Abstract
High levels of hyaluronan (ΗΑ), a major extracellular matrix (ECM) glycosaminoglycan, have been correlated with poor clinical outcome in several malignancies, including breast cancer. The high and low molecular weight HΑ forms exert diverse biological functions. Depending on their molecular size, ΗΑ forms either promote or attenuate signaling cascades that regulate cancer progression. In order to evaluate the effects of different ΗΑ forms on breast cancer cells' behavior, ΗΑ fragments of defined molecular size were synthesized. Breast cancer cells of different estrogen receptor (ER) status – the low metastatic, ERα-positive MCF-7 epithelial cells and the highly aggressive, ERβ-positive MDA-MB-231 mesenchymal cells – were evaluated following treatment with HA fragments. Scanning electron microscopy revealed that HA fragments critically affect the morphology of breast cancer cells in a molecular-size dependent mode. Moreover, the ΗΑ fragments affect cell functional properties, the expression of major ECM mediators and epithelial-to-mesenchymal transition (ΕΜΤ) markers. Notably, treatment with 200 kDa ΗΑ increased the expression levels of the epithelial marker Ε-cadherin and reduced the expression levels of HA synthase 2 and mesenchymal markers, like fibronectin and snail2/slug. These novel data suggest that the effects of HA in breast cancer cells depend on the molecular size and the ER status. An in-depth understanding on the mechanistic basis of these effects may contribute on the development of novel therapeutic strategies for the pharmacological targeting of aggressive breast cancer.
Abbreviations: HA, hyaluronan or hyaluronic acid; HMW HA, high molecular weight hyaluronan; LMW HA, low molecular weight hyaluronan; o-HA, hyaluronan oligomers; s-HA, sulfated hyaluronan; BTH, bovine testes hyaluronidase; ER, estrogen receptor; EMT, epithelial-to-mesenchymal transition; ECM, extracellular matrix; MMPs, matrix metalloproteinases; MET, mesenchymal-to-epithelial transition; HAS, hyaluronan synthase; HYAL, hyaluronidase; SDC, syndecan; TIMPs, tissue inhibitors of metalloproteinases; tPA, tissue plasminogen activator; uPA, urokinase plasminogen activator; SEM, scanning electron microscopy
Keywords: Hyaluronan, Breast cancer, Epithelial-to-mesenchymal transition, Estrogen receptors, CD44, Scanning electron microscopy
Highlights
-
•
HA fragments affect breast cancer cell phenotype, functional properties and matrix composition
-
•
Τhe effects of HA in breast cancer cells depends on the molecular size and the estrogen receptor status
-
•
200 kDa HA fragment evokes mesenchymal-to-epithelial transition and expression of epithelial markers
-
•
Targeting HA is a promising tool for novel pharmaceutical approaches in breast cancer
Introduction
Breast cancer, a heterogeneous and complex malignancy is the second most commonly reported type of cancer worldwide [1,2]. Estrogens and their receptors (ERα and ERβ) play pivotal roles in breast cancer development and progression. ERs are principal signaling molecules that regulate the expression of several biological effectors that modulate breast cancer properties and progression [[3], [4], [5], [6], [7]]. The majority of breast cancer cells are ERα positive, which have been correlated with epithelial characteristics and a low metastatic potential [[8], [9], [10]]. Epithelial-to-mesenchymal transition (EMT) is a crucial step for breast cancer invasion and metastasis [1]. Mesenchymal markers, such as snail2/slug, fibronectin and zeb1/zeb2 are overexpressed, whereas the epithelial marker E-cadherin is downregulated. As a result, the cell-to-cell junctions are lost, cells lose their polarity and their migratory capacity is increased. In addition, the expression of several matrix metalloproteinases (MMPs), which are critical mediators of the extracellular matrix (ECM), is altered. Under certain conditions, the opposite process, i.e. mesenchymal-to-epithelial transition (MET) is also possible [11,12].
ECM is a well-organized, multitasking, three-dimensional network, providing a physical scaffold for the cells and regulates their growth, differentiation and migration. ECM consists of a variety of macromolecules, such as collagen, proteoglycans and glycosaminoglycans [[13], [14], [15], [16]]. Hyaluronan (HA) is a linear glycosaminoglycan composed of disaccharide repeating units of d-glucuronic acid (GlcA) and N-acetyl-d-glucosamine (GlcNAc). Despite its simple structure, HA plays a crucial role in several biological processes, such as tissue hydration and maintenance of the viscoelasticity of liquid connective tissues, whereas it has been correlated with tumor development, fibrotic wound healing and inflammation [17,18]. HA is synthesized by three enzymes, the HA synthases (HASs): HAS1, HAS2 and HAS3. The latter is the most active of the three enzymes and can produce high molecular weight HA (HMW HA) and low molecular weight HA (LMW HA). HAS2 produces HMW HA and it is responsible for the increased HA synthesis as stress response. HAS1 is able to synthesize LMW HA and HMW HA and its upregulation has been correlated to inflammation [19]. The enzymes that are responsible for HA degradation are HYAL 1–4, HYALP and PH20. HYAL 2 is located at the cell surface and hydrolyzes HMW HA to LMW HA (approximately 20 kDa). HYAL 1 is located at the lysosomes and hydrolyzes LMW HA to HA oligomers [13,19,20]. HA binds to several receptors initiating a variety of biological responses. The most well-known HA receptors are CD44 and receptor for HA-mediated motility, RHAMM. CD44 regulates a plethora of cell–cell and cell–matrix interactions as well as cell survival and proliferation. It is overexpressed in several cancer types; thus, it is considered as a desirable therapeutic target [[21], [22], [23], [24]].
HA exhibits different biological properties and functions depending on its molecular size. HMW HA (1000–6000 kDa) is able to regulate cellular behavior in a different mode of action as compared to LMW HA (10–250 kDa) and HA oligomers (o-HA). HMW HA seems to promote anti-inflammatory, anti-proliferative and anti-angiogenic effects. It is also a key regulator of wound healing and embryogenesis [19,[25], [26], [27]]. On the other hand, HA fragments (2–10 disaccharides) initiate signaling cascades that lead to a variety of cellular responses, such as inflammation, endothelial cell proliferation and angiogenesis [[28], [29], [30], [31], [32]]. Even though increased levels of endogenous HA have been correlated to several types of cancer, LMW HA (100–300 kDa) and o-HA (3–12 disaccharides) are able to attenuate signaling pathways that regulate cancer cell proliferation, migration and metastasis [[33], [34], [35], [36], [37], [38], [39], [40], [41]]. This phenomenon could possibly occur due to the ability of HA fractions to compete with the endogenous HA for binding to HA receptors. These observations indicate that HA effects on cancer cells are size-dependent [26,[42], [43], [44], [45], [46]]. It also indicates that HA fractions could be used as therapeutics depending on their molecular size. Several studies have proven that LMW HA and o-HA are able to attenuate cell growth, migration and invasion [33,38,45,47] and can sensitize cancer cells to chemotherapy as well [34,35].
The aim of this study was therefore to evaluate the biological effects on breast cancer cells with different ER status. For this purpose, HA fractions of defined molecular size were synthesized, characterized for their molecular size and used to treat two breast cancer cell lines of different ER status; the low invasive MCF-7 and the highly aggressive MDA-MB-231 cells. Here, we demonstrate that HA fractions are able to regulate breast cancer cells' functional properties and affect their morphology depending on their molecular size. HA can also affect EMT and modify the expression of a variety of genes, such as MMPs, syndecan-4 and heparanase, in a molecular size-dependent mode. The obtained data suggest that the molecular size of HA is a critical regulator of breast cancer cells behavior opening a new area for intensive research for future pharmacological targeting approaches in mammary cancer.
Results
HA of different molecular size affects breast cancer cells' functional properties
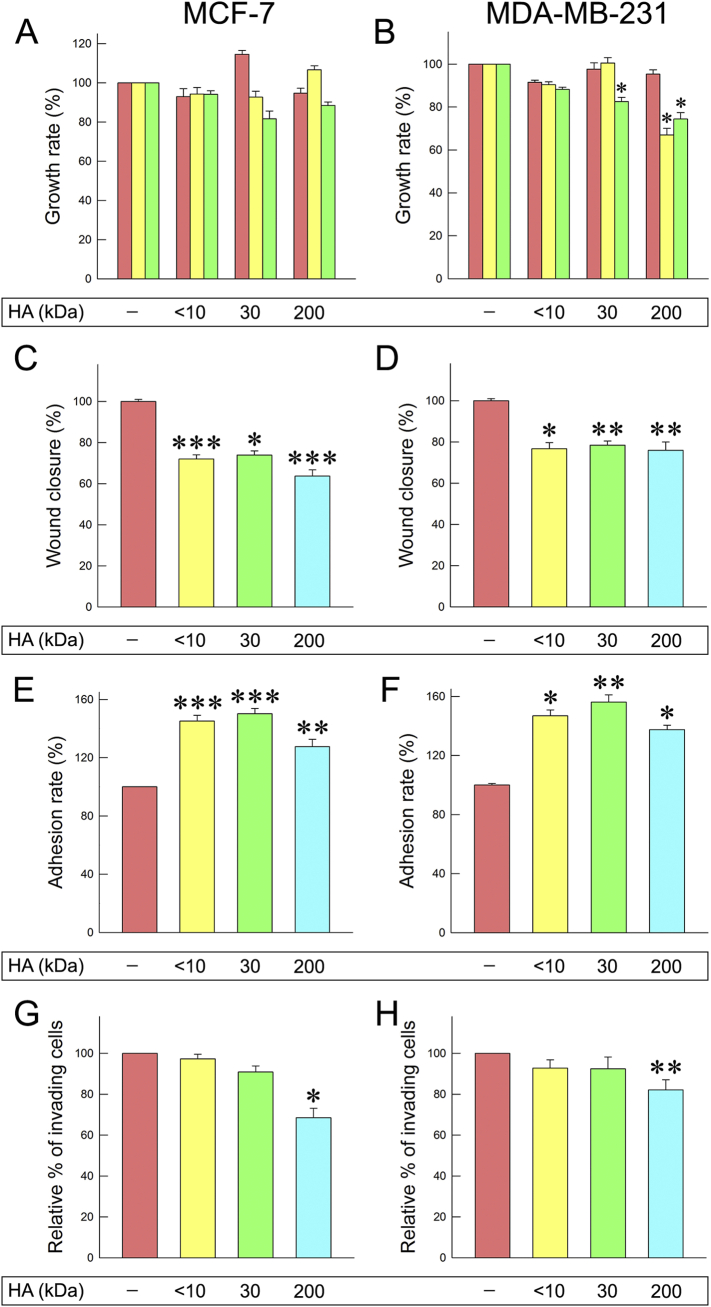
HA fractions of different molecular size were synthesized (supplementary Table 1) in order to evaluate their biological effects in two breast cancer cell lines with different ER status and aggressiveness: MCF-7 and MDA-MB-231. First, the effect of HA fractions on the proliferation of breast cancer cells was studied. Three different HA concentrations (12.5, 25, 200 μg/mL) were tested. The obtained results demonstrate that the HA fragments of 30 and 200 kDa reduced cell proliferation at the higher concentrations tested for both cell lines. According to the pilot experiments, the functional properties were further examined for all three fractions, <10, 30 and 200 kDa (provided by Fidia Farmaceutici S.p.A.) at a final concentration of 200 μg/mL. At a next step, we determined whether HA fractions could affect cell migratory potential by conducting wound healing assay. Notably, all of the HA fractions tested could decrease the migration (from 22% to 37%) of both cell lines (Fig. 1C and D).
Fig. 1.
Effects of different HA molecular sizes on breast cancer cells. (A, B) Cell proliferation after 24 h of HA treatment (concentrations: 12.5 μg/mL, 25 μg/mL and 200 μg/mL). (C, D) Cell migration after 24 h of HA treatment at a final concentration of 200 μg/mL. (E, F) Cell adhesion on collagen type I. (G, H) Cell invasion after 24 h of HA treatment at a final concentration of 200 μg/mL. Each experiment was conducted three times. Each bar represents mean ± S.D. values from triplicate samples. Asterisks (*), (**) indicate statistically significant differences (p < 0.05 and p < 0.01, respectively).
Cancer cells in order to invade and metastasize alter cell-cell and cell-matrix interactions [11]. Therefore, the next step was to evaluate the effects on cancer cell adhesion ability and invasiveness. The obtained data demonstrated that the three HA fractions tested significantly increased cell adhesiveness on collagen type I. MCF-7 adhesion ability increased by 45%, 50% and 27% following a 24 h treatment with <10, 30 and 200 kDa HA fractions, respectively (Fig. 1E). As shown in Fig. 1F, MDA-MB-231 adhesion ability was also increased (46%, 56% and 37%, respectively). These results could be correlated with the HA-mediated decreased migratory potential of the cells. Evaluation of cancer cells invasiveness showed that only the 200 kDa HA fraction significantly decreased the invasiveness of both cell lines in collagen type I gels (Fig. 1G and H).
HA fragments competition with endogenous HA affects cell migration
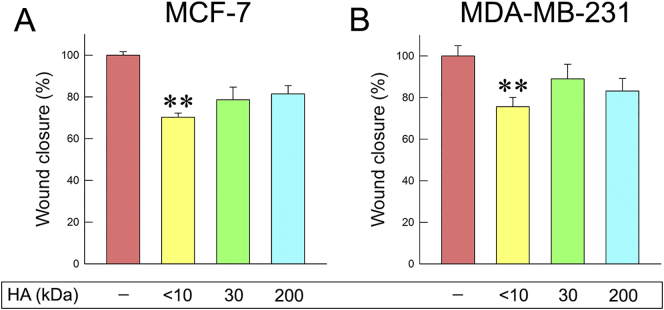
The results described above could possibly occur due to competitive effects between the HA fragments and endogenous HA. In order to investigate this possibility, breast cancer cells were treated with hyaluronidase at a final concentration of 1 U/mL for 1 h to remove pericellular HA coating. Following treatment with hyaluronidase, 200 kDa HA was added, since it affected the cellular functional properties the most. Wound healing assay was conducted for both cell lines tested. According to the obtained data even though 200 kDa could reduce cellular migration, pretreatment with hyaluronidase attenuated this effect (Fig. 2). Notably, similar results were observed in both cell lines although MCF-7 cells produce low amounts of HA (Fig. 2A). Consequently, the effects of the HA fragments on cell migration partly occur due to competition with endogenous HA.
