Abstract
The microenvironment contributes to the excessive connective tissue deposition that characterizes fibrosis. Members of the CCN family of matricellular proteins are secreted by fibroblasts into the fibrotic microenvironment; however, the role of endogenous CCN1 in skin fibrosis is unknown. Mice harboring a fibroblast-specific deletion for CCN1 were used to assess if CCN1 contributes to dermal homeostasis, wound healing, and skin fibrosis. Mice with a fibroblast-specific CCN1 deletion showed progressive skin thinning and reduced accumulation of type I collagen; however, the overall mechanical property of skin (Young's modulus) was not significantly reduced. Real time-polymerase chain reaction analysis revealed that CCN1-deficient skin displayed reduced expression of mRNAs encoding enzymes that promote collagen stability (including prolyl-4-hydroxylase and PLOD2), although expression of COL1A1 mRNA was unaltered. CCN1-deficent skin showed reduced hydroxyproline levels. Electron microscopy revealed that collagen fibers were disorganized in CCN1-deficient skin. CCN1-deficient mice were resistant to bleomycin-induced skin fibrosis, as visualized by reduced collagen accumulation and skin thickness suggesting that deposition/accumulation of collagen is impaired in the absence of CCN1. Conversely, CCN1-deficient mice showed unaltered wound closure kinetics, suggesting de novo collagen production in response to injury did not require CCN1. In response to either wounding or bleomycin, induction of α-smooth muscle actin-positive myofibroblasts was unaffected by loss of CCN1. CCN1 protein was overexpressed by dermal fibroblasts isolated from lesional (i.e., fibrotic) areas of patients with early onset diffuse scleroderma. Thus, CCN1 expression by fibroblasts, being essential for skin fibrosis, is a viable anti-fibrotic target.
Keywords: Fibrosis, CCN family, CCN1, cyr61, Matricellular proteins, Connective tissue, Dermis, Scleroderma
Highlights
-
•
The role of endogenous CCN1 in skin biology is largely unknown
-
•
Fibroblast-specific deletion CCN1 causes thinner skin and misaligned collagen
-
•
CCN1-deficient mice were resistant to bleomycin-induced skin fibrosis
-
•
Wound healing closure kinetics was unaffected by loss of CCN1
-
•
CCN1 may be as a target for anti-fibrotic therapy
Introduction
Fibrotic diseases, including diffuse cutaneous systemic sclerosis (dcSSc), are characterized by excessive deposition of extracellular matrix (ECM) and account for ~45% of the deaths and health care costs in the developed world [1,2]. Targeting proteins secreted into the microenvironment may represent novel anti-fibrotic therapeutic strategies [3,4].
The CCN family of matricellular proteins, secreted by fibroblasts into the microenvironment, are potential anti-fibrotic targets [5,6]. In vitro, CCN family members have minimal activity, but function as proadhesive signaling modifiers [7]. In vivo, CCN proteins act as a “centralized coordination network” and, by interacting with extracellular ligands, integrate signals emanating from multiple sources to affect organ development, homeostasis and disease [7]. Binding sites for integrins, heparin sulfate proteoglycans, TrkA, fibronectin and other proteins decorate CCN molecules [[7], [8], [9]]. Thus, although examining the effect of mutations impairing the ability of individual CCN proteins to interact with individual binding partners is informative [10], elucidating the function of CCN proteins requires using knockout animal models.
CCN2 (connective tissue growth factor) was proposed as a mediator of fibrosis ~20 years ago [11]. Conditional knockout and neutralizing antibody approaches have shown that CCN2 contributes to fibrosis [[12], [13], [14], [15]]. CCN2 promotes the differentiation of pericyte-like progenitor cells into myofibroblasts, a feature that appears to contribute to fibrosis but not to cutaneous tissue repair [12,16]. Importantly, an anti-CCN2 strategy (FG-3019) is under clinical evaluation as a treatment for idiopathic pulmonary fibrosis [17,18].
After CCN2, CCN1 (cyr61) is the second most-studied CCN family member. Both CCN1 and CCN2 are induced in fibroblasts by TGFβ1 via ALK5/NOX1/4, and are upregulated in response to mechanotransduction by the Hippo/YAP pathway [[19], [20], [21], [22]]. CCN1, like all CCN family members, is composed of an N–terminal signal peptide followed by 4 conserved structural domains, namely: the insulin-like growth factor binding protein (IGFBP) domain, the von Willebrand factor type C (VWC) domain, the thrombospondin type 1 (TSP1) domain, and the C–terminal (CT) heparin-binding domain [6,7]. Mice with a global CCN1 knockout die in utero due to severe angiogenic defects [23]; however, the role of intact, endogenous CCN1 protein postnatally is almost wholly unknown. To begin to address this gap in our knowledge, herein we use a conditional knockout strategy to delete CCN1 specifically in fibroblasts postnatally [24]. Our data generate novel insights into the role of CCN1 in skin homeostasis, repair and fibrosis and also suggest CCN1 may be an anti-fibrotic target.
Results
Loss of CCN1 expression by fibroblasts results in reduced and misaligned collagen accumulation in skin
To assess the role of CCN1 in dermal fibroblasts, we used a conditional knockout strategy, employing mice expressing a tamoxifen-dependent cre recombinase expressed under the control of a fibroblast-specific Col1A2 promoter/enhancer. We have previously used these mice to delete genes specifically in fibroblasts [12,24,25,29]. Three weeks postnatally, ccn1f/f mice hemizygous for COL1A2-CRE(ER)T were treated with tamoxifen or corn oil to generate mice deleted or not for CCN1 in fibroblasts (as described in Methods, these mice are subsequently described as CCN1−/− or CCN1f/f, respectively). Deletion of CCN1 was verified by genotyping, extraction of dermal fibroblasts for qPCR analysis and subjecting dermal fibroblasts treated with or without TGFβ1, which induces CCN1 protein expression [19] to Western blot analysis with an anti-CCN1 antibody (Fig. 1A–C). ~4 weeks post-deletion, CCN1-deficient skin began to show macroscopic evidence of thinning (Fig. 1D); indeed, histological analysis of skin tissue verified that loss of CCN1 resulted in thinner skin (p = 0.0005) (Fig. 1E). However, intriguingly, loss of CCN1 from fibroblasts did not impair the mechanical properties (Young's modulus) of skin; in fact, the Young's modulus of skin was somewhat increased in the absence of CCN1 (Fig. 1F). Analysis of tissue sections stained with DAPI revealed that loss of CCN1 caused an increase (p = 0.0013) in the number of cells per unit area (Fig. 1G). Conversely, proliferation of fibroblasts, as assessed using an anti-PCNA antibody, and apoptosis (as observed by TUNEL) was not impaired in CCN1-deficient skin (Fig. 1H, I). These data suggest that the phenotype of CCN1-deficent skin resulted from decreased stability/impaired organization of ECM acausing a compressed dermis.
Fig. 1.
Loss of CCN1 in fibroblasts in mice leads to decreased skin thickness. A) Genotyping showing loss of CCN1 expression in fibroblasts two weeks post-tamoxifen injections. Fibroblasts extracted from skin of mice show loss of CCN1 as revealed by analysis of B) mRNA (P = 0.0122; Graph shows mean fold change +/− SD; Student t-test; n = 6); and C) protein (representative image, n = 3) expression. D) In mice, 4 weeks post-injection of tamoxifen (i.e., at 8 weeks of age), loss of CCN1 results in translucent skin. (Representative image; n = 4). E) Dermal layer of skin is significantly thinner (p = 0.0005) in CCN1−/− mice as revealed by H&E staining. (Graph shows mean dermal thickness +/− SD; Student's t-test; n = 4). F) There is an increase in Young's modulus (stiffness) that is increased (p = 0.0354, n = 4) in the absence of CCN1 but is unlikely to be of biological significance and G) more nuclei present (p = 0.0013) per unit area of skin in CCN1−/−, compared to CCN1f/f, mice as shown by DAPI staining of. (Representative images shown; Graph shows mean cells per 120,000 μm2 +/− SD; Student's t-test; n = 5). H) No alterations were observed in the proliferation marker PCNA. (Representative images shown; Graph represents mean percentage of cells PCNA positive +/− SD; Student's t-test; n = 5). I) Tunel assay showed no alteration in apoptosis. (Representative images shown; n = 6).
