Abstract
The COVID-19 pandemic has ravaged the globe, and its causative agent, SARS-CoV-2, continues to rage. Prospects of ending this pandemic rest on the development of effective interventions. Single and combination monoclonal antibody (mAb) therapeutics have received emergency use authorization1–3, with more in the pipeline4–7. Furthermore, multiple vaccine constructs have shown promise8, including two with ~95% protective efficacy against COVID-199,10. However, these interventions were directed toward the initial SARS-CoV-2 that emerged in 2019. The recent emergence of new SARS-CoV-2 variants B.1.1.7 in the UK11 and B.1.351 in South Africa12 is of concern because of their purported ease of transmission and extensive mutations in the spike protein. We now report that B.1.1.7 is refractory to neutralization by most mAbs to the N-terminal domain (NTD) of spike and relatively resistant to a few mAbs to the receptor-binding domain (RBD). It is not more resistant to convalescent plasma or vaccinee sera. Findings on B.1.351 are more worrisome in that this variant is not only refractory to neutralization by most NTD mAbs but also by multiple individual mAbs to the receptor-binding motif on RBD, largely due to an E484K mutation. Moreover, B.1.351 is markedly more resistant to neutralization by convalescent plasma (9.4 fold) and vaccinee sera (10.3–12.4 fold). B.1.351 and emergent variants13,14 with similar spike mutations present new challenges for mAb therapy and threaten the protective efficacy of current vaccines.
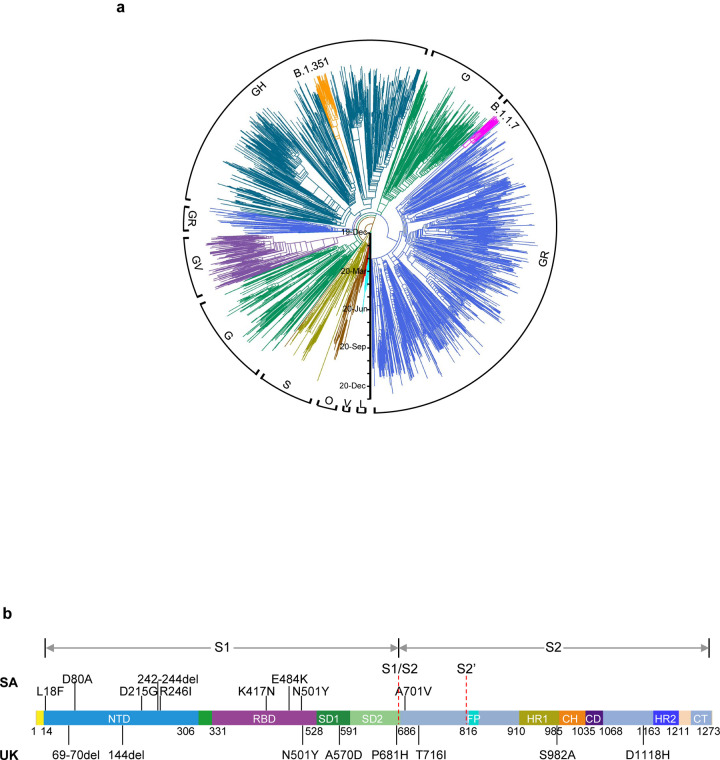
Considerable SARS-CoV-2 evolution has occurred since its initial emergence, including variants with a D614G mutation15 that have become dominant. Viruses with this mutation alone do not appear to be antigenically distinct, however16. SARS-CoV-2 B.1.1.7, also known as 501Y.V1 in the GR clade (Fig. 1a), emerged in September 2020 in South East England and rapidly became the dominant variant in the UK, possibly due to its enhanced transmissibility11. This strain has now spread to over 50 countries, and there are indications that it may be more virulent17. B.1.1.7 contains 8 spike mutations in addition to D614G, including two deletions (69–70del & 144del) in NTD, one mutation (N501Y) in RBD, and one mutation (P681H) near the furin cleavage site (Fig. 1b). SARS-CoV-2 B.1.351, also known as 501Y.V2 in the GH clade (Fig. 1a), emerged in late 2020 in Eastern Cape, South Africa (SA)12. This variant has since become dominant locally, raising the specter that it too has enhanced transmissibility. B.1.351 contains 9 spike mutations in addition to D614G, including a cluster of mutations (e.g., 242–244del & R246I) in NTD, three mutations (K417N, E484K, & N501Y) in RBD, and one mutation (A701V) near the furin cleavage site (Fig. 1b). There is a growing concern that these new variants could impair the efficacy of current mAb therapies or vaccines, because many of the mutations reside in the antigenic supersite in NTD18,19 or in the ACE2-binding site (also known as the receptor-binding motif—RBM) that is a major target of potent virus-neutralizing antibodies. We therefore addressed this concern by assessing the susceptibility of authentic B.1.1.7 and B.1.351 viruses to neutralization by 30 mAbs, 20 convalescent plasma, and 22 vaccinee sera. In addition, we created VSV-based SARS-CoV-2 pseudoviruses that contain each of the individual mutations as well as one with all 8 mutations of the B.1.1.7 variant (UKΔ8) and another with all 9 mutations of the B.1.351 variant (SAΔ9). A total of 18 mutant pseudoviruses were made as previously described20,21, and each was found to have a robust titer (Extended Data Fig. 1) adequate for neutralization studies.