Fig. 2.
Effects of 200 kDa HA on cell migration after treatment with hyaluronidase. (A) MCF-7 cell migration after hyaluronidase treatment (1 h) and 200 kDa HA treatment (24 h) at a final concentration of 200 μg/mL. (B) MDA-MB-231 cell migration after 1 h of hyaluronidase treatment and 24 h of 200 kDa HA treatment at a final concentration of 200 μg/mL.
Evaluation of the HA size-dependent functional properties
The observed effects of the various HA molecular sizes on the functional properties of both cancer cell lines raised the question whether their morphology is also affected. Moreover, we found of importance to evaluate the invasiveness of cancer cells using a matrix mimetic environment. For this purpose, breast cancer cells were cultured on Matrigel and were analyzed using scanning electron microscopy (SEM). The patterns of lamellipodia and filopodia, known to contribute to migration, invasion and metastasis of cancer cells [48,49], were also evaluated by SEM.
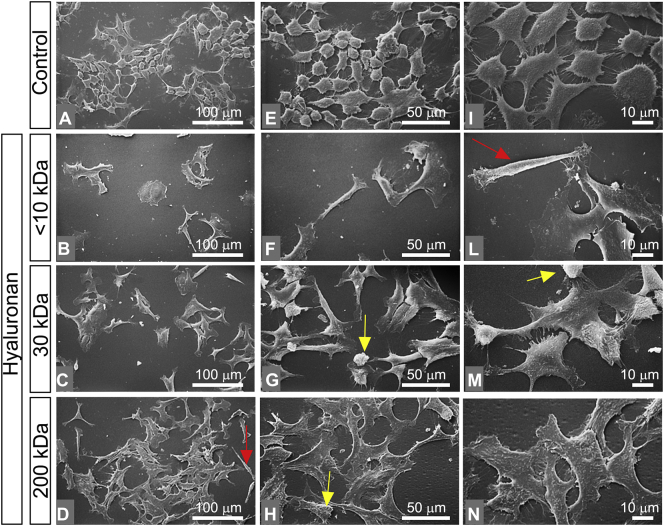
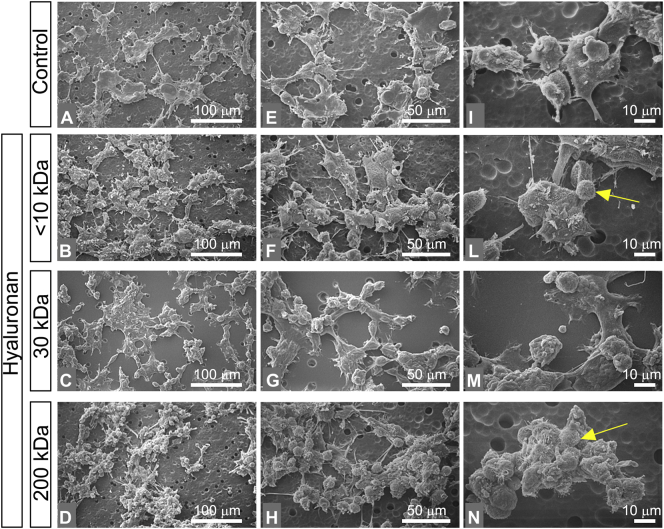
Almost all of MCF-7 cells (control group in culture flasks) showed an ovoidal flattened shape with few and short cytoplasmic microvilli and no microvesicles (epithelial phenotype). Very rare globular and elongated fusiform cells were detectable. Even though all cells created cell-cell contacts, gaps between adjacent cells were still observable. Isolated cells were not observed (Fig. 3A, E, I). Cells treated with <10 kDa HA mainly included isolated groups of less than ten large stem-like shaped cells, with few microvilli and no microvesicles. Rare isolated flattened ovoidal cells were observed (Fig. 3B and F). Isolated fusiform/elongated cells comparable to mesenchymal shaped cells were also detectable (Fig. 3F and L). MCF-7 cells treated with 30 kDa HA showed groups of many polygonal flattened cells with cell-cell contacts, filopodia, few microvilli and no microvesicles (Fig. 3C, G, M). Rare elongated cells and very few globular cells were also detectable. All cells exhibited few microvilli and no microvesicles (Fig. 3G, M). Most of MCF-7 cells treated with 200 kDa HA were extremely flattened polygonal cells, with very few microvilli and no microvesicles. Very rare stem-like cells and elongated ones were also present (Fig. 3D, H, N). The flattened polygonal cells had a tight cell-cell contact that makes it difficult to distinguish the interface between the adjacent cells (Fig. 3N).
Fig. 3.
Impact of HA molecular sizes in morphology of MCF-7 cells. (A, E, I) MCF-7 cultured in flasks. Cells show an ovoidal flattened shape with few and short cytoplasmic microvilli and no microvesicles (epithelial phenotype) (bar 0.1 mm for A, E and 10 μm for I). Very rare globular and elongated fusiform cells are also detectable (A, E). Even though all cells create cell-cell contacts by thin cytoplasmic protrusions, gaps between adjacent cells are observable (I). No isolated cells are present. (B, F, L) MCF-7 cells treated with <10 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for B, F and 10 μm for L). The culture mainly includes isolated groups of less than ten large stem-like cells with few microvilli and no microvesicles. (F) An isolated flattened ovoidal cell is observed in the center. (L) An isolated fusiform/elongated cell comparable to mesenchymal shaped cells is visible (red arrow). (C, G, M) MCF-7 cells treated with 30 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for C, G and 10 μm for M). The culture includes many grouped polygonal flattened cells with cell-cell contacts, filopodia, few microvilli and no microvesicles. Some elongated cells and very few globular cells are also detectable. All these cells exhibit few microvilli (yellow arrow) and no microvesicles (G, M). (D, H, N) MCF-7 cells treated with 200 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for D, H and 10 μm for N). Most of the cells appear with extremely thin flattened polygonal pattern with very few microvilli (yellow arrow) and no microvesicles. Very rare stem like cells and elongated ones (red arrow) are also present (D, H). When the very thin and flattened polygonal cells exhibit tight cell-cell contacts, it is difficult to distinguish the interface between the adjacent cells (N). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
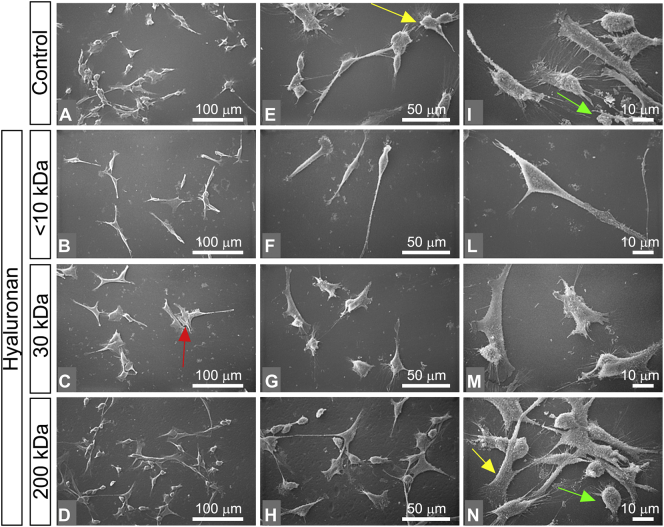
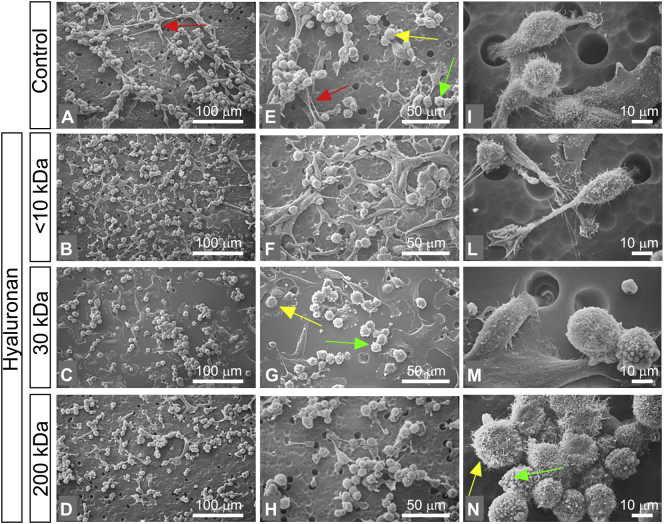
In contrast to MCF-7, MDA-MB-231 cells (control group in culture flasks) appeared as isolated cells with no cell-cell contacts. Most of the cells showed globular and spindle-like shapes (mesenchymal phenotype). All cells exhibited both cytoplasmic microvilli and microvesicles which were particularly distributed on the surface of the globular shaped cells (Fig. 4A, E, I). When the cells were treated with <10 kDa HA, the cell culture included isolated and very elongated spindle-like shaped cells, which showed very long filopodia (mesenchymal phenotype) and very rare cell-cell contacts (Fig. 4B, F, L). Most of the MDA-MB-231 cells treated with 30 kDa HA appeared in groups of three to five stem-like cells with cell-cell contacts, very few isolated globular shaped cells and rare spindle-like shaped cells (Fig. 4C, G, M). MDA-MB-231 cells treated with 200 kDa HA included only a few spindle-like cells, more numerous stem like cells and globular cells. Cell-cell contacts were more frequently observed compared to those of the previous groups and the number of globular shaped cells was significantly increased compared to the control cells (Fig. 4D, H, N). Microvilli on flattened cells and microvesicles on the globular ones were still observable (Fig. 4N).
Fig. 4.
Impact of HA molecular sizes in morphology of MDA-MB-231 cells. (A, E, I) MDA-MB-231 cultured in flasks appear as isolated cells with no cell-cell contacts or in groups of few cells (bar 0.1 mm for A, E and 10 μm for I). Most of the cells show globular and spindle-like shape (mesenchymal phenotype). Rare polygonal flattened cells are observed. All cells show both cytoplasmic microvilli (yellow arrow) and microvesicles (green arrow), which are particularly located on the surface of the globular shaped cells. (B, F, L) MDA-MB-231 cells treated with <10 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for B, F and 10 μm for L). Most of the cells are isolated and appear like very elongated spindle-like shaped cells showing very long filopodia (mesenchymal phenotype) and very rare cell-cell contacts. (C, G, M) MDA-MB-231 cells treated with 30 kD HA at a final concentration of 200 μg/mL (bar 0.1 mm for C, G and 10 μm for M). The cells appear in groups of three to five stem-like cells (red arrow) with cell-cell contacts. Isolated globular shaped cells and rare spindle like shaped cells are observable. (D, H, N) MDA-MB-231 cells treated with 200 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for D, H and 10 μm for N). The cultures include few spindle-like cells, more numerous stem like cells and globular cells. Cell-cell contacts are frequently observable if compared to those of the previous groups and the globular shaped cells are increased in comparison to the control group. Microvilli (yellow arrow) on flattened cells and microvesicles (green arrow) on the globular ones are observable (N). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Cultures of MCF-7 cells on Millipore filter coated by Matrigel (control group) showed groups of flattened polygonal cells with short filopodia and globular cells with no microvesicles. Many cell-cell contacts among all cells were detectable (Fig. 5A, E, I). MCF-7 cells treated with <10 kDa HA showed the same globular cells with few microvilli and no microvesicles however the number of the flattened polygonal cells was increased compared to the control group. Most of the cells exhibited cell-cell contacts and groups (Fig. 5B, F, L). The MCF-7 cells treated with 30 kDa HA appeared also as flattened polygonal cells. A slight increase in the number of the globular ones, growing on the flattened cells, if compared to the control group, was observed. All cells were grouped and demonstrated cell-cell contacts (Fig. 5C, G and M). The MCF-7 cells treated with 200 kDa HA included few flattened polygonal cells and many globular cells as compared to all of the previous groups. All cells exhibited cell-cell contacts, few microvilli and no microvesicles (Fig. 5D, H, N).
Fig. 5.
Impact of HA molecular sizes in morphology of MCF-7 cells in Millipore filter coated with Matrigel. (A, E, I) MCF-7 cells control group on Matrigel coated Millipore filters (bar 0.1 mm for A, E and 10 μm for I). The cultures include groups of flattened polygonal cells with short filopodia and globular cells with no microvesicles growing also on the flattened ones. Many cell-cell contacts among all cells are observed. (B, F, L) MCF-7 cells treated with <10 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for B, F and 10 μm for L). The cultures include groups of globular cells with few microvilli (yellow arrow) and no microvesicles. In addition, a higher number of flattened polygonal cells in comparison to the control group are observed. Most of the cells are grouped and show cell-cell contact. (C, G, M) MCF-7 cells treated with 30 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for C, G and 10 μm for M). The cultures include flattened polygonal cells and a slight increase in number of the globular ones in comparison to the control group is observed. The globular cells are growing on the flattened ones. All the cells are grouped and create cell-cell contacts. (D, H, N) MCF-7 cells treated with 200 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for D, H and 10 μm for N). The cultures include few flattened polygonal cells and many globular cells in comparison to all cells of the previous groups. The numerous globular cells grow also on the flattened ones. All cells show cell-cell contacts, few microvilli (yellow arrow) and no microvesicles. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Most of the MDA-MB-231 cells cultured on a Millipore filter coated by Matrigel (control group) appeared grouped and formed cell-cell contacts. Globular cells with microvilli and microvesicles were more numerous than spindle-like cells (Fig. 6A, E, I). The cells after treatment with <10 kDa HA exhibited the same globular and spindle-like shape with the control group. However, the mesenchymal cells were increased in number in comparison to the globular ones. Most of cells formed cell-cell contacts and many isolated cells passing through the pores of Matrigel-coated Millipore were also observed (Fig. 6B, F, L). The cultures of MDA-MB-231 cells that were treated with 30 kDa HA included very thin and flattened polygonal cells with no filopodia, very few microvilli and no microvesicles. In these cell cultures, globular cells with microvilli and microvesicles were also detectable. Cell-cell contacts were observed among all the cells (Fig. 6C, G, M). Most of the MDA-MB-231 cultures treated with 200 kDa HA included globular cells with microvilli, microvesicles and a few elongated cells (Fig. 6D, H). Most of the cells exhibited cell-cell contacts even though very few isolated cells were detectable (Fig. 6N).
