.A., Horwitz A.F. Integrin-ligand binding properties govern cell migration speed through cell-substratum adhesiveness. Nature. 1997;385(6616):537–540. doi: 10.1038/385537a0. [DOI] [PubMed] [Google Scholar]
- 45.Couchman J.R., Rees D.A. The behaviour of fibroblasts migrating from chick heart explants: changes in adhesion, locomotion and growth, and in the distribution of actomyosin and fibronectin. Journal of Cell Science. 1979;39:149–165. doi: 10.1242/jcs.39.1.149. [DOI] [PubMed] [Google Scholar]
- 46.Hynes R.O. Integrins: bidirectional, allosteric signaling machines. Cell. 2002;110(6):673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 47.Laukaitis C.M., Webb D.J., Donais K., Horwitz A.F. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. The Journal of Cell Biology. 2001;153(7):1427–1440. doi: 10.1083/jcb.153.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ryan M.J., Johnson G., Kirk J., Fuerstenberg S.M., Zager R.A., Torok-Storb B. HK-2: an immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney International. 1994;45(1):48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 49.Wilm M., Shevchenko A., Houthaeve T., Breit S., Schweigerer L., Fotsis T., Mann M. Femtomole sequencing of proteins from polyacrylamide gels by nano-electrospray mass spectrometry. Nature. 1996;379(6564):466–469. doi: 10.1038/379466a0. [DOI] [PubMed] [Google Scholar]
- 50.Sap K.A., Bezstarosti K., Dekkers D.H.W., Voets O., Demmers J.A.A. Quantitative proteomics reveals extensive changes in the ubiquitinome after perturbation of the proteasome by targeted dsRNA-mediated subunit knockdown in Drosophila. Journal of Proteome Research. 2017;16(8):2848–2862. doi: 10.1021/acs.jproteome.7b00156. [DOI] [PubMed] [Google Scholar]
- 51.Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nature Biotechnology. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material