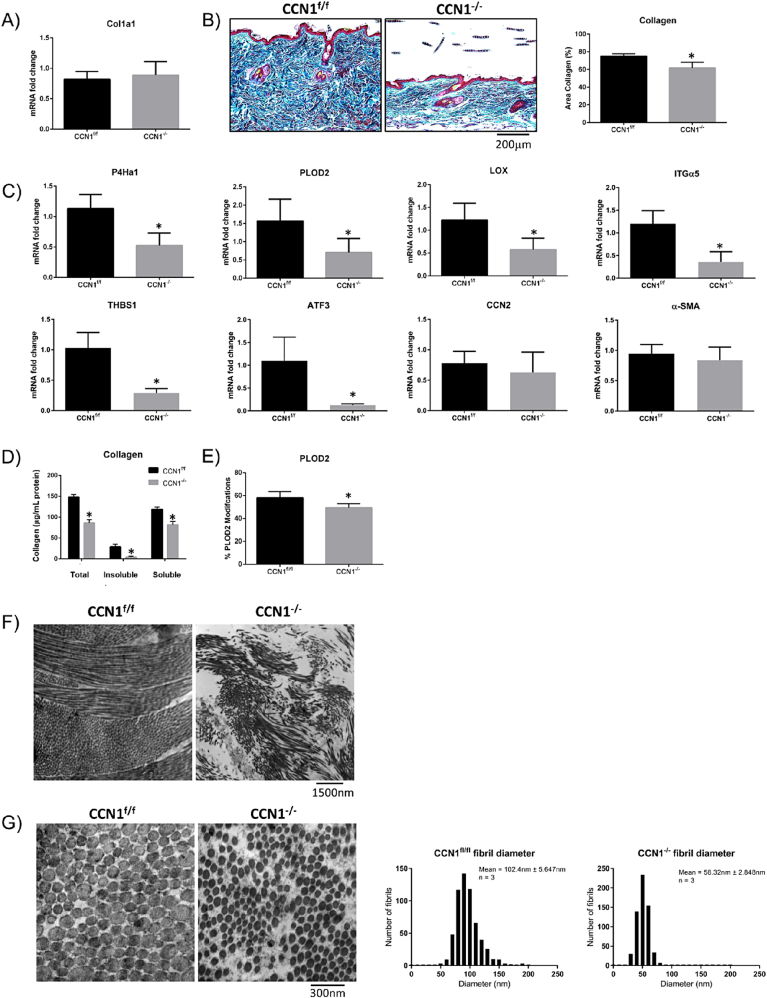
Our data suggested that the skin thinning phenotype in CCN1-deficient skin occurred due to impaired collagen accumulation. Although real-time PCR (RTPCR) analysis of RNA extracted from total skin revealed that COL1A1 mRNA was not reduced in the CCN2-deficient skin; trichrome staining of tissue sections from control and CCN1-deficient skin revealed less collagen accumulation in the mice deleted for CCN1 in fibroblasts (Fig. 2A, B). RTPCR analysis of RNA from CCN1-deficient and control skin revealed reduced expression of mRNAs encoding genes involved with collagen synthesis/stability including: PLOD2, lysyl oxidase (LOX) and prolyl-4-hydroxylase (P4H) [[30], [31], [32], [33]] (Fig. 2C). Supporting these data, analysis of protein extracted from CCN1-deficient and control skin revealed evidence of reduced collagen accumulation, as determined by hydroxyproline content (Fig. 2D). Mass spectrophotometry detected a decreased abundance of PLOD2-modified lysine residues (i.e. PLOD2-generated hydroxylysine residues) in type I collagen-derived tryptic peptides isolated from CCN1-deficient skin (Fig. 2E). Examples of deconvoluted mass spectrophotometry data showing examples of PLOD2-modified or unmodified lysine residues of Col1a2 are shown in Supplemental Fig. 1. Closer examination, using electron microscopy, showed disorganization of collagen matrix in CCN1-deficient skin indicating impaired collagen alignment (Fig. 2F). The fibril diameter was also significantly smaller (p = 0.0022) with larger spaces between collagen fibrils (Fig. 2G), further supporting that collagen accumulation/stability was impaired in the absence of CCN1 expression by fibroblasts.
Fig. 2.
Loss of CCN1 from fibroblasts resulted in decreased in collagen stability, but no difference in collagen mRNA expression. A) RNA from skin tissue showed no changes in collagen type I mRNA expression. (Graph represents mean fold change +/− SD; Student's t-test; n = 4). B) Skin tissue showed a significant decrease in collagen (p = 0.0014) shown by trichrome staining. (Representative images shown; Graph shows mean area of collagen present in images +/− SD; Student's t-test; n = 4). C) Skin tissue from CCN1-deficient skin showed a significant decrease in expression of mRNAs encoding cross-linking enzymes P4Ha1 (P = 0.0009), PLOD2 (P = 0.0115) and LOX (P = 0.0046), and mRNAs encoding the ECM-promoting proteins integrin alpha5 (ITGα5, p = 0.0002), thrombospondin-1 (THBS1, p < 0.0001) and ATF3 (p = 0.0325). There were no changes observed in the fibrotic markers CCN2 and α-smooth muscle actin (α-SMA) in CCN1-deficient skin tissue. (Graphs represent mean mRNA fold change +/− SD; Student's t-test; n = 4). Note: total skin tissue was examined. D) Significantly less total (p > 0.0001), insoluble (p = 0.0003) and soluble (p = 0.0003) collagen were present in the skin of mice lacking CCN1 as shown by a hydroxyproline assay. (Graph represents mean collagen content (ug/mg) +/− SD; Student's t-tests; n = 4). E) Proteomics using mass spectrometry revealed a significant decrease in PLOD2 (p = 0.0364)-generated collagen crosslinks in tissue of CCN1−/− mice. (Graph represents mean modifications +/− SD; Student's t-test; n = 4). F.G) electron microscopy of sections of CCN1f/f and CCN1−/− skin. Images show a significant decrease (p = 0.0022) in collagen fibril diameter in the CCN1−/− mouse dermis. Inset shows mean fibril diameter +/− SD; Student's t-test (n = 3).
Further indication that loss of CCN1 expression by skin resulted in impaired ECM organization was provided by genome-wide expression profiling of CCN1-deficient and control skin (Table 1, Supplemental data). We found decreased mRNA expression of genes encoding proteins involving cell adhesion to extracellular matrix and transcription factors involved with ECM gene expression, notably those of the AP-1 gene family. We used real-time PCR analysis to confirm that integrin alpha 5, thrombospondin-1 and activating transcription factor (ATF-3) mRNA expression was reduced in CCN1-deficient skin (Fig. 2C). Integrin alpha 5 is an integrin subunit that binds fibronectin, thrombospondin-1 promotes ECM homeostasis, and ATF3 enhances the profibrotic effects of TGFbeta and contributes to bleomycin-induced skin fibrosis [[34], [35], [36]]. Of note, neither CCN2 nor alpha-smooth muscle actin (αSMA, ACTA2) mRNA expression was reduced in CCN1-deficient skin (Fig. 2C).
Table 1.
Genome-wide expression profiling of RNA from skin of CCN1-deficient and control skin reveals that clusters involved with cell migration, adhesion and transcription are affected by loss of CCN1 (fold change reflects decrease in expression in response to loss of CCN1). For entire lists of mRNAs affected greater or equal to 1.5-fold by loss of CCN1, please see Supplemental information.
| ID | Gene name | Fold change |
|---|---|---|
| Cell migration cluster | ||
| NM_010415 | Heparin-binding EGF-like growth factor | −2.2867 |
| NM_001139509 | Nuclear receptor subfamily 4, group A, member 2 | −1.92135 |
| NM_013749 | Tumor necrosis factor receptor superfamily, member 12a | −1.8584 |
| Cell adhesion cluster | ||
| NM_133743 | Ly6/Plaur domain containing 3 | −1.52012 |
| NM_010516 | Cysteine rich protein 61 | −1.60192 |
| NM_053110 | Glycoprotein (transmembrane) nmb | −1.60465 |
| NM_010577 | Integrin alpha 5 (fibronectin receptor alpha) | −1.57323 |
| NM_001081445 | Neural cell adhesion molecule 1 | −1.53712 |
| NM_009398 | Similar to TNF-stimulated gene 6 protein; tumor necrosis factor alpha induced protein 6 | −2.20598 |
| NM_011580 | Thrombospondin 1; similar to thrombospondin 1 | −2.18805 |
| NM_013749 | Tumor necrosis factor receptor superfamily, member 12a | −1.8584 |
| Regulation of transcription cluster | ||
| NM_008036 | FBJ osteosarcoma oncogene B | −2.38818 |
| NM_007498 | Activating transcription factor 3 | −2.07292 |
| NM_153287 | Cysteine-serine-rich nuclear protein 1 | −1.72143 |
| NM_007950 | Epiregulin | −2.12905 |
| NM_010235 | Fos-like antigen 1 | −2.6796 |
| NM_010212 | Four and a half LIM domains 2 | −1.5412 |
| NM_010849 | Myelocytomatosis oncogene | −1.58666 |
| NM_008719 | Neuronal PAS domain protein 2 | −1.64826 |
| NM_010444 | Nuclear receptor subfamily 4, group A, member 1 | −2.583 |
| NM_001139509 | Nuclear receptor subfamily 4, group A, member 2 | −1.92135 |
| NM_027787 | Regulatory factor X, 2 (influences HLA class II expression) | −2.14973 |
| NM_017373 | Similar to NFIL3/E4BP4 transcription factor; nuclear factor, interleukin 3, regulated | −1.66494 |
Loss of CCN1 expression results in resistance to bleomycin-induced skin fibrosis.