Fig. 6.
Impact of HA molecular sizes in morphology of MDA-MB-231 cells in Millipore filter coated with Matrigel. (A, E, I) MDA-MB-231 cells control group on Matrigel coated Millipore filters (bar 0.1 mm for A, E and 10 μm for I). The cells appear grouped with cell-cell contacts. Globular cells with microvilli (yellow arrow) and microvesicles (green arrow) are more numerous than spindle like shaped cells (red arrows) (mesenchymal phenotype). (B, F, L) MDA-MB-231 cells treated with <10 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for B, F and 10 μm for L). The cultures include globular and spindle like shaped cells. The elongated mesenchymal-like cells are more numerous than the globular ones (B, F). Most of the cells show cell-cell contacts. However, many isolated cells passing through Millipore pores are also visible (L). (C, G, M) MDA-MB-231 cells treated with 30 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for C, G and 10 μm for M). Very thin and flattened polygonal cells with no filopodia, very few microvilli and no microvesicles are observed. On this culture, globular cells with microvilli (yellow arrow) and microvesicles (green arrow) are also detectable. Cell-cell contacts are observed among all the cells. (D, H, N) MDA-MB-231 cells treated with 200 kDa HA at a final concentration of 200 μg/mL (bar 0.1 mm for D, H and 10 μm for N). The cultures include globular cells with microvilli, microvesicles and fewer elongated cells (D, H). Most of the cells show cell-cell contacts even though very few isolated cells are present. Grouped globular cells with microvilli (yellow arrow) and microvesicles (green arrow) are also visible (N). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
HA fractions evokes size-dependent changes in the expression of EMT markers
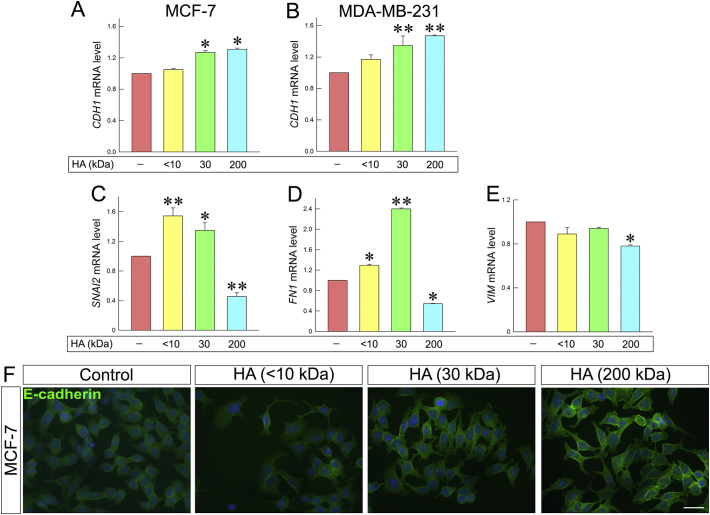
During EMT process, the expression patterns of characteristic molecules are altered. E-cadherin is an epithelial marker, which is suppressed during EMT process, while mesenchymal markers, such as fibronectin and snail2/slug are overexpressed [50]. Notably, following treatment with HA, E-cadherin expression was increased in both cell lines (Fig. 7A and B). Treatment of MCF-7 cells with 30 and 200 kDa HA resulted in increased expression of E-cadherin (ca. 27% and 31%), as compared to control cells. These results also confirmed by immunofluorescence (Fig. 7F). More interestingly, the expression levels of E-cadherin in MDA-MB-231 cells were increased upon treatment with 30 kDa and 200 kDa HA (ca. 34% and 47%). MDA-MB-231 control cells exhibit mesenchymal phenotype and express low levels of E-cadherin. Considering the upregulation of E-cadherin after HA treatment, the expression levels of major mesenchymal markers were evaluated. Although the expressions of snail2/slug and fibronectin were increased with <10 and 30 kDa HA (Fig. 7C and D), the treatment with 200 kDa HA downregulated the expression levels of the two EMT markers (ca. 46% and 55%), demonstrating the crucial role of this HA molecular size to affect EMT. In addition, the expression of vimentin (Fig. 7E) was also reduced with 200 kDa HA (ca. 22%).
Fig. 7.
HA fragments mediate the expression of major EMT markers. Quantitative RT-PCR analysis of E-cadherin mRNA levels in MCF-7 (A) and MDA-MB-231 cells (B) following a 24 h HA treatment at a final concentration of 200 μg/mL. (C, D) Expression of the mesenchymal marker snail2/slug, fibronectin and vimentin in MDA-MB-231 cells. Asterisks (*), (**) indicate statistically significant differences (p < 0.05 and p < 0.01, respectively); (F) Immunofluorescence analysis for E-cadherin (green). Cells were treated with HA fragments of different molecular size for 24 h. Nuclei are shown in blue (DAPI). Scale bars ~20 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
HA modulates the expression of HASs, CD44 and ECM regulators
Taking into consideration that breast cancer cells functional properties and morphology are affected upon treatment with HA of various molecular sizes, we prompted to further examine whether these changes could be correlated to modifications of the expression patterns of ECM effectors, such as HA synthases (HAS2 and 3) and MMPs, known to play crucial role for cell survival, motility and EMT.
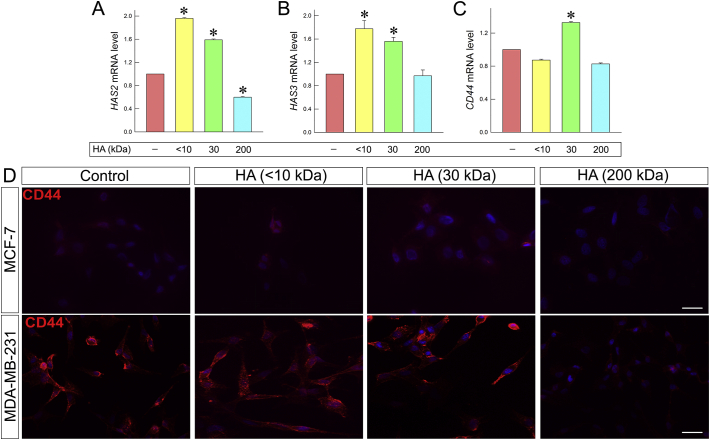
Elevated expression of HAS2 has been correlated to breast cancer cells invasiveness, EMT, increased migratory ability and metastasis [[51], [52], [53], [54], [55], [56], [57], [58]], whereas downregulation of HAS2 has been correlated to less aggressive cellular behavior [[59], [60], [61]]. HAS2 is the major HAS expressed in MDA-MB-231 cells [62,63] and therefore the effects of the HA fractions were evaluated in this cell line. Interestingly, HAS2 expression in MDA-MB-231 cells was increased following treatment with <10 and 30 kDa HA (ca. 96% and 60%, respectively). However, as shown in Fig. 8A, HAS2 expression was remarkably decreased (ca. 40%) upon treatment with the 200 kDa HA. HAS3 is overexpressed in a variety of tumors [18,19]. A similar pattern was obtained for the mRNA level of HAS3 in the MDA-MB-231 cells with an increment upon <10 and 30 kDa HA treatment (ca. 78% and 56%) as compared to control (Fig. 8B). The obtained data clearly suggest that the different molecular sizes of HA play crucial role in the expression of HAS2 and -3. It is worth noticing that the 200 kDa HA fraction has a beneficial anticancer effect for the highly aggressive MDA-MB-231 cells as it suppressed the expression of the HAS2, which in turn has been correlated with the invasiveness of these cells.
Fig. 8.
HA fragments effect on HA synthesis and HA receptor CD44. (A) Real-time PCR analysis of HAS2 mRNA levels in MDA-MB-231 cells after 24 h of HA treatment. (B) Real-time PCR analysis of HAS3 mRNA levels in MDA-MB-231 cells after 24 h of HA treatment. (C) Real-time PCR analysis of CD44 total mRNA levels in MDA-MB-231 cells after 24 h of HA treatment. Asterisks (*), (**) indicate statistically significant differences (p < 0.05 and p < 0.01, respectively); (D) Immunofluorescence analysis for CD44 (red). Cells were treated with HA fragments of different molecular size at final concentration of 200 μg/mL for 24 h. Nuclei are shown in blue (DAPI). Scale bars ~20 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
CD44 is one of the major HA receptors. CD44 interactions with pericellular HA lead to breast cancer cells migration, invasion, EMT and metastasis [[64], [65], [66], [67]]. The disruption of these interactions could inhibit the above processes [68,69]. Immunofluorescence revealed that upon HA treatment CD44 signal is attenuated in cellular protrusions as compared to the control cells. This is more profound after treatment with 200 kDa HA in MDA-MB-231 cells that express high levels of CD44. Moreover, CD44 signal is slightly reduced (Fig. 8D). This result is well correlated with the morphological changes observed in the cells after the addition of 200 kDa HA. On the other hand, the signal seems to be slightly increased following treatment with 30 kDa HA in both cell lines. These data are also confirmed by qPCR (Fig. 8C). The expression of CD44 was increased after treatment with 30 kDa HA and slightly decreased after treatment with 200 kDa HA (ca. 30% and 18% respectively).
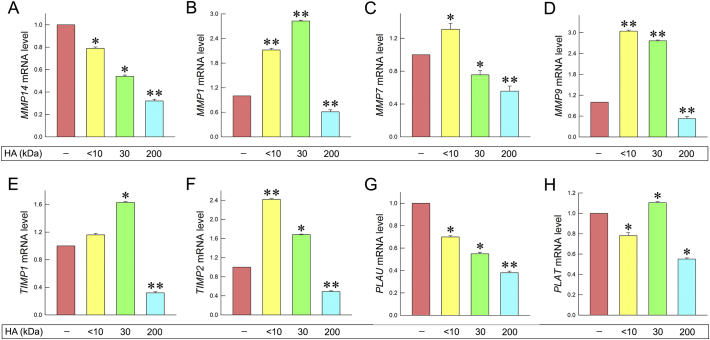
MMPs are major ECM regulators, since they degrade the matrix macromolecules thus altering the cell-cell or cell-matrix interactions. These changes could promote cell migration, invasion and angiogenesis [11,70]. Generally, MMPs and their natural inhibitors, tissue inhibitors of metalloproteinases (TIMPs), are upregulated in breast cancer [[71], [72], [73]]. MMPs, like MMP9, are able to form complexes with HA receptor, CD44, triggering signaling pathways that increase cancer cells invasiveness. Moreover, endogenous HA-CD44 interactions lead to upregulation of MMPs expression, like MT1-MMP promoting cells invasion [74,75]. Thus, as next step we evaluated the effects of HA fractions on MMPs and TIMPs expression in the highly aggressive MDA-MB-231 cells. Notably, MT1-MMP (MMP14) expression was decreased upon treatment with 30 and 200 kDa (ca. 46% and 68%, respectively) (Fig. 9A). Although the expression of MMP1 increased with <10 and 30 kDa (Fig. 9B), it was decreased following treatment with 200 kDa HA (ca. 40%). The expression of MMP7 increased after treatment with <10 kDa HA (ca. 31%), whereas it decreased after treatment with 30 and 200 kDa HA (ca. 25% and 45%, respectively) (Fig. 9C). Concerning the MMP9 expression, although an increment was noted with <10 and 30 kDa HA, as shown in Fig. 9D, it was downregulated after treatment with 200 kDa HA (ca. 47%).
Fig. 9.
HA fragments modify the mRNA expression profiles of major ECM regulators in breast cancer cells. (A) Quantitative RT-PCR analysis of MT1-MMP mRNA levels of MDA-MB-231 cells after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. (B) qRT-PCR analysis of MMP1 mRNA levels of MDA-MB-231 cells after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. (C) qRT-PCR analysis of MMP7 mRNA levels of MDA-MB-231 cells after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. (D) qRT-PCR analysis of MMP9 mRNA levels of MDA-MB-231 cells after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. (E) qRT-PCR analysis of TIMP1 mRNA levels of MDA-MB-231 cells after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. (F) qRT-PCR analysis of TIMP2 mRNA levels of MDA-MB-231 cells after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. (G) qRT-PCR analysis of uPA (PLAU) mRNA levels of MDA-MB-231 after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. (H) qRT-PCR analysis of tPA (PLAT) mRNA levels of MDA-MB-231 after treatment with HA fragments at a final concentration of 200 μg/mL for 24 h. Asterisks (*), (**) indicate statistically significant differences (p < 0.05 and p < 0.01, respectively).
In addition, the expression levels of TIMPs were evaluated in MDA-MB-231 cells. The expression levels of TIMP1 were increased after treatment with 30 kDa HA (ca. 63%), while suppressed after treatment with 200 kDa HA (ca. 68%) (Fig. 9E). The expression levels of TIMP2 of these cells were increased after treatment with <10 and 30 kDa HA (ca. 2-fold and 68% respectively). However, the mRNA levels of TIMP2 were downregulated after treatment with 200 kDa HA (ca. 51%), as shown in Fig. 9F.
