Rüthnick et al. decipher how the assembly of the daughter spindle pole body (dSPB) works on a molecular level. Recruitment of the proteins Spc29 and Spc42 to the N-terminus of the bridge protein Sfi1 facilitated by centrin initiates dSPB assembly.
Abstract
The spindle pole body (SPB) provides microtubule-organizing functions in yeast and duplicates exactly once per cell cycle. The first step in SPB duplication is the half-bridge to bridge conversion via the antiparallel dimerization of the centrin (Cdc31)-binding protein Sfi1 in anaphase. The bridge, which is anchored to the old SPB on the proximal end, exposes free Sfi1 N-termini (N-Sfi1) at its distal end. These free N-Sfi1 promote in G1 the assembly of the daughter SPB (dSPB) in a yet unclear manner. This study shows that N-Sfi1 including the first three Cdc31 binding sites interacts with the SPB components Spc29 and Spc42, triggering the assembly of the dSPB. Cdc31 binding to N-Sfi1 promotes Spc29 recruitment and is essential for satellite formation. Furthermore, phosphorylation of N-Sfi1 has an inhibitory effect and delays dSPB biogenesis until G1. Taking these data together, we provide an understanding of the initial steps in SPB assembly and describe a new function of Cdc31 in the recruitment of dSPB components.
Introduction
In the yeast Saccharomyces cerevisiae, the spindle pole body (SPB) functions as a microtubule (MT)-organizing center similar to the centrosome in animal cells (Jaspersen and Winey, 2004; Rüthnick and Schiebel, 2018). The SPB is a large multilayered structure composed of at least 16 proteins that is embedded in the nuclear envelope (NE) throughout the cell cycle. This embedding is a reflection of the closed mitosis in yeast and enables the SPB to organize simultaneously nuclear and cytoplasmic MTs (Byers and Goetsch, 1975). Nuclear MTs segregate the duplicated chromosomes in mitosis, while the cytoplasmic MTs contact the cell cortex and align the mitotic spindle along the mother–bud axis.
The SPB, as well as the human centrosome, duplicates exactly once per cell cycle (Byers and Goetsch, 1975; Rüthnick and Schiebel, 2016). This tight regulation is essential for the viability of yeast cells. SPB duplication occurs at an SPB substructure that, depending on its length, is called the “half bridge” or “bridge” (Byers and Goetsch, 1975; Rüthnick and Schiebel, 2016). The half bridge is a one-sided attachment to the SPB central plaque and is layered on both sides of the NE (Byers and Goetsch, 1975). The cytoplasmic side of the half bridge consists of parallel bundles of elongated Sfi1 proteins organized in the same orientation (Kilmartin, 2003; Li et al., 2006). All Sfi1 N-termini (N-Sfi1) are embedded in the SPB core, while the Sfi1 C-termini (C-Sfi1) are positioned away from the SPB. The protein Kar1 with its C-terminal membrane anchor attaches the Sfi1 layer to the NE (Seybold et al., 2015; Vallen et al., 1992). Each Sfi1 molecule binds up to 21 centrin (Cdc31 in yeast) molecules through conserved binding sites along the central α-helical region of Sfi1. Cdc31 stabilizes the elongated structure of Sfi1 but also promotes lateral interactions between Sfi1 molecules of the half bridge (Kilmartin, 2003; Seybold et al., 2015). The nuclear side of the half bridge mainly consists of the SUN (Sad1p/UNC-84) domain protein Mps3 that has an unclear function in SPB duplication (Friederichs et al., 2011; Hagan and Yanagida, 1995; Jaspersen et al., 2002).
Cyclin-dependent kinase 1 (Cdk1) phosphorylates six sites within C-Sfi1 and thereby prevents C-tail to C-tail dimerization (Avena et al., 2014; Elserafy et al., 2014). With anaphase onset, the phosphatase Cdc14 becomes released from the nucleolus and dephosphorylates Cdk1 phosphorylation sites in C-Sfi1 (Avena et al., 2014; Elserafy et al., 2014; Visintin et al., 1998). This triggers antiparallel C-tail to C-tail dimerization of Sfi1 in anaphase, converting the half bridge into the nearly twice as long bridge structure (Kilmartin, 2003). This dimerization also exposes free N-Sfi1 at the bridge distal end that are believed to promote the assembly of an SPB precursor on the cytoplasmic side of the NE, known as the “satellite” (Byers and Goetsch, 1975; Rüthnick and Schiebel, 2016). The satellite consists mainly of the central plaque proteins SPB component 29 (Spc29) and 42 (Spc42; Adams and Kilmartin, 1999; Burns et al., 2015). How these proteins become recruited to N-Sfi1 and the regulation of this interaction are currently unclear.
After formation in early G1, the satellite expands with rising Cdk1 kinase activity into the duplication plaque. The duplication plaque inserts next to the mother SPB while still being attached to the parental structure via the bridge (Adams and Kilmartin, 1999; Byers and Goetsch, 1975). A nuclear pore complex, together with a number of integral membrane and membrane-associated proteins that become recruited to the NE insertion site of the new SPB, assists duplication plaque insertion (Chen et al., 2019; Rüthnick et al., 2017). After or during NE insertion, the inner plaque assembles from within the nucleus (Elliott et al., 1999; Knop and Schiebel, 1998). As a final step of the duplication process, the SPB bridge separates in S phase through phosphorylation of C-Sfi1 and the joint activities of the kinesin-5 motor proteins Kip1 and Cin8 that push the SPBs apart (Avena et al., 2014; Elserafy et al., 2014; Leary et al., 2019; Saunders and Hoyt, 1992). The mother and daughter SPBs carry a half bridge each in late S phase until the upcoming anaphase.
The role of Sfi1 in SPB duplication is conserved in Schizosaccharomyces pombe. Similarly, in this organism, the half-bridge–like assembly of parallel Sfi1 molecules duplicates into a bridge at the end of mitosis by C-tail to C-tail Sfi1 interactions (Bouhlel et al., 2015). Recently, a function of human Sfi1 (hSfi1) in early steps of centriole duplication has been described (Kodani et al., 2019). hSfi1 promotes centriole duplication by recruiting the ubiquitin-deconjugating enzyme USP9X that stabilizes the centrosomal protein STIL (SCL/TAL-interrupting locus). Whether such a recruiting function is conserved in yeast and whether also C-tail dimerization of hSfi1 is required for its centriole-promoting functions are open questions.
How free N-Sfi1 at the distal end of the bridge contribute to the assembly of the satellite is not understood. In this study, we used mutations in SFI1 that impair SPB duplication and biochemical experiments to show that N-Sfi1 can recruit Spc29. This process is mediated via the binding of Cdc31 to the first three N-terminal binding sites in N-Sfi1. Also, Spc42 directly interacts with Sfi1. Furthermore, we provide evidence that polo-like kinase Cdc5 prevents the assembly of the satellite in anaphase due to inhibitory phosphorylations in N-Sfi1. This together explains how bridge formation in anaphase is linked to the assembly of the satellite in G1 and how SPB and bridge components are interacting.
Results
Binding of the satellite proteins Spc29 and Spc42 to Sfi1 requires N-Sfi1 and Cdc31 binding sites
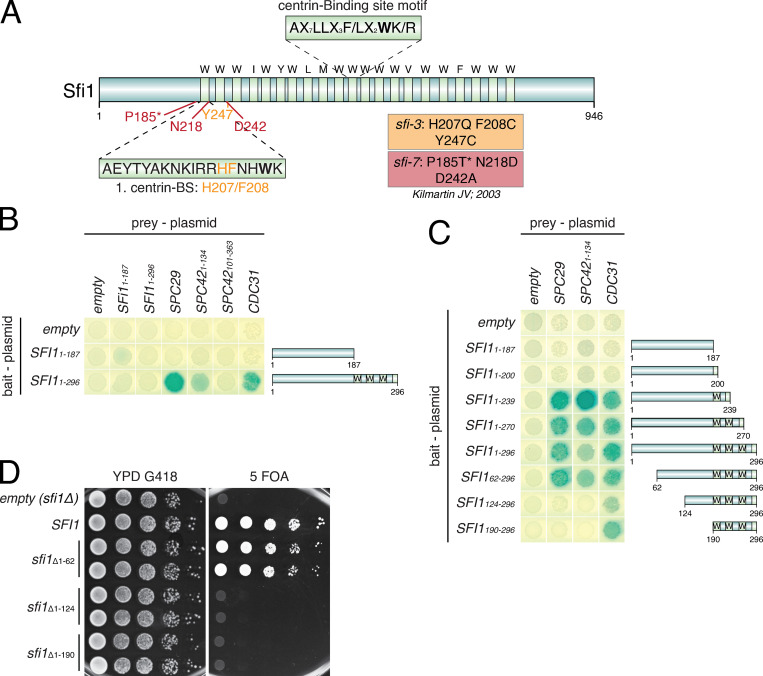
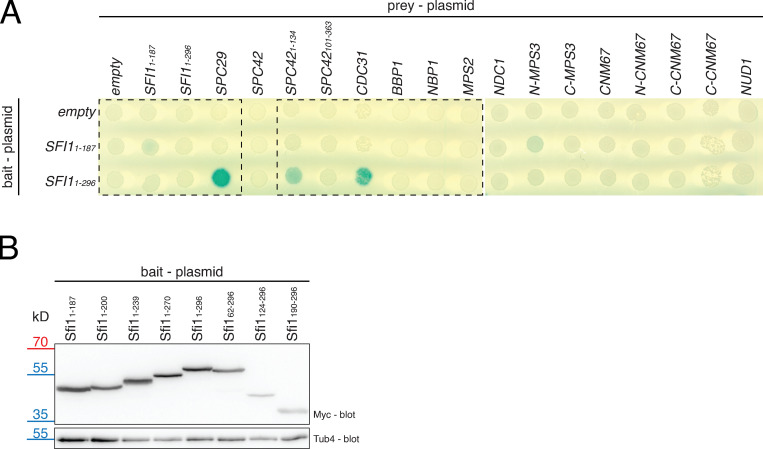
Sfi1 is an elongated protein with an N-terminal extension of 187 amino acids and 21 putative Cdc31-binding sites in its center (Fig. 1 A; Cdc31-binding sites are marked by light green shades) and a C-terminal extension of 148 amino acids. Previously, it was shown that the sfi1-3 and sfi1-7 alleles with mutations within the first three N-terminal Cdc31-binding sites (Fig. 1 A) affect SPB duplication (Kilmartin, 2003). Because these mutations in sfi1-3 and sfi-7 are quite distant from the C-Sfi1 that is essential for bridge assembly (Elserafy et al., 2014), the N-Sfi1 might have a novel role in SPB duplication, perhaps in the formation of the satellite through protein recruitment. To test this notion, we performed a yeast two-hybrid (Y2H) screen with all SPB core proteins, including Spc29 and Spc42, the two main satellite components, as prey constructs and the N-Sfi1 as bait (Fig. 1 B; Fig. S1 A; Adams and Kilmartin, 1999; Burns et al., 2015). One N-Sfi1 version contained only the N-terminal region without Cdc31-binding sites (Sfi11–187), while the other included the first three Cdc31-binding sites (Sfi11–296) and therefore spanned the region that carried the mutations in sfi-3 and sfi1-7 (Kilmartin, 2003; Li et al., 2006). The Sfi11–187 failed to interact with any of the prey proteins. Sfi11–296 strikingly showed Y2H interactions (blue color) in combination with Spc29, the N-terminus of Spc42 (Spc421–134; N-Spc42), and Cdc31. In contrast, the C-terminus of Spc42 (Spc42101–363) failed to interact with Sfi11–296 (Fig. 1 B; and Fig. S1 A), although it showed strong Y2H interactions with the SPB component Cnm67 (Elliott et al., 1999), indicating that it is functional per se. Thus, the N-Sfi1 including the first Cdc31-binding sites interacts with the SPB components Spc29 and N-Spc42.