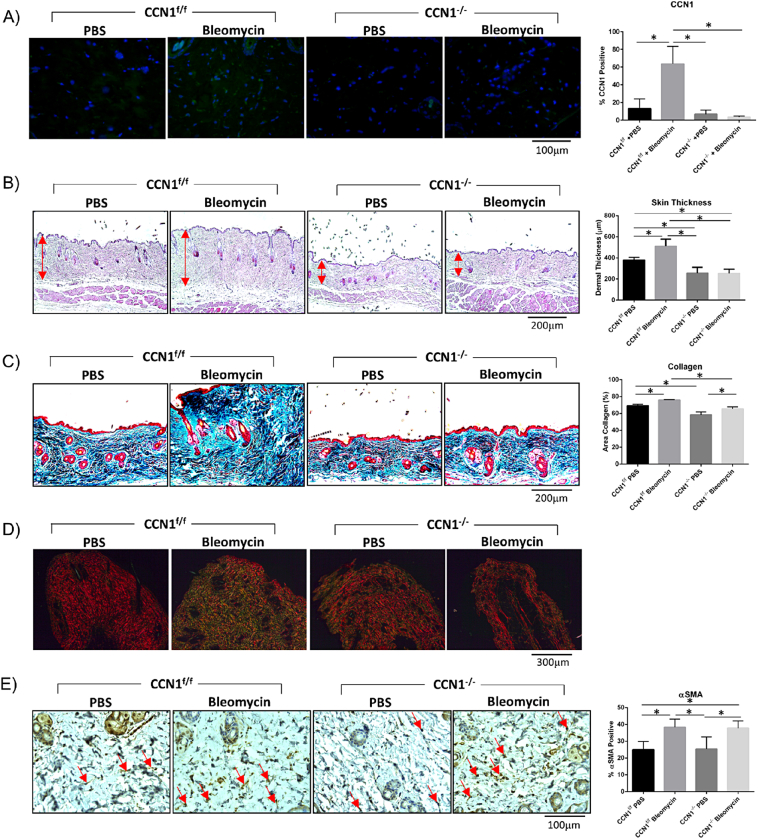
As our preliminary data suggested that accumulation/stability of collagen is impaired in mice deleted for CCN1 in fibroblasts, we evaluated whether CCN1 expression by fibroblasts was required for skin fibrogenesis. To test this hypothesis, we subjected mice, deleted or not for CCN1 in fibroblasts, to the bleomycin model of skin fibrosis. Two weeks post-deletion, mice were injected every day for 28 days with either the pro-inflammatory agent bleomycin, to induce fibrosis, or vehicle (PBS) control. We found that CCN1 protein was induced in response to bleomycin in control, but not CCN1-deficient, mice (Fig. 3A). [Our detection methods were insufficiently sensitive to detect basal CCN1 protein expression in skin, presumably also reflecting that, despite its contribution to aspects of dermal homeostasis (Fig. 1, Fig. 2), basal expression of CCN1 is minimal in skin.] Although, in control mice, bleomycin caused an increase in skin thickness and collagen accumulation (as visualized by hematoxylin and eosin staining and Trichrome staining, respectively), mice deficient in CCN1 were relatively resistant to the profibrotic effects of bleomycin (Fig. 3B, C). Similar results were obtained using Sirius red staining of tissue sections (Fig. 3D). Using this method, in polarized light, the thickest collagen fibers appear yellow/orange, whereas newly formed fibers are green. We found that accumulation of both thick and newly formed fibers in response to bleomycin was essentially absent in CCN1-deficient mice compared to control mice.
Fig. 3.
Loss of CCN1 impaired induction of collagen production and skin thickness in response to bleomycin-induced skin fibrosis. However, myofibroblasts differentiation still occurred in CCN1-deficient skin. A) An increase in CCN1 protein expression was observed in skin sections of wild-type mice subjected to bleomycin-induced skin fibrosis (p < 0.0001); CCN1−/− mice showed no CCN1 staining, as observed by staining with an anti-CCN1 antibody, as described in Methods. (Representative images shown; Graphs represent mean CCN1 positive cells +/− SD; one-way ANOVA; n = 5). B) Bleomycin-induced skin thickness (p = 0.0042) is not observed in CCN1−/− mice. Skin is significantly thinner in PBS CCN1−/− (p = 0.0175) and Bleomycin CCN1−/− (p = 0.0082) compared to PBS CCN1f/f. (Representative images shown; Graph shows mean skin thickness +/-SD; one-way ANOVA; n = 4). C) Trichrome images show CCN1−/− mice do not get an over-production of collagen in response to bleomycin induced fibrosis (p = 0.0228). (Representative images shown; Graph represents mean area collagen in images +/− SD; one-way ANOVA; n = 4). D) Birefringence analyses of picroSirius stained skin sections indicate that CCN1−/− does not display the excessive collagen fiber size and density seen in regular bleomycin-fibrosis. (Representative images shown; n = 6). E) Dermal increase in αSMA (p = 0.0037) seen in bleomycin-induced fibrosis is still observed in CCN1−/− dermis (p = 0.0071), compared to control PBS mice. (Representative images shown; Graphs represent mean αSMA positive cells +/− SD; one-way ANOVA; n = 5).
Intriguingly, myofibroblast induction was observed in both control and CCN1-deficient mice (Fig. 3E), suggesting that CCN1 was essential for collagen synthesis and deposition in response to bleomycin, but not for myofibroblast differentiation.
Loss of CCN1 expression by fibroblasts does not impair cutaneous wound closure kinetics
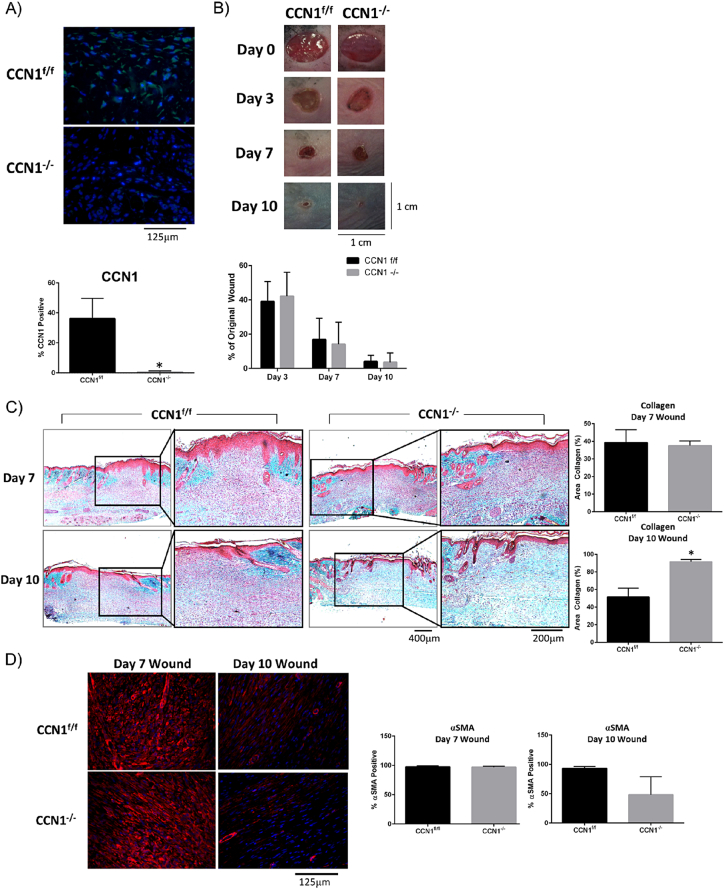
Fibrosis has been considered in the past to represent a persistent tissue repair program. To investigate this issue and to test if CCN1 might represent a specific antifibrotic target, we assessed if CCN1 was also required for cutaneous tissue repair, which involves the de novo synthesis of collagen in response to injury. To test if CCN1 was required for de novo collagen protein synthesis in response to wounding and if the mechanisms underlying normal tissue repair and fibrosis might differ in their requirements for endogenous CCN1, we assessed if loss of CCN1 expression by fibroblasts was impaired cutaneous tissue repair. To test this hypothesis, we subjected mice deleted or not for CCN1 to the dermal punch model of wounding. Staining with anti-CCN1 antibody revealed induction of CCN1 in wounds of control, but not CCN1-deficient, mice (Fig. 4A). Morphometric analysis revealed that wound closure kinetics was not impaired in CCN1-deficient skin (Fig. 4B). Intriguingly, trichrome staining revealed that de novo collagen synthesis was not impaired in the CCN1-deficient wounds; in fact, wound resolution, as observed by more blue-staining collagen in day 10 wounds, appeared to be improved in the absence of CCN1 (Fig. 4C). Additionally, αSMA expression was still induced in Day 7 wounds supporting he notion that CCN1 is not responsible for myofibroblast induction (Fig. 4D). At Day 10 there was a trend toward less αSMA present in the wound bed, consistent with the idea wound resolution was enhanced in the absence of CCN1 expression by fibroblasts. These data suggest that loss of CCN1 expression by fibroblasts does not significantly impair de novo protein synthesis in response cutaneous wound healing.
