Abstract
One year in the coronavirus disease 2019 (COVID-19) pandemic, the first vaccines are being rolled out under emergency use authorizations. It is of great concern that newly emerging variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) can escape antibody-mediated protection induced by previous infection or vaccination through mutations in the spike protein. The glutamate (E) to Lysine (K) substitution at position 484 (E484K) in the receptor binding domain (RBD) of the spike protein is present in the rapidly spreading variants of concern belonging to the B.1.351 and P.1 lineages. We performed in vitro microneutralization assays with both the USA-WA1/2020 virus and a recombinant (r)SARS-CoV-2 virus that is identical to USA-WA1/2020 except for the E484K mutation introduced in the spike RBD. We selected 34 sera from study participants based on their SARS-CoV-2 spike ELISA antibody titer (negative [N=4] versus weak [N=8], moderate [N=11] or strong positive [N=11]). In addition, we included sera from five individuals who received two doses of the Pfizer SARS-CoV-2 vaccine BNT162b2. Serum neutralization efficiency was lower against the E484K rSARS-CoV-2 (vaccination samples: 3.4 fold; convalescent low IgG: 2.4 fold, moderate IgG: 4.2 fold and high IgG: 2.6 fold) compared to USA-WA1/2020. For some of the convalescent donor sera with low or moderate IgG against the SARS-CoV-2 spike, the drop in neutralization efficiency resulted in neutralization ID50 values similar to negative control samples, with low or even absence of neutralization of the E484K rSARS-CoV-2. However, human sera with high neutralization titers against the USA-WA1/2020 strain were still able to neutralize the E484K rSARS-CoV-2. Therefore, it is important to aim for the highest titers possible induced by vaccination to enhance protection against newly emerging SARS-CoV-2 variants. Two vaccine doses may be needed for induction of high antibody titers against SARS-CoV-2. Postponing the second vaccination is suggested by some public health authorities in order to provide more individuals with a primer vaccination. Our data suggests that this may leave vaccinees less protected against newly emerging variants.
Introduction
One year in the coronavirus disease 2019 (COVID-19) pandemic, the first vaccines are being rolled out under emergency use authorizations. Recently, rapidly spreading variants of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, have been reported. It is of great concern that these newly emerging variants can escape neutralizing antibodies induced by previous infection and/or vaccination through mutations in the spike (S) protein, including the receptor binding domain (RBD), a target for neutralizing antibodies. We and others have previously reported that the asparagine (N) to tyrosine (Y) substitution at position 501 (N501Y), present in variants of concern belonging to the B.1.1.7, B.1.351 and P.1 lineages, does not seem to affect in vitro neutralization of SARS-CoV-2 viruses by human sera from convalescent or vaccinated human donors. However, there remains concern about additional substitutions like E484K present in B.1.351 and P.1 lineages allowing escape from neutralizing antibodies (1–4), thereby potentially rendering vaccine-induced immunity less protective.
In order to investigate the impact of the E484K mutation in the neutralizing activity of SARS-CoV-2 specific antisera, we performed in vitro microneutralization assays with both the USA-WA1/2020 virus and a recombinant (r)SARS-CoV-2 virus that is identical to USA-WA1/2020 except for the E484K mutation introduced in the spike RBD.
The E484K mutant rSARS-CoV-2 was generated using previously described reverse genetics based on the use of a bacterial artificial chromosome (BAC) (5–7). The USA-WA1/2020 reflects SARS-CoV-2 strains that circulated in the early phase of the COVID-19 pandemic. A total of 34 sera were selected from study participants based on their SARS-CoV-2 S enzyme linked immunosorbent assay (ELISA) antibody titer (negative [N=4] versus weak [N=8], moderate [N=11] or strong positive [N=11]). In addition, we included sera from five individuals who received two doses of the Pfizer SARS-CoV-2 vaccine BNT162b2 (V1-V5). Demographics and available metadata for each participant is summarized in Supplementary Table 1. We performed all experiments in a blinded manner. The same sera have been tested for neutralization studies with a N501Y SARS-CoV-2 variant in our recent report (8).
Results
Sera from vaccinated donors gave high neutralization titers, similar to convalescent samples with the highest neutralization titers. Serum neutralization efficiency was lower against the E484K rSARS-CoV-2 (vaccination samples: 3.4 fold; convalescent low IgG: 2.4 fold, moderate IgG: 4.2 fold and high IgG: 2.6 fold based on geometric means) which was significantly different for the convalescent sera (see Figure 1), suggesting that the single E484K mutation in the RBD affects binding by serum polyclonal neutralizing antibodies from both convalescent and vaccinated donors. In the case of convalescent donor sera with low or moderate IgG against SARS-CoV-2 S protein, the drop in neutralization efficiency could result in neutralization ID50 values similar to negative control samples, resulting in low or even absence of neutralization of the E484K recombinant virus by those sera.
Figure 1.
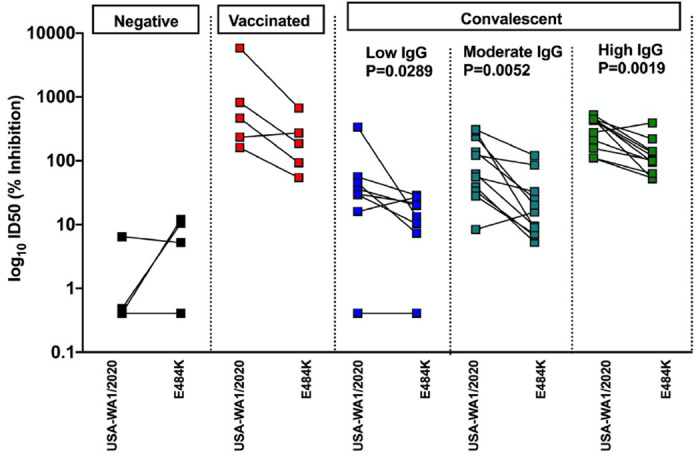
Human convalescent and post-vaccination sera neutralize E484K rSARS-CoV-2 less efficient than USA-WA1/2020 in an in vitro microneutralization assay. Convalescent sera are subdivided in low, moderate and high IgG classes based on anti-spike ELISA titers. Two-sided Mann Whitney-U tests were performed to calculate statistical differences.