Plasminogen activation system also plays an important role in ECM remodeling and cancer cells migration, invasion and metastasis [[76], [77], [78]]. Endogenous HA-CD44 interactions promote the expression of plasminogen activation system molecules leading to the increment of cancer cells invasiveness [74]. Plasminogen activation system molecules are overexpressed at high invasive MDA-MB-231 cells [76]. For this purpose, the HA fractions effects were evaluated on the expression of these molecules (Fig. 9G and H). Notably, urokinase plasminogen activator (uPA/PLAU) expression levels were downregulated after HA treatment (ca. 30%, 45% and 62%). Furthermore, the mRNA levels of tissue plasminogen activator (tPA/PLAT) were increased after treatment with 30 kDa HA (ca. 10%), while its gene expression was downregulated after treatment with 200 kDa (ca. 45%).
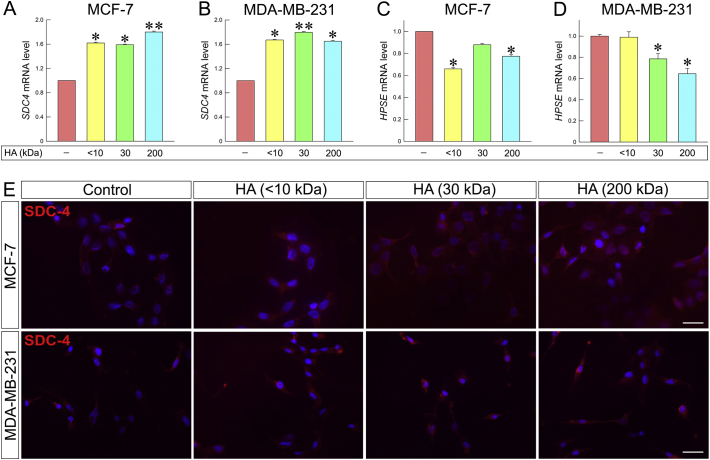
As mentioned above, breast cancer cell behavior depends on their microenvironment. ECM components are able to modify cellular properties, influence cell signaling and enhance or inhibit cancer progression. Syndecans (SDCs) are cell surface heparan sulfate proteoglycans that play pivotal role to the regulation of cellular responses. SDC-4 is ubiquitously expressed. Elevated levels of SDC-4 in breast cancer are correlated to ERs and PgR expression. SDC-4 promotes cell adhesion and inhibits the cellular invasion into collagen as well [[79], [80], [81], [82], [83]]. Heparanase is a matrix degradation enzyme that is involved in the shedding of SDCs. Elevated levels of heparanase are associated with tumor aggressiveness, angiogenesis and metastasis [[84], [85], [86], [87]]. Notably, as shown in Fig. 10A and B, treatment with all HA fractions of different molecular sizes increased the expression of SDC-4 in both cell lines (62%, 59% and 80% in MCF-7 and 67%, 80% and 65% in MDA-MB-231 cells following treatments with <10, 30 and 200 kDa HA, respectively). The above results are also confirmed by immunofluorescence (Fig. 10E). On the other hand, the expression of heparanase decreased in both cells lines (Fig. 10C and D).
Fig. 10.
HA fragments change the expression patterns of SDC-4 and heparanase. (A) qRT-PCR analysis of SDC-4 mRNA levels of MCF-7 after 24 h of HA treatment at a final concentration of 200 μg/mL. (B) qRT-PCR analysis of SDC-4 mRNA levels of MDA-MB-231 cells after 24 h of HA treatment at a final concentration of 200 μg/mL. (C) qRT-PCR analysis of heparanase mRNA levels of MCF-7 cells after 24 h of HA treatment at a final concentration of 200 μg/mL. (D) qRT-PCR analysis of heparanase mRNA levels of MDA-MB-231 cells after 24 h of HA treatment at a final concentration of 200 μg/mL. Asterisks (*), (**) indicate statistically significant differences (p < 0.05 and p < 0.01, respectively). (E) Immunofluorescence analysis for SDC-4 (red). Cells were treated with HA fragments of different molecular size at final concentration of 200 μg/mL for 24 h. Nuclei are shown in blue (DAPI). Scale bars ~20 μm. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Discussion
HA is one of the major ECM components, which plays pivotal role in several biological processes. Recent reports demonstrated that HA molecular size is a key regulator of its properties and functions [26,88,89]. HA binding to its receptor, CD44, triggers various signaling pathways promoting cancer cells proliferation, migration and invasiveness [[64], [65], [66],[90], [91], [92], [93]]. On the other hand, the interruption of HA-CD44 interactions can attenuate these signaling cascades [37,38,[94], [95], [96]]. Several studies indicated that HA fragments, depending on their molecular size are able to compete with the endogenous HA for binding to the HA receptors [26,45,88]. As a result they are able to inhibit the signaling pathways that are initiated by the HA binding to CD44 [42,44,89]. This ability of the HA fractions has been proven to reduce the aggressiveness of various types of cancer cells [[33], [34], [35], [36],39,40,47,97].
In the present study, HA fragments of defined molecular size were synthesized. The effects of HA fragments were further evaluated on breast cancer cells with different ER status; the epithelial ERα-positive, MCF-7, and the aggressive ERβ-positive, MDA-MB-231 breast cancer cells. HA fragments of <10, 30 and 200 kDa were evaluated in terms of their effects on breast cancer cells' functional properties, morphology and gene expression.
The obtained data demonstrated that the adhesion of breast cancer cells on collagen type I was increased after HA treatment (Fig. 10A and B). These results may be correlated to the increased expression levels of SDC-4 that were observed following HA treatment. SDCs play crucial role in cellular growth, spreading, adhesion and signaling by acting as co-receptors with integrins and cell-cell adhesion molecules such collagen type I and fibronectin [98,99]. SDC-4 in particular has been correlated with better cancer prognosis and it is mainly present and concentrated in focal adhesions [79,81,82,100].
Our data revealed that among the tested HA fragments, the 200 kDa HA fraction exhibited significant effects on breast cancer cells. Even though all the synthesized fractions were able to inhibit breast cancer cell migration, only this fraction was able to inhibit cellular invasion and evoked significant morphological changes in both cell lines leading to a less aggressive phenotype. This phenomenon may depend on different signaling pathways and the partial competition with endogenous HA for CD44 binding.
In breast cancer, MMPs and TIMPs are overexpressed. Elevated levels of MMPs are associated to poor prognosis, EMT, cancer cells' invasion and aggressiveness [11,72,74,[101], [102], [103], [104], [105], [106], [107], [108], [109]]. CD44 interactions with the endogenous HA upregulate the expression of MMPs like MT1-MMP, which is necessary for MMPs activation. MMP9 forms complexes with CD44 leading to cellular invasion [110,111]. Furthermore, MT1-MMP and MMP9 are associated with the formation of invadopodia. Several studies have proven that the downregulation of these molecules expression can reduce migration, invasion and angiogenesis [[112], [113], [114], [115], [116], [117]]. Treatment with 200 kDa HA could reduce invasiveness and cellular protrusions evoking morphological changes by significantly decreasing the expression levels of MT1-MMP, MMP1, MMP7 and MMP9. In addition, the expression levels of TIMP2 which activates MMP2 promoting cellular aggressiveness, were downregulated upon treatment [[118], [119], [120], [121]].
The morphological changes that were triggered by the addition of 200 kDa HA are supported by differentiated mRNA levels of major EMT markers in both cell lines. E-cadherin is the main epithelial marker that is normally expressed in very low levels in mesenchymal type cells like MDA-MB-231 [12,122,123]. Notably, our data indicate that the mRNA levels of E-cadherin were increased in both cell lines. On the contrary, mesenchymal markers that are upregulated during EMT process and highly expressed in MDA-MB-231 [1,12,50] were downregulated.
Moreover, treatment with this 200 kDa HA led to significant changes of the expression profile of important ECM regulators in MDA-MB-231. The expression levels of HAS2 are significantly upregulated in breast cancer cases and are associated with bad prognosis [[51], [52], [53], [54], [55], [56],124,125], whereas their downregulation leads to the reduction of cancer cells' aggressiveness [[59], [60], [61]]. Our data indicate that after treatment, the levels of HAS2 expression were significantly downregulated (Fig. 8A). This observation can be correlated to the reduction of MDA-MB-231 aggressiveness. Furthermore, the immunofluorescence data demonstrate that after the treatment CD44 signal was attenuated, in particular in MDA-MB-231 protrusions. CD44-HA interactions lead to cancer cells proliferation, migration, invasiveness and chemotherapy resistance [65,67,90,[126], [127], [128], [129], [130], [131]].
Plasminogen activation system is essential for ECM remodeling. It is also correlated to breast cancer cells invasion and metastasis. Expression of uPA has been associated with ERK expression [[76], [77], [78],132,133] and can be triggered by HA-CD44 interactions [74]. Our data demonstrate that the expression levels of uPA and tPA in MDA-MB-231 were downregulated after treatment (Fig. 9G and H). Furthermore, the expression levels of heparanase that are highly correlated with increased cancer cells aggressiveness and bad prognosis [84,86,134] were decreased in both cell lines (Fig. 10C and D).
In conclusion, our findings demonstrate that the HA fragments could regulate mammary cancer cells functional properties, morphology and gene expression. Treatment with 200 kDa HA not only downregulated the expression levels of molecules that are correlated to cellular aggressiveness but also reduced breast cancer cell invasion, proliferation and migration and led to a less aggressive phenotype. The exact mechanism by which the HA fragments drive these actions is still to be elucidated. However, our novel data indicate that 200 kDa HA fragment could be useful in the development of therapeutic strategies for aggressive mammary cancer.
Materials and methods
Synthesis of HA fractions
LMW HA fraction (30 kDa) was obtained by chemical hydrolysis. Solution of 15 mg/mL of 200 kDa HANa (Fidia Farmaceutici S.p.A.) in 0.1 M NaOH was prepared. The mixture was split into two batches (A-B) and stirred at 45 °C for 44 h (batch A) and 66 h (batch B). Then, the solutions were transferred into new flasks and 1.0 M HCl was added dropwise for pH neutralization (final pH = 7.0 ± 0.5). Milli-Q water (15% of the total solvent amount) and NaCl (0.55 g for 100 mL of solvent) were added. Then, both the solutions were precipitated with two volumes of cold ethanol 96% v/v. Afterwards the solutions were filtered, washed three times with EtOH/water 9:1 and vacuum dried overnight. The products were dissolved in PBS (phosphate buffer saline) 0.1 M, pH = 7, at a concentration of 30 mg/mL, then dialyzed (membrane cut-off: 20 kDa) against PBS for 2 days and against milli-Q water for 1 day. The final step was the freeze-drying. Viscotek analysis was carried out in order to evaluate the MW of each fraction (Table 1). Batch B (30 kDa HA) was used for biological evaluation.
HA oligomer (<10 kDa) was obtained by enzymatic hydrolysis. 200 kDa HANa (Fidia Farmaceutici S.p.A.) was added to 0.1 M acetate buffer pH = 5 (final HA concentration: 20 mg/mL), and stirred until total HA dissolution. Afterwards, bovine testes hyaluronidase – BTH (Fidia Farmaceutici S.p.A., 800 U/mg, 55 kDa, code: 3506500) was added, under stirring at 37 °C. The reaction was split into two parts (batches C and D). Batch C was terminated after 72 h: in order to deactivate the enzyme, the solution was heated at 98 °C for 10 min under stirring. The solution was ultra-filtrated using a 30 kDa cut-off cassette (Millipore), in order to obtain products with a MW < 30 kDa. Following, the solution was ultra-filtrated using a 10 kDa cut-off cassette, in order to separate two polymer fractions, one with MW up to 10 kDa (10–30 kDa HA) and a second one with MW < 10 kDa (<10 kDa HA). Finally, the samples were freeze-dried. The <10 kDa fraction was used for biological evaluation.
Cell cultures and reagents
MCF-7 (ERα-positive, low metastatic potential) and MDA-MB-231 (ERβ-positive, high metastatic potential) breast cancer cells were obtained from the American Type Culture Collection (ATCC). Breast cancer cell lines were cultured in complete medium [Dulbecco's Modified Eagle's Medium (DMEM, LMD1110/500, Biosera)] supplemented with 10% fetal bovine serum (FBS, FB-1000/500, Biosera), a cocktail of antimicrobial agents (100 IU/mL penicillin, 100 μg/mL streptomycin, 10 μg/mL gentamycin sulfate and 2.5 μg/mL amphotericin B), 1.0 mM sodium pyruvate and 2 mM l-glutamine. All of the above were obtained from Biosera LTD. (Courtaboeuf Cedex, France). The cell lines were routinely cultured at 37 °C, in 95% humidified atmosphere air in 5% CO2. When the cell confluency was approximately 80–85%, cells were harvested with trypsin-EDTA 1× in PBS (LM-T1706/500, Biosera). All experiments were conducted in serum-free conditions. The cytostatic agent cytarabine was purchased from Sigma Chemical Co. (St Louis, MO, USA). 200 kDa HA was supplied by Fidia Farmaceutici S.p.A. All the other HA fractions were synthesized at Fidia Farmaceutici S.p.A. (Abano Terme, Italy) as described above. All other chemicals used were of the best commercially available grade.
Proliferation assay
MCF-7 and MDA-MB-231 cells were seeded on 96-well plates at a density of 7500 and 5000 cells per well, respectively. The cells were incubated in complete medium for 24 h and were serum starved overnight. Then, serum-free medium containing the HA fractions at the following concentrations: 12.5 μg/mL, 25 μg/mL, 50 μg/mL, 100 μg/mL and 200 μg/mL, was added and the cells were incubated for 24 h. In order to evaluate the HA fractions effect on breast cancer cells proliferation, a crystal violet assay was used, as described in previous study [135]. Briefly, cells were washed twice with PBS and stained with 0.5% w/v crystal violet solution in 20% methanol/distilled water. The cells were incubated with the dye at 37 °C for 20 min with 150 oscillations on a bench rocker. Then, the staining solution was removed and the cells were washed three times with distilled water. The cells were left at room temperature overnight in order to dry. The next day, methanol was added in order to retrieve the dye that had been bounded to the cells. For this purpose, the plate was incubated at 37 °C for 20 min with 150 oscillations on a bench rocker. After incubation, the optical density of each well was measured at 570 nm (TECAN photometer).