Figure 1.
N-Sfi1 interacts with Spc29 and N-Spc42. (A) Schematic representation of Sfi1. All 21 conserved Cdc31-binding sites (the key W, I, Y, L, and F residues in the Cdc31-binding motifs are marked by light green shades) are shown, as well as the mutations identified in the sfi1-3 and sfi-7 mutants as described elsewhere (Kilmartin, 2003). The asterisk in P185T* indicates the P185T mutation in sfi1-7 that was identified by sequencing of the original sfi1-7 plasmid. (B and C) Y2H interaction studies after X-Gal overlay. (D) Representative image of a drop test (10-fold dilutions) of N-terminally deleted Sfi1-truncation mutants analyzed in a SFI1-shuffle strain and incubated at 30°C for 2.5 d. BS, binding site; YPD, yeast peptone dextrose.
Figure S1.
Y2H interaction study of N-Sfi1. (A) Uncut Y2H assay with all analyzed SPB components. Parts are shown in Fig. 1 B. The full-length Spc42 Y2H construct did not interact with Sfi11–187 and Sfi11–296, because this construct is nonfunctional, since it does not show a positive interaction with Spc29 and Cnm67 (Elliott et al., 1999). (B) Immunoblot to confirm the expression of all Sfi1 constructs analyzed in Fig. 1 C and Fig. 3 B. Tub4 is the loading control.
We next asked about the minimal requirements of the N-Sfi1 for the binding to Spc29 and N-Spc42. At least one functional Cdc31-binding site of Sfi1 (Sfi11–239) was required for the interaction with Spc29, N-Spc42, and Cdc31 (Fig. 1 C). Removal of the first 62 N-terminal amino acids from Sfi11–296 had no impact on the interaction with Spc29, N-Spc42, and Cdc31. However, further truncations in the N-Sfi1 (Sfi1124–296 or Sfi1190–296) abolished interaction with Spc29 and N-Spc42 without affecting Cdc31 binding (Fig. 1 C). All Sfi1 constructs were expressed in yeast, as indicated by an immunoblot for the myc-tagged bait proteins (Fig. S1 B). Taken together, the amino acids 62–239, including the first Cdc31-binding site, are important for the interaction with Spc29 and N-Spc42.
Because SPB duplication is essential for the viability of yeast cells, we expected that interfering with regions of Sfi1 that mediate binding to Spc29 and Spc42 should cause lethality. Sfi1 fragments that are not important for these interactions, such as the N-terminal 62 amino acids, may not have a strong impact on the viability of cells. To test this hypothesis, we constructed SFI1 alleles with N-terminal truncations and tested their ability to support growth of an sfi1Δ strain in a plasmid shuffle complementation experiment. Indeed, cells expressing the sfi1Δ1–62 allele were viable on 5′-fluoroorotic acid (5-FOA) plates, which select for cells that have lost the URA3-based WT SFI1 plasmid but still express sfi1Δ1–62. In contrast, cells only expressing the sfi1Δ1–124 or sfi1Δ1–190 deletions were not viable (Fig. 1 D). These data indicate that the N-terminal region of Sfi1 encompassing amino acids 62–239, including the first Cdc31-binding site, bind the satellite components Spc29 and Spc42.
Complex formation of recombinant N-Sfi1, Cdc31, and Spc29
To validate the Y2H data and to test whether these interactions were direct (Fig. 1, B and C), we coexpressed different versions of N-SFI1 together with CDC31 and six histidine-tagged SPC29 (SPC29-6His) in Escherichia coli. We chose E. coli for these experiments because of the tight attachment of Sfi1 to the SPB in yeast cells, which makes extraction difficult (Elliott et al., 1999). Furthermore, because E. coli does not encode for SPB components, interactions were most likely direct and not mediated by other proteins in the system. After coexpression, we immunoprecipitated Flag-tagged N-Sfi1 and analyzed for coelution with Spc29-6His and Cdc31. Cdc31 coimmunoprecipitated with Sfi11–239, Sfi11–270, and Sfi11–296 but not with Sfi11–187, as indicated by the Cdc31 immunoblot (Fig. 2 A, top) and the Coomassie blue–stained gel (Fig. 2 A, bottom). The amount of Cdc31 that bound to the Sfi1 fragments correlated with the number of Cdc31-binding sites (Fig. 2 B).
Figure 2.
N-Sfi1 directly interacts with Spc29. (A) FLAG-IP experiment of N-Sfi1-FLAG constructs with zero, one, two, and three Cdc31-binding sites coexpressed with Cdc31 and Spc29-6His in E. coli. Proteins were analyzed by Coomassie blue staining (common band at 20 kD in the IP lanes represents the light chain of the FLAG antibody) and immunoblotting with the indicated antibodies. The asterisk indicates the position of the enriched Sfi1-FLAG constructs. (B) Quantifications of the Coomassie blue–stained gel (as shown in A) to determine the Cdc31/Sfi1-FLAG ratio. n = 2. (C) FLAG-IP as described for A with the exception that Sfi1-FLAG fragments were also expressed without Cdc31. (D) Quantification of Spc29/Sfi11–296 ratio from the immunoblots in C. n = 3. Error bars are SD. P value (0.005) was derived by using a two-tailed t test. (E) FLAG-IP with N-terminally shortened Sfi1-FLAG constructs coexpressed with Cdc31 and Spc29-6His analyzed by immunoblotting and Coomassie staining. The asterisk indicates the position of the Sfi1-FLAG constructs. Common band at 20 kD in the IP lanes represents the light chain of the FLAG antibody. (F) Quantification of the ratio between Spc29 and Sfi1 from the Coomassie blue–stained gel (as shown in E). n = 3. Error bars are SD. P value (0.024) was derived by using a two-tailed t test. Shape symbols in B, D, and F represent the individual datapoints of each experiment.
Spc29 was only weakly detected in the Sfi11–239-FLAG immunoprecipitation (IP) by anti-Spc29 antibodies (Fig. 2 A). In contrast, Sfi11–270 and Sfi11–296, with two and three Cdc31 binding sites, respectively, associated more strongly with Spc29. Binding of Spc29 to Sfi11–296 was already detected by Coomassie blue staining (Fig. 2 A). The relatively weak interaction between Spc29 and Sfi11–239 in this in vitro system compared with the Y2H analysis reflects either the higher stringency of the biochemical approach that discriminates binding differences better than the Y2H or the lack of modifications in the E. coli–expressed proteins that, in yeast, possibly enhance the interaction efficiency. In summary, this experiment suggests direct binding of Spc29 to the N-Sfi1–Cdc31 complex.
We next addressed whether binding of Spc29 to N-Sfi1 requires the presence of Cdc31. For this, we coexpressed N-Sfi1–FLAG constructs and Spc29-6His with and without Cdc31 in E. coli (Fig. 2 C). In general, N-Sfi1–FLAG expression was always weaker in the absence of Cdc31 than when it was coexpressed with Cdc31 (Fig. 2 C; compare INPUT lanes with and without Cdc31). Although Sfi11–296 was only weakly expressed in the absence of Cdc31, in the anti-FLAG IP, Sfi11–296 and the Sfi11–296–Cdc31 complex were both present at similar levels (Fig. 2 C, FLAG-IP lanes in the righthand panel). Sfi11–296–Cdc31 efficiently coimmunoprecipitated Spc29, whereas there was only a very weak interaction with Sfi11–296 alone (Fig. 2, C and D). Thus, Cdc31 promotes binding of Spc29 to Sfi11–296.
Using the E. coli system, we further evaluated the importance of regions within the N-terminus of Sfi1 for Spc29 recruitment. Sfi11–296, Sfi162–296, and Sfi1190–296 were expressed together with Cdc31 in E. coli. We also expressed FLAG-tagged Sfi1124–296 and alternatively Sfi1127–296; however, for unknown reasons, both constructs were not expressed efficiently in E. coli. Consistent with the Y2H data (Fig. 1 C), Sfi162–296 interacted with Cdc31 and Spc29 (Fig. 2 E). Interestingly, Spc29 repeatedly bound with greater efficiency to Sfi162–296 than to Sfi11–296, as judged from the immunoblot analysis and the quantification of the Coomassie blue–stained gel (Fig. 2, E and F). This stronger binding suggests a potential negative role of the N-terminal 62 amino acids of Sfi1 for Spc29 binding. As expected from the Y2H experiment, Sfi1190–296 failed to interact with Spc29 (Fig. 2 E). However, we noticed that Cdc31 binding to Sfi1190–296 was reduced, which may indicate a folding problem of E. coli–expressed Sfi1190–296.
Together, Spc29 binds directly to N-Sfi1, depending on the recruitment of Cdc31. The first 62 amino acids of Sfi1 are not required for Spc29 binding, but instead have a negative impact on the Spc29 recruitment.
