Abstract
Mesenchymal stem cells (MSCs) are highly responsive to cues in the microenvironment (niche) that must be recapitulated ex vivo to study their authentic behavior. In this study, we hypothesized that native bone marrow (BM)- and adipose (AD)-derived extracellular matrices (ECM) were unique in their ability to control MSC behavior. To test this, we compared proliferation and differentiation of bone marrow (BM)-derived MSCs when maintained on native decellularized ECM produced by BM versus AD stromal cells (i.e. BM- versus AD-ECM).
We found that both ECMs contained similar types of collagens but differed in the relative abundance of each. Type VI collagen was the most abundant (≈60% of the total collagen present), while type I was the next most abundant at ≈30%. These two types of collagen were found in nearly equal proportions in both ECMs. In contrast, type XII collagen was almost exclusively found in AD-ECM, while types IV and V were only found in BM-ECM. Physically and mechanically, BM-ECM was rougher and stiffer, but less adhesive, than AD-ECM.
During 14 days in culture, both ECMs supported BM-MSC proliferation better than tissue culture plastic (TCP), although MSC-related surface marker expression remained relatively high on all three culture surfaces. BM-MSCs cultured in osteogenic (OS) differentiation media on BM-ECM displayed a significant increase in calcium deposition in the matrix, indicative of osteogenesis, while BM-MSCs cultured on AD-ECM in the presence of adipogenic (AP) differentiation media showed a significant increase in Oil Red O staining, indicative of adipogenesis. Further, culture on BM-ECM significantly increased BM-MSC-responsiveness to rhBMP-2 (an osteogenic inducer), while culture on AD-ECM enhanced responsiveness to rosiglitazone (an adipogenic inducer).
These findings support our hypothesis and indicate that BM- and AD-ECMs retain unique elements, characteristic of their tissue-specific microenvironment (niche), which promote retention of MSC differentiation state (i.e. “stemness”) during expansion and direct cell response to lineage-specific inducers. This study provides a new paradigm for precisely controlling MSC fate to a desired cell lineage for tissue-specific cell-based therapies.
Keywords: Cell culture, Mesenchymal stem cells, Native extracellular matrix, Tissue-specific, Microenvironment, Differentiation
Highlights
-
•
Tissue culture plastic is an inadequate surface for studying cell behavior in vitro.
-
•
Cell-derived ECMs have been shown to recapitulate the stem cell microenvironment.
-
•
These ECMs direct MSC response to lineage-specific induction media and factors.
-
•
This novel culture system facilitates modeling of physiologic cell behavior in vitro.
Introduction
The use of human mesenchymal stem cells (MSCs) for tissue engineering and cell-based therapeutic applications requires the expansion of large numbers of multipotent cells whose differentiation along a specific cell lineage can be precisely controlled. To obtain sufficient numbers of cells, conventional expansion methods require multiple, long-term subcultures, during which MSCs often enter premature senescence or undergo spontaneous differentiation to undesired cell lineages [[1], [2], [3]]. In order to retain the phenotype and differentiation potential of MSCs, they must be expanded in a culture environment which mimics the native MSC niche to control two fundamental aspects of stem cell-fate: self-renewal and differentiation potential [[4], [5], [6]].
Stem cell fate is regulated by both the intrinsic properties of the cells and the extrinsic cues contained within the stem cell niche. A major component of this niche (or microenvironment) is the extracellular matrix (ECM) which plays a multitude of roles that affect the resident stem cell population, such as modulation of growth factor activity, mediation of mechano-transduction effects, and cell-to-cell interactions. In particular, the ECM anchors and organizes the cells and provides both mechanical and physical cues, as well as a reservoir of structural/matricellular biomolecules and soluble factors. A number of groups have reported the results of studies where an individual component or property of the ECM (biomolecular composition, architecture, mechanical and electrostatic properties) has been experimentally modeled to gain insight into the cell-ECM interactions occurring in the niche [[7], [8], [9], [10]]. These studies have clearly demonstrated the importance of individual biochemical, architectural and mechanical properties in the ECM, but the appropriate combinations of intrinsic and extrinsic cues required to recapitulate the native stem cell niche ex vivo and effectively direct cell behavior, remain to be defined.
The current study extends our earlier observations which showed that BM- and AD-ECMs constitute unique tissue-specific microenvironments, containing different protein compositions, architectures, and mechanical properties, that influence the behavior of MSCs [4,11]. Here, we test the hypothesis that the ECM, produced by BM- and AD-derived stromal cells, is capable of maintaining MSC differentiation state (“stemness”) during expansion and directing MSC differentiation to the lineage of the stromal cells that synthesized the ECM.
Results
ECM produced by stromal cells from bone marrow and adipose tissues displayed differences in relative matrix protein abundance, topography, and physical properties
Stromal cells from bone marrow and adipose tissues were used to prepare BM- and AD-ECM, respectively, and their protein composition and physical/architectural properties compared. Proteomic analysis showed that the two ECMs shared 50% of their protein components (Fig. 1A). Based on protein classification, collagens and glycoproteins represented the two main protein classes found, followed by proteoglycans and ECM-associated components.
Fig. 1.
Proteomic analysis of BM- and AD-ECM by mass spectrometry. (A) Number of shared (common) or unique proteins found in BM- and AD-ECM by proteomic analysis. (B) Proteins common to both BM- and AD-ECM were identified and relative abundance determined. The data shown in the tables have been segregated into protein classes/types and represent the average of separate analyses of ECMs produced by 4 donors (±95% confidence interval). For each ECM, the average amount of each protein (i.e. number of spectra) was calculated as a percent relative to the total number of spectra identified in each donor's ECM. (C) Proteins unique to either BM- or AD-ECM were identified and relative abundance determined. The data shown in the tables were calculated as described in (B).
Of the collagens present, type VI collagen was the most abundant, followed by type I collagen (Figs. 1B, 1C, and 2A). While type VI collagen was present in nearly equivalent quantities in both ECMs and comprised approximately 60% of the total collagen content, type I collagen represented about 30% of the collagen content in BM-ECM and 36% in AD-ECM (Fig. 2A). In contrast, type XII collagen accounted for almost 12% of the collagen content in AD-ECM, but was entirely absent in BM-ECM (Figs. 1C and 2A), while types IV and V were unique to BM-ECM. Type I and XII collagens, in particular, are known to affect fibril orientation and tissue mechanics [12] and may, in part, account for the differences observed in structure and function of the two matrices.
Fig. 2.
Characterization of the biochemical and physical properties of the BM- and AD-ECMs using proteomic analysis and atomic force microscopy. (A) Relative abundance of types I, IV, V, VI, and XII collagen in BM- and AD-ECM based on mass spectrometric analyses. (B) Representative atomic force micrographs showing differences in topography, adhesion, and stiffness of the BM- and AD-ECMs. Each image represents a 40 × 40 μm region of the matrix. (C) Adhesion and stiffness (elastic modulus) of BM- and AD-ECM as measured by atomic force microscopy. Each data point represents one randomly selected independent measurement. The error bars represent the 95% confidence interval; the box and diamond represent the median of the samples.