Conclusions
These data indicate that the E484K mutation present in circulating SARS-CoV-2 strains that belong to the B.1.351 and P.1 lineages reduces the neutralizing activity of human polyclonal sera induced in convalescent (infected with previous strains) and vaccinated individuals. The significant impact of a single point mutation in the neutralizing activity of polyclonal sera highlights the need for the rapid characterization of SARS-CoV-2 variants. However, human sera with high neutralization titers against the USA-WA1/2020 strain were still able to neutralize the E484K rSARS-CoV-2. Therefore, it is important to aim for the highest titers possible induced by vaccination, as this should enhance the chances for protection even in the case of antigenic drift of circulating SARS-CoV-2 strains. Currently deployed SARS-CoV-2 vaccines are recommended as a prime-boost regimen. Because of vaccine shortage and relatively strong seroconversion being observed after a single dose, some public health authorities recommended to postpone the second booster vaccination in order to be able to provide more individuals with a first primer vaccination. This will result in lower neutralizing antibody titers. Our data show that this may be problematic in the context of newly emerging SARS-CoV-2 variants, as it may leave some vaccinees unprotected. It is currently unknown which neutralization titer correlates with (full) protection, and to what extent immune mechanisms beyond direct virus neutralization contribute to protection, especially for specific target groups with comorbidities that are currently being prioritized for vaccination.
Viruses that belong to the B.1.351 and P.1 lineages have originally been described in the Republic of South Africa and Brazil, but are now reported on multiple continents already. Therefore, while it is premature to update vaccines based on these lineages, it is important that the worldwide vaccination effort will aim at fully vaccinating as many people as possible using vaccination strategies that result in induction of high neutralizing antibody titers.
Supplementary Material
Acknowledgements
PVI study group (in alphabetical order): H. Alshammary, A. Amoako, M. Awawda, K. Beach, C. M. Bermúdez-González, R. Chernet, L. Eaker, E. Ferreri, D. Floda, C. Gleason, Dr. G. Kleiner, D. Jurczyszak, J. Matthews, W. Mendez, Dr. LCF Mulder, K. Russo, A. Salimbangon, Dr. M. Saksena, A. Shin, L. Sominsky and K. Srivastava.
We thank all the study participants for their continued support of COVID19 research. This research was partly funded by CRIP (Center for Research for Influenza Pathogenesis), a NIAID supported Center of Excellence for Influenza Research and Surveillance (CEIRS, contract # HHSN272201400008C); by NCI grant U54CA260560; by the generous support of the JPB Foundation and the Open Philanthropy Project (research grant 2020-215611 (5384); and by anonymous donors to A.G-S. Work on SARS-CoV-2 in the Krammer and Simon laboratories was funded by the Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051. Research in Martinez-Sobrido laboratory was partially funded by the New York Influenza Center of Excellence (NYICE), a member of the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Department of Health and Human Services, Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract No. HHSN272201400005C (NYICE).
Conflicts of interest
The García-Sastre Laboratory has received research support from Pfizer, Senhwa Biosciences and 7Hills Pharma. Adolfo García-Sastre has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Accurius and Esperovax. The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays and NDV-based SARS-CoV-2 vaccines which list Florian Krammer as co-inventor. Daniel Stadlbauer and Viviana Simon are also listed on the serological assay patent application as co-inventors. Florian Krammer has consulted for Merck and Pfizer (before 2020), and is currently consulting for Seqirus and Avimex. The Krammer laboratory is also collaborating with Pfizer on animal models of SARS-CoV-2.
References
- 1.Weisblum Y, Schmidt F, Zhang F, DaSilva J, Poston D, Lorenzi JC, et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife [Internet]. [cited 2020 Dec 22];9. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7723407/ [DOI] [PMC free article] [PubMed]
- 2.Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, et al. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv. 2021. January 4;2020.12.31.425021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. bioRxiv. 2021. January 19;2021.01.15.426911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu K, Werner AP, Moliva JI, Koch M, Choi A, Stewart-Jones GBE, et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv. 2021. January 25;2021.01.25.427948. [Google Scholar]
- 5.Chiem K, Morales Vasquez D, Park J-G, Platt RN, Anderson T, Walter MR, et al. Generation and Characterization of recombinant SARS-CoV-2 expressing reporter genes. J Virol. 2021. January 11; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiem K, Ye C, Martinez-Sobrido L. Generation of Recombinant SARS-CoV-2 Using a Bacterial Artificial Chromosome. Curr Protoc Microbiol [Internet]. 2020. December [cited 2021 Jan 25];59(1). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7646048/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ye C, Chiem K, Park J-G, Oladunni F, Platt RN, Anderson T, et al. Rescue of SARS-CoV-2 from a Single Bacterial Artificial Chromosome. mBio [Internet]. 2020. September 25 [cited 2021 Jan 25];11(5). Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7520601/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rathnasinghe R, Jangra S, Cupic A, Martinez-Romero C, Mulder LCF, Kehrer T, et al. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv. 2021. January 20;2021.01.19.21249592. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


