Abstract
Lung fibrosis is characterized by excessive deposition of extracellular matrix (ECM), in particular collagens, by fibroblasts in the interstitium. Transforming growth factor-β1 (TGF-β1) alters the expression of many extracellular matrix (ECM) components produced by fibroblasts, but such changes in ECM composition as well as modulation of collagen post-translational modification (PTM) levels have not been comprehensively investigated. Here, we performed mass spectrometry (MS)-based proteomics analyses to assess changes in the ECM deposited by cultured lung fibroblasts from idiopathic pulmonary fibrosis (IPF) patients upon stimulation with transforming growth factor β1 (TGF-β1). In addition to the ECM changes commonly associated with lung fibrosis, MS-based label-free quantification revealed profound effects on enzymes involved in ECM crosslinking and turnover as well as multiple positive and negative feedback mechanisms of TGF-β1 signaling. Notably, the ECM changes observed in this in vitro model correlated significantly with ECM changes observed in patient samples. Because collagens are subject to multiple PTMs with major implications in disease, we implemented a new bioinformatic platform to analyze MS data that allows for the comprehensive mapping and site-specific quantitation of collagen PTMs in crude ECM preparations. These analyses yielded a comprehensive map of prolyl and lysyl hydroxylations as well as lysyl glycosylations for 15 collagen chains. In addition, site-specific PTM analysis revealed novel sites of prolyl-3-hydroxylation and lysyl glycosylation in type I collagen. Interestingly, the results show, for the first time, that TGF-β1 can modulate prolyl-3-hydroxylation and glycosylation in a site-specific manner. Taken together, this proof of concept study not only reveals unanticipated TGF-β1 mediated regulation of collagen PTMs and other ECM components but also lays the foundation for dissecting their key roles in health and disease.
The proteomic data has been deposited to the ProteomeXchange Consortium via the MassIVE partner repository with the data set identifier MSV000082958.
Abbreviations: AGC, automatic gain control; ANXA11, annexin A11; BGN, biglycan; COL1A1, collagen-I alpha 1 chain; DCN, decorin; ECM, extracellular matrix; FN1, fibronectin 1; G-HyK, galactosylhydroxylysine; GG-HyK, glucosylgalactosylhydroxylysine; HyK, hydroxylysine; HyP, hydroxyproline; 3-HyP, 3-hydroxyproline; 4-HyP, 4-hydroxyproline; ILD, interstitial lung disease; IPF, idiopathic pulmonary fibrosis; LH, lysyl hydroxylase; LOX(L), lysyl oxidase(-like); LTBP2, latent-transforming growth factor β -binding protein 2; PAI1, plasminogen activator inhibitor 1; PCA, principal component analysis; P3H, prolyl-3-hydroxylase; P4H, prolyl-4-hydroxylase; PLOD (LH), procollagen-lysine,2-oxoglutarate 5-dioxygenases (lysyl hydroxylases); PTM, post-translational modification; SEMA7A, semaphorin 7a; α-SMA, α-smooth muscle actin; TGF-β, transforming growth factor β; TGM2, transglutaminase 1; VCAN, versican; Xaa, Xaa position in the Gly-Xaa-Yaa repeat in triple-helical collagen; Yaa, Yaa position in the Gly-Xaa-Yaa repeat in triple-helical collagen
Keywords: Pulmonary fibrosis, Collagen, Extracellular matrix, Transforming growth factor-β, Collagen post-translational modifications, Prolyl hydroxylation, Lysyl hydroxylation, Lysyl glycosylation
Highlights
-
•
Quantitative proteomics of TGF-β-induced changes in ECM composition and collagen PTM in pulmonary fibroblasts
-
•
TGF-β promotes crosslinking and turnover as well as complex feedback mechanisms that alter fibroblast ECM homeostasis.
-
•
A novel bioinformatic workflow for MS data analysis enabled global mapping and quantitation of known and novel collagen PTMs
-
•
Quantitative assessment of prolyl-3-hydroxylation site occupancy and lysine-O-glycosylation microheterogeneity
-
•
TGF-β1 modulates collagen PTMs in a site-specific manner that may favor collagen accumulation in lung fibrosis
1. Introduction
Fibrosis is a major cause of mortality and morbidity and has been estimated to contribute to at least one third of all deaths worldwide and even 45% of deaths in the developed world [1,2]. Idiopathic pulmonary fibrosis (IPF) is one of the most progressive types of organ fibrosis, with a five-year survival rate of 30–50%, poorly understood etiology, and few treatment options [3].
Organ fibrosis is thought to be initiated by repeated or chronic epithelial injury. The current belief is that damaged epithelial cells induce an aberrant and unresolved wound repair process by activating fibroblasts via various profibrotic cues. Upon injury, epithelial cells activate the profibrotic cytokine transforming growth factor β (TGF-β) from latent complexes in the extracellular matrix (ECM) [[4], [5], [6]] and release TGF-β-containing exosomes [7]. TGF-β is well-established as a central driver of fibrogenesis through induction of expression and deposition of major ECM components like type I collagen and fibronectin by fibroblasts [1,[8], [9], [10]]. Furthermore, fibrotic ECM as synthesized by IPF fibroblasts provides profibrotic cues resulting in even more ECM synthesis, i.e. a positive feedback loop that may underlie the highly progressive character of the disease [[11], [12], [13], [14], [15]]. However, to the best of our knowledge, no study has taken an unbiased, global proteomic approach to comprehensively quantify the changes in ECM proteins and their modifications synthesized by IPF fibroblasts upon TGF-β1 stimulation.
Constituting between 30% and 70% of ECM protein in all tissue types, collagen clearly is one of the main components of the ECM [16]. Collagen biosynthesis and deposition is highly upregulated in fibrotic disease, leading to impaired ECM homeostasis and scar formation [[17], [18], [19]]. Importantly, all collagens are subject to various intracellular post-translational modifications (PTMs) as well as extracellular maturation steps [20,21]. Evidence has emerged that strongly argues for an important impact of collagen PTMs on protein-protein or protein-cell interactions [[22], [23], [24], [25], [26], [27], [28]], which can be highly relevant for disease. For instance, collagen prolyl-3-hydroxylation, a comparatively rare collagen PTM, has been shown to affect binding to the proteoglycan decorin, an extracellular regulator of TGF-β activity in the context of osteogenesis imperfecta [27]. Causal associations have been established for prolyl-3-hydroxylase 1 (P3H1, LEPRE1) gene mutations with osteogenesis imperfecta [29,30], for prolyl-3-hydroxylase 2 (P3H2, LEPREL1) gene mutations with myopia and several other eye defects [31,32], and for prolyl-3-hydroxylase 3 (P3H3, LEPREL2) gene mutations with Ehlers-Dalnos syndrome type VIA [33]. Although collagen PTMs have been studied by targeted mass spectrometry as well as computational prediction methods [34,35], they have neither been described in depth, nor in the context of lung fibrosis [36,37]. Therefore, strategies enabling global assessment and characterization of collagen PTMs are likely to provide important insight about their biological function.
In this work, we used mass spectrometry-based quantitative analysis together with immunoblotting and quantitative real time-PCR (qRT-PCR) to fully characterize and validate TGF-β1 mediated changes in ECM proteins deposited by IPF patient-derived lung fibroblasts. In addition, we developed a new MS data analysis workflow that enabled global identification and label-free quantitation of site-specific changes of hydroxylation and glycosylation of collagens in response to TGF-β1 treatment. The findings validate many of the known pleiotropic functions that characterize TGF-β1 as a profibrotic cytokine, and reveal an unanticipated role of TGF-β1 in site-specific regulation of collagen PTMs that may favor ECM accumulation and alter cell-matrix interactions in lung fibrosis.
2. Results and discussion
2.1. Primary human lung fibroblast culture is suitable to explore TGF-β1-mediated changes in ECM
In previous studies, we have established a reliable in vitro system of lung fibrosis that allows for the study of ECM composition and collagen biosynthesis and secretion [9,38,39]. To this end, primary human lung fibroblasts, for example fibroblasts isolated from IPF patients, are cultured in presence of 2-phosphoascorbate to allow for maximal collagen production [9,39] in absence and presence of concentrations of TGF-β1 (2 ng/ml) that mimic a profibrotic environment. Notably, our previous studies using identical experimental conditions have shown that control and IPF fibroblasts respond similarly to TGF-β1 stimulation as judged by expression of collagens, collagen biosynthetic enzymes, other ECM proteins, and collagen secretion [9,38,39]. As the availability of fibroblasts derived from healthy donors is limited, we have chosen to quantify ECM changes induced by TGF-β1 stimulation of IPF fibroblasts. As we previously showed [9,39], TGF-β1 consistently induced myofibroblast differentiation, type I collagen and fibronectin synthesis in these cells (Suppl. Fig. S1A–D). At the same time TGF-β1 downregulated miR-29b, a central negative regulator of ECM expression (Suppl. Fig. S1E) [11,12,40,41]. Collectively, these results suggested that this cell culture system is suitable to explore TGF-β1-mediated changes in ECM and collagen PTMs in a global proteomic approach.
2.2. ECM composition changed upon treatment of IPF fibroblasts with TGF-β1
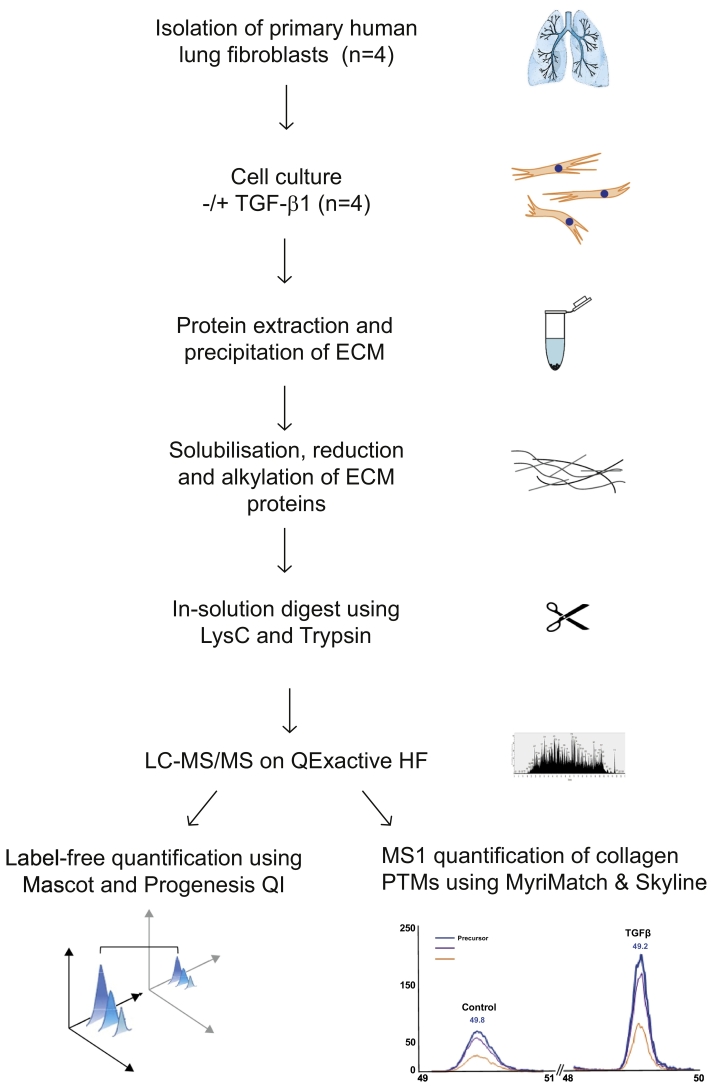
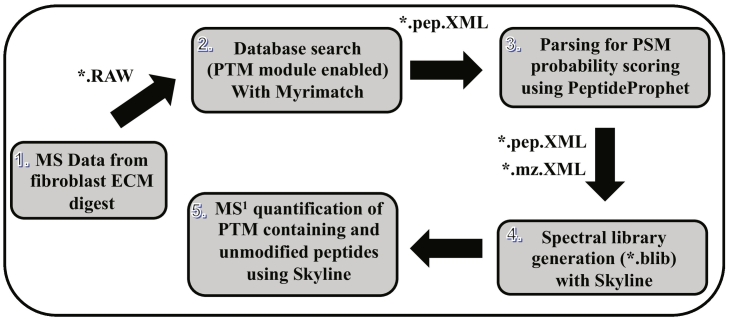
ECM enrichment was achieved by detergent-based decellularization of control and TGF-β1 stimulated fibroblast monolayer cultures. The insoluble pellet was solubilized in guanidine-containing buffer before reduction, alkylation, and digestion of protein samples. Peptides were analyzed by liquid-chromatography/tandem mass spectrometry (LC-MS/MS) followed by label-free quantitative analysis of MS data to reveal changes in fibroblast ECM upon TGF-β1 stimulation (Fig. 1). The same raw MS files were also subjected to an MS1 analysis using Skyline software to assess changes in collagen PTM.
Fig. 1.
Overview of experimental approach.
In total, we consistently detected 149 matrisomal proteins synthesized by IPF fibroblasts. Of these, 115 proteins were quantified with at least two unique peptides, including 58 core matrisome proteins and 57 matrisome-associated proteins, using the matrisome classification developed by Naba et al. [42] (Fig. 2A). In terms of coverage, this is superior to many other in vitro studies where ECM proteins synthesized by fibroblasts were analyzed following various enrichment protocols, including acetone-precipitation of cell culture supernatants [43] or decellularization of cell culture dishes [[44], [45], [46]], sometimes followed by resolubilization with 8 M urea [45] or on-plate enzymatic digestion [46]. In summary, the proteomics analyses suggest that our detergent-based ECM enrichment procedure (Fig. 1) yields excellent coverage of ECM and ECM-associated proteins (Fig. 2A).
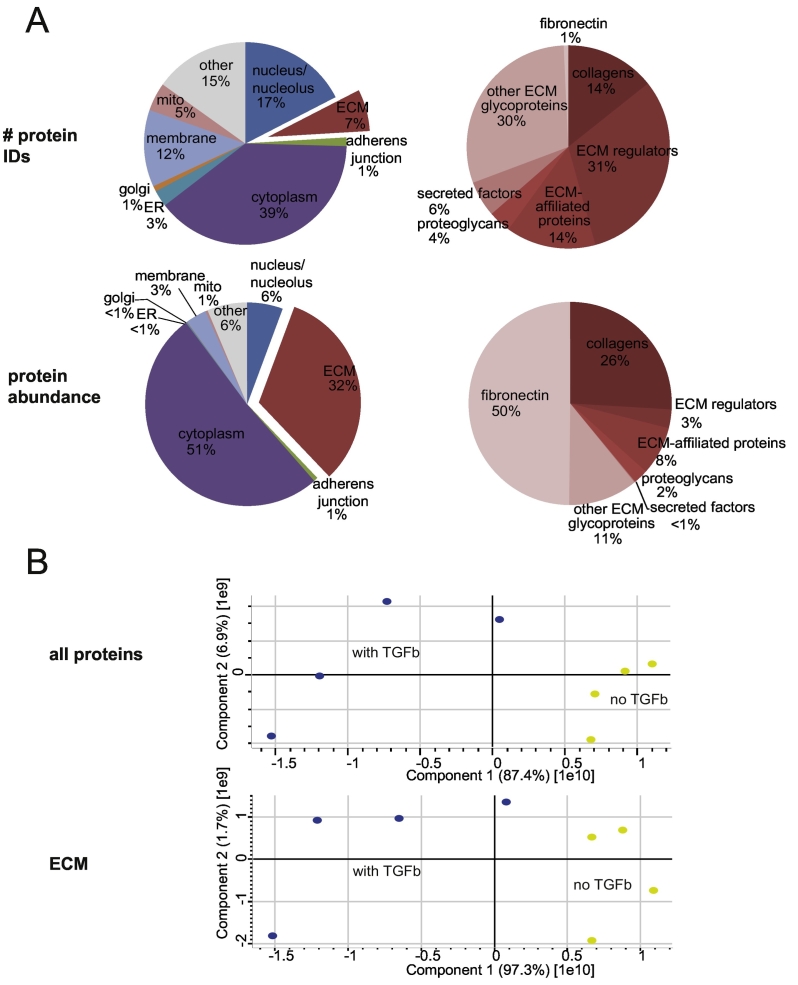
Fig. 2.
(A) Relative contribution of ECM proteins to the total number of protein IDs classified by cellular localization (upper panel, left) and distribution of ECM protein IDs according to matrisome classification [42] (upper panel, right); relative contribution of ECM proteins in terms of total protein abundance (lower panel, left) and distribution of ECM protein abundance according to matrisome classification [42] (lower panel, right). (B) Principal component analysis of control (yellow) and TGF-β1-treated (blue) distinct biological replicates of all protein IDs (top) and ECM proteins subset (bottom)
The proportion of matrisome proteins in comparison to the overall detected protein number, was 7% in terms of individual protein identifications (IDs). However, the proportion rose to 32% when based on summed protein abundances, indicating a significant enrichment of ECM proteins (Fig. 2A). As it has been observed in other studies using detergent-mediated decellularization, cytoplasmic proteins constituted the main part of identified proteins [47]. The predominant presence of cytoskeletal proteins such as myosin and actin suggests that chaotropic action of detergents is not strong enough to disrupt the interactions between the ECM and cellular proteins including plasma membrane integrins and associated cytoskeletal proteins.
Matrisome-associated proteins including ECM regulators such as collagen biosynthetic enzymes as prolyl-4- and lysyl hydroxylases (e.g. P4HA1, P4HA2, PLOD1, PLOD2, PLOD3) and ECM-affiliated proteins like membrane-associated annexins (proteins of the ANXA family, Figs. 2A and 3) were also detected in the ECM produced by primary culture of fibroblasts. A recent study comparing ECM produced by mesenchymal stem cells and fibroblasts in vitro, in which decellularization was followed by ECM protein solubilization with 8 M urea, identified 38 to 45 core matrisome proteins and 16 to 26 matrisome-associated proteins [45], suggesting that our ECM isolation protocol is sufficiently mild and sensitive to allow for the identification of ECM resident proteins even those weakly bound to the ECM core proteins.