In vitro wound healing assay
MCF-7 and MDA-MB-231 cells were seeded on 12-well plates at a density of 30 × 104 and 25 × 104 cells per well, respectively. Cells were cultured in complete medium for 24 h and serum starved overnight. The next day, the cell layer was wounded by scratching with a sterile 100 μL pipette tip. The cell layer was washed twice with PBS in order to remove the detached cells. Then serum-free medium containing the cytostatic cytarabine (10 μΜ) was added, as to minimize possible contribution of cell proliferation, and it remained until the end of experiment. After 40 min of incubation, the HA fractions were added at a final concentration of 200 μg/mL. The wound closure was monitored at 0 and 24 h using a digital camera connected to a phase-contrast microscope. In order to evaluate the HA fractions effect on breast cancer cells migration, the wound surface area was quantified by image analysis (Image J 1.50b Launcher Symmetry Software).
Cell adhesion assay
Breast cancer cells were seeded in 6-well plates. MCF-7 were seeded at a density of 30 × 104 cells per well and MDA-MB-231 were seeded at a density of 25 × 104 cells per well. Cells were cultured in complete medium for 24 h; afterwards, they were serum starved overnight. The next day, HA fractions were added for 24 h at a final concentration 200 μg/mL in serum-free medium. Cells were then harvested using 4 mM EDTA in PBS, centrifuged and resuspended in serum-free medium containing 0.1% BSA. Then, the cells were counted and seeded in 96-well plates that were pre-coated with 40 μg/mL collagen type I in PBS, at 4 °C overnight. MCF-7 were seeded at a density of 2 × 104 cells per well. MDA-MB-231 were seeded at a density of 1 × 104 cells per well. The next day the solution was removed, and the plate was washed twice with PBS and blocked with 1% BSA in PBS for 30 min. After seeding, the cells were incubated at 37 °C for 30 min in order to adhere to the surface. Then the cells were washed twice with PBS in order to remove the non-adherent ones. The adherent cells were stained with 0.5% w/v crystal violet in 20% methanol/distilled water and incubated for 20 min at 37 °C, at 150 oscillations on a bench rocker. The optical density of each well was measured following the crystal violet assay, as described above.
Collagen type I invasion assay
In order to evaluate the HA fractions effect on breast cancer cells invasive capacity, a collagen type I invasion assay was used, as described in previous study [136]. Briefly, breast cancer cells were cultured in complete medium for 24 h and then serum starved overnight. A collagen type I solution was prepared as following: 5 volumes of CMF-HBSS were mixed with 2.65 volumes of complete medium, 1 volume of MEM 10×, 1 volume of 0.25 M NaHCO3, 0.3 volumes of 1.0 M NaOH and 4 volumes of collagen type I (stock concentration 5 mg/mL). The final concentration of collagen type I was 1 mg/mL. The solution was spread homogeneously in 12-well plates and the plates were left to gelify at 37 °C, 5% CO2 for 1 h. Then, breast cancer cells were seeded in the plates at a density of 6 × 104 cells per well and the HA fractions were added at a final concentration of 200 μg/mL. Cells were incubated for 24 h. Afterwards, images were taken using a digital camera connected to a phase-contrast microscope.
In vitro wound healing assay after hyaluronidase treatment
MCF-7 and MDA-MB-231 cells were seeded on 12-well plates at a density of 30 × 104 and 25 × 104 cells per well, respectively. Cells were cultured in complete medium for 24 h and serum starved overnight. The next day the cell layer was wounded by scratching with a sterile 100 μL pipette tip. The cell layer was washed twice with PBS in order to remove the detached cells. Then hyaluronidase from Streptomyces hyalurolyticus (Sigma, H1136-1AMP) was added in serum free-medium at a final concentration of 1 U/mL and the cells were incubated at 37 °C, 5% CO2 for 1 h. Following the medium was removed, fresh serum-free medium containing the cytostatic cytarabine (10 μΜ) was added and the cells were incubated for 40 min. Afterwards, the HA fraction was added at a final concentration of 200 μg/mL. The wound closure was monitored at 0 and 24 h using a digital camera connected to a phase-contrast microscope. In order to evaluate the HA fraction effect on breast cancer cells migration, the wound surface area was quantified by image analysis (Image J 1.50b Launcher Symmetry Software).
Scanning electron microscopy
MCF-7 and MDA-MB-231 breast cancer cells were previously treated with HA fractions of different molecular size (<10 kDa, 30 kDa and 200 kDa) and then cultured for 5 h on polystyrene flasks in order to obtain different HA treated groups of cells. Untreated MCF-7 and MDA-MB-231 breast cancer cells were used as control groups. MCF-7 and MDA-MB-231 breast cancer cells were also cultured for 5 h on a “Isopore Membrane Filter” with pore size of 8.0 μm (Millipore, Milan, Italy) coated by 50 μL of Matrigel solution (0.18 μg/μL) (BD Biosciences, Milan, Italy) as control groups. Furthermore, MCF-7 and MDA-MB-231 breast cancer cells were first treated with HA fragments of different molecular size (<10 kDa, 30 kDa and 200 kDa) and then cultured for 5 h on Millipore filter plus Matrigel to collect different HA treated groups of breast cancer cells.
All of the MCF-7 and MDA-MB-231 cells cultured on flasks and on Millipore filter coated by 50 μL of Matrigel solution were fixed in a Karnovsky's solution for 20 min. The flasks and the Millipore filters with adhering cells were rinsed three times with 0.1% cacodylate buffer, dehydrated with increasing concentrations of ethanol, and finally dehydrated with hexamethyldisilazane (Sigma-Aldrich Inc.) for 15 min. Portions of the flasks and the hole Millipore filters were mounted on proper stubs, coated with a 5 nm palladium gold film (Emitech 550 sputter-coater) to be observed under a scanning electron microscope (SEM) (Philips 515, Eindhoven, the Netherlands) operating in secondary-electron mode.
RNA isolation, cDNA synthesis and real-time qPCR
Cells were cultured in petri dishes at a density of 80 × 104 cells for 24 h and were serum starved overnight. HA fractions were added to the cultures at a final concentration 200 μg/mL and incubated for 24 h. Cells were collected, RNA isolation was conducted using the NucleoSpin® RNA II Kit (Macherey-Nagel, Duren, Germany). In order to quantify the isolated RNA, the absorbance of each sample was measured at 260 nm. For the cDNA synthesis, the PrimeScript™ 1st strand cDNA synthesis kit perfect real time (Takara Bio Inc., Japan) was used. Real-time PCR was conducted using KAPA Taq ReadyMix DNA Polymerase (KAPA BIOSYSTEMS, Wilmington, Massachusetts). All the experiments were carried out according to the manufacturer's instructions. Real-time qPCR analysis was conducted in 20 μL reaction mixture. The amplification was performed utilizing Rotor Gene Q (Qiagen, USA). All the reactions were performed in triplicate and a standard curve was always included for each pair of primers for assay validation. In addition, a melting curve analysis was always performed for detecting the SYBR Green-based objective amplicon. To provide quantification, the point of product accumulation in the early logarithmic phase of the amplification plot was defined by assigning a fluorescence threshold above the background, defined as the threshold cycle (Ct) number. Relative expression of different gene transcripts was calculated by the ΔΔCt method. The Ct of any gene of interest was normalized to the Ct of the normalizer (GAPDH). Fold changes (arbitrary units) were determined as 2−ΔΔCt. Genes of interest and the primers that were used are presented in Table 2.
Immunofluorescence
Cells were seeded on glass cover slips inside a 12-well cell culture plate; MCF-7 at a density of 10 × 104 cells per well and MDA-MB-231 at a density of 5 × 104 cells per well. Cells were cultured in complete medium for 24 h and then serum starved overnight. The next day the HA fractions were added at a final concentration of 200 μg/mL and the cells were incubated for 24 h. Cells were washed twice with 1× PBS and fixed in 4% paraformaldehyde in 1× PBS. For E-cadherin staining, the cells were fixed in cold methanol for 5 min and cold acetone for 5 min. Then, cells were washed three times with 1× PBS-Tween 0.01% and permeabilized with 0.05% Triton X-100/PBS-Tween 0.01%. Cells were washed again with PBS-Tween 0.01% and the specimens were blocked with 5% BSA in PBS-Tween 0.01% for 1 h. Coverslips were stained with the primary antibodies in 1% BSA/PBS-Tween 0.01%, overnight at 4 °C. The appropriate secondary antibodies in 1% BSA/PBS-Tween 0.01% were used for 1 h in the dark, and the coverslips were mounted with DAPI on microscope slides. Stained slides were observed using a fluorescent phase contrast microscope (OLYMPUS CKX41, QImaging Micro Publisher 3.3RTV) through a 60× objective. The following antibodies were used: E-cadherin (Takara, ECCD-2, 1:200), Hermes-3 (1 μg/mL), SDC-4(sc-12766, 1:50), Alexa Fluor-594 or -488 anti-mouse (Biotium, PSF006, 1:2000).
Statistical analysis
Reported values are expressed as mean ± standard deviation (SD) of experiments in triplicate. Statistically significant differences were evaluated using the analysis of variance (ANOVA) test and were considered statistically significant at the level of at least p ≤ 0.05. Statistical analysis and graphs were made using GraphPad Prism 6 (GraphPad Software).
Funding information
This work was supported by the EU Horizon 2020 project RISE-2014, action No. 645756 “GLYCANC – Matrix glycans as multifunctional pathogenesis factors and therapeutic targets in cancer”. AGT was supported by General Secretariat for Research and Technology (GSRT) & Hellenic Foundation of Research & Innovation (H.F.R.I.). ZP was supported by the project ArthroMicroPerMed (MIS 5033644) which is implemented under the NSRF 2014–2020 and co-financed by Greece and the ΕU.
Author contributions
AGT performed the main experimental part and prepared the manuscript draft and the figures, ZP contributed to the manuscript writing, editing, data demonstration and supervised the experiments, CB organized, performed and supervised part of the cell culture experiments, RB designed and supervised the HA synthesis experiments, IC performed part of the experiments, VM performed the 3D cultures experiments, MO designed and supervised the 3D cultures experiments, MF performed SEM image analysis and interpretation and contributed to design and manuscript writing and editing, GD supervised part of the experiments and contributed to data demonstration and manuscript editing, NKK had the overall design and supervision of the experiments, demonstration of the data, writing, editing and submission of the manuscript. All authors reviewed the manuscript.
Declaration of Competing Interest
Carlo Barbera, Mauro Pavan and Devis Galesso were, at the time of the study, full-time employees at Fidia Farmaceutici S.p.A.
Acknowledgments
We wish to thank Dr. Spyros Skandalis (University of Patras) for kindly providing primers and antibodies, Dr. Mauro Pavan (Fidia S.p.A.) for HPLC-MS spectrometry analysis and Viscotek analysis, Mr. Gianfranco Filippini DipSA (University of Bologna) for technical assistance during samples preparation for SEM, Professor Renato Iozzo and Dr. Simone Buraschi (Thomas Jefferson University) for valuable advices and helpful discussion. Dr. Ilaria Caon was a PhD student of the “Biotechnology, Biosciences and Surgical Technology” course at Università degli studi dell'Insubria.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mbplus.2019.100008.