Spc42 binds directly to N-Sfi1
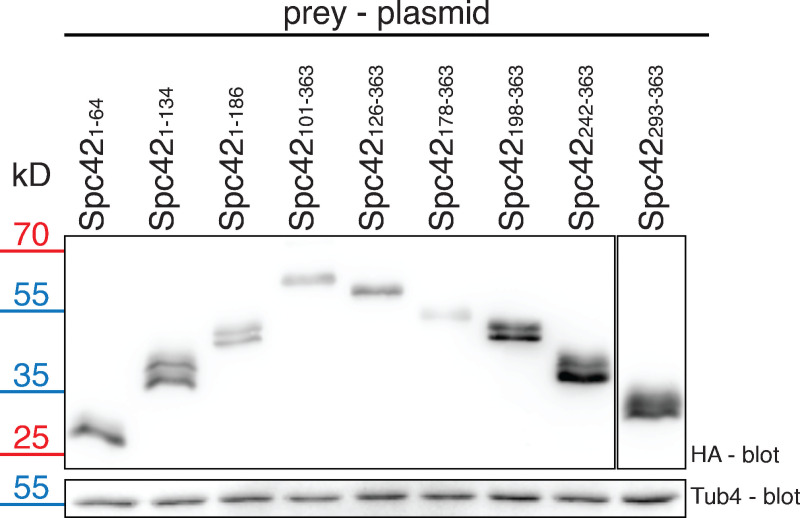
N-Spc42 has the ability to interact with N-Sfi1 (Fig. 1 B). To confirm this binding and to further map interaction, we tested Spc42 subfragments as prey (Fig. 3 A) together with different N-Sfi1 bait constructs. The N-terminal 1–134 amino acids of Spc42 were adequate for the binding to N-Sfi1 constructs containing at least one Cdc31-binding site (Sfi11–239). Spc421–64 and the C-Spc42 fragments were insufficient to mediate binding to N-Sfi1 fragments in this assay. As for Spc29 (Fig. 1 C), the N-terminal 62 amino acids of Sfi1 were not essential for Spc421–134 binding (Fig. 3 B). We confirmed that all HA-tagged prey proteins were expressed (Fig. S2).
Figure 3.
N-Spc42 directly interacts with Sfi1. (A) Schematic representation of the SPB protein Spc42. (B) Y2H analysis of the interactions between Spc42 and Sfi1 after X-Gal overlay. (C) FLAG-IP experiment of Sfi11–296-FLAG/Cdc31 with the indicated 3K2N-Spc42-6His constructs expressed separately in E. coli followed by mixing. Protein interaction was analyzed by immunoblotting and Coomassie blue staining. Asterisks indicate the position of Spc42 constructs in the Coomassie blue stain gel. (D) FLAG-IP of Sfi11–296-FLAG/Cdc31 with 3K2N-Spc421–186-6His in the presence or absence of Spc29 (performed by mixing of Sfi11–296-FLAG/Cdc31, 3K2N-Spc421–186-6His, and Spc29-6His extracts). Protein interaction was analyzed by immunoblotting. (E) Quantification of the ratio between Spc42 and Sfi11–296/Cdc31 with and without Spc29 from immunoblots in D. n = 3. The result of a two-tailed t test was NS. Shape symbols represent individual datapoints from each experiment. CC, coiled-coil domain.
Figure S2.
Immunoblot analysis to confirm the expression of all SPC42 constructs analyzed in Fig. 3 B. The Spc42 double bands are explained by phosphorylation of the protein (Donaldson and Kilmartin, 1996). Tub4 serves as a loading control.
Expression of 6His-tagged SPC42 fragments together with N-SFI11–296-FLAG and CDC31 in E. coli, followed by IP experiments, confirmed these results. For this experiment, we used Spc42-6His fragments that contained the sigma-54–dependent transcriptional regulator domain from Chlorobium tepidum (3K2N) at the N-terminus (3K2N-Spc42-6His). 3K2N increases the solubility of the Spc42 constructs without changing the binding properties of the Spc42 moiety (Drennan et al., 2019). In this setup, the FLAG-tagged, Sfi11–296/Cdc31–containing E. coli extract was mixed with extracts containing 3K2N-Spc42-6His constructs in order to ensure equal input of Sfi11–296/Cdc31 followed by FLAG IP. Constant input in case of 3K2N-Spc42-6His and Sfi11–296/Cdc31 coexpression was difficult to achieve because of toxic effects for the E. coli cells in some combinations. Analysis of the FLAG immunoprecipitates by Coomassie blue staining of the gel and immunoblot analysis indicated that Spc421–134 and Spc421–186 but not the C-terminal Spc42101–363 and Spc42126–363 fragments bound to Sfi11–296/Cdc31 (Fig. 3 C), which is consistent with the Y2H data (Fig. 3 A). These data together suggest that the N-Spc42 binds to N-Sfi1.
We next tested whether addition of Spc29 to the extracts containing N-Sfi1/Cdc31 and 3K2N-Spc421–186-6His would increase the binding efficiency of N-Spc42 to N-Sfi1/Cdc31. As shown in Fig. 3, D and E, the presence of Spc29 did not increase N-Spc42 binding to N-Sfi1/Cdc31, indicating that Spc42 and Spc29 do not bind cooperatively to N-Sfi1/Cdc31 in vitro. In summary, we can conclude that N-Spc42 directly interacts with the N-terminus of Sfi1 independent of Spc29.
Mutations in sfi1-3 and sfi1-7 affect binding of Spc29 and Spc42
The conditional lethal sfi1-3 and sfi1-7 alleles carry mutations in the N-terminal region of Sfi1 (Fig. 1 A; Kilmartin, 2003), which is important for Spc29/N-Spc42 binding (Figs. 1, 2, and 3). Mutations in sfi1-3 and sfi1-7 could therefore interfere with Spc29 and Spc42 binding to Sfi1. However, because the molecular defects of sfi1-3 and sfi1-7 were never analyzed in detail, proof for this possibility is missing. To test this model, we first mapped the mutations in sfi1-3 and sfi1-7 that contributed to the growth defect and therefore may interfere with the binding of Spc29 and Spc42. Only the combined presence of all three published mutations (H207Q, F208C, and Y247C) in sfi1-3 caused a conditional lethal growth defect at 37°C (Fig. 4 A; Kilmartin, 2003). Sequencing of the original sfi1-7 plasmid identified, besides the two published mutations, N218D and D242A, one additional mutation, P185T. Surprisingly, cells with sfi1N218D D242A grew as WT SFI1 cells at 37°C (Fig. 4 B). In contrast, sfi1P185T cells showed a conditional lethal growth defect at 37°C (Fig. 4 B). The further addition of N218D and D242A mutations enhanced the growth defect of sfi1P185T cells (sfi1P185T N218D D242A in Fig. 4 B; 23°C). Thus, it is predominately the P185T mutation preceding the first Cdc31-binding site (Fig. 1 A) that is responsible for the growth defect of sfi1-7 cells.
Figure 4.
N-terminal sfi1 mutants are deficient in SPB duplication. (A and B) Growth test of the indicated SFI1 mutants. Yeast cells were serially diluted 1:10 and spotted onto the indicated plates. Plates were incubated for 2.5 d at 37°C or for 4 d at 23°C. (C and D) Y2H analysis of N-Sfi1 constructs carrying the indicated mutations after X-Gal overlay. (E and F) Microscopic phenotype analysis of the indicated sfi1 mutants 4 h after the release from the G1 block and the temperature shift to 37°C. Cells incubated at 23°C served as a control. Graph (left) shows the average of 3 independent experiments (n = 3) with >100 cells per cell type analyzed for each time. Error bars indicate SD. Of note, whether multiple Spc42-yeGFP signals in sfi1-3 cells reflect SPBs or only Spc42 assemblies is presently unclear. Exemplary fluorescence images of the analyzed phenotypes are shown on the right. Scale bars: 5 µm. (G and H) EM micrographs of the indicated SFI1 mutants 4 h after the release from G1 block and the temperature shift. Cartoons illustrate the SPB phenotype. Scale bars: 150 nm. B, bridge; cMTs, cytoplasmic microtubules; nMTs, nuclear microtubules; YPD, yeast peptone dextrose.
Using the Y2H assay to test whether mutations in sfi1-3 or sfi1-7 interfere with Spc29 and Spc42 recruitment, we analyzed binding of the N-Sfi1F208C, N-Sfi1Y247C, N-Sfi1H207C F208C, and N-Sfi1H207C F208C Y247C from sfi1-3 and N-Sfi1P185T, N-Sfi1N218D D242A, and N-Sfi1P185T N218D D242A from sfi1-7 to Cdc31, Spc29, and N-Spc42. All N-Sfi11–296 mutant constructs interacted with Cdc31 (Fig. 4, C and D). Interestingly, Sfi11–296H207Q F208C Y247C (sfi1-3) failed to interact with N-Spc42 and Spc29, while Sfi11–296F208C and Sfi11-296H207Q F208C primarily showed impaired N-Spc42 binding with only a mild impact on Spc29, suggesting different binding requirements for Spc29 and Spc42 (Fig. 4 C). We next analyzed the impact of the mutations present in sfi1-7 on Spc29/Spc42 binding. Consistent with the lack of a growth phenotype of sfi1N218D D242A cells (Fig. 4 B), the Sfi11–296N218D D242A construct had no obvious impact on the binding of Cdc31, Spc29, and N-Spc42 (Fig. 4 D). However, the N-Sfi11–296P185T mutation alone or in combination with N218D and D242A abolished the interaction with N-Spc42 and reduced the binding to Spc29 dramatically (Fig. 4 D). These data together suggest that mutations in sfi1-3 and sfi1-7 impair the interactions with Spc29 and Spc42, but not with Cdc31. The defective interactions are probably causing the growth defect of the sfi1-3 and sfi1-7 mutant cells.