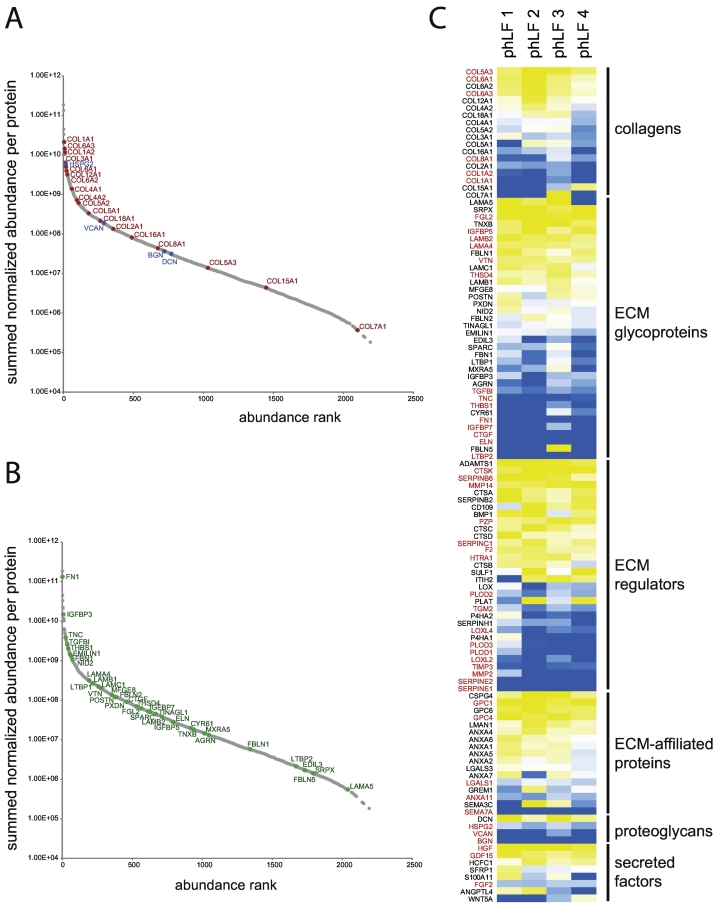
Fig. 3.
Relative protein abundance in IPF fibroblast ECM and their regulation by TGF-β1. (A) Summed normalized protein abundances for collagens and proteoglycans. (B) Summed normalized protein abundances for ECM glycoproteins. (C) Heat map showing fold-change of ECM proteins in response to TGF-β1 treatment. Levels of proteins depicted in red font were significantly changed (p < 0.05).
To characterize the differences between ECM proteins derived from TGF-β1 treated and control ECM samples, we performed principal component analysis (PCA). Fig. 2B shows that principal component 1 (PC1, x-axis), which separates untreated from TGF-β1-treated samples and thus reflects a TGF-β1 signature, was by far the largest source of variability in the data. PC1 accounted for 87% of data variation, when all proteins, and even for 97%, when only ECM proteins were included into the analysis. This observation underlines the particular importance of TGF-β1 in the context of ECM protein expression and demonstrates that the TGF-β1 effect largely overruled other sources of variability, implying that for example donor-derived variability played a negligible role in this data set. In agreement, ECM proteins known to be induced by TGF-β1 in lung fibroblasts, including representatives of core matrisome and matrisome-associated proteins, were consistently upregulated in our study, as e.g. both collagen type I chains (COL1A1, COL1A2) [9,48], fibronectin 1 (FN1) [9], the proteoglycan biglycan (BGN) [[49], [50], [51]], as well as intra- and extracellular ECM regulators like collagen lysyl hydroxylases (PLOD1, PLOD2, PLOD3) [52,53], plasminogen activator inhibitor 1 (PAI1, gene name SERPINE1) [9], and latent-transforming growth factor β -binding protein 2 (LTBP2) [54] (Fig. 3).
2.3. TGF-β1 stimulation modulated IPF fibroblast expression of several collagens, but maintained collagen heterotrimer chain stoichiometries
Primary IPF fibroblasts expressed all major fibrillar collagen alpha chains, namely type I (COL1A1, COL1A2), type II (COL2A1), type III (COL3A1), and type V (COL5A1, COL5A2, COL5A3, Fig. 3). Consistent with being a minor component of collagen fibrils, type V collagen was considerably less abundant than type I and type III collagens (Fig. 3A) [55]. Except for type II collagen, all of these have been reported to be expressed and upregulated by TGF-β1 in lung fibroblasts, including our own studies [9,39,48,56,57]. In contrast, collagen type II is considered a classical cartilage collagen and has not previously been described to be expressed by lung fibroblasts [58]. Furthermore, of all fibrillar collagens, we only observed consistent and significant upregulation of both type I collagen alpha chains (COL1A1 and COL1A2), whereas type III collagen (COL3A1) and the type V collagen alpha chains (COL5A1 and COL5A2) were only upregulated in three or two of the four primary fibroblast lines, respectively (Fig. 3B). At first sight, this is in contrast to our previously published results on type III and V collagen expression and secretion, but may simply reflect lower statistical power of the current analysis, as we have used more fibroblast lines (n = 8) in our previous studies [9,39]. However, it is also conceivable, that upregulation of collagen types III and V protein expression and secretion may not always result in higher deposition of these collagens in the ECM.
Also the beaded-filament-forming type VI collagen belonged to the collagens with highest abundance (Fig. 3A). Type VI collagen is currently emerging as an important regulator of fibroblast migration [38,59]. Interestingly, contrary to type I collagen, both COL6A1 and COL6A3 were significantly downregulated in response to TGF-β1 and COL6A2 showed a similar trend (p = 0.063, Fig. 3B). This is in agreement with our recent study [38] as well as studies on normal skin development and fibrosis which showed that expression of type VI collagen was not coordinated with expression of type I collagen [60]. Our study suggests that TGF-β1 may be the regulatory factor underlying this phenomenon.
Two fibril-associated collagens with interrupted triple helices (FACITs) were detected in IPF fibroblasts, namely type XII (COL12A1) and type XVI collagen (COL16A1). Collagens belonging to FACITs have been described to play important roles in tissue plasticity by binding to the surface of fibrillar collagens, to proteoglycans, and to integrins among other ECM proteins [[61], [62], [63], [64], [65], [66], [67]]. In agreement, type XII collagen has been observed to localize to fibrotic regions in skin and lung, including in IPF [68,69]. However, while TGF-β has been described to upregulate COL12A1 expression in cultured equine tenocytes [70], we did not observe significant regulation in our approach. For COL16A1, moderate to two-fold upregulation by TGF-β1 was evident for three out of four fibroblast lines, just failing significance (p = 0.065). Positive correlations between TGF-β levels and COL16A1 expression in human left ventricle tissue samples from dilated cardiomyopathy patients have been reported, providing indirect support of TGF-β-induced expression of COL16A1 [71]. Furthermore, in the context of inflammatory bowel disease, COL16A1 synthesis is elevated in myofibroblasts and likely increases focal adhesion stability [72]. Our results indicate, that type XVI collagen may also be involved in lung fibroblast migration and tissue remodeling.
While expression of fibrillar and fibril-associated collagens is well-described for lung fibroblasts, the expression of network-forming collagens, with collagen IV being the most prominent representative, is typically attributed to epithelial cells [[73], [74], [75]]. Here, we show that the α1 and α2 chains of collagen IV are expressed in relatively high abundance by IPF fibroblasts, but not significantly altered in response to TGF-β1 (Fig. 3A, B). In contrast, a much less well-studied network-forming collagen, namely collagen VIII, was moderately expressed, but significantly upregulated by TGF-β1 (Fig. 3A, B). Collagen VIII is described as a major component of subendothelial membranes [76,77], but recently, Seet et al. reported that COL8A1 expression is increased in a mouse model of conjunctival fibrosis where collagen VIII colocalizes with type I collagen in scar tissue [78]. Interestingly, the study by Seet et al. evidenced an increase of both collagen VIII chains (COL8A1 and COL8A2) on transcript, but not on protein level, and concluded that collagen VIII may be strongly regulated at the post-translational level. Here, we detected only COL8A1 in the ECM and observed similar upregulation at transcript and protein level (Figs. 3B, 4C), demonstrating that COL8A1 is not strongly regulated at the post-translational level in lung fibroblasts treated with TGF-β1 in vitro. However, as Seet et al. suggested, collagen VIII may still be degraded in the ECM by neutrophil elastase in vivo. To our knowledge, upregulation of COL8A1 by TGF-β1 has not been described previously for fibroblasts, but TGF-β1 has been reported to upregulate COL8A1 in a renal carcinoma cell line [79] and there is supporting circumstantial evidence in the context of dilated cardiomyopathy [71].
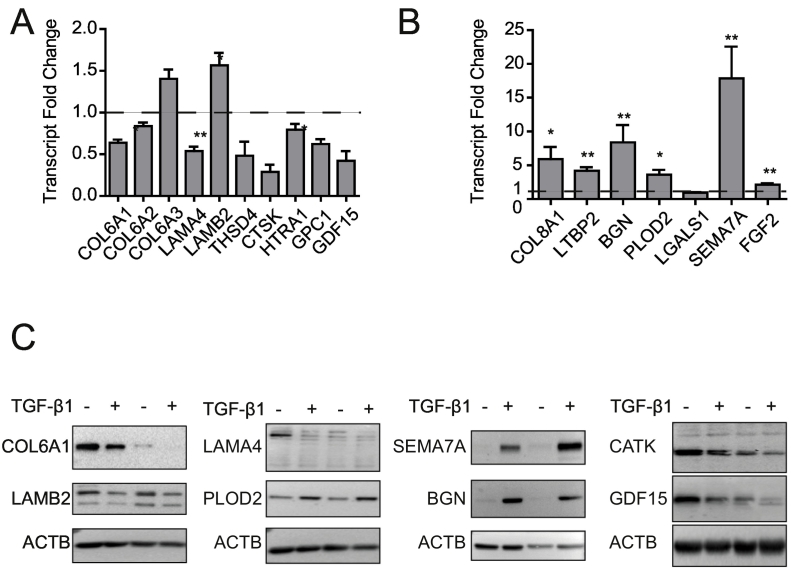
Fig. 4.
Transcriptional and post-translational regulation of ECM and ECM-associated proteins in TGF-β1 treated IPF fibroblasts. Fold-change derived from qRT-PCR analyses of transcripts of proteins quantified as decreased (A) and increased (B) by label-free proteomics profiling of ECM derived from TGF-β1-treated IPF fibroblast (Fig. 3B). Each bar represents the fold-change in TGF-β1-treated samples. Data is plotted as mean ± SD (n = 4 for each transcript) using GraphPad Prism 5. Significant changes are denoted by single asterisk (*p < 0.05) and double asterisks (**p < 0.01) as determined by the two-tailed paired t-test. (C) Western blot analyses of representative ECM and ECM-associated proteins in cell lysates (≈intracellular) from control and TGF-β1-treated IPF fibroblast. Two independently derived primary fibroblast lines are shown.
Also COL18A1, a ubiquitous basement membrane component expressed by many cell types, was detected at relatively high abundance in IPF fibroblast ECM, while we detected type VII (COL7A1) and type XV (COL15A1) collagens at comparatively low abundance (Fig. 3A), the latter a close structural homologue of type XVIII collagen. To our knowledge, the function of these three collagens in the lung is unknown and their expression by lung fibroblasts has not been described previously. Increased levels of COL18A1, including increased plasma levels of endostatin, a proteolytically derived fragment with anti-angiogenic properties, has been linked to kidney fibrosis, both in patient samples as well as in experimental models of kidney disease [[80], [81], [82], [83], [84], [85], [86], [87]]. COL15A1 plays an important role in the organization of fibrillar matrices in the heart [88] and is enriched in interstitial renal fibrosis [89] and the bleomycin-induced model of lung fibrosis [90]. Type VII collagen is established as the major component of anchoring fibrils of the dermal-epidermal adhesion in skin [91,92] and is upregulated in skin of systemic sclerosis patients [93]. COL7A1 and COL15A1 have been reported to be expressed and upregulated by TGF-β1 in human dermal fibroblasts [94,95]. Here, we observed increased levels of COL7A1 and COL15A1 in response to TGF-β1 in the ECM of three out of four IPF-patient derived fibroblast lines, indicating individual differences between patients. In contrast, levels of COL18A1 were largely unchanged (Fig. 3B).
Type I collagen typically consists of two α1(I) chains and one α2(I) chains, but also the homotrimeric form, which is resistant to collagenase-mediated degradation [[96], [97], [98]], has been described in the context of several pathologies including fibrosis [[99], [100], [101]]. Therefore, we assessed collagen chain stoichiometries for type I collagen as well as type IV, V, VI, and VIII collagen, for which the final triple helix can be assembled from two to six different chains. Judging from abundance of these collagens and what is deposited for their heterotrimerization pattern in the UniProt database, our results suggest that the following major collagen trimers are formed by lung fibroblasts: α12α2(I), α12α2(IV), α12α2(V), α1α2α3(VI), α13(VIII). In order to assess whether collagen trimerization is changed upon TGF-β1 treatment, we determined chain stoichiometries for these collagens under both conditions relative to the respective α1 chain and found that these were largely unchanged (Table 1).
Table 1.
Collagen chain stoichiometries for type I, IV, V, and VI collagens.
| Collagen type | Detected chains | Control | TGF-β1 |
|---|---|---|---|
| I | COL1A1 - α1(I) | 1 | 1 |
| COL1A2 - α2(I) | 0.62 ± 0.22 | 0.56 ± 0.15 | |
| IV | COL4A1 - α1(IV) | 1 | 1 |
| COL4A2 - α2(IV) | 0.56 ± 0.22 | 0.48 ± 0.20 | |
| V | COL5A1 - α1(V) | 1 | 1 |
| COL5A2 - α2(V) | 1.77 ± 0.57 | 1.74 ± 0.25 | |
| COL5A3 - α3(V) | 0.04 ± 0.02 | 0.02 ± 0.01 | |
| VI | COL6A1 - α1(VI) | 1 | 1 |
| COL6A2 - α2(VI) | 0.64 ± 0.29 | 0.67 ± 0.19 | |
| COL6A3 - α3(VI) | 0.84 ± 0.37 | 0.89 ± 0.25 |
Chain stoichiometries for type I and IV collagens were in good agreement with the major heterotrimeric form α12α2(I) and α12α2(IV), respectively (Table 1). Surprisingly, for type V collagen we detected more α2 chain relative to α1 chain, which indicates that either other heterotrimeric forms exist than the known α12α2(V) and α1α2α3(V) triple helical molecules [102,103] or that the α2(V) chain engages in hitherto undescribed heterotypic collagen molecules, as has been described for α1(XI) and α2(V) [104]. COL11A1, however, was only detected with one unique peptide (and therefore not included in our analysis), which indicates that α1(XI) is too low-abundant to interact with all α2(V) not engaged with α1(V). For type VI collagen, notably, the comparably low levels of the α2(VI) chain do not agree well with α1α2α3(VI) as the only heterotrimeric form and argue for the additional existence of an α12α3(VI) form and/or of non-triple-helical α1(VI) collagen, which has been described recently [105].
2.4. IPF fibroblasts express a specific selection of laminin chains, which are decreased in response to TGF-β1
Laminins, in particular the chains LAMA4 (α4), LAMA5 (α5), LAMB1 (β1), LAMB2 (β2), and LAMC1 (ɣ1), were abundantly expressed by IPF fibroblasts (Fig. 3C). We predicted the possible heterotrimeric laminin assemblies present in the ECM synthesized by pulmonary fibroblasts using the normalized relative abundance of identified laminin chains depicted in Fig. 3C. Thus, out of the 16 laminin heterotrimers isolated or experimentally predicted thus far [106], our data suggests the following laminin heterotrimers (given in order of decreasing abundance): α4β1ɣ1 (411), α4β2ɣ1 (421), α5β1ɣ1 (511); α5β2ɣ1 (521). As α4 is detected at much higher abundance than α5, we conclude that the α4-containing heterotrimers represent by far the major laminin molecules in IPF fibroblast-derived ECM. Laminins are large molecular weight ECM glycoproteins that are one of the main components of basement membranes where they serve as ligands to cellular receptors, a cell-matrix interaction important for signaling and anchoring of cells [106]. Even though laminins are typically considered to be almost exclusively expressed by epithelial cells, the contributions of other cell types to the pool of laminin chains within the basement membrane have been poorly described. However, consistent with our findings, previous studies have suggested that pulmonary fibroblasts synthesize laminins [107,108]. The synthesis of basement membrane proteins by non-epithelial cells does not seem to be restricted to the lung since studies using skin, gut, and cornea fibroblasts have also shown the synthesis of collagen IV and laminins contributing to basement membrane assembly [[109], [110], [111], [112]].
Interestingly, we observed moderate (up to −1.4-fold) but consistent downregulation by TGF-β1 of all four major laminins, which reached significance for LAMA4 and LAMB2, but just failed to do so for LAMC1 (p = 0.0929) and LAMB1 (p = 0.0628, Fig. 3B). Notably, laminin, in contrast to fibronectin and type I collagen, has been shown to suppress TGF-β1-induced myofibroblast differentiation in 2D culture [113]. Downregulation of laminin levels may thus represent a contributing mechanism by which TGF-β1 drives myofibroblast differentiation.
2.5. TGF-β1 regulates ECM proteins both transcriptionally as well as post-translationally
For a number of selected targets, including representatives of all matrisome compartments, we assessed whether TGF-β1-mediated changes were consistent at both transcript and soluble protein (≈intracellular) levels (Fig. 4). Expression of most regulated matrisome proteins was altered similarly on transcript and soluble protein levels. Regarding the downregulated proteins, two exceptions clearly stood out, namely COL6A3 and LAMB2. Both proteins are components of heterotrimeric protein complexes, namely type VI collagen (mostly α1α2α3) and laminins (e.g. α4β2ɣ1 and α5β2ɣ1), and at least one other component of the same heterotrimer was downregulated on transcript level. For type VI collagen, the 3 chains (COL6A1, COL6A2, and COL6A1) were decreased in our proteomics studies (Fig. 3B). We validated the result for COL6A1 by Western blot (Fig. 4B). However, qRT-PCR data showed a different pattern where COL6A1 and COL6A2 were downregulated, but in contrast COL6A3 was upregulated (Fig. 4A). The inconsistency between proteomics and qRT-PCR data suggests that type VI collagen synthesis is regulated post-transcriptionally. Interestingly, studies in mice have shown that deletion of the gene encoding the α1(VI) chain (COL6A1) results in complete absence of extracellular type VI collagen, suggesting that regulation of type VI collagen chain expression is not coordinated at the transcript level, but rather at the level of triple helix formation and secretion, which depends on the presence of the α1(VI) chain and the correct chain stoichiometry. Similarly, downregulation of only one laminin chain transcript (LAMA4 coding for the alpha-4 chain, Fig. 4A) of the three necessary for the heterotrimer leads to overall lower laminin levels in the soluble protein fraction (Fig. 4A, LAMA4 and LAMB2) and in the ECM (Fig. 3B). Surprisingly, the LAMB2 transcript was upregulated, suggesting that regulation of laminin expression by TGF-β1 is not coordinated on transcript level. As to the upregulated proteins, all of the selected targets except for Galectin 1 (LGALS1), were consistently upregulated on transcript and protein level, with biglycan (BGN) and semaphorin 7A (SEMA7A) showing the highest level of regulation (Fig. 4B, C).
2.6. TGF-β1 altered the expression of novel and known ECM-associated proteins including several crosslinking enzymes
Several of the matrisome proteins identified in this study have been previously described as TGF-β1 targets in human lung fibroblasts, e.g. COL1A1, COL1A2, tenascin C (TNC), fibronectin (FN1), lysyl hydroxylase 1 (PLOD1), lysyl oxidase like-2 (LOXL2), the proteoglycans BGN, VCAN, HSPG2 (also termed perlecan) and fibroblast growth factor 2 (FGF2), validating the overall proteomics approach [9,39,48,56,57,[114], [115], [116], [117], [118]]. However, in addition to COL2A1, COL8A1, and COL16A1 discussed above, our proteomics study identified other novel TGF-β1 targets in the lung fibroblast ECM. For instance, the ECM-affiliated protein ANXA11 has to our knowledge not been described as a TGF-β1 target.