Fig. 1 |. Emerging SARS-CoV-2 variants identified in the United Kingdom and South Africa.
a, Phylogenetic tree of SARS-CoV-2 variants, with B.1.351 and B.1.1.7 highlighted. b, Mutations in the viral spike identified in B.1.351 (SA) and B.1.1.7 (UK) in addition to D614G.
Monoclonal antibodies
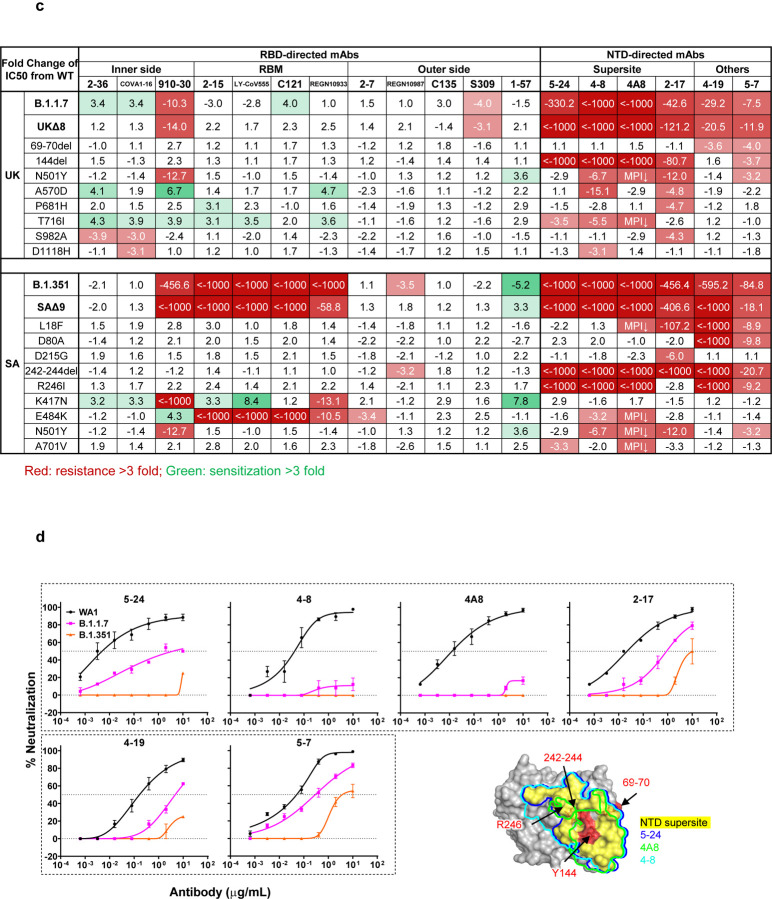
We first assayed the neutralizing activity of 12 RBD mAbs against authentic B.1.1.7 and B.1.351 viruses, as compared to the original SARS-CoV-2 strain (WT), in Vero E6 cells as previously described20,21. Three mAbs target the “inner side”, four target RBM, and five target the “outer side”. The footprints of these mAbs on RBD are shown in Fig. 2a, and their neutralization profiles are shown in Fig. 2b. For neutralization of B.1.1.7, only the activities of 910–3022 and S3095 are significantly impaired. For neutralization of B.1.351, however, the activities of 910–30, 2–1520, LY-CoV555 (bamlanivimab)1,23, C12124, and REGN10933 (casirivimab)2 are completely or markedly abolished. The four mAbs that target RBM are among the most potent SARS-CoV-2-neutralizing antibodies in clinical use or development. Note that mAbs directed to lower aspects of the “inner side” (2–3620 & COVA1–1625,26) or to the “outer side” retain their activities against B.1.351, including 2–720, REGN10987 (imdevimab)2, C13524, and S309 that are in clinical use or development. The results on the neutralization of B.1.1.7 and B.1.351 by these 12 mAbs are summarized in Fig. 2c as fold increase or decrease in IC50 neutralization titers relative to the WT. To understand the specific spike mutations responsible for the observed changes, we also tested the same panel of mAbs against pseudoviruses UKΔ8 and SAΔ9, as well as those containing only a single mutation found in B.1.1.7 or B.1.351. The results are displayed, among others, in Extended Data Fig. 2 and summarized in Fig. 2c. There is general agreement for results between B.1.1.7 and UKΔ8, as well as between B.1.351 and SAΔ9. Against B.1.1.7, the decreased activity of 910–30 is mediated by N501Y, whereas the slightly impaired activity of S309 is unexplained. Against B.1.351, the complete loss of activity of 2–15, LY-CoV555, and C121 is mediated by E484K; the complete loss for 910–30 is mediated by K417N; and the marked reduction for REGN10933 is mediated by K417N and E484K, as has been reported27. A structural explanation on how E484K disrupts the binding of 2–15, LY-CoV555, and REGN10933 is presented in Extended Data Fig. 3a.