Fig. 4.
Loss of CCN1 does not impair the kinetics of cutaneous wound healing. Mice were subjected to the dermal punch (6 mm) model of tissue repair. A) anti-CCN1 antibody staining confirms absence of CCN1 in fibroblasts of CCN1-deficient mice (p = 0.0008). (Representative images shown; n = 5). B) Images show no changes in the kinetics of wound closure between CCN1f/f and CCN1−/− mice. (Graphs show mean percentage of original wound size +/− SD; two-way ANOVA; n = 40 wounds). C) Trichrome staining on day 7 and day 10 wounds from CCN1f/f and CCN1−/− mice, show at day 10 significantly more (p = 0.0027) collagen deposited in the dermis in the CCN1−/− mice compared to CCN1f/f mice. (Representative images shown; Graphs represent percentage of tissue stain blue for collagen; n = 3). D) Staining of day 7 wounds with anti-αSMA antibody show no changes between CCN1f/f and CCN1−/−, however in Day 10 wounds there is a trend toward less αSMA in CCN1−/− mice. (Representative images, Graphs show mean percent of cells stained +/− SD, Student's t-test, n = 3).
CCN1 is overexpressed by dermal fibroblasts cultured from fibrotic lesions of patients with early onset (<2 years) diffuse cutaneous SSc (dcSSc)
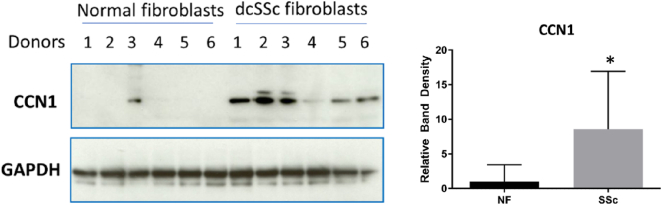
To begin to provide a clinical context for our studies, we cultured dermal fibroblasts from lesional areas of patients with early onset (<2 years) dcSSc that were diagnosed according to EULAR/ACR criteria as having skin fibrosis [37]. Protein extracts were prepared and subjected to Western blot analysis with an anti-CCN1 antibody. Protein extracts were simultaneously prepared from dermal fibroblasts isolated from age-, sex- and site-matched healthy controls. Although there was variation in protein expression among samples, we found that CCN1 protein was significantly overexpressed (p = 0.0087) in early onset dcSSc dermal fibroblasts (Fig. 5).
Fig. 5.
Early onset dcSSc dermal fibroblasts express higher levels of CCN1 compared to control dermal fibroblasts. Biopsies were taken from six patients with diffuse cutaneous systemic sclerosis (dcSSc fibroblasts) and six age- site- and sex-matched healthy controls (normal fibroblasts) (donors 1 through 6, as indicated), fibroblasts cultured and protein analysis via western blot performed. A significant (p = 0.0087) increase in CCN1 protein expression was observed in dcSSc dermal fibroblasts, compared to control dermal fibroblasts. Graph represents mean band density of CCN1 compared to GAPDH control; a Mann Whitney test was performed as protein expression among human samples is highly variable. We observed expected donor-to-donor variation in protein expression data within both normal and SSc groups. The patients all qualified for immediate classification as having SSc due to fibrosis in the forearm, as described by EULAR/ACR diagnosis criteria [37].
Collectively, our data support the notion that endogenous CCN1 expression by fibroblasts contributes to collagen stability in skin and that CCN1 may be a novel, specific anti-fibrotic target.
Discussion
The CCN family of matricellular proteins mediate interactions among cells, extracellular matrix and growth factors and cytokines, are highly spatiotemporally regulated and contribute to development, tissue homeostasis, cancers and fibrosis [[6], [7], [8], [9]]. Due to their context-dependent effects, and their ability to interact with a myriad of proteins, establishing a specific mechanism of CCN action has been elusive [[6], [7], [8], [9], [10]]. To understand the actual function of CCN molecules in vivo, the use of genetic (e.g., conditional knockout) models is essential.
The primary sources of CCN proteins in vivo are mesenchymal cells [[6], [7], [8], [9]]. CCN1 is also expressed in endothelial cells, and is induced in sites undergoing inflammation and repair [38]. In this report, we use fibroblast-specific CCN1 knockout mice to report the first detailed analysis of the role of endogenous CCN1 protein in dermal fibroblasts. CCN1 has a context-dependent role in collagen synthesis and accumulation in skin. Loss of CCN1 protein, although not appreciably affecting type I collagen mRNA expression, results in decreased mRNA expression of prolyl-4-hydroxylase, lysyl oxidase and PLOD2, which generate proteins that promote collagen crosslinking/stabilization [[29], [30], [31], [32], [33]]. Expression of these mRNAs was decreased but not abolished; collagen alignment was impaired in CCN1-deficient skin and resembled that of aged skin. This loss was insufficient to impair the basal mechanical properties of uninjured skin. Similarly, wound closure kinetics was unaltered in CCN1-deficient skin. However, trichrome staining of tissue sections of day 10 wounds revealed that wounds of CCN1-deficient mice showed increased resolution, consistent with a previous study that showed mice harboring a mutated CCN1 protein displayed faster wound resolution in granulation tissue [39]. Overall, our date revealed that loss of CCN1 expression by fibroblasts does not impair matrix stiffness or wound healing; i.e., CCN1 may represent a novel, specific anti-fibrotic target. These data also emphasize the notion that differences exist between the fundamental mechanisms underlying normal tissue repair and fibrosis.
Loss of CCN1 expression by skin fibroblasts caused resistance to bleomycin-induced skin fibrosis. Loss of CCN1, however, but did not impair myofibroblast induction, perhaps related to the observation that CCN1 was not involved with maintaining skin stiffness, a feature required for myofibroblast differentiation. Instead, loss of CCN1 impaired new collagen synthesis and remodeling in response to bleomycin. In skin, fibroblast-specific expression of endogenous CCN2 and CCN1 appear to promote different profibrotic activities; CCN2 promotes myofibroblast differentiation [17], conversely CCN1 appears to promote matrix organization/stability/alignment. Indeed, the primary role of CCN1 in vivo may be as a chaperone to help organize fibrillar collagen. Consequently, it may be useful therapeutically to block both CCN2 and CCN1, for example, by treating with the antifibrotic protein CCN3 [5]. Bleomycin-induced fibrosis is in widespread use an appropriate, initial proof-of-concept model for evaluating possible targets for antifibrotic therapy in inflammatory-driven conditions like SSc. In the future, it will be interesting to evaluate whether CCN1 is required for skin fibrosis in other models, e.g., the TSK mouse [40].
Previous experiments examining the potential role of CCN1 in fibrosis have yielded somewhat contradictory results, emphasizing the context-dependent roles of CCN proteins [[6], [7], [8], [9], [10]]. Nonetheless, the preponderance of the available data suggests that CCN1 is overexpressed in conditions of inflammation, fibrosis and injury [38]. In one study, CCN1 was found to be upregulated in humans with chronic obstructive pulmonary disease and in animal models of lung fibrosis; in this report, overexpression of CCN1 exacerbated lung injury [41]. CCN1 blockade was found to be protective against fibrosis in a model of ischemic kidney injury and in the unilateral ureter obstruction model of kidney fibrosis [42,43]. In liver fibrosis, CCN1 expression is also increased; adenovirus-mediated overexpression of CCN1 in hepatic stellate cells suppressed fibrogenic responses due to increased apoptosis caused by endoplasmic reticulum stress and an unfolded protein response [44]. It must be stated, however, that hepatocyte-specific deletion of endogenous CCN1 exacerbated liver fibrosis [45] although, given the extreme context-dependent effects of CCN proteins, it may be that endogenous CCN1 would have a different effect on experimental liver fibrosis if deleted in hepatic stellate cells.
In terms of SSc, one study used microarray analysis to show that CCN1 mRNA expression was elevated in clinically affected (fibrotic) skin [46]. Consistent with this latter report, elevated levels of CCN1 protein were seen in serum of SSc patients [47]. It is noteworthy that, in our current study, although overall expression of CCN1 protein was increased in early onset dcSSc fibroblasts, the expression of CCN1 varied among samples. Intriguingly, studies focusing on patients with later stages of SSc (i.e., not early onset patients with active fibrosis but those with severe digital ulcers) showed reduced CCN1 levels in endothelial cells and circulation of patients, consistent with the notion that CCN1 expression is reduced in avascular tissue [40,48,49]. These results reflect that CCN1 expression is controlled by many, often opposing, parameters [40]; transcription of CCN1 is directly induced in response to mechanical loading through YAP/TAZ, but is epigenetically suppressed in SSc patients with digital ulcers via fli-1 and histone deacetylase 5 [40,[48], [49], [50]]. In this regard, CCN1 expression has been shown to correlate with the induction of profibrotic inflammatory responses, in a wide variety of indications [38,42,43,47,51,52]. Thus, CCN1 protein may be induced in the early, fibrotic phase of SSc, which is characterized by the presence of mechanical tension, but may be suppressed in patients with severe vascular damage. In conditions of vascular damage, addition of exogenous CCN1 may be advantageous, due to its potent proangiogenic activity [53]. In a study, published after this manuscript was submitted for review, involving cells derived from patients with vascular disease, overexpression of CCN1 in SSc dermal fibroblasts caused apoptosis and reduced protein synthesis including that of collagen [53]; however, as discussed above, this phenomenon has been shown to arise due to an accumulation of unprocessed CCN1 resulting in an unfolded protein response [44]. That said, exogenous recombinant CCN1 has been shown to slow a fibrotic response in response to wound healing through the stress-related generation of reactive oxygen species [39].