Failure of Sfi1 to bind to Spc29 and Spc42 should affect not bridge formation but instead the assembly of the satellite. By analyzing the phenotype of sfi1 cells that were synchronized with the mating pheromone α-factor before the release from the cell cycle block and incubation at the restrictive temperature (37°C), we tested this notion. At 37°C, more than half of the sfi1-3 cells arrested as large-budded cells with a single Spc42–yeast-enhanced GFP (yeGFP) signal reflecting the SPB (Fig. 4 E). About three-fourths of sfi1P185T cells accumulated with a large bud and a single Spc42-tdTomato SPB signal (Fig. 4 F). The single Spc42 signal phenotype of large-budded mitotic cells is explained either by two side-by-side SPBs or by an unduplicated SPB. To discriminate between both possibilities, we analyzed synchronized sfi1-3 and sfi1P185T cells incubated at 37°C by thin-section EM. Large-budded sfi1-3 carried a single SPB with a bridge but lacking a satellite (Fig. 4 G, SPB in one section and bridge in the next section; Fig. S3 A). Quantification of the EM images showed a nearly WT SPB size (Fig. S3 D) and that this single SPB in the sfi1-3 cells was connected to a bridge with a length of 113 nm (Fig. S3 E). A similar “single SPB with bridge” phenotype was observed in sfi1P185T mutant cells (Figs. 4 H and S3 E). However, in contrast to sfi1-3 cells, the single SPB of sfi1P185T cells was enlarged in size (Fig. S3 D). Finally, to show that our EM analysis is capable of visualizing the satellite and that synchronized WT cells incubated at 37°C carry mostly a half bridge, we performed EM analysis with WT cells. α-Factor arrested G1 cells contained a single SPB that was attached to a satellite-bearing bridge (Fig. S3 B). When α-factor–arrested WT cells were released from the G1 block at 37°C, we detected two SPBs of normal size (Bullitt et al., 1997) that were associated with a short half bridge lacking a satellite (Fig. S3, C–E).
Figure S3.
EM analysis of the SPB fate in WT and N-terminal Sfi1 mutants. (A) Additional EM analysis of synchronized sfi1H207Q F208C Y247C (sfi1-3) cells as in Fig. 4 G. (B) EM analysis of α-factor–arrested WT cells. (C) EM analysis of large-budded WT cells for 4 h at 37°C after release from α-factor. Cartoons in A–C illustrate the SPB phenotype. (D and E) Quantification of the SPB size (D) with n ≥ 13 and the half-bridge/bridge length of the indicated cell types from A–C and Fig. 4, G and H, with n ≥ 14. P values (< 0.0001 and n.s., D; < 0.0001, E) were derived by using a two-tailed t test. B, bridge; cMTs, cytoplasmic microtubules; HB, half bridge; nMTs, nuclear microtubules. Scale bar: 150 nm.
This together indicates that preventing binding of Spc29 and Spc42 to N-Sfi1 blocks SPB duplication at the level of satellite formation without affecting bridge assembly.
Importance of the Cdc31-binding sites in N-Sfi1 for SPB duplication
Our in vitro experiments indicate the importance of Cdc31-binding sites 1–3 in N-Sfi1 for the recruitment of Spc29. The Sfi11–239 with only one Cdc31-binding site bound Spc29 less efficiently than Sfi11–296 with three binding sites (Fig. 2 A). Cdc31-binding sites in the N-Sfi1 could function jointly in Spc29 and Spc42 recruitment, or, alternatively, only one Cdc31-binding site could be part of the Spc29/Spc42 binding region. To test these possibilities, we replaced the highly conserved tryptophan (see Fig. 1 A) in the first, second, and third Cdc31-binding sites with alanine in order to interfere with Cdc31 binding. Surprisingly, the W211A mutation in Sfi11–239 had little impact on Cdc31 binding in the Y2H system (Fig. S4 A). To further disrupt Cdc31 binding, we chose to mutate the phenylalanine at position −3 in respect to tryptophan (see Fig. 1 A). On the basis of structural predictions, this F208 could also be part of the interaction surface with Cdc31, and because it was mutated in sfi1-3 to cysteine (F208C), we decided to stay with this mutation (Li et al., 2006). F208C W211A in N-Sfi11–239 completely abolished the interaction with Cdc31 (Fig. S4 A). In addition, inactivation of all three Cdc31-binding sites (F208C W211A, F237C W240A, and F268C W271A) completely impaired binding of Cdc31 to Sfi11–296 (three Cdc31-binding sites).
Figure S4.
Additional analysis of sfi1Cdc31-binding site mutants. (A) Y2H study of Cdc31 with all Cdc31-binding site mutants in different N-Sfi1 constructs after X-Gal overlay. Partially, the same results as in Fig. 5 are shown here. (B) Drop test analysis of the indicated tryptophan mutants in SFI1 in a plasmid shuffle experiment. “Empty (sfi1∆)” indicates the sfi1Δ cells with the empty control plasmid pRS305K. Plates were incubated as described in Fig. 5 B. Cartoon illustrates which Cdc31-binding sites (marked with light green shades) are affected (marked in red). (C) Additional EM images for sfi1W211A W271A cells after 3 h at 37°C (as in Fig. 5 E). Cartoons illustrate the SPB phenotype. B, bridge; cMTs, cytoplasmic microtubules; nMTs, nuclear microtubules. YPD, yeast peptone dextrose.
With this knowledge, we inactivated the first, second, and third Cdc31-binding sites in Sfi11–296 (three Cdc31-binding sites) and analyzed the impact of these mutations on Cdc31, Spc29, and N-Spc42 binding in a Y2H assay. Inactivation of the first (F208C W211A) Cdc31-binding site completely abolished N-Spc42 interaction with little effect on Spc29 (Fig. 5 A). Cdc31 still bound to Sfi11–296F208C W211A through the second and third binding sites. The same consequence on Spc29 and N-Spc42 interactions as for N-Sfi11–296F208C W211A was observed when the third Cdc31-binding site was impaired (F268C W271A), while interference with the second Cdc31-binding sites in Sfi11–296 (F237C W240A) had no impact on the binding of Spc29 and N-Spc42 (Fig. 5 A). This analysis suggests that the second Cdc31-binding site in Sfi1 does not have a crucial role for Spc29 and Spc42 binding as long as the first and third binding sites are intact.
Figure 5.
Role of the N-terminal Cdc31-binding sites of Sfi1 for the SPB assembly. (A) Y2H analysis of tryptophan and phenylalanine mutants affecting the Cdc31-binding sites in Sfi1 after X-Gal overlay. Fig. S4 A shows the same Cdc31 Y2H analysis supplemented with further mutants of Sfi1. (B) Drop test of the SFI1 mutants in an SFI1-shuffle strain. 10-fold serial dilutions of cells on yeast peptone dextrose (YPD) G418 and 5-FOA plates were incubated for 2.5 d at 37°C and for 4 d at 23°C. Empty (sfi1∆) indicates the sfi1Δ cells with the empty control plasmid pRS305K. Cartoons on the right illustrate which Cdc31-binding site (Cdc31-binding sites are marked with light green shades; mutation is in the site marked with red) was mutated. (C) sfi1W211A W271A mutant phenotypes 3 h after temperature shift to 37°C or growth at 23°C. Left: Quantification of the cell cycle phases. Graph illustrates average of 3 independent experiments (n = 3) with >100 cells analyzed each time. Error bars are SD. Right: Exemplary fluorescence images. G1, S/M, and anaphase are sfi1W211A W271A cells grown at 23°C; mitotic arrest is an sfi1W211A W271A cell incubated at 37°C. (D) Quantification of the Spc42-yeGFP signal of the cells in C. Graph shows one representative result, which was confirmed in 3 independent experiments with n > 100 per strain and experiment. Average and SD are given. P value (< 0.0001) was derived by using a two-tailed t test. (E) EM micrographs of sfi1W211A W271A cells after 3 h at 23°C and 37°C are shown. See Fig. S4 C for additional EM images. Cartoons illustrate the SPB phenotype. (F and G) Quantification of the EM images for the SPB size and bridge length, respectively. For each cell type and temperature, n ≥ 17 for the SPB size and n ≥ 11 for the length of the bridge. Average and SD are given. P values (<0.0007, F; < 0.0001, G) were derived by using a two-tailed t test. B, bridge; cMTs, cytoplasmic microtubules; HB, half bridge; nMTs, nuclear microtubules; RFI, relative fluorescence intensity.
We next combined mutations in different Cdc31-binding sites of N-Sfi1 in order to test for cooperation. Double mutations in the first and second (W211A W240A) and the first and third (W211A W271A) Cdc31-binding sites of Sfi11–296 impaired binding of Spc29 and N-Spc42. Because the single W211A and W271A mutations only affected Spc42 binding with no influence on Spc29 (Fig. 5 A), the failure of the double mutants to interact with Spc29 suggests a cooperative impact of these two Cdc31 sites on Spc29 interaction (Fig. 5 A). In contrast, the W240A W271A (second and third) combination behaved as W271A and only affected N-Spc42 binding.
To verify the model of assisted Spc29 and Spc42 binding to Sfi1 by specific Cdc31-binding sites, we analyzed the growth behavior of sfi1 mutants carrying mutations in the first, second, and third Cdc31-binding sites of Sfi1. sfi1W211A and sfi1F208C W211A (first Cdc31-binding site) showed a reduction in the growth of cells at 37°C (Fig. 5 B, top and middle). sfi1W240A and sfi1F237C W240A (second) had no impact on cell growth (Fig. 5 B). sfi1W271A alone had no effect, while sfi1F268C W271A (third) cells were inviable at 23°C and 37°C (Fig. 5 B). Consistent with the Y2H experiment (Fig. 5 A), this result emphasizes the importance of the first Cdc31-binding site and, even more, the third Cdc31-binding site for the function of Sfi1.
We next addressed the impact of Cdc31-binding site cooperation on cell viability. The combination of W211A W240A (mutations in first and second binding sites) and W240A W271A (second and third binding sites) was lethal for cells (Fig. 5 B, bottom), while the single mutants had no or little growth influence compared with WT (Fig. 5 B, top). sfi1W211A W271A cells (first and third binding sites) showed a conditional lethal growth defect at 37°C (Fig. 5 B). The strong impact of W240A W271A on cell growth was surprising because the Y2H (Fig. 5 A) detected Spc29 binding. We propose that the Spc29 interaction with N-Sfi1 is not productive for satellite formation when the second and third Cdc31-binding sites are partially defective. This analysis suggests that the partial inactivation of two Cdc31-binding sites in the N-terminus of Sfi1 enhances the growth defect of cells.