Fig. 2 |. Susceptibility of B.1.1.7 and B.1.351 to neutralization by mAbs.
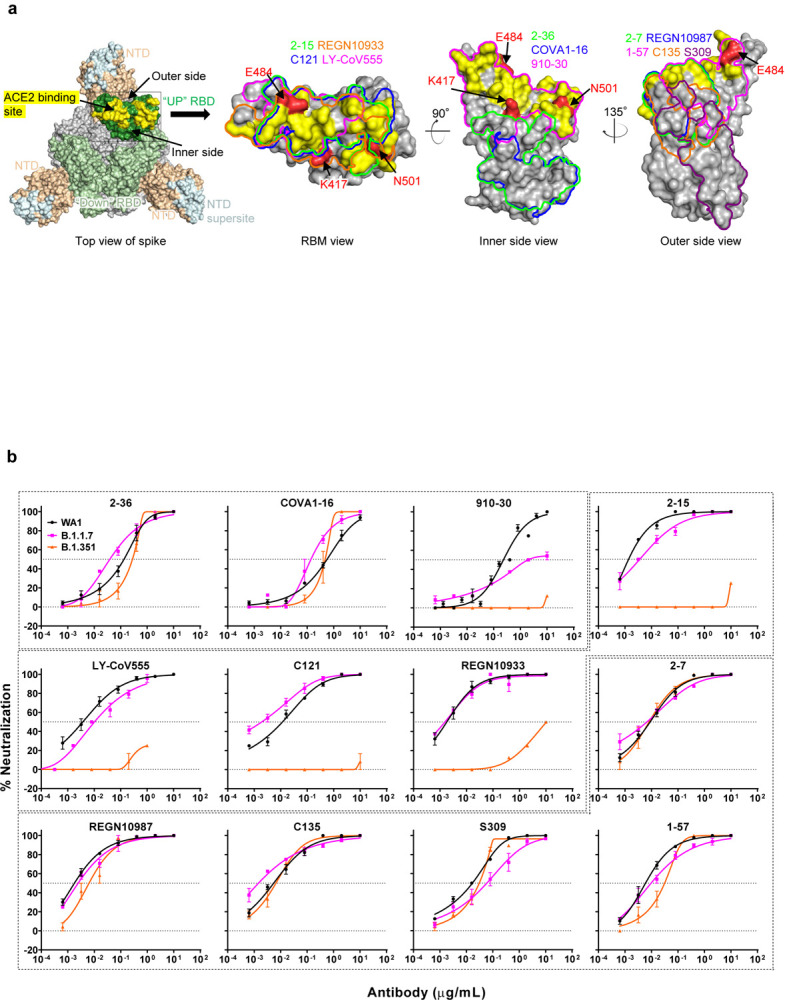
a, Footprints of neutralizing mAbs on the RBD. Left panel, top view of SARS-COV-2 spike with one RBD in the “up” conformation (pdb: 6zgg). RBD and NTD are colored green and peach, respectively. The positions of ‘inner’ and ‘outer’ sides are indicated on the “up” RBD with the ACE2-binding site colored yellow. The three panels to the right show the antibody footprints on RBD. b, Neutralization of B.1.1.7, B.1.351, and WT viruses by select RBD mAbs. c, Fold increase or decrease in IC50 of neutralizing mAbs against B.1.1.7 and B.1.351, as well as UKΔ8, SAΔ9, and single-mutation pseudoviruses, relative to WT, presented as a heatmap with darker colors implying greater change. MPI↓ denotes that maximum percent inhibition is substantially reduced, confounding IC50 calculations. d, Neutralization of B.1.1.7, B.1.351, and WT viruses by NTD-directed mAbs, the footprints of which are delineated by the color tracings in the insert. e, Changes in neutralization IC50 of authorized or investigational therapeutic mAbs against B.1.1.7, B.1.351, WT (WA1) viruses as well as UKΔ8, SAΔ9, and WT (D614G) pseudoviruses. Data in b and d are mean ± SEM of technical triplicates, and represent one of two independent experiments.
We also assessed the neutralizing activity of six NTD mAbs against B.1.1.7, B.1.351, and WT viruses. Both B.1.1.7 and B.1.351 are profoundly resistant to neutralization by our antibodies 5–24 and 4–820, as well as by 4A828, all of which target the antigenic supersite in NTD18 (Insert in Fig. 2d). The activities of 2–17, 4–19, and 5–720 are variably impaired, particularly against B.1.351. To understand the specific mutations responsible for the observed changes, we then tested these mAbs against pseudoviruses containing only a single mutation found in B.1.1.7 or B.1.351 (Extended Data Fig. 2). The results are summarized in Fig. 2c as fold increase or decrease relative to the WT (D614G). It is evident that the resistance of B.1.1.7 to most NTD mAbs is largely conferred by 144del, whereas the resistance of B.1.351 is largely conferred by 242–244del and/or R246I. Amino-acid residues 144, 242–244, and 246 all fall within the NTD supersite18,19 (Insert in Fig. 2d; details in Extended Data Fig. 3b).