In interpreting these studies, it is important to remember that the biological roles of CCN proteins can vary and are highly context dependent [7]. Specifically, the biological role of CCN1 may differ based on the cell type examined, the clinical stage of the disease, the levels of CCN1 protein present or, critically, whether the effects of endogenous or exogenously added CCN1 protein are examined. It is imperative to emphasize that, in our current study, we are examining the role of endogenous CCN1 expression by fibroblasts on skin biology. In any event, that CCN1 appears to have context-dependent roles suggests that caution be exercised regarding the potential future use of CCN1 as a therapeutic target and that each indication should be considered separately.
In summation, CCN1 behaves like a typical matricellular protein in that it acts in a context-dependent fashion, emphasizing the necessity of studying the role of CCN1 in as close to an in vivo context as possible. Additional work needs to be conducted to show if endogenous CCN1 contributes to pathological fibrosis in humans. Nonetheless, our data suggest that further efforts aimed at developing anti-CCN1 strategies to treat fibrosis are warranted.
Methods
Generation of fibroblast-specific CCN1 conditional knock-out mice
Mice hemizygous for a tamoxifen-dependent cre recombinase expressed under the control of a fibroblast-specific Col1a2 promoter/enhancer [Col1A2-Cre(ER)T)] that were also ccn1f/f were created and genotyped as previously described [24]. Male mice (3 weeks old) were injected intraperitoneally with tamoxifen (10 mg/mL) or corn oil (vehicle control) every day for 5 days to generate mice deleted or not for CCN1. In this manuscript, mice not deleted for ccn1 in fibroblasts are designated CCN1f/f whereas mice deleted for ccn1 are designated CCN1−/−.
Confirmation of CCN1 expression and deletion in fibroblasts
Back skin from CCN1f/f and CCN1−/− mice was placed in 10% FBS/DMEM with 2 mg/ml collagenase type II (3 h, 37 °C). Connective tissue was removed and placed into 10% FBS/DMEM. Cell debris was removed by centrifugation (1 min, 500 rpm). The supernatant was filtered (70 μm) and cell pelleted. Cell pellets were cultured in 10% FBS/DMEM (Invitrogen) until 70% confluent, serum-starved overnight in 0.5% FBS/DMEM and treated +/-TGFβ1 (4 ng/mL, 24 h, R and D Systems). Protein was harvested by cell lysis in RIPA buffer; protein (50 μg) was subjected to SDS-PAGE and Western blot analysis with anti-CCN1 (Sc-13,100, 1:800) antibody and HRP-conjugated antibody. Chemiluminescent substrate (Pierce) was added (5 min); X-Ray images were taken. For RNA analysis, cells were similarly treated; however, Trizol (Invitrogen) was used to collect RNA. RNA was used (40 ng/well) for RT-PCR analysis of to detect CCN1 expression using the delta-delta CT method using 18S RNA as an internal control [19,22]. Student's t-test was used for statistical analysis.
Culturing of scleroderma fibroblasts
As previously described [22,54,55], scleroderma dermal fibroblasts were obtained by explant culture from punch biopsies (4 mm) taken from affected forearms of patients with early onset diffuse cutaneous SSc (dcSSc) (patients 2 years from the first non-Raynaud's manifestation of SSc) and the EULAR/ACR criteria for automatic SSc classification due to skin fibrosis [37]. These criteria have been used at the Royal Free Centre for Rheumatology for classification of patients as SSc since 2013. Normal fibroblasts (NF) were obtained from age- gender- and site-matched healthy individuals. Cells, acquired under informed consent with the approval of the institutional review board of the Royal Free Hospital, were cultured and treated as described above; cells were not exposed to TGFβ1. Protein samples were harvested at passage 4.
Bleomycin-induced skin fibrosis mouse model
In this manuscript, mice not deleted for Ccn1 in fibroblasts [24] are designated CCN1f/f whereas mice deleted for Ccn1 are designated CCN1−/−. After confirming deletion of ccn1, mice were separated into four groups: PBS CCN1f/f, Bleomycin CCN1f/f, PBS CCN1−/− and Bleomycin CCN1−/−. Each day for 28 days, PBS groups were injected subcutaneously with 0.1 mL PBS; bleomycin groups were injected with 0.1 ml (0.1 units) bleomycin [12]. Then, mice were sacrificed and skin collected.
Wound healing mouse model
Mice were subjected to 6 mm punch biopsies, four wounds/mouse [24,25]. Northern Eclipse software was used to measure the size of wounds at day 0, 3, 7 and 10 post-wounding, and for determining the wound size as a percentage of original wound size (two-way ANOVA). Tissue was collected from mice at Day 3, 7 and 10 for histological analysis.
Electron microscopy
Skin from CCN1f/f and CCN1−/− mice was harvested and cut into 0.5–1.0 mm wedges, then fixed in 2.5% glutaraldehyde in cacodylate buffer overnight. Tissues were oriented and immobilized using mPREP system and workbench (Microscopy Innovations; Marshfield, WI). \All subsequent steps were conducted in mPREM system. Tissue was fixed/stained in 1% osmium tetroxide in cacodylate buffer (0.1 M, pH 7.2) for 1 h on ice, then rinsed 5 times in ddH2O. Tissue was stained in 1% Tannic acid in cacodylate buffer for 2 h on ice, then again in fresh 1% Tannic acid in cacodylate buffer for 2 more hours on ice. Samples were rinsed 5 times in ddH2O, then incubated in 1% uranyl acetate at 4 °C overnight in the dark. Samples were rinsed 5 times in ddH2O before being dehydrated in successive baths of 20%, 50%, 70%, 90%, 95%, and 3 times in 100% acetone. After dehydration, samples were infiltrated overnight by mixtures of 1 part Epon-Araldite resin:2 parts acetone followed by 1 part resin:1 part acetone, followed by 2 parts resin:1 part acetone, followed by 100% resin. Samples were embedded in 100% resin, which was polymerized in 60 °C oven for 5 days. Embedded samples were sectioned to a thickness of 70-80 nm on the Ultracut E (Reichert-Jung) and collected on nickel grids. Grids were stained with 1% uranyl acetate for 15 min, rinsed 24 times before staining with Reynolds' lead citrate. Grids were rinsed 24 more times before being dried. Images were obtained on a Philips CM10 transmission electron microscope at 60 kV.
Histological analysis
Tissue samples were collected as described above, fixed overnight in 4% paraformaldehyde (Sigma) at 4 °C, processed, embedded in paraffin wax blocks, and sectioned (5 μm thickness). Skin tissue was stained with hematoxylin and eosin, and skin thickness assessed using northern eclipse software. Three measurements were taken from each image; three images were taken/skin section (Zeiss, Northern Eclipse). Student's t-test or a one-way ANOVA was performed. Skin sections were stained for Masson's trichrome to examine collagen content. Three images/sections were taken (Zeiss). Using ImageJ, percent of the dermal area stained with alanine blue was determined. Student's t-test or a one-way ANOVA was performed for statistical analysis. Skin tissue was stained with picrosirius red for collagen and subjected to plane polarized light to assess collagen birefringence and organization, as previously described [26].
Immunohistochemical analysis
Immunohistological analysis of tissue sections (5 μm) was performed essentially as previously described [12,24] using anti-PCNA (ab2426, 1:500), anti-CCN1 (ABC102, 1:500) and anti-αSMA (ab5694; 1:400), appropriate secondary antibody containing fluorophore, and mounting media containing DAPI (Vector Laboratories). Images were taken using Zeiss microscope and camera and percent positive cells were calculated using image J. Number of nuclei were also counted using DAPI stained images to determine cell number using Image J. Student's t-test or a one-way ANOVA was performed. Alternatively, Vector Laboratories ABC-HRP staining kit (PK-4001) was used with a DAB enzyme substrate (SK-4105) for colorimetric analyses using anti-αSMA (ab5694; 1:400) antibody and a biotinylated secondary antibody was added for 30 min, followed by a 30 min incubation with ABC reagent followed by counterstaining with hematoxylin for tissue architecture. Images were taken using Zeiss microscope and camera and percent positive cells were calculated using image J. A one-way ANOVA was performed for statistical analysis.
