Abstract
Coronaviruses (CoVs) cause diseases such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and coronavirus disease 2019 (COVID-19). Therefore, this study was conducted to combat major CoVs via a trivalent subunit vaccine, which was engineered by implementing sequences of spike (S) protein, nucleocapsid (N), envelope (E), membrane (M) protein, non-structural protein (nsp) 3, and nsp8 antigens. The CTL, HTL, MHC I, and IFN-γ epitopes were predicted via CTLPRED, IEDB, and IFN epitope servers, respectively. Also, to stimulate strong helper T lymphocytes (HTLs) responses, Pan HLA DR-binding epitope (PADRE) was used. Also, for boosting the immune response, β-defensin 2 was added to the construct as an adjuvant. Furthermore, TAT was applied to the vaccine to facilitate the intracellular delivery. Finally, TAT, adjuvant, PADRE, and selected epitopes were appropriately assembled. Based on the predicted epitopes, a trivalent multi-epitope vaccine with a molecular weight of 74.8 kDa was constructed. Further analyses predicted the molecule to be a strong antigen, and a non-allergenic and soluble protein. Secondary and tertiary structures were predicted. Additionally, analyses validated the stability of the proposed vaccine. Molecular docking and molecular dynamics simulation (MDS) showed binding affinity and stability of the vaccine-TLR3 complex was favorable. The predicted epitopes demonstrated a strong potential to stimulate T and B-cell mediated immune responses. Furthermore, codon optimization and in silico cloning guaranteed increased expression. In summary, investigations demonstrated that this next-generation approach might provide a new horizon for the development of a highly immunogenic vaccine against SARS‐CoV, MERS‐CoV, and SARS-CoV-2.
Communicated by Ramaswamy H. Sarma
Keywords: Immunoinformatics, MERS‐CoV, SARS‐CoV, SARS-CoV-2, Subunit vaccine
Graphical Abstract
1. Introduction
Coronaviruses (CoVs) are positive-strand viruses from the family Coronaviridae, which is named after its crown-like shape (Tortorici & Veesler, 2019). CoVs have caused major outbreaks in recent years (Azhar et al., 2016; Habibzadeh & Stoneman, 2020). Various human CoVs have spread globally, causing mostly mild illness, while others are highly pathogenic. The first severe acute respiratory syndrome (SARS)-CoV emerged in china in 2001, and resulted in a global outbreak. However, this outbreak was successfully contained. In 2012, another CoV variant named Middle East respiratory syndrome (MERS)-CoV emerged in Jeddah, Saudi Arabia. However, this outbreak never diminished perfectly, and hospitals and community outbreaks continued to occur annually (Fehr & Perlman, 2015). Recently, SARS-CoV-2 initiated an outbreak that turned into a pandemic, raising global consternation. This newly surfaced virus that caused a new disease named coronavirus disease (COVID-19), has a considerably lower mortality rate. However, the rapid transmission of the virus has turned COVID-19 into a fatal pandemic, which indicates a raising need for novel treatments and preventive methods.
Numerous vaccine and drug design efforts have been targeted on CoVs. Some vaccine design efforts against SARS-CoV-2 have progressed into phase I clinical trials for SARS-CoV-2 in the USA (NCT04283461) and China (NCT04313127). However, results of preventive and therapeutic approaches targeted at CoVs, in particular, vaccines have not been adequately successful. Several aspects of CoVs may highlight the importance of prevention. Remarkably, various subtypes of CoVs such as MERS-CoV, which has caused past outbreaks, have remained relevant (Kleine-Weber et al., 2018).
CoV proteins can be classified into two subgroups, structural and non-structural proteins (nsps). The genome of CoVs is surrounded by structural proteins comprising spike (S) protein, nucleocapsid (N), envelope (E), and membrane (M) proteins (Rappuoli et al., 2016). S protein performs various functions and mediates the cellular entrance of CoVs. After attachment of the S protein to its receptor, CoVs can enter the cytoplasm. Protease breakdown of S protein is required for virus-cell fusion and the entry of genomic RNA (Dawson et al., 2019; Rappuoli et al., 2016; Zumla et al., 2015). The S protein of SARS-CoV-2 is highly similar to SARS-CoV. Also, polyclonal antibodies for SARS-CoV suppress spike-regulated entrance of SARS-CoV-2 (Walls et al., 2020). HCoV-NL63, a virus that only causes mild symptoms, also has S protein, and recruits the angiotensin-converting enzyme 2 (ACE2) for entering the host cell, while showing a very low pathogenicity. This indicates that additional factors are responsible for the pathogenicity of the major CoVs involved in significant outbreaks. (Hofmann et al., 2005). A high similarity was determined for N protein of SARS-CoV, SARS-CoV-2, and MERS-CoV, while it was significantly different from other CoVs that mainly cause mild illness. Thus, the N protein has been suggested as a promising candidate for cross-protective vaccine design. (Ong et al., 2020). The E and M proteins have been suggested as vaccine candidates in reverse vaccinology (RV) assessments. The E and M proteins showed key roles in the viral assembly of CoVs. Further, the N protein is essential for viral RNA synthesis. Deletion of the E protein inhibits the virulence of CoVs, and some researches have been directed at the efficacy of live attenuated vaccines that comprise recombinant SARS-CoV or MERS-CoV with a mutated E protein (Shang et al., 2020). Also, M protein has been utilized for DNA vaccine construction against SARS-CoV (Shi et al., 2006). The implementation of non-structural proteins (nsp), a part of ORF1a polyprotein, may result in additional protection. The nsp3 is the second-highest pathogenic factor of SARS-CoV-2 protein. Most vaccines contain an attenuated or inactivated whole virus and structural proteins. However, such vaccines could not give complete protection, and whole vaccines may cause side effects. Therefore, next-generation vaccines are to be developed.
Taken together, an efficient multipotent preventive vaccine against various subclasses of rapidly emerging human CoVs remains to be discovered. This is highly important because of the recent CoV outbreak, which has raised an urgent need for the development of vaccine research. For this purpose, bioinformatics analyses have been utilized to aid the vaccine design process. Hence, we utilized in silico methods to design a trivalent novel cross-protective multi-epitope vaccine against SARS-CoV, MERS-CoV, and SARS-CoV-2 via spike (S) protein, nucleocapsid (N), envelope (E), membrane (M) protein, nsp3, and nsp8 antigens.
2. Material and methods
2.1. Retrieval of protein sequences
The critical immunogenic proteins of the SARS-CoV-2, SARS-CoV and MERS-CoV. were evaluated and six proteins were selected and the amino acid sequence of nsp3 (Accession no. YP_009725299.1, ACZ72252.1:819-2740, and YP_009047231.1), nsp8 (Accession no. YP_009725304.1, ACZ72252.1:3920-4117, and YP_009047236.1), E protein (Accession no. QIH45055.1, ABI96961.1and ATG84893.1), M protein (Accession no. QII57163.1, P59596.1, and AXN73531.1), S protein (Accession no. QII57161.1, A.1, and AXN73514.1), and N protein (Accession no. QIH45060.1, P59595.1, and ATG84895.1) were retrieved from the Universal Protein Resource (UniProt) (http://www.uniprot.org) and the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/protein/). The sequences were then saved in FASTA format.
2.2. Cytotoxic T lymphocytes (CTL) and IFN-γ inducing epitopes prediction
Choosing CTLs epitopes is one of the most important elements of vaccine development. This is due to that CTLs kill virus-infected or damaged cells by recognizing the epitopes presented by MHC-I. Indeed, the initial step of stimulating the immune system is presenting an antigen on the MHC-I. CTLPred (http://crdd.osdd.net/raghava/ctlpred/) is a freely accessible web server for the prediction of CTL epitopes. This server is based on artificial neural network (ANN), support vector machine (SVM), consensus, and combined prediction methods.
2.3. Helper T-lymphocyte (HTL) epitope prediction and IFN-γ inducing epitopes prediction
IEDB prediction tool (http://tools.immuneepitope.org/mhcii/) was employed to predict helper T cell (HTL) epitopes via the selected antigens. Each selected peptide was a 15mer. The epitopes were predicted for human HLA-II alleles as target species. We used the IEDB recommended method to achieve the best possible result. Besides, IFN-γ plays an essential role in the innate and adaptive immune response by stimulating natural killer cells and macrophages, and provides an elevated response to MHC antigens. Also, IFN-γ epitope server (http://crdd.osdd.net/raghava/ifnepitope/scan.php) was used to predict IFN-positive sequences in the selected antigens (Dhanda et al., 2013; Webb & Sali, 2016).
2.4. Trivalent subunit multi-epitope vaccine construct and gene optimization
The N-terminal sequence of the subunit vaccine starts with the TAT peptide of HIV as cell-penetrating peptides (CPPs) and is followed by the β-defensin 2 as an adjuvant. Following this step, PADRE was utilized as helper T lymphocyte epitope. Then, HTL and CTL selected epitopes were added sequentially for each SARS-CoV-2, SARS-CoV, and MERS-CoV protein, respectively. The AAY, GPGPG, EAAAK, KK, and GGGS amino acids sequences were used as suitable linkers to attach the selected epitopes and achieve a multi-epitope vaccine consisting of an antigenic part of a pathogenic organism to elicit an immunological response. Finally, the DNA sequence of the subunit vaccine and its adaptation to E. coli codon usage were obtained from the JCAT server (http://www.jcat.de).
2.5. Population coverage
IEDB population coverage tool (http://tools.iedb.org/population/) was used to determine the population coverage of the final construct within the global population. Epitope/MHC restriction data were loaded on IEDB and prediction calculations were performed for MHC-I and MHC-II separate epitopes.
2.6. Physico-chemical parameters analysis
The codon usage was analyzed by Java Codon Adaptation Tool (JCat).The Mfold server (http://unafold.rna.albany.edu/) was used to predict the secondary structure of RNA. Mfold online server predicts the true positive base pairs and minimum ΔG for structures via thermodynamics methods. Besides, the physical and chemical vaccine parameters were discovered. Some physicochemical characteristics of the designed structure, including amino acid composition, instability index, molecular weight, PI value, in vitro and in vivo estimated half-life, aliphatic index, extinction coefficient, and grand average of hydropathicity (GRAVY) were evaluated using ProtParam online server (https://web.expasy.org/protparam/).