In the second most abundant class of proteins, fibronectin (FN1) was the most abundant glycoprotein in both ECMs, but tenascin C (TNC), transforming growth factor-beta-induced protein (TGFBI), and thrombospondin-1 (THBS1) were also found in both ECMs but at much lower levels and in slightly different amounts (Fig. 1B). In contrast, there were several glycoproteins that were unique to specific ECMs (Fig. 1C). Three glycoproteins, known to be involved in promoting osteogenesis, were found exclusively in BM-ECM (periostin (POSTN), fibrillin-1 (FBN1), and cysteine-rich angiogenic inducer 61 [CYR61]), while three other proteins were found in AD-ECM but only one is known to play a role in adipogenesis (fibulin-2 [FBLN2]).
Several proteoglycans were shared by both ECMs, but heparan sulfate proteoglycan-2 (HSPG2) was most abundant in BM-ECM, while biglycan (BGN) was most abundant in AD-ECM (Fig. 1B). Other proteoglycans (VCAN, HAPLN1, and DCN) were also found in the two ECMs but in low amounts. In contrast, one proteoglycan was uniquely found, in very low abundance, in each ECM (i.e., chondroitin sulfate proteoglycan-4 [CSPG4] in BM-ECM and lumican [LUM] in AD-ECM).
Three ECM-associated components were identified in both ECMs and found in similar abundance (annexin A2 [ANXA2], semaphorin 7A [SEMA7A], and peroxidasin [PXDN]) (Fig. 1B). In contrast, each ECM (Fig. 1C) contained proteins that were unique and present in very low abundance (nidogen-2 [NID2], insulin-like growth factor binding protein-7 [IGFBP7], and gremlin-1 [GREM1] in BM-ECM and galectin-1/lectin, galactoside-binding, soluble, 1 [LGALS1] in AD-ECM).
The two ECMs displayed vastly different structures and physical properties when analyzed with atomic force microscopy (AFM). BM-ECM has a relatively rough topography and consists of highly-organized and aligned fibrils when compared to AD-ECM (Fig. 2B); this can be seen in the AFM topographical maps. ECM stiffness, measured with AFM methods in the current study, is similar to what we reported previously using rheometry [4]. BM-ECM is roughly an order of magnitude stiffer than AD-ECM (P < 0.001, Fig. 2C). Lastly, we performed adhesion surface mapping studies for the two ECMs (Fig. 2C) and found that BM-ECM was one-third as adhesive as AD-ECM (P < 0.0001). The effect of this property, or related properties (e.g. surface energy/wettability), on cell function is particularly difficult to ascertain as any differences are likely tied to changes in the biochemical composition or organization of the matrix. Still, they each represent another relatively subtle microenvironmental variable that may have substantial effects on matrix properties.
Both BM- and AD-ECMs have similar effects on BM-MSC proliferation and retention of the MSC immunophenotype
To evaluate the effect of culture surface on cell proliferation, BM-MSCs were seeded onto standard TCP and the two ECMs (BM- and AD-ECM) at the same seeding density and cultured for 5, 7, and 14 days. At each time point, cells were released from the surface, counted and immunophenotyped. After 5 days in culture, cell density was significantly higher on the two ECMs compared to TCP (Fig. 3A). Cell counts on day 7 showed that BM-MSCs continued to proliferate and cell density on the two ECMs was higher than on TCP. However, cultures on AD-ECM had a cell density that was slightly lower than on BM-ECM. With continued culture to 14 days, cells on the two ECMs continued to display a significantly higher cell density than those on TCP.
Fig. 3.
BM-MSC proliferation and cell surface marker expression after culture for up to 14 days on TCP and BM- and AD-ECMs. (A) BM-MSCs were seeded onto TCP and BM- and AD-ECM culture surfaces and grown for 5, 7 and 14 days. At each time point, cells were detached from the substrate, stained with trypan blue, and counted. The data are presented as the average (±95% confidence interval) number of cells/cm2 in 3 independent experiments. *P < 0.05, vs. TCP; +P < 0.05, vs. AD-ECM. (B) Flow cytometry histograms of stem cell marker expression by BM-MSCs after 14 days in culture on TCP, BM-ECM, or AD-ECM. The data include standard surface markers for BM-MSCs (e.g. CD73, CD90, CD105 and CD146) and the embryonic stem cell marker SSEA-4. For each surface marker, the percent positive cells are shown in blue and the isotype control is shown in red. While the data are for cells from one donor cultured on TCP and the two ECMs for 14 days, similar results were obtained for cells from 3 additional donors cultured on the same surfaces for 5, 7, and 14 days.
Uniformly high expression of MSC surface markers (CD73, CD90, CD105, and CD146) was maintained through 14 days in culture on all surfaces. SSEA-4, which is an important marker of uncommitted MSCs [13] and is related to dividing cells [14], was typically higher on the two ECMs than TCP at all timepoints. This expression pattern was uniformly observed with BM-MSCs from four individual donors. Flow cytometry histograms from a representative experiment showed typical expression of MSC surface markers on day 14 of culture on all substrates (Fig. 3B).
Tissue-specific ECM maintains BM-MSC differentiation state and directs cell response to lineage-specific differentiation media
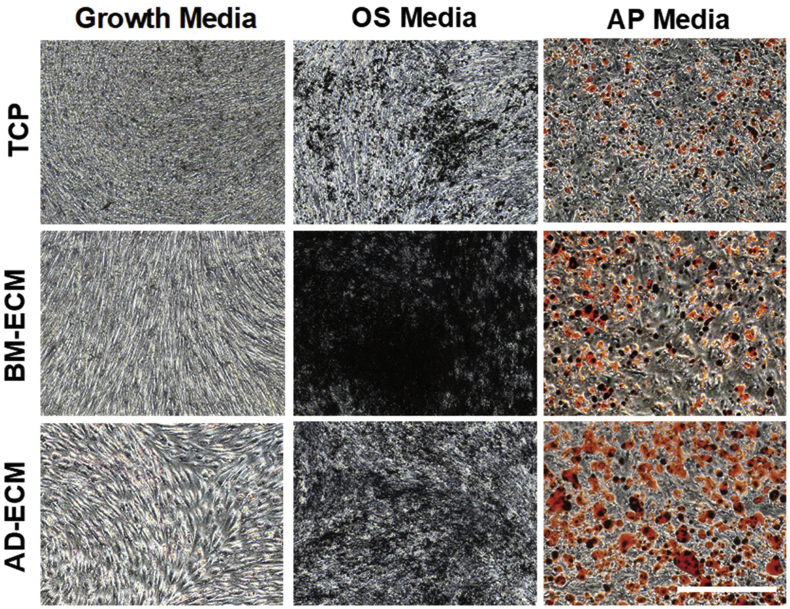
BM-MSCs were cultured for 7 days in growth media on either TCP or one of the ECMs (BM- and AD-ECM) and then switched to osteogenic (OS) or adipogenic (AP) differentiation media for an additional 7 days. Cultures of BM-MSCs, maintained on BM-ECM and treated with OS media, contained regions of strong von Kossa-positive staining, suggesting extensive calcification associated with osteogenic differentiation (Fig. 4). OS-treated cells that had been cultured on either TCP or AD-ECM also contained regions of von Kossa positive staining, but calcification was not as intense as found on the BM-ECM cultures. Similarly, Oil Red O staining suggested that BM-MSCs cultured on AD-ECM in the presence of AP media accumulated the greatest number of lipid droplets (Fig. 4).