Furthermore, we find that TGF-β1 increases expression of more proteins involved in enzymatic collagen crosslinking than previously described, including lysyl oxidases LOXL2 and LOXL4, lysyl hydroxylases PLOD1, PLOD2, and PLOD3, and, finally transglutaminase 2 (TGM2) in primary human lung fibroblasts. LOXL2 has been described as TGF-β1 target in the context of glaucoma and hepatocellular carcinoma [119,120] and is upregulated in liver [121], heart [122], kidney [123,124] and lung fibrosis [125,126]. Our findings suggest that increased extracellular LOXL2 in lung fibrosis may be derived from lung fibroblasts stimulated by TGF-β1 (Fig. 3B). Surprisingly, we did not detect LOXL1. Judging from the mouse homologues LOXL1 and LOXL2, these lysyl oxidases have very similar detergent solubility profiles [90] and hence, if LOXL1 was expressed by lung fibroblasts on our model, we should have readily detected it with our ECM enrichment protocol. Therefore, this may indicate that increased levels of LOXL1 in IPF, as recently reported by Tjin et al. [127], are generated by another cell type than lung fibroblasts. LOXL4, in contrast, is a comparatively unexplored member of the lysyl oxidase family. Evidence from the literature suggests that LOXL4 contributes to ECM remodeling in cancer and wound healing [128,129] and upregulation of LOXL4 by TGF-β1 has been reported in trabecular meshwork and aortic endothelial cells [120,130]. Here, we provide the first evidence, that LOXL4 also may play a role in fibrotic lung remodeling.
Hydroxylation of lysine residues in collagen chains occurs in the endoplasmic reticulum and is catalyzed by lysyl hydroxylases (LH, also referred to as procollagen-lysine,2-oxoglutarate 5-dioxygenase, PLOD). Hydroxylysines (HyK) play an important role in collagen secretion and assembly as they are substrates for further modification including O-glycosylation and lysyl oxidase (LOX)-mediated crosslinking. Notably, the chemical nature of mature collagen crosslinks (lysyl pyrrole, hydroxylysyl pyrrole, hydroxylysyl pyridinoline, or lysylpyridinoline) is determined by the extent of hydroxylation of the lysines involved in crosslinking [131]. Interestingly, PLOD2 (LH2) is increased in cancer-associated fibroblasts which leads to increased hydroxylation of key lysine residues leading to the formation of more stable pyridinoline-derived crosslinks which are thought to contribute to the increased stiffness of tumor stroma [132]. All three LH (PLOD1, PLOD2, and PLOD3), have been reported to be upregulated in fibroblasts by TGF-β1 on transcript level before [115], a finding which we confirm for PLOD2 (Fig. 4B) and extend for all three LH on protein level (Figs. 3B, 4C).
Finally, we observed that TGM2 is significantly upregulated by TGF-β1 in primary IPF fibroblasts. TGM2 catalyzes the reaction between glutamyl and lysyl residues of substrate proteins both in the cytosol and in the extracellular space, leading to intracellular or extracellular crosslinking of proteins via γ-glutamyl-lysyl crosslinks. In the extracellular space, in contrast to lysyl oxidase activity, TGM2-mediated crosslinking is not restricted to collagens, but has been reported for fibronectin, integrins, and heparan sulfate-bearing proteoglycans such as syndecan-4. Except for a recent study which showed that TGM2 is elevated in chronic obstructive pulmonary disease (COPD) [133], TGM2 has not received much attention in the context of lung pathology. Nevertheless, evidence from other organs suggest an important role in fibrosis: For instance, recently Wen Z. et al. found that TGM2 regulates liver fibrosis via positive feedback regulation of TLR4 signaling [134] and it has been reported that TGF-β1 increases TGM2 in apoptotic mouse thymocytes [135], in keratinocytes and in oral fibroblasts [136].
2.7. TGF-β1 induced matrisome changes indicate both positive and negative feedback mechanisms and highlight the dynamics and complexity of ECM turnover
Highlighting the dynamic nature of TGF-β1 effects on human lung fibroblasts, our results suggest several positive and negative feedback effects on TGF-β1 activity. For instance, upregulation of THBS1 and downregulation of HTRA1 are likely to potentiate TGF-β1 effects: THBS-1 is responsible for much of the activation of latent TGF-β1 in vivo [137,138], and the serine protease HTRA1 inhibits TGF-β1 signaling by cleavage of its receptors [139,140]. In contrast, the drastic upregulation (>10-fold) of latent-transforming growth factor β-binding protein 2 (LTBP2) clearly represents a negative feedback mechanism to increase scavenging of endogenously produced TGF-β1 in presence of abundant active extracellular TGF-β1. Also downregulation of thrombin (F2), which has been shown to stimulate TGF-β secretion in cultured human epithelial cells [141], may contribute to decreased synthesis of endogenous TGF-β1.
In addition to increased crosslinking of collagen and other ECM proteins discussed above, changes in expression levels of several ECM-degrading proteins indicate a profound effect of TGF-β1 on ECM turnover. For instance, we show that cathepsin K, the most efficient protease for the degradation of fibrillar collagen [142,143], is significantly decreased by TGF-β1 in human lung fibroblasts (Fig. 3, Fig. 4). Furthermore, increased SERPINE1 (plasminogen activator inhibitor 1, PAI-1) blocks plasmin-dependent activation of MMPs and thus protects ECM proteins from proteolytic degradation [144]. Matrix metalloproteinase 2 (MMP2), also increased in our study, degrades elastin and the collagen types IV and V, all three ECM proteins abundantly expressed by human lung fibroblasts in the present study (Fig. 3) [145]. In contrast, MMP14, which was significantly decreased, degrades fibronectin (FN1), tenascin C (TNC), perlecan (HSPG2), type I and type III collagen [146], again all ECM proteins detected in abundance in our approach. In addition, simultaneous upregulation of TIMP3, a matrix metalloproteinase inhibitor, adds another level of complexity to altered fibroblast ECM turnover in response to TGF-β1. Overall, the results indicate that TGF-β1 drives a substantial part of the impaired protease/anti-protease balance in pulmonary fibrosis [147].
2.8. ECM changes in the described in vitro model show significant correlation with ECM changes in human lung fibrosis
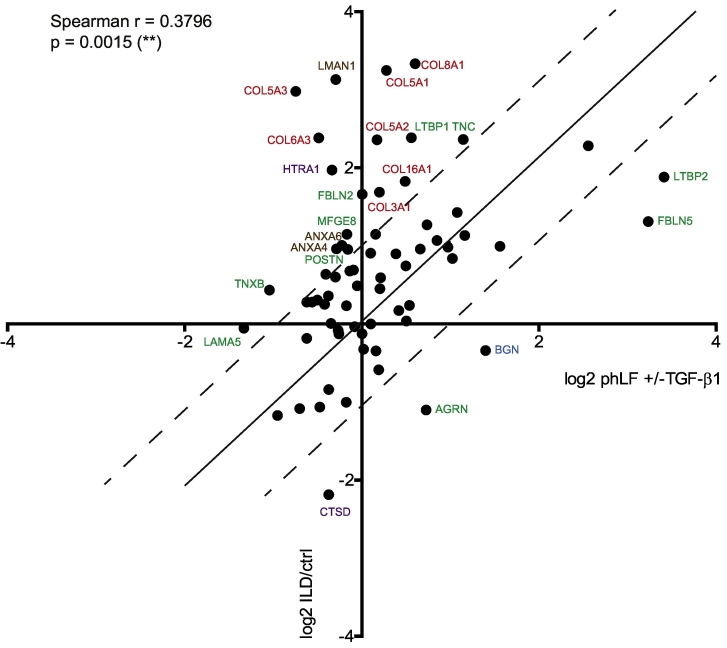
A recent study by Rosmark et al. elegantly demonstrated that lung fibroblast production of some ECM proteins, in particular proteoglycans, differs when cells are cultured on native decellularized tissue scaffolds as compared to conventional monolayer culture [148]. Even if the aim of this study was not to explore ECM changes in the context of fibrosis, the reported findings point out that the use of a two-dimensional fibroblast culture model may not recapitulate all ECM changes observed in vivo in the context of a native scaffold. Therefore, we investigated this issue by performing a correlation analysis of ECM changes between our in vitro results from cultured fibroblasts and publicly available data set from human lung fibrosis reported by Schiller et al. [149]. In the latter, a proteomic approach was used to compare several subtypes of interstitial lung disease (ILD) including IPF to healthy control tissue. Similar to our study, all samples used in this report represented end-stage lung fibrosis and had also been obtained from the CPC bioArchive. The authors used quantitative detergent-based solubility profiling to fractionate proteins according to their solubility. Hence, for correlation, we directly compared our guanidine-solubilized data set with the corresponding insoluble protein data set enriched for ECM components and found a moderate (Spearman r = 0.38), but significant correlation (Fig. 5). Notably, for decorin (DCN) and versican (VCAN), two proteoglycans expression of which was found to depend on native scaffolds [148], the observed effects in our TGF-β1-driven in vitro model of lung fibrosis reflected the changes in clinical lung fibrosis well (DCN expression not significantly altered, VCAN expression significantly increased, see Suppl. Table S3). These results confirm our approach based on the concept that TGF-β1-driven changes of fibroblast-produced ECM describe a significant part of overall ECM changes in lung fibrosis. Nevertheless, we found a number of proteins without correlation or even anti-correlation (Fig. 5). There may be several reasons for the observed outliers. First, addition of TGF-β1 alone may not be sufficient to induce some of the ECM components in lung fibroblasts. For instance, COL5A1 and COL5A2 are not significantly changed in our in vitro model, but substantially altered in ILD. As it has been shown that collagen V secretion is additionally regulated by platelet-derived growth factor (PDGF) signaling, lack of PDGF in our culture system may explain some of the discrepancies observed [150]. Second, in human ILD, other cell types than fibroblasts may be responsible for the observed ECM changes. For example, in the normal lung, fibulin-2 (FBLN2) and lactadherin (MFGE8) are both mainly expressed by epithelial cells [151,152]. Third, the discrepancies may also result from the different extraction protocols for ECM components. However, similar correlation analysis between ECM data from the murine model for lung fibrosis [90] and human lung fibrosis [149] showed that there was no significant correlation between these two in vivo derived data sets, even though the ECM extraction protocols used were identical (Supplementary Fig. S2B). Notably, these results also demonstrate that, in terms of ECM changes, our in vitro model correlates better with human lung fibrosis data than the most commonly used mouse model.
Fig. 5.
Correlation analysis of the here presented data set with a publicly available ECM data set from human lung fibrosis samples. Names are given for proteins that deviate by a log2 ratio > 1 from the perfect correlation. Collagens are given in red, ECM glycoproteins in green, proteoglycans in blue, ECM regulators in purple, ECM-affiliated proteins in brown, and secreted factors in black.
The ECM data derived from the mouse model of bleomycin-induced lung fibrosis neither correlated significantly with our in vitro model, nor with human ILD (Supplementary Fig. S2). It is highly likely that this reflects the transient nature of bleomycin-induced lung fibrosis in the mouse where the fibrotic response peaks two weeks after administration of bleomycin followed by at least partial resolution of fibrosis within the subsequent six weeks [9,90]. Interestingly, many striking discrepancies observed in the mouse model correspond to ECM regulators like metalloproteinases (MMP14, ADAMTS1), cathepsins (CTSA, CTSB, CTSD) and extracellular sulfatase SULF1 (Supplementary Fig. S2). Expression of these ECM components may therefore protect from further progression or contribute to resolution of lung fibrosis in the mouse model.
2.9. Global PTM characterization of ECM collagens identifies novel prolyl-3-hydroxylation and lysyl glycosylation sites
Collagen PTMs such as lysine and proline hydroxylation as well as lysine glycosylation have been shown to play an important role in fibril assembly, cell-matrix interactions and tissue development. Collagen PTM characterization by common mass spectrometry-based proteomics workflows is challenging because enzymatic digestion generates long peptide sequences harboring a high number of different PTMs present at different levels of occupancy. We have previously developed an optimized bioinformatics platform to characterize PTMs of purified collagen IV [21]. However, characterization and quantitation of collagen PTMs in rather crude ECM preparations (without any biochemical purification) has remained a daunting task. Here, we have implemented an optimized high-resolution global MS workflow to characterize the PTMs of all the different collagens present in ECM produced by the primary cultures of lung fibroblasts isolated from IPF patients (Fig. 1, Fig. 6).
Fig. 6.
Bioinformatics pipeline for the global identification and quantitation of site-specific PTM in collagens. Thermo .raw MS/MS files were initially searched with Myrimatch to identify the proteins present in the fibroblast ECM. The subset of identified proteins were used as database to perform a second search in which sequence motifs were defined for the identification of site-specific PTM in collagens present in ECM. The resulting *.pep.XML output files from Myrimatch were parsed by PeptideProphet to compute the probability score (0–1) for each peptide spectrum match (PSM). The PeptideProphet parsed .pep.XML output file was imported into Skyline along with all the raw MS/MS files in *.mz.XML format (converted from *.RAW by MSConvert) to generate the spectral library (.blib). This spectral library was used for the targeted extraction of all the PTM modified and unmodified peptide species for each specific site. MS1 area was computed for each peptide for different samples from Skyline.
For the initial global characterization of site-specific hydroxylation and glycosylation of proline and lysines in the different collagen types identified in the tryptic digests of the ECM produced by IPF fibroblasts, we took advantage of the unique ‘motif search feature’ embedded within MyriMatch to search the MS data generated by the Q-Exactive HF mass spectrometer [21]. We have identified a total of 18 different collagen chains from the lung fibroblast ECM (Fig. 3). Table 2 summarizes all the identified sites of modification in 15 different alpha chains (out of 18) including 69 3-HyP, 42 HyK, 49 glucosylgalactosyl-hydroxylysine (GG-HyK) and 4 galactosyl-hydroxylysine (G-HyK) sites. Notably, site-specific PTMs were identified in COL1A1 as well as COL5A3 which differ in as much as 3 order of magnitude, showcasing the wide dynamic range capabilities and high sensitivity of our workflow (Fig. 3A and Table 2).
Table 2.
Mass spectrometry-based identification of site-specific hydroxylation and glycosylation of different collagens identified in the ECM of human lung fibroblast.
| Collagen chain | 3-HyP Sites (Gly-HyP-HyP) | Glucosylgalactosyl-hydroxylysine | Galactosyl-hydroxylysine | Hydroxylysine |
|---|---|---|---|---|
| COL1A1 | P426, P555, P567, P690, P771,P885, P894, P897, P1119, P1122, P1164, P1179 | K265, K277, K448, K520, K586, K862 | K277, K448, K520, K586 | K265, K277, K286, K352, K397, K442, K505, K538, K586, K742, K781, K826, K862, K934 |
| COL1A2 | P101, P419, P593, P782, P797, P806, P809, P902 | K189 | – | K189, K198, K354, K498, K621, K846 |
| COL2A1 | P1096 | – | – | K608, K848 |
| COL3A1 | P940, P1147, P1162 | K584, K662, K740, K743, K779, K932 | – | K108, K251, K350, K461, K503, K584, K629, K779, K860, K923 |
| COL4A1 | P372, P478, P647, P786, P803, P1424, P1436 | K295, K298, K322, K343, K361, K497, K527 | – | – |
| COL4A2 | P197, P424, P733 | K225, K409, K453, K456, K883, K1106, K1175, K1247, K1373 | – | K597, K907 |
| COL5A1 | P500, P516, P575, P599, P761, P770, P779, P833, P992, P995, P1322 | K582, K774, K1038, K1074, K1311 | – | K963, K1125, K1326, K1473 |
| COL5A2 | P136, P409, P442, P448, P658, P919, P1177, P1198 | K320, K386, K554, K785 | – | K431, K860 |
| COL5A3 | P576, P1113 | K502, K1132 | – | – |
| COL6A1 | P582, P588 | K546 | – | – |
| COL6A2 | – | K545 | – | – |
| COL6A3 | – | K2052, K2103, K2170, K2209, K2212 | – | K2121 |
| COL12A1 | P2775, P2831, P2837, P2855, P2858, P2976, P3022, P3025 | – | – | – |
| COL16A1 | P1402 | K658, K1116 | – | K1245 |
| COL18A1 | P862, P868, P871, P907, P910 | – | – | – |
Although collagen V is a minor component of heterotypical collagen fibrils, comprehensive maps of proline and lysine hydroxylation have been generated by mass spectrometry analyses of purified bovine COL5A1 and COL5A2 chains [153]. As shown in Table 2, our analyses not only identified a large number of prolyl-3-hydroxylation sites in human COL5A1 and COL5A2 chains, but also new 3-hydroxyproline (3-HyP576, 3-HyP1113) and glucosyl-galactosyl-hydroxylysine (GG-HyK502 and GG-HyK1132) sites in COL5A3.
We also identified 3 sites (P940, P1147, and P1162) of prolyl-3-hydroxylation in COL3A1. Interestingly, 3-hydroxylation of evolutionarily conserved P1162 was previously reported in many mammals, but not in human COL3A1 [154]. We detected 3-HyP in this site, indicating not only superior sensitivity of our mass spectrometry method but also evidencing that prolyl-3-hydroxylation of this site is evolutionarily conserved.
Although collagen IV is not the major collagen type expressed by lung fibroblasts, our analyses yielded similar results for sites of 3-HyP and GG-HyK to those reported in our previous studies with the purified proteins [21]. Furthermore, many other new sites of prolyl-3-hydroxylation were identified for COL6A1, COL12A1, COL16A1 and COL18A1 (Table 2), indicating that our analyses of ECM preparations present an advantage because these insoluble collagens are not easy to purify and thus unlikely to be analyzed by common proteomics platforms. Taken together, these results show that our proteomics platform enabled the simultaneous identification and mapping of site-specific PTM in many different collagens found in fibroblast ECM.