To exclude the possibility that mutations in two Cdc31-binding sites of Sfi1 would generally cause lethality, we changed tryptophan or corresponding residues in Cdc31-binding sites 4–9 as single and double mutations. All single mutants grew at 23°C and 37°C. Double mutants affecting two Cdc31-binding sites were viable, except for sfi1W327 W377A (fifth and seventh binding sites; Fig. S4 B). This result indicates that the three Cdc31-binding sites in N-Sfi1 are particularly important for the function of Sfi1 because combining two tryptophan mutations in binding site 1, 2, or 3 impaired viability, whereas the growth defect caused by combined mutations in sites 4–9 was mostly negligible.
We took advantage of the temperature sensitivity of the sfi1W211A W271A cells for phenotype analysis. Considering the impact of these two mutations on Spc29 and Spc42 binding to N-Sfi1 (Fig. 5 A), we might expect that sfi1W211A W271A cells assemble a bridge but then fail in the recruitment of satellite components. Indeed, most sfi1W211A W271A cells arrested after 3 h at 37°C as large-budded cells with a single Spc42-yeGFP SPB signal (Fig. 5 C, sfi1W211A W271A cell with mitotic arrest at 37°C on the right; G1, S/M, and anaphase sfi1W211A W271A cells on the left grew at 23°C). Quantification of the Spc42-yeGFP signal in sfi1W211A W271A cells indicated an increase in the relative fluorescence intensity, which is consistent with either two side-by-side SPBs or the enlargement of the single SPB (Fig. 5 D). EM analysis of sfi1W211A W271A cells grown at 23°C revealed SPBs of normal size with an attached half bridge (Fig. 5 E, cell 1 and cell 2; Fig. 5, F and G). At the restrictive temperature (37°C), however, large-budded sfi1W211A W271A cells had only one slightly enlarged SPB (Fig. 5, E and F; Fig. S4 C) that carried a bridge without a satellite (Fig. 5, E and G; Fig. S4 C). This together supports the model that impairment of the N-terminal Cdc31-binding sites in Sfi1 is not important for bridge assembly but plays a role in the assembly of the satellite.
Protein kinase–induced block of satellite formation in anaphase
The bridge assembles in anaphase, while the satellite components Spc29 and Spc42 are not recruited before G1 (Burns et al., 2015). What delays the assembly of the satellite? In vitro phosphorylation analysis of purified GST-Sfi1 combined with mass spectrometry identified five sites that became phosphorylated by polo-like kinase Cdc5 but not by the kinase-dead variant of Cdc5, Mps1 kinase, the aurora kinase Ipl1, and mitotic Cdk1–Clb2 complex (Fig. 6 A). Three of these Cdc5 phosphosites have been described to be subject to phosphorylation in vivo (Keck et al., 2011; Fig. 6 A).
Figure 6.
Role of phosphorylation sites in the N-Sfi1. (A) Summary of phosphosites identified in vitro in purified GST-Sfi1 incubated with Cdc5 [a kinase-dead variant of Cdc5 [Cdc5-KD]), Mps1, Cdk1-Clb2, and Ipl1 and subsequent mass spectrometric analysis. Comparison with the phosphorylation sites identified in vivo as published elsewhere (Keck et al., 2011). (B) Drop test analysis of the indicated phosphoinhibitory (A) and phosphomimetic (D) mutants of SFI1 in an SFI1-shuffle experiment. Empty (sfi1∆) indicates the sfi1Δ cells with the empty control plasmid pRS305K. Plates were incubated as described in Fig. 5 B. (C) Phenotypic analysis of sfi1S10A T11A S42A and sfi1S10D T11D S42D mutants together with a WT SFI1 as described in Materials and methods. Top: Graph illustrates the average of 3 independent experiments (n = 3) with >100 large-budded cells analyzed each time. Error bars indicate SD. Bottom: Exemplary fluorescence images of the quantified phenotypes are shown. Scale bar: 5 µm. (D) EM micrographs of sfi1S10D T11D S42D cells 4 h after release from G1 block and temperature shift to 37°C. Cartoons illustrate the SPB phenotype. (E and F) Quantification of the EM images for the SPB size and bridge length, respectively, with n ≥ 9 for the SPB size and n = 6 for the length of the bridge per cell type. “Single SPB” indicates sfi1S10D T11D S42D cells that are large budded but contain only one SPB by EM. “Two SPBs” indicate large-budded sfi1S10D T11D S42D cells with two SPBs. Average and SD are given. P values (< 0.039, E; < 0.0001 ,F) were derived by using a two-tailed t test. B, bridge; cMTs, cytoplasmic microtubules; HB, half bridge; nMTs, nuclear microtubules; YPD, yeast peptone dextrose.
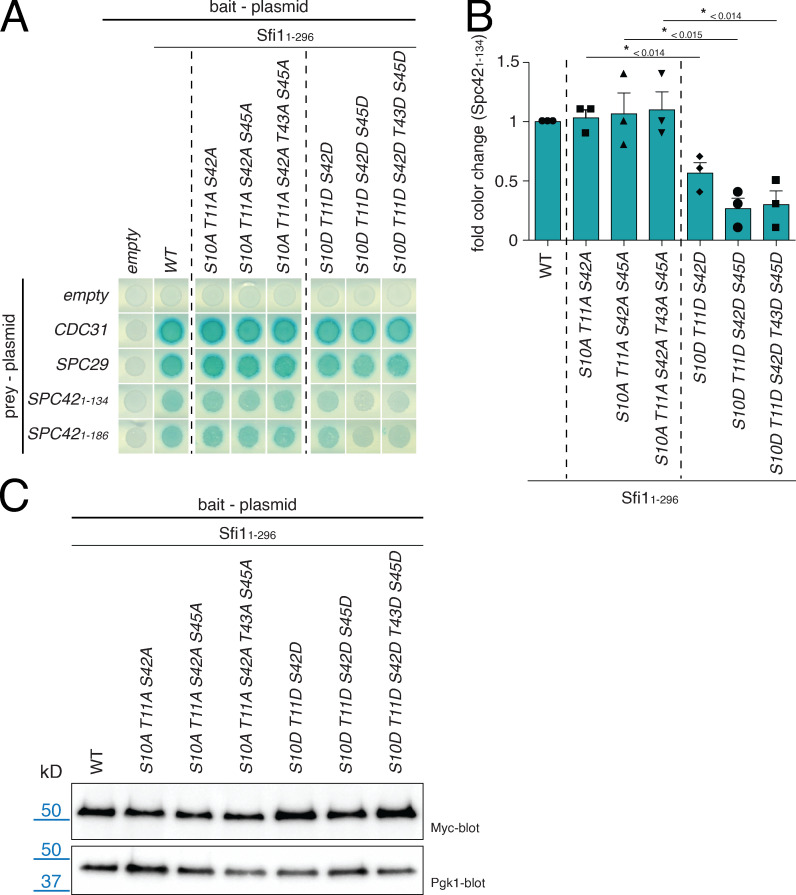
We tested the possibility that this phosphoregulation delays Spc29 and Spc42 binding to N-Sfi1 by introducing phosphoinhibitory (serine/threonine to alanine) and phosphomimetic mutations (serine/threonine to aspartic acid) in Sfi1. Phosphoinhibitory SFI1 mutants had little impact on the growth of cells at 23°C and 37°C (Fig. 6 B, A mutants). In contrast, the combination of at least three phosphomimetic SFI1 mutations (sfi1S10D T11D S42D) slowed growth at 23°C and impaired growth at 37°C (Fig. 6 B, D mutants). The addition of the S45D mutation further enhanced this growth defect (Fig. 6 B).
What could be the molecular defect of the phosphomimetic SFI1 mutations? Using the Y2H system, we detected a reduced interaction of phosphomimetic SFI1 D mutants (Fig. S5, A and B; e.g., Sfi11–296S10D T11D S42D) with N-Spc42 without affecting Spc29 or Cdc31 interaction (Fig. S5 A). Such reduced binding was not observed for the phosphoinhibitory A mutants (Fig. S5, A and B). As a control, we established that all Sfi1 Y2H constructs were expressed similarly in yeast cells (Fig. S5 C). This suggests that the phosphorylation of N-Sfi1 affects interaction with Spc42.
Figure S5.
Y2H interaction study of sfi1Cdc5 mutants. (A) Y2H analysis of the sfi1S10D T11D S42D mutations on the interaction with N-Spc42 after X-Gal overlay. (B) Quantification of A. Blue color of the cell patches in A was quantified and normalized to WT. n = 3. Error bars are SD. P values are indicated in the figure and were derived by using a two-tailed t test. Shape symbols represent individual data points from each experiment. (C) Immunoblot analysis to check for the expression of the SFI11–296 Y2H bait constructs from A. Pgk1 is the loading control.
The phenotype of sfi1S10D T11D S42D and sfi1S10A T11A S42A cells (three phosphosites that were confirmed in vivo and in vitro) was analyzed at 37°C. While the A mutant behaved similar to the WT, D mutant cells arrested in the cell cycle with a large bud, a phenotype that is typical for spindle assembly checkpoint maintenance (Hoyt et al., 1991; Li and Murray, 1991). About 60% of large-budded sfi1S10D T11D S42D cells carried only one Spc42-tdTomato SPB signal (Fig. 6 C). Analysis of large-budded sfi1S10D T11D S42D cells by EM detected cells with a single enlarged SPB (Fig. 6 D, left panel; and Fig. 6 E) that carried a full bridge structure but lacked a satellite (Fig. 6 D, left panel; and Fig. 6 F). In contrast, when large-budded sfi1S10D T11D S42D cells contained two SPBs, these SPBs were associated with a half bridge (Fig. 6 D, right panel; and Fig. 6 F). This analysis suggests that phosphorylation of N-Sfi1 inhibits satellite assembly despite successful bridge formation.
Discussion
The yeast SPB is a relatively simple MT-organizing center that consists of only 16 proteins (Seybold and Schiebel, 2013). As is true of the human centrosomes, it has the essential feature of duplication once per cell cycle (Rüthnick and Schiebel, 2016). The small number of SPB components and the comprehensive understanding of the yeast cell cycle make it an ideal model to study the conserved mechanism of how the cell cycle machinery restricts duplication of a proteinaceous structure to one event per cell cycle.