2.7. Prediction of solubility, antigenicity, and allergenicity
SOLpro online server (http://scratch.proteomics.ics.uci.edu) was utilized to predict the propensity of the protein to be soluble upon overexpression in Escherichia Coli. This server uses a two-stage SVM architecture method to evaluate the protein solubility based on multiple representations of the primary sequence. Besides, the prediction of allergenicity with high precision was discovered by AlgPred (http://crdd.osdd.net/raghava/algpred/). Furthermore, the antigenicity of the proposed vaccine was assessed using another online server VaxiJen v2.0 (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html). The accuracy of this server varies from 70% to 89% depending on the organism.
2.8. Prediction of secondary and tertiary structures
The secondary structure of the predicted vaccine was predicted using GOR IV and PSIPRED online servers (http://expasy.org/tool/gor4.html and http://bioinf.cs. ucl.ac.uk/psipred/), respectively. The percentage of the helices (H), strands (S), and coils (C) were determined. Homology modeling of the final protein construct was performed by GalaxyTBM (http://galaxy.seoklab.org/index.html). GalaxyTBM is a template-based tertiary structure prediction online server, and predicts protein structure from the sequence when experimental structures of homologous proteins are available as templates, and refines loop or terminus regions by ab initio modeling.
2.9. Refinement and validation of the 3 D modeled structure
The tertiary structure of the final vaccine construct produced by the RaptorX server depending on the degree of similarity between available template structure options and the target protein. Therefore, the GalaxyRefine server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) was used to further improve the accuracy of the modeled structure. The CASP10 assessed the GalaxyRefine server and reported that GalaxyRefine is one of the best-performing methods for improving the local structure quality. Also, SAVES v5.0 server (http://servicesn.mbi.ucla.edu/PROCHECK/) was employed to validate the refined model and design a Ramachandran plot for our structure.
2.10. Prediction of intrinsic protein disorder
DisEMBL 1.5 (http://dis.embl.de/) and IUPred (http://iupred.enzim.hu/pred.php) were utilized to predict intrinsic protein disorder and unstructured regions within a protein. DisEMBL is a public online server for predicting disorder in proteins. Also, DISOPRED3 was employed to predict essential protein disorder and unstructured regions in our proposed vaccine.
2.11. Prediction of conformational and linear B-cell epitopes
We utilized Discotope 2.0 server (http://www.cbs.dtu.dk/services/DiscoTope/) to predict conformational epitopes for the modeled vaccine. The Discotope 2.0 server was used for the estimation of surface accessibility in terms of contact numbers and a new epitope propensity amino acid score to predict discontinuous B cell epitopes from the tertiary structure. The final scores were determined by combining the propensity scores of residues in spatial proximity and contact numbers. Also, ElliPro online server (http://tools.iedb.org/ellipro/) was employed to predict linear and discontinuous B-cell epitopes. ElliPro classifies the conformational epitopes based on protrusion index, and clusters them based on distance R (Å), while the Discotope method utilizes a calculation of surface accessibility and epitope propensity amino acid score. Finally, conformational epitopes with the highest scores were selected.
2.12. Disulfide bond engineering
Improving the stability of the modeled vaccine, an important step in the vaccine design process, was completed by an approach to disulfide bond formation. Disulfide bonds provide a well-defined geometric conformation and significant stability for the protein structure. Disulfide engineering was done by Disulfide by design v2.0 (DbD2) to design novel disulfide bonds in the vaccine protein construct (Craig & Dombkowski, 2013).
2.13. Molecular docking between vaccine construct and TLRs
Molecular Docking was performed to perceive the interaction between the receptors (TLR-3, TLR4, and TLR8) and the ligand (the final vaccine construct) at their stable form and to check the binding affinity between them. The protein-protein docking was also performed by the Hdock online server (http://hdock.phys.hust.edu.cn/). HDOCK is a docking server that efficiently integrates various components, comprising sequence search, template selection, and model building. The predictive power of the HDOCK server can also be improved by ranking the template-based models. Moreover, the center of extremely populated clusters and the lowest binding energy of the receptor-ligand complexes were predicted. After several rotations of the ligand, the docked complex was selected based on the lowest binding energy. Moreover, re-affirmation was performed by the PatchDock online server (https://bioinfo3d.cs.tau.ac.il/PatchDock/) to evaluate the interaction between the designed vaccine construct and the receptors. This server produces the output based on three algorithms: filtering and scoring, molecular shape representation, and surface patch matching. The output composed of rank, global energy, atomic contact energy, transformations, and PDB file for the complex.
2.14. Molecular dynamics simulation study
The molecular dynamics (MD) simulation technique was carried out by the utilization of GROningen MAchine for Chemical Simulations (single-precision, version 2020.3) (GROMACS) (Van Der Spoel et al., 2005). The vaccine underwent a brief simulation, which was followed by last-frame extraction. This was then taken to the next MD step. Then, we simulated the TLR3 (as the main involved TLR in virus infections)-vaccine in a blood-like environment. VMD (Humphrey et al., 1996), and UCSF chimera softwares were utilized to check MD process. OPLS-AA force field was used to make the topology of proteins (Sambasivarao & Acevedo, 2009). We conducted the minimization step through the steepest descent algorithm in order to obtain a convergence Fmax aim of less than 1000 KJ.mol−1.nm−1. To avoid atoms from exceeding the box, periodic boundary conditions (PBC) by XYZ coordinates were considered. Afterwards, two equilibration stages comprising of NVT (for 100 ps), followed by NPT (for 100 ps) were carried out. We used the Brendsen pressure coupling approach with a reference pressure of 1 bar, and set the temperature to 300°K. Last, the final MD simulation was performed for 10 ns. System specifications for simulation were Intel Core i7 4.2 Ghz and AMD Radeon Pro 580 (8 Gb VRAM). We also performed the Molecular Mechanics Poisson-Boltzmann Surface Area (MM-PBSA) analyses. The sets of structures were acquired with MD or Monte Carlo methods. Of note, the collections of structures should be stored in the format of an Amber trajectory file. The MM-PBSA package has been provided for the GROMACS package (https://rashmikumari.github.io/g_mmpbsa/) as well. The MMPBSA/GBSA method combines the molecular mechanical energies with the continuum solvent approaches. MM-PBSA analyses were conducted for the equilibrated stage of the MD run. Then, SASA, electrostatic, WCA, SAV, and Van der Waal energies were reported for the contact area of the complex. We used UCSF Chimera with default contact setting and H-bond criteria to index all atoms involved in the contact area in TLR3-Vaccine complex. The contact criteria find atoms with a VDW overlap of at least −0.4 angstroms. The indexed atoms were grouped for both TLR3 and the vaccine construct, and used as an input for MMPBSA analysis.
2.15. In silico cloning
The optimized sequence of the proposed vaccine was reversely transcribed, and to ensure the restriction of cloning, XhoI and Not I restriction sites were added to the N- and C-terminals of sequence, respectively. Then, we used the Snap-Gene restriction cloning module to insert the optimized sequence into the PET21b (+) vector between defined restriction sites.
2.16. Immune simulation
The C-ImmSim online server (http://150.146.2.1/C-IMMSIM/index.php) was used to determine the immune response profile of the constructed vaccine (Rapin et al., 2010). C-ImmSim simulates three sections that represent three different anatomical regions in mammals, including the bone marrow, the tertiary lymphatic organ, and the thymus. This is an important step for the simulation of the immune response, because it determines immunogenicity. The binding of the epitope, which is the immunogenic part of an invading pathogen, together with activation and cooperation from T helper cells, is required to trigger an immune response in the affected host.
3. Results
3.1. Primary analysis of the retrieved sequences
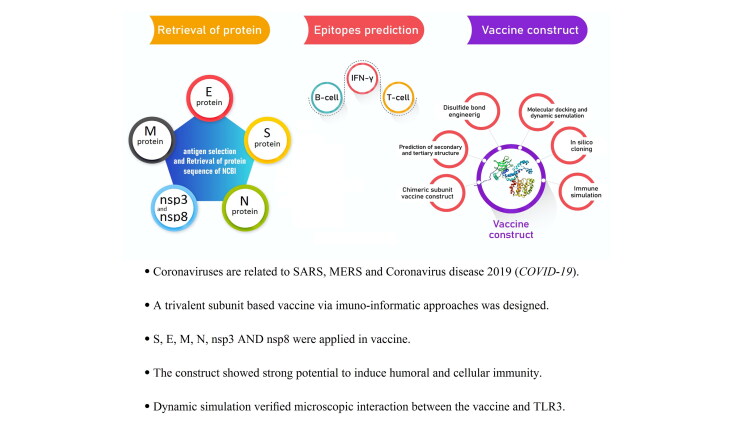
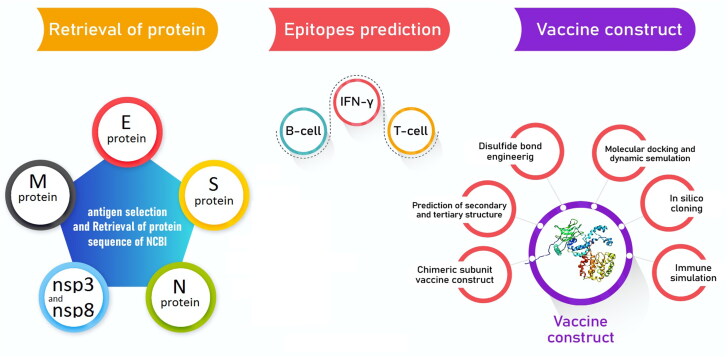
In this study, spike (S) protein, nucleocapsid (N), envelope (E), membrane (M) protein, nsp3 and nsp8 antigens (totally 6 protein) were used for bioinformatics analyses, as well as detecting CTL, HTL, IFN-γ, and MHC I binder's epitopes. The highly conserved regions of β-defensin 2 were employed as an adjuvant because the vaccine requires a strong induction of T cell immune responses. The signal sequences were excluded from all constructed structures. The graphical abstract which represents the overall procedures of vaccine design is shown in Figure 1.