Fig. 4.
Von Kossa and Oil Red O staining of BM-MSC cultures maintained on either TCP, BM-ECM, or AD-ECM in growth media or osteogenic (OS) or adipogenic (AP) induction media. BM-MSCs were cultured for 7 days in growth media, followed by culture for an additional 7 days in either OS or AP induction media or growth media (control). Cells were imaged on day 14 of culture with brightfield microscopy at 4× magnification (bar in figure = 500 μm).
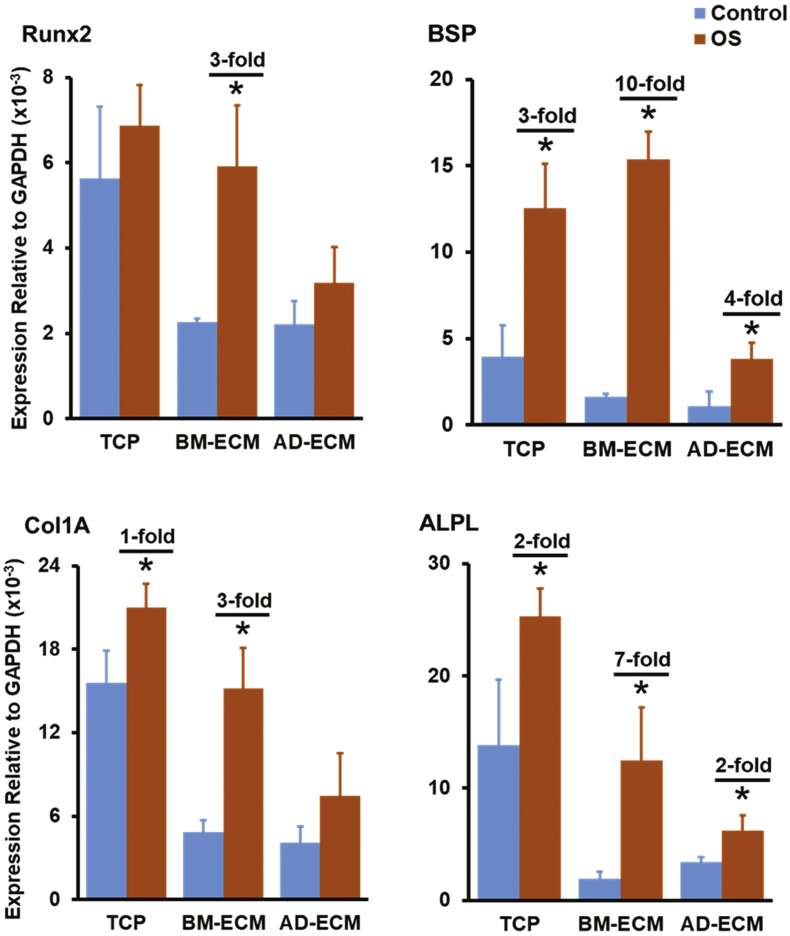
The effect of differentiation media on the cells was assessed by measuring the expression of transcripts widely associated with osteogenic or adipogenic differentiation. BM-MSCs maintained on BM-ECM, but not TCP or AD-ECM, in OS media displayed the largest and most significant increases in the expression of osteogenic genes: runt-related transcription factor 2 (Runx2; 3-fold), bone sialoprotein (BSP; 10-fold), collagen type I (Col1A; 3-fold) and alkaline phosphatase (ALPL; 7-fold) (Fig. 5). Similarly, BM-MSCs maintained on AD-ECM in the presence of AP media displayed the largest increase in the expression of adipogenic genes: peroxisome proliferator-activated receptor gamma (PPARγ; 14-fold), CCAAT/enhancer-binding protein alpha (CEBPα; 88-fold), and lipoprotein lipase (LPL; 19,292-fold), while cells cultured on TCP and BM-ECM Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7 also showed significant fold-increases but to a smaller degree.
Fig. 5.
Tissue-specific ECM directs the differentiation of BM-MSCs in response to osteogenic (OS) induction media. Expression of osteogenesis-associated transcripts by BM-MSCs cultured on TCP, BM-ECM, or AD-ECM in growth (=control) or OS induction media is shown. Transcript levels are reported relative to GAPDH expression; each experiment was performed in triplicate and repeated three times. *P < 0.05, vs. untreated control on a particular culture surface. When a significant difference was observed, the fold-increase over control is shown.
Fig. 6.
Tissue-specific ECM directs the differentiation of BM-MSCs in response to adipogenic (AD) induction media. Expression of adipogenesis-associated transcripts by BM-MSCs cultured on TCP, BM-ECM, or AD-ECM in growth (=control) or AP induction media is shown. Transcript levels are reported relative to GAPDH expression; each experiment was performed in triplicate and repeated three times. *P < 0.05, vs. untreated control on a particular culture surface. When a significant difference was observed, the fold-increase over control is shown.
Fig. 7.