Appendix A. Supplementary data
Supplementary tables
References
- 1.Jo S.J., Park P., Cha H., Ahn S.G., Kim M.J., Koo J.S., Jeong J., Park J.H., Dong S.M., Lee M. Cellular inhibitor of apoptosis protein 2 promotes the epithelial- mesenchymal transition in triple-negative breast cancer cells through activation of the AKT signaling pathway. Oncotarget. 2017;8:78781–78795. doi: 10.18632/oncotarget.20227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mook S., Van'T Veer L.J., Rutgers E.J., Ravdin P.M., Van De Velde A.O., Van Leeuwen F.E., Visser O., Schmidt M.K. Independent prognostic value of screen detection in invasive breast cancer. Journal of the National Cancer Institute. 2011;103:585–597. doi: 10.1093/jnci/djr043. [DOI] [PubMed] [Google Scholar]
- 3.Björnström L., Sjöberg M. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Molecular Endocrinology. 2005;19:833–842. doi: 10.1210/me.2004-0486. [DOI] [PubMed] [Google Scholar]
- 4.Tsonis A.I., Afratis N., Gialeli C., Ellina M.-I., Piperigkou Z., Skandalis S.S., Theocharis A.D., Tzanakakis G.N., Karamanos N.K. Evaluation of the coordinated actions of estrogen receptors with epidermal growth factor receptor and insulin-like growth factor receptor in the expression of cell surface heparan sulfate proteoglycans and cell motility in breast cancer cells. The FEBS Journal. 2013;280:2248–2259. doi: 10.1111/febs.12162. [DOI] [PubMed] [Google Scholar]
- 5.Yaşar P., Ayaz G., User S.D., Güpür G., Muyan M. Molecular mechanism of estrogen–estrogen receptor signaling. Reproductive Medicine and Biology. 2017;16:4–20. doi: 10.1002/rmb2.12006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levin E.R. Cellular functions of plasma membrane estrogen receptors. Steroids. 2002;67:471–475. doi: 10.1016/S0039-128X(01)00179-9. [DOI] [PubMed] [Google Scholar]
- 7.Piperigkou Z., Franchi M., Götte M., Karamanos N.K. Estrogen receptor beta as epigenetic mediator of miR-10b and miR-145 in mammary cancer. Matrix Biology. 2017;64:94–111. doi: 10.1016/j.matbio.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Bouris P., Skandalis S.S., Piperigkou Z., Afratis N., Karamanou K., Aletras A.J., Moustakas A., Theocharis A.D., Karamanos N.K. Estrogen receptor alpha mediates epithelial to mesenchymal transition, expression of specific matrix effectors and functional properties of breast cancer cells. Matrix Biology. 2015;43:42–60. doi: 10.1016/j.matbio.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 9.Grober O.M.V., Mutarelli M., Giurato G., Ravo M., Cicatiello L., De Filippo M.R., Ferraro L., Nassa G., Papa M.F., Paris O., Tarallo R., Luo S., Schroth G.P., Benes V., Weisz A. Global analysis of estrogen receptor beta binding to breast cancer cell genome reveals an extensive interplay with estrogen receptor alpha for target gene regulation. BMC Genomics. 2011;12 doi: 10.1186/1471-2164-12-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Piperigkou Z., Bouris P., Onisto M., Franchi M., Kletsas D., Theocharis A.D., Karamanos N.K. Estrogen receptor beta modulates breast cancer cells functional properties, signaling and expression of matrix molecules. Matrix Biology. 2016;56:4–23. doi: 10.1016/j.matbio.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Gialeli C., Theocharis A.D., Karamanos N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. The FEBS Journal. 2011;278:16–27. doi: 10.1111/j.1742-4658.2010.07919.x. [DOI] [PubMed] [Google Scholar]
- 12.Sciacovelli M., Frezza C. Metabolic reprogramming and epithelial-to-mesenchymal transition in cancer. The FEBS Journal. 2017;284:3132–3144. doi: 10.1111/febs.14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Theocharis A.D., Skandalis S.S., Gialeli C., Karamanos N.K. Extracellular matrix structure. Advanced Drug Delivery Reviews. 2016;97:4–27. doi: 10.1016/j.addr.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 14.Karamanos N.K., Piperigkou Z., Theocharis A.D., Watanabe H., Franchi M., Baud S., Brézillon S., Götte M., Passi A., Vigetti D., Ricard-Blum S., Sanderson R.D., Neill T., Iozzo R.V. Proteoglycan Chemical Diversity Drives Multifunctional Cell Regulation and Therapeutics. Chemical Reviews. 2018;118(18):9152–9232. doi: 10.1021/acs.chemrev.8b00354. [DOI] [PubMed] [Google Scholar]
- 15.Iozzo R.V., Gubbiotti M.A. Extracellular matrix: the driving force of mammalian diseases. Matrix Biology. 2018;71–72:1–9. doi: 10.1016/j.matbio.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pozzi A., Yurchenco P.D., Iozzo R.V. The nature and biology of basement membranes. Matrix Biology. 2017;57–58:1–11. doi: 10.1016/j.matbio.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Necas J., Bartosikova L., Brauner P., Kolar J. Hyaluronic acid (hyaluronan): a review. Veterinární Medicína. 2008;53:397–411. doi: 10.17221/1930-VETMED. [DOI] [Google Scholar]
- 18.Oikari S., Kettunen T., Tiainen S., Häyrinen J., Masarwah A., Sudah M., Sutela A., Vanninen R., Tammi M., Auvinen P. UDP-sugar accumulation drives hyaluronan synthesis in breast cancer. Matrix Biology. 2018;67:63–74. doi: 10.1016/j.matbio.2017.12.015. [DOI] [PubMed] [Google Scholar]
- 19.Bohaumilitzky L., Huber A.-K., Stork E.M., Wengert S., Woelfl F., Boehm H. A trickster in disguise: hyaluronan's ambivalent roles in the matrix. Frontiers in Oncology. 2017;7:242. doi: 10.3389/fonc.2017.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McAtee C.O., Booth C., Elowsky C., Zhao L., Payne J., Fangman T., Caplan S., Henry M.D., Simpson M.A. Prostate tumor cell exosomes containing hyaluronidase Hyal1 stimulate prostate stromal cell motility by engagement of FAK-mediated integrin signaling. Matrix Biology. 2018;78-79:165–179. doi: 10.1016/j.matbio.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heldin P., Karousou E., Bernert B., Porsch H., Nishitsuka K., Skandalis S. Importance of hyaluronan-CD44 interactions in inflammation and tumorigenesis. Connective Tissue Research. 2008;49:215–218. doi: 10.1080/03008200802143323. [DOI] [PubMed] [Google Scholar]
- 22.Misra S., Heldin P., Hascall V.C., Karamanos N.K., Skandalis S.S., Markwald R.R., Ghatak S. Hyaluronan-CD44 interactions as potential targets for cancer therapy. The FEBS Journal. 2011;278:1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misra S., Hascall V.C., Markwald R.R., Ghatak S. Interactions between hyaluronan and its receptors (CD44, RHAMM) regulate the activities of inflammation and cancer. Frontiers in Immunology. 2015;6 doi: 10.3389/fimmu.2015.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arasu U.T., Kärnä R., Härkönen K., Oikari S., Koistinen A., Kröger H., Qu C., Lammi M.J., Rilla K. Human mesenchymal stem cells secrete hyaluronan-coated extracellular vesicles. Matrix Biology. 2017 doi: 10.1016/j.matbio.2017.05.001. [DOI] [PubMed] [Google Scholar]
- 25.Stern R., Asari A.A., Sugahara K.N. Hyaluronan fragments: an information-rich system. European Journal of Cell Biology. 2006;85:699–715. doi: 10.1016/j.ejcb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 26.Yang C., Cao M., Liu H., He Y., Xu J., Du Y., Liu Y., Wang W., Cui L., Hu J., Gao F. The high and low molecular weight forms of hyaluronan have distinct effects on CD44 clustering. The Journal of Biological Chemistry. 2012;287:43094–43107. doi: 10.1074/jbc.M112.349209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tavianatou A.G., Caon I., Franchi M., Piperigkou Z., Galesso D. Hyaluronan: molecular size-dependent signaling and biological functions in inflammation and cancer. The FEBS Journal. 2019:1–26. doi: 10.1111/febs.14777. [DOI] [PubMed] [Google Scholar]
- 28.Gao F., Yang C.X., Mo W., Liu Y.W., He Y.Q. Hyaluronan oligosaccharides are potential stimulators to angiogenesis via RHAMM mediated signal pathway in wound healing. Clinical and Investigative Medicine. 2008;31:106–116. doi: 10.25011/cim.v31i3.3467. [DOI] [PubMed] [Google Scholar]
- 29.Slevin M., Kumar S., Gaffney J. Angiogenic oligosaccharides of hyaluronan induce multiple signaling pathways affecting vascular endothelial cell mitogenic and wound healing responses. The Journal of Biological Chemistry. 2002;277:41046–41059. doi: 10.1074/jbc.M109443200. [DOI] [PubMed] [Google Scholar]
- 30.Slevin M., Krupinski J., Gaffney J., Matou S., West D., Delisser H., Savani R.C., Kumar S. Hyaluronan-mediated angiogenesis in vascular disease: uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biology. 2007;26:58–68. doi: 10.1016/j.matbio.2006.08.261. [DOI] [PubMed] [Google Scholar]
- 31.Pardue E.L., Ibrahim S., Ramamurthi A. Role of hyaluronan in angiogenesis and its utility to angiogenic tissue engineering. Organogenesis. 2008;4:203–214. doi: 10.4161/org.4.4.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hauser-Kawaguchi A., Luyt L.G., Turley E. Design of peptide mimetics to block pro-inflammatory functions of HA fragments. Matrix Biology. 2018;78-79:346–456. doi: 10.1016/j.matbio.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 33.Urakawa H., Nishida Y., Knudson W., Knudson C.B., Arai E., Kozawa E., Futamura N., Wasa J., Ishiguro N. Therapeutic potential of hyaluronan oligosaccharides for bone metastasis of breast cancer. Journal of Orthopaedic Research. 2012;30:662–672. doi: 10.1002/jor.21557. [DOI] [PubMed] [Google Scholar]
- 34.Russo R.I.C., García M.G., Alaniz L., Blanco G., Alvarez E., Hajos S.E. Hyaluronan oligosaccharides sensitize lymphoma resistant cell lines to vincristine by modulating P-glycoprotein activity and PI3K/Akt pathway. International Journal of Cancer. 2008;122:1012–1018. doi: 10.1002/ijc.23122. [DOI] [PubMed] [Google Scholar]
- 35.Lompardía S.L., Díaz M., Papademetrio D.L., Mascaró M., Pibuel M., Álvarez E., Hajos S.E. Hyaluronan oligomers sensitize chronic myeloid leukemia cell lines to the effect of imatinib. Glycobiology. 2016;26:343–352. doi: 10.1093/glycob/cwv107. [DOI] [PubMed] [Google Scholar]
- 36.Alaniz L., Rizzo M., Malvicini M., Jaunarena J., Avella D., Atorrasagasti C., Aquino J.B., Garcia M., Matar P., Silva M., Mazzolini G. Low molecular weight hyaluronan inhibits colorectal carcinoma growth by decreasing tumor cell proliferation and stimulating immune response. Cancer Letters. 2009;278:9–16. doi: 10.1016/j.canlet.2008.12.029. [DOI] [PubMed] [Google Scholar]
- 37.Slomiany M.G., Dai L., Tolliver L.B., Grass G.D., Zeng Y., Toole B.P. Inhibition of functional hyaluronan-CD44 interactions in CD133-positive primary human ovarian carcinoma cells by small hyaluronan oligosaccharides. Clinical Cancer Research. 2009;15:7593–7601. doi: 10.1158/1078-0432.CCR-09-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slomiany M.G., Dai L., Bomar P.A., Knackstedt T.J., Kranc D.A., Tolliver L., Maria B.L., Toole B.P. Abrogating drug resistance in malignant peripheral nerve sheath tumors by disrupting hyaluronan-CD44 interactions with small hyaluronan oligosaccharides. Cancer Research. 2009;69:4992–4998. doi: 10.1158/0008-5472.CAN-09-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng C., Toole B.P., Kinney S.D., Kuo J.W., Stamenkovic I. Inhibition of tumor growth in vivo by hyaluronan oligomers. International Journal of Cancer. 1998;77:396–401. doi: 10.1002/(SICI)1097-0215(19980729)77:3<396::AID-IJC15>3.0.CO;2-6. (pii) [DOI] [PubMed] [Google Scholar]
- 40.Ward J.A., Huang L., Guo H., Ghatak S., Toole B.P. Perturbation of hyaluronan interactions inhibits malignant properties of glioma cells. The American Journal of Pathology. 2003;162:1403–1409. doi: 10.1016/S0002-9440(10)64273-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tammi M.I., Oikari S., Pasonen-Seppänen S., Rilla K., Auvinen P., Tammi R.H. Activated hyaluronan metabolism in the tumor matrix — causes and consequences. Matrix Biology. 2018;78-79:147–164. doi: 10.1016/j.matbio.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 42.Ghatak S., Misra S., Toole B.P. Hyaluronan oligosaccharides inhibit anchorage-independent growth of tumor cells by suppressing the phosphoinositide 3-kinase/Akt cell survival pathway. The Journal of Biological Chemistry. 2002;277:38013–38020. doi: 10.1074/jbc.M202404200. [DOI] [PubMed] [Google Scholar]
- 43.Fieber C. Hyaluronan-oligosaccharide-induced transcription of metalloproteases. Journal of Cell Science. 2004;117:359–367. doi: 10.1242/jcs.00831. [DOI] [PubMed] [Google Scholar]
- 44.Ghatak S., Misra S., Toole B.P. Hyaluronan constitutively regulates ErbB2 phosphorylation and signaling complex formation in carcinoma cells. The Journal of Biological Chemistry. 2005;280:8875–8883. doi: 10.1074/jbc.M410882200. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Y. fang, Qiao S. pei, Shi S. liang, Yao L. fen, Hou X. lu, Li C. feng, Lin F.H., Guo K., Acharya A., Chen X. biao, Nie Y., Tian W. ming. Modulating three-dimensional microenvironment with hyaluronan of different molecular weights alters breast cancer cell invasion behavior. ACS Applied Materials and Interfaces. 2017;9:9327–9338. doi: 10.1021/acsami.6b15187. [DOI] [PubMed] [Google Scholar]
- 46.Tolg C., Yuan H., Flynn S.M., Basu K., Ma J., Tse K.C.K., Kowalska B., Vulkanesku D., Cowman M.K., McCarthy J.B., Turley E.A. Hyaluronan modulates growth factor induced mammary gland branching in a size dependent manner. Matrix Biology. 2017;63:117–132. doi: 10.1016/j.matbio.2017.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Toole B.P., Ghatak S., Misra S. Hyaluronan oligosaccharides as a potential anticancer therapeutic. Current Pharmaceutical Biotechnology. 2008;9:249–252. doi: 10.2174/138920108785161569. [DOI] [PubMed] [Google Scholar]