Figure 1.
Flowchart summarizing the different steps of a trivalent subunit vaccine CoV design.
3.2. Cytotoxic T lymphocyte (CTL) prediction
CTL epitopes were selected according to the highest specificity and lowest sensitivity for their adaptive immune receptor. The top three epitopes with the highest score were chosen from spike S, N, E, M, nsp3, and nsp8 antigens to get a total of 18 final CTL epitopes (Table 1). On the other hand, we utilized the Immune Epitope Database (IEDB) tool to predict MHC-I peptide binders to human alleles HLA-B*57:01, HLA-A*26:01, HLA-A*11:01, HLA-B*35:01, HLA-A*03:01, HLA-B*15:01, HLA-A*01:01, HLA-A*11:01, HLA-A*30:02, HLA-A*23:01, HLA-B*38:01, HLA-B*07:02, HLA-C*07:01, HLA-A*03:01, HLA-A*25:01, HLA-A*31:01, and HLA-A*29:02. Finally, CTL epitopes were analyzed and IFN-γ inducing epitopes were applied to the final construct.
Table 1.
Predicted CTL epitopes with MHC I binding epitopes against human emerging pathogenic SARS-CoV, MERS-CoV, and SARS-CoV-2.
| Selected Antigen | Organism | Position | CTL epitope | MHC-I epitope | Allele | Percentile rank |
|---|---|---|---|---|---|---|
| nsp3 | SARS-CoV-2 | 829-840 | LNHTKKWKY | MSALNHTKKWKY | HLA-B*57:01 | 0.26 |
| SARS-CoV | 1184-1195 | FEVLAVEDT | FEVLAVEDTQGM | HLA-A*26:01 | 0.62 | |
| MERS-CoV | 391-402 | SFDYLIREA | AVSFDYLIREAK | HLA-A*11:01 | 0.32 | |
| nsp8 | SARS-CoV-2 | 12-23 | YAAFATAQE | YAAFATAQEAYE | HLA-B*35:01 | 0.21 |
| SARS-CoV | 35-46 | KKLKKSLNV | LKKLKKSLNVAK | HLA-A*03:01 | 2.10 | |
| MERS-CoV | 39-50 | AVNIAKNAY | LQKAVNIAKNAY | HLA-B*15:01 | 0.52 | |
| S protein | SARS-CoV-2 | 27-38 | YTNSFTRGV | AYTNSFTRGVYY | HLA-A*01:01 | 0.04 |
| SARS-CoV | 454-465 | ISNVPFSPD | DISNVPFSPDGK | HLA-A*11:01 | 1.40 | |
| MERS-CoV | 303-314 | IQSDRKAWA | IQSDRKAWAAFY | HLA-A*01:01 | 0.11 | |
| M protein | SARS-CoV-2 | 185-196 | RVAGDSGFA | QRVAGDSGFAAY | HLA-A*30:02 | 0.06 |
| SARS-CoV | 53-64 | LWLLWPVTL | LWLLWPVTLACF | HLA-A*23:01 | 0.32 | |
| MERS-CoV | 152-163 | HFGACDYDR | GMHFGACDYDRL | HLA-B*38:01 | 0.49 | |
| N protein | SARS-CoV-2 | 45-56 | NNTASWFTA | LPNNTASWFTAL | HLA-B*07:02 | 0.40 |
| SARS-CoV | 62-73 | EELRFPRGQ | KEELRFPRGQGV | HLA-C*07:01 | 1.40 | |
| MERS-CoV | 249-260 | HKRTSTKSF | KMRHKRTSTKSF | HLA-A*03:01 | 0.80 | |
| Eprotein | SARS-CoV-2 | 33-44 | TALRLCAYC | ILTALRLCAYCC | HLA-A*25:01 | 1.40 |
| SARS-CoV | 50-61 | VKPTVYVYS | SLVKPTVYVYSR | HLA-A*31:01 | 0.87 | |
| MERS-CoV | 49-60 | VQPALYLYN | TLLVQPALYLYN | HLA-A*29:02 | 1.30 |
3.3. HTL epitope prediction
HTL epitope prediction server for human HLA-II alleles was done with the help of the IEDB online server, including HLA-DPA1*01:03, HLA-DPB1*02:01, HLA-DQA1*01:01, HLA-DQB1*05:01, HLA-DRB1*09:01, HLA-DRB1*13:02, HLA-DRB1*13:21, HLA-DRB1*04:26, HLA-DRB1*13:02, HLA-DRB1*01:01, HLA-DQA1*05:01, HLA-DQB1*02:01, HLA-DPB1*02:01, HLA-DRB1*09:01, HLA-DRB1*11:14, HLA-DQA1*01:02, HLA-DQB1*06:02, HLA-DPA1*03:01, HLA-DPB1*04:02, HLA-DPA1*03:01, HLA-DPB1*04:02, HLA-DRB1*07:03 and HLA-DRB1*01:01 for antigens. Finally, 18 epitopes were selected from the predicted HTL epitopes based on their lowest percentile rank (Table 2). The lowest percentile rank reflects their binding affinity level for the MHC-II receptor. In the end, HTL epitopes were analyzed and IFN-γ inducing epitopes were predicted.
Table 2.
Predicted MHC II epitopes against emerging human pathogenic CoVs, SARS-CoV, MERS-CoV, and SARS-CoV-2 antigens.
| Antigen | Organism | Allele | Position | HTL epitopes |
Percentile rank | IFN-γ |
|---|---|---|---|---|---|---|
| nsp3 | SARS-CoV-2 | HLA-DPA1*01:03 HLA-DPB1*02:01 |
1512-1526 | AYILFTRFFYVLGLA | 0.01 | + |
| SARS-CoV | HLA-DQA1*01:01 HLA-DQB1*05:01 |
1517-1531 | FISNSWLMWFIISIV | 0.01 | – | |
| MERS-CoV | HLA-DRB1*09:01 | 1473-1487 | DWRSYNYAVSSAFWL | 0.01 | + | |
| nsp8 | SARS-CoV-2 | HLA-DRB1*13:02 | 184-198 | LIVTALRANSAVKLQ | 0.09 | + |
| SARS-CoV | HLA-DRB1*13:21 | 86-100 | AMQTMLFTMLRKLDN | 0.16 | – | |
| MERS-CoV | HLA-DRB1*04:26 | 178-192 | ENLTWPLVLECTRAS | 0.23 | – | |
| S Protein | SARS-CoV-2 | HLA-DRB1*13:02 | 113-127 | KTQSLLIVNNATNVV | 0.01 | – |
| SARS-CoV | HLA-DRB1*01:01 | 499-513 | LSFELLNAPATVCGP | 0.01 | – | |
| MERS-CoV | HLA-DQA1*05:01 HLA-DQB1*02:01 |
239-253 | FMYTYNITEDEILEW | 0.03 | – | |
| M Protein | SARS-CoV-2 | HLA-DPA1*01:03 HLA-DPB1*02:01 |
89-103 | GLMWLSYFIASFRLF | 0.05 | – |
| SARS-CoV | HLA-DRB1*09:01 | 173-187 | RTLSYYKLGASQRVG | 0.07 | + | |
| MERS-CoV | HLA-DRB1*11:14 | 8-62 | FKMFVLWLLWPSSMA | 0.06 | – | |
| N Protein | SARS-CoV-2 | HLA-DQA1*01:02 HLA-DQB1*06:02 |
150-164 | NPANNAAIVLQLPQG | 0.03 | – |
| SARS-CoV | HLA-DRB1*09:01 | 306-320 | AQFAPSASAFFGMSR | 0.01 | + | |
| MERS-CoV | HLA-DRB1*07:03 | 328-342 | FLRYSGAIKLDPKNP | 0.04 | – | |
| E Protein | SARS-CoV-2 | HLA-DPA1*03:01 HLA-DPB1*04:02 |
18-32 | LLFLAFVVFLLVTLA | 0.02 | – |
| SARS-CoV | HLA-DPA1*03:01 HLA-DPB1*04:02 |
15-29 | NSVLLFLAFVVFLLV | 0.06 | – | |
| MERS-CoV | HLA-DRB1*01:01 | 46-60 | GFNTLLVQPALYLYN | 0.03 | – |
3.4. Construction of the subunit vaccine
To design the final vaccine construct, the top-scored epitopes of CTL and HTL were assembled using the HEYGAEALERAG linker. To facilitate cell penetration of the proposed vaccine, the TAT peptide of HIV (GRKKRRQRRRPPQ) was added to the N-terminal of the construct. In addition, for increasing the immunogenicity of the vaccine, a 41 amino acid-long sequence of β-defensin 2 was added after TAT peptide as an adjuvant and was followed by the PADRE sequence (AGLFQRHGEGTKATVGEPV). The TAT peptide and β-defensin 2 were linked to the PADRE sequence through GPGPG and EAAAK linkers, respectively. The order of sequences in the final construct is summarized in Figure 2.
Figure 2.
Schematic representation of the trivalent subunit SARS-CoV vaccine construct. This vaccine construct covering of TAT sequence, β-defencin and PADRE at N-terminal that linked to HTL and CTL epitopes with appropriate linker, and terminated with His tag in C-terminal.
3.5. Population coverage
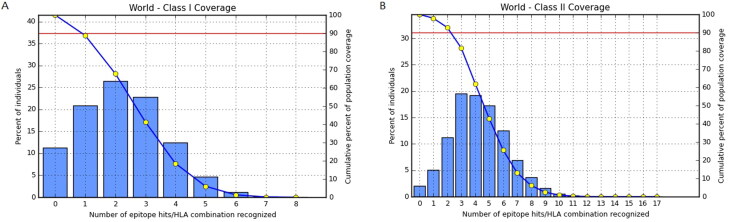
The prediction of MHC-I and MHC-II based coverage of the applied epitopes in the constructed vaccine was performed by IEDB analysis of resources for the world population. As SARS-CoV-2 is global pandemic, worldwide analysis has been accomplished. The world population coverage of MHC-I and MHC-II was found to be 88.77%, and 97.91%, respectively, that showed in Figure 3.