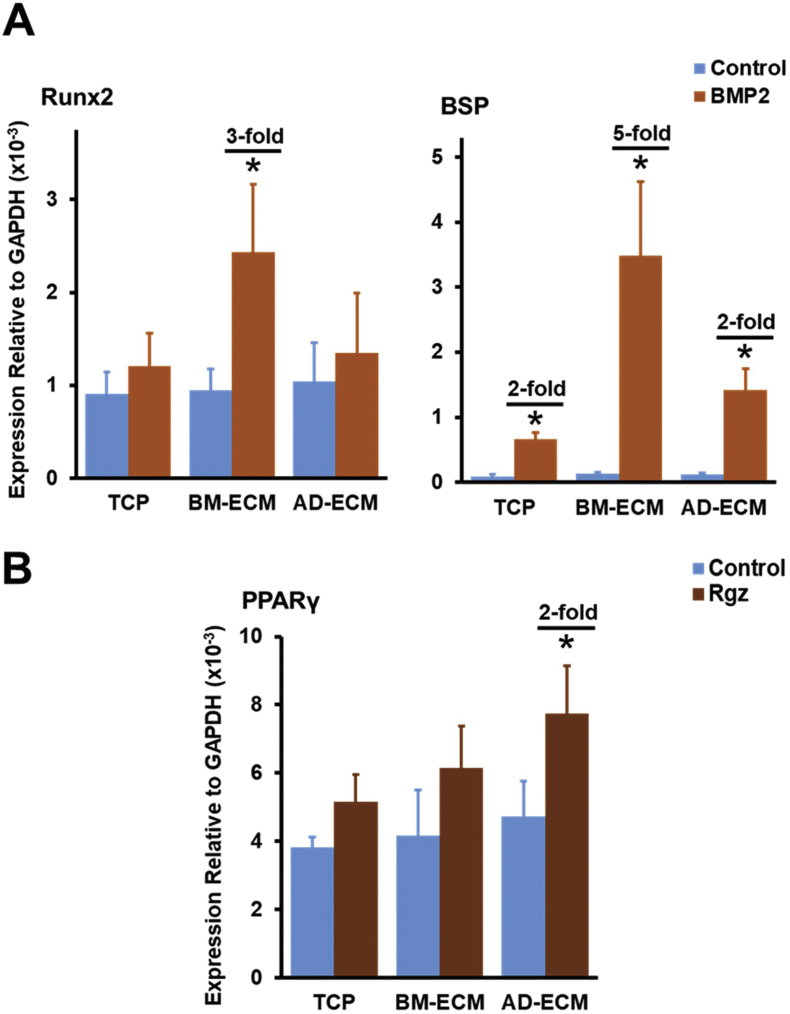
Gene expression of BM-MSCs in response to specific inducers of osteogenesis (BMP-2) and adipogenesis (rosiglitazone, Rgz) is increased by culture on tissue-specific ECMs. BM-MSCs were cultured in growth media on TCP or either BM- or AD-ECM for 7 days, switched to low serum-containing media for 24 h, and then treated with BMP-2 (200 ng/ml) or Rgz (1.9 μg/ml) for 48 h. (A) Cell response to BMP-2 was assessed by measuring the expression of transcripts associated with osteogenesis (e.g. Runx2 and BSP) using RT-PCR. (B) Cell response to Rgz was assessed by measuring the expression of transcripts associated with adipogenesis (e.g. PPARγ) using RT-PCR. In both panels, transcript levels are reported relative to GAPDH expression; each experiment was performed in triplicate and repeated three times. *P < 0.05, vs. untreated control on a particular culture surface. When a significant difference was observed, the fold-increase over control is shown.
Response of BM-MSCs to specific inducers of osteogenesis (BMP-2) and adipogenesis (Rgz) is increased by culture on tissue-specific ECM
For these experiments, BM-MSCs were cultured in growth media for 7 days on either TCP or one of the ECMs (BM- and AD-ECM), switched to low serum-containing growth media for 24 h, and then treated for an additional 48 h with the same media supplemented with either BMP-2 or Rgz, to induce osteogenesis or adipogenesis, respectively. BM-MSC response to these inducers was then assessed by measuring changes in the expression of osteogenesis- or adipogenesis-related mRNA transcripts using quantitative RT-PCR (Fig. 7). The data showed that cells maintained on BM-ECM and treated with BMP-2 exhibited a significant increase in Runx2 (3-fold) and BSP (5-fold) expression over controls, while cells cultured on TCP or AD-ECM failed to display an increase with BMP-2 treatment or the increase was significant but greatly diminished (Fig. 7A). Similarly, only BM-MSCs maintained on AD-ECM demonstrated responsiveness to Rgz as measured by a significant 2-fold increase in PPARγ expression (Fig. 7B).
Discussion
During routine expansion on TCP, MSCs lose their regenerative capacity with multiple passages due to cellular senescence, spontaneous differentiation, or other types of phenotypic drift [1]. This results in a loss of MSC differentiation state (i.e. “stemness”) and overall diminished capacity to respond to differentiation inducers and/or cues, leading to inconsistent outcomes [13,14].
Previously, our group described a native decellularized BM-ECM culture system that promoted the rapid proliferation of BM-MSCs, as well as retention of their stem cell properties (“stemness”), with multiple passages relative to cells cultured on TCP [5,11]. To understand the mechanisms involved, we investigated the characteristics of BM-ECM and compared its biochemical, architectural and mechanical attributes to another ECM produced by adipose-derived stromal cells (AD-ECM) [4].
The protein composition of both BM- and AD-ECMs was determined using mass spectrometry to identify any potential key components that may be different between the two ECMs. The studies revealed that the two ECMs were quite similar, with the shared number of proteins accounting for approximately 50% of the total number identified in both ECMs. Of the proteins found uniquely in one of the ECMs, many were associated with a particular cell lineage. For example, two glycoproteins, periostin and cysteine-rich angiogenic inducer 61 (Cyr61), found exclusively in BM-ECM, are strongly implicated in bone formation and maintenance (Fig. 1C). Periostin upregulates MSC osteogenesis via the Wnt/β-catenin pathway [15], while bone-specific, conditional knockout of Cyr61 reduces osteocyte/osteoblast activity, substantially reducing bone mineral density [16]. Similarly, fibulin 2 has been shown to be enriched in adipocyte ECM [17]. These findings support our assertion that ECMs, produced by BM- or AD-derived stromal cells ex vivo, retain the characteristics of their tissue of origin and suggest a mechanism whereby they may direct cell lineage-specific differentiation of MSCs in culture.
Interestingly, type VI collagen was the predominant collagen in both ECMs and several studies have reported that this collagen plays a direct role in regulating stem cell fate [[18], [19], [20]]. Andersen et al. used immunohistochemical approaches to demonstrate a shift in type VI collagen localization with MSC differentiation. In undifferentiated cells, type VI collagen was localized exclusively to the cytoplasm and then became redistributed to the cell membrane and peri-cellular matrix with differentiation to the osteoblast or adipocyte cell lineage [21]. More recently, Urciuolo et al. demonstrated that type VI collagen plays a key role in creating the muscle satellite cell niche; in type VI collagen knock-out mice muscle regenerative capacity was reduced along with impaired satellite cell self-renewal [22]. With engraftment of wild-type fibroblasts, type VI collagen production resumed, along with the return of satellite cell function, to near-normal levels. These observations suggest an important role for type VI collagen in directing stem cell behavior and provide support for our hypothesis that BM- and AD-ECMs replicate the MSC tissue-specific microenvironment present in bone marrow and adipose tissue.