Tissue RNA extraction and real time polymerase chain reaction (PCR)
Tissue was homogenized in 1 ml of trizol and a trizol-chloroform extraction was performed. TAQman PCR was performed on 40 ng of mRNA to examine expression of Ccn1, Col1a1, Lox, P4Ha1 and Plod2. Experiments were run in triplicate, delta-delta CT method was used for analysis using 18S RNA was used as an internal control. Student's t-tests were used for statistical analysis.
Hydroxyproline analysis
Soluble collagen was extracted using 0.5 M acetic acid, sonicated, and rotated at 4 °C overnight. Samples were centrifuged at high speed for 45 min. The supernatant and precipitate were collected separately, the former was used to detect soluble collagen using QuickZyme soluble collagen assay (QZBcol1) and the latter for insoluble collagen using total collagen kit from QuickZyme (QZBtotcol1). The combination of soluble and insoluble accounted for the total. Student t-tests were used for statistical analysis.
Tunel assay
Paraffin embedded skin samples were sectioned 5 μm thick and analyzed for apoptosis using Trevigen Tunnel assay kit (4810-30-K). Control slides were treated with TACS-nuclease to break DNA for positive control. The kit uses biotinylated nucleotides that integrate into the tissue and are detected using streptavidin-horseradish peroxidase, and colorimetric substrates diaminobenzidine (DAB). Sections were counterstained with hematoxylin and imaged using Zeiss microscope and camera.
Mechanical testing of skin
Full thickness skin was excised from four mice per group (CCN1f/f and CCN1−/−) and placed on a rigid support surface. Compressive stiffness (Young's modulus) was measured by indentation with the epidermis oriented upwards. Uni-axial compression tests were performed with a mechanical tester (Mach-1v500css, Biomomentum Inc., Canada) equipped with vertically loaded spherical indenter with 1 mm tip diameter at a ramping velocity of 0.05 mm/s. Four different positions were measured per sample. Slopes of the resulting force-distance curves were fitted and the Young's modulus was calculated using a Herz model, using the instrument's analysis software (Biomomentum Inc). Data were statistically compared and analyzed using Student's t-test followed by Tukey HSD post-hoc analysis (using Prism, Graph pad) and represented as mean ± standard error (N = 4).
Proteomic analysis
Skin from CCN1f/f and CCN1−/− mice was harvested, minced, and protein was suspended in 8 M urea and 10 mM DTT, diluted eight-fold with 50 mM NH4HCO3, pH 7.8, and subjected to tryptic digestion [18 h, 37 °C with =2% (w/w) sequencing-grade trypsin (Promega, Madison, WI)]. Peptide separation and mass spectrometric analyses were carried out with a nano-HPLC Proxeon (Thermo Scientific, San Jose, CA, USA) and a capillary fused silica column (75 μm × 10 cm, Pico Tip™ EMITTER, New Objective, Woburn, MA) packed in-house with C18 resin of 3 μm spherical beads and 20 nm pore size (Michrom BioResources, Auburn, CA) linked to a mass spectrometer (LTQ-Velos, Thermo Scientific, San Jose, CA) using an electrospray ionization in a survey scan in the range of m/z values 390–2000 tandem MS/MS. Samples were dried and re-suspended in 20 μl of 97.5% H2O/2.4% acetonitrile/0.1% formic acid and subjected to reversed-phase nLC-ESI-MS/MS with a linear 85-minute gradient ranging from 5% to 100% of solvent (99.9% acetonitrile/0.1% formic acid) at a flow rate of 200 nl/min with a maximum pressure of 280 bar. Electrospray voltage and the temperature of the ion transfer capillary were 1.9 kV and 250 °C respectively. Each survey scan (MS) was followed by automated sequential selection of seven peptides for CID, with dynamic exclusion of the previously selected ions. Obtained spectra were searched against mouse collagen protein databases (Swiss Prot and TrEMBL, Swiss Institute of Bioinformatics, Geneva, Switzerland, http://ca.expasy.org/sprot/) using SEQUEST algorithm (Proteome Discoverer 1.3;Thermo Scientific, San Jose, CA). Results were filtered for a False Discovery rate of 1% employing a decoy search strategy utilizing a reverse database. A protein which passes beyond the inclusion of the filter arrangement at least in three different MS analyzes of the same group in a total of four analyzes by MS group. For qualitative proteome, three MS raw files from each pooled group were analyzed (SIEVE Version 2.0 Thermo Scientific, San Jose, CA, USA). Signal processing was performed in a total of 6 MS raw files. The SIEVE experimental workflow was defined as “Two Sample Differential Analysis” where two classes of samples are compared to each other. Peptides were grouped, and a protein ratio and p-value were calculated. Lysyl hydroxylase 2 (LH2, encoded by PLOD2) modifies X-Lys-Gly, X-Lys-Ala, and X-Lys-Ser sequences in type I collagen [27] (PMID: 17289364). In our samples, X-Lys-Ala was not detected; X-Lys-Ser was minimally detected. Extracted ion chromatographs (EIC) of peptide ions containing different degrees of hydroxylation were generated as previously described [28].
Genome-wide expression profiling
Microarray analysis was performed, by the London Regional Genomics Center, essentially as described (Affymetrix GeneChip Mouse Gene 2.0 ST arrays, Santa Clara, CA) followed by cluster analysis [DAVID v6.7 (https://david.ncifcrf.gov/)] [[56], [57], [58], [59]]. Experiments were performed twice; the data set after average fold increase values were calculated are in Supplementary information.
The following are the supplementary data related to this article.
Sample deconvoluted collision-induced dissociation spectrum corresponding to tryptic peptide GPKGENGIVGPTGSVGAAGPSGPNGPPGPVGSR derived from mouse type I collagen (Col1a2) that is hydroxylated (TOP PANEL, Hyl) or not (BOTTOM PANEL, Lys) at the lysine residue.
Gene array data: mRNAs increased in CCN1-deficient skin
Gene array data: mRNAs decreased in CCN1-deficient skin
Author contributions
KQ analysis of data, conducting and designing experiments, data interpretation, writing of the manuscript.
RJS obtaining clinical samples.
SL, AP, JH analysis of data, conducting and designing experiments, data interpretation.
DN analysis of data, conducting experiments.
AL, DO'G, BH, XS, RJS experimental supervision, data interpretation, writing of the manuscript.
Ethical approval
All procedures approving animal and human materials were approved by the appropriate boards at the University of Western Ontario and the Royal Free Hospital.
Declaration of Competing Interest
AL is a shareholder of FibroGen. FibroGen had no input into the design or interpretation of the experiments, or in the writing of the manuscript.
Acknowledgements
Electron microscopy was performed with the expert assistance of Richard Gardiner, Karen Nygard with and Reza Khazaee (Integrated Microscopy Unit, Biotron, University of Western Ontario). We thank David Carter (London Regional Genomics Center, London, ON) for expert assistance regarding genome-wide expression profiling. AL was funded by the CIHR (MOP-77603 and MOP-119410), NSERC (CPG-146479), the Scleroderma Society of Ontario and the Arthritis Society. AP was funded by the Joint Motion Program.