Figure 3.
Worldwide population coverage of selected epitopes based on (A) MHC-I and (B) MHC-II binding alleles.
3.6. Optimized codon and mRNA structure of the chimeric vaccine
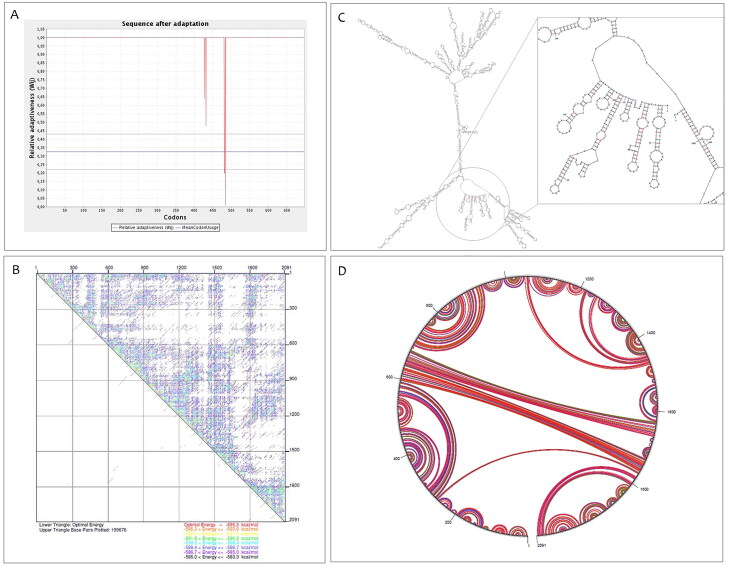
We use the JCAT online server to obtain the codon-optimized DNA sequence of the proposed vaccine to verify for cloning and expressing in E. coli strain K12 as a host. The optimized codon sequences had 2091 nucleotides long, GC-content reached 52.9% and its codon adaptation index was raised to 0.98. The graphical illustration of the optimized gene was shown in Figure 4A. Besides, the mfold server was used to get free energy related to the whole mRNA structure and its 5′ end. Its details are shown in Table 3. As shown in Figure 3B, C, and D, the minimum free energy of the predicted secondary RNA structure was ΔG = −595.30 kcal/mol at 37 °C without hairpin or pseudoknot at the 5′ side.
Figure 4.
Analysis of RNA structures. (A) Graphical view of codon usage in the optimized gene. (B) The energy dot plot for the predicted mRNA. (C) The predicted of RNA secondary structure has no hairpin and pseudo knot at 5′site of mRNA. (D) A circle graph is one way to display base pairs of RNA structure.
Table 3.
Free energy details related to 5′ end of recombinant gene mRNA structure was predicted by mfold web server.
| Structural element | Free energy (kcal/mol) | Base pair |
|---|---|---|
| External loop | −8.70 | 27 ss bases & 8 closing helices. |
| Stack | −2.90 | External closing pair is G1780-C2089 |
| Stack | −2.90 | External closing pair is G1783-C2086 |
| Stack | −1.80 | External closing pair is U1785-A2084 |
| Helix | −18.60 | 9 base pairs. |
| Stack | −2.30 | External closing pair is G1804-C2074 |
| Interior loop | 0.80 | External closing pair is G1798-C2080 |
3.7. Evaluation of allergenicity, antigenicity, physiochemical parameters, and solubility of the vaccine construct
Investigating allergenicity of the proposed vaccine is a crucial consideration to avoid allergic reactions. The resulting output of the Algpred online server determined the non-allergenic behavior of the vaccine constructs with a score of −0.59 (Threshold=-0.4). Also, the antigenicity of the construct was predicted by the VaxiJen v2.0 online server. It confirmed that the protein was a probable antigen with an overall prediction score of antigen of 0.4996. Furthermore, the physicochemical parameters were evaluated, and the molecular weight of the vaccine protein was found to be 74.8 kDa which is good to support the antigenic nature of the vaccine construct. The predicted instability index of the proposed vaccine was 39.09, which is below 40, hence, classifying the protein as stable. The isoelectric point (pI) of the construct was 9.30 and showed the basic nature of the vaccine.
The estimated half-life (in vitro) in mammalian reticulocytes was found to be 30 h, while in vivo studies in Yeast and E. coli showed the interval was longer than 20 and 10 h, respectively. In addition, the aliphatic index was 80.26, which determines the thermostable nature of the proposed vaccine. Also, the grand average of hydropathicity was obtained to be 0.063, which proves the hydrophilic nature of the constructed vaccine. The value for the solubility tendency of the proposed vaccine upon overexpression in E. coli host was 0.55.
3.8. Secondary and tertiary structure of subunit vaccine
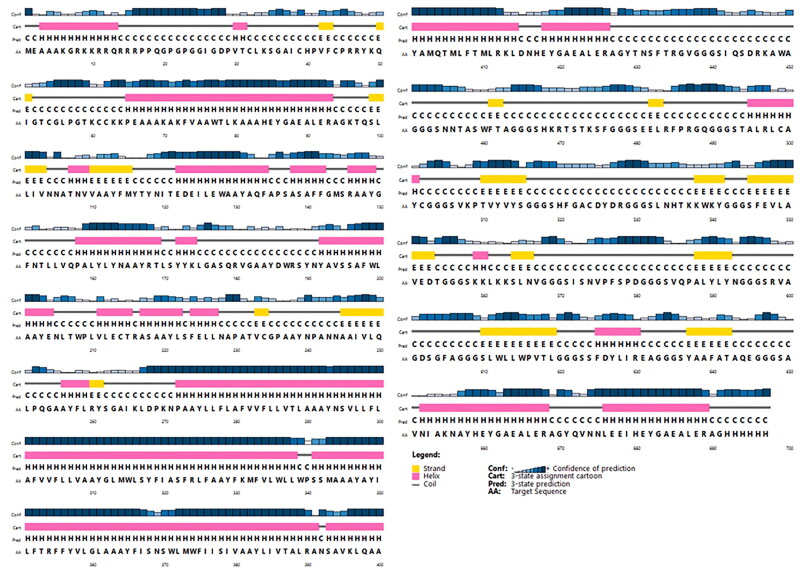
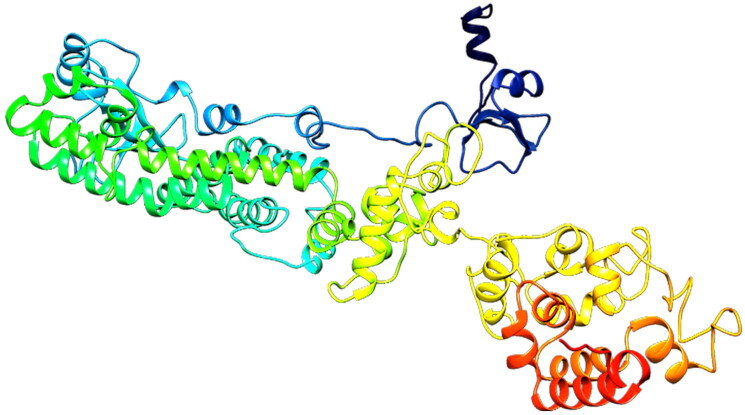
PSIPRED server was employed to predict the secondary structure of the proposed vaccine (Figure 5). This server showed that the vaccine was made of 43.76% alpha-helix, 16.93% extended strand, and 39.31% random coil. The tertiary structure was predicted as three domain structures using the galaxyWEB web server that is a template-based protein structure modeling web server that resulted in a modeled protein structure (Figure 6). The 1FD3_C, 5MKE_A, and 5MKF_A were used as the best-suited templates to model the tertiary structure. All 697 amino acid residues were modeled, while the 70-92, 681-688, and 690-697 amino acids of the vaccine were unreliable local regions.
Figure 5.
PSIPRED graphical results from secondary structure prediction of vaccine. The PSIPRED results indicated that constructed vaccine included 43.76% alpha helix (H), 16.93% extended strand, and 39.31% random coil.
Figure 6.
Predicted 3D structure of constructed vaccine.
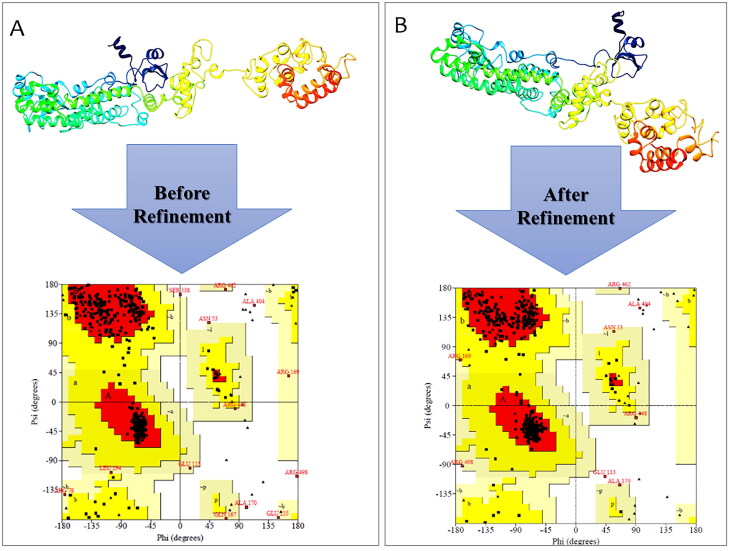
3.9. Refinement and validation of the 3D structure
The generated 3 D structure was submitted for the refinement process to the GalaxyRefine online server to enhance the quality of predicted 3 D modeled structure beyond the accuracy. Furthermore, the refined model which was received from GalaxyRefine was analyzed by the Ramachandran plot. Validation of the 3 D structure by Ramachandran plot demonstrated 540 (92%), 37 (6.3%), and 9(1.5%) of residues were placed in favored, additionally allowed and disallowed regions, before conducting the refinement, respectively (Figure 7A). In the refined model, 550 (93.7%), 26 (4.4%), and 7 (1.2%) of residues were placed in favored, additionally allowed and disallowed regions, respectively (Figure 7B).