While type VI collagen was the most abundant collagen in both ECMs and present in nearly the same amounts (~60%), type I collagen was the second most abundant collagen and present in both ECMs as well (~30%). Type XII collagen was only found in AD-ECM, while types IV and V collagen accounted for a minor percentage of only BM-ECM. Type I collagen is a widely distributed fibrillar collagen that is present at high levels in bone and skin [[23], [24], [25], [26]]. In contrast, type XII collagen is less common and is a member of the FACIT (or fibril-associated collagens with interrupted triple-helices) family of collagens which function to stabilize the fibrillar collagen network by forming bridges across fibrils [12,27]. Taking into consideration the architectural and mechanical differences we identified previously between BM- and AD-ECM [4], the highly-organized, relatively high-stiffness properties of BM-ECM (Fig. 2B and C) may be partially attributed to the abundance of type I and VI collagens [28]. In contrast, the relatively disorganized architecture and low-stiffness of AD-ECM may be due to the considerable percentage (~12%) of type XII collagen relative to type I [29]. These differences may be important in understanding how the ECM regulates cell behavior since changes in the elasticity of culture surfaces has been shown to influence MSC differentiation [30]. Together, these matrix properties (protein composition, architecture, physical properties) provide clues as to the nature of the microenvironment (niche) experienced by the cells in each tissue but fall short of providing any mechanistic information [31,32].
Previously, we showed that different cues displayed by these two ECMs (i.e., physical/mechanical, architectural) influenced discrete cellular behaviors such as proliferation, spreading morphology (circularity), and calcium/lipid deposition [4]. Based on these results and the proteomic analyses above, we hypothesized that native ECM plays a tissue-specific role in directing MSC differentiation to the lineage of the stromal cells that produced it. To test this, we compared the effect of culture on TCP and the two ECMs for up to 14 days on BM-MSC proliferation and cell surface marker expression. In addition to confirming our previously reported finding that the proliferation of BM-MSCs maintained on either ECM was superior to that of cells maintained on TCP, the current study finds that BM-MSCs cultured on the ECMs for up to 14 days displayed better retention of the MSC phenotype (i.e. higher expression of SSEA-4) than cells maintained on TCP. SSEA-4 is an important marker of uncommitted MSCs [13] and is related to dividing cells [14]. Despite the high density achieved on day 14 of culture, MSCs expanded on BM- and AD-ECM demonstrated the highest SSEA-4 expression compared to TCP. These results suggest that while both ECMs promote MSC proliferation, relative to TCP, they may also protect the “primitive” phenotype of BM-MSCs and restrain their tendency towards phenotypic drift and spontaneous differentiation.
We compared the capacity of TCP and the BM- and AD-ECMs to direct tissue-specific differentiation of BM-MSCs by culturing the cells on the three substrates in the presence of osteogenic (OS) or adipogenic (AP) differentiation media. In these studies, the greatest amount of mineral formation (i.e. von Kossa-positive staining for calcium deposition) was observed in cultures on BM-ECM maintained in OS media, while lipid-deposition (i.e. Oil Red O staining) was highest in cultures on AD-ECM in AP differentiation media (Fig. 4). In contrast, mineral and lipid formation did not occur, or occurred to a much smaller degree, in cultures maintained on BM- or AD-ECM as compared to TCP in growth media (control), suggesting that BM-MSCs cultured on the ECMs were able to retain their undifferentiated state until presented with lineage-specific differentiation media and, at that time, MSC differentiation favored the stromal cell lineage responsible for producing the tissue-specific ECM. These observations were further confirmed at the gene expression level by studies showing that both BM- and AD-ECM contain tissue-specific cues which direct and augment the differentiation of MSCs when cultured under osteogenic or adipogenic induction conditions, respectively (Fig. 5, Fig. 6).
To more precisely assess the tissue-specific role of the ECM in modulating the responsiveness of BM-MSCs to inducers of differentiation, we treated BM-MSCs with BMP-2 (a known specific inducer of osteogenesis) or rosiglitazone (a known specific inducer of adipogenesis) during culture on TCP or the two ECMs (Fig. 7). In this experiment, the concentration of BMP-2 (200 ng/ml) was selected so that it was just high enough to stimulate a change in gene expression in BM-MSCs maintained on BM-ECM, but not TCP. Indeed, BM-MSCs maintained on BM-ECM exhibited a significant increase in Runx2 (3-fold) and BSP (5-fold) expression with BMP-2 treatment compared to AD-ECM and TCP. Conversely, only BM-MSCs cultured on AD-ECM demonstrated a significant response to induction with rosiglitazone (2-fold increase in PPARγ expression). These data suggest that BM- and AD-ECM have the ability to increase the sensitivity of resident cells to lineage-specific inducers (e.g. BMP-2 for osteogenesis on BM-ECM). While studies have shown that certain ECM components, such as specific matricellular proteins, regulate the interaction of cells and soluble differentiation factors, the precise mechanism(s) by which BM- and AD-ECM direct stem cell fate remain to be elucidated.
Although many biochemical, architectural, and physical/mechanical attributes of culture surfaces have been shown to individually influence MSC self-renewal and differentiation, it is a much greater challenge to investigate how these parameters work together, as in the native stem cell niche in vivo. Here we report that tissue-specific ECMs, assembled by stromal cells ex vivo, provide a useful approach for modeling the complexity of the stem cell niche in culture. Specifically, this study provides evidence that bone marrow- and adipose-derived ECMs perform two very important roles. First, they increase the sensitivity of MSCs to tissue-specific growth factors and second they promote MSC self-renewal and maintenance of phenotype in the absence of differentiation inducers.
Based on what has been learned from native ECMs to date, the stem cell niche is undoubtedly a highly complex system, featuring interrelated signaling cues which are observed as distinct architectural, mechanical and biochemical properties, but whose discrete role in cell signaling is exceedingly difficult to investigate. We assert that the ECMs described herein provide a method for re-creating much of the complexity of the native stem cell niche ex vivo, and perhaps offer a new approach for dissecting out the roles of its manifold components. In the future, it will be necessary to develop collections of more defined tissue-specific ECMs which contain appropriate combinations of biological contexts to accurately replicate how the cells behave in the body. Defined, tissue-specific ECMs may help overcome a number of current obstacles in cell-based therapeutics and tissue engineering by providing a unique environment for both efficient stem cell expansion, without loss of differentiation state (i.e. “stemness”), and retention of tissue-specific differentiation capacity.
Tissue-specific matrices may also be useful for increasing our understanding of the mechanisms used by the ECM to regulate homeostasis in various tissues and organs [33]. Such ex vivo models might also be used as important tools for investigating the role of the ECM in the pathophysiology of fibrotic disease, cancer and aging [34,35]. The ability to study these ECMs directly may yield important insights into the cause of degenerative disease and provide clues, leading to new therapeutic approaches, which reverse pathologic changes by targeting the ECM.
Materials and methods
Preparation of BM- and AD-MSCs
Human bone marrow (BM), collected from healthy donors (20–25 years old) after obtaining informed consent, was purchased from Lonza Group Ltd. (Walkersville, MD, see company website for IRB information). Fresh unprocessed BM samples, containing BM-MSCs, were seeded into tissue culture plastic (TCP) vessels at 5 × 105 cells/cm2 and cultured as previously described [4] to assess cell behavior and prepare BM-ECM as described below. AD-MSCs were also obtained from Lonza Group Ltd. (Walkersville, MD) and used to prepare AD-ECM according to methods described previously [4].