It was proposed that the exposure of free N-Sfi1 triggers the assembly of the daughter SPB precursor, the satellite, at the distal end of the bridge (Kilmartin, 2003). However, little experimental evidence for such a function of N-Sfi1 has been provided. Here, we show by in vivo and in vitro approaches that N-Sfi1 can recruit the satellite components Spc29 and Spc42 to the distal end of the bridge. Biochemical data suggest direct binding of Spc29 and Spc42 to N-Sfi1. Both binding sites are probably distinct, as supported by the analysis of sfi1 mutants. sfi1P185T and sfi1F208C only moderately affected Spc29 binding but completely abolished the interaction with Spc42. This stronger impact on Spc42 binding was also observed for the sfi1W211A and sfi1W271A mutants. Although recombinant Spc29 and Spc42 proteins bind independently from each other to N-Sfi1/Cdc31 and no enhancement of N-Spc42 binding by addition of Spc29 was observed, this finding does not exclude the possibility that, during SPB duplication in the natural cellular environment, both interacting proteins become recruited to N-Sfi1/Cdc31 in a cooperative manner (Elliott et al., 1999). The importance of Spc29 and Spc42 binding to N-Sfi1/Cdc31 for satellite formation is emphasized by the phenotypes of sfi1-3, sfi1-7, sfi1W211A W271A, and sfi1S10D T11D S42D cells. All mutant cells fail completely or partly in SPB duplication despite bridge assembly due to their deficiency to bind Spc29 and/or Spc42, and therefore satellite assembly is not initiated.
The yeast centrin Cdc31 has an essential role in SPB duplication. Conditional lethal cdc31(ts) cells arrest in mitosis with an enlarged SPB that does not carry a half bridge or bridge (Baum et al., 1986). The lateral cross-linking activity of Cdc31 on Sfi1 molecules within the bridge that is defective in cdc31(ts) mutant cells explains this phenotype (Seybold et al., 2015). The data of this study suggest an additional, novel function of Cdc31 in the recruitment of Spc29 and Spc42 to N-Sfi1 and therefore in satellite formation. As shown by in vitro binding analysis, binding of Spc29 to N-Sfi1 strongly requires the presence of Cdc31 (Fig. 2 C). In addition, sfi1 mutations that impair Cdc31 binding also disrupt Spc42 and Spc29 interaction with N-Sfi1 (Fig. 5). This raises the question how Cdc31 contributes to the recruitment of Spc29 and Spc42 to N-Sfi1. The Y2H data indicate a complex interplay of different Cdc31-binding sites in N-Sfi1 and emphasize the special importance of the first and third Cdc31-binding sites for the recruitment of Spc29 and Spc42 (Fig. 5). However, neither in Y2H (Fig. 3 B) nor in co-IP experiments we performed for this study did we find evidence for a direct interaction of Cdc31 with either Spc29 or N-Spc42. Nevertheless, Cdc31 bound to N-Sfi1 could promote Spc29 and Spc42 binding by assisting the recruitment of these proteins to N-Sfi1, by stabilizing the interactions of the tetrameric Spc29–Spc42–N-Sfi1–Cdc31 complex, or indirectly by stabilizing a binding site in N-Sfi1 and therefore providing an interaction platform for Spc29 and Spc42. Structural analysis of N-Sfi1 with bound Cdc31, Spc29, and Spc42 will be needed to solve this open question.
Puzzlingly, while bridge formation occurs in anaphase, the assembly of the satellite does not happen before G1 phase (Burns et al., 2015; Elserafy et al., 2014). This indicates that a mechanism in addition to the exposure of free N-Sfi1 regulates the assembly of the satellite. Because the deletion of the N-terminal 62 amino acids in Sfi1 enhances binding of Spc29 (Fig. 2, E and F), we tested the possibility that this Sfi1 region functions as such a regulatory element. Phosphorylation analysis suggests that polo-like kinase Cdc5 has the ability to phosphorylate several residues in this N-terminal region of Sfi1. These phosphorylations of N-Sfi1 have a negative impact on the interaction with Spc42 in the Y2H system without affecting Spc29 (Fig. S5, A and B). However, it is still possible that, within the bridge environment at the SPB, where ∼80 similarly positioned N-Sfi1 become phosphorylated (Seybold et al., 2015), this combined phosphorylation load influences Spc29 binding. In addition, phosphomimetic sfi1S10D T11D S42D mutant cells assemble a bridge at the restrictive temperature but then fail in satellite formation (Fig. 5). This together strongly supports the notion that phosphorylation of N-Sfi1 by Cdc5 delays satellite formation until the decline of Cdc5 activity with mitotic exit (Charles et al., 1998) and is consistent with structured illumination microscopy data showing that Spc29 and Spc42 only become recruited to the distal end of the bridge with mitotic exit in G1 (Burns et al., 2015).
Data provided in this article also suggest that inhibition of free N-Sfi1 by Cdc5 phosphorylation is not essential for the viability of cells, as indicated by the relatively normal growth of the phosphoinhibitory sfi1S10A T11A S42A (A mutant) and the sfi1Δ1–62 mutant cells. Thus, N-Sfi1 phosphorylation by Cdc5 is probably a safeguard mechanism that only becomes essential under disturbing conditions, such as during prolonged mitotic arrest or if an unknown compensation mechanism fails. Future studies will clarify under what circumstances N-Sfi1 regulation by Cdc5 becomes essential.
On the basis of our data, we now can close the gap between bridge assembly in early anaphase and satellite biogenesis in G1. The exposed free N-Sfi1 are first blocked by Cdc5 phosphorylation in mitosis. The decline of Cdc5 kinase activity with mitotic exit (Charles et al., 1998) allows binding of Spc29 and Spc42 to the distal end of the bridge in early G1. Cdk1 kinase activity that promotes Spc42 polymerization into a 2D crystal then most likely triggers Spc42 layer expansion into a duplication plaque after START of the cell cycle (Bullitt et al., 1997; Jaspersen et al., 2004). Mps1 kinase that phosphorylates Spc29 and Spc42 at multiple sites is also involved in this step (Araki et al., 2010; Jones et al., 2018). Finally, with the help of a recruited nuclear pore complex and the SPB insertion network consisting of the proteins Bbp1, Mps2, Ndc1, and Nbp1, the duplication plaque becomes inserted into the double membrane of the NE (Araki et al., 2006; Chen et al., 2019; de la Cruz Muñoz-Centeno et al., 1999; Rüthnick et al., 2017; Schramm et al., 2000; Winey et al., 1993).
Sfi1 proteins are encoded by the genomes of yeast to human cells. However, the conserved region is restricted to the >20 central centrin-binding sites (Rüthnick and Schiebel, 2016). Because Cdc31 provides lateral interactions between Sfi1/Cdc31 molecules (Seybold et al., 2015), the common function of the central Sfi1/centrin region could be the formation of parallel protein arrays. The N-terminal region of Sfi1 homologues is not conserved. However, this is also not the case for Spc29 and Spc42. An SPC29 orthologue is not encoded by the S. pombe genome (Bestul et al., 2017). S. pombe Pcp89 that fulfills the function of Spc42 is not related to Spc42 at the amino acid level (Rosenberg et al., 2006). It was suggested that human Cep57 might play an analogous role to Spc42/Pcp89 at centrioles (Bestul et al., 2017). It will be interesting to test whether these functionally related Spc42/Pcp89/Cep57 proteins interact with the various N-Sfi1/centrin complexes whose amino acid sequences are now adapted to the organism-specific protein of interaction.
Taken together, this study describes the missing link between SPB bridge formation in anaphase and the assembly of the daughter SPB precursor, the satellite, at the distal end of the bridge in G1. In addition, it suggests a model for the function of the divergent N-Sfi1.
Materials and methods
Yeast strains, plasmids, and culture conditions
All strains used in this study are listed in Table S1. Endogenous gene tagging and deletions were constructed via PCR-based methods (Janke et al., 2004). Point mutations in SFI1 were generated in a pRS305K-SFI1 plasmid via a two-step overlap mutagenesis PCR (Francis et al., 2017). Primers that were used for mutagenesis are listed in Table S1. Single integrations of these plasmids into the genome were achieved by double digest of the plasmid with DraIII-HF and ZraI (Taxis and Knop, 2006), subsequent transformation into a SFI1-shuffle strain, and selection for Geneticin (G418)-resistant clones. The integration of the plasmid fragment was confirmed via colony PCR. To analyze the growth behavior of the mutants, 10-fold serial diluted drop tests were performed on yeast peptone dextrose plates with G418 and synthetic complete (SC) media plates with 5-FOA. The latter ones allowed counterselection for strains that spontaneously lost the URA3-based pRS316-SFI1 rescue plasmid. Yeast cells were generally grown in selective synthetic defined or SC media supplemented with 2% glucose or 3% raffinose. To induce the expression of proteins under the Gal1 promoter (e.g., to test the expression of the Y2H proteins), galactose was added to a final concentration of 2%. Alkaline lysis and TCA precipitations were used to prepare yeast cell extracts (Janke et al., 2004).
For the analysis of the temperature-sensitive mutants, cells were precultured at 23°C, synchronized in G1 by the addition of α-factor (10 µg/ml) for 2.5 h, and released back into the cell cycle by a wash and medium change to α-factor–free medium. One half of the culture was incubated at 23°C, and the other half was shifted to 37°C. Cells were subjected directly to live-cell microscopy or fixed for 10 min in 4% PFA/2% sucrose for later microscopic analysis.
Y2H
All indicated gene fragments were cloned into pMM5 and pMM6 (Schramm et al., 2001) and transformed in SGY37 and YPH500, respectively. Transformants were incubated in selective media. On the next day, the density was adjusted to OD600 1, and mating was performed in a 96-well plate in yeast extract, peptone, adenine, and dextrose overnight at 30°C. On the next day, the cells were resuspended carefully and plated on double-selective SC plates lacking histidine and leucine. The plates were incubated again at 30°C for 30 h before the X-Gal overlay was performed (Schramm et al., 2001). Change of color was carefully documented every hour, and figures usually show the Y2H result after 4–5 h. For the quantification of the color change, the images were further analyzed with the open-source ImageJ software (National Institutes of Health), and GraphPad Prism software was implemented for statistical analysis.