Figure 7.
Validation of 3D modeled structure. (A) Ramachandran plot of the initial model showed 92%, 6.3%, and 1.5% of residues were located in favored, allowed and outlier regions, respectively. (B) In the refined model 93.7%, 4.4% and 1.2% of residues were located in favored, allowed and outlier regions, respectively.
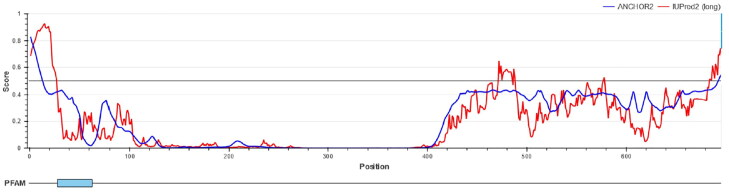
3.10. Prediction of intrinsic protein disorder
DisEMBL online server identified and annotated disordered regions. Amino acids in the input sequence were considered disordered by remark-465 definition in the following regions: 1-23, 464-478, 689-697. Based on IUPred results, amino acids were considered disordered when the confidence score was higher than 0.5. Disordered regions were also presented in Figure 8.
Figure 8.
Intrinsically disorder regions. Amino acids in the input sequence were considered disordered when the red line is above the confidence score is higher than 0.5.
3.11. Predicted B-cell epitopes
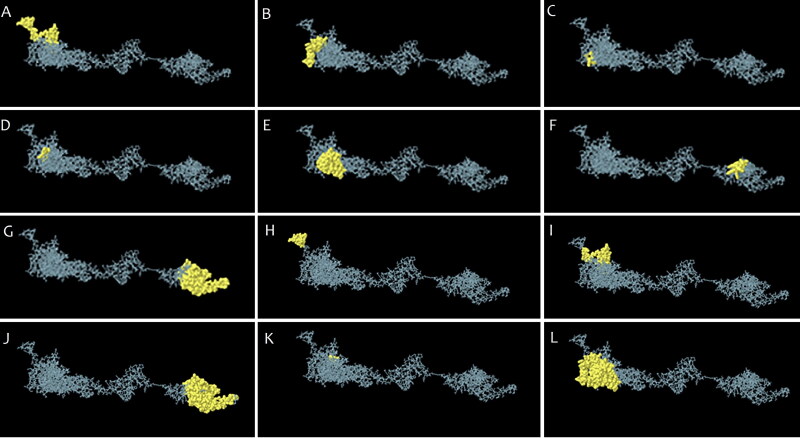
Ellipro and the Discotope online servers were utilized to predict the linear B-cell and discontinuous B cell epitopes, respectively. The linear B-cell and discontinuous B cell epitopes on the 3D structure are shown in Figures 9A–G and 9H–L, respectively. Moreover, epitopes predicted in the modeled vaccine according to various parameters were listed in Table 4.
Figure 9.
The B-cell epitopes. (A-G) Linear and (H-L) discontinuous B cell epitopes on the 3 D structure.
Table 4.
B cell epitopes predicted in constructed vaccine based on different parameters.
| Prediction parameter | Epitope sequence |
|---|---|
| Hydrophilicity | AKGRKKRRQRRR, QGPGPGG, ERAGKTQS, DNHEYGA, GGSIQSD, AGGGSNNTAS, TAGGGSHKRTSTKS, PRGQGGGSTA, GACDYDRGGGS, AVEDTGGGSKK, SPDGGGSVQ, YNGGGSRVAGDS, REAGGGSY, ATAQEGGGSA |
| Flexibility | EAAAKGRKKRRQRRRPP, EALERAGKT, GSIQSDRKAWAGGGSNNT, FTAGGGSHKRTSTKSFGGGS, LRFPRGQGGG, AYCGGGS, YVYSGGG, CDYDRGGG, LAVEDTGGGSKKLK, VPFSPDGG, YLYNGGGS, PVTLGGGSS, LIREAGG, ATAQEGGG |
| Accessibility | AAAKGRKKRRQRRRPPQGPGP, FCPRRYKQIGT, TKCCKKPEAAAK, LERAGKTQS, YTYNITEDE, YNAAYRTLSYYKLGASQR, AYDWRSYNYAV, AYNPANNA, IKLDPKNPAAY, MLRKLDNHEYGAE, ERAGYTNSFTR, SIQSDRKAWA, GGSHKRTSTKSF, SEELRFPRGQG, SLNHTKKWKYGG, GGSKKLKKSLN, AKNAYHEYGAE, YQVNNLEE |
| Turns | YNPANNAA, KLDNHEY, GGSNNTAS, |
| Exposed Surface | AAKGRKKRRQRRRPPQG, CPRRYKQIG, TKCCKKPE, IKLDPKNP, RKLDNHE, HKRTSTKS, NHTKKWKYG, GSKKLKKSLN |
| Polarity | AAKGRKKRRQRRRPPQG, FCPRRYKQIG, GTKCCKKPEA, KAAAHEYGAEALERAGKTQ, TEDEILEW, LRKLDNHEYGAEA, GGGSHKRTSTKS, GGSEELRFPRG, SLNHTKKWKYG, GSKKLKKSLN, AKNAYHEYGAEA, NNLEEIHEYGAEALERAGHHHHHH |
| Antigenic Propensity | IGDPVTCLKSG, ICHPVFCPRRY, LPGTKCC, TQSLLIVN, FNTLLVQP, TLSYYKLG, LTWPLVLECTR, YLSFELL, IVLQLPQG, FVVFLLVTL, YNSVLLFL, FVVFLLVA, YFKMFVLWLLWPS, TRFFYVLGL, WFIISIV, SVKPTVYVYSGG, SLWLLWPVTLG |
3.12. Vaccine protein disulfide engineering
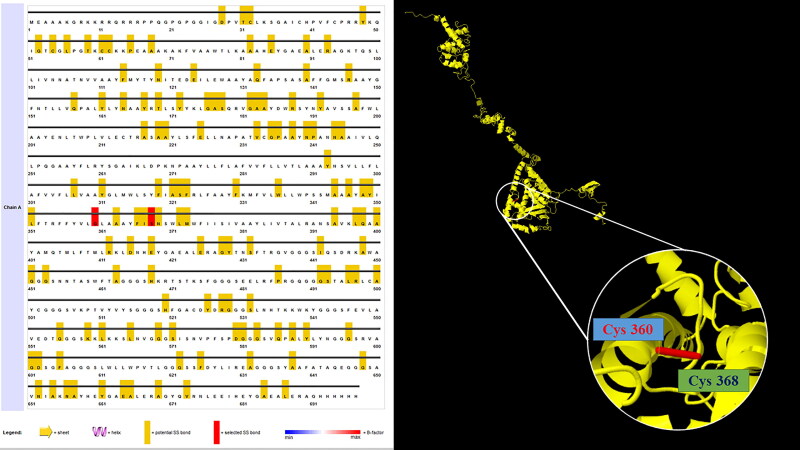
Disulfide engineering leads to an increase in vaccine stability through mutating the residues present in high mobility areas with cysteine. 76 residue pairs were found to have the ability of disulfide bond formation. But, when all these residue pairs were further analyzed for other parameters like energy on chi3 value and B-factor, only 1 pair of residues provided the disulfide bond formation. Those residue pairs were GLY360-SER368 and ASP580- GLY601 (Figure 10). These two residues were mutated to cysteine. The energy value was analyzed for the residue screening was less than 2.2 and the value of chi3 was between −87 to +97.
Figure 10.
Disulfide engineering to improve protein stability. Two pairs of mutated residues were selected based on their energy, chi3 value, and B-factor.
3.13. Molecular docking of vaccine construct with the TLR-3, TLR-4 and TLR-8
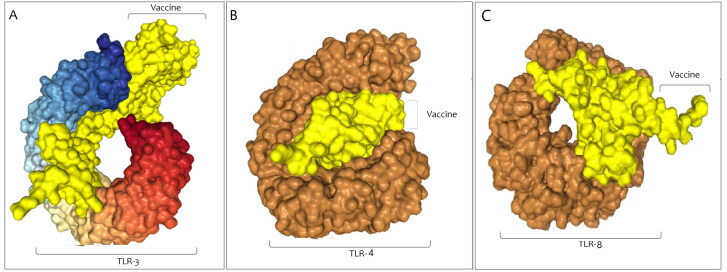
Protein-protein docking was done with the help of the Hdock online server between the final vaccine construct as a ligand and the TLR-3, TLR-4, and TLR-8 as receptors. We procured 20 models and the model with the lowest binding energy was selected. The Docking score for the best acceptable model of vaccine construct with TLR-3 was −326.86, whereas the ligand RMSD (Å) was 205.96. PatchDock server was used to re-affirm the docking between the final vaccine construct and TLR-3. The top 20 models were provided by the server and the model with the top-ranked and lowest binding energy was chosen.The obtained output encompasses the score (17080), Area (2493.80), ACE (408.17), and transformation (-2.83 − 0.57 − 1.83 − 49.29 3.75 160.90). The Docking score for the best acceptable model of the vaccine construct and TLR4 was −357.83, whereas the ligand RMSD (Å) was 245.12. Also, the docking score for the best acceptable model for docking among vaccine construct and TLR8 was −336.95, whereas the ligand RMSD (Å) was 292.88. The docked complexes for TLR3, TLR4, and TLR8 that was achieved from the server has been shown in Figure 11A–C, respectively.
Figure 11.
Docking complex of vaccine construct with TLRs. (A) The result was obtained from protein-protein docking with the help of the Hdock for TLR3, (B) TLR4, and (C) TLR8.