Preparation of cell-free BM- and AD-ECM
BM- and AD-ECM were prepared as described previously [[2], [3], [4], [5]]. Briefly, BM- or AD-derived stromal cells were seeded into TCP vessels. At confluence, cells were induced to synthesize ECM. Afterwards, cells were removed by treatment with Triton X-100 (Millipore-Sigma), a non-denaturing detergent. The resulting cell-free ECM was washed thoroughly and stored dry at 4 °C for no more than 1 year. Prior to use, ECM was rehydrated for 1 h with PBS at 37 °C.
Assessment of cell proliferation and immunophenotyping
Cell proliferation was determined as previously described [2,4,6]. Briefly, BM-MSCs (P3) were seeded at 5 × 103 cells/cm2 on BM-ECM, AD-ECM, or TCP and cultured for 5, 7 and 14 days in α-MEM (Life Technologies, Grand Island, NY) containing 2 mM glutamine, penicillin/streptomycin, and 15% fetal bovine serum (FBS) (Atlanta Biologicals, Lawrenceville, GA) (=growth media). At harvest, cells were released from the culture surfaces, using trypsin for cultures on TCP and collagenase for cultures on ECM, and then counted using a hemocytometer after trypan blue staining.
After cell counting, BM-MSC surface marker expression was measured using flow cytometry. Mouse anti-human non-conjugated antibodies (IgG1, IgG3, CD34, CD73, CD90, CD105, CD146 and SSEA-4) were purchased from BD Biosciences (San Jose, CA, U.S.A.). Single cell suspensions (1 × 105/100 μl) were incubated at 4 °C for 1 h with primary antibody (10 μg/ml), washed twice with staining buffer (PBS + 5% FBS + 0.01% sodium azide), and then incubated with FITC-conjugated goat anti-mouse IgG at 4 °C for 30 min. Cells were subsequently washed two times with staining buffer and then immediately analyzed using a BD Bioscience LSRII flow cytometer running FACSDiva software. Alternatively, cells were fixed with 1% paraformaldehyde, followed by analysis within 72 h. Data were analyzed and figures created using FlowJo software. At least 10,000 events were measured in each sample and the percent positive cells (relative to isotype control) determined.
Compositional analysis of ECM using mass spectrometry
Extraction of proteins from the ECM for mass spectrometric (MS) analysis involved a more comprehensive approach than described in our previous report [4] and reflected further refinement of the method adapted from [36]. ECM proteins were isolated by physical agitation and sonication (2 times) for 10 min in PBS at room temperature. The ECM pellet was treated with Protein Extraction Reagent Type 4 (Millipore-Sigma) and sonicated on ice for 30 min, followed by vacuum drying. The dried ECM was reconstituted in Ready Prep 2-D Rehydration Sample Buffer (Bio-Rad) and treated with repeated cycles of sonication on ice and vortexing at 32 °C to disperse the pellet. The sample was then subjected to two freeze-thaw cycles in liquid nitrogen, before centrifugation at 21,000g for 10 min. The resulting supernatant (supernatant #1) was collected and adjusted to a final urea concentration of 8 M, while the pellet was dissolved in 100 μl DMSO, sonicated for 30 min on ice, and re-centrifuged at 21,000g for 10 min. The resulting supernatant (supernatant #2) was made up to 8 M urea, the two supernatants combined, and then vortexed to produce supernatant #3. A 200 μl aliquot of supernatant #3 was then combined with 1 ml acetone, mixed by vortexing, and stored overnight at 4 °C. Following a brief centrifugation, the acetone supernatant was removed, and the pellet allowed air-drying at room temperature. Dried pellets were reconstituted in 50 μl 2× SDS buffer and boiled at 100 °C for 5 min before loading onto an SDS-PAGE gel.
Protein separation was performed using standard (one-dimension) SDS-PAGE and Coomassie blue staining. Proteins were released from discrete serial sections of each lane of the gel by in situ digestion with trypsin (Promega). Digests were analyzed using capillary HPLC-electrospray ionization tandem mass spectrometry (HPLC-ESI-MS/MS) on a Thermo Fisher LTQ fitted with a New Objective PicoView 550 nanospray interface. On-line HPLC separation of the digests was accomplished with an Eksigent NanoLC micro HPLC. A mass spectral scan strategy was used in which a survey scan was acquired followed by data-dependent collision-induced dissociation (CID) spectra of the seven most intense ions in the survey scan. Mascot (Matrix Science) was used to search the mass spectra searched against the SwissProt database. Methionine oxidation was considered as a variable modification. Cross correlation of the Mascot results with X! Tandem and determination of protein and peptide identity probabilities were accomplished by Scaffold (Proteome Software). Protein identifications were accepted using the following criteria: minimum number of peptides, 2; peptide probability, ≥95%; protein probability, ≥99%.
In order to investigate the reproducibility and overall consistency of the matrices, compositional data from freshly-prepared ECMs was pooled with results originally reported in Marinkovic et al. [4] and Ragelle et al. [37] (accessible via the PRIDE proteomics database of the European Bioinformatics Institute (EMBL-EBI) with identifiers PXD005521 and https://doi.org/10.6019/PXD005521). The combined data were re-analyzed to reflect BM- and AD-ECM composition produced under similar conditions by stromal cells derived from a total of four individual donors for each matrix. Proteomic data not only included extracellular matrix proteins, but also traces of proteins previously reported or known to be present in cells or the cell membrane. For tabulating the proteins found in the ECMs, these trace amounts of protein were manually excluded and not included in Fig. 1.
Physical and structural characterization of ECM by atomic force microscopy
Topography, adhesion, and stiffness of the matrices were determined using a Nanoscope Catalyst (Bruker) atomic force microscope (AFM), mounted on a Nikon Ti inverted epifluorescence microscope, in PeakForce Quantitative Nanomechanical Mapping (PF-QNM) mode. The probes and matrices were immersed in PBS. At least 5, randomly selected, 100x100μm fields were scanned for each group.
Assessment of BM-MSC response to osteogenic or adipogenic induction media
BM-MSCs (P3) were seeded onto the three culture surfaces (BM-ECM, AD-ECM and TCP) at 5 × 103 cells/cm2 and cultured in growth media. After 7 days, the cells were switched to osteogenic (OS) or adipogenic (AP) induction media and the cultures continued for an additional 7 days. OS induction media contained growth media supplemented with 100 nM dexamethasone, 10 mM β-glycerophosphate, and 50 μM L-ascorbate-2-phosphate. Alternatively, AP induction media consisted of DMEM containing 10% FBS, supplemented with 5 mM 3-isobutyl-L-methylxanthine (IBMX), 1 mM indomethacin, 1 μM dexamethasone, and 10 μM insulin. All OS and AP induction media supplements were purchased from Millipore Sigma (St. Louis, MO, USA). The effect of culture on the three different culture surfaces in the presence of OS or AP induction media was evaluated by the staining and gene expression studies described below.
Von Kossa and Oil Red O staining after culture on TCP, BM- and AD-ECM
After culture, cells were fixed with 10% formalin for 1 h at room temperature. Calcium deposition, an index of mineralization, was assessed by von Kossa staining (1% silver nitrate) of the cultures overnight and then washing with 5% sodium thiosulfate. To assess lipid deposition, the cultures were stained with Oil Red O (0.5% in 2-propanol) overnight and then washed extensively with distilled water to remove unbound dye. Representative areas of the stained cultures were photographed at 4× magnification using a brightfield microscope.
Quantification of BM-MSC gene expression with culture on TCP, BM- and AD-ECM
Cells were rinsed with cold PBS and total RNA isolated using the Trizol reagent (Life Technologies, Carlsbad, CA). cDNA was reverse-transcribed from the extracted RNA (2 μg) using a High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA, USA). Transcripts of interest, as well as that of the housekeeping gene (GAPDH), were amplified from the cDNA by real-time PCR using TaqMan Universal PCR Master Mix and Assay-on-Demand or Assay-by-Design primer/probe sets (Applied Biosystems). Amplification and detection were carried out with an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems). Gene expression was quantified by subtracting the GAPDH threshold cycle (Ct) value from the Ct value of the gene of interest and expressed as 2−ΔCt as described by the manufacturer's protocol.
Assessment of BM-MSC-responsiveness to treatment with specific inducers of osteogenesis (rhBMP-2) and adipogenesis (Rgz)
BM-MSCs (P3) were seeded onto the three different culture surfaces (BM-ECM, AD-ECM, and TCP) at 5 × 103 cells/cm2 and cultured in growth media for 7 days. On day 7, the media were switched to growth media containing 2% FBS and the cultures continued overnight. On day 8, 200 ng/ml rhBMP-2 (Life Technologies, Frederick, MD) or 1.9 μg/ml Rgz (Millipore Sigma, St. Louis, MO) was added to the media and the cultures continued for an additional 48 h. Changes in transcripts in response to BMP-2 or Rgz treatment were measured using RT-PCR as described above.
Statistical interpretation of the data
Student's t-test was used to determine significance and included a Bonferroni correction to account for multiple comparisons (α = 0.0167). At a minimum, replicates were performed in triplicate and experiments repeated three times. All error bars depicted in the figures reflect the 95% confidence interval for each data set.
Declaration of competing interest
Dr. Chen is a Board member and shareholder in StemBioSys, Inc. (San Antonio, TX). Dr. Travis Block is currently an employee of StemBioSys, Inc. (San Antonio, TX) and receives a salary. All other authors have no financial or competing interests to declare.
Acknowledgments
Acknowledgements
This study was supported by a VA Merit Review (1I01BX002145-01) to Dr. Chen and a National Institutes of Health/NIDCR grant (DE025286) to Dr. Yeh. Dr. Marinkovic was supported by a NIH-NCATS TL1 Translational Science Training grant (TL1 TR001119) and NIH-NIDCR F31 National Research Service Award (F31 DE02668). Mass spectrometry analyses were conducted at the UTHSCSA Institutional Mass Spectrometry Laboratory which is supported in part by an NIH shared instrumentation grant (1 S10 RR021160-01) to Dr. Susan T. Weintraub who is the Director of the Mass Spectrometry Core Laboratory. The expert technical assistance of Kevin Hakala, M.S. and Sammy Pardo, B.A. in performing the mass spectrometry studies is greatly appreciated.
Authors' contributions
MM: designed and performed experiments, analyzed data, prepared figures/tables, prepared first draft of the manuscript; TJB, RR, ONT, and AOG: performed experiments, acquired data; DDD, CKY: performed data analysis and interpretation, wrote/edited manuscript; XDC: conceived/designed the study, obtained financial support, analyzed/interpreted the data, wrote/edited manuscript; All authors have reviewed and approved the final manuscript.
Contributor Information
Milos Marinkovic, Email: marinkovic@uthscsa.edu.
Olivia N. Tran, Email: trano@livemail.uthscsa.edu.
Rubie Rakian, Email: rakianr@livemail.uthscsa.edu.
Aaron O. Gonzalez, Email: gonzaleza33@livemail.uthscsa.edu.
David D. Dean, Email: deand@uthscsa.edu.
Chih-Ko Yeh, Email: yeh@uthscsa.edu.
Xiao-Dong Chen, Email: chenx4@uthscsa.edu.
References
- 1.Mosna F., Sensebé L., Krampera M. Human bone marrow and adipose tissue mesenchymal stem cells: a user’s guide. Stem Cells Dev. 2010;19:1449–1470. doi: 10.1089/scd.2010.0140. [DOI] [PubMed] [Google Scholar]
- 2.Chen X.-D., Dusevich V., Feng J.Q., Manolagas S.C., Jilka R.L. Extracellular matrix made by bone marrow cells facilitates expansion of marrow-derived mesenchymal progenitor cells and prevents their differentiation into osteoblasts. J. Bone Miner. Res. 2007;22:1943–1956. doi: 10.1359/jbmr.070725. [DOI] [PubMed] [Google Scholar]
- 3.Lai Y., Sun Y., Skinner C.M., Son E.L., Lu Z., Tuan R.S. Reconstitution of marrow-derived extracellular matrix ex vivo: a robust culture system for expanding large-scale highly functional human mesenchymal stem cells. Stem Cells Dev. 2010;19:1095–1107. doi: 10.1089/scd.2009.0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marinkovic M., Block T.J., Rakian R., Li Q., Wang E., Reilly M.A. One size does not fit all: developing a cell-specific niche for in vitro study of cell behavior. Matrix Biol. 2016;52:426–441. doi: 10.1016/j.matbio.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen X. Extracellular matrix provides an optimal niche for the maintenance and propagation of mesenchymal stem cells. Birth defects Res. Part C. 2010;90:45–54. doi: 10.1002/bdrc.20171. [DOI] [PubMed] [Google Scholar]
- 6.Block T.J., Marinkovic M., Tran O.N., Gonzalez A.O., Marshall A., Dean D.D. Restoring the quantity and quality of elderly human mesenchymal stem cells for autologous cell-based therapies. Stem Cell Res Ther. 2017;8:239. doi: 10.1186/s13287-017-0688-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy W.L., McDevitt T.C., Engler A.J. Materials as stem cell regulators. Nat. Mater. 2014;13:547–557. doi: 10.1038/nmat3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalby M.J., Gadegaard N., Tare R., Andar A., Riehle M.O., Herzyk P. The control of human mesenchymal cell differentiation using nanoscale symmetry and disorder. Nat. Mater. 2007;6:997–1003. doi: 10.1038/nmat2013. [DOI] [PubMed] [Google Scholar]