We next tested the neutralizing activity of 12 additional RBD mAbs, including ones from our own collection (1–20, 4–20, 2–4, 2–43, 2–30, & 2–38)20 as well as CB6 (etesevimab)3,6, COV2–2196 & COV2–21307, Brii-196 & Brii-1984, and REGN10985. The results against B.1.1.7, B.1.351, and WT are highlighted in Extended Data Fig. 4a, and the detailed findings against the single-mutation pseudoviruses are shown in Extended Data Fig. 2. The fold changes in neutralization IC50 titers relative to the WT are tabulated in Extended Data Fig. 4b. Here, we only comment on results for mAbs in clinical development. The activity of CB6 is rendered inactive against B.1.351 because of K417N. Brii-196 and COV2–2130 are essentially unaffected by the new variants; the activities of Brii-198 and COV2–2196 are diminished 14.6 fold and 6.3 fold, respectively, against B.1.351 but not against B.1.1.7.
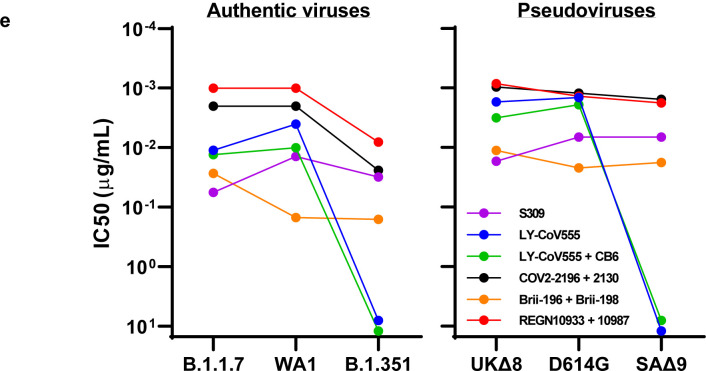
Lastly, we examined, in a single experiment, the neutralizing activity of mAb therapies in clinical use or under clinical investigation against B.1.1.7, B.1.351, and WT viruses, as well as against UKΔ8, SAΔ9, and WT pseudoviruses. The results for single mAb LY-CoV555 and S309, as well as for combination regimens REGN10933+REGN10987, LY-CoV555+CB6, Brii-196+Brii-198, and COV2–2196+COV2–2130, are shown in Extended Data Fig. 5 and summarized in Fig. 2e. Note that LY-CoV555, alone or in combination with CB6, is no longer able to neutralize B.1.351. While REGN10933+REGN10987 and COV2–2196+COV2–2130 are seemingly unaffected against variant pseudoviruses, there are noticeable decreases in their activity against B.1.351 authentic virus. Although S309 and the Brii-196+Brii-198 combination are not significantly impaired, their potencies are noticeably lower (Fig. 2e). These findings suggest that antibody treatment of this virus might need to be modified in localities where B.1.351 and related variants13,14 are prevalent, and highlight the importance of combination antibody therapy to address the expanding antigenic diversity of SARS-CoV-2.
Convalescent plasma
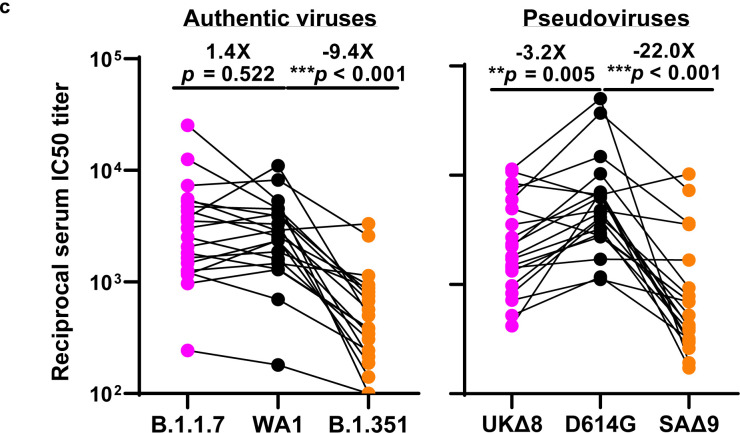
We obtained convalescent plasma from 20 patients more than one month after documented SARS-CoV-2 infection in the Spring of 2020. Each plasma sample was then assayed for neutralization against B.1.1.7, B.1.351, and WT viruses. Fig. 3a shows that most (16 of 20) plasma samples lost >2.5-fold neutralizing activity against B.1.351, while maintaining activity against B.1.1.7. Only plasma from P7, P10, P18, and P20 retain neutralizing activities similar to those against the WT. These results are summarized as fold increase or decrease in plasma neutralization IC50 titers in Fig. 3b. Furthermore, the magnitude of the drop in plasma neutralization is better seen in Fig. 3c, showing no loss of activity against B.1.1.7 but substantial loss against B.1.351 (9.4 fold).
Fig. 3 |. B.1.351 is more resistant to neutralization by convalescent plasma from patients.