References
- 1.Borthwick L.A., Wynn T.A., Fisher A.J. (2013) Cytokine mediated tissue fibrosis. Biochimica et Biophysica Acta. 2013;1832:1049–1060. doi: 10.1016/j.bbadis.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denton C.P., Khanna D. (2017) Systemic sclerosis. Lancet. 2017;390:1685–1699. doi: 10.1016/S0140-6736(17)30933-9. [DOI] [PubMed] [Google Scholar]
- 3.Schulz J.N., Plomann M., Sengle G., Gullberg D., Krieg T., Eckes B. New developments on skin fibrosis - essential signals emanating from the extracellular matrix for the control of myofibroblasts. Matrix Biology. 2018;68-69:522–532. doi: 10.1016/j.matbio.2018.01.025. [DOI] [PubMed] [Google Scholar]
- 4.Schoenherr C., Frame M.C., Byron A. Trafficking of adhesion and growth factor receptors and their effector kinases. Annual Review of Cell and Developmental Biology. 2018;34:29–58. doi: 10.1146/annurev-cellbio-100617-062559. [DOI] [PubMed] [Google Scholar]
- 5.Riser B.L., Barnes J.L., Varani J. Balanced regulation of the CCN family of matricellular proteins: a novel approach to the prevention and treatment of fibrosis and cancer. Journal of Cell Communication and Signaling. 2015;9:327–339. doi: 10.1007/s12079-015-0309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jun J.I., Lau L.F. Taking aim at the extracellular matrix: CCN proteins as emerging therapeutic targets. Nature Reviews. Drug Discovery. 2011;10:945–963. doi: 10.1038/nrd3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perbal B. The concept of the CCN protein family revisited: a centralized coordination network. Journal of Cell Communication and Signaling. 2018;12:3–12. doi: 10.1007/s12079-018-0455-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lau L.F. Cell surface receptors for CCN proteins. Journal of Cell Communication and Signaling. 2016;10:121–127. doi: 10.1007/s12079-016-0324-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takigawa M. An early history of CCN2/CTGF research: the road to CCN2 via hcs24, ctgf, ecogenin, and regenerin. Journal of Cell Communication and Signaling. 2018;12:253–264. doi: 10.1007/s12079-017-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lau L.F. CCN1/CYR61: the very model of a modern matricellular protein. Cellular and Molecular Life Sciences. 2011;68:3149–3163. doi: 10.1007/s00018-011-0778-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dammeier J., Brauchle M., Falk W., Grotendorst G.R., Werner S. Connective tissue growth factor: a novel regulator of mucosal repair and fibrosis in inflammatory bowel disease? The International Journal of Biochemistry & Cell Biology. 1998;30:909–922. doi: 10.1016/s1357-2725(98)00046-6. [DOI] [PubMed] [Google Scholar]
- 12.Liu S., Shi-wen X., Abraham D.J., Leask A. CCN2 is required for bleomycin-induced skin fibrosis in mice. Arthritis and Rheumatism. 2011;63:239–246. doi: 10.1002/art.30074. [DOI] [PubMed] [Google Scholar]
- 13.Makino K., Makino T., Stawski L., Lipson K.E., Leask A., Trojanowska M. Anti-connective tissue growth factor (CTGF/CCN2) monoclonal antibody attenuates skin fibrosis in mice models of systemic sclerosis. Arthritis Research & Therapy. 2017;19:134. doi: 10.1186/s13075-017-1356-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kinashi H., Falke L.L., Nguyen T.Q., Bovenschen N., Aten J., Leask A., Ito Y., Goldschmeding R. Connective tissue growth factor regulates fibrosis-associated renal lymphangiogenesis. Kidney International. 2017;92:850–863. doi: 10.1016/j.kint.2017.03.029. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez D., Rebolledo D.L., Correa L.M. The inhibition of CTGF/CCN2 activity improves muscle and locomotor function in a murine ALS model. Human Molecular Genetics. 2018;27:2913–2926. doi: 10.1093/hmg/ddy204. [DOI] [PubMed] [Google Scholar]
- 16.Tsang M., Leask A. CCN2 is required for recruitment of Sox2-expressing cells during cutaneous tissue repair. Journal of Cell Communication and Signaling. 2015;9:341–346. doi: 10.1007/s12079-014-0245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leask A. CCN2: a novel, specific and valid target for anti-fibrotic drug intervention. Expert Opinion on Therapeutic Targets. 2013;17:1067–1071. doi: 10.1517/14728222.2013.812074. [DOI] [PubMed] [Google Scholar]
- 18.Raghu G., Scholand M.B., de Andrade J., Lancaster L., Mageto Y., Goldin J., Brown K.K., Flaherty K.R., Wencel M., Wanger J., Neff T., Valone F., Stauffer J., Porter S. FG-3019 anti-connective tissue growth factor monoclonal antibody: results of an open-label clinical trial in idiopathic pulmonary fibrosis. The European Respiratory Journal. 2016;47:1481–1491. doi: 10.1183/13993003.01030-2015. [DOI] [PubMed] [Google Scholar]
- 19.Thompson K., Murphy-Marshman H., Leask A. ALK5 inhibition blocks TGFbeta-induced CCN1 expression in human foreskin fibroblasts. Journal of Cell Communication and Signaling. 2014;8:59–63. doi: 10.1007/s12079-014-0229-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leask A., Holmes A., Black C.M., Abraham D.J. Connective tissue growth factor gene regulation. Requirements for its induction by transforming growth factor-beta 2 in fibroblasts. The Journal of Biological Chemistry. 2003;278:13008–13015. doi: 10.1074/jbc.M210366200. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y., Huang T., Cheng A.S. The TEAD family and its oncogenic role in promoting tumorigenesis. International Journal of Molecular Sciences. 2016;17 doi: 10.3390/ijms17010138. (pii: E138) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy-Marshman H., Quensel K., Shi-Wen X. Antioxidants and NOX1/NOX4 inhibition blocks TGFβ1-induced CCN2 and α-SMA expression in dermal and gingival fibroblasts. PLoS One. 2017;12 doi: 10.1371/journal.pone.0186740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mo F.E., Muntean A.G., Chen C.C. CYR61 (CCN1) is essential for placental development and vascular integrity. Molecular and Cellular Biology. 2002;22:8709–8720. doi: 10.1128/MCB.22.24.8709-8720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu S., Thompson K., Leask A. CCN2 expression by fibroblasts is not required for cutaneous tissue repair. Wound Repair and Regeneration. 2014;22:119–124. doi: 10.1111/wrr.12131. [DOI] [PubMed] [Google Scholar]
- 25.Liu S., Xu S.W., Blumbach K., Eastwood M., Denton C.P., Eckes B., Krieg T., Abraham D.J., Leask A. Expression of integrin beta1 by fibroblasts is required for tissue repair in vivo. Journal of Cell Science. 2010;123:3674–3682. doi: 10.1242/jcs.070672. [DOI] [PubMed] [Google Scholar]
- 26.Rittié L. Method for picrosirius red-polarization detection of collagen fibers in tissue sections. Methods in Molecular Biology. 2017;1627:395–407. doi: 10.1007/978-1-4939-7113-8_26. [DOI] [PubMed] [Google Scholar]
- 27.van der Slot A.J., Zuurmond A.M., Bardoel A.F., Wijmenga C., Pruijs H.E., Sillence D.O., Brinckmann J., Abraham D.J., Black C.M., Verzijl N., DeGroot J., Hanemaaijer R., TeKoppele J.M., Huizinga T.W., Bank R.A. Identification of PLOD2 as telopeptide lysyl hydroxylase, an important enzyme in fibrosis. The Journal of Biological Chemistry. 2003;278:40967–40972. doi: 10.1074/jbc.M307380200. [DOI] [PubMed] [Google Scholar]
- 28.Perdivara I., Yamauchi M., Tomer K.B. Molecular characterization of collagen hydroxylysine o-glycosylation by mass spectrometry: current status. Australian Journal of Chemistry. 2013;66:760–769. doi: 10.1071/CH13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S., Kapoor M., Denton C.P., Abraham D.J., Leask A. Loss of beta1 integrin in mouse fibroblasts results in resistance to skin scleroderma in a mouse model. Arthritis and Rheumatism. 2009;60:2817–2821. doi: 10.1002/art.24801. [DOI] [PubMed] [Google Scholar]
- 30.Yeowell H.N., Marshall M.K., Walker L.C. Regulation of lysyl oxidase mRNA in dermal fibroblasts from normal donors and patients with inherited connective tissue disorders. Archives of Biochemistry and Biophysics. 1994;308:299–305. doi: 10.1006/abbi.1994.1042. [DOI] [PubMed] [Google Scholar]
- 31.Yeowell H.N., Murad S., Pinnell S.R. Hydralazine differentially increases mRNAs for the alpha and beta subunits of prolyl 4-hydroxylase whereas it decreases pro alpha 1(I) collagen mRNAs in human skin fibroblasts. Archives of Biochemistry and Biophysics. 1991;289:399–404. doi: 10.1016/0003-9861(91)90430-q. [DOI] [PubMed] [Google Scholar]
- 32.Myllyharju J. Prolyl 4-hydroxylases, the key enzymes of collagen biosynthesis. Matrix Biology. 2003;22:15–24. doi: 10.1016/s0945-053x(03)00006-4. [DOI] [PubMed] [Google Scholar]
- 33.van der Slot A.J., Zuurmond A.M., van den Bogaerdt A.J., Ulrich M.M., Middelkoop E., Boers W., Karel Ronday H., DeGroot J., Huizinga T.W., Bank R.A. Increased formation of pyridinoline cross-links due to higher telopeptide lysyl hydroxylase levels is a general fibrotic phenomenon. Matrix Biology. 2004;23:251–257. doi: 10.1016/j.matbio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 34.Epstein Shochet G., Brook E., Israeli-Shani L., Edelstein E., Shitrit D. Fibroblast paracrine TNF-α signaling elevates integrin A5 expression in idiopathic pulmonary fibrosis (IPF) Respiratory Research. 2017 Jun 19;18(1):122. doi: 10.1186/s12931-017-0606-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosini S., Pugh N., Bonna A.M., DJS Hulmes, Farndale R.W., Adams J.C. Thrombospondin-1 promotes matrix homeostasis by interacting with collagen and lysyl oxidase precursors and collagen cross-linking sites. Science Signaling. 2018 May 29;11(532) doi: 10.1126/scisignal.aar2566. (pii: eaar2566) [DOI] [PubMed] [Google Scholar]