- 9.Lutolf M.P., Gilbert P.M., Blau H.M. Designing materials to direct stem-cell fate. Nature. 2009;462:433–441. doi: 10.1038/nature08602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lecuit T., Lenne P.F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 2007;8:633–644. doi: 10.1038/nrm2222. [DOI] [PubMed] [Google Scholar]
- 11.Rakian R., Block T.J., Johnson S.M., Marinkovic M., Wu J., Dai Q. Native extracellular matrix preserves mesenchymal stem cell “stemness” and differentiation potential under serum-free culture conditions. Stem Cell Res Ther. 2015;6:235. doi: 10.1186/s13287-015-0235-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiquet M., Birk D.E., Bönnemann C.G., Koch M. Collagen XII: protecting bone and muscle integrity by organizing collagen fibrils. Int. J. Biochem. Cell Biol. 2014;53:51–54. doi: 10.1016/j.biocel.2014.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv F.-J., Tuan R.S., Cheung K.M.C., Leung V.Y.L. Concise review: the surface markers and identity of human mesenchymal stem cells. Stem Cells. 2014;32:1408–1419. doi: 10.1002/stem.1681. [DOI] [PubMed] [Google Scholar]
- 14.Gang E.J., Bosnakovski D., Figueiredo C.A., Visser J.W., Perlingeiro R.C.R. SSEA-4 identifies mesenchymal stem cells from bone marrow. Blood. 2007;109:1743–1751. doi: 10.1182/blood-2005-11-010504. [DOI] [PubMed] [Google Scholar]
- 15.Zhang F., Luo K., Rong Z., Wang Z., Luo F., Zhang Z. Periostin upregulates Wnt/β-catenin signaling to promote the osteogenesis of CTLA4-modified human bone marrow-mesenchymal stem cells. Sci. Rep. 2017;7:1–10. doi: 10.1038/srep41634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao G., Huang B.-L., Rigueur D., Wang W., Bhoot C., Charles K.R. CYR61/CCN1 regulates sclerostin levels and bone maintenance. J. Bone Miner. Res. 2018;33:1076–1089. doi: 10.1002/jbmr.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariman E.C.M., Wang P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 2010;67:1277–1292. doi: 10.1007/s00018-010-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan T., Muise E.S., Iyengar P., Wang Z.V., Chandalia M., Abate N. Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol. Cell. Biol. 2009;29:1575–1591. doi: 10.1128/MCB.01300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gattazzo F., Urciuolo A., Bonaldo P. Extracellular matrix: a dynamic microenvironment for stem cell niche. Biochim. Biophys. Acta. 2014;1840:2506–2519. doi: 10.1016/j.bbagen.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alexeev V., Arita M., Donahue A., Bonaldo P., Chu M.-L., Igoucheva O. Human adipose-derived stem cell transplantation as a potential therapy for collagen VI-related congenital muscular dystrophy. Stem Cell Res Ther. 2014;5:21. doi: 10.1186/scrt411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andersen D.C., Kortesidis A., Zannettino A.C.W., Kratchmorova I., Chen L., Jensen O.N. Development of novel monoclonal antibodies that define differentiation stages of human stromal (mesenchymal) stem cells. Mol. Cells. 2011;32:133–142. doi: 10.1007/s10059-011-2277-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urciuolo A., Quarta M., Morbidoni V., Gattazzo F., Molon S., Grumati P. Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun. 2013;4:1964–1977. doi: 10.1038/ncomms2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon M.Y. Extracellular matrix of the marrow microenvironment. Br. J. Haematol. 1988;70:1–4. doi: 10.1111/j.1365-2141.1988.tb02425.x. [DOI] [PubMed] [Google Scholar]
- 24.Calvi L.M., Adams G.B., Weibrecht K.W., Weber J.M., Olson D.P., Knight M.C. Osteoblastic cells regulate the haematopoietic stem cell niche. Nature. 2003;425:841–846. doi: 10.1038/nature02040. [DOI] [PubMed] [Google Scholar]
- 25.Gronthos S., Simmons P.J., Graves S.E., Robey P.G. Integrin-mediated interactions between human bone marrow stromal precursor cells and the extracellular matrix. Bone. 2001;28:174–181. doi: 10.1016/s8756-3282(00)00424-5. [DOI] [PubMed] [Google Scholar]
- 26.Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., MacArthur B.D., Lira S.A. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwal P., Zwolanek D., Keene D.R., Schulz J.-N., Blumbach K., Heinegard D. Collagen XII and XIV, new partners of cartilage oligomeric matrix protein in the skin extracellular matrix suprastructure. J. Biol. Chem. 2012;287:22549–22559. doi: 10.1074/jbc.M111.335935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reese S.P., Underwood C.J., Weiss J.A. Effects of decorin proteoglycan on fibrillogenesis, ultrastructure, and mechanics of type I collagen gels. Matrix Biol. 2013;32:414–423. doi: 10.1016/j.matbio.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mariman E.C.M., Wang P. Adipocyte extracellular matrix composition, dynamics and role in obesity. Cell. Mol. Life Sci. 2010;67:1277–1292. doi: 10.1007/s00018-010-0263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Engler A.J., Sen S., Sweeney H.L., Discher D.E. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 31.Prewitz M.C., Seib F.P., von Bonin M., Friedrichs J., Stissel A., Niehage C. Tightly anchored tissue-mimetic matrices as instructive stem cell microenvironments. Nat. Methods. 2013;10:788–794. doi: 10.1038/nmeth.2523. [DOI] [PubMed] [Google Scholar]
- 32.Prewitz M.C., Stissel A., Friedrichs J., Traber N., Vogler S., Bornhauser M., Werner C. Extracellular matrix deposition of bone marrow stroma enhanced by macromolecular crowding. Biomaterials. 2015;73:60–69. doi: 10.1016/j.biomaterials.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Karamanos N.K., Theocharis A.D., Neill T., Iozzo R.V. Matrix modeling and remodeling: a biological interplay regulating tissue homeostasis and diseases. Matrix Biol. 2019;75–76:1–11. doi: 10.1016/j.matbio.2018.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaefer L. Decoding fibrosis: mechanisms and translational aspects. Matrix Biol. 2018;68–69:1–7. doi: 10.1016/j.matbio.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Iozzo R.V., Gubbiotti M.A. Extracellular matrix: the driving force of mammalian diseases. Matrix Biol. 2018;71–72:1–9. doi: 10.1016/j.matbio.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Castro Brás L.E., Ramirez T.A., DeLeon-Rennell K.Y., Chiao Y.A., Ma Y., Dai Q. Texas 3-step decellularization protocol: looking at the cardiac extracellular matrix. J. Proteome. 2013;86:43–52. doi: 10.1016/j.jprot.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ragelle H., Naba A., Larson B.L., Zhou F., Prijić M., Whittaker C.A. Comprehensive proteomic characterization of stem cell-derived extracellular matrices. Biomaterials. 2017;128:147–159. doi: 10.1016/j.biomaterials.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]