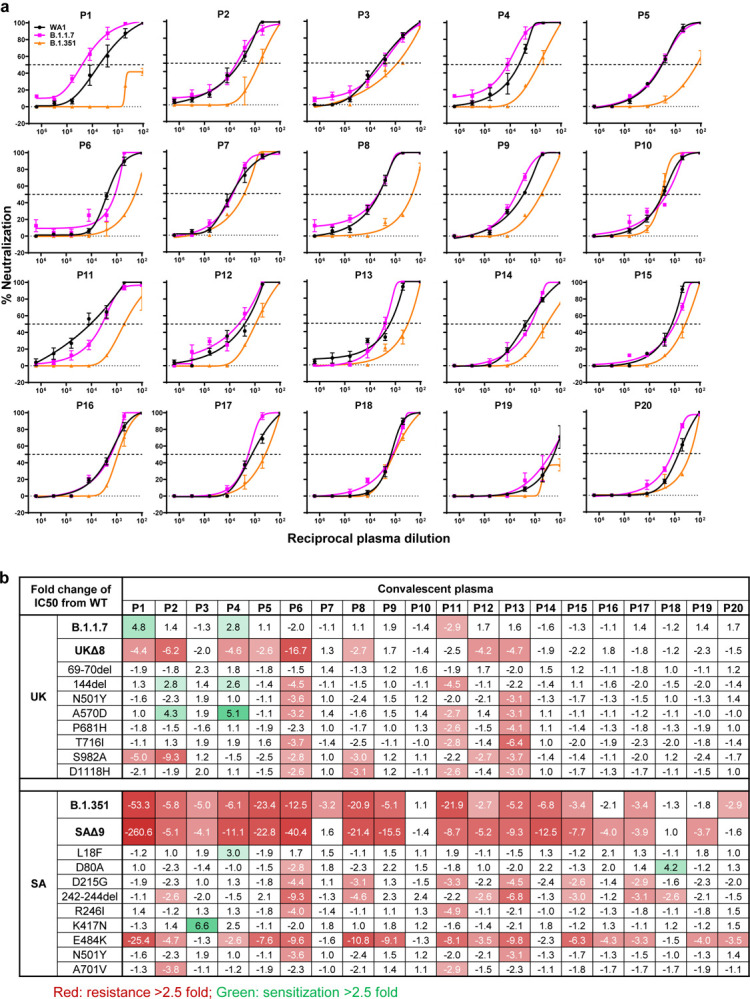
a, Neutralization results for 20 convalescent plasma samples (P1–P20) against B.1.1.7, B.1.351, and WT viruses. Data represent mean ± SEM of technical triplicates. b, Fold increase or decrease in neutralization IC50 of B.1.1.7 and B.1.351, as well as UKΔ8, SAΔ9, and single-mutation pseudoviruses, relative to the WT presented as a heatmap with darker colors implying greater change. c, Change in reciprocal plasma neutralization IC50 values of convalescent plasma against B.1.1.7 and B.1.351, as well as UKΔ8 and SAΔ9, relative to the WT. Mean fold changes in IC50 values relative to the WT are written above the p values. Statistical analysis was performed using a Wilcoxon matched-pairs signed rank test. Two-tailed p-values are reported.
Every plasma sample was also tested against each mutant pseudovirus, and those findings are shown in Extended Data Fig. 6 and summarized in Figs. 3b & 3c. Eight samples show >2.5-fold decrease in neutralizing activity against UKΔ8, in contrast to the results for B.1.1.7 neutralization. These discrepant results highlight our previous observation20 that pseudovirus neutralization does not always faithfully recapitulate live virus neutralization. The loss of plasma neutralizing activity against B.1.351 could be largely attributed to E484K (Fig. 3b), which has been shown to attenuate the neutralizing activity of convalescent sera29. Our findings here suggests that this RBM mutation is situated in an immunodominant epitope for most infected persons. It is also interesting to note that cases such as P7, P10, and P18 have neutralizing antibodies that are essentially unperturbed by the multitude of spike mutations found in these two new variants (Fig. 3b). A detailed analysis of their antibody repertoire against the viral spike could be informative.