3.14. Molecular dynamics simulation and advanced binding results
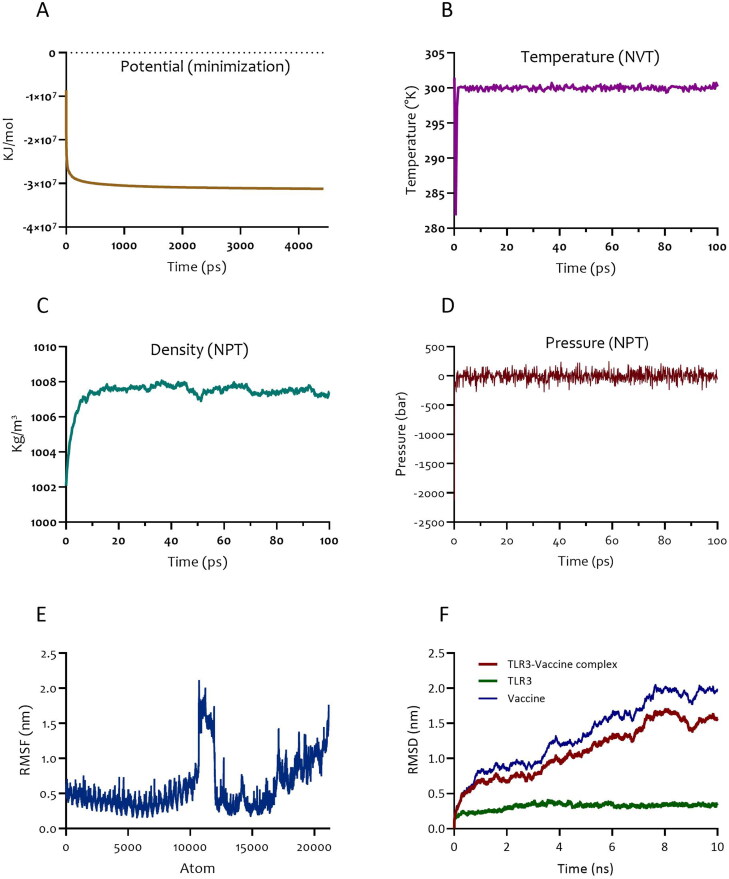
The selected docked complex (TLR3-vaccine) was taken into MD simulation. The system was minimized and reached Fmax < 1000 KJ.mol−1.nm−1 (Figure 12A). The temperature progression curve indicated that the system reached 300 K very rapidly. The temperature remained constant for the rest of the 100 ps equilibration period (Figure 12B). Density showed a stable value of approximately 1008 after 10 ps (Figure 12C). The pressure progression curve validated that equilibration was also favorable. Indeed, after 20 ps, the pressure did not deviate from the fixed value of 0 bar (Figure 12D). The output of MD simulation was obtained as Root Mean Square Fluctuation (RMSF) of all amino acid residues and Root Mean Square Deviation (RMSD) of TLR3-vaccine construct backbone. Root Mean Square Fluctuation (RMSF) plot for the side chain of the docked TLR3-vaccine complex did not show significant fluctuations, confirming the protein did not unfold nor did the construct denature (Figure 12E). The RMSD of the TLR3-vaccine complex reflected its stability over a 10 ns timescale. The deviation started with 0.2 nm and reached a stable value of about 1.7 nm after 8 ns. From this point, RMSD values remained stable (Figure 12F). The stability of the complex during the MD run was also validated by visualization of the MD trajectory. Finally, MM-PBSA analysis of the equilibrated interval of the MD simulation for the contact area showed van der Waal energy = −399.247 ± 0.755 kJ/mol, electrostattic energy = −239.238 ± 1.310 kJ/mol, SASA energy = −78.710 ± 0.126 kJ/mol, SAV energy = 0.000 ± 0.000 kJ/mol, and WCA energy = 0.000 ± 0.000 kJ/mol. Collectively, these outputs indicate additional favorable aspects of the atoms involved in the complex contact area, and further validate our MD results.
Figure 12.
Molecular dynamics simulation of the TLR-vaccine complex. (A) Potential progression curve of TLR3 and vaccine construct. (B) Temperature progression curve of TLR3- vaccine complex. (C) Density curve and (D) pressure progression curve of TLR3-vaccine indicating pressure fluctuation reached equilibration phase over 100 ps. (E) RMSF representation of the docked complex protein side chains. (F) RMSD representation of the docked complex protein backbone consists of TLR3as a receptor and vaccine as a ligand.
3.15. In silico cloning
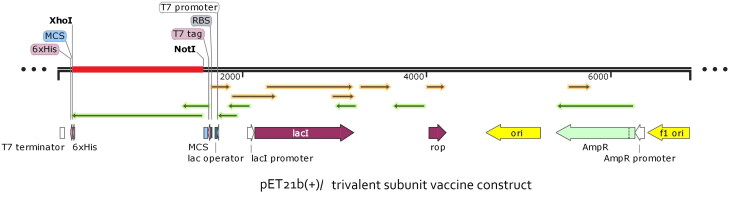
To examine the expression of the recommended vaccine into a vector, cloning was done by utilizing the SnapGene server. As the expression system of the human and E. coli K12 differ from each other and the codon requires the adaptation as per the host expression system, the codon sequence that was optimized by JCat was reverse transcribed and modified as per the E. coli K12. The restriction enzyme sites Not I and Xho I were introduced at the N and C-terminal, respectively. The adapted codon was entered into the pET21b (+) vector and a recombinant plasmid with the length of 7514 base pair was obtained as displayed in Figure 13.
Figure 13.
In silico cloning to express final vaccine construct in E. coli. The vaccine construct gene (red) was inserted into the pET21b (+) vector (black) between Not I and Xho I restriction sites to create a recombinant vector.
3.16. Immune simulation
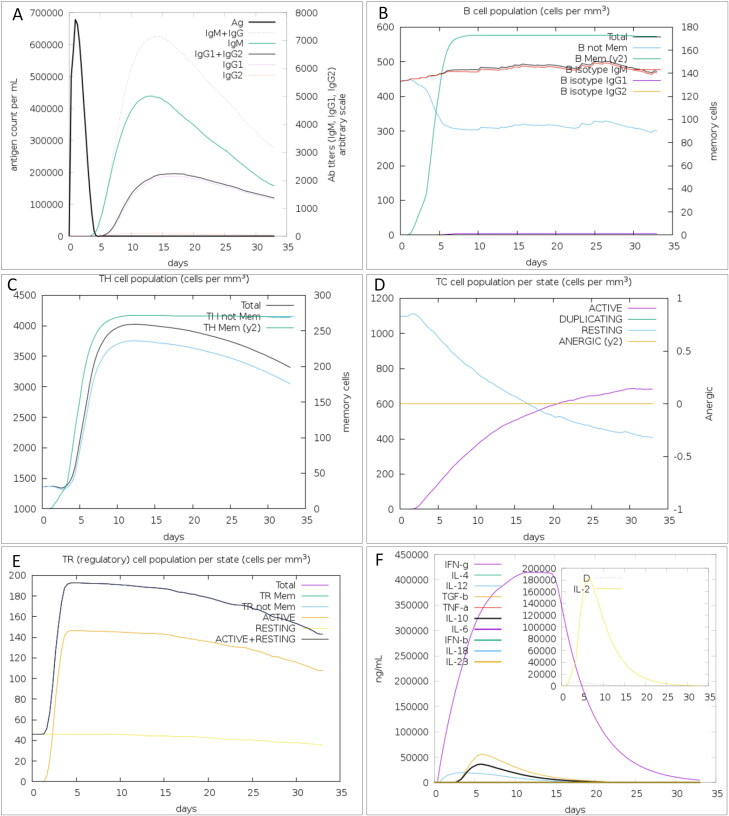
C-ImmSim server results revealed a noticeable increase in the generation of immune responses. The humoral immune response was identified by high levels of IgM along with IgG + IgM (Figure 14A). Estimation of the B cell population uncovered an increase in B memory cells and B-cell isotype IgM with a corresponding reduction in antigen concentration (Figure 14B). Besides, a high response was observed in the TH1 and TC cell populations with corresponding memory development (Figure 10C and D). This profile identified the development of immune memory and consequently enhanced clearance of the antigen upon subsequent exposures. In contrast, regulatory T cells were estimated to be at a low level (Figure 14E). Moreover, these predictions were consistent with the induced level of IFN-γ produced after immunization with the proposed vaccine (Figure 14F).
Figure 14.
In silico immune simulation with the trivalent subunit vaccine. (A) Immunoglobulin production in response to antigen, (B) B-cell populations after exposure to antigen (C), The evolution of T-helper, (D) T-cytotoxic, and (E) T-regulatory cell populations per state after antigen injection. (F) Level of cytokines induced by the vaccine. The insert plot shows the IL-2 level with the Simpson index, D indicated by the dotted line.
4. Discussion
Designing novel vaccines for preventing the ever-rising global burden of disease is an important issue that cannot be ignored. CoVs are a large family of zoonotic viruses that cause illnesses ranging from the common cold to more severe diseases such as SARS-CoV, MERS-CoV, and SARS-CoV-2 (Hui & IAzharE, 2020; Lu et al., 2020). While many CoV vaccine studies targeting various structural proteins were conducted, most of these endeavors finally stopped soon after the outbreak of SARS and MERS (Ong et al., 2020).
In the present study, we designed a multi-epitope vaccine with both structural proteins and nsps against severe human CoVs (e.g. SARS-CoV-2, SARS-CoV, and MERS-CoV) via immunoinformatics strategies utilizing various in silico tools.