- 36.Mallano T., Palumbo-Zerr K., Zerr P., Ramming A., Zeller B., Beyer C., Dees C., Huang J., Hai T., Distler O., Schett G., Distler J.H. Activating transcription factor 3 regulates canonical TGFβ signalling in systemic sclerosis. Annals of the Rheumatic Diseases. 2016 Mar;75(3):586–592. doi: 10.1136/annrheumdis-2014-206214. [DOI] [PubMed] [Google Scholar]
- 37.van den Hoogen F., Khanna D., Fransen J., Johnson S.R., Baron M., Tyndall A., Matucci-Cerinic M., Naden R.P., Medsger T.A., Jr., Carreira P.E., Riemekasten G., Clements P.J., Denton C.P., Distler O., Allanore Y., Furst D.E., Gabrielli A., Mayes M.D., van Laar J.M., Seibold J.R., Czirjak L., Steen V.D., Inanc M., Kowal-Bielecka O., Müller-Ladner U., Valentini G., Veale D.J., Vonk M.C., Walker U.A., Chung L., Collier D.H., Csuka M.E., Fessler B.J., Guiducci S., Herrick A., Hsu V.M., Jimenez S., Kahaleh B., Merkel P.A., Sierakowski S., Silver R.M., Simms R.W., Varga J., Pope J.E. 2013 classification criteria for systemic sclerosis: an American College of Rheumatology/European League against Rheumatism collaborative initiative. Arthritis and Rheumatism. 2013;65 doi: 10.1002/art.38098. (2737-274) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krupska I., Bruford E.A., Chaqour B. Eyeing the Cyr61/CTGF/NOV (CCN) group of genes in development and diseases: highlights of their structural likenesses and functional dissimilarities. Human Genomics. 2015;23(9):24. doi: 10.1186/s40246-015-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jun J.I., Lau L.F. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nature Cell Biology. 2010;12:676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marangoni R.G., Varga J., Tourtellotte W.G. Animal models of scleroderma: recent progress. Current Opinion in Rheumatology. 2016;28:561–570. doi: 10.1097/BOR.0000000000000331. [DOI] [PubMed] [Google Scholar]
- 41.Grazioli S., Gil S., An D., Kajikawa O., Farnand A.W., Hanson J.F., Birkland T., Chen P., Duffield J., Schnapp L.M., Altemeier W.A., Matute-Bello G. CYR61 (CCN1) overexpression induces lung injury in mice. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2015;308:L759–L765. doi: 10.1152/ajplung.00190.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai C.F., Lin S.L., Chiang W.C., Chen Y.M., Wu V.C., Young G.H., Ko W.J., Kuo M.L., Tsai T.J., Wu K.D., Lai C.F., Lin S.L., Chiang W.C., Chen Y.M., Wu V.C., Young G.H., Ko W.J., Kuo M.L., Tsai T.J., Wu K.D. Blockade of cysteine-rich protein 61 attenuates renal inflammation and fibrosis after ischemic kidney injury. American Journal of Physiology. Renal Physiology. 2014;307(5):F581–F592. doi: 10.1152/ajprenal.00670.2013. [DOI] [PubMed] [Google Scholar]
- 43.Lai C.F., Chen Y.M., Chiang W.C., Lin S.L., Kuo M.L., Tsai T.J. Cysteine-rich protein 61 plays a proinflammatory role in obstructive kidney fibrosis. PLoS One. 2013;8(2) doi: 10.1371/journal.pone.0056481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borkham-Kamphorst E., Steffen B.T., Van de Leur E., Tihaa L., Haas U., Woitok M.M., Meurer S.K., Weiskirchen R. Adenoviral CCN gene transfers induce in vitro and in vivo endoplasmic reticulum stress and unfolded protein response. Biochimica et Biophysica Acta. 2016;1863:2604–2612. doi: 10.1016/j.bbamcr.2016.07.006. [DOI] [PubMed] [Google Scholar]
- 45.Kim K.H., Chen C.C., Monzon R.I., Lau L.F. Matricellular protein CCN1 promotes regression of liver fibrosis through induction of cellular senescence in hepatic myofibroblasts. Molecular and Cellular Biology. 2013;33:2078–2090. doi: 10.1128/MCB.00049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gardner H., Shearstone J.R., Bandaru R., Crowell T., Lynes M., Trojanowska M., Pannu J., Smith E., Jablonska S., Blaszczyk M., Tan F.K., Mayes M.D. Gene profiling of scleroderma skin reveals robust signatures of disease that are imperfectly reflected in the transcript profiles of explanted fibroblasts. Arthritis and Rheumatism. 2006;54:1961–1973. doi: 10.1002/art.21894. [DOI] [PubMed] [Google Scholar]
- 47.Lin J., Li N., Chen H., Liu C., Yang B., Ou Q. Serum Cyr61 is associated with clinical disease activity and inflammation in patients with systemic lupus erythematosus. Medicine (Baltimore) 2015;94 doi: 10.1097/MD.0000000000000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saigusa R., Asano Y., Taniguchi T., Yamashita T., Takahashi T., Ichimura Y., Toyama T., Tamaki Z., Tada Y., Sugaya M., Kadono T., Sato S. A possible contribution of endothelial CCN1 downregulation due to Fli1 deficiency to the development of digital ulcers in systemic sclerosis. Experimental Dermatology. 2015;24:127–132. doi: 10.1111/exd.12602. [DOI] [PubMed] [Google Scholar]
- 49.Tsou P.S., Wren J.D., Amin M.A., Schiopu E., Fox D.A., Khanna D., Sawalha A.H. Histone deacetylase 5 Is overexpressed in scleroderma endothelial cells and impairs angiogenesis via repression of proangiogenic factors. Arthritis and Rheumatism. 2016;68:2975–2985. doi: 10.1002/art.39828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quan T., Xu Y., Qin Z., Robichaud P., Betcher S., Calderone K., He T., Johnson T.M., Voorhees J.J., Fisher G.J. Elevated YAP and its downstream targets CCN1 and CCN2 in basal cell carcinoma: impact on keratinocyte proliferation and stromal cell activation. The American Journal of Pathology. 2014;184:937–943. doi: 10.1016/j.ajpath.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu P., G& Ma, Li N. The profile of Cyr61 expression data correlate to the skin inflammation in psoriasis. Data in Brief. 2016;2016(10):487–491. doi: 10.1016/j.dib.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y., Zhang J., Zhou Z., Wu P., Huo R., Wang B., Shen Z., Li H., Zhai T., Shen B., Chen X., Li N. CCN1, a pro-inflammatory factor, aggravates psoriasis skin lesions by promoting keratinocyte activation. The Journal of Investigative Dermatology. 2015;135:2666–2675. doi: 10.1038/jid.2015.231. [DOI] [PubMed] [Google Scholar]
- 53.Tsou P.S., Khanna D., Sawalha A.H. Identifying CYR61 as a potential anti-fibrotic and pro-angiogenic mediator in scleroderma. Arthritis and Rheumatism. 2019 doi: 10.1002/art.40890. Mar 18. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shiwen X., Stratton R., Nikitorowicz-Buniak J., Ahmed-Abdi B., Ponticos M., Denton C., Abraham D., Takahashi A., Suki B., Layne M.D., Lafyatis R., Smith B.D. A role of myocardin related transcription factor-A (MRTF-A) in scleroderma related fibrosis. PLoS One. 2015;10(5) doi: 10.1371/journal.pone.0126015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denton C.P., Ong V.H., Xu S., Chen-Harris H., Modrusan Z., Lafyatis R., Khanna D., Jahreis A., Siegel J., Sornasse T. Therapeutic interleukin-6 blockade reverses transforming growth factor-beta pathway activation in dermal fibroblasts: insights from the faSScinate clinical trial in systemic sclerosis. Annals of the Rheumatic Diseases. 2018;77(9):1362–1371. doi: 10.1136/annrheumdis-2018-213031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shi-wen X., Parapuram S.K., Pala D., Chen Y., Carter D.E., Eastwood M., Denton C.P., Abraham D.J., Leask A. Requirement of transforming growth factor beta-activated kinase 1 for transforming growth factor beta-induced alpha-smooth muscle actin expression and extracellular matrix contraction in fibroblasts. Arthritis and Rheumatism. 2009;60:234–241. doi: 10.1002/art.24223. [DOI] [PubMed] [Google Scholar]
- 57.Guo F., Carter D.E., Leask A. Mechanical tension increases CCN2/CTGF expression and proliferation in gingival fibroblasts via a TGFbeta-dependent mechanism. PLoS One. 2011;6 doi: 10.1371/journal.pone.0019756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen S., McLean S., Carter D.E., Leask A. The gene expression profile induced by Wnt 3a in NIH 3T3 fibroblasts. Journal of Cell Communication and Signaling. 2007;1:175–183. doi: 10.1007/s12079-007-0015-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuk H., Hutchenreuther J., Murphy-Marshman H., Carter D., Leask A. 5Z-7-Oxozeanol inhibits the effects of tgfbeta1 on human gingival fibroblasts. PLoS One. 2015;10 doi: 10.1371/journal.pone.0123689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sample deconvoluted collision-induced dissociation spectrum corresponding to tryptic peptide GPKGENGIVGPTGSVGAAGPSGPNGPPGPVGSR derived from mouse type I collagen (Col1a2) that is hydroxylated (TOP PANEL, Hyl) or not (BOTTOM PANEL, Lys) at the lysine residue.
Gene array data: mRNAs increased in CCN1-deficient skin
Gene array data: mRNAs decreased in CCN1-deficient skin