Vaccinee Sera
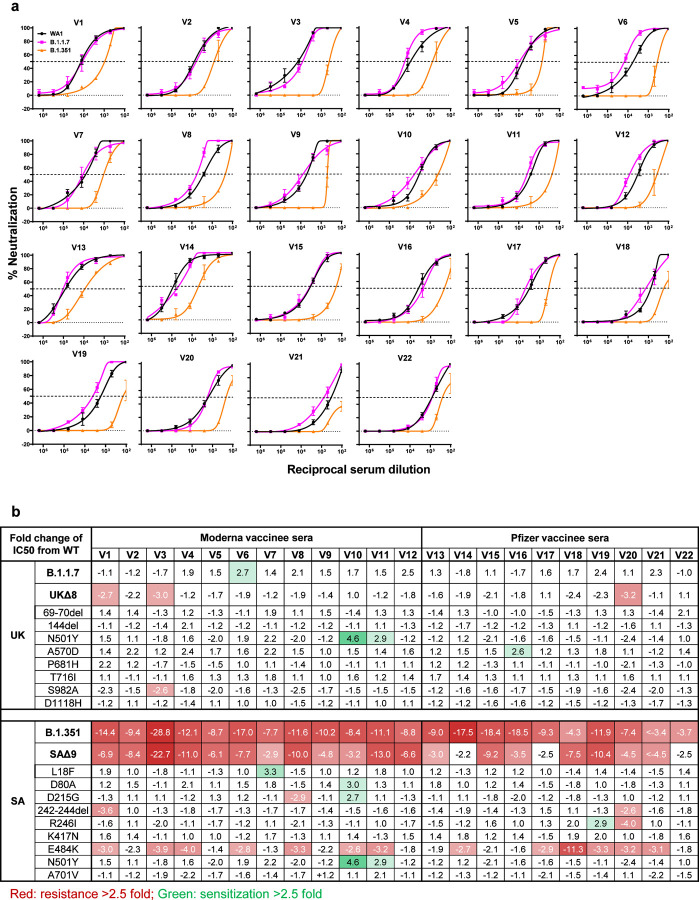
Sera were obtained from 12 participants of a Phase 1 clinical trial of Moderna SARS-Co-2 mRNA-1273 Vaccine9 conducted at the NIH. These volunteers received two immunizations with the vaccine (100 μg) on days 0 and 28, and blood was collected on day 43. Additional vaccinee sera were obtained from 10 individuals who received the Pfizer BNT162b2 Covid-19 Vaccine10 under emergency use authorization at the clinical dose on days 0 and 21. Blood was collected on day 28 or later.
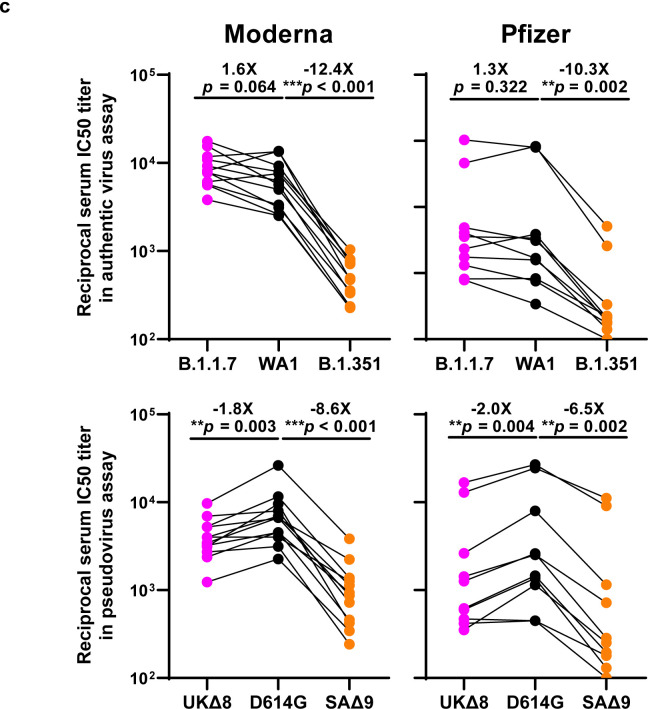
Each vaccinee serum sample was assayed for neutralization against B.1.1.7, B.1.351, and WT viruses. Fig. 4a shows no loss of neutralizing activity against B.1.1.7, whereas every sample lost activity against B.1.351. These results are quantified and tabulated as fold increase or decrease in neutralization IC50 titers in Fig. 4b, and the extent of the decline in neutralization activity is more evident in Fig. 4c. Overall, the neutralizing activity against B.1.1.7 was essentially unchanged, but significantly lower against B.1.351 (12.4 fold, Moderna; 10.3 fold, Pfizer).
Fig. 4 |. B.1.351 is more resistant to neutralization by vaccinee sera.
a, Neutralization profiles for 22 serum samples obtained from persons who received SARS-CoV-2 vaccine made by Moderna (V1-V12) or Pfizer (V13-V22) against B.1.1.7, B.1.351, and WT viruses. Data are mean ± SEM of technical triplicates, and represent one of two independent experiments. b, Fold change in serum neutralization IC50 of B.1.1.7 and B.1.351, as well as UKΔ8, SAΔ9, and single-mutation pseudoviruses, relative to the WT, presented as a heatmap with darker colors implying greater change. c, Change in reciprocal serum IC50 values for Moderna and Pfizer vaccinees against B.1.1.7 and B.1.351, as well as UKΔ8 and SAΔ9, relative to the WT. Mean fold change in IC50 relative to the WT is written above the p values. Statistical analysis was performed using a Wilcoxon matched-pairs signed rank test. Two-tailed p-values are reported.
Every vaccinee serum was also tested against each mutant pseudovirus, and the results are presented in Extended Data Fig. 7 and summarized in Figs. 4b & 4c. No single mutation in B.1.1.7 has an appreciable impact on the neutralizing activity of vaccinee sera. The loss of neutralizing activity against SAΔ9 is largely consistent with the loss in B.1.351 live virus neutralization. A major contributor to the neutralization resistance of this variant virus appears to be E484K (Fig. 4b), indicating that this RBM mutation is situated in an immunodominant epitope recognized by all vaccinees studied.