Fluorescence light microscopy
A DeltaVision RT system (Olympus IX71 based; Applied Precision Ltd.) equipped with a Photometrics CoolSnap HQ camera (Roper Scientific), a 100×/1.4-NA Super-Plan Apochromat oil objective (Olympus), a four-color Standard Insight SSI module light source, a workstation with a CentOS operating system, and softWorx software (Applied Precision Ltd.) was used for imaging at room temperature (23°C). Cells were placed on a microscopic slide and kept in their growth media (live cells) or PBS (fixed cells) for the time of imaging. For all fluorescence signal quantification experiments, all imaging was conducted with the same exposure and illumination settings on living cells to allow the direct comparison of the results. For the quantification of the signal intensity, the integrated density (IntDen) of the SPB (Spc42-yeGFP) in the brightest stack was measured with a 5 × 5 pixel-square and a 7 × 7 pixel-square for background correction. The following formula was used to calculate the relative fluorescence intensity: RFI = IntDen5×5 – {[IntDen7×7 – IntDen5×5] × [area5×5/(area7×7 – area5×5)]}. Image processing and phenotypic analysis were performed with ImageJ software. Quantifications were performed three times and analyzed with GraphPad Prism software. A combined graph is shown in Fig. 4, E and F; Fig. 5, C and D; and Fig. 6 C.
EM
Yeast cells were prepared for EM analysis as described previously (Rüthnick et al., 2017). In short, cells were high-pressure frozen with an HPM010 (Abra-Fluid) after collection via filtration, then freeze substituted using the EM-AFS2 device (Leica Microsystems), and 70-nm serial sections were prepared on a Reichert Ultracut S Microtome (Leica Instruments) and post-stained with 2% uranyl acetate and lead citrate. Sections were scanned by using a Jeol JE-1400 (JEOL Ltd.) operating at 80 kV and equipped with a 4k × 4k digital camera (F416; TVIPS). Micrographs were further processed using ImageJ software, and statistical analysis was performed with GraphPad Prism software.
Protein purification, in vitro kinase assay, and mass spectrometry
These methods were performed as described before (Elserafy et al., 2014). In short, all kinases were purified from yeast cells (Geymonat et al., 2007; Ubersax et al., 2003), while GST-Sfi1 was purified from E. coli by using GST-Sepharose beads (Macherey-Nagel). Subsequently, the in vitro kinase reaction was performed in the presence of 5 µCi of γ-32P-ATP (0.05 nM). Samples were prepared for mass spectrometry, where peptides were analyzed by liquid chromatography–tandem mass spectrometry (Orbitrap Elite; Thermo Fisher Scientific). The data were processed using Proteome Discoverer software (version 1.4; Thermo Fisher Scientific). Phosphorylation site localization was performed on the Mascot results using PhosphoRS.
IP
E. coli BL21-CodonPlus was (co)transformed with the indicated plasmids. The protein expression was induced with 0.5 mM IPTG at 37°C for 2–3 h or, in the case of the 3K2N-Spc42-6His constructs, with 1 mM IPTG overnight at 16°C (Drennan et al., 2019). The bacteria were harvested by centrifugation, and the pellet was snap frozen in liquid nitrogen and stored at −80°C for further processing. The pellet was thawed on ice, resuspended in 1.5 ml of FLAG-IP-lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM PMSF, Complete Mini Protease Inhibitor Cocktail; Merck), and sonicated on ice until 90% of the cells had burst. Cell lysate was supplemented with 0.5% Triton X-100, 250 U Benzonase Nuclease (EMD Millipore), and incubated for 1 h, rotating at 4°C. The soluble proteins were separated from the cell debris by centrifugation (21,000 g for 15 min) and incubated with previously equilibrated anti-FLAG M2 agarose or magnetic beads (Merck) for 3–4 h, rotating at 4°C. Sfi1, Cdc31, and Spc29 were coexpressed at 37°C (Fig. 2). The Spc42 fusion constructs were most soluble when expressed at 16°C. To combine them with the N-Sfi1/Cdc31 and Spc29 constructs that were expressed separately, we mixed the proteins in experiments (Fig. 3, C and D). The anti-FLAG beads were incubated with E. coli extracts containing Sfi1-FLAG, Sfi1-FLAG/Cdc31, Spc42, or Spc29 constructs. The bound proteins were washed five times with lysis buffer and eluted with 100 µl of 4× Laemmli buffer. The proteins were separated by SDS-PAGE and analyzed with Coomassie blue staining or transferred to 0.22-µm nitrocellulose membrane (Bio-Rad Laboratories) for Western blotting. After staining of the membrane with primary and secondary antibodies (see next section), the signals were detected with the LAS-3000 imaging system (Fujifilm). For quantifications, the scanned gels or membranes were further analyzed with ImageJ software, and statistical analysis was performed with GraphPad Prism software.
Antibodies
Antibodies used in this study were as follows: rabbit anti-Spc42 (Western blot, 1:1,000) and rabbit anti-Spc29 (Western blot, 1:500; Elliott et al., 1999), mouse anti-FLAG (Western blot, 1:1,000, F1804; Cell Signaling Technology), rabbit anti-FLAG (Western blot, 1:1,000, 20543-1-AP; ProteinTech), goat anti-Cdc31 (Western blot, 1:500; Spang et al., 1993), mouse anti-Penta-His (Western blot, 1:1,000, 34660; Qiagen), mouse monoclonal anti-His-HRP (Western blot, 1:2,500, HRP-66005; ProteinTech), mouse monoclonal antipolyhistidine (Western blot, 1:3,000, H1029; Merck), rat anti-HA (Western blot, 1:1,000, 1867423; Merck), mouse anti-Myc (Western blot, 1:1,000, OP10; Cell Signaling Technology), rabbit anti-Tub4 (Western blot, 1:500; Geissler et al., 1996). Corresponding secondary HRP-tagged antibodies were mostly purchased from Jackson ImmunoResearch Laboratories: donkey anti-rabbit HRP (1:5,000; 711-035-152), anti-mouse HRP (1:10,000; 715-035-151), and rabbit anti-goat HRP (1:10,000; 305-035-045). Furthermore, goat anti-rat HRP (1:5,000, A10549; Molecular Probes) and anti-mouse Ig HRP (1:1,000, 18-8817-33; Rockland Immunochemicals) antibodies were used.
Online supplemental material
Fig. S1 shows the full initial Y2H screen of Fig. 1 B and the expression of the Sfi1-Y2H bait constructs. Fig. S2 confirms expression of the Spc42-Y2H bait constructs. Fig. S3 displays EM images and quantifications supplementary to Fig. 4. In Fig. S4, Y2H analysis for the interaction of Cdc31 with all sfi1W/F mutants is shown together with the growth assay for mutations in SFI1 that affect the more central Cdc31-binding sites and EM micrographs for sfi1W211A W271A (supplemental to Fig. 5 E). Fig. S5 shows the interaction of the different sfi1Cdc5 mutants and confirms their expression from the Y2H bait plasmid. Table S1 lists yeast strains, plasmids, and primers used in this study.
Supplementary Material
lists all yeast strains, describing their genotype, and lists all plasmids used in this study.
Acknowledgments
We are thankful to the Center for Molecular Biology Mass Spectrometry Facility for analyzing phosphorylated proteins and the Electron Microscopy Core Facility of Heidelberg University for their support. We thank U. Jäkle for excellent technical support and Dr. Hans Hombauer (University of Heidelberg, Heidelberg, Germany) for sharing several reagents and expertise. We thank Dr. J. Kilmartin (Emeritus of Medical Research Council Laboratory of Molecular Biology, Cambridge, UK) for sending us plasmids carrying sfi1-3 and sfi1-7.
This work was supported by Deutsche Forschungsgemeinschaft grant Schi 295/5-3.
The authors declare no competing financial interests.
Author contributions: Conceptualization, methodology, project administration, supervision, and writing the original draft were accomplished by E. Schiebel and D. Rüthnick. Further on, D. Rüthnick was responsible for data curation, formal analysis, and visualization. The investigation was carried out by D. Rüthnick, J. Vitale, and A. Neuner (precisely all EM experiments). E. Schiebel ensured funding acquisition and provision of all resources.