- 48.Machesky L.M. Lamellipodia and filopodia in metastasis and invasion. FEBS Letters. 2008;582:2102–2111. doi: 10.1016/j.febslet.2008.03.039. [DOI] [PubMed] [Google Scholar]
- 49.Jacquemet G., Hamidi H., Ivaska J. Filopodia in cell adhesion, 3D migration and cancer cell invasion. Current Opinion in Cell Biology. 2015;36:23–31. doi: 10.1016/j.ceb.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 50.Medici D., Hay E.D., Olsen B.R. Snail and slug promote epithelial-mesenchymal transition through β-catenin–T-cell factor-4-dependent expression of transforming growth factor-β3. Molecular Biology of the Cell. 2008;19:4875–4887. doi: 10.1091/mbc.e08-05-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vanneste M., Hanoux V., Bouakka M., Bonnamy P.J. Hyaluronate synthase-2 overexpression alters estrogen dependence and induces histone deacetylase inhibitor-like effects on ER-driven genes in MCF7 breast tumor cells. Molecular and Cellular Endocrinology. 2017;444:48–58. doi: 10.1016/j.mce.2017.01.046. [DOI] [PubMed] [Google Scholar]
- 52.Preca B., Bajdak K., Mock K., Lehmann W., Sundararajan V., Bronsert P., Matzge-ogi A. Full-Text. 2017;8:11530–11543. doi: 10.18632/oncotarget.14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li P., Xiang T., Li H., Li Q., Yang B., Huang J., Zhang X., Shi Y., Tan J., Ren G. Hyaluronan synthase 2 overexpression is correlated with the tumorigenesis and metastasis of human breast cancer. International Journal of Clinical and Experimental Pathology. 2015;8:12101–12114. [PMC free article] [PubMed] [Google Scholar]
- 54.Okuda H., Kobayashi A., Xia B., Watabe M., Pai S.K., Hirota S., Xing F., Liu W., Pandey P.R., Fukuda K., Modur V., Ghosh A., Wilber A., Watabe K. Hyaluronan synthase HAS2 promotes tumor progression in bone by stimulating the interaction of breast cancer stem-like cells with macrophages and stromal cells. Cancer Research. 2012;72:537–547. doi: 10.1158/0008-5472.CAN-11-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernert B., Porsch H., Heldin P. Hyaluronan synthase 2 (HAS2) promotes breast cancer cell invasion by suppression of tissue metalloproteinase inhibitor 1 (TIMP-1) The Journal of Biological Chemistry. 2011;286:42349–42359. doi: 10.1074/jbc.M111.278598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zoltan-Jones A., Huang L., Ghatak S., Toole B.P. Elevated hyaluronan production induces mesenchymal and transformed properties in epithelial cells. The Journal of Biological Chemistry. 2003;278:45801–45810. doi: 10.1074/jbc.M308168200. [DOI] [PubMed] [Google Scholar]
- 57.Kolliopoulos C., Lin C.Y., Heldin C.H., Moustakas A., Heldin P. Has2 natural antisense RNA and Hmga2 promote Has2 expression during TGFβ-induced EMT in breast cancer. Matrix Biology. 2018 doi: 10.1016/j.matbio.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 58.Heldin P., Lin C.Y., Kolliopoulos C., Chen Y.H., Skandalis S.S. Regulation of hyaluronan biosynthesis and clinical impact of excessive hyaluronan production. Matrix Biology. 2018 doi: 10.1016/j.matbio.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 59.Li Y., Li L., Brown T.J., Heldin P. Silencing of hyaluronan synthase 2 suppresses the malignant phenotype of invasive breast cancer cells. International Journal of Cancer. 2007;120:2557–2567. doi: 10.1002/ijc.22550. [DOI] [PubMed] [Google Scholar]
- 60.Karalis T.T., Heldin P., Vynios D.H., Neill T., Buraschi S., Iozzo R.V., Karamanos N.K., Skandalis S.S. Tumor-suppressive functions of 4-MU on breast cancer cells of different ER status: regulation of hyaluronan/HAS2/CD44 and specific matrix effectors. Matrix Biology. 2018;78-79:118–138. doi: 10.1016/j.matbio.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 61.Udabage L., Brownlee G.R., Waltham M., Blick T., Walker E.C., Heldin P., Nilsson S.K., Thompson E.W., Brown T.J. Antisense-mediated suppression of hyaluronan synthase 2 inhibits the tumorigenesis and progression of breast cancer. Cancer Research. 2005;65:6139–6150. doi: 10.1158/0008-5472.CAN-04-1622. doi:65/14/6139 [pii] [DOI] [PubMed] [Google Scholar]
- 62.Udabage L., Brownlee G.R., Nilsson S.K., Brown T.J. The over-expression of HAS2, Hyal-2 and CD44 is implicated in the invasiveness of breast cancer. Experimental Cell Research. 2005;310:205–217. doi: 10.1016/j.yexcr.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 63.Heldin P., Basu K., Olofsson B., Porsch H., Kozlova I., Kahata K. Deregulation of hyaluronan synthesis, degradation and binding promotes breast cancer. Journal of Biochemistry. 2013;154:395–408. doi: 10.1093/jb/mvt085. [DOI] [PubMed] [Google Scholar]
- 64.Hamilton S.R., Fard S.F., Paiwand F.F., Tolg C., Veiseh M., Wang C., McCarthy J.B., Bissell M.J., Koropatnick J., Turley E.A. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. The Journal of Biological Chemistry. 2007;282:16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bourguignon L.Y.W., Wong G., Earle C., Krueger K., Spevak C.C. Hyaluronan-CD44 interaction promotes c-Src-mediated twist signaling, microRNA-10b expression, and RhoA/RhoC up-regulation, leading to rho-kinase-associated cytoskeleton activation and breast tumor cell invasion. The Journal of Biological Chemistry. 2010;285:36721–36735. doi: 10.1074/jbc.M110.162305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bourguignon L.Y.W., Spevak C.C., Wong G., Xia W., Gilad E. Hyaluronan-CD44 interaction with protein kinase Cε promotes oncogenic signaling by the stem cell marker nanog and the production of microRNA-21, leading to down-regulation of the tumor suppressor protein PDCD4, anti-apoptosis, and chemotherapy resistance. The Journal of Biological Chemistry. 2009;284:26533–26546. doi: 10.1074/jbc.M109.027466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hiscox S., Baruha B., Smith C., Bellerby R., Goddard L., Jordan N., Poghosyan Z., Nicholson R.I., Barrett-Lee P., Gee J. Overexpression of CD44 accompanies acquired tamoxifen resistance in MCF7 cells and augments their sensitivity to the stromal factors, heregulin and hyaluronan. BMC Cancer. 2012;12:1. doi: 10.1186/1471-2407-12-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peterson R.M., Yu Q., Stamenkovic I., Toole B.P. Perturbation of hyaluronan interactions by soluble CD44 inhibits growth of murine mammary carcinoma cells in ascites. The American Journal of Pathology. 2000;156:2159–2167. doi: 10.1016/S0002-9440(10)65086-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Toole B.P. Hyaluronan-CD44 interactions in cancer: paradoxes and possibilities. Clinical Cancer Research. 2009;15:7462–7468. doi: 10.1158/1078-0432.CCR-09-0479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang K., Wu F., Seo B.R., Fischbach C., Chen W., Hsu L., Gourdon D. HHS Public Access. 2018:86–95. doi: 10.1016/j.matbio.2016.08.001.Breast. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Benson C.S., Babu S.D., Radhakrishna S., Selvamurugan N., Sankar B.R. Expression of matrix metalloproteinases in human breast cancer tissues. Disease Markers. 2013;34:395–405. doi: 10.3233/DMA-130986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Figueira R.C.S., Gomes L.R., Neto J.S., Silva F.C., Silva I.D.C.G., Sogayar M.C. Correlation between MMPs and their inhibitors in breast cancer tumor tissue specimens and in cell lines with different metastatic potential. BMC Cancer. 2009;9:1–11. doi: 10.1186/1471-2407-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen W., Zhou S., Mao L., Zhang H., Sun D., Zhang J., Li J., Tang J. hai. Crosstalk between TGF-β signaling and miRNAs in breast cancer metastasis. Tumor Biology. 2016;37:10011–10019. doi: 10.1007/s13277-016-5060-8. [DOI] [PubMed] [Google Scholar]
- 74.Montgomery N., Hill A., McFarlane S., Neisen J., O'Grady A., Conlon S., Jirstrom K., Kay E.W., Waugh D.J.J. CD44 enhances invasion of basal-like breast cancer cells by upregulating serine protease and collagen-degrading enzymatic expression and activity. Breast Cancer Research. 2012;14:R84. doi: 10.1186/bcr3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bourguignon L.Y.W. CD44-mediated oncogenic signaling and cytoskeleton activation during mammary tumor progression. Journal of Mammary Gland Biology and Neoplasia. 2001;6:287–297. doi: 10.1023/A:1011371523994. [DOI] [PubMed] [Google Scholar]
- 76.Holst-Hansen C., Johannessen B., Høyer-Hansen G., Rømer J., Ellis V., Brünner N. Urokinase-type plasminogen activation in three human breast cancer cell lines correlates with their in vitro invasiveness. Clinical & Experimental Metastasis. 1996;14:297–307. doi: 10.1007/BF00053903. http://www.ncbi.nlm.nih.gov/pubmed/8674284 [DOI] [PubMed] [Google Scholar]
- 77.Lang D.S., Marwitz S., Heilenkötter U., Schumm W., Behrens O., Simon R., Reck M., Vollmer E., Goldmann T. Transforming growth factor-beta signaling leads to uPA/PAI-1 activation and metastasis: a study on human breast cancer tissues. Pathology Oncology Research. 2014;20:727–732. doi: 10.1007/s12253-014-9753-2. [DOI] [PubMed] [Google Scholar]
- 78.Wolff C., Malinowsky K., Berg D., Schragner K., Schuster T., Walch A., Bronger H., Höfler H., Becker K.-F. Signalling networks associated with urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in breast cancer tissues: new insights from protein microarray analysis. The Journal of Pathology. 2011;223:54–63. doi: 10.1002/path.2791. [DOI] [PubMed] [Google Scholar]
- 79.Vicente C.M., Aparecida D., Sartorio P.V., Silva T.D., Saad S.S., Nader H.B., Forones N.M., Toma L. Heparan sulfate proteoglycans in human colorectal cancer. Analytical Cellular Pathology (Amsterdam) 2018;2018 doi: 10.1155/2018/8389595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barbouri D., Afratis N., Gialeli C., Vynios D.H., Theocharis A.D., Karamanos N.K. Syndecans as modulators and potential pharmacological targets in cancer progression. Frontiers in Oncology. 2014;4:1–11. doi: 10.3389/fonc.2014.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lim C.C., Multhaupt H.A.B., Couchman J.R. Cell surface heparan sulfate proteoglycans control adhesion and invasion of breast carcinoma cells. Molecular Cancer. 2015;14:1–18. doi: 10.1186/s12943-014-0279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Woods A., Couchman J.R. Syndecan 4 heparan sulfate proteoglycan is a selectively enriched and widespread focal adhesion component. Molecular Biology of the Cell. 1994;5:183–192. doi: 10.1091/mbc.5.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Afratis N.A., Bouris P., Skandalis S.S., Multhaupt H.A., Couchman J.R., Theocharis A.D., Karamanos N.K. IGF-IR cooperates with ERα to inhibit breast cancer cell aggressiveness by regulating the expression and localisation of ECM molecules. Scientific Reports. 2017;7:1–12. doi: 10.1038/srep40138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Purushothaman A., Hurst D.R., Pisano C., Mizumoto S., Sugahara K., Sanderson R.D. Heparanase-mediated loss of nuclear syndecan-1 enhances histone acetyltransferase (HAT) activity to promote expression of genes that drive an aggressive tumor phenotype. The Journal of Biological Chemistry. 2011;286:30377–30383. doi: 10.1074/jbc.M111.254789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kelly T., Suva L.J., Huang Y., MacLeod V., Miao H.Q., Walker R.C., Sanderson R.D. Expression of heparanase by primary breast tumors promotes bone resorption in the absence of detectable bone metastases. Cancer Research. 2005;65:5778–5784. doi: 10.1158/0008-5472.CAN-05-0749. [DOI] [PubMed] [Google Scholar]
- 86.Reiland J., Sanderson R.D., Waguespack M., Barker S.A., Long R., Carson D.D., Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: functional implications for tumor cell invasion. The Journal of Biological Chemistry. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- 87.Theocharis A.D., Karamanos N.K. Proteoglycans remodeling in cancer: underlying molecular mechanisms. Matrix Biology. 2017;75-76:220–259. doi: 10.1016/j.matbio.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 88.Fuchs K., Hippe A., Schmaus A., Homey B., Sleeman J.P., Orian-Rousseau V. Opposing effects of high-and low-molecular weight hyaluronan on CXCL12-induced CXCR4 signaling depend on CD44. Cell Death & Disease. 2013;4:e819-11. doi: 10.1038/cddis.2013.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kothapalli D., Flowers J., Xu T., Puré E., Assoian R.K. Differential activation of ERK and Rac mediates the proliferative and anti-proliferative effects of hyaluronan and CD44. The Journal of Biological Chemistry. 2008;283:31823–31829. doi: 10.1074/jbc.M802934200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bourguignon L.Y.W., Peyrollier K., Xia W., Gilad E. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. The Journal of Biological Chemistry. 2008;283:17635–17651. doi: 10.1074/jbc.M800109200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herrera-Gayol A., Jothy S. Effects of hyaluronan on the invasive properties of human breast cancer cells in vitro. International Journal of Experimental Pathology. 2001;82:193–200. doi: 10.1046/j.1365-2613.2001.00196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McFarlane S., Coulter J.A., Tibbits P., O'Grady A., McFarlane C., Montgomery N., Hill A., McCarthy H.O., Young L.S., Kay E.W., Isacke C.M., Waugh D.J.J. CD44 increases the efficiency of distant metastasis of breast cancer. Oncotarget. 2015;6:11465–11476. doi: 10.18632/oncotarget.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tzircotis G. Chemotaxis towards hyaluronan is dependent on CD44 expression and modulated by cell type variation in CD44-hyaluronan binding. Journal of Cell Science. 2005;118:5119–5128. doi: 10.1242/jcs.02629. [DOI] [PubMed] [Google Scholar]