2.10. Comprehensive mapping of PTMs in COL1A1 from the ECM produced by IPF fibroblasts
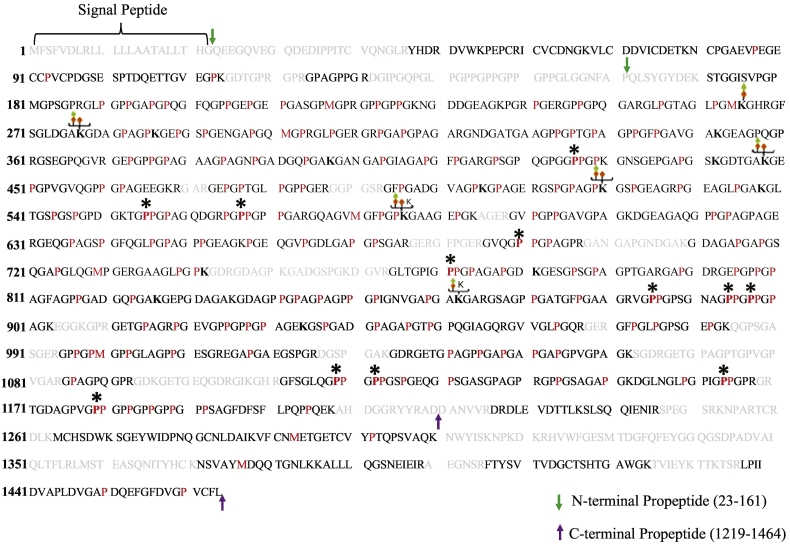
To evaluate the sensitivity and specificity of our global MS workflow for mapping and quantifying collagens PTMs from the unfractionated ECM, we focussed on COL1A1 as a model protein because it is the most abundant component of lung fibroblast ECM. In addition, type I collagen, purified from different tissues and species, has been extensively characterized by classical biochemical and mass spectrometry methods [33,[154], [155], [156], [157], [158], [159]]. In Fig. 7 we present a comprehensive map of prolyl/lysyl-hydroxylation and lysyl-glycosylation of COL1A1 produced by our global analysis of ECM generated by primary human lung fibroblasts. Our optimized bioinformatics workflow resulted in ~85% sequence coverage for processed COL1A1 molecule. Interestingly, a few peptides from the N- and C-terminal propeptides were also detected (Fig. 7), evidencing the dynamics of the collagen fibril assembly process. A total of 109 4-HyP sites were mapped to the Yaa position of the classical 4-hydroxylation motif (Gly-Xaa-HyP) within COL1A1. In sum, our results indicate that about 95% of the proline residues present in the Yaa position of “Gly-Xaa-Yaa” motif in processed COL1A1 molecule are 4-hydroxylated, as evidenced by previous Edman microsequencing analyses [160]. Although the total number of prolyl-4-hydroxylation sites in control (n = 106) and TGF-β1 treated (n = 106) lung fibroblasts was identical, a small number of these (<3%) were mapped to specific sites within the COL1A1 of each sample group.
Fig. 7.
Comprehensive map of hydroxylation and O-linked glycosylation sites in human COL1A1. Lung fibroblast ECM was used to generate Collagen I PTM maps. MS identified collagen peptide sequences are shown in black which together represent an overall sequence coverage of 85% for the processed form of COL1A1. Signal peptide encompassing amino acid residues 1–22 are indicated. The green and purple arrows delimit the N-terminal (23–161) and C-terminal (1219–1464) pro-peptide sequences, respectively. Sequences not identified in this study are colored gray. “P” indicates 4-hydroxyproline occurring in the Yaa position of Gly-Xaa-Yaa motif, and a bold “P*” indicates 3-hydroxyproline in the Xaa position of Gly-Xaa-HyP motif. In addition, hydroxylation in red “P” residues occurring in the Xaa position followed by either Ala, Val, Met, Arg, Asp, or Glu in the Yaa position of Gly-Xaa-Yaa motif is reported but cannot be defined as either 3- or 4-HyP solely on the current MS/MS strategy [21]. Green and orange diamonds denote glucosyl and galactosyl sugar moieties attached to hydroxylysine (bold “K”). A summary of the PTMs is presented in Table 2, and PSMs for O-glycosylated lysine and 3-hydroxyproline sites are provided in Supplementary Figs. S3–S19. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In addition to 4-HyP, we also investigated the occurrence of 3-HyP sites which have also been considered to provide structural stability to collagen molecules and affect binding affinities to other ECM proteins [22,27,28]. We identified a total of 12 hydroxylated prolines (P426, P555, P567, P690, P771, P885, P894, P897, P1119, P1122, P1164, P1179) in the Xaa position of the classical (Gly-HyP-HyP) motif (Fig. 8A and B, Supplementary Figs. S3–S9). We assigned these as 3-hydroxyprolines (3-HyP) as previous experimental evidence has shown that they are hydroxylated in the third position of the 5-membered nitrogen containing ring [160]. Interestingly, nine (P426, P555, P567, P690, P771, P894, P897, P1119, P1122) of these 3-HyP sites (manually inspected as shown in Supplementary Figs. S3–S9), have not been reported for human COL1A1. Our analyses detected hydroxylation of P885 (referred to as P707 for processed COL1A1), P1164 (referred to as P986 for processed COL1A1) and the C-terminal (GPP)n cluster to be 3-hydroxylated in COL1A1 [155,156,161]. The signal to noise ratio of the mass spectra corresponding to this modified peptide was very low and we have identified only P1179 to be 3-hydroxylated in the entire (GPP)n cluster. It has been shown that the occurrence of 3-prolyl hydroxylation in the COL1A1 (GPP)n cluster is tissue specific [155] and it seems that COL1A1 produced by human fibroblasts lacks or displays a very low level of 3-prolyl-hydroxylation in this cluster.
Fig. 8.
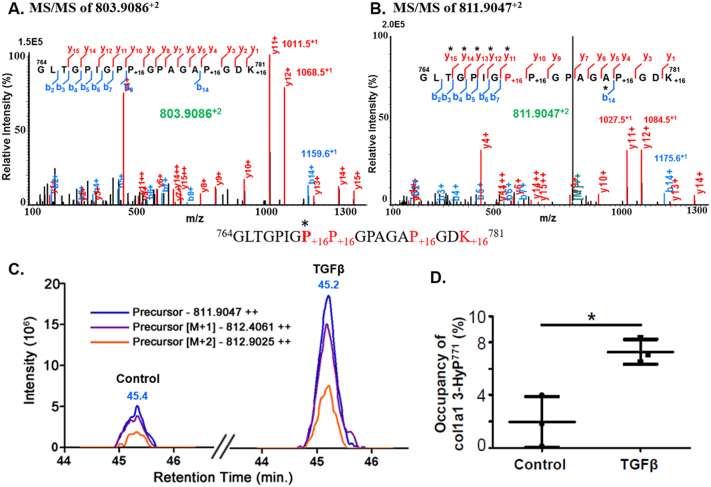
Quantitation of site-specific hydroxylation occupancy of P771 in COL1A1 from control and TGF-β1 treated human lung fibroblasts. (A, B) MS/MS spectra of peptide 764GLTGPIGPP+16GPAGAP+16GDK+16781 in which Pro771 is either unmodified (m/z 803.9086+2) or 3-hydroxylated (m/z 811.9047+2). (C) Skyline MS1 quantitation showing the extracted ion chromatograms for the 3-HyP771-containing peptide 764GLTGPIGP+16P+16GPAGAP+16GDK+16781 from the ECM digest of control and TGF-β1 treated human fibroblasts. (D) Determination of 3-hydroxylation occupancy (%) for P771 calculated from the relative abundances of unmodified and 3-hydroxylated peptides obtained by Skyline MS1 quantitation of control and TGF-β1 samples. MS1 area for 3-HyP771 modified peptide was normalized by dividing the area of modified peptide to the summed area of all the unmodified and modified peptide “764GLTGPIGPPGPAGAPGDK781” species. The occupancy (%) of 3-HyP771 in COL1A1 was increased about 3.7 fold in TGF-β1 treated fibroblast ECM (Table 3). Each dot and square represents the measurement of 3-HyP771 occupancy in each control and TGF-β treated samples. Data is plotted as mean ± SEM (n = 3 in each group) using GraphPad Prism 5. The increased hydroxylation occupancy for Pro771 in TGF-β1 treated samples was significant (*p < 0.05, two-tailed unpaired ‘t’-test).
2.11. Site-specific quantitation of prolyl-3-hydroxylation and lysine glycosylation in COL1A1
Recent studies have shown that mutations in members of the prolyl 3-hydroxylase family such as LEPRE1 (P3H1) and LEPREL2 (P3H3) are associated with osteogenesis imperfecta which results in absence of 3-prolyl-hydroxylation at a single site (P1164) of COL1A1 [30,162]. Thus, quantifying the occupancy (fraction of COL1A1 molecules containing 3-HyP at a specific site) of different 3-HyP sites holds premier importance in health and disease. Here, we have optimized and validated an MS1 based quantitative pipeline for the relative quantitation of post-translationally modified peptides between two or more different conditions across different samples (Fig. 5). The results showed that with the exception of one 3-HyP site, overall occupancies are <10% and did not change significantly upon TGF-β1 treatment (Table 3). Notably, the occupancy of 3-HyP771 site significantly increased 3.7 fold in the TGF-β1 treated samples (Fig. 9, Table 3). While the biological significance of this finding remains unclear, it is conceivable that increased prolyl-3-hydroxylation of this site may contribute to proteolytic resistance of collagen fibrils. In analogy to the mechanism of triple-helix stabilization by 4-HyP [34], the additional hydroxyl group in 3-HyP771 may favor the formation of a water bridge with a carbonyl group of an adjacent triple-helixal molecule. Furthermore, increased 3-hydroxylation of Pro771 may also alter specific collagen interactions with other ECM proteins or collagen receptors on adherent cells. Interestingly, although the 3-HyP1164 site (referred to as P986 for processed COL1A1) has been previously shown to be fully hydroxylated in COL1A1 from mouse skin fibroblasts [162], our results indicate that this 3-HyP site is hydroxylated at a level of ~43% in both control and TGF-β1 treated human lung fibroblast ECM, suggesting that the occupancy of this site could vary in a tissue- as well as species-specific manner.
Table 3.
Quantitative occupancy (%) of 3-hydroxyproline (3-HyP) sites identified in COL1A1 from lung fibroblast ECM. Results are expressed as mean ± SEM (n = 3 in each group). Significant statistical differences were estimated by two-tailed unpaired ‘t’-test (*p < 0.05, ns > 0.05).
| COL1A1 3-HyP site1 |
Occupancy (%) of 3-HyP sites |
|
|---|---|---|
| Control | TGF-β1 | |
| P426 | 1.5 ± 0.5 | 2.8 ± 0.8ns |
| P555,567 | 3.0 ± 1.5 | 1.6 ± 0.19ns |
| P690 | 1.6 ± 0.6 | 1.2 ± 0.08ns |
| P771 | 1.9 ± 1.1 | 7.2 ± 0.5* |
| P885,894,897 | 0.6 ± 0.4 | 0.8 ± 0.4ns |
| P1119,1122 | 9.4 ± 4.9 | 11.9 ± 1.2ns |
| P1164 | 42.7 ± 11 | 41.4 ± 2.2ns |
Numbering of collagen alpha 1 chain (I) corresponds to that of the translated protein product starting from the signal peptide.
Fig. 9.
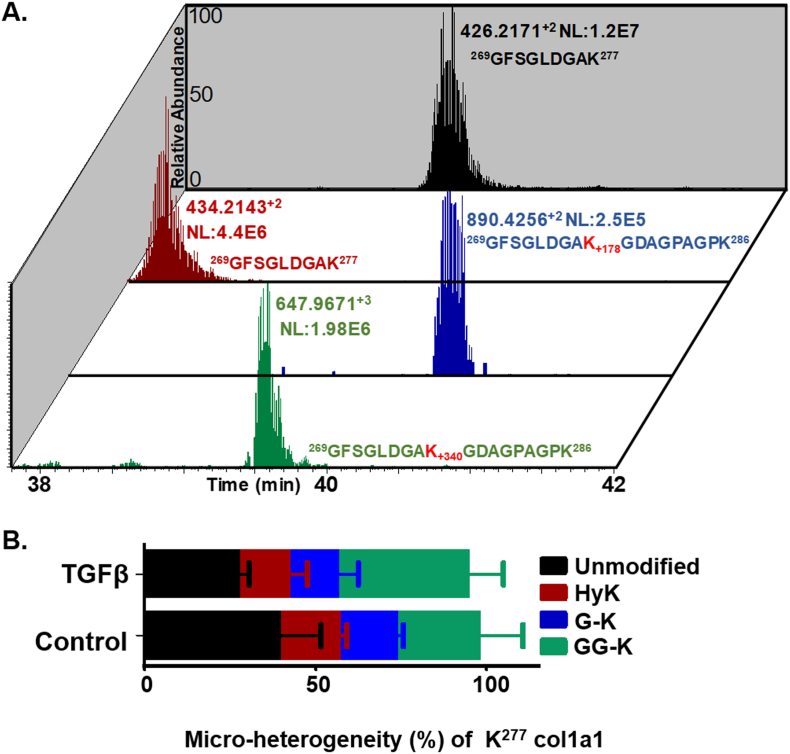
Skyline quantitation of micro-heterogeneity of the K277 site in COL1A1 from human lung fibroblasts. (A) Chromatographic elution profiles of unmodified (K), hydroxylysine (HyK), galactosyl-hydroxylysine (G-HyK), and glucosylgalactosyl-hydroxylysine (GG-HyK) modified (K277) peptide in human COL1A1 from lung fibroblast ECM digests. (B) Graphical representation of Skyline quantitation of micro-heterogenic distribution of unmodified K277 (black), HyK277 (maroon), G-HyK277 (blue) and GG-HyK277 (green) species in COL1A1 from the ECM digest of control and TGF-β1 treated human fibroblasts. The different colors in bar represent the occupancy of different forms at K277 site in COL1A1 with mean ± SEM (n = 3 in each group) (see Table 4).
In addition to 3-HyP found within the classical “Gly-3HyP-4HyP” motif, we also identified 37 prolyl-hydroxylation sites at the ‘Xaa’ position of “Gly-HyP-[Ala/Met/Ser/Gln/Lys/Thr/Iso]” motif in COL1A1. Some of these exemptions including “Gly-HyP-[Ala/Val/Gln/HyK]” have already been reported by mass spectrometry studies of type II, IV and V collagens [21,153,163]. Our results expand the list of exemptions to the classical Gly-3HyP-4HyP motif and further confirm that hydroxylation of Pro residues in the Xaa position is not as restricted to the presence of a 4HyP in the Yaa position as previously thought.
Glycosylation has been shown to be important for many structural and functional properties of collagen molecules including stability, proteolytic degradation [157], fibril formation [164], affinity to cell-surface receptors [[165], [166], [167]], and collagen crosslinking [24,158,159]. We have successfully employed our global PTM analyses platform to quantify microheterogeneity of the O-glycosylation sites in COL1A1. For the purpose of our analyses, we define microheterogeneity as the distribution of occupancy between the different possible modification states of lysine residues including unmodified lysine (K), hydroxylated lysine (HyK), galactosyl-hydroxylysine (G-HyK) and glucosylgalactosyl-hydroxylysine (GG-HyK) at any given specific site. Overall our analyses revealed a total of 14 HyK sites, 9 of which did not carry any of the 2 possible glycosylation moieties (Table 2). We also identified a total of 6 O-glycosylation sites in COL1A1, where 4 of these (K277, K448, K520, K586) were heterogenous containing either galactosyl or glucosylgalactosyl moieties (Table 4, Supplementary Figs. S10–S19).
Table 4.
Quantitative micro-heterogenic occupancy (%) of O-glycosylated lysine sites identified in COL1A1 from lung fibroblast ECM. Results are expressed as mean ± SEM (n = 3 in each group). Statistically significant differences were estimated by two-tailed unpaired ‘t’-test (*p < 0.05, ns > 0.05). N/D, not detected; HyK, hydroxylysine; G-HyK, galactosylhydroxy-lysine; GG-HyK, glucosylgalactosylhydroxylysine.
| COL1A1 Lysine site1 |
Modification | Micro-heterogeneity (%) |
|
|---|---|---|---|
| Control | TGF-β1 | ||
| K277 | K | 39.7 ± 8.2 | 29.8 ± 3.4ns |
| HyK | 17.9 ± 1.1 | 12.1 ± 2.2ns | |
| G-HyK | 16.7 ± 0.8 | 14.2 ± 3.9ns | |
| GG-HyK | 25.5 ± 7.2 | 43.7 ± 6.6ns | |
| K448 | K | N/D | N/D |
| HyK | N/D | N/D | |
| G-HyK | 30.5 ± 11.6 | 31 ± 0.8ns | |
| GG-HyK | 69.5 ± 11.5 | 69 ± 0.8ns | |
| K520 | K | N/D | N/D |
| HyK | N/D | N/D | |
| G-HyK | 56.5 ± 21 | 34 ± 9.5ns | |
| GG-HyK | 43.5 ± 21 | 66 ± 9.5ns | |
| K586 | K | 28 ± 9.3 | 13.6 ± 7.1ns |
| HyK | 3.6 ± 1.1 | 1.1 ± 0.17 ns | |
| G-HyK | 29.4 ± 7.4 | 29.5 ± 6.7ns | |
| GG-HyK | 38.9 ± 12.5 | 55.7 ± 13.3ns | |
Numbering of collagen alpha 1 chain (I) corresponds to that of the translated protein product starting from the signal peptide.
Since our proteomic analyses showed upregulation of PLOD1, PLOD2 and PLOD3 in primary lung fibroblasts upon TGF-β treatment, we investigated the microheterogeneity of the glycosylation sites in COL1A1, by integrating Skyline MS1 level quantitation to our database search results obtained with MyriMatch (Table 4). Fig. 8 shows an example of the chromatographic elution profile of the different peptides taken into account to determine microheterogeneity at K277 site. As expected the hydroxylysine containing peptide 269GFSGLDGAHyK277 elutes almost 2 min before the unmodified version of the peptide (269GFSGLDGAK277). Similarly, the glycosylated forms of HyK277 (G-HyK and GG-HyK) are not cleaved by trypsin and thus are found in longer peptide sequence (269GFSGLDGAKGDAGPAGPK286). As expected the peptide containing GG-HyK277 elutes from the C18 column before the G-HyK277 containing peptide. Using Skyline to quantify the areas of each peptide, we determined the microheterogeneity of K277 in both control and TGF-β1 treated samples. Interestingly, our analyses revealed a reduction in hydroxylation and galactosylation of K277 levels upon TGF-β1 treatment, which correlates with a corresponding increase of glucosylation of K277 (GG-K277). Using this strategy, we also found K448 and K520 to be completely (100%) glycosylated. The microheterogenic distribution of glucosylation of K520 and K586 showed an increasing trend upon TGF-β1 treatment, but they were not statistically significant (Table 4). Such microheterogenic differences for these specific sites may play important role in the progression and development of fibrosis.
2.12. Conclusions
In conclusion, we report the first comprehensive proteomic analysis of TGF-β1-induced changes in lung fibroblast-synthesized ECM. The results correlate significantly with ECM changes found in human lung fibrosis and reveal profound and highly dynamic effects of TGF-β1 on ECM composition and structure. Analysis of collagen PTMs in crude ECM preparations from cultured fibroblasts serves as a proof of concept demonstrating that our proteomics platform is sensitive, specific and yields comparable or better results than those obtained with purified collagen preparations. In addition, our results from global identification and quantification of collagen revealed novel sites of prolyl hydroxylation and lysyl glycosylation. The occupancy of one of these sites was shown to be regulated by TGF-β1 and it will be interesting to investigate the impact of this modification in the rate of collagen deposition in fibrosis. Because purification of collagens is not required, our platform will facilitate the quantitation of site specific collagen modifications to address key questions about their role in development and progression of fibrotic disease.
3. Experimental procedures
3.1. Human lung material, isolation and culture of primary human lung fibroblasts
The study was carried out with four biological replicates. Primary human lung fibroblasts (phLF) were isolated from human lung explant material, representing end-stage disease, of four IPF patients obtained from the BioArchive CPC-M for lung diseases at the Comprehensive Pneumology Center (CPC Munich, Germany). Patients' characteristics were as follows: Three men, one woman; mean age, 55 ± 7; mean forced expiratory volume in 1 s (FEV1), 2.0 ± 0.9 l; mean forced vital capacity (FVC), 56 ± 21% FVCpredicted; mean diffusing capacity of the lungs for carbon monoxide (DLCO), 2.5 ± 0.3 mmol CO∗min−1∗kPa−1. The study was approved by the local ethics committee of the Ludwig-Maximilians University of Munich, Germany, and all participants had given written informed consent. Isolation and culture of phLF was performed as described previously [9,39].
3.2. Treatment of phLF with TGF-β1
Cells were seeded at a density of 20,000–25,000 cells/cm2. Twenty-four hours later starvation in Dulbecco's modified Eagle medium/F-12 including 0.5% fetal bovine serum and 0.1 mM 2-phospho-l-ascorbic acid was performed for 24 h. Then, cells were treated with 2 ng/ml TGF-β1 (R&D Systems, Minneapolis, MN) in starvation medium for another 48 h, followed by harvesting for RNA and protein analysis. Unless stated otherwise, all data is derived from four independent experiments using independently derived fibroblast lines.
We chose a TGF-β1 concentration of 2 ng/ml because our previous studies demonstrated that this concentration is suitable to mimic a myofibroblast phenotype in vitro [9,38,39]. Indeed, 2 ng/ml TGF-β1 reproducibly and robustly induces a myofibroblast phenotype and increases collagen synthesis and secretion in primary human lung fibroblasts (see Supplementary Fig. 1A–D).
3.3. RNA isolation and Real-Time Quantitative Reverse-Transcriptase PCR (qRT-PCR) analysis
RNA isolation and qRT-PCR analysis was performed as described previously [9,39]. Primers were obtained from MWG Eurofins (Ebersberg, Germany) and are given in Supplementary Table S1.
3.4. Western blot analyses
Western Blot analysis were performed essentially as described previously [9,39]: Cells were scraped into Radio-Immunoprecipitation Assay (RIPA) buffer (50 mM Tris HCl pH 7.4, 150 mM NaCl, 1% Triton X100, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS) containing a protease inhibitor and a phosphatase inhibitor cocktail (both Roche), incubated 30 min, followed by centrifugation for 20 min at 13,000 rpm and 4 °C to clarify the lysates and obtain the ECM pellet used for proteomics analyses. Protein concentration of the supernatant was determined using the Pierce BCA Protein Assay (Thermo Fisher Scientific; Waltham, USA). Samples were denatured in Laemmli buffer (65 mM Tris-HCl pH 6.8, 10% glycerol, 2% SDS, 0.01% bromophenol blue, 100 mM DTT) and proteins resolved by SDS-PAGE. Proteins were then transferred to polyvinylidene difluoride (PVDF) membranes. Nonspecific binding to membrane was blocked with 5% milk in TBS-T (0.1% Tween 20, TBS). Membranes were shortly rinsed and washed three times for 5 min in TBS-T and incubated with primary antibody (see Supplementary Table S2 for details on the specific antibodies) overnight at 4 °C. After washing with TBS-T, membranes were incubated with secondary antibody for 1 h at room temperature. Blots were rinsed with TBS-T and visualized with the enhanced chemiluminescence (ECL) system (Thermo Fisher Scientific, Waltham, MA, USA) and analyzed with the ChemiDocXRS+ imaging system (Bio-Rad, Munich, Germany).
Primary antibodies used in this study are given in Supplementary Table S2. HRP-linked and fluorescent labeled secondary antibodies were purchased from GE Healthcare Life Sciences (Freiburg, Germany).
3.5. Denaturation, reduction and alkylation of ECM proteins
For each of the four IPF fibroblast lines used, the ECM pellets from three confluent wells of 6-well culture plates were solubilized in 6 M guanidinium-hydrochloride, 100 mM Tris/HCl, pH 8.5 and denatured by heating at 100 °C for 10 min, disulfide bridges in ECM proteins were reduced by adding dithiothreitol to a final concentration of 25 mM and incubation at 56 °C for 30 min. Sulfhydryl groups were alkylated by adding iodoacetamide to a final concentration of 50 mM and incubated for 45 min at RT in the dark. Alkylated ECM proteins were precipitated with 9 parts of ice-cold ethanol at −20 °C overnight. Precipitated ECM proteins were collected by centrifugation for 30 min at 4 °C. ECM pellet was resuspended in 50 mM ammonium bicarbonate, pH 7.8 before proteolytic digestions.
3.6. LysC-trypsin digestion
ECM proteins were digested in-solution with LysC (Wako Chemicals, USA) at 1:20 ratio for 4 h at 37 °C prior to addition of trypsin (Promega) at 1:20 ratio at 37 °C for overnight (18 h). After that, additional trypsin was added at 1:50 ratio and further incubated at 37 °C for 4 h. Trypsin was deactivated by heating at 100 °C for 10 min. Samples were acidified with trifluoroacetic acid and stored at −20 °C before mass spectrometry analyses.
3.7. Mass spectrometry
LC-MS/MS analysis was performed as described previously on a Q-Exactive HF mass spectrometer online coupled to an Ultimate 3000 nano-RSLC (Thermo Scientific) [168]. Briefly, approximately 0.5 μg of ECM protein sample was loaded onto the trap column and after 5 min eluted and separated on the C18 analytical column (custom made Acquity UPLC M-class HSST3 column, 1.8 μm particles, 25 cm length, Waters) by a 90 min non-linear acetonitrile gradient. MS spectra were recorded at a resolution of 60,000 with an automatic gain control (AGC) target of 3 × 106 and a maximum injection time of 50 ms from 300 to 1500 m/z. From the MS scan, the 10 most abundant peptide ions were selected for fragmentation via higher-energy collisional dissociation with a normalized collision energy of 27, an isolation window of 1.6 m/z, and a dynamic exclusion of 30 s. MS/MS spectra were recorded at a resolution of 15,000 with an AGC target of 105 and a maximum injection time of 50 ms. Intensity threshold was set to 1 × 104 and unassigned charges, and ions with charges of +1 and >+8 were excluded.
3.8. Label-free proteomic analysis
The acquired spectra were loaded to the Progenesis QI proteomics software (version 3.0, Nonlinear Dynamics) for label-free peptide quantification and analyzed as described previously [[168], [169], [170]]. Total ion current (TIC) were used to normalize control versus TGF-β1-treated data sets reflecting the general difference in ECM deposition per cell line. On average, the normalization values used were 1.29 ± 0.08, indicating that in total there is only about 29% more ECM deposition with TGF-β1 and analyzing the raw abundances yielded similar results (not shown). Peptide identification was done with the Mascot search engine (version 2.5.1) in the Swissprot human protein database (Release 2017_02, 11,329,970 residues, 20,194 sequences). Search parameters used were: 10 ppm peptide mass tolerance and 20 mmu fragment mass tolerance, one missed cleavage allowed, carbamidomethylation of cysteine was set as fixed modification, methionine and proline oxidation and asparagine or glutamine deamidation were allowed as variable modifications. A Mascot-integrated decoy database search calculated a false discovery rate of 0.47% when the search was performed using Mascot percolator score and a significance threshold of 0.05. Peptide assignments were reimported into the Progenesis QI software, and the abundances of all unique peptides allocated to each protein were summed up. Resulting normalized protein abundances were used for calculation of fold-changes and a repeated-measures ANOVA.
3.9. Protein classification
Protein classification was done with Matrisome annotator for ECM proteins [171] and with DAVID for non-ECM proteins [172].
3.10. Stoichiometry calculations
Stoichiometries of collagen chains were calculated using the iBAQ method by dividing summed normalized intensities per protein per sample by the number of observable peptides for each protein [[173], [174], [175]].
3.11. Correlation analysis with in vivo lung fibrosis data sets
Protein abundance ratios and significance values of the insoluble fractions for the mouse model of bleomycin-induced lung fibrosis (day 14) were extracted from Schiller et al. [90]. For correlation with human lung fibrosis, protein abundance values from Schiller et al. [149] (interstitial lung disease patients vs. healthy controls) were used for calculation of fold changes and significance values by unpaired t-test after imputation of missing values in Perseus software [176]. Ratios calculated with imputed values are given in Supplementary Table S3, but the corresponding log2 ratios were omitted from the correlation analysis to avoid imputation-based errors.
3.12. Analysis of collagen PTMs
Thermo (.raw) files were independently searched by MyriMatch algorithm (located at: https://svn.code.sf.net/p/proteowizard/code/trunk/pwiz/pwiz_tools/Bumbershoot/myrimatch/doc/index.html) [177] as described previously [21]. Files were initially searched against a human database (Uniprot, 20,232 entries), which also included the reverse sequences for each protein to compute the false discovery rate with a maximum of two missed cleavages with a minimum peptide length of 5 residues and a maximum length of 35 residues. The search was configured to identify peptides with fixed carbamidomethylation of cysteine (+57.021) and variable oxidation of methionine (+15.9949). Precursor and fragment mass tolerances were set to 10 and 20 ppm, respectively. The .pepXML files obtained from each search were imported into IDPicker (version 3.1) for the parsimonious grouping of identified proteins at a 0% protein global FDR [178]. A subset fasta database was exported from the identified proteins through IDPicker. The .raw files were further searched against this subset FASTA database allowing for a maximum of three missed cleavages and expanding the search space with the addition of dynamic modifications (proline hydroxylation, lysine glycosylation, etc.) up to 10 per peptide. We employed a very unique feature of MyriMatch, which allows the use of a limited set of regular expressions to consider dynamic PTMs linked to primary structure peptide motifs found in collagens. We have considered ‘Yaa’ to be a 4-HyP site in the ‘Gly-Xaa-Yaa’ collagenous sequence. Only in the presence of 4-HyP site in ‘Yaa’ position, it is likely that prolyl 3-hydroxylation can occur at the ‘Xaa’ position. For lysine hydroxylation and subsequent glycosylation ‘Gly-Xaa-K’ motif was considered. To estimate FDRs, each sequence of the database was reversed and concatenated to the database and <1% FDR was considered for PSM, peptide and protein inferences using IDPicker. Candidate peptides were required to feature trypsin cleavages at both ends. Resulting PSMs were manually inspected, and MS2 spectra were examined to confirm correct assignment of PTMs. In addition, known peptides of COL1A1 containing tissue specific 3-HyP cluster near the c-terminal were searched using slight changes in the search strategy by increasing the maximum peptide length from 35 to 50 and including maximum 15 dynamic modifications per peptide. Because the length of tryptic peptides generated in this region were >35 and possibly harbor >10 PTMs, further search space expansion strategy was undertaken.
3.13. In-house pipeline for relative quantitation of collagen PTMs
To quantitate the post-translationally modified peptides from COL1A1 we have plugged in open-source softwares for proteomics analyses with our in-house PTM detection workflow. PeptideProphet, a part of the trans-proteomic tandem pipeline, was used to assign peptide to spectrum matches (PSM) and correlating probability scores of post translationally modified peptides identified with the MyriMatch search algorithm. To do this, initially the MyriMatch output file extension was renamed from “.pepXML” to “.pep.XML” for proper compatibility with the PeptideProphet software. As the MyriMatch search was performed against a database concocted with reverse decoy sequences, accurate mass and non-parametric modeling was employed. The resulting .pep.XML file and converted .mzXML from Thermo MS .RAW files were used for construction of the spectral library in Skyline (Daily-64 bit, 4.1.1.18118) with a cut-off score of 0.95 using DDA with MS1 filtering workflow. Full-scan settings were configured using the Orbitrap mass analyzer with a resolving power of 60,000 at 400 m/z including 3 isotope peaks with precursor charge ranging from +2 to +7. Only scans with 1 min of MS/MS ID's were used for retention time filtering. Maximum of two missed cleavages were considered in the peptide settings tab of Skyline before importing the raw MS/MS data files in .mzXML format (Fig. 7). Tryptic peptides containing proline or lysine residues, their unmodified, hydroxylated and/or glycosylated forms were identified by IDPicker with manual inspection of the MS/MS spectra. Skyline was used for targeted extraction of 3-HyP and/or the various forms of glycosylated hydroxylysine containing peptide species. MS1 peak area was computed from Skyline for different peptide species containing the site of modification. The relative quantitation (% occupancy of 3-HyP sites and micro- heterogeneity of glycosylated lysine sites) were calculated by dividing the total ion current calculated for each species by the sum of the ion current of all observed species containing that particular modified site [179]. Comparison of different 3-HyP and glycosylated lysine sites occurring in COL1A1 between control and TGF-β1 treated samples was performed using two-tailed unpaired ‘t’ test by GraphPad Prism 5.
The following are the supplementary data related to this article.
Supplementary material: Supplementary Tables S1 and S2; Supplementary Figures S1-S19
Table of quantified ECM proteins with respective average ratios withTGFb/noTGFb and significance values, in comparison to ratios and significance values Bleo/PBS and ILD/control extracted from Schiller et al. 2015 and 2017.
Ratios >1 are given in blue, ratios <1 are given in yellow. Significance values <0.05 are given in red. ILD/control ratios obtained after imputation of missing values are marked by +. Proteins that could not be quantified in the respective data set are marked with ND.
Acknowledgments
Acknowledgements
We thank Dr. David Tabb (Stellenbosch University, SA), Dr. David Shteynberg and the entire Skyline team (Institute for Systems Biology, Seattle, WA) for their valuable inputs in order to build PTM quantitation pipeline at different times using Myrimatch, Peptideprophet, and Skyline. We are also grateful to Herbert Schiller for help with the mouse and human fibrosis ECM data sets and valuable discussions, and Elisabeth Hennen and Fabian Gruhn for excellent technical assistance. We gratefully acknowledge the provision of human biomaterial and clinical data from the CPC-M bioArchive and its partners at the Asklepios Biobank Gauting, the Klinikum der Universität München, and the Ludwig-Maximilians-Universität München.
This work was supported in part by the Friedrich-Baur-Stiftung (Reg.-Nr. 51/16, grant to CASW), the Deutsche Forschungsgemeinschaft (DFG) within the Research Training Group DFG GRK2338, the Helmholtz Association, the German Center for Lung Research (DZL), and by NIH grants DK099467, DK103067 and DK065138. M.A. is supported by T32DK007569.
Author contributions statement
C.A.S.-W. and R.M.V. conceived and designed the study. J.M.-P., T.B., L.K., D.R., and M.A., performed experiments and acquired data. J.M.-P., T.B., J.B., S.E., O.E., S.M.H., R.M.V. and C.A.S.-W. analyzed and interpreted the data. J.M.-P., T.B., R.M.V. and C.A.S.-W. prepared the manuscript for submission. All authors have read and approved the final manuscript.
Contributor Information
Juliane Merl-Pham, Email: juliane.merl@helmholtz-muenchen.de.
Trayambak Basak, Email: trayambak.basak@vanderbilt.edu.
Larissa Knüppel, Email: larissa.knueppel@helmholtz-muenchen.de.
Deepak Ramanujam, Email: deepak.ramanujam@tum.de.
Mark Athanason, Email: mark.athanason@vanderbilt.edu.
Jürgen Behr, Email: juergen.behr@med.uni-muenchen.de.
Stefan Engelhardt, Email: stefan.engelhardt@tum.de.
Oliver Eickelberg, Email: oliver.eickelberg@ucdenver.edu.
Stefanie M. Hauck, Email: hauck@helmholtz-muenchen.de.
Roberto Vanacore, Email: roberto.vanacore@vanderbilt.edu.
Claudia A. Staab-Weijnitz, Email: staab-weijnitz@helmholtz-muenchen.de.
References
- 1.Wynn T.A., Ramalingam T.R. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nature Medicine. 2012;18(7):1028–1040. doi: 10.1038/nm.2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. Zeisberg, R. Kalluri, Cellular mechanisms of tissue fibrosis. 1. Common and organ-specific mechanisms associated with tissue fibrosis, Am. J. Physiol. Cell Physiol. 304(3) (2013) C216–25. [DOI] [PMC free article] [PubMed]
- 3.Kim D.S., Collard H.R., King T.E., Jr. Classification and natural history of the idiopathic interstitial pneumonias. Proceedings of the American Thoracic Society. 2006;3(4):285–292. doi: 10.1513/pats.200601-005TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatler A.L., Jenkins G. TGF-beta activation and lung fibrosis. Proceedings of the American Thoracic Society. 2012;9(3):130–136. doi: 10.1513/pats.201201-003AW. [DOI] [PubMed] [Google Scholar]
- 5.D. Sheppard, Epithelial-mesenchymal interactions in fibrosis and repair. Transforming growth factor-beta activation by epithelial cells and fibroblasts, Ann. Am. Thorac. Soc. 12 Suppl 1 (2015) S21–3. [DOI] [PMC free article] [PubMed]
- 6.Henderson N.C., Arnold T.D., Katamura Y., Giacomini M.M., Rodriguez J.D., McCarty J.H., Pellicoro A., Raschperger E., Betsholtz C., Ruminski P.G., Griggs D.W., Prinsen M.J., Maher J.J., Iredale J.P., Lacy-Hulbert A., Adams R.H., Sheppard D. Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nature Medicine. 2013;19(12):1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borges F.T., Melo S.A., Ozdemir B.C., Kato N., Revuelta I., Miller C.A., Gattone V.H., 2nd, LeBleu V.S., Kalluri R. TGF-beta1-containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. Journal of the American Society of Nephrology. 2013;24(3):385–392. doi: 10.1681/ASN.2012101031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aschner Y., Downey G.P. Transforming growth factor-beta: master regulator of the respiratory system in health and disease. American Journal of Respiratory Cell and Molecular Biology. 2016;54(5):647–655. doi: 10.1165/rcmb.2015-0391TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staab-Weijnitz C.A., Fernandez I.E., Knüppel L., Maul J., Heinzelmann K., Juan-Guardela B.M., Hennen E., Preissler G., Winter H., Neurohr C., Hatz R., Lindner M., Behr J., Kaminski N., Eickelberg O. FK506-binding protein 10 is a potential novel drug target for idiopathic pulmonary fibrosis. American Journal of Respiratory and Critical Care Medicine. 2015;192(4):455–467. doi: 10.1164/rccm.201412-2233OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Györfi A.H., Matei A.E., Distler J.H.W. Targeting TGF-beta signaling for the treatment of fibrosis. Matrix Biology. 2018;68-69:8–27. doi: 10.1016/j.matbio.2017.12.016. [DOI] [PubMed] [Google Scholar]
- 11.Parker M.W., Rossi D., Peterson M., Smith K., Sikstrom K., White E.S., Connett J.E., Henke C.A., Larsson O., Bitterman P.B. Fibrotic extracellular matrix activates a profibrotic positive feedback loop. The Journal of Clinical Investigation. 2014;124(4):1622–1635. doi: 10.1172/JCI71386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Y. Zhou, J.C. Horowitz, A. Naba, N. Ambalavanan, K. Atabai, J. Balestrini, P.B. Bitterman, R.A. Corley, B.S. Ding, A.J. Engler, K.C. Hansen, J.S. Hagood, F. Kheradmand, Q.S. Lin, E. Neptune, L. Niklason, L.A. Ortiz, W.C. Parks, D.J. Tschumperlin, E.S. White, H.A. Chapman, V.J. Thannickal, Extracellular matrix in lung development, homeostasis and disease, Matrix Biol. 73(77–104) (2018). [DOI] [PMC free article] [PubMed]
- 13.Walraven M., Hinz B. Therapeutic approaches to control tissue repair and fibrosis: extracellular matrix as a game changer. Matrix Biology. 2018;71-72:205–224. doi: 10.1016/j.matbio.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 14.Hewlett J.C., Kropski J.A., Blackwell T.S. Idiopathic pulmonary fibrosis: epithelial-mesenchymal interactions and emerging therapeutic targets. Matrix Biology. 2018;71-72:112–127. doi: 10.1016/j.matbio.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu B.H., Madison M.C., Corry D., Kheradmand F. Matrix remodeling in chronic lung diseases. Matrix Biology. 2018;73:52–63. doi: 10.1016/j.matbio.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beachley V.Z., Wolf M.T., Sadtler K., Manda S.S., Jacobs H., Blatchley M.R., Bader J.S., Pandey A., Pardoll D., Elisseeff J.H. Tissue matrix arrays for high-throughput screening and systems analysis of cell function. Nature Methods. 2015;12(12):1197–1204. doi: 10.1038/nmeth.3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Selman, T.E. King, A. Pardo, S. American Thoracic, S. European Respiratory, P. American College of Chest, Idiopathic pulmonary fibrosis: prevailing and evolving hypotheses about its pathogenesis and implications for therapy, Ann. Intern. Med. 134(2) (2001) 136–51. [DOI] [PubMed]
- 18.M. Selman, A. Pardo, Revealing the pathogenic and aging-related mechanisms of the enigmatic idiopathic pulmonary fibrosis. an integral model, Am. J. Respir. Crit. Care Med. 189(10) (2014) 1161–72. [DOI] [PubMed]
- 19.Wolters P.J., Collard H.R., Jones K.D. Pathogenesis of idiopathic pulmonary fibrosis. Annual Review of Pathology. 2014;9:157–179. doi: 10.1146/annurev-pathol-012513-104706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishikawa Y., Bachinger H.P. A molecular ensemble in the rER for procollagen maturation. Biochimica et Biophysica Acta. 2013;1833(11):2479–2491. doi: 10.1016/j.bbamcr.2013.04.008. [DOI] [PubMed] [Google Scholar]
- 21.Basak T., Vega-Montoto L., Zimmerman L.J., Tabb D.L., Hudson B.G., Vanacore R.M. Comprehensive characterization of glycosylation and hydroxylation of basement membrane collagen IV by high-resolution mass spectrometry. Journal of Proteome Research. 2016;15(1):245–258. doi: 10.1021/acs.jproteome.5b00767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.E. Pokidysheva, S. Boudko, J. Vranka, K. Zientek, K. Maddox, M. Moser, R. Fassler, J. Ware, H.P. Bachinger, Biological role of prolyl 3-hydroxylation in type IV collagen, Proceedings of the National Academy of Sciences of the United States of America 111(1) (2014) 161–6. [DOI] [PMC free article] [PubMed]
- 23.Liao H., Zakhaleva J., Chen W. Cells and tissue interactions with glycated collagen and their relevance to delayed diabetic wound healing. Biomaterials. 2009;30(9):1689–1696. doi: 10.1016/j.biomaterials.2008.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jürgensen H.J., Madsen D.H., Ingvarsen S., Melander M.C., Gardsvoll H., Patthy L., Engelholm L.H., Behrendt N. A novel functional role of collagen glycosylation: interaction with the endocytic collagen receptor uparap/ENDO180. The Journal of Biological Chemistry. 2011;286(37):32736–32748. doi: 10.1074/jbc.M111.266692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ryan B.J., Nissim A., Winyard P.G. Oxidative post-translational modifications and their involvement in the pathogenesis of autoimmune diseases. Redox Biology. 2014;2:715–724. doi: 10.1016/j.redox.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bartling B., Desole M., Rohrbach S., Silber R.E., Simm A. Age-associated changes of extracellular matrix collagen impair lung cancer cell migration. The FASEB Journal. 2009;23(5):1510–1520. doi: 10.1096/fj.08-122648. [DOI] [PubMed] [Google Scholar]
- 27.Grafe I., Yang T., Alexander S., Homan E.P., Lietman C., Jiang M.M., Bertin T., Munivez E., Chen Y., Dawson B., Ishikawa Y., Weis M.A., Sampath T.K., Ambrose C., Eyre D., Bachinger H.P., Lee B. Excessive transforming growth factor-beta signaling is a common mechanism in osteogenesis imperfecta. Nature Medicine. 2014;20(6):670–675. doi: 10.1038/nm.3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery N.T., Zientek K.D., Pokidysheva E.N., Bachinger H.P. Post-translational modification of type IV collagen with 3-hydroxyproline affects its interactions with glycoprotein VI and nidogens 1 and 2. The Journal of Biological Chemistry. 2018;293(16):5987–5999. doi: 10.1074/jbc.RA117.000406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldridge D., Schwarze U., Morello R., Lennington J., Bertin T.K., Pace J.M., Pepin M.G., Weis M., Eyre D.R., Walsh J., Lambert D., Green A., Robinson H., Michelson M., Houge G., Lindman C., Martin J., Ward J., Lemyre E., Mitchell J.J., Krakow D., Rimoin D.L., Cohn D.H., Byers P.H., Lee B. CRTAP and LEPRE1 mutations in recessive osteogenesis imperfecta. Human Mutation. 2008;29(12):1435–1442. doi: 10.1002/humu.20799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cabral W.A., Chang W., Barnes A.M., Weis M., Scott M.A., Leikin S., Makareeva E., Kuznetsova N.V., Rosenbaum K.N., Tifft C.J., Bulas D.I., Kozma C., Smith P.A., Eyre D.R., Marini J.C. Prolyl 3-hydroxylase 1 deficiency causes a recessive metabolic bone disorder resembling lethal/severe osteogenesis imperfecta. Nature Genetics. 2007;39(3):359–365. doi: 10.1038/ng1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mordechai S., Gradstein L., Pasanen A., Ofir R., El Amour K., Levy J., Belfair N., Lifshitz T., Joshua S., Narkis G., Elbedour K., Myllyharju J., Birk O.S. High myopia caused by a mutation in LEPREL1, encoding prolyl 3-hydroxylase 2. American Journal of Human Genetics. 2011;89(3):438–445. doi: 10.1016/j.ajhg.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo H., Tong P., Peng Y., Wang T., Liu Y., Chen J., Li Y., Tian Q., Hu Y., Zheng Y., Xiao L., Xiong W., Pan Q., Hu Z., Xia K. Homozygous loss-of-function mutation of the LEPREL1 gene causes severe non-syndromic high myopia with early-onset cataract. Clinical Genetics. 2014;86(6):575–579. doi: 10.1111/cge.12309. [DOI] [PubMed] [Google Scholar]
- 33.Hudson D.M., Weis M., Rai J., Joeng K.S., Dimori M., Lee B.H., Morello R., Eyre D.R. P3h3-null and Sc65-null mice phenocopy the collagen lysine under-hydroxylation and cross-linking abnormality of Ehlers-Danlos syndrome type VIA. The Journal of Biological Chemistry. 2017;292(9):3877–3887. doi: 10.1074/jbc.M116.762245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hudson D.M., Eyre D.R. Collagen prolyl 3-hydroxylation: a major role for a minor post-translational modification? Connective Tissue Research. 2013;54(4–5):245–251. doi: 10.3109/03008207.2013.800867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z.R. Predict collagen hydroxyproline sites using support vector machines. Journal of Computational Biology. 2009;16(5):691–702. doi: 10.1089/cmb.2008.0167. [DOI] [PubMed] [Google Scholar]
- 36.Ricard-Blum S., Baffet G., Theret N. Molecular and tissue alterations of collagens in fibrosis. Matrix Biology. 2018;68-69:122–149. doi: 10.1016/j.matbio.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 37.Yu G., Ibarra G.H., Kaminski N. Fibrosis: lessons from OMICS analyses of the human lung. Matrix Biology. 2018;68-69:422–434. doi: 10.1016/j.matbio.2018.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knüppel L., Heinzelmann K., Lindner M., Hatz R., Behr J., Eickelberg O., Staab-Weijnitz C.A. FK506-binding protein 10 (FKBP10) regulates lung fibroblast migration via collagen VI synthesis. Respiratory Research. 2018;19(1):67. doi: 10.1186/s12931-018-0768-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.L. Knüppel, Y. Ishikawa, M. Aichler, K. Heinzelmann, R. Hatz, J. Behr, A. Walch, H.P. Bächinger, O. Eickelberg, C.A. Staab-Weijnitz, A novel antifibrotic mechanism of nintedanib and pirfenidone. Inhibition of collagen fibril assembly, Am. J. Respir. Cell Mol. Biol. 57(1) (2017) 77–90. [DOI] [PubMed]
- 40.Montgomery R.L., Yu G., Latimer P.A., Stack C., Robinson K., Dalby C.M., Kaminski N., van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Molecular Medicine. 2014;6(10):1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.L. Cushing, P.P. Kuang, J. Qian, F. Shao, J. Wu, F. Little, V.J. Thannickal, W.V. Cardoso, J. Lu, miR-29 is a major regulator of genes associated with pulmonary fibrosis, Am. J. Respir. Cell Mol. Biol. 45(2) (2011) 287–94. [DOI] [PMC free article] [PubMed]
- 42.Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., Hynes R.O. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Molecular & Cellular Proteomics. 2012;11(4):M111–014647. doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma L., Gan C., Huang Y., Wang Y., Luo G., Wu J. Comparative proteomic analysis of extracellular matrix proteins secreted by hypertrophic scar with normal skin fibroblasts. Burns & Trauma. 2014;2(2):76–83. doi: 10.4103/2321-3868.130191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang K.E., Kwon J., Rhim J.H., Choi J.S., Kim S.I., Lee S.H., Park J., Jang I.S. Differential expression of extracellular matrix proteins in senescent and young human fibroblasts: a comparative proteomics and microarray study. Molecules and Cells. 2011;32(1):99–106. doi: 10.1007/s10059-011-0064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ragelle H., Naba A., Larson B.L., Zhou F., Prijic M., Whittaker C.A., Del Rosario A., Langer R., Hynes R.O., Anderson D.G. Comprehensive proteomic characterization of stem cell-derived extracellular matrices. Biomaterials. 2017;128:147–159. doi: 10.1016/j.biomaterials.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pflieger D., Chabane S., Gaillard O., Bernard B.A., Ducoroy P., Rossier J., Vinh J. Comparative proteomic analysis of extracellular matrix proteins secreted by two types of skin fibroblasts. Proteomics. 2006;6(21):5868–5879. doi: 10.1002/pmic.200402040. [DOI] [PubMed] [Google Scholar]
- 47.Lennon R., Byron A., Humphries J.D., Randles M.J., Carisey A., Murphy S., Knight D., Brenchley P.E., Zent R., Humphries M.J. Global analysis reveals the complexity of the human glomerular extracellular matrix. Journal of the American Society of Nephrology. 2014;25(5):939–951. doi: 10.1681/ASN.2013030233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kurundkar A.R., Kurundkar D., Rangarajan S., Locy M.L., Zhou Y., Liu R.M., Zmijewski J., Thannickal V.J. The matricellular protein CCN1 enhances TGF-beta1/SMAD3-dependent profibrotic signaling in fibroblasts and contributes to fibrogenic responses to lung injury. The FASEB Journal. 2016;30(6):2135–2150. doi: 10.1096/fj.201500173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Andersson-Sjoland A., Thiman L., Nihlberg K., Hallgren O., Rolandsson S., Skog I., Mared L., Hansson L., Eriksson L., Bjermer L., Westergren-Thorsson G. Fibroblast phenotypes and their activity are changed in the wound healing process after lung transplantation. The Journal of Heart and Lung Transplantation. 2011;30(8):945–954. doi: 10.1016/j.healun.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Todorova L., Bjermer L., Westergren-Thorsson G., Miller-Larsson A. TGFbeta-induced matrix production by bronchial fibroblasts in asthma: budesonide and formoterol effects. Respiratory Medicine. 2011;105(9):1296–1307. doi: 10.1016/j.rmed.2011.03.020. [DOI] [PubMed] [Google Scholar]
- 51.Venkatesan N., Roughley P.J., Ludwig M.S. Proteoglycan expression in bleomycin lung fibroblasts: role of transforming growth factor-beta(1) and interferon-gamma. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2002;283(4):L806–L814. doi: 10.1152/ajplung.00061.2002. [DOI] [PubMed] [Google Scholar]
- 52.Remst D.F., Blaney Davidson E.N., Vitters E.L., R.A. Bank, van den Berg W.B., van der Kraan P.M. TGF-ss induces Lysyl hydroxylase 2b in human synovial osteoarthritic fibroblasts through ALK5 signaling. Cell and Tissue Research. 2014;355(1):163–171. doi: 10.1007/s00441-013-1740-5. [DOI] [PubMed] [Google Scholar]
- 53.Knippenberg M., Helder M.N., Doulabi B.Z., R.A. Bank, Wuisman P.I., Klein-Nulend J. Differential effects of bone morphogenetic protein-2 and transforming growth factor-beta1 on gene expression of collagen-modifying enzymes in human adipose tissue-derived mesenchymal stem cells. Tissue Engineering. 2009;15(8):2213–2225. doi: 10.1089/ten.tea.2007.0184. [DOI] [PubMed] [Google Scholar]
- 54.Ahmed W., Kucich U., Abrams W., Bashir M., Rosenbloom J., Segade F., Mecham R., Rosenbloom J. Signaling pathway by which TGF-beta1 increases expression of latent TGF-beta binding protein-2 at the transcriptional level. Connective Tissue Research. 1998;37(3–4):263–276. doi: 10.3109/03008209809002444. [DOI] [PubMed] [Google Scholar]
- 55.Wenstrup R.J., Florer J.B., Brunskill E.W., Bell S.M., Chervoneva I., Birk D.E. Type V collagen controls the initiation of collagen fibril assembly. The Journal of Biological Chemistry. 2004;279(51):53331–53337. doi: 10.1074/jbc.M409622200. [DOI] [PubMed] [Google Scholar]
- 56.Mia M.M., Boersema M., R.A. Bank Interleukin-1beta attenuates myofibroblast formation and extracellular matrix production in dermal and lung fibroblasts exposed to transforming growth factor-beta1. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0091559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mak K.M., Png C.Y., Lee D.J. Type V collagen in health, disease, and fibrosis. The Anatomical Record. 2016;299(5):613–629. doi: 10.1002/ar.23330. [DOI] [PubMed] [Google Scholar]
- 58.Kadler K.E., Baldock C., Bella J., Boot-Handford R.P. Collagens at a glance. Journal of Cell Science. 2007;120(Pt 12):1955–1958. doi: 10.1242/jcs.03453. [DOI] [PubMed] [Google Scholar]
- 59.Theocharidis G., Drymoussi Z., Kao A.P., Barber A.H., Lee D.A., Braun K.M., Connelly J.T. Type VI collagen regulates dermal matrix assembly and fibroblast motility. The Journal of Investigative Dermatology. 2016;136(1):74–83. doi: 10.1038/JID.2015.352. [DOI] [PubMed] [Google Scholar]
- 60.Pablos J.L., Everett E.T., Harley R., LeRoy E.C., Norris J.S. Transforming growth factor-beta 1 and collagen gene expression during postnatal skin development and fibrosis in the tight-skin mouse. Laboratory Investigation. 1995;72(6):670–678. [PubMed] [Google Scholar]
- 61.Keene D.R., Lunstrum G.P., Morris N.P., Stoddard D.W., Burgeson R.E. Two type XII-like collagens localize to the surface of banded collagen fibrils. The Journal of Cell Biology. 1991;113(4):971–978. doi: 10.1083/jcb.113.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown J.C., Mann K., Wiedemann H., Timpl R. Structure and binding properties of collagen type XIV isolated from human placenta. The Journal of Cell Biology. 1993;120(2):557–567. doi: 10.1083/jcb.120.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Font B., Aubert-Foucher E., Goldschmidt D., Eichenberger D., van der Rest M. Binding of collagen XIV with the dermatan sulfate side chain of decorin. The Journal of Biological Chemistry. 1993;268(33):25015–25018. [PubMed] [Google Scholar]