Discussion
Both SARS-CoV-2 variants B.1.1.7 and B.1.351 are raising concerns not only because of their increase transmissibility but also because of their extensive mutations in spike that could lead to antigenic changes detrimental to mAb therapies and vaccine protection. It is of equal concern that another variant known as P.1 or 501Y.V3 is increasing rapidly in Brazil and spreading far beyond13,14. P.1 contains three mutations (K417T, E484K, and N501Y) at the same RBD residues as B.1.351. Much of our findings on B.1.351 would likely be similar for this emergent variant. N501Y is shared among viruses in these three lineages; while this mutation may confer enhanced binding to ACE230, its antigenic impact is limited to a few mAbs (Fig. 2c & Extended Data Fig. 4b), with no pronounced effects on the neutralizing activity of convalescent plasma or vaccinee sera (Figs. 3b & 4b), as others are reporting31–33.
Our findings have relevance to the use of mAb to treat or prevent SARS-CoV-2. Both B.1.1.7 and B.1.351 are resistant to neutralization by mAbs directed to the NTD supersite (Figs. 2c, 2d, & Extended Data Fig. 3b). More importantly, B.1.351 is resistant to a major group of potent mAbs that target the RBM, including three regimens authorized for emergency use (Fig. 2c). LY-CoV555 alone and in combination with CB6 are inactive against B.1.351, and the activity of REGN10933 is impaired (Fig. 2b) while its combination with REGN10987 retains much of the activity (Fig. 2e). Several other mAbs in development are similarly impaired (Figs. 2c, 2e, & Extended Data Fig. 4b) against this variant. Decisions on the use of these mAbs will depend heavily on the local prevalence of B.1.351 or variants with an E484K mutation, thus highlighting the importance of viral genomic surveillance worldwide and proactive development of next-generation antibody therapeutics, including combinations that target antigenically distinct epitopes.
Convalescent plasma from patients infected with SARS-CoV-2 from early in the pandemic show no significant change in neutralizing activity against B.1.1.7, but the diminution against B.1.351 is remarkable (Figs. 3b &3c). This relative resistance is largely due to E484K, a mutation shared by B.1.351 and P.112–14. Again, in areas where such viruses are common, one would have a concern about re-infection, as other studies are also suggesting34,35. This apprehension is heightened by the recent observation from the Novavax vaccine trial in South Africa that placebo recipients with prior SARS-CoV-2 infection were not protected against a subsequent exposure to B.1.35136,37. Even more disturbing is the situation in Manaus, Brazil where a second wave of infection due to P.1 is sweeping through a population that was already 76% seropositive due to prior infection in the Spring of 202038.
As for the ramifications of our findings for the protective efficacy of current SARS-CoV-2 vaccines, the neutralizing activity of vaccinee sera against B.1.1.7 is largely intact and no adverse impact on current vaccines is expected (Fig. 4c), consistent with conclusions being reached by others33,39,40. On the other hand, the loss of 10.3–12.4 fold in activity against B.1.351 is larger than results being reported using mutant pseudoviruses33,41,42 or live virus43. Taken together, the overall findings are worrisome, particularly in light of recent reports that both Novavax and Johnson & Johnson vaccines showed a substantial drop in efficacy in South Africa36,37,44.
The recent emergence of B.1.1.7, B.1.351, and P.1 marks the beginning of SARS-CoV-2 antigenic drift. This conclusion is supported by data presented herein, illustrating how so many of these spike changes conferred resistance to antibody neutralization, and by studies reporting similar spike mutations selected by antibody pressure in vitro or in vivo45–49. Mutationally, this virus is traveling in a direction that could ultimately lead to escape from our current therapeutic and prophylactic interventions directed to the viral spike. If the rampant spread of the virus continues and more critical mutations accumulate, then we may be condemned to chasing after the evolving SARS-CoV-2 continually, as we have long done for influenza virus. Such considerations require that we stop virus transmission as quickly as is feasible, by redoubling our mitigation measures and by expediting vaccine rollout.
Supplementary Material
References
- 1.Chen P. et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N Engl J Med 384, 229–237 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hansen J. et al. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science 369, 1010–1014 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gottlieb R. L. et al. Effect of Bamlanivimab as Monotherapy or in Combination With Etesevimab on Viral Load in Patients With Mild to Moderate COVID-19: A Randomized Clinical Trial. JAMA (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju B. et al. Human neutralizing antibodies elicited by SARS-CoV-2 infection. Nature 584, 115–119 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Pinto D. et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature 583, 290–295 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Shi R. et al. A human neutralizing antibody targets the receptor-binding site of SARS-CoV-2. Nature 584, 120–124 (2020). [DOI] [PubMed] [Google Scholar]
- 7.Zost S. J. et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature 584, 443–449 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krammer F. SARS-CoV-2 vaccines in development. Nature 586, 516–527 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Anderson E. J. et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N Engl J Med 383, 2427–2438 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polack F. P. et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med 383, 2603–2615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rambaut A, et al. Preliminary genomic characterisation of an emergent SARSCoV-2 lineage in the UK defined by a novel set of spike mutations. https://virological.org/t/preliminarygenomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-ofspike-mutations/563. (2020).
- 12.Tegally H. et al. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv, 2020.2012.2021.20248640 (2020). [Google Scholar]
- 13.Faria NR, et al. Genomic characterisation of an emergent SARS-CoV-2 lineage in Manaus: preliminary findings. https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings/586. (2021).
- 14.Naveca F, et al. Phylogenetic relationship of SARS-CoV-2 sequences from Amazonas with emerging Brazilian variants harboring mutations E484K and N501Y in the Spike protein. https://virological.org/t/phylogenetic-relationship-of-sars-cov-2-sequences-from-amazonas-with-emerging-brazilian-variants-harboring-mutations-e484k-and-n501y-in-the-spike-protein/585. (2021).
- 15.Korber B. et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 182, 812–827 e819 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hou Y. J. et al. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science 370, 1464–1468 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies N. G. et al. Increased hazard of death in community-tested cases of SARS-CoV-2 Variant of Concern 202012/01. medRxiv, 2021.2002.2001.21250959 (2021). [Google Scholar]
- 18.Cerutti G. et al. Potent SARS-CoV-2 Neutralizing Antibodies Directed Against Spike N-Terminal Domain Target a Single Supersite. bioRxiv, 2021.2001.2010.426120 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCallum M. et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. bioRxiv, 2021.2001.2014.426475 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L. et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 584, 450–456 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Wang P. et al. SARS-CoV-2 neutralizing antibody responses are more robust in patients with severe disease. Emerg Microbes Infect 9, 2091–2093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Banach B. B. et al. Paired heavy and light chain signatures contribute to potent SARS-CoV-2 neutralization in public antibody responses. bioRxiv, 2020.2012.2031.424987 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones B. E. et al. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv (2020). [Google Scholar]
- 24.Robbiani D. F. et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature 584, 437–442 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brouwer P. J. M. et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science 369, 643-+ (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu H. et al. Cross-Neutralization of a SARS-CoV-2 Antibody to a Functionally Conserved Site Is Mediated by Avidity. Immunity 53, 1272–1280 e1275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Starr T. N. et al. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science, eabf9302 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chi X. et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science 369, 650–655 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greaney A. J. et al. Comprehensive mapping of mutations to the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human serum antibodies. bioRxiv, 2020.2012.2031.425021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Starr T. N. et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell 182, 1295–1310 e1220 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xie X. et al. Neutralization of SARS-CoV-2 spike 69/70 deletion, E484K and N501Y variants by BNT162b2 vaccine-elicited sera. Nature Medicine (2021). [DOI] [PubMed] [Google Scholar]
- 32.Rees-Spear C. et al. The impact of Spike mutations on SARS-CoV-2 neutralization. bioRxiv, 2021.2001.2015.426849 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu K. et al. mRNA-1273 vaccine induces neutralizing antibodies against spike mutants from global SARS-CoV-2 variants. bioRxiv, 2021.2001.2025.427948 (2021). [Google Scholar]
- 34.Cele S. et al. Escape of SARS-CoV-2 501Y.V2 variants from neutralization by convalescent plasma. medRxiv, 2021.2001.2026.21250224 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wibmer C. K. et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv, 2021.2001.2018.427166 (2021). [DOI] [PubMed] [Google Scholar]
- 36.Wadman M. & Cohen J. Novavax vaccine delivers 89% efficacy against COVID-19 in U.K.—but is less potent in South Africa, https://www.sciencemag.org/news/2021/01/novavax-vaccine-delivers-89-efficacy-against-covid-19-uk-less-potent-south-africa. (2021).
- 37.Callaway E. & Mallapaty S. Novavax offers first evidence that COVID vaccines protect people against variants. https://www.nature.com/articles/d41586-021-00268-9. (2021). [DOI] [PubMed]
- 38.Sabino E. C. et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet 397, 452–455 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collier D. et al. Impact of SARS-CoV-2 B.1.1.7 Spike variant on neutralisation potency of sera from individuals vaccinated with Pfizer vaccine BNT162b2. medRxiv, 2021.2001.2019.21249840 (2021). [Google Scholar]
- 40.Muik A. et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. bioRxiv, 2021.2001.2018.426984 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Z. et al. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tada T. et al. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv, 2021.2002.2005.430003 (2021). [Google Scholar]
- 43.Huang B. et al. Neutralization of SARS-CoV-2 VOC 501Y.V2 by human antisera elicited by both inactivated BBIBP-CorV and recombinant dimeric RBD ZF2001 vaccines. bioRxiv, 2021.2002.2001.429069 (2021). [Google Scholar]
- 44.Haseltine WA. Novavax And Johnson & Johnson’s Vaccines Are Less Effective Against The UK And South African Variants. https://www.forbes.com/sites/williamhaseltine/2021/02/01/novavax-and-johnson--johnsons-vaccines-are-less-effective-against-the-uk-and-south-african-variants/?sh=545908868e4f. (2021).
- 45.Baum A. et al. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science 369, 1014–1018 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weisblum Y. et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kemp S. A. et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McCarthy K. R. et al. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science, eabf6950 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andreano E. et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv, 2020.2012.2028.424451 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.