Advancement in genomics has altered the procedures and concepts of vaccine design (Ahluwalia et al., 2017). Reverse vaccinology, a novel method to merge immunogenetics and immunogenomics via immunoinformatic has been widely used to introduce a novel vaccine. The most important benefit of RV in the context of SARS-CoV-2 is the quick identification of favorable protein vaccines, while traditional vaccinology may take decades (Srivastava et al., 2018, 2019, 2020). Thus, in this study, we developed a novel trivalent subunit vaccine from immune-dominant peptides of spike (S) protein, nucleocapsid (N), envelope (E), membrane (M) protein, nsp3, and nsp8 antigens. In our suggested vaccine construct, the CTL, MHC-I, MHC-II epitopes were predicted and used. Cytotoxic CD8 T lymphocytes (CTL) limit the spread of infectious pathogens via recognition and elimination of the infected cells or secreting particular antiviral cytokines (Garcia et al., 1999; Sharmeen et al., 2012). Thus, T-cell epitope-based vaccination plays a unique role against infectious pathogens such as viruses by eliciting a potent immune response (Shrestha & Diamond, 2004). HTL responses exert a critical role in the induction of both humoral and cellular immune responses. Therefore, HTL epitopes are believed to be an important part of prophylactic and therapeutic vaccines (Alexander et al., 1998). Furthermore, once the development of the novel multi-epitope vaccine begins, there will be great concern about its immunogenicity. Therefore, adjuvants were established and attached to vaccine construction to enhance its efficacy. Recently, the role of TLRs in the relation between the innate and adaptive immunity has been highlighted in vaccine design. TLRs are a subgroup of pattern recognition receptors which are present on antigen-presenting cells (APCs), which activating via an appropriate TLR agonist will elicit cellular and humoral reactions as well as innate responses (Dowling & Mansell, 2016; Steinhagen et al., 2011). There are multiple ways that induce TLRs. For example, β-defensin 2 which induces signal transduction via TLR-4 regulates adaptive immunity during infection (Amelin et al., 2002). Self-destructive signaling which eliminates activated APCs, and thus modifies the adaptive immunity is also possible via utilizing β-defensin 2 (Biragyn et al., 2008).
In our vaccine construct, human β-defensin 2 serves as an adjuvant which promotes the expression of primary immune-stimulatory and antiviral compounds (Kim et al., 2018). In addition, to increase the quality of vaccine delivery, a cell-penetrating peptide, namely TAT, from HIV was engaged to involve T cells via APCs (Pouniotis et al., 2016). Furthermore, the PADRE T helper epitope can bind to a prominent number of HLA-DR types with an affinity spanning from intermediate to high (Feng et al., 2014). Also, the YQVNNLEEI epitope located in a conserved region of EBOV NP was added to the C terminal of the vaccine construct. A recent study showed this epitope contains a protective CTL epitope against COVID-19 (Herst et al., 2020).
Allergenicity is one of the main problems in vaccine development. Many vaccines stimulate the immune system, and may lead to allergic reactions (McKeever et al., 2004). Based on the WHO/FAO, if a sequence shows at least six contiguous amino acids to identified allergens over a window of 80 amino acids (0.35% sequence identity), it is considered as potentially allergenic. The result of the allergenicity assessment showed that the vaccine was not allergenic. The subunit chimeric vaccine included 697 residues. Restriction sites, instability elements, and all the cis-acting sites which significantly interfere with cloning were omitted from the construct. Moreover, codon optimization was performed to facilitate the high-level expression of the multi-epitope vaccine in E. coli and improve translational and transcriptional output. mRNA secondary structure is an essential parameter in the expression of subunit vaccines. The outputs from mRNA prediction by Mfold online server showed that the mRNA to be sufficiently stable for an impressive translation in the host. Besides, physico-chemical factors analysis via ProtParam online server showed the amino acid sequence of the subunit vaccine contained 575 residues and an average molecular weight (MW) of 65,378.43 D which represents a good antigenic nature (antigens with a MW of <5–10 KDa are considered as poorly immunogenic) (Foroutan et al., 2018; Timmerman, 2009). The predicted instability index of the proposed vaccine was 39.09, which was below 40, and hence classified the protein as stable. The isoelectric point of the construct was 9.30 (pI) and showed the basic nature of the vaccine. The estimated half-life (in vitro) in mammalian reticulocytes was found to be 30 hr. Moreover, in in vivo studies in Yeast and E. coli, it was found to be more than 20 and 10 hr, respectively. In addition, the aliphatic index was 80.26, which determined the thermostable nature of the proposed vaccine, and the Grand average of hydropathicity was 0.063, which proved the hydrophilic nature of the constructed vaccine (Validi et al., 2018). Physicochemical parameters showed this subunit vaccine to be an appropriate candidate for molecular biology-based issues. Also, the tendency of vaccine solubility upon overexpression in E. coli was computed 0.95%, showing an admissible percentage of solubility in an overexpressed state.
On the other hand, outputs from the primary model with the Galaxyweb server showed that the Galaxyweb program model was applied to generate the primary 3 D structure of the protein vaccine. Obtained results showed 43.76% alpha-helix, 16.93% extended strand, and 39.31% random coil structure existed in the tertiary model. After the refinement, the predicted structure showed a high-quality tertiary model.
The analysis of all discontinuous and continuous B-cell epitopes showed that the identified epitopes on the protein surface could interact comfortably with antibodies, and were commonly flexible. Furthermore, DisEMBL online server was utilized to specify unstructured regions and disorders in the subunit vaccine. Obtained results indicated that disorder regions were sited among positions 1-23, 464-478, and 689-697. Then, the docking process between TLR3, TLR4, and TLR8 and the designed vaccine was accomplished and the foremost docked models were selected and investigated via MD simulation to compute the stability of the vaccine–protein complex. Outputs of MD simulation supported the subunit vaccine-TLR3 interactions. The interactions between the two molecules were modified during the MD simulation. The MD simulation results confirmed that the proposed novel vaccine was a robust vaccine target, and the equilibrated system reached a favorably stable state without decomposition or severe fluctuation.
In the case of conventional vaccines, some recent clinical trial studies resolved safety concerns while corroborating conventional methods for vaccine development (Tameris et al., 2013). Interestingly, it has been indicated that when the cellular lines employed for the generation of the vaccine may be contaminated via microorganisms that do not produce any cytopathic effect while undergoing replication along with the vaccine virus (Committee, 2003; Merten, 2002). These contaminations perhaps prove dangerous for the host, especially when the quality control process fails. However, the recombinant multi-epitope vaccine does not carry any risk of the emergence of virulence property as it solely contains the antigenic sections (not the entire virus itself). On the other hand, C-ImmSim server results revealed a noticeable increase in the generation of immune responses. Moreover, several instruments have been employed in the current study to accredit the non-allergic and nontoxic behavior of the designed vaccine construct. However, the predicted bioinformatics results were based on the various investigations of sequences and different immune servers. We suggest that future wet lab-based analyses should be employed in in vivo models for the experimental validation of our subunit vaccine are warranted.
5. Conclusion
In the current study, we developed an efficient trivalent subunit vaccine against CoVs via different in silico servers. The physico-chemical properties and allergenicity of vaccine were found to be very satisfactory for carrier protein development. The secondary and tertiary structure prediction of the vaccine gave a clear view of the structural properties. Molecular docking and dynamics simulation validated the strong interaction and stability of vaccine with TLRs. Immunoinformatics analysis indicated this vaccine was able to induce both humoral and cellular immune responses effectively due to the presence of several epitopes from a combination of antigens, PADRE, and an adjuvant. Thus, this subunit vaccine can conceivably be utilized for prophylactic aims against emerging human pathogenic CoVs such as SARS-CoV, MERS-CoV, and SARS-CoV-2.
Acknowledgements
We thank Student Research Committee, Babol University of Medical Sciences, and all staff at Immunoregulation Research Center, Health Research Institute, Babol University of Medical Sciences.
Funding Statement
The Deputy for Research and Technology of Babol University of Medical Sciences supported this study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Availability of data and materials
All data and materials of this work are available from the corresponding author on reasonable request.