- 64.Giry-Lozinguez C., Aubert-Foucher E., Penin F., Deleage G., Dublet B., van der Rest M. Identification and characterization of a heparin binding site within the NC1 domain of chicken collagen XIV. Matrix Biology. 1998;17(2):145–149. doi: 10.1016/s0945-053x(98)90027-0. [DOI] [PubMed] [Google Scholar]
- 65.Kassner A., Hansen U., Miosge N., Reinhardt D.P., Aigner T., Bruckner-Tuderman L., Bruckner P., Grassel S. Discrete integration of collagen XVI into tissue-specific collagen fibrils or beaded microfibrils. Matrix Biology. 2003;22(2):131–143. doi: 10.1016/s0945-053x(03)00008-8. [DOI] [PubMed] [Google Scholar]
- 66.Kassner A., Tiedemann K., Notbohm H., Ludwig T., Morgelin M., Reinhardt D.P., Chu M.L., Bruckner P., Grassel S. Molecular structure and interaction of recombinant human type XVI collagen. Journal of Molecular Biology. 2004;339(4):835–853. doi: 10.1016/j.jmb.2004.03.042. [DOI] [PubMed] [Google Scholar]
- 67.Eble J.A., Kassner A., Niland S., Morgelin M., Grifka J., Grassel S. Collagen XVI harbors an integrin alpha1 beta1 recognition site in its C-terminal domains. The Journal of Biological Chemistry. 2006;281(35):25745–25756. doi: 10.1074/jbc.M509942200. [DOI] [PubMed] [Google Scholar]
- 68.Moinzadeh P., Agarwal P., Bloch W., Orteu C., Hunzelmann N., Eckes B., Krieg T. Systemic sclerosis with multiple nodules: characterization of the extracellular matrix. Archives of Dermatological Research. 2013;305(7):645–652. doi: 10.1007/s00403-013-1383-0. [DOI] [PubMed] [Google Scholar]
- 69.Tzortzaki E.G., Koutsopoulos A.V., Dambaki K.I., Lambiri I., Plataki M., Gordon M.K., Gerecke D.R., Siafakas N.M. Active remodeling in idiopathic interstitial pneumonias: evaluation of collagen types XII and XIV. The Journal of Histochemistry and Cytochemistry. 2006;54(6):693–700. doi: 10.1369/jhc.5A6835.2006. [DOI] [PubMed] [Google Scholar]
- 70.Arai K., Kasashima Y., Kobayashi A., Kuwano A., Yoshihara T. TGF-beta alters collagen XII and XIV mRNA levels in cultured equine tenocytes. Matrix Biology. 2002;21(3):243–250. doi: 10.1016/s0945-053x(02)00005-7. [DOI] [PubMed] [Google Scholar]
- 71.Gil-Cayuela C., Rosello L.E., Ortega A., Tarazon E., Trivino J.C., Martinez-Dolz L., Gonzalez-Juanatey J.R., Lago F., Portoles M., Rivera M. New altered non-fibrillar collagens in human dilated cardiomyopathy: role in the remodeling process. PLoS One. 2016;11(12) doi: 10.1371/journal.pone.0168130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ratzinger S., Eble J.A., Pasoldt A., Opolka A., Rogler G., Grifka J., Grassel S. Collagen XVI induces formation of focal contacts on intestinal myofibroblasts isolated from the normal and inflamed intestinal tract. Matrix Biology. 2010;29(3):177–193. doi: 10.1016/j.matbio.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 73.Sage H., Farin F.M., Striker G.E., Fisher A.B. Granular pneumocytes in primary culture secrete several major components of the extracellular matrix. Biochemistry. 1983;22(9):2148–2155. doi: 10.1021/bi00278a015. [DOI] [PubMed] [Google Scholar]
- 74.Crouch E.C., Moxley M.A., Longmore W. Synthesis of collagenous proteins by pulmonary type II epithelial cells. The American Review of Respiratory Disease. 1987;135(5):1118–1123. doi: 10.1164/arrd.1987.135.5.1118. [DOI] [PubMed] [Google Scholar]
- 75.Abrahamson D.R., Hudson B.G., Stroganova L., Borza D.B., John P.L. St. Cellular origins of type IV collagen networks in developing glomeruli. Journal of the American Society of Nephrology. 2009;20(7):1471–1479. doi: 10.1681/ASN.2008101086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sawada H., Konomi H., Hirosawa K. Characterization of the collagen in the hexagonal lattice of Descemet's membrane: its relation to type VIII collagen. The Journal of Cell Biology. 1990;110(1):219–227. doi: 10.1083/jcb.110.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kittelberger R., Davis P.F., Greenhill N.S. Immunolocalization of type VIII collagen in vascular tissue. Biochemical and Biophysical Research Communications. 1989;159(2):414–419. doi: 10.1016/0006-291x(89)90007-7. [DOI] [PubMed] [Google Scholar]
- 78.Seet L.F., Toh L.Z., Chu S.W.L., Finger S.N., Chua J.L.L., Wong T.T. Upregulation of distinct collagen transcripts in post-surgery scar tissue: a study of conjunctival fibrosis. Disease Models & Mechanisms. 2017;10(6):751–760. doi: 10.1242/dmm.028555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Boguslawska J., Kedzierska H., Poplawski P., Rybicka B., Tanski Z., Piekielko-Witkowska A. Expression of genes involved in cellular adhesion and extracellular matrix remodeling correlates with poor survival of patients with renal cancer. The Journal of Urology. 2016;195(6):1892–1902. doi: 10.1016/j.juro.2015.11.050. [DOI] [PubMed] [Google Scholar]
- 80.Hamano Y., Okude T., Shirai R., Sato I., Kimura R., Ogawa M., Ueda Y., Yokosuka O., Kalluri R., Ueda S. Lack of collagen XVIII/endostatin exacerbates immune-mediated glomerulonephritis. Journal of the American Society of Nephrology. 2010;21(9):1445–1455. doi: 10.1681/ASN.2009050492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Charytan D.M., Padera R., Helfand A.M., Zeisberg M., Xu X., Liu X., Himmelfarb J., Cinelli A., Kalluri R., Zeisberg E.M. Increased concentration of circulating angiogenesis and nitric oxide inhibitors induces endothelial to mesenchymal transition and myocardial fibrosis in patients with chronic kidney disease. International Journal of Cardiology. 2014;176(1):99–109. doi: 10.1016/j.ijcard.2014.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen J., Hamm L.L., Mohler E.R., Hudaihed A., Arora R., Chen C.S., Liu Y., Browne G., Mills K.T., Kleinpeter M.A., Simon E.E., Rifai N., Klag M.J., He J. Interrelationship of multiple endothelial dysfunction biomarkers with chronic kidney disease. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ruge T., Carlsson A.C., Larsson T.E., Carrero J.J., Larsson A., Lind L., Arnlov J. Endostatin level is associated with kidney injury in the elderly: findings from two community-based cohorts. American Journal of Nephrology. 2014;40(5):417–424. doi: 10.1159/000369076. [DOI] [PubMed] [Google Scholar]
- 84.Rydzewska-Rosolowska A., Borawski J., Mysliwiec M. High plasma endostatin level unaffected by low-molecular weight heparin in hemodialysis patients—a preliminary report. Advances in Medical Sciences. 2009;54(2):199–202. doi: 10.2478/v10039-009-0030-7. [DOI] [PubMed] [Google Scholar]
- 85.Stoessel A., Paliege A., Theilig F., Addabbo F., Ratliff B., Waschke J., Patschan D., Goligorsky M.S., Bachmann S. Indolent course of tubulointerstitial disease in a mouse model of subpressor, low-dose nitric oxide synthase inhibition. American Journal of Physiology. Renal Physiology. 2008;295(3):F717–F725. doi: 10.1152/ajprenal.00071.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maciel T.T., Coutinho E.L., Soares D., Achar E., Schor N., Bellini M.H. Endostatin, an antiangiogenic protein, is expressed in the unilateral ureteral obstruction mice model. Journal of Nephrology. 2008;21(5):753–760. [PubMed] [Google Scholar]
- 87.Heljasvaara R., Aikio M., Ruotsalainen H., Pihlajaniemi T. Collagen XVIII in tissue homeostasis and dysregulation - lessons learned from model organisms and human patients. Matrix Biology. 2017;57-58:55–75. doi: 10.1016/j.matbio.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 88.Rasi K., Piuhola J., Czabanka M., Sormunen R., Ilves M., Leskinen H., Rysa J., Kerkela R., Janmey P., Heljasvaara R., Peuhkurinen K., Vuolteenaho O., Ruskoaho H., Vajkoczy P., Pihlajaniemi T., Eklund L. Collagen XV is necessary for modeling of the extracellular matrix and its deficiency predisposes to cardiomyopathy. Circ. Res. 2010;107(10):1241–1252. doi: 10.1161/CIRCRESAHA.110.222133. [DOI] [PubMed] [Google Scholar]
- 89.Hagg P.M., Hagg P.O., Peltonen S., Autio-Harmainen H., Pihlajaniemi T. Location of type XV collagen in human tissues and its accumulation in the interstitial matrix of the fibrotic kidney. The American Journal of Pathology. 1997;150(6):2075–2086. [PMC free article] [PubMed] [Google Scholar]
- 90.Schiller H.B., Fernandez I.E., Burgstaller G., Schaab C., Scheltema R.A., Schwarzmayr T., Strom T.M., Eickelberg O., Mann M. Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Molecular Systems Biology. 2015;11(7):819. doi: 10.15252/msb.20156123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chung H.J., Uitto J. Type VII collagen: the anchoring fibril protein at fault in dystrophic epidermolysis bullosa. Dermatologic Clinics. 2010;28(1):93–105. doi: 10.1016/j.det.2009.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Uitto J., Chung-Honet L.C., Christiano A.M. Molecular biology and pathology of type VII collagen. Experimental Dermatology. 1992;1(1):2–11. doi: 10.1111/j.1600-0625.1992.tb00065.x. [DOI] [PubMed] [Google Scholar]
- 93.L. Rudnicka, J. Varga, A.M. Christiano, R.V. Iozzo, S.A. Jimenez, J. Uitto, Elevated expression of type VII collagen in the skin of patients with systemic sclerosis. Regulation by transforming growth factor-beta, J. Clin. Invest. 93(4) (1994) 1709–15. [DOI] [PMC free article] [PubMed]
- 94.Vindevoghel L., Kon A., Lechleider R.J., Uitto J., Roberts A.B., Mauviel A. Smad-dependent transcriptional activation of human type VII collagen gene (COL7A1) promoter by transforming growth factor-beta. The Journal of Biological Chemistry. 1998;273(21):13053–13057. doi: 10.1074/jbc.273.21.13053. [DOI] [PubMed] [Google Scholar]
- 95.Kivirikko S., Mauviel A., Pihlajaniemi T., Uitto J. Cytokine modulation of type XV collagen gene expression in human dermal fibroblast cultures. Experimental Dermatology. 1999;8(5):407–412. doi: 10.1111/j.1600-0625.1999.tb00390.x. [DOI] [PubMed] [Google Scholar]
- 96.Han S., Makareeva E., Kuznetsova N.V., DeRidder A.M., Sutter M.B., Losert W., Phillips C.L., Visse R., Nagase H., Leikin S. Molecular mechanism of type I collagen homotrimer resistance to mammalian collagenases. The Journal of Biological Chemistry. 2010;285(29):22276–22281. doi: 10.1074/jbc.M110.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kuznetsova N.V., McBride D.J., Leikin S. Changes in thermal stability and microunfolding pattern of collagen helix resulting from the loss of alpha2(I) chain in osteogenesis imperfecta murine. Journal of Molecular Biology. 2003;331(1):191–200. doi: 10.1016/s0022-2836(03)00715-0. [DOI] [PubMed] [Google Scholar]
- 98.Sharma U., Carrique L., Vadon-Le Goff S., Mariano N., Georges R.N., Delolme F., Koivunen P., Myllyharju J., Moali C., Aghajari N., Hulmes D.J. Structural basis of homo- and heterotrimerization of collagen I. Nature Communications. 2017;8:14671. doi: 10.1038/ncomms14671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rojkind M., Giambrone M.A., Biempica L. Collagen types in normal and cirrhotic liver. Gastroenterology. 1979;76(4):710–719. [PubMed] [Google Scholar]
- 100.Kilian O., Pfeil U., Wenisch S., Heiss C., Kraus R., Schnettler R. Enhanced alpha 1(I) mRNA expression in frozen shoulder and dupuytren tissue. European Journal of Medical Research. 2007;12(12):585–590. [PubMed] [Google Scholar]
- 101.Roberts-Pilgrim A.M., Makareeva E., Myles M.H., Besch-Williford C.L., Brodeur A.C., Walker A.L., Leikin S., Franklin C.L., Phillips C.L. Deficient degradation of homotrimeric type I collagen, alpha1(I)3 glomerulopathy in oim mice. Molecular Genetics and Metabolism. 2011;104(3):373–382. doi: 10.1016/j.ymgme.2011.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hashimoto Y., Shieh T.Y., Aoyama H., Izawa Y., Hayakawa T. Isolation and characterization of type V collagen from human post-burn granulation tissues. The Journal of Investigative Dermatology. 1986;87(4):540–543. doi: 10.1111/1523-1747.ep12455645. [DOI] [PubMed] [Google Scholar]
- 103.C. Niyibizi, P.P. Fietzek, M. van der Rest, Human placenta type V collagens. Evidence for the existence of an alpha 1(V) alpha 2(V) alpha 3(V) collagen molecule, J. Biol. Chem. 259(22) (1984) 14170–4. [PubMed]
- 104.Kleman J.P., Hartmann D.J., Ramirez F., van der Rest M. The human rhabdomyosarcoma cell line A204 lays down a highly insoluble matrix composed mainly of alpha 1 type-XI and alpha 2 type-V collagen chains. European Journal of Biochemistry. 1992;210(1):329–335. doi: 10.1111/j.1432-1033.1992.tb17425.x. [DOI] [PubMed] [Google Scholar]
- 105.Sato T., Takano R., Tokunaka K., Saiga K., Tomura A., Sugihara H., Hayashi T., Imamura Y., Morita M. Type VI collagen alpha1 chain polypeptide in non-triple helical form is an alternative gene product of COL6A1. Journal of Biochemistry. 2018;164(2):173–181. doi: 10.1093/jb/mvy040. [DOI] [PubMed] [Google Scholar]
- 106.Aumailley M. The laminin family. Cell Adhesion & Migration. 2013;7(1):48–55. doi: 10.4161/cam.22826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Torry D.J., Richards C.D., Podor T.J., Gauldie J. Anchorage-independent colony growth of pulmonary fibroblasts derived from fibrotic human lung tissue. The Journal of Clinical Investigation. 1994;93(4):1525–1532. doi: 10.1172/JCI117131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Pierce R.A., Griffin G.L., Mudd M.S., Moxley M.A., Longmore W.J., Sanes J.R., Miner J.H., Senior R.M. Expression of laminin alpha3, alpha4, and alpha5 chains by alveolar epithelial cells and fibroblasts. American Journal of Respiratory Cell and Molecular Biology. 1998;19(2):237–244. doi: 10.1165/ajrcmb.19.2.3087. [DOI] [PubMed] [Google Scholar]
- 109.Marinkovich M.P., Keene D.R., Rimberg C.S., Burgeson R.E. Cellular origin of the dermal-epidermal basement membrane. Developmental Dynamics. 1993;197(4):255–267. doi: 10.1002/aja.1001970404. [DOI] [PubMed] [Google Scholar]
- 110.Simon-Assmann P., Bouziges F., Arnold C., Haffen K., Kedinger M. Epithelial-mesenchymal interactions in the production of basement membrane components in the gut. Development. 1988;102(2):339–347. doi: 10.1242/dev.102.2.339. [DOI] [PubMed] [Google Scholar]
- 111.Hassell J.R., Schrecengost P.K., Rada J.A., SundarRaj N., Sossi G., Thoft R.A. Biosynthesis of stromal matrix proteoglycans and basement membrane components by human corneal fibroblasts. Investigative Ophthalmology & Visual Science. 1992;33(3):547–557. [PubMed] [Google Scholar]
- 112.Wilson S.E., Marino G.K., Torricelli A.A.M., Medeiros C.S. Injury and defective regeneration of the epithelial basement membrane in corneal fibrosis: a paradigm for fibrosis in other organs? Matrix Biology. 2017;64:17–26. doi: 10.1016/j.matbio.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Thannickal V.J., Lee D.Y., White E.S., Cui Z., Larios J.M., Chacon R., Horowitz J.C., Day R.M., Thomas P.E. Myofibroblast differentiation by transforming growth factor-beta1 is dependent on cell adhesion and integrin signaling via focal adhesion kinase. The Journal of Biological Chemistry. 2003;278(14):12384–12389. doi: 10.1074/jbc.M208544200. [DOI] [PubMed] [Google Scholar]
- 114.Estany S., Vicens-Zygmunt V., Llatjos R., Montes A., Penin R., Escobar I., Xaubet A., Santos S., Manresa F., Dorca J., Molina-Molina M. Lung fibrotic tenascin-C upregulation is associated with other extracellular matrix proteins and induced by TGFbeta1. BMC Pulmonary Medicine. 2014;14:120. doi: 10.1186/1471-2466-14-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mia M.M., R.A. Bank The IkappaB kinase inhibitor ACHP strongly attenuates TGFbeta1-induced myofibroblast formation and collagen synthesis. Journal of Cellular and Molecular Medicine. 2015;19(12):2780–2792. doi: 10.1111/jcmm.12661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wei Y., Kim T.J., Peng D.H., Duan D., Gibbons D.L., Yamauchi M., Jackson J.R., Le Saux C.J., Calhoun C., Peters J., Derynck R., Backes B.J., Chapman H.A. Fibroblast-specific inhibition of TGF-beta1 signaling attenuates lung and tumor fibrosis. The Journal of Clinical Investigation. 2017;127(10):3675–3688. doi: 10.1172/JCI94624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hallgren O., Nihlberg K., Dahlback M., Bjermer L., Eriksson L.T., Erjefalt J.S., Lofdahl C.G., Westergren-Thorsson G. Altered fibroblast proteoglycan production in COPD. Respiratory Research. 2010;11:55. doi: 10.1186/1465-9921-11-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Goldsmith K.T., Gammon R.B., Garver R.I., Jr. Modulation of bFGF in lung fibroblasts by TGF-beta and PDGF. The American Journal of Physiology. 1991;261(6 Pt 1):L378–L385. doi: 10.1152/ajplung.1991.261.6.L378. [DOI] [PubMed] [Google Scholar]
- 119.Ezzoukhry Z., Henriet E., Piquet L., Boye K., Bioulac-Sage P., Balabaud C., Couchy G., Zucman-Rossi J., Moreau V., Saltel F. TGF-beta1 promotes linear invadosome formation in hepatocellular carcinoma cells, through DDR1 up-regulation and collagen I cross-linking. European Journal of Cell Biology. 2016;95(11):503–512. doi: 10.1016/j.ejcb.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 120.Sethi A., Mao W., Wordinger R.J., Clark A.F. Transforming growth factor-beta induces extracellular matrix protein cross-linking lysyl oxidase (LOX) genes in human trabecular meshwork cells. Investigative Ophthalmology & Visual Science. 2011;52(8):5240–5250. doi: 10.1167/iovs.11-7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ikenaga N., Peng Z.W., Vaid K.A., Liu S.B., Yoshida S., Sverdlov D.Y., Mikels-Vigdal A., Smith V., Schuppan D., Popov Y.V. Selective targeting of lysyl oxidase-like 2 (LOXL2) suppresses hepatic fibrosis progression and accelerates its reversal. Gut. 2017;66(9):1697–1708. doi: 10.1136/gutjnl-2016-312473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang J., Savvatis K., Kang J.S., Fan P., Zhong H., Schwartz K., Barry V., Mikels-Vigdal A., Karpinski S., Kornyeyev D., Adamkewicz J., Feng X., Zhou Q., Shang C., Kumar P., Phan D., Kasner M., Lopez B., Diez J., Wright K.C., Kovacs R.L., Chen P.S., Quertermous T., Smith V., Yao L., Tschope C., Chang C.P. Targeting LOXL2 for cardiac interstitial fibrosis and heart failure treatment. Nature Communications. 2016;7:13710. doi: 10.1038/ncomms13710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Stangenberg S., Saad S., Schilter H.C., Zaky A., Gill A., Pollock C.A., Wong M.G. Lysyl oxidase-like 2 inhibition ameliorates glomerulosclerosis and albuminuria in diabetic nephropathy. Scientific Reports. 2018;8(1):9423. doi: 10.1038/s41598-018-27462-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Cosgrove D., Dufek B., Meehan D.T., Delimont D., Hartnett M., Samuelson G., Gratton M.A., Phillips G., MacKenna D.A., Bain G. Lysyl oxidase like-2 contributes to renal fibrosis in Col4alpha3/Alport mice. Kidney International. 2018;94(2):303–314. doi: 10.1016/j.kint.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barry-Hamilton V., Spangler R., Marshall D., McCauley S., Rodriguez H.M., Oyasu M., Mikels A., Vaysberg M., Ghermazien H., Wai C., Garcia C.A., Velayo A.C., Jorgensen B., Biermann D., Tsai D., Green J., Zaffryar-Eilot S., Holzer A., Ogg S., Thai D., Neufeld G., Van Vlasselaer P., Smith V. Allosteric inhibition of lysyl oxidase-like-2 impedes the development of a pathologic microenvironment. Nature Medicine. 2010;16(9):1009–1017. doi: 10.1038/nm.2208. [DOI] [PubMed] [Google Scholar]