References
- Adams, I.R., and Kilmartin J.V.. 1999. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 145:809–823. 10.1083/jcb.145.4.809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, Y., Lau C.K., Maekawa H., Jaspersen S.L., Giddings T.H. Jr., Schiebel E., and Winey M.. 2006. The Saccharomyces cerevisiae spindle pole body (SPB) component Nbp1p is required for SPB membrane insertion and interacts with the integral membrane proteins Ndc1p and Mps2p. Mol. Biol. Cell. 17:1959–1970. 10.1091/mbc.e05-07-0668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, Y., Gombos L., Migueleti S.P.S., Sivashanmugam L., Antony C., and Schiebel E.. 2010. N-terminal regions of Mps1 kinase determine functional bifurcation. J. Cell Biol. 189:41–56. 10.1083/jcb.200910027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avena, J.S., Burns S., Yu Z., Ebmeier C.C., Old W.M., Jaspersen S.L., and Winey M.. 2014. Licensing of yeast centrosome duplication requires phosphoregulation of sfi1. PLoS Genet. 10:e1004666. 10.1371/journal.pgen.1004666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum, P., Furlong C., and Byers B.. 1986. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc. Natl. Acad. Sci. USA. 83:5512–5516. 10.1073/pnas.83.15.5512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestul, A.J., Yu Z., Unruh J.R., and Jaspersen S.L.. 2017. Molecular model of fission yeast centrosome assembly determined by superresolution imaging. J. Cell Biol. 216:2409–2424. 10.1083/jcb.201701041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhlel, I.B., Ohta M., Mayeux A., Bordes N., Dingli F., Boulanger J., Velve Casquillas G., Loew D., Tran P.T., Sato M., et al. 2015. Cell cycle control of spindle pole body duplication and splitting by Sfi1 and Cdc31 in fission yeast. J. Cell Sci. 128:1481–1493. 10.1242/jcs.159657 [DOI] [PubMed] [Google Scholar]
- Bullitt, E., Rout M.P., Kilmartin J.V., and Akey C.W.. 1997. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 89:1077–1086. 10.1016/S0092-8674(00)80295-0 [DOI] [PubMed] [Google Scholar]
- Burns, S., Avena J.S., Unruh J.R., Yu Z., Smith S.E., Slaughter B.D., Winey M., and Jaspersen S.L.. 2015. Structured illumination with particle averaging reveals novel roles for yeast centrosome components during duplication. eLife. 4:e08586. 10.7554/eLife.08586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers, B., and Goetsch L.. 1975. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 124:511–523. 10.1128/JB.124.1.511-523.1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles, J.F., Jaspersen S.L., Tinker-Kulberg R.L., Hwang L., Szidon A., and Morgan D.O.. 1998. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 8:497–507. 10.1016/S0960-9822(98)70201-5 [DOI] [PubMed] [Google Scholar]
- Chen, J., Gardner J.M., Yu Z., Smith S.E., McKinney S., Slaughter B.D., Unruh J.R., and Jaspersen S.L.. 2019. Yeast centrosome components form a noncanonical LINC complex at the nuclear envelope insertion site. J. Cell Biol. 218:1478–1490. 10.1083/jcb.201809045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz Muñoz-Centeno, M., McBratney S., Monterrosa A., Byers B., Mann C., and Winey M.. 1999. Saccharomyces cerevisiae MPS2 encodes a membrane protein localized at the spindle pole body and the nuclear envelope. Mol. Biol. Cell. 10:2393–2406. 10.1091/mbc.10.7.2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson, A.D., and Kilmartin J.V.. 1996. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) with an essential function during SPB duplication. J. Cell Biol. 132:887–901. 10.1083/jcb.132.5.887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drennan, A.C., Krishna S., Seeger M.A., Andreas M.P., Gardner J.M., Sether E.K.R., Jaspersen S.L., and Rayment I.. 2019. Structure and function of Spc42 coiled-coils in yeast centrosome assembly and duplication. Mol. Biol. Cell. 30:1505–1522. 10.1091/mbc.E19-03-0167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott, S., Knop M., Schlenstedt G., and Schiebel E.. 1999. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA. 96:6205–6210. 10.1073/pnas.96.11.6205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elserafy, M., Šarić M., Neuner A., Lin T.C., Zhang W., Seybold C., Sivashanmugam L., and Schiebel E.. 2014. Molecular mechanisms that restrict yeast centrosome duplication to one event per cell cycle. Curr. Biol. 24:1456–1466. 10.1016/j.cub.2014.05.032 [DOI] [PubMed] [Google Scholar]
- Francis, M.S., Amer A.A., Milton D.L., and Costa T.R.. 2017. Site-directed mutagenesis and its application in studying the interactions of T3S components. Methods Mol. Biol. 1531:11–31. 10.1007/978-1-4939-6649-3_2 [DOI] [PubMed] [Google Scholar]
- Friederichs, J.M., Ghosh S., Smoyer C.J., McCroskey S., Miller B.D., Weaver K.J., Delventhal K.M., Unruh J., Slaughter B.D., and Jaspersen S.L.. 2011. The SUN protein Mps3 is required for spindle pole body insertion into the nuclear membrane and nuclear envelope homeostasis. PLoS Genet. 7:e1002365. 10.1371/journal.pgen.1002365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler, S., Pereira G., Spang A., Knop M., Souès S., Kilmartin J., and Schiebel E.. 1996. The spindle pole body component Spc98p interacts with the γ-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 15:3899–3911. 10.1002/j.1460-2075.1996.tb00764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geymonat, M., Spanos A., and Sedgwick S.G.. 2007. A Saccharomyces cerevisiae autoselection system for optimised recombinant protein expression. Gene. 399:120–128. 10.1016/j.gene.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Hagan, I., and Yanagida M.. 1995. The product of the spindle formation gene sad1+ associates with the fission yeast spindle pole body and is essential for viability. J. Cell Biol. 129:1033–1047. 10.1083/jcb.129.4.1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt, M.A., Totis L., and Roberts B.T.. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517. 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- Janke, C., Magiera M.M., Rathfelder N., Taxis C., Reber S., Maekawa H., Moreno-Borchart A., Doenges G., Schwob E., Schiebel E., et al. 2004. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast. 21:947–962. 10.1002/yea.1142 [DOI] [PubMed] [Google Scholar]
- Jaspersen, S.L., and Winey M.. 2004. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 20:1–28. 10.1146/annurev.cellbio.20.022003.114106 [DOI] [PubMed] [Google Scholar]
- Jaspersen, S.L., Giddings T.H.J. Jr., and Winey M.. 2002. Mps3p is a novel component of the yeast spindle pole body that interacts with the yeast centrin homologue Cdc31p. J. Cell Biol. 159:945–956. 10.1083/jcb.200208169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen, S.L., Huneycutt B.J., Giddings T.H. Jr., Resing K.A., Ahn N.G., and Winey M.. 2004. Cdc28/Cdk1 regulates spindle pole body duplication through phosphorylation of Spc42 and Mps1. Dev. Cell. 7:263–274. 10.1016/j.devcel.2004.07.006 [DOI] [PubMed] [Google Scholar]
- Jones, M.H., O’Toole E.T., Fabritius A.S., Muller E.G., Meehl J.B., Jaspersen S.L., and Winey M.. 2018. Key phosphorylation events in Spc29 and Spc42 guide multiple steps of yeast centrosome duplication. Mol. Biol. Cell. 29:2280–2291. 10.1091/mbc.E18-05-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck, J.M., Jones M.H., Wong C.C., Binkley J., Chen D., Jaspersen S.L., Holinger E.P., Xu T., Niepel M., Rout M.P., et al. 2011. A cell cycle phosphoproteome of the yeast centrosome. Science. 332:1557–1561. 10.1126/science.1205193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin, J.V. 2003. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J. Cell Biol. 162:1211–1221. 10.1083/jcb.200307064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop, M., and Schiebel E.. 1998. Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J. 17:3952–3967. 10.1093/emboj/17.14.3952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani, A., Moyer T., Chen A., Holland A., Walsh C.A., and Reiter J.F.. 2019. SFI1 promotes centriole duplication by recruiting USP9X to stabilize the microcephaly protein STIL. J. Cell Biol. 218:2185–2197. 10.1083/jcb.201803041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary, A., Sim S., Nazarova E., Shulist K., Genthial R., Yang S.K., Bui K.H., Francois P., and Vogel J.. 2019. Successive kinesin-5 microtubule crosslinking and sliding promote fast, irreversible formation of a stereotyped bipolar spindle. Curr. Biol. 29:3825–3837.e3. 10.1016/j.cub.2019.09.030 [DOI] [PubMed] [Google Scholar]
- Li, R., and Murray A.W.. 1991. Feedback control of mitosis in budding yeast. Cell. 66:519–531. 10.1016/0092-8674(81)90015-5 [DOI] [PubMed] [Google Scholar]
- Li, S., Sandercock A.M., Conduit P., Robinson C.V., Williams R.L., and Kilmartin J.V.. 2006. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 173:867–877. 10.1083/jcb.200603153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg, J.A., Tomlin G.C., McDonald W.H., Snydsman B.E., Muller E.G., Yates J.R. III, and Gould K.L.. 2006. Ppc89 links multiple proteins, including the septation initiation network, to the core of the fission yeast spindle-pole body. Mol. Biol. Cell. 17:3793–3805. 10.1091/mbc.e06-01-0039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüthnick, D., and Schiebel E.. 2016. Duplication of the yeast spindle pole body once per cell cycle. Mol. Cell. Biol. 36:1324–1331. 10.1128/MCB.00048-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüthnick, D., and Schiebel E.. 2018. Duplication and nuclear envelope insertion of the yeast microtubule organizing centre, the spindle pole body. Cells. 7:42. 10.3390/cells7050042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüthnick, D., Neuner A., Dietrich F., Kirrmaier D., Engel U., Knop M., and Schiebel E.. 2017. Characterization of spindle pole body duplication reveals a regulatory role for nuclear pore complexes. J. Cell Biol. 216:2425–2442. 10.1083/jcb.201612129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, W.S., and Hoyt M.A.. 1992. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 70:451–458. 10.1016/0092-8674(92)90169-D [DOI] [PubMed] [Google Scholar]
- Schramm, C., Elliott S., Shevchenko A., Shevchenko A., and Schiebel E.. 2000. The Bbp1p-Mps2p complex connects the SPB to the nuclear envelope and is essential for SPB duplication. EMBO J. 19:421–433. 10.1093/emboj/19.3.421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm, C., Janke C., and Schiebel E.. 2001. Molecular dissection of yeast spindle pole bodies by two hybrid, in vitro binding, and co-purification. Methods Cell Biol. 67:71–94. 10.1016/S0091-679X(01)67006-7 [DOI] [PubMed] [Google Scholar]
- Seybold, C., and Schiebel E.. 2013. Spindle pole bodies. Curr. Biol. 23:R858–R860. 10.1016/j.cub.2013.07.024 [DOI] [PubMed] [Google Scholar]
- Seybold, C., Elserafy M., Rüthnick D., Ozboyaci M., Neuner A., Flottmann B., Heilemann M., Wade R.C., and Schiebel E.. 2015. Kar1 binding to Sfi1 C-terminal regions anchors the SPB bridge to the nuclear envelope. J. Cell Biol. 209:843–861. 10.1083/jcb.201412050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang, A., Courtney I., Fackler U., Matzner M., and Schiebel E.. 1993. The calcium-binding protein cell division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J. Cell Biol. 123:405–416. 10.1083/jcb.123.2.405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis, C., and Knop M.. 2006. System of centromeric, episomal, and integrative vectors based on drug resistance markers for Saccharomyces cerevisiae. Biotechniques. 40:73–78. 10.2144/000112040 [DOI] [PubMed] [Google Scholar]
- Ubersax, J.A., Woodbury E.L., Quang P.N., Paraz M., Blethrow J.D., Shah K., Shokat K.M., and Morgan D.O.. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature. 425:859–864. 10.1038/nature02062 [DOI] [PubMed] [Google Scholar]
- Vallen, E.A., Hiller M.A., Scherson T.Y., and Rose M.D.. 1992. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J. Cell Biol. 117:1277–1287. 10.1083/jcb.117.6.1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin, R., Craig K., Hwang E.S., Prinz S., Tyers M., and Amon A.. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol. Cell. 2:709–718. 10.1016/S1097-2765(00)80286-5 [DOI] [PubMed] [Google Scholar]
- Winey, M., Hoyt M.A., Chan C., Goetsch L., Botstein D., and Byers B.. 1993. NDC1: a nuclear periphery component required for yeast spindle pole body duplication. J. Cell Biol. 122:743–751. 10.1083/jcb.122.4.743 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists all yeast strains, describing their genotype, and lists all plasmids used in this study.