Authors’ contributions
A.R., M.B., K.S., S.M., and H.R.N. conceptualized the study and prepared the initial draft. MD simulations and visualized results conducted by K.S. H.R.N. supervised the project, critically appraised the manuscript, and prepared the final draft
References
- Ahluwalia, P. K., Pandey, R. K., Sehajpal, P. K., & Prajapati, V. K. (2017). Perturbed microRNA expression by Mycobacterium tuberculosis promotes macrophage polarization leading to pro-survival foam cell. Frontiers in Immunology, 8, 107. 10.3389/fimmu.2017.00107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander, J., Fikes, J., Hoffman, S., Franke, E., Sacci, J., Appella, E., Chisari, F. V., Guidotti, L. G., Chesnut, R. W., Livingston, B., & Sette, A. (1998). The optimization of helper T lymphocyte (HTL) function in vaccine development. Immunologic Research, 18(2), 79–92. 10.1007/BF02788751 [DOI] [PubMed] [Google Scholar]
- Amelin, Y., Krot, A. N., Hutcheon, I. D., & Ulyanov, A. A. (2002). Lead isotopic ages of chondrules and calcium-aluminum-rich inclusions. Science (New York, N.Y.).), 297(5587), 1678–1683. 10.1126/science.1073950 [DOI] [PubMed] [Google Scholar]
- Azhar, E. I., Lanini, S., Ippolito, G., & Zumla, A. (2016). The Middle East respiratory syndrome coronavirus–a continuing risk to global health security. In Emerging and re-emerging viral infections. Advances in Experimental Medicine and Biology, 972, 49–60. 10.1007/5584_2016_133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biragyn, A., Coscia, M., Nagashima, K., Sanford, M., Young, H. A., & Olkhanud, P. (2008). Murine beta-defensin 2 promotes TLR-4/MyD88-mediated and NF-kappaB-dependent atypical death of APCs via activation of TNFR2. Journal of Leukocyte Biology, 83(4), 998–1008. 10.1189/jlb.1007700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee, I. S. R. (2003). Immunization safety review: Vaccinations and sudden unexpected death in infancy. National Academies Press. [PubMed] [Google Scholar]
- Craig, D. B., & Dombkowski, A. A. (2013). Disulfide by design 2.0: A web-based tool for disulfide engineering in proteins. BMC Bioinformatics, 14, 346. 10.1186/1471-2105-14-346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson, P., Malik, M. R., Parvez, F., & Morse, S. S. (2019). What have we learned about Middle East respiratory syndrome coronavirus emergence in humans? A systematic literature review. Vector-Borne and Zoonotic Diseases, 19(3), 174–192. 10.1089/vbz.2017.2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda, S. K., Vir, P., & Raghava, G. P. (2013). Designing of interferon-gamma inducing MHC class-II binders. Biology Direct, 8(1), 30. 10.1186/1745-6150-8-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling, J. K., & Mansell, A. (2016). Toll-like receptors: The swiss army knife of immunity and vaccine development. Clinical & Translational Immunology, 5(5), e85. 10.1038/cti.2016.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr, A. R., & Perlman, S. (2015). Coronaviruses: An Overview of their replication and pathogenesis. In Maier H., Bickerton E., Britton P. (Eds.), Coronaviruses. Methods in Molecular Biology, vol. 1282. New York, NY: Humana Press. 10.1007/978-1-4939-2438-7_1 [DOI] [PMC free article] [PubMed]
- Feng, G.-D., Xue, X.-C., Gao, M.-L., Wang, X.-F., Shu, Z., Mu, N., Gao, Y., Wang, Z.-L., Hao, Q., Li, W.-N., Li, M., Zhang, C., Zhang, W., & Zhang, Y.-Q. (2014). Therapeutic effects of PADRE-BAFF autovaccine on rat adjuvant arthritis. BioMed Research International, 2014, 854954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foroutan, M., Ghaffarifar, F., Sharifi, Z., Dalimi, A., & Pirestani, M. (2018). Bioinformatics analysis of ROP8 protein to improve vaccine design against Toxoplasma gondii. Infection, Genetics and Evolution, 62, 193–204. 10.1016/j.meegid.2018.04.033 [DOI] [PubMed] [Google Scholar]
- Garcia, K. C., Teyton, L., & Wilson, I. A. (1999). Structural basis of T cell recognition. Annual Review of Immunology, 17(1), 369–397. 10.1146/annurev.immunol.17.1.369 [DOI] [PubMed] [Google Scholar]
- Habibzadeh, P., & Stoneman, E. K. (2020, April). The novel coronavirus: A bird's eye view. The International Journal of Occupational and Environmental Medicine, 11(2), 65–71. 10.15171/ijoem.2020.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herst, C. V., Burkholz, S., Sidney, J., Sette, A., Harris, P. E., Massey, S., Brasel, T., Cunha-Neto, E., Rosa, D. S., Chao, W. C. H., Carback, R., Hodge, T., Wang, L., Ciotlos, S., Lloyd, P., & Rubsamen, R. (2020). An effective CTL peptide vaccine for ebola zaire based on survivors’ CD8+ targeting of a particular nucleocapsid protein epitope with potential implications for COVID-19 vaccine design. Vaccine, 38(28), 4464–4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann, H., Pyrc, K., van der Hoek, L., Geier, M., Berkhout, B., & Pöhlmann, S. (2005). Human coronavirus NL63 employs the severe acute respiratory syndrome coronavirus receptor for cellular entry. Proceedings of the National Academy of Sciences, 102(22), 7988–7993. 10.1073/pnas.0409465102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, D., & IAzharE, M. T. (2020). Thecontinuing2019-nCoVep-idemic threat of novel coronaviruses to global health-the latest 2019 novel coronavirus outbreak in Wuhan, China. International Journal of Infectious Diseases, 91, 264–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey, W., Dalke, A., & Schulten, K. (1996). VMD: Visual molecular dynamics. Journal of Molecular Graphics, 14(1), 33–38. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- Kim, J., Yang, Y. L., Jang, S.-H., & Jang, Y.-S. (2018). Human β-defensin 2 plays a regulatory role in innate antiviral immunity and is capable of potentiating the induction of antigen-specific immunity. Virology Journal, 15(1), 124. 10.1186/s12985-018-1035-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Weber, H., Elzayat, M. T., Wang, L., Graham, B. S., Müller, M. A., Drosten, C., Pöhlmann, S., & Hoffmann, M. (2018). Mutations in the spike protein of MERS-CoV transmitted in Korea increase resistance towards antibody-mediated neutralization. Journal of Virology, 93(2), e01381-18. 10.1128/JVI.01381-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H., Stratton, C. W., & Tang, Y. W. (2020). Outbreak of pneumonia of unknown etiology in Wuhan China: The mystery and the miracle. Journal of Medical Virology, 92(4), 401–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeever, T. M., Lewis, S. A., Smith, C., & Hubbard, R. (2004). Vaccination and allergic disease: A birth cohort study. American Journal of Public Health, 94(6), 985–989. 10.2105/ajph.94.6.985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merten, O.-W. (2002). Virus contaminations of cell cultures - A biotechnological view. Cytotechnology, 39(2), 91–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong, E., Wong, M. U., Huffman, A., & He, Y. (2020). COVID-19 coronavirus vaccine design using reverse vaccinology and machine learning. Frontiers in Immunology,11,1581. 10.3389/fimmu.2020.01581 [DOI] [PMC free article] [PubMed]
- Pouniotis, D., Tang, C.-K., Apostolopoulos, V., & Pietersz, G. (2016). Vaccine delivery by penetratin: Mechanism of antigen presentation by dendritic cells. Immunologic Research, 64(4), 887–900. 10.1007/s12026-016-8799-5 [DOI] [PubMed] [Google Scholar]
- Rapin, N., Lund, O., Bernaschi, M., & Castiglione, F. (2010). Computational immunology meets bioinformatics: The use of prediction tools for molecular binding in the simulation of the immune system. PLoS One, 5(4), e9862. 10.1371/journal.pone.0009862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli, R., Bottomley, M. J., D’Oro, U., Finco, O., & De Gregorio, E. (2016). Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. Journal of Experimental Medicine, 213(4), 469–481. 10.1084/jem.20151960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivarao, S. V., & Acevedo, O. (2009). Development of OPLS-AA force field parameters for 68 unique ionic liquids. Journal of Chemical Theory and Computation, 5(4), 1038–1050. 10.1021/ct900009a [DOI] [PubMed] [Google Scholar]
- Shang, W., Yang, Y., Rao, Y., & Rao, X. (2020). The outbreak of SARS-CoV-2 pneumonia calls for viral vaccines . npj Vaccines, 5(1), 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharmeen, R., Hossain, M. N., Rahman, M. M., Foysal, M. J., & Miah, M. F. (2012). In-vitro antibacterial activity of herbal aqueous extract against multi-drug resistant Klebsiella sp. isolated from human clinical samples. International Current Pharmaceutical Journal, 1(6), 133–137. 10.3329/icpj.v1i6.10534 [DOI] [Google Scholar]
- Shi, S.-Q., Peng, J.-P., Li, Y.-C., Qin, C., Liang, G.-D., Xu, L., Yang, Y., Wang, J.-L., & Sun, Q.-H. (2006). The expression of membrane protein augments the specific responses induced by SARS-CoV nucleocapsid DNA immunization. Molecular Immunology, 43(11), 1791–1798. 10.1016/j.molimm.2005.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha, B., & Diamond, M. S. (2004). Role of CD8+ T cells in control of West Nile virus infection. + T cells in control of West Nile virus infection. Journal of Virology, 78(15), 8312–8321. 10.1128/JVI.78.15.8312-8321.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, S., Kamthania, M., Kumar Pandey, R., Kumar Saxena, A., Saxena, V., Kumar Singh, S., Kumar Sharma, R., & Sharma, N. (2019). Design of novel multi-epitope vaccines against severe acute respiratory syndrome validated through multistage molecular interaction and dynamics. Journal of Biomolecular Structure & Dynamics, 37(16), 4345–4360. 10.1080/07391102.2018.1548977 [DOI] [PubMed] [Google Scholar]
- Srivastava, S., Kamthania, M., Singh, S., Saxena, A. K., & Sharma, N. (2018). Structural basis of development of multi-epitope vaccine against Middle East respiratory syndrome using in silico approach. Infection and Drug Resistance, 11, 2377–2391. 10.2147/IDR.S175114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava, S., Verma, S., Kamthania, M., Kaur, R., Badyal, R. K., Saxena, A. K., Shin, H.-J., Kolbe, M., & Pandey, K. C. (2020). Structural basis for designing multiepitope vaccines against COVID-19 infection: In silico vaccine design and validation. JMIR Bioinformatics and Biotechnology, 1(1), e19371. 10.2196/19371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhagen, F., Kinjo, T., Bode, C., & Klinman, D. M. (2011). TLR-based immune adjuvants. Vaccine, 29(17), 3341–3355. 10.1016/j.vaccine.2010.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tameris, M. D., Hatherill, M., Landry, B. S., Scriba, T. J., Snowden, M. A., Lockhart, S., Shea, J. E., McClain, J. B., Hussey, G. D., Hanekom, W. A., Mahomed, H., & McShane, H. (2013). Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: A randomised, placebo-controlled phase 2b trial. The Lancet, 381(9871), 1021–1028. 10.1016/S0140-6736(13)60177-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman, J. M. (2009). Carrier protein conjugate vaccines: The “missing link” to improved antibody and CTL responses? Human Vaccines, 5(3), 181–183. 10.4161/hv.5.3.7476 [DOI] [PubMed] [Google Scholar]
- Tortorici, M. A., & Veesler, D. (2019). Structural insights into coronavirus entry. Advances in virus research, 105, 93–116. 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Validi, M., Karkhah, A., Prajapati, V. K., & Nouri, H. R. (2018). Immuno-informatics based approaches to design a novel multi epitope-based vaccine for immune response reinforcement against Leptospirosis. Molecular Immunology, 104, 128–138. 10.1016/j.molimm.2018.11.005 [DOI] [PubMed] [Google Scholar]
- Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718. 10.1002/jcc.20291 [DOI] [PubMed] [Google Scholar]
- Walls, A. C., Park, Y.-J., Tortorici, M. A., Wall, A., McGuire, A. T., & Veesler, D. (2020). Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell, 183(6), 1735. 10.1016/j.cell.2020.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb, B., & Sali, A. (2016). Comparative protein structure modeling using MODELLER. Current Protocols in Bioinformatics, 54(1), 1-37. 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumla, A., Hui, D. S., & Perlman, S. (2015). Middle East respiratory syndrome. The Lancet, 386(9997), 995–1007. 10.1016/S0140-6736(15)60454-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials of this work are available from the corresponding author on reasonable request.