- 126.Aumiller V., Strobel B., Romeike M., Schuler M., Stierstorfer B.E., Kreuz S. Comparative analysis of lysyl oxidase (like) family members in pulmonary fibrosis. Scientific Reports. 2017;7(1):149. doi: 10.1038/s41598-017-00270-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tjin G., White E.S., Faiz A., Sicard D., Tschumperlin D.J., Mahar A., Kable E.P.W., Burgess J.K. Lysyl oxidases regulate fibrillar collagen remodelling in idiopathic pulmonary fibrosis. Disease Models & Mechanisms. 2017;10(11):1301–1312. doi: 10.1242/dmm.030114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Choi S.K., Kim H.S., Jin T., Moon W.K. LOXL4 knockdown enhances tumor growth and lung metastasis through collagen-dependent extracellular matrix changes in triple-negative breast cancer. Oncotarget. 2017;8(7):11977–11989. doi: 10.18632/oncotarget.14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.M. Rouzaire, A. Comptour, C. Belville, D. Bouvier, G. Clairefond, F. Ponelle, V. Sapin, D. Gallot, L. Blanchon, All-trans retinoic acid promotes wound healing of primary amniocytes through the induction of LOXL4, a member of the lysyl oxidase family, Int. J. Biochem. Cell Biol. 81(Pt A) (2016) 10–19. [DOI] [PubMed]
- 130.Busnadiego O., Gonzalez-Santamaria J., Lagares D., Guinea-Viniegra J., Pichol-Thievend C., Muller L., Rodriguez-Pascual F. LOXL4 is induced by transforming growth factor beta1 through Smad and JunB/Fra2 and contributes to vascular matrix remodeling. Molecular and Cellular Biology. 2013;33(12):2388–2401. doi: 10.1128/MCB.00036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Eyre D.R., Weis M.A., Wu J.J. Advances in collagen cross-link analysis. Methods. 2008;45(1):65–74. doi: 10.1016/j.ymeth.2008.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.D. Pankova, Y. Chen, M. Terajima, M.J. Schliekelman, B.N. Baird, M. Fahrenholtz, L. Sun, B.J. Gill, T.J. Vadakkan, M.P. Kim, Y.H. Ahn, J.D. Roybal, X. Liu, E.R. Parra Cuentas, J. Rodriguez, Wistuba, II, C.J. Creighton, D.L. Gibbons, J.M. Hicks, M.E. Dickinson, J.L. West, K.J. Grande-Allen, S.M. Hanash, M. Yamauchi, J.M. Kurie, Cancer-associated fibroblasts induce a collagen cross-link switch in tumor stroma, Mol. Cancer Res. 14(3) (2016) 287–95. [DOI] [PMC free article] [PubMed]
- 133.Ohlmeier S., Nieminen P., Gao J., Kanerva T., Ronty M., Toljamo T., Bergmann U., Mazur W., Pulkkinen V. Lung tissue proteomics identifies elevated transglutaminase 2 levels in stable chronic obstructive pulmonary disease. American Journal of Physiology. Lung Cellular and Molecular Physiology. 2016;310(11):L1155–L1165. doi: 10.1152/ajplung.00021.2016. [DOI] [PubMed] [Google Scholar]
- 134.Wen Z., Ji X., Tang J., Lin G., Xiao L., Liang C., Wang M., Su F., Ferrandon D., Li Z. Positive feedback regulation between transglutaminase 2 and toll-like receptor 4 signaling in hepatic stellate cells correlates with liver fibrosis post Schistosoma japonicum infection. Frontiers in Immunology. 2017;8:1808. doi: 10.3389/fimmu.2017.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sandor K., Daniel B., Kiss B., Kovacs F., Szondy Z. Transcriptional control of transglutaminase 2 expression in mouse apoptotic thymocytes. Biochimica et Biophysica Acta. 2016;1859(8):964–974. doi: 10.1016/j.bbagrm.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 136.Khan I., Agarwal P., Thangjam G.S., Radhesh R., Rao S.G., Kondaiah P. Role of TGF-beta and BMP7 in the pathogenesis of oral submucous fibrosis. Growth Factors. 2011;29(4):119–127. doi: 10.3109/08977194.2011.582839. [DOI] [PubMed] [Google Scholar]
- 137.Crawford S.E., Stellmach V., Murphy-Ullrich J.E., Ribeiro S.M., Lawler J., Hynes R.O., Boivin G.P., Bouck N. Thrombospondin-1 is a major activator of TGF-beta1 in vivo. Cell. 1998;93(7):1159–1170. doi: 10.1016/s0092-8674(00)81460-9. [DOI] [PubMed] [Google Scholar]
- 138.Murphy-Ullrich J.E., Suto M.J. Thrombospondin-1 regulation of latent TGF-beta activation: a therapeutic target for fibrotic disease. Matrix Biology. 2017;68-69:28–43. doi: 10.1016/j.matbio.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Oka C., Tsujimoto R., Kajikawa M., Koshiba-Takeuchi K., Ina J., Yano M., Tsuchiya A., Ueta Y., Soma A., Kanda H., Matsumoto M., Kawaichi M. HtrA1 serine protease inhibits signaling mediated by Tgfbeta family proteins. Development. 2004;131(5):1041–1053. doi: 10.1242/dev.00999. [DOI] [PubMed] [Google Scholar]
- 140.Graham J.R., Chamberland A., Lin Q., Li X.J., Dai D., Zeng W., Ryan M.S., Rivera-Bermudez M.A., Flannery C.R., Yang Z. Serine protease HTRA1 antagonizes transforming growth factor-beta signaling by cleaving its receptors and loss of HTRA1 in vivo enhances bone formation. PLoS One. 2013;8(9) doi: 10.1371/journal.pone.0074094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Shirato K., Osawa H., Kaizuka M., Nakamura N., Sugawara T., Nakamura M., Tamura M., Yamabe H., Okumura K. Thrombin stimulates production of fibronectin by human proximal tubular epithelial cells via a transforming growth factor-beta-dependent mechanism. Nephrology, Dialysis, Transplantation. 2003;18(11):2248–2254. doi: 10.1093/ndt/gfg398. [DOI] [PubMed] [Google Scholar]
- 142.Panwar P., Du X., Sharma V., Lamour G., Castro M., Li H., Bromme D. Effects of cysteine proteases on the structural and mechanical properties of collagen fibers. The Journal of Biological Chemistry. 2013;288(8):5940–5950. doi: 10.1074/jbc.M112.419689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Aguda A.H., Panwar P., Du X., Nguyen N.T., Brayer G.D., Bromme D. Structural basis of collagen fiber degradation by cathepsin K. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(49):17474–17479. doi: 10.1073/pnas.1414126111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ghosh A.K., Vaughan D.E. PAI-1 in tissue fibrosis. Journal of Cellular Physiology. 2012;227(2):493–507. doi: 10.1002/jcp.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Van Doren S.R. Matrix metalloproteinase interactions with collagen and elastin. Matrix Biology. 2015;44-46:224–231. doi: 10.1016/j.matbio.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.d'Ortho M.P., Will H., Atkinson S., Butler G., Messent A., Gavrilovic J., Smith B., Timpl R., Zardi L., Murphy G. Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. European Journal of Biochemistry. 1997;250(3):751–757. doi: 10.1111/j.1432-1033.1997.00751.x. [DOI] [PubMed] [Google Scholar]
- 147.Menou A., Duitman J., Crestani B. The impaired proteases and anti-proteases balance in Idiopathic Pulmonary Fibrosis. Matrix Biology. 2018;68-69:382–403. doi: 10.1016/j.matbio.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 148.Rosmark O., Ahrman E., Muller C., Elowsson Rendin L., Eriksson L., Malmstrom A., Hallgren O., Larsson-Callerfelt A.K., Westergren-Thorsson G., Malmstrom J. Quantifying extracellular matrix turnover in human lung scaffold cultures. Scientific Reports. 2018;8(1):5409. doi: 10.1038/s41598-018-23702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Schiller H.B., Mayr C.H., Leuschner G., Strunz M., Staab-Weijnitz C., Preisendorfer S., Eckes B., Moinzadeh P., Krieg T., Schwartz D.A., Hatz R.A., Behr J., Mann M., Eickelberg O. Deep proteome profiling reveals common prevalence of MZB1-positive plasma B cells in human lung and skin fibrosis. American Journal of Respiratory and Critical Care Medicine. 2017;196(10):1298–1310. doi: 10.1164/rccm.201611-2263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Heinzelmann K., Noskovicova N., Merl-Pham J., Preissler G., Winter H., Lindner M., Hatz R., Hauck S.M., Behr J., Eickelberg O. Surface proteome analysis identifies platelet derived growth factor receptor-alpha as a critical mediator of transforming growth factor-beta-induced collagen secretion. The International Journal of Biochemistry & Cell Biology. 2016;74:44–59. doi: 10.1016/j.biocel.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 151.B.N. Baird, M.J. Schliekelman, Y.H. Ahn, Y. Chen, J.D. Roybal, B.J. Gill, D.K. Mishra, B. Erez, M. O'Reilly, Y. Yang, M. Patel, X. Liu, N. Thilaganathan, I.V. Larina, M.E. Dickinson, J.L. West, D.L. Gibbons, D.D. Liu, M.P. Kim, J.M. Hicks, Wistuba, II, S.M. Hanash, J.M. Kurie, Fibulin-2 is a driver of malignant progression in lung adenocarcinoma, PLoS One 8(6) (2013) e67054. [DOI] [PMC free article] [PubMed]
- 152.Heinbockel L., Marwitz S., Schromm A.B., Watz H., Kugler C., Ammerpohl O., Schnepf K., Rabe K.F., Droemann D., Goldmann T. Identification of novel target genes in human lung tissue involved in chronic obstructive pulmonary disease. International Journal of Chronic Obstructive Pulmonary Disease. 2018;13:2255–2259. doi: 10.2147/COPD.S161958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Yang C., Park A.C., Davis N.A., Russell J.D., Kim B., Brand D.D., Lawrence M.J., Ge Y., Westphall M.S., Coon J.J., Greenspan D.S. Comprehensive mass spectrometric mapping of the hydroxylated amino acid residues of the alpha1(V) collagen chain. The Journal of Biological Chemistry. 2012;287(48):40598–40610. doi: 10.1074/jbc.M112.406850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Weis M.A., Hudson D.M., Kim L., Scott M., Wu J.J., Eyre D.R. Location of 3-hydroxyproline residues in collagen types I, II, III, and V/XI implies a role in fibril supramolecular assembly. The Journal of Biological Chemistry. 2010;285(4):2580–2590. doi: 10.1074/jbc.M109.068726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Eyre D.R., Weis M., Hudson D.M., Wu J.J., Kim L. A novel 3-hydroxyproline (3Hyp)-rich motif marks the triple-helical C terminus of tendon type I collagen. The Journal of Biological Chemistry. 2011;286(10):7732–7736. doi: 10.1074/jbc.C110.195768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Hudson D.M., Garibov M., Dixon D.R., Popowics T., Eyre D.R. Distinct post-translational features of type I collagen are conserved in mouse and human periodontal ligament. Journal of Periodontal Research. 2017;52(6):1042–1049. doi: 10.1111/jre.12475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Perdivara I., Yamauchi M., Tomer K.B. Molecular characterization of collagen hydroxylysine O-glycosylation by mass spectrometry: current status. Australian Journal of Chemistry. 2013;66(7):760–769. doi: 10.1071/CH13174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Terajima M., Perdivara I., Sricholpech M., Deguchi Y., Pleshko N., Tomer K.B., Yamauchi M. Glycosylation and cross-linking in bone type I collagen. The Journal of Biological Chemistry. 2014;289(33):22636–22647. doi: 10.1074/jbc.M113.528513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sricholpech M., Perdivara I., Yokoyama M., Nagaoka H., Terajima M., Tomer K.B., Yamauchi M. Lysyl hydroxylase 3-mediated glucosylation in type I collagen: molecular loci and biological significance. The Journal of Biological Chemistry. 2012;287(27):22998–23009. doi: 10.1074/jbc.M112.343954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Fietzek P.P., Kuhn K. Information contained in the amino acid sequence of the alpha1(I)-chain of collagen and its consequences upon the formation of the triple helix, of fibrils and crosslinks. Molecular and Cellular Biochemistry. 1975;8(3):141–157. doi: 10.1007/BF01792765. [DOI] [PubMed] [Google Scholar]
- 161.Hudson D.M., Werther R., Weis M., Wu J.J., Eyre D.R. Evolutionary origins of C-terminal (GPP)n 3-hydroxyproline formation in vertebrate tendon collagen. PLoS One. 2014;9(4) doi: 10.1371/journal.pone.0093467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Morello R., Bertin T.K., Chen Y., Hicks J., Tonachini L., Monticone M., Castagnola P., Rauch F., Glorieux F.H., Vranka J., Bachinger H.P., Pace J.M., Schwarze U., Byers P.H., Weis M., Fernandes R.J., Eyre D.R., Yao Z., Boyce B.F., Lee B. CRTAP is required for prolyl 3-hydroxylation and mutations cause recessive osteogenesis imperfecta. Cell. 2006;127(2):291–304. doi: 10.1016/j.cell.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 163.Song E., Mechref Y. LC-MS/MS identification of the O-glycosylation and hydroxylation of amino acid residues of collagen alpha-1 (II) chain from bovine cartilage. Journal of Proteome Research. 2013;12(8):3599–3609. doi: 10.1021/pr400101t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yamauchi M., Sricholpech M. Lysine post-translational modifications of collagen. Essays in Biochemistry. 2012;52:113–133. doi: 10.1042/bse0520113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Bhadriraju K., Chung K.H., Spurlin T.A., Haynes R.J., Elliott J.T., Plant A.L. The relative roles of collagen adhesive receptor DDR2 activation and matrix stiffness on the downregulation of focal adhesion kinase in vascular smooth muscle cells. Biomaterials. 2009;30(35):6687–6694. doi: 10.1016/j.biomaterials.2009.08.036. [DOI] [PubMed] [Google Scholar]
- 166.Vogel W., Gish G.D., Alves F., Pawson T. The discoidin domain receptor tyrosine kinases are activated by collagen. Molecular Cell. 1997;1(1):13–23. doi: 10.1016/s1097-2765(00)80003-9. [DOI] [PubMed] [Google Scholar]
- 167.Lauer-Fields J.L., Malkar N.B., Richet G., Drauz K., Fields G.B. Melanoma cell CD44 interaction with the alpha 1(IV)1263-1277 region from basement membrane collagen is modulated by ligand glycosylation. The Journal of Biological Chemistry. 2003;278(16):14321–14330. doi: 10.1074/jbc.M212246200. [DOI] [PubMed] [Google Scholar]
- 168.Philipp J., Azimzadeh O., Subramanian V., Merl-Pham J., Lowe D., Hladik D., Erbeldinger N., Ktitareva S., Fournier C., Atkinson M.J., Raj K., Tapio S. Radiation-induced endothelial inflammation is transferred via the secretome to recipient cells in a STAT-mediated process. Journal of Proteome Research. 2017;16(10):3903–3916. doi: 10.1021/acs.jproteome.7b00536. [DOI] [PubMed] [Google Scholar]
- 169.Hauck S.M., Dietter J., Kramer R.L., Hofmaier F., Zipplies J.K., Amann B., Feuchtinger A., Deeg C.A., Ueffing M. Deciphering membrane-associated molecular processes in target tissue of autoimmune uveitis by label-free quantitative mass spectrometry. Molecular & Cellular Proteomics. 2010;9(10):2292–2305. doi: 10.1074/mcp.M110.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Merl J., Ueffing M., Hauck S.M., von Toerne C. Direct comparison of MS-based label-free and SILAC quantitative proteome profiling strategies in primary retinal Muller cells. Proteomics. 2012;12(12):1902–1911. doi: 10.1002/pmic.201100549. [DOI] [PubMed] [Google Scholar]
- 171.Naba A., Pearce O.M.T., Del Rosario A., Ma D., Ding H., Rajeeve V., Cutillas P.R., Balkwill F.R., Hynes R.O. Characterization of the extracellular matrix of normal and diseased tissues using proteomics. Journal of Proteome Research. 2017;16(8):3083–3091. doi: 10.1021/acs.jproteome.7b00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.W. Huang da, B.T. Sherman, R. Stephens, M.W. Baseler, H.C. Lane, R.A. Lempicki, DAVID gene ID conversion tool, Bioinformation 2(10) (2008) 428–30. [DOI] [PMC free article] [PubMed]
- 173.Fabre B., Lambour T., Garrigues L., Ducoux-Petit M., Amalric F., Monsarrat B., Burlet-Schiltz O., Bousquet-Dubouch M.P. Label-free quantitative proteomics reveals the dynamics of proteasome complexes composition and stoichiometry in a wide range of human cell lines. Journal of Proteome Research. 2014;13(6):3027–3037. doi: 10.1021/pr500193k. [DOI] [PubMed] [Google Scholar]
- 174.Krey J.F., Wilmarth P.A., Shin J.B., Klimek J., Sherman N.E., Jeffery E.D., Choi D., David L.L., Barr-Gillespie P.G. Accurate label-free protein quantitation with high- and low-resolution mass spectrometers. Journal of Proteome Research. 2014;13(2):1034–1044. doi: 10.1021/pr401017h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Schwanhäusser B., Busse D., Li N., Dittmar G., Schuchhardt J., Wolf J., Chen W., Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 176.Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nature Protocols. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 177.Tabb D.L., Fernando C.G., Chambers M.C. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. Journal of Proteome Research. 2007;6(2):654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Z.Q. Ma, S. Dasari, M.C. Chambers, M.D. Litton, S.M. Sobecki, L.J. Zimmerman, P.J. Halvey, B. Schilling, P.M. Drake, B.W. Gibson, D.L. Tabb, IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering, J. Proteome Res. 8(8) (2009) 3872–81. [DOI] [PMC free article] [PubMed]
- 179.Cabral W.A., Perdivara I., Weis M., Terajima M., Blissett A.R., Chang W., Perosky J.E., Makareeva E.N., Mertz E.L., Leikin S., Tomer K.B., Kozloff K.M., Eyre D.R., Yamauchi M., Marini J.C. Abnormal type I collagen post-translational modification and crosslinking in a cyclophilin B KO mouse model of recessive osteogenesis imperfecta. PLoS Genetics. 2014;10(6) doi: 10.1371/journal.pgen.1004465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material: Supplementary Tables S1 and S2; Supplementary Figures S1-S19
Table of quantified ECM proteins with respective average ratios withTGFb/noTGFb and significance values, in comparison to ratios and significance values Bleo/PBS and ILD/control extracted from Schiller et al. 2015 and 2017.
Ratios >1 are given in blue, ratios <1 are given in yellow. Significance values <0.05 are given in red. ILD/control ratios obtained after imputation of missing values are marked by +. Proteins that could not be quantified in the respective data set are marked with ND.