- 94.Misra S., Obeid L.M., Hannun Y.A., Minamisawa S., Berger F.G., Markwald R.R., Toole B.P., Ghatak S. Hyaluronan constitutively regulates activation of COX-2-mediated cell survival activity in intestinal epithelial and colon carcinoma cells. The Journal of Biological Chemistry. 2008;283:14335–14344. doi: 10.1074/jbc.M703811200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Toole B.P., Wight T.N., Tammi M.I. Hyaluronan-cell interactions in cancer and vascular disease. The Journal of Biological Chemistry. 2002;277:4593–4596. doi: 10.1074/jbc.R100039200. [DOI] [PubMed] [Google Scholar]
- 96.Toole B.P., Slomiany M.G. Hyaluronan: a constitutive regulator of chemoresistance and malignancy in cancer cells. Seminars in Cancer Biology. 2008;18:244–250. doi: 10.1016/j.semcancer.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hosono K., Nishida Y., Knudson W., Knudson C.B., Naruse T., Suzuki Y., Ishiguro N. Hyaluronan oligosaccharides inhibit tumorigenicity of osteosarcoma cell lines MG-63 and LM-8 in vitro and in vivo via perturbation of hyaluronan-rich pericellular matrix of the cells. The American Journal of Pathology. 2007;171:274–286. doi: 10.2353/ajpath.2007.060828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Theocharis A.D., Skandalis S.S., Neill T., Multhaupt H.A.B., Hubo M., Frey H., Gopal S., Gomes A., Afratis N., Lim H.C., Couchman J.R., Filmus J., Ralph S., Schaefer L., Iozzo R.V., Karamanos N.K. Insights into the key roles of proteoglycans in breast cancer biology and translational medicine. Biochimica et Biophysica Acta, Reviews on Cancer. 2015;1855:276–300. doi: 10.1016/j.bbcan.2015.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rodríguez Rodríguez A.C., Francisco Ortega M.I. Estudio de los picos tallados de la época preeuropea de Gran Canaria. Un ejemplo de especialización en el trabajo a partir de las evidencias recuperadas en la cantera de molinos de Montaña Quemada. Complutum. 2012;23:77–97. doi: 10.5209/rev-CMPL.2012.v23.n1.39532. [DOI] [Google Scholar]
- 100.Malavaki C.J., Roussidis A.E., Gialeli C., Kletsas D., Tsegenidis T., Theocharis A.D., Tzanakakis G.N., Karamanos N.K. Imatinib as a key inhibitor of the platelet-derived growth factor receptor mediated expression of cell surface heparan sulfate proteoglycans and functional properties of breast cancer cells. The FEBS Journal. 2013 doi: 10.1111/febs.12163. [DOI] [PubMed] [Google Scholar]
- 101.Huang J., Ang L., Liu M.Q., Hu H.G., Wang J., Zou Q., Zhao Y., Zheng L., Zhao M., Wu Z.S. Serum and tissue expression of gelatinase and twist in breast cancer. European Review for Medical and Pharmacological Sciences. 2014;18:2662–2669. [PubMed] [Google Scholar]
- 102.Fernandez-Garcia B., Eiró N., Marín L., González-Reyes S., González L.O., Lamelas M.L., Vizoso F.J. Expression and prognostic significance of fibronectin and matrix metalloproteases in breast cancer metastasis. Histopathology. 2014;64:512–522. doi: 10.1111/his.12300. [DOI] [PubMed] [Google Scholar]
- 103.Radisky E.S., Radisky D.C. Matrix metalloproteinases as breast cancer drivers and therapeutic targets. Frontiers in Bioscience (Landmark Edition) 2015;20:1144–1163. doi: 10.1016/j.ygyno.2014.12.035.Pharmacologic. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Merdad A., Karim S., Schulten H.-J., Dallol A., Buhmeida A., Al-Thubaity F., Gari M.A., Chaudhary A.G., Abuzenadah A.M., Al-Qahtani M.H. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer: MMP-9 as a potential biomarker for cancer invasion and metastasis. Anticancer Research. 2014;34:1355–1366. http://ar.iiarjournals.org/content/34/3/1355.long [PubMed] [Google Scholar]
- 105.Kim G.E., Lee J.S., Choi Y.D., Lee K.H., Lee J.H., Nam J.H., Choi C., Kim S.S., Park M.H., Yoon J.H., Kweon S.S. Expression of matrix metalloproteinases and their inhibitors in different immunohistochemical-based molecular subtypes of breast cancer. BMC Cancer. 2014;14:959. doi: 10.1186/1471-2407-14-959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Heo D.S., Choi H., Yeom M.Y., Song B.J., Oh S.J. Serum levels of matrix metalloproteinase-9 predict lymph node metastasis in breast cancer patients. Oncology Reports. 2014;31:1567–1572. doi: 10.3892/or.2014.3001. [DOI] [PubMed] [Google Scholar]
- 107.Ren F., Tang R., Zhang X., Madushi W.M., Luo D., Dang Y., Li Z., Wei K., Chen G., Ahmad A. Overexpression of MMP family members functions as prognostic biomarker for breast cancer patients: a systematic review and meta-analysis. PLoS One. 2015;10:1–18. doi: 10.1371/journal.pone.0135544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kuo Y.C., Su C.H., Liu C.Y., Chen T.H., Chen C.P., Wang H.S. Transforming growth factor-?? Induces CD44 cleavage that promotes migration of MDA-MB-435s cells through the up-regulation of membrane type 1-matrix metalloproteinase. International Journal of Cancer. 2009;124:2568–2576. doi: 10.1002/ijc.24263. [DOI] [PubMed] [Google Scholar]
- 109.Köhrmann A., Kammerer U., Kapp M., Dietl J., Anacker J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: new findings and review of the literature. BMC Cancer. 2009;9:1–20. doi: 10.1186/1471-2407-9-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bourguignon L.Y.W., Gunja-smith Z., Iida N., Zhu H.B., Young L.J.T., Muller W.J., Cardiff R.D. Cytoskeleton-mediated tumor cell migration and matrix metalloproteinase (MMP-9) association in metastatic breast cancer cells. Journal of Cellular Physiology. 1998;215:206–215. doi: 10.1002/(SICI)1097-4652(199807)176:1<206::AID-JCP22>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 111.Montgomery N., Hill A., McFarlane S., Neisen J., O'Grady A., Conlon S., Jirstrom K., Kay E.W., Waugh D. CD44 enhances invasion of basal-like breast cancer cells by up-regulating serine protease and collagen-degrading enzymatic expression and activity. Breast Cancer Research. 2012:1–19. doi: 10.1186/bcr3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fan S.H., Wang Y.Y., Lu J., Zheng Y.L., Wu D.M., Zhang Z.F., Shan Q., Hu B., Li M.Q., Cheng W. CERS2 suppresses tumor cell invasion and is associated with decreased V-ATPase and MMP-2/MMP-9 activities in breast cancer. Journal of Cellular Biochemistry. 2015;116:502–513. doi: 10.1002/jcb.24978. [DOI] [PubMed] [Google Scholar]
- 113.Li J., Qiu D.-M., Chen S.-H., Cao S.-P., Xia X.-L. Suppression of human breast cancer cell metastasis by coptisine in vitro. Asian Pacific Journal of Cancer Prevention. 2014;15:5747–5751. doi: 10.7314/APJCP.2014.15.14.5747. [DOI] [PubMed] [Google Scholar]
- 114.Li J., Zhang J., Wang Y., Liang X., Wusiman Z., Yin Y., Shen Q. Synergistic inhibition of migration and invasion of breast cancer cells by dual docetaxel/quercetin-loaded nanoparticles via Akt/MMP-9 pathway. International Journal of Pharmaceutics. 2017;523:300–309. doi: 10.1016/j.ijpharm.2017.03.040. [DOI] [PubMed] [Google Scholar]
- 115.Latocha M., Płonka J., Kusmierz D., Jurzak M., Polaniak R., Nowosad A. Transcripional activity of genes encoding mmps and timps in breast cancer cells treated by genistein and in normal cancer-associated fibroblasts — in vitro studies. Acta Poloniae Pharmaceutica. Drug Research. 2014;71:1095–1102. [PubMed] [Google Scholar]
- 116.Lv Y., Zhao X., Zhu L., Li S., Xiao Q., He W., Yin L. Targeting intracellular MMPs efficiently inhibits tumor metastasis and angiogenesis. Theranostics. 2018;8:2830–2845. doi: 10.7150/thno.23209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gialeli C., Viola M., Barbouri D., Kletsas D., Passi A., Karamanos N.K. Dynamic interplay between breast cancer cells and normal endothelium mediates the expression of matrix macromolecules, proteasome activity and functional properties of endothelial cells. Biochimica et Biophysica Acta - General Subjects. 2014;1840:2549–2559. doi: 10.1016/j.bbagen.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 118.Ingvarsen S., Madsen D.H., Hillig T., Lund L.R., Holmbeck K., Behrendt N., Engelholm L.H. Dimerization of endogenous MT1-MMP is a regulatory step in the activation of the 72-kDa gelatinase MMP-2 on fibroblasts and fibrosarcoma cells. Biological Chemistry. 2008;389:943–953. doi: 10.1515/BC.2008.097. [DOI] [PubMed] [Google Scholar]
- 119.Li Z., Takino T., Endo Y., Sato H. Activation of MMP-9 by membrane type-1 MMP/MMP-2 axis stimulates tumor metastasis. Cancer Science. 2017;108:347–353. doi: 10.1111/cas.13134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kazes I., Elalamy I., Sraer J.D., Hatmi M., Nguyen G. Platelet release of trimolecular complex components MT1-MMP/TIMP2/MMP2: involvement in MMP2 activation and platelet aggregation. Blood. 2000;96:3064–3069. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=11049985&retmode=ref&cmd=prlinks%5Cnpapers2://publication/uuid/44FE6805-2C07-40BC-A29F-32400C673BD8 [PubMed] [Google Scholar]
- 121.Kim H.J., Il Park C., Park B.W., De Lee H., Jung W.H. Expression of MT-1 MMP, MMP2, MMP9 and TIMP2 mRNAs in ductal carcinoma in situ and invasive ductal carcinoma of the breast. Yonsei Medical Journal. 2006;47:333–342. doi: 10.3349/ymj.2006.47.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Thiery J.P. Epithelial–mesenchymal transitions in tumour progression. Nature Reviews. Cancer. 2002;2:442–454. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 123.Gritsenko P.G., Ilina O., Friedl P. Interstitial guidance of cancer invasion. The Journal of Pathology. 2012;226:185–199. doi: 10.1002/path.3031. [DOI] [PubMed] [Google Scholar]
- 124.Auvinen P., Rilla K., Tumelius R., Tammi M., Sironen R., Soini Y., Kosma V.M., Mannermaa A., Viikari J., Tammi R. Hyaluronan synthases (HAS1-3) in stromal and malignant cells correlate with breast cancer grade and predict patient survival. Breast Cancer Research and Treatment. 2014;143:277–286. doi: 10.1007/s10549-013-2804-7. [DOI] [PubMed] [Google Scholar]
- 125.Vigetti D., Viola M., Karousou E., De Luca G., Passi A. Metabolic control of hyaluronan synthases. Matrix Biology. 2014 doi: 10.1016/j.matbio.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 126.Hanagiri T., Shinohara S., Takenaka M., Shigematsu Y., Yasuda M., Shimokawa H., Nagata Y., Nakagawa M., Uramoto H., So T., Tanaka F. Effects of hyaluronic acid and CD44 interaction on the proliferation and invasiveness of malignant pleural mesothelioma. Tumour Biology. 2012;33:2135–2141. doi: 10.1007/s13277-012-0473-5. [DOI] [PubMed] [Google Scholar]
- 127.Draffin J.E., McFarlane S., Hill A., Johnston P.G., Waugh D.J. CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Research. 2004;64:5702–5711. doi: 10.1158/0008-5472.CAN-04-0389\r64/16/5702. (pii) [DOI] [PubMed] [Google Scholar]
- 128.Fang X.J., Jiang H., Zhao X.P., Jiang W.M. The role of a new CD44st in increasing the invasion capability of the human breast cancer cell line MCF-7. BMC Cancer. 2011;11:290. doi: 10.1186/1471-2407-11-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Smith S.M., Lyu Y.L., Cai L. NF-κB affects proliferation and invasiveness of breast cancer cells by regulating CD44 expression. PLoS One. 2014;9 doi: 10.1371/journal.pone.0106966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zhang C., Xu Y., Hao Q., Wang S., Li H., Li J., Gao Y., Li M., Li W., Xue X., Wu S., Zhang Y., Zhang W. FOXP3 suppresses breast cancer metastasis through downregulation of CD44. International Journal of Cancer. 2015;137:1279–1290. doi: 10.1002/ijc.29482. [DOI] [PubMed] [Google Scholar]
- 131.Fang X.J., Jiang H., Zhu Y.Q., Zhang L.Y., Fan Q.H., Tian Y. Doxorubicin induces drug resistance and expression of the novel CD44st via NF-κB in human breast cancer MCF-7 cells. Oncology Reports. 2014;31:2735–2742. doi: 10.3892/or.2014.3131. [DOI] [PubMed] [Google Scholar]
- 132.Andreasen P.A., Kjoller L., Christensen L., Duffy M.J. The urokinase-type plasminogen activator system in cancer metastasis: a review. International Journal of Cancer. 1997;72:1–22. doi: 10.1002/(SICI)1097-0215(19970703)72:1<1::AID-IJC1>3.0.CO;2-Z. (pii) [DOI] [PubMed] [Google Scholar]
- 133.Dutta S., Bandyopadhyay C., Bottero V., Veettil M.V., Wilson L., Pins M.R., Johnson K.E., Warshall C., Chandran B. Angiogenin interacts with the plasminogen activation system at the cell surface of breast cancer cells to regulate plasmin formation and cell migration. Molecular Oncology. 2014;8:483–507. doi: 10.1016/j.molonc.2013.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Afratis N., Gialeli C., Nikitovic D., Tsegenidis T., Karousou E., Theocharis A.D., Pavão M.S., Tzanakakis G.N., Karamanos N.K. Glycosaminoglycans: key players in cancer cell biology and treatment. The FEBS Journal. 2012;279:1177–1197. doi: 10.1111/j.1742-4658.2012.08529.x. [DOI] [PubMed] [Google Scholar]
- 135.Feoktistova M., Geserick P., Leverkus M. Crystal violet assay for determining viability of cultured cells. Cold Spring Harbor Protocols. 2016 doi: 10.1101/pdb.prot087379. [DOI] [PubMed] [Google Scholar]
- 136.De Wever O., Hendrix A., De Boeck A., Westbroek W., Braems G., Emami S., Sabbah M., Gespach C., Bracke M. Modeling and quantification of cancer cell invasion through collagen type I matrices. The International Journal of Developmental Biology. 2010;54:887–896. doi: 10.1387/ijdb.092948ow. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables