Thousands of variants are associated with genetic cardiomyopathies; however, the mechanisms driving these progressive, heterogeneous diseases are not well understood. Functional molecular studies are necessary for both mechanistically understanding disease pathogenesis and realizing the promise of precision medicine–based therapeutics.
Abstract
Genetic cardiomyopathies have been studied for decades, and it has become increasingly clear that these progressive diseases are more complex than originally thought. These complexities can be seen both in the molecular etiologies of these disorders and in the clinical phenotypes observed in patients. While these disorders can be caused by mutations in cardiac genes, including ones encoding sarcomeric proteins, the disease presentation varies depending on the patient mutation, where mutations even within the same gene can cause divergent phenotypes. Moreover, it is challenging to connect the mutation-induced molecular insult that drives the disease pathogenesis with the various compensatory and maladaptive pathways that are activated during the course of the subsequent progressive, pathogenic cardiac remodeling. These inherent complexities have frustrated our ability to understand and develop broadly effective treatments for these disorders. It has been proposed that it might be possible to improve patient outcomes by adopting a precision medicine approach. Here, we lay out a practical framework for such an approach, where patient subpopulations are binned based on common underlying biophysical mechanisms that drive the molecular disease pathogenesis, and we propose that this function-based approach will enable the development of targeted therapeutics that ameliorate these effects. We highlight several mutations to illustrate the need for mechanistic molecular experiments that span organizational and temporal scales, and we describe recent advances in the development of novel therapeutics based on functional targets. Finally, we describe many of the outstanding questions for the field and how fundamental mechanistic studies, informed by our more nuanced understanding of the clinical disorders, will play a central role in realizing the potential of precision medicine for genetic cardiomyopathies.
Introduction
Genetic cardiomyopathies encompass a broad array of progressive cardiac disorders that result in complex patterns of pathogenic ventricular and atrial remodeling (Lorenzini et al., 2020). The most common subclasses, hypertrophic (HCM) and dilated (DCM) cardiomyopathies, are characterized by hypertrophy of the left ventricular wall and dilation of the left ventricular chamber, respectively. The classification of these differing remodeling patterns has long been based on noninvasive diagnostic criteria (e.g., echocardiography) of often end-stage disease. Clinical symptoms and outcomes are largely defined by the functional effects on myocardial performance, as dictated by both the observed patterns of morphologic change and the timing of disease onset. While the precise disease frequency rates are somewhat controversial, extensive clinical data support a worldwide frequency of ∼1/300–1/500 individuals for HCM and ∼1/250 for DCM (Semsarian et al., 2015; McNally and Mestroni, 2017). In 1990, cardiac myosin (MYH7) was the first gene linked to HCM, and most of the subsequently identified genes associated with HCM encode sarcomeric proteins (Geisterfer-Lowrance et al., 1990). While the genetic basis of DCM is broader and more complex, a recent study identified 12 genes robustly associated with DCM, including a majority of sarcomeric proteins (Mazzarotto et al., 2020b).
Despite an extensive array of studies over the last ∼30 yr encompassing biophysical, cellular, animal model, and, more recently, patient tissue methodologies, the current approaches to community-based patient management for cardiomyopathies remains largely unchanged since the era preceding genetic linkage. At present, medical therapy is largely palliative and reserved for patients with symptoms that occur later in the pathogenic process after irreversible cardiac remodeling has occurred (Gersh et al., 2011). This lack of translational insight is humbling, given our relatively sophisticated understanding of the structure and function of myocellular processes and the proteins encoded by the genes linked to cardiomyopathies.
As we have learned more about these progressive disorders, it has become apparent that they are more complex in their molecular etiologies and clinical presentation than generally appreciated. This complexity has limited our understanding of the molecular and cellular effectors that drive disease pathogenesis and frustrated the development of effective therapeutics. Overcoming these enduring limitations will require updated, nuanced models based on experimentally derived insights into the molecular mechanisms that drive this most biophysical of disorders, coupled with a more modern and robust understanding of the variability of clinical expression. The goals of this review are to (1) highlight recent advances in our basic science and clinical understanding of these diseases, and (2) provide a roadmap forward for leveraging basic science discoveries to make translational advances for these complex disorders.
Cardiomyopathies have variable and complex clinical presentations
To begin to overcome these limitations, it is important to acknowledge the current challenges in full. At the most basic level, the primary question to be addressed is deceptively simple: How does mutation of a given protein result in a given morphological–clinical phenotype? Our increasing understanding of the genetics, molecular underpinnings, and clinical phenotypes of cardiomyopathies have demonstrated that these disorders are anything but simple.
As of 2020, we have identified thousands of mutations linked to cardiomyopathies. With the recent development of large and curated genomic data sets, it has become clear that simply assigning pathogenicity to a given gene variant in the absence of robust genetic linkage is highly imperfect (Mazzarotto et al., 2020a). Connecting genetic variants with disease phenotype is not only a central challenge in clinical medicine, but this issue also greatly impacts basic science. Many basic science studies of variants have assumed rigorous linkage to disease when the variant might not be causative. This is most commonly observed with variants that have only been observed in a single individual or family, and this issue is compounded by the lack of precise clinical phenotyping in many of the submissions to large databases. While the use of such rare variants to probe basic biology and structure can be informative, translational insights and inferences about disease mechanism drawn from such approaches are limited. Both genotype and phenotype must be rigorously determined and validated in order to generate clinically relevant and actionable mechanistic information.
Our understanding of the phenotype component of this equation has also vastly expanded over the past decade. The terms HCM and DCM are historic, representing descriptive terminology originally derived from autopsy studies or echocardiography (Teare, 1958; Frank and Braunwald, 1968). For example, HCM is classically defined as a diagnosis of exclusion of other potential causes or “unexplained left ventricular hypertrophy.” In the modern, postgenetics era, these terms are imprecise and used by clinicians only in the broadest sense. Recent data from highly curated clinical data sets have validated what clinicians have long known—that genetic cardiomyopathies, especially HCM, are protean in their morphologic manifestations. For example, the most important clinical question regarding a patient diagnosed with HCM is whether they exhibit obstructive (hypertrophic obstructive cardiomyopathy; ∼60%) or nonobstructive (∼40%) physiology, largely determined from the hemodynamic and morphologic profiles of the left ventricular outflow tract. This classification determines both the treatment approaches and likely outcomes (Neubauer et al., 2019). Other impactful morphologic observations that determine management include the distribution of left ventricular hypertrophy (e.g., apical versus concentric), fibrotic burden, degree of diastolic impairment, atrial remodeling, and potential mitral valve involvement. Thus, the robust linkage of genotype to phenotype in HCM or DCM requires more precise characterization of patient-specific phenotypes to reflect the reality of clinical management.
The pathologic remodeling in HCM and DCM is progressive. At birth, patients have the mutation yet typically do not show signs of remodeling (genotype+/phenotype−); however, many patients progress and develop remodeling and subsequent disease symptoms (phenotype+/genotype+). Disease severity is closely correlated to the time of onset, with worse outcomes correlated to younger age of diagnosis. Patterns of pathogenic remodeling are often nonlinear, with unpredictable zones of accelerated or quiescent remodeling. Moreover, patients with HCM can transition to DCM, which is predictive of poor outcomes in sarcomere gene–positive patients (Helms and Day, 2016; Ho et al., 2018).
There are currently no definitive cures for cardiomyopathies. Current therapy is largely dependent on the contractile state of the heart. In systolic heart failure (frequently seen with DCM), also known as heart failure with reduced ejection fraction, the heart shows reduced contractile function during systole. Treatment of heart failure with reduced ejection fraction, which is comprehensive and a highly active research area, focuses on supporting hemodynamics and preventing additional adverse remodeling (e.g., β-blockers, angiotensin-converting enzyme inhibitors, angiotensin receptor blockers, and, recently, neprilysin inhibitors; Yancy et al., 2017). The efficacy of these approaches in patients with genetic DCM is highly variable and often insufficient to avoid progression to end-stage disease. Pediatric patients with DCM frequently do not respond well to these therapeutics (Kantor et al., 2010). Later-stage therapies focus on direct contractile support via left ventricular assist devices or heart transplantation.
In contrast, systolic performance is often preserved or supranormal in patients with HCM, where the patterns of ventricular remodeling are near protean compared with DCM. HCM patients often demonstrate diastolic dysfunction, which impacts diastolic filling, and their hearts operate at elevated pressures, similar to heart failure with preserved ejection fraction, for which there are currently no effective therapies. Current guidelines support therapy only in patients with overt symptoms and are mostly limited to therapeutics that improve hemodynamics in obstructive patients (e.g., β-blockers, nondihydropyridine calcium channel blockers, and disopyramide; Gersh et al., 2011). These palliative approaches are effective in a subset of patients for improving exercise tolerance and quality of life, but do not address the underlying disease mechanism. For the significant cohort of patients with HCM without obstruction, no specific therapies are effective (Day, 2019). As such, new approaches are needed to identify therapeutics that improve outcomes.
Cardiomyopathies are excellent candidates for a precision medicine approach
Clearly, the field is challenged with highly complex disorders defined by large degrees-of-freedom issues with respect to both sides of the genotype–phenotype linkage. Addressing this challenge will require new approaches that leverage growing data sets with more refined and precise views of the clinical disorders. Rather than considering HCM and DCM as monolithic entities, translational advances will likely come from a precision medicine approach of dividing patients into rational subsets with therapeutics tailored to each subset.
We propose defining subsets based on the primary molecular mechanisms that drive the disease pathogenesis. This requires identifying common structural–functional groups or biophysical bins, rather than the current approaches that either focus on individual mutations in isolation or that do not consider genetic information. This is important for many reasons. First, in the era of next-generation sequencing and the rapid expansion of patient genotyping, the number of potentially pathogenic mutations identified is rising quickly, and when coupled to the complexity of patient phenotypes, the ability to link genotype to phenotype such that genetic information can be used in patient management remains elusive. Genetic screening is primarily used to identify genotype-positive family members for additional screening. This limitation remains a central concern of modern clinical management. The ability to stratify or place newly identified mutations or variants into functional bins based on biophysical mechanisms would provide a robust framework both for clinical prediction and rational therapeutic targeting.
Second, given that the cardiac sarcomere is a near-crystalline array of proteins that form highly evolved and allosterically regulated macromolecular complexes, the identification of disease-causing mechanisms must consider both inter- and intramolecular interactions that may occur at a significant distance from the mutational site. Not surprisingly, this comprehensive molecular phenotyping has revealed that the “one gene, one mechanism” or even “one mutation, one mechanism” assumptions are overly simplistic since a given mutation can affect multiple functional units of macromolecular complexes.
Finally, the initial myocellular response(s) to perturbations in sarcomeric function are likely compensatory, involving a myriad of signaling pathways affecting metabolism/energetics, calcium homeostasis, and proteasomal degradation, to name but a few (Spirito et al., 1985; Javadpour et al., 2003; Day, 2013; Helms et al., 2016; Lehman et al., 2019). The coupling of robust functional bins to early phases of the molecular response would facilitate targeting either the primary biophysical defect and/or the initial downstream signaling pathways, and this could potentially alter the natural history of pathogenic cardiac remodeling in these disorders. Taken together, this overall approach would be predicted to transcend the simple notions that HCM and DCM represent monolithic disorders and enable the field to leverage the overall pathogenic complexity of these disorders to develop new panels of mechanistically driven therapeutic options.
The molecular and cellular manifestations of cardiomyopathies are complex, and they evolve with disease progression
It can be challenging to decipher the initial molecular insult that drives the disease pathogenesis from other cellular dysfunctions. Genome-wide association studies and targeted next-generation sequencing of patients have revealed thousands of mutations in genes primarily expressed in cardiomyocytes (McNally and Mestroni, 2017; Ho et al., 2018; Fatkin et al., 2019). These genes can be broadly characterized into those encoding proteins involved in sarcomeric contraction (e.g., MYH7, TNNT2, MYBPC3, and TTN), mechanotransduction (e.g., LMNA and DES), excitation-contraction coupling and calcium handling (e.g., PLN, SERCA2A, and SCN5A), metabolism (e.g., TAZ), and organizing cytoskeletal architecture (e.g., ACTN1 and VCL). Thus, mutations in genes involved in different pathways can lead to convergent cellular manifestations and ventricular remodeling phenotypes. For example, a primary biophysical effect of phospholamban mutations—a protein that regulates sarcoplasmic calcium uptake via SERCA2a inhibition—is altered intramyocellular calcium handling; however, the resulting calcium dysregulation will impact both contractility and metabolism due to the increased energetic cost of sequestering calcium. Likewise, altered myofilament contractility due to mutations in cardiac troponin T can affect cellular mechanotransduction (Clippinger et al., 2019), leading to downstream changes in the expression of calcium-handling genes (Chandra et al., 2001; Montgomery et al., 2001).
Moreover, despite similarities observed in later stage, symptomatic disease, independent genetic etiologies can cause different mutation-induced changes in molecular and cellular function early in the disease. The initial mutation-induced perturbations in function act as molecular stress tests that trigger subsequent responses by the cell to maintain homeostatic and functional mechanisms. These responses can vary between mutations and they define the nature and magnitude of early, often compensatory pathways driving myocardial remodeling. These molecular and cellular signatures are highly dynamic and vary over the often-extended time course of the disease.
Initial molecular insults affect protein biochemistry and biophysics
Mechanistic studies have highlighted the need to reevaluate some of the earlier models for organizing mutations. These studies have repeatedly revealed that it is impossible to classify mutations based on the affected gene alone. For example, numerous variants in cardiac myosin (MYH7) have been identified, some of which cause HCM, some of which cause DCM, and some of which are benign (Alamo et al., 2017). Similarly, it is not possible to classify the diseases as arising from either a gain or loss of molecular function. For example, mutations in titin (TTN) and myosin-binding protein C (MyBPC; MYBPC3 gene) can both cause loss of protein function (Andersen et al., 2004; Hinson et al., 2015; Helms et al., 2020a); however, titin mutations typically cause DCM, while MyBPC mutations typically cause HCM. Moreover, it has been proposed that HCM is caused by mutations that result in molecular hypercontractility and DCM results from hypocontractility; however, many mutations associated with cardiomyopathies, such as those in lamin A/C—the second-most frequently mutated gene in DCM—or desmin play no direct role in contractility. Finally, patients with HCM can transition to DCM with the activation of maladaptive pathways. Taken together, it has become clear that understanding the disease pathogenesis will require the development of models that better capture these subtleties.
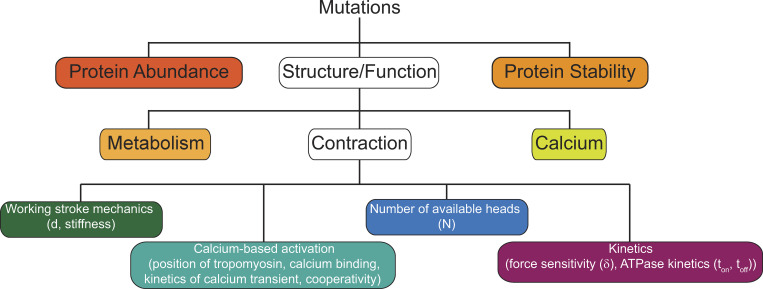
We propose classifying patient mutations into bins based on the initial biophysical insult driving the disease pathogenesis at the molecular scale and the resultant functional effects (Fig. 1). For each bin, therapeutics can be designed to ameliorate the underlying biophysical defect. This is a central point since, given the relative infrequency of individual mutations, therapeutics targeting independent mutations are not feasible. Instead, robust mutational binning based on the initial molecular insult can identify subsets of mutations that would be predicted to respond similarly to a given therapy. While these proposed biophysical bins are not mutually exclusive, they provide a useful organizational framework for organizing mutations.
Figure 1.
Mutations can be grouped into bins based on the biophysical consequences of the initial molecular insult that drives the disease pathogenesis. Colored boxes represent potential biophysical bins for organizing mutations. White boxes represent broad bins that we subdivided into more specialized bins to describe the effects of sarcomeric mutations. While these bins are neither mutually exclusive nor exhaustive, they provide a useful framework for classifying patient subpopulations and for identifying biophysical parameters that can be targeted pharmacologically for the development of precision therapeutics for these subpopulations.
The molecular effects of mutations can be divided into three broad categories: protein abundance, stability, and function; however, these categories are not mutually exclusive. For example, while DCM-causing dystrophin mutations affect its function in vitro, the protein is degraded in the cell, leading to reduced abundance (Kerr et al., 2001). As such, it is necessary to consider the molecular effects of mutations both in vitro and in vivo, especially in the development of new therapeutics. A compound that restores activity to a protein that is degraded in the cell would not be useful, and, similarly, a compound that increases protein abundance would not be useful if the protein is nonfunctional. In the following section, we describe potential bins and provide specific examples of mutations that fall within these bins.
Mutations affecting protein abundance
Some mutations affect protein abundance, leading to a net haploinsufficiency. Haploinsufficiency can arise from nonsense-mediated decay of mRNA, degradation of proteins that fail quality control, or changes in protein structure that affect its integration into stable macromolecular complexes. For example, titin, the most frequently mutated gene in DCM, is a large protein with multiple globular domains that connects the thick filament to the Z-disc, and it is a major source of muscle passive tension. Many titin mutations are frameshift or nonsense mutations that lead to protein degradation (Hinson et al., 2015). Similarly, in Duchenne’s muscular dystrophy, DCM is a common late-stage manifestation. The disorder is frequently caused by frameshift mutations in dystrophin (DMD), which anchors the sarcomere to the extracellular environment via the sarcolemma. These frameshift mutations lead to protein degradation, and many patients have no detectable expression of dystrophin (Kerr et al., 2001). This leads to repeated sarcolemmal damage with eventual myocyte loss and extensive replacement fibrosis.
Haploinsufficiency due to protein loss can also lead to HCM. For example, one of the most commonly mutated proteins in HCM is MyBPC. MyBPC lies along the thick filament and its N-terminal region can bind to the thin filament in a phosphorylation-dependent manner (Shaffer et al., 2009). It has been proposed that MyBPC plays a role in regulating both thin and thick filament activation (Mun et al., 2014; McNamara et al., 2019; Toepfer et al., 2019). Many MyBPC HCM mutations are frameshift or nonsense mutations that lead to a depletion of MyBPC and dysregulation of contractility (Helms et al., 2020b). Deletion of MYBPC3 causes HCM (Harris et al., 2002).
Mutations affecting protein stability
Some cardiomyopathy-causing mutations affect the stability of proteins, which can lead to changes in protein function and/or abundance. One example of a cardiomyopathy caused by changes in noncardiac protein stability is cardiac amyloidosis (Bart et al., 2020). This disorder presents as infiltrative cardiomyopathy, resulting in a loss of ventricular compliance. It can be familial or arise from clonal mutations in plasma cells. In the most common form, light chain amyloidosis, mutations in Ig light chains in clonal plasma cells lead to protein instability and the formation of insoluble amyloid. Similarly, inherited mutations in liver-produced transthyretin (TTR) can cause amyloid formation and cardiac deposition. The exact mechanism by which these amyloid plaques cause disease is an active field of research.
Cardiomyopathic remodeling can also be caused by mutations that affect the stability of macromolecular complexes. For example, there are disease-causing mutations in the coiled-coil cardiac myosin tail/rod domain that affect the assembly and stability of the thick filament (Blair et al., 2002). It is important to note that the stability of macromolecular complexes depends on the Kd of the components as well as the local cellular protein concentration, which can vary throughout the cell. For example, within the sarcomere, there are very high concentrations of troponin (∼0.15 mM) and actin (∼1 mM) due to crystalline packing of thick and thin filament latices. Outside the sarcomere, the effective concentration of troponin is very small. The Kd for troponin binding to the thin filament in solution is ∼50 µM (Gangadharan et al., 2017). Given the high concentration of actin and troponin within the sarcomere, all of the troponin will be bound to the thin filament. In this case, subtle mutation-induced changes in the Kd would be predicted to have little impact on the steady-state fraction of troponin bound to the thin filament. That being said, subtle changes in affinity can provide insights into mutation-induced structural changes.
Mutations affecting protein function
The majority of cardiomyopathy-causing mutations are single point mutations. Some of these mutations occur in introns where they can affect gene splicing, leading to altered protein abundance or function. Other mutations occur in exons, resulting in amino acid substitutions. Many of these mutations directly affect protein function, such as point mutations in the myosin head domain that affect the kinetics of its ATPase cycle (Sommese et al., 2013; Liu et al., 2018; Adhikari et al., 2019; Sarkar et al., 2020) and mutations within troponin and tropomyosin that affect calcium-based thin filament regulation (Tardiff et al., 1999; Barrick et al., 2019; Clippinger et al., 2019; Clippinger et al., 2020 Preprint; Ezekian et al., 2020). Other mutations can affect the mechanical properties of proteins. For example, mutations in the myosin light chain binding domain can potentially affect the stiffness of the lever arm, impacting myosin’s ability to efficiently generate force (Greenberg et al., 2010). Other point mutations affect posttranslational modifications that regulate protein activity, such as phosphorylation of troponin I (Memo et al., 2013).
It remains challenging to predict functional consequences solely from the mutation’s location in the molecule’s three-dimensional structure. Crystal structures capture a single, static, typically low-energy conformation of a protein, and, therefore, higher energy, functionally relevant conformations and transition states are frequently not captured. Proteins and protein complexes are complicated machines that can be functionally regulated by allosteric interactions through the molecule, and, therefore, point mutations distal from the protein active site can affect the conformational dynamics of side chains at the active site. Therefore, not all functionally relevant mutations occur at active sites. For example, some mutations in the myosin converter domain occur far from the active site and affect the ATPase cycling kinetics (Gunther et al., 2019; Vera et al., 2019). The recent use of computational modeling coupled with high-resolution in vitro experiments is expanding our understanding of these processes and the effects of disease-causing mutations and genetic modifiers (Guinto et al., 2007; Manning et al., 2012; Williams et al., 2016; McConnell et al., 2017; Porter et al., 2020). We anticipate that these tools will greatly enhance our understanding of mutations at the molecular/atomic scale (Manning et al., 2012; Williams et al., 2016; Porter et al., 2020).
Classic genetic cardiomyopathies are caused by sarcomeric mutations
To illustrate how functional binning of initial molecular insults could be accomplished, we will discuss targetable biophysical bins for sarcomeric protein mutations (Fig. 1). HCM patients with known pathogenic mutations in sarcomeric proteins have worse outcomes than do patients with variants in nonsarcomeric genes or genotype-negative patients (Ho et al., 2018). In general, HCM is typically associated with diastolic dysfunction, and DCM is typically associated with systolic dysfunction in patients; however, the mutation’s effects on molecular contractility are not necessarily the same as those observed in symptomatic patient hearts. The initial insult causes the activation of adaptive and maladaptive pathways, and, therefore, it can be masked by secondary adaptations associated with disease progression. Understanding the molecular basis of sarcomeric mutations necessitates a careful examination of their impact across multiple levels of biologic complexity and experimental resolution.
The sarcomere is the fundamental unit of cardiac contraction, where force and power are generated by the collective motion of myosin molecular motors interacting with the thin filament. Myosins must generate sufficient power during systole to pump blood throughout the body and then stop generating power during diastolic filling. The physiologic state of the heart and hemodynamic demand vary with disease and patient activity, and, therefore, multiple complex mechanisms have evolved to regulate power output. In the next sections, we review the molecular mechanisms of muscle contraction and describe targetable biophysical bins based on these mechanisms.
Calcium-based regulation of muscle contraction
Cardiomyocyte contraction is initiated by depolarization of the sarcolemma, which leads to the activation of voltage-gated ion channels, including L-type calcium channels. Local calcium influx and activation of the ryanodine receptor on the sarcoplasmic reticulum follows, initiating calcium-induced calcium release. This causes the levels of intracellular calcium to rise, activating the thin filament and initiating sarcomeric contraction. At the end of the contraction, calcium is removed from the cytoplasm via the sarcoendoplasmic reticulum calcium ATPase pump and the sodium-calcium exchanger, facilitating muscle relaxation. The time scale of muscle activation depends on the amplitude and kinetics of the calcium transient.
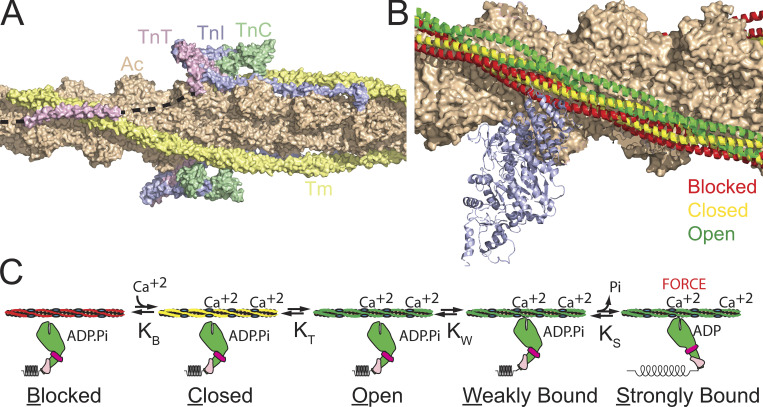
Force is generated by myosins pulling on thin filaments consisting of actin, troponin, and tropomyosin (Fig. 2). During diastole, when the intracellular calcium levels in the cell are low, tropomyosin lies along the myosin strong-binding sites on actin, sterically inhibiting force generation (blocked state of tropomyosin; McKillop and Geeves, 1993; Lehman et al., 1994). When calcium enters the sarcomere, it binds to troponin C, leading to an axial shift in the tropomyosin positioning on actin to the closed state. This partially exposes the myosin strong-binding sites on actin. Myosin strong binding to the thin filament pushes tropomyosin into the open state, which opens multiple adjacent myosin-binding sites, leading to cooperative recruitment of myosin cross-bridges. Therefore, calcium-based regulation depends on calcium, myosin binding, and the coupling between troponin and tropomyosin.
Figure 2.
Regulation of contraction by the thin filament. (A) The thin filament, consisting of actin (Ac; peach), tropomyosin (Tm; yellow), troponin I (TnI; blue), troponin C (TnC; green), and troponin T (TnT; pink) regulates calcium-dependent interactions between myosin and the thin filament. Black dashed line shows regions of troponin T that were not resolved in the structure. Structure is from PDB accession no. 6KN7. (B) Tropomyosin can lie in three positions along the thin filament, blocked (red), closed (yellow), and open (green). When tropomyosin lies in the blocked position, it sterically blocks the strong binding of myosin (blue ribbon structure). When tropomyosin is pushed into the open position by myosin binding, it opens adjacent myosin-binding sites, leading to cooperative recruitment of additional myosin cross-bridges. Based on PDB accession nos. 6KN7 (blocked), 6KN8 (closed), and 4A7L (open, myosin bound). (C) Cartoon of thin filament regulation. Calcium binding to the thin filament causes tropomyosin to shift to the closed position. The tropomyosin can then either thermally diffuse or be pushed into the open position by myosin binding. Myosin initially binds weakly to the thin filament, and then strongly. Upon the transition to myosin strong binding of the thin filament, myosin releases phosphate and undergoes its power stroke, generating force. KB, KT, KW, and KS are equilibrium constants between states.
Bins: Kinetics of the calcium transient, calcium binding to troponin, tropomyosin positioning along actin, calcium dissociation kinetics, cooperativity of activation.
Mechanics of the myosin working stroke
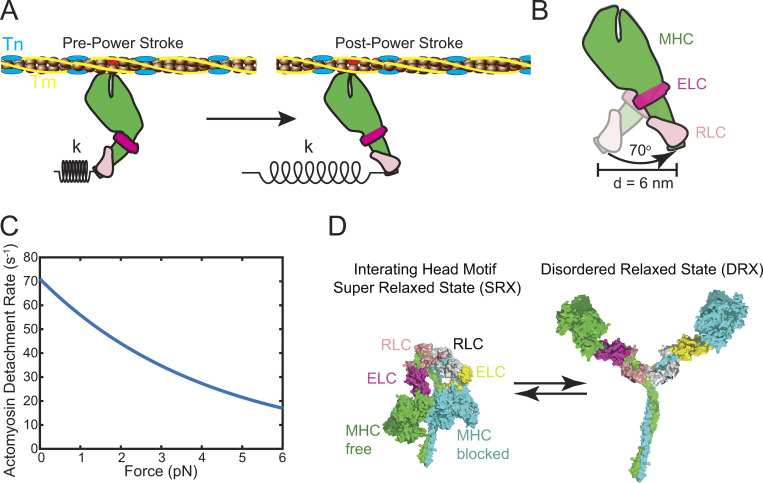
Cardiac myosin has a core structure that is conserved with other myosins (Preller and Manstein, 2012; Winkelmann et al., 2015). It is a hexameric complex comprised of two myosin heavy chains (MHCs), two regulatory light chains (RLCs), and two essential light chains (ELCs). The two MHCs dimerize via a long coiled-coil tail that facilitates oligomerization into the thick filament. The head domain of the MHC contains the sites of ATP hydrolysis and actin binding. Small conformational changes at the nucleotide binding site associated with nucleotide binding, hydrolysis, and release cause kinking of the relay helix and rotation of the converter domain (Fig. 3). The converter domain acts as a fulcrum, and its rotation is amplified into a larger displacement by the lever arm composed of the light chain binding domain (Rayment et al., 1993b). This rotation is known as the powerstroke, and the lever arm rotates by ∼70° to generate a unitary displacement (or working stroke), d, of ∼6 nm (Dominguez et al., 1998; Fig. 3 B). The force generated is proportional to both the unitary displacement and the stiffness of the myosin cross-bridge. Thus, if a given mutation affects the mechanics of the working stroke, by either changing the angular swing or the stiffness of the lever arm, this can affect myosin force production.
Figure 3.
The myosin motor drives cardiac contraction. (A) The myosin motor generates force when it transitions from the pre–power stroke state to the post–power stroke state. This force generates a displacement, d, and the transmission of this force depends on the stiffness of the myosin, k. Troponin (Tn; blue); tropomyosin (Tm; yellow); MHC (green); ELC (magenta); RLC (pink). (B) The transition from the pre–power stroke state causes the light chain binding domain to rotate by 70°, generating a displacement, d, of ∼6 nm. (C) Force slows the rate of actomyosin detachment according to Eq. 1. Mutation or drug-induced changes in the load dependence of the rate of detachment affects the speed of shortening (Eq. 2), the myosin duty ratio (Eq. 3), force generation (Eq. 4), and power output (Eq. 5). (D) Myosin can form an autoinhibited state, known as the interacting heads motif. The SRX state is likely related to the formation of the interacting heads motif, where one myosin head, the blocked head, binds to the coiled-coil S2 region, and the other head, the free head, forms interactions with the blocked head. The interacting heads motif is regulated by several mechanisms, including mechanical stretch, RLC phosphorylation, and interactions between myosin and MyBPC. Relief of the autoinhibition causes the myosin to adopt a disordered relaxed state, where the myosin heads can interact with activated thin filaments. Interacting head motif is based on PDB accession no. 5TBY.
Bins: Mechanics of the working stroke.
Kinetics of the myosin enzymatic cycle
Myosin generates force in an ATP-dependent manner (Lymn and Taylor, 1971; De La Cruz and Ostap, 2004). In the absence of nucleotide, myosin strongly binds actin. ATP binding causes rapid dissociation of actin and myosin. Myosin then hydrolyzes ATP and it adopts a primed “pre–power stroke conformation” (Dominguez et al., 1998). Upon strong binding to actin, phosphate is released and the myosin undergoes its power stroke, generating force. ADP is then released, generating an additional small displacement (Greenberg et al., 2014) and the cycle resets. The rates and equilibrium constants that define these biochemical transitions are major determinants of the rates of muscle shortening (see below). The slowest transition in the ATPase cycle is the rate of myosin strong binding to actin and the subsequent phosphate release (∼10 s−1 at 20°C). The unloaded muscle shortening speed is limited by the rate of ADP release (Bárány, 1967; Siemankowski et al., 1985) from actomyosin (∼70 s−1 at 20°C; Deacon et al., 2012).
The rates of individual biochemical transitions can be affected by mechanical forces, and this can affect both the muscle-shortening speed and force production (Spudich, 2014; Greenberg et al., 2016). Force slows myosin’s ATPase kinetics (Hill, 1938), and this slowing contributes to the Fenn effect (Fenn, 1923). The rate of a transition in the presence of force, k(F), can be calculated as (Bell, 1978):
| (1) |
where ko is the rate of the transition in the absence of force, kBT is the thermal energy, F is the force on the molecule, and δ is the distance to the transition state, where a larger δ represents a greater force sensitivity. In the case of cardiac muscle, force slows the rate of ADP release (Nyitrai and Geeves, 2004; Takagi et al., 2006), the transition that sets the rate of unloaded muscle shortening (Siemankowski et al., 1985), and δ equals ∼1 nm (Greenberg et al., 2014; Sung et al., 2015; Fig. 3 C).
Bins: Myosin kinetics, load dependence of kinetics.
Myosin thick filament–based regulation of muscle contraction
Cardiac contractility can also be regulated at the level of myosins within the thick filament. Intact striated muscle has a population of myosin heads that hydrolyze ATP slower than the rate seen using purified proteins (Stewart et al., 2010; Hooijman et al., 2011). This led to the hypothesis that striated muscle myosin can adopt a super-relaxed state (SRX). Based on EM reconstructions of smooth and skeletal muscle thick filaments (Woodhead et al., 2005; Alamo et al., 2016), it was proposed that the SRX represents an autoinhibitory state of myosin, where the two myosin heads interact both with each other and the S2 region (Fig. 3 D). In this state, the myosin heads form an interacting heads motif that sequesters them to the thick filament backbone and prevents binding to the thin filament. The heads can be released from the thick filament backbone by phosphorylation of the RLC (Yang et al., 1998) or via a mechanosensitive mechanism following mechanical stretch (Linari et al., 2015). Once the heads release from the thick filament backbone, they adopt a disordered relaxed state where they can bind the thin filament. There is emerging evidence that the SRX is regulated by MyBPC, potentially through phosphorylation of the N-terminal region (Nag et al., 2017; McNamara et al., 2019). Thus, muscle contraction can be regulated by modulating the number of myosin heads available to interact with the thin filament (N).
Bins: Number of myosin heads available to bind the thin filament.
Cardiomyopathies are complex, dynamic disorders whereby changes in the molecular and cellular environment drives pathogenic cardiac remodeling
Each of the biophysical bins described above have multiple levels of regulation that can be tuned in both health and disease. These adaptive and maladaptive processes are highly dynamic, and they can affect these parameters differently as the disease progresses. One prominent experimental challenge is that many studies provide a snapshot of the disease at a single time point; however, it is important to consider the time-dependent nature of the disorders. As cardiomyopathic remodeling progresses, changes in both tissue structure and cellular function are often observed. At the molecular scale, there are changes in protein phosphorylation patterns (e.g., RLCs, troponin I, MyBPC) that tune the biophysical properties of contraction (Nakano et al., 2019; Tucholski et al., 2020). There are also functionally significant shifts in protein isoform expression, with a shift toward a more fetal gene expression pattern in the later stages of disease (Yin et al., 2015). Moreover, modification of proteins, such as methylglyoxal modification of contractile proteins in patients with diabetes (Papadaki et al., 2018), can affect their biophysical properties. Thus, when designing rigorous experiments, the question of which precise phenotype and which phase of the disorder is being modeled is paramount for determining translational insights. Of note, many pathways activated in disease are not unique to genetic cardiomyopathies, but are shared, in part, with other causes of pathogenic remodeling, including myocardial infarction, aortic stenosis, chemotherapy-induced cardiomyopathy, diabetic cardiomyopathy, and chronic hypertension. Since not all causes of cardiac remodeling are similar in presentation or treatability, it is important to understand both the initial molecular insults that drive early myocellular remodeling and subsequent downstream compensatory responses, as these prepathogenetic states are potential therapeutic targets.
How biophysical parameters relate to muscle shortening, force production, and power output
The biophysical parameters described above can be combined to describe the rate of muscle shortening, force production, and power output. The muscle shortening rate depends on both the kinetics and mechanics of the myosin working stroke. An unloaded or lightly loaded muscle will shorten at its maximal speed if at least one cross-bridge is attached to the thin filament at any given time. Under these circumstances, the shortening rate, V(F), is limited by the rate of actomyosin dissociation (Huxley, 1990):
| (2) |
where d is the unitary step size and ton(F) is the amount of time that actin and myosin are attached (typically set by the ADP release rate). Perturbations that affect either parameter will affect the shortening speed. It should be noted that there are alternative models describing the unloaded shortening speed as being limited by attachment-limited kinetics that use similar biophysical bins (Brizendine et al., 2015). At higher loads, the speed depends on the number of force-generating cross-bridges, and expressions have been derived that use similar biophysical bins to define the loaded speed (Baker et al., 2002).
The force produced by a muscle will depend on several parameters. One parameter is the duty ratio, r(F), which defines the fraction of myosin’s biochemical cycle spent attached to actin:
| (3) |
where ton(F) is the amount of time that myosin spends attached to actin, tcycle(F) is the time that it takes for myosin to complete one ATPase cycle (i.e., the inverse of the ATPase rate), and toff(F) is the amount of time that myosin spends detached from actin (typically set by the rate of myosin strong binding and subsequent phosphate release; Muretta et al., 2015; Woody et al., 2019). Therefore, mutations affecting the rates of individual biochemical transitions can potentially affect the duty ratio. Since these rates depend on force (Eq. 1), mutation-induced changes in myosin’s force-sensing properties can affect the duty ratio (Liu et al., 2018). Moreover, the effective duty ratio depends on calcium-based regulation, since at low calcium, myosin is inhibited from interacting with the thin filament (McKillop and Geeves, 1993).
The force generated by an ensemble of motors in muscle, Fens, is given by (Spudich, 2014)
| (4) |
where N is the number of myosin heads available to interact with the thin filament; Funi is the unitary force of a single myosin, which depends on the working stroke mechanics; and r(F) is the load-dependent duty ratio. Changes in any of these parameters will affect force production.
The power output, P(F), is given by the product of force and velocity:
| (5) |
And, therefore, it depends on the composite effects of the biophysical bins.
Mechanobiology links aberrant mechanical forces with biologic adaptation
As discussed above, many cardiomyopathy mutations occur in sarcomeric genes and thus affect contractility. It has been challenging to connect altered cardiac contraction with the disease progression which manifests itself over the course of decades; however, the development of a hypertrophic or dilated phenotype can be predicted based on the integral of the tension transient (Davis et al., 2016). Therefore, the initial molecular insult of altered contractility can activate downstream adaptive and maladaptive pathways, including changes in metabolism, calcium handling, gene expression, and protein posttranslational modifications.
The cell must sense altered mechanics and respond via activation of mechanobiological signaling pathways. These changes can vary over length and time scales (Iskratsch et al., 2014). For example, short-term changes in cellular function can occur via rapid phosphorylation of proteins, whereas longer-term changes can occur via altered transcription or tissue-scale remodeling (DuFort et al., 2011). Mechanobiological signaling pathways have been thoroughly described in several cell types, and determining their exact molecular mediators in the heart remains an active field of research (Majkut et al., 2014; Boothe et al., 2016; Chiou et al., 2016; Cho et al., 2019).
Mechanotransduction in cardiomyocytes can occur via several pathways (Iskratsch et al., 2014; Pruitt et al., 2014). Forces outside of the cell are transmitted into the cell via linkages between the cell and the extracellular matrix (e.g., dystroglycan complex) or between cells (e.g., intercalated discs). These forces can be directly transmitted to the nucleus through cytoskeletal elements or indirectly via force-induced unfolding of signaling molecules (Hu et al., 2017). Alternatively, forces at the membrane can open mechanosensitive ion channels (e.g., transient receptor potential channels), leading to ion influx and the activation of secondary messenger pathways (e.g., calcium–calcineurin). The reader is referred to several reviews on cardiomyocyte mechanosensing (Smith et al., 2017; Nakamura and Sadoshima, 2018; Saucerman et al., 2019).
While we have focused on sarcomeric gene mutations, there are other genes beyond the sarcomere linked to cardiomyopathies. Many of these genes lie along canonical mechanotransduction pathways, including dystrophin (DMD), desmin (DES), and lamin A/C (LMNA; Cho et al., 2016). In fact, lamin A is one of the most frequently mutated genes associated with DCM. Lamin intermediate filaments in the nucleus form lamin-associated domains (LADs) that bind to chromatin (van Steensel and Belmont, 2017). These LADs are regions of low transcriptional activity, and mechanical forces can cause the disruption of these domains (Janota et al., 2020). A recent study examining an LMNA DCM mutation in stem cell–derived cardiomyocytes showed changes in LAD topology, leading to altered gene expression (Lee et al., 2019).
Recent work demonstrated that cardiomyocyte microtubules may play a role in mechanotransduction (Caporizzo et al., 2019). Microtubules stretch perpendicular to and are mechanically coupled to the Z-discs, likely through interactions between detyrosinated tubulin and desmin (Robison et al., 2016). These interactions contribute to the viscoelastic properties of the muscle and are disrupted in patients with heart failure (Chen et al., 2018; Caporizzo et al., 2020). The microtubule network also plays a role in regulating the production of reactive oxygen species in response to stretch. Disruption of this network leads to aberrant X-ROS signaling, which increases the frequency of proarrhythmogenic calcium sparks (Kerr et al., 2015). This provides a mechanism by which disrupted mechanics can lead to calcium dysregulation, independent of changes in gene expression.
Mechanotransduction also plays a central role in sarcomeric formation and maintenance (Chopra et al., 2018). Disruption of mechanotransduction in stem cell–derived cardiomyocytes by knockout of vinculin or inhibition of myosin contraction by blebbistatin leads to sarcomeric disassembly (Chopra et al., 2018). Moreover, troponin T, which regulates cardiac force generation, is necessary for proper sarcomere assembly in several model systems (Ahmad et al., 2008; Nishii et al., 2008; Ferrante et al., 2011). Taken together, these results demonstrate that mutation-induced changes in molecular tension or mechanotransduction could affect sarcomeric organization.
There is emerging evidence that the primary disruption of mechanotransduction plays a role in cardiomyopathies. DCM-linked mutations in sarcomeric proteins, including titin and troponin T, cause sarcomeric disarray in stem cell–derived cardiomyocytes (Chopra et al., 2018; Clippinger et al., 2019; Dai et al., 2020). Our recent work demonstrated that the DCM mutation, ΔK210, in troponin T reduces molecular and cellular force generation and alters cardiomyocyte mechanosensing (Clippinger et al., 2019). We cultured human pluripotent stem cell–derived cardiomyocytes on substrates of different stiffness. While the WT cells showed robust sarcomeric organization over a range of stiffnesses, ΔK210 cells showed sarcomeric disarray on stiff substrates, but not on substrates matching the stiffness of the healthy heart. Thus, mutation of sarcomeric proteins can affect not only cellular contraction, but also mechanotransduction. Moreover, our results suggest that fibrosis-induced stiffening of the heart could contribute to disease progression.
The role of altered mechanical forces and mechanobiology in disease likely extends beyond cardiomyocytes. There are multiple cell types in the heart, including macrophages, fibroblasts, endothelial cells, and pericytes that likely play critical roles in disease pathogenesis. Importantly, myocardial tissue in both HCM and DCM exhibits varying degrees of fibrosis, which can act as a substrate for arrhythmias. Fibrosis, in part, is due to activation of quiescent fibroblasts to become myofibroblasts (Nakamura and Sadoshima, 2018). Myofibroblasts show increased contractility and increased deposition of extracellular matrix due to the activation of the TGF-β pathway (Davis and Molkentin, 2014). Mechanical forces can promote the myofibroblast transition, and, therefore, mutation-induced altered cardiac contractility can affect processes beyond cardiomyocytes.
Examples of well-characterized sarcomeric mutations
There have been many excellent mechanistic studies of sarcomeric cardiomyopathy mutations, and it is impossible to cover them all. Here, we focus on two well-studied mutations that highlight the complex disease pathogenesis and the need to examine mutations over several scales of organization.
R403Q in MYH7 (MHC) causes HCM
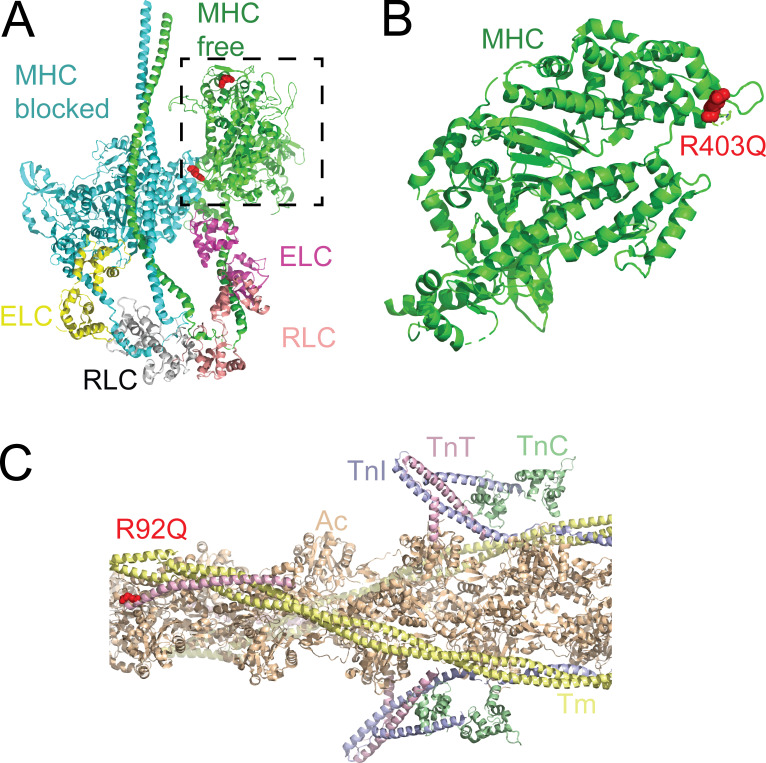
R403Q, the first mutation associated with HCM (Geisterfer-Lowrance et al., 1990), is one of the most studied mutations. The mutation results in a charge reversal, suggesting that it could affect electrostatic interactions between residue 403 and binding partners (Fig. 4). R403 is located in the MHC cardiomyopathy loop (Rayment et al., 1993a), which derives its name from the multiple cardiomyopathy-causing mutations found there (Preller and Manstein, 2012).
Figure 4.
Examples of well-studied mutations. (A) R403Q in the MHC (MYH7) causes HCM. Structural model of the myosin interacting heads motif (PDB accession no. 5TBY). The MHCs and their associated ELCs and RLCs fold back to form an autoinhibited structure. R403 is shown in red. R403 on the blocked head sits at the interface formed with the free head. Dashed box highlights the myosin head domain in B. (B) Structure of the myosin head domain (rotated from the dashed box in A). R403 lies in the cardiomyopathy loop in the myosin head that forms part of the actin binding interface. (C) R92Q in troponin T (TNNT2) causes HCM. The thin filament, consisting of actin (Ac; peach), tropomyosin (Tm; yellow), troponin I (TnI; blue), troponin C (TnC; green), and troponin T (TnT; pink) regulates calcium-dependent interactions between myosin and the thin filament. R92Q lies near the region of troponin T that binds the overlap region between two tropomyosin molecules. Based on PDB accession no. 6KN8.
Studying the initial molecular insult driving R403Q pathogenesis has been challenging, in part, due to difficulties in establishing model systems that faithfully recapitulate the human disease phenotype. Early studies were conducted with myosin extracted from patient samples; however, some studies showed decreased contractile function (e.g., reduced sliding speed [Cuda et al., 1997] and tension [Lankford et al., 1995]), while others showed increased contractility (e.g., increased actomyosin detachment kinetics; Palmiter et al., 2000). These experiments were confounded by the limited availability of human tissue and challenges with preserving the myosin.
A major advance was the development of an R403Q transgenic mouse that expresses abundant amounts of control and mutant protein (Geisterfer-Lowrance et al., 1996); however, mouse ventricles primarily express α-cardiac MHC (MYH6), not the β-isoform (MYH7) expressed in human ventricles. The α-isoform has faster motility and ATPase kinetics and lower force than the β-isoform (VanBuren et al., 1995; Alpert et al., 2002; Deacon et al., 2012) and, as such, the use of α-cardiac myosin may not reproduce the human phenotype. Biochemical studies of murine R403Q α-cardiac isoform showed an increase in motility, ensemble force, and ATPase kinetics. Based on these results, it was proposed that R403Q causes molecular hypercontractility (Tyska et al., 2000; Debold et al., 2007). Studies using transgenic mice and rabbits showed different molecular defects with the β-isoform (Lowey, 2002; Lowey et al., 2008; Lowey et al., 2013; Lowey et al., 2018).
Expressing recombinant human β-isoform MHC has been challenging since it cannot be expressed by using the baculovirus/Sf9 system typically used to express myosin proteins due to the lack of proper chaperones (Srikakulam and Winkelmann, 2004). However, it is possible to express MHC fragments of recombinant human cardiac myosin in C2C12 cells, which contain muscle specific chaperones (Deacon et al., 2012), and this system has been used to generate recombinant human R403Q. Unexpectedly, human R403Q myosin showed reduced intrinsic force, unchanged size of the working stroke, decreased ensemble force, increased motile speed with unregulated actin, and decreased motile speed with regulated thin filaments (Nag et al., 2015).
The finding of reduced contractility with human R403Q motor domains was inconsistent with the model that HCM mutations cause molecular hypercontractility; however, myosin operates in the sarcomeric macromolecular complex. Spudich (2015) noted that the myosin head has a domain enriched with charged amino acids that he dubbed the myosin mesa, and he proposed that the mesa participates in protein–protein interactions. Many cardiomyopathy-causing mutations, including R403, are located along the mesa (Alamo et al., 2017). Structural models have suggested that the mesa helps form the SRX and that it interacts with MyBPC (Nag et al., 2017; Trivedi et al., 2018). In the interacting head configuration, R403 on the blocked head likely interacts with the free head, and R403 of the free head interacts with MyBPC (Sarkar et al., 2020). Consistent with this model, biochemical experiments and x-ray fiber diffraction studies demonstrated that R403Q destabilizes the SRX (Anderson et al., 2018; Sarkar et al., 2020) and reduces binding to MyBPC (Sarkar et al., 2020). Based on these results, it was proposed that the R403Q mutation destabilizes the SRX, leading to hypercontractility by increasing the number of available myosin heads. This hypercontractility is independent of changes in the intrinsic force-generating or ATPase properties of individual motors. These results highlight the need to consider the molecular effects of mutations in systems with biophysical properties similar to the human isoforms, as well as the importance of studying mutations over several scales of organization.
R92Q in TNNT2 (troponin T) causes HCM
The R92 residue in troponin T has been proposed as a hot spot for HCM mutations (Forissier et al., 1996). To date, three separate mutations (R92Q/L/W) have been identified at this site with different clinical presentations, depending on the mutation. Structurally, troponin T is part of the complex that regulates calcium-dependent interactions between myosin and the thin filament (Fig. 4). Troponin is a trimeric protein composed of troponin C, which binds calcium; troponin I, which inhibits the movement of tropomyosin in the absence of calcium; and troponin T, which anchors the troponin complex to tropomyosin and plays a role in thin filament inhibition (Tobacman et al., 2002; Madan et al., 2020). The troponin complex core domain was crystalized in 2003 (Takeda et al., 2003); however, the majority of troponin T, including R92, was not resolved, likely because this region is intrinsically disordered in solution. Recently, a high-resolution cryo-EM structure of the thin filament provided near-atomic resolution structures of actin, tropomyosin, and most of the troponin complex (Yamada et al., 2020), and the position of troponin T was further refined by computational docking (Pavadai et al., 2020). The majority of troponin T forms an elongated structure, and R92 is located near the region of troponin T that interacts with tropomyosin. Whole-atom molecular dynamics simulations of fully regulated thin filaments revealed that the R92L mutation increases the distance between troponin T and tropomyosin, potentially reducing the affinity and coupling between tropomyosin and troponin T (McConnell et al., 2017). These simulations also revealed that mutations at R92 cause allosteric changes in troponin C that potentially affect calcium-binding dynamics (Williams et al., 2016).
The molecular consequences of R92Q have been studied in both in vitro and in vivo systems. The earliest studies involved overexpressing R92Q troponin T in mouse, rat, cat, and quail cells (Marian et al., 1997; Morimoto et al., 1998; Sweeney et al., 1998). In all of these studies, the R92Q protein integrated into sarcomeres, suggesting that the primary effect of the mutation is disrupted protein function. Furthermore, there were several studies in which recombinant troponin was exchanged into muscle fibers (Yanaga et al., 1999; Szczesna et al., 2000), and the majority, but not all (Sweeney et al., 1998; Rust et al., 1999), of these studies demonstrated that the mutation shifts the force–calcium curve toward submaximal calcium activation (Morimoto et al., 1998; Yanaga et al., 1999; Szczesna et al., 2000). Similar shifts were also seen by using recombinant proteins (Robinson et al., 2007; Messer et al., 2016; Clippinger et al., 2020 Preprint). A shift toward submaximal calcium activation would be expected to cause hypercontractility during a calcium transient. Some (Robinson et al., 2007), but not all (Liu et al., 2012; Clippinger et al., 2020 Preprint), of these studies showed changes in calcium binding to troponin C.
A major step forward was the generation of the R92Q transgenic mouse (Tardiff et al., 1999), which recapitulates several features of the disease phenotype, including increased ventricular mass, diastolic dysfunction, cellular fibrosis, and myocyte disarray. The degree of cardiac dysfunction depends on the expression levels of mutant protein. Muscle fibers from these mice showed a shift toward submaximal calcium activation (Chandra et al., 2001) and energetic abnormalities (Javadpour et al., 2003). There were important sex-based differences with the mice, with males having worse outcomes (Maass et al., 2004). Importantly, the R92Q disease phenotype in the mouse depends on the MHC isoform (He et al., 2007; Rice et al., 2010; Ford et al., 2012). The R92Q mutation with the β-isoform MHC background showed differences in calcium handling, cross-bridge kinetics, energetics, and diastolic dysfunction. This result highlights the importance of using humanized protein isoforms in disease modeling.
Three models have been proposed for the initial insult of the R92Q mutation: altered cross-bridge kinetics (Ford et al., 2012), altered calcium homeostasis due to changed myofilament buffering (Robinson et al., 2007; Robinson et al., 2018), and mutation-induced changes in tropomyosin positioning (McConnell et al., 2017). Our recent biochemical work demonstrated that R92Q affects the stability of tropomyosin’s blocked state without affecting calcium binding or cross-bridge kinetics (Clippinger et al., 2020 Preprint). Moreover, R92Q mutation reduces the affinity of troponin for tropomyosin (Gangadharan et al., 2017). These results are consistent with molecular dynamics simulations showing that mutation of R92 affects the coupling between tropomyosin and troponin (McConnell et al., 2017).
Development of therapeutics for cardiomyopathies
There is an outstanding need to develop therapeutics for HCM and DCM that improve outcomes and patients’ quality of life. Here, we discuss two compounds in clinical trials that were rationally designed to target myosin-based contractility, rather than traditional therapeutic pathways. We are still at an early stage of understanding of how these modulators can be used therapeutically.
Omecamtiv mecarbil (OM) as a treatment for systolic dysfunction
OM was identified in high-throughput screens for compounds that increased the steady-state, actin-activated myosin ATPase rate (i.e., myosin activators; Morgan et al., 2010; Malik et al., 2011). OM has selectivity for human ventricular myosin over fast skeletal or smooth muscle isoforms and it has a Kd of 1.6 µM for ventricular myosin. This compound increases the steady-state, actin-activated myosin ATPase rate by accelerating the rates of phosphate release and ATP hydrolysis. As such, this compound was expected to increase the myosin duty ratio (Eq. 3) and, thus, the ensemble force production (Eq. 4). When added to rat ventricular cardiomyocytes, OM increased the rate and extent of muscle shortening without affecting the amplitude or kinetics of the calcium transient.
Recent experiments have shown that the molecular mechanism of OM is more complex than originally appreciated. The first evidence of this was seen in studies with regulated thin filaments (Malik et al., 2011), where, under fully activating calcium concentrations, the ATPase rate was lower with OM, not faster as would be expected for a myosin activator. The ATPase rate was higher only at intermediate calcium concentrations (Kieu et al., 2019), suggesting that the primary mechanism of action may involve activating the thin filament. In vitro motility assays demonstrated that OM causes a 14-fold, dose-dependent reduction in the speed of myosin movement (Liu et al., 2015; Swenson et al., 2017). A reduction in speed could come from decreasing the unitary displacement or increasing ton (Eq. 2). The rates of ADP release and ATP-induced actomyosin dissociation, which typically set ton, were unchanged by the addition of OM (Liu et al., 2015), suggesting that the drug decreases the working stroke and/or changes the biochemical pathway.
Optical trapping experiments demonstrated that OM causes a dose-dependent increase in the fraction of myosin cross-bridges that bind the thin filament without actively generating a working stroke (Woody et al., 2018). Moreover, the measured attachment durations show that OM-bound cross-bridges have a slower dissociation rate, suggesting that OM causes the myosin to enter a noncanonical biochemical pathway. A similar result was seen by using transient time-resolved FRET, which showed that OM decreases the rate of the powerstroke and causes the myosin to enter a noncanonical biochemical pathway (Rohde et al., 2017). Taken together, the measured speed in the in vitro motility assay decreases because the number of cross-bridges that generate a productive displacement decreases, independent of the accelerated attachment kinetics seen in the ATPase measurements. OM binds to the myosin converter domain, near the junction between the head and lever arm domains (Malik et al., 2011; Winkelmann et al., 2015; Planelles-Herrero et al., 2017). Thus, the effects on myosin ATPase kinetics occur allosterically. It is possible that OM’s binding near the converter domain helps to uncouple lever arm rotation from the ATPase kinetics. Taken together, these results strongly suggest that OM is not a pure myosin activator as initially proposed.
If OM is not a myosin activator, how does it increase contractility? The answer likely lies in the effects on thin filament activation. As described above, thin filament activation depends both on calcium and myosin cross-bridge binding (McKillop and Geeves, 1993). OM, by increasing the rate of cross-bridge association and prolonging the time that the cross-bridge remains attached to the thin filament, likely increases thin filament activation by moving tropomyosin and exposing additional myosin-binding sites on actin (Woody et al., 2018). This explains the shift toward submaximal calcium activation seen in the steady-state ATPase rate with regulated thin filaments. At low levels of OM, the minority population of slowly cycling, OM-bound cross-bridges would tend to recruit more force-generating cross-bridges to the thin filament. At high levels of OM, most cross-bridges would bind OM, which inhibits the working stroke, and thus the muscle force would decrease. This effect—activation at low levels of OM and reduced contractility at high concentrations of OM—has been modeled computationally (Woody et al., 2018) and observed in muscle fibers (Nagy et al., 2015). Therefore, OM is better characterized as a thin filament activator rather than a myosin activator.
The mechanism of OM likely extends beyond its effects on single myosin motor domains. OM binds near the interface adopted in the interacting head motif. X-ray diffraction studies of muscle fibers demonstrated that OM disrupts the SRX, increasing the number of heads available to interact with the thin filament (Kampourakis et al., 2018), potentially increasing force and power generation (Eq. 4). This effect on SRX formation is nonexclusive from the effects on thin filament activation.
Taken together, OM likely acts through a different mechanism than initially envisioned. The two effects of thin filament activation and SRX destabilization would increase myosin force production and systolic function. These predictions were largely supported by the results of the phase II trial (COSMIC-HF, NCT01786512), a short-term, randomized trial designed to assess left ventricular morphology and systolic performance in patients with New York Heart Association class II and III systolic heart failure (Teerlink et al., 2016). Both safety and increased systolic function (systolic ejection time), decreased left ventricular end-systolic and end-diastolic diameter, decreased heart rate, and decreased plasma NT-proBNP were observed. This was followed by the first phase III outcomes trial (GALACTIC-HF, NTC02929329) designed to assess whether OM, when added to the standard of care, was well tolerated and superior to placebo in reducing the risk of cardiovascular death or heart failure in patients with chronic systolic heart failure (Teerlink et al., 2020). This was a large, well-designed multinational trial with 8,526 patients randomized to either placebo or three different oral doses of OM (25 mg, 37.5 mg, or 50 mg, twice daily). While the primary composite outcome—a small but significant decrease in the incidence of heart failure or death from cardiovascular causes—was met, none of the secondary outcomes, including cardiovascular death, improvement in the Kansas City Cardiomyopathy Questionnaire, first heart failure hospitalization, or death from any cause were achieved. While the apparent discordance between the results of the two trials, whereby clear short-term functional and structural improvement (COSMIC-HF) did not translate to the important clinical outcome of cardiovascular death (GALACTIC-HF) was surprising en face, the complexity and heterogeneity of the clinical syndrome is a likely contributor. Further subgroup analysis may identify specific patient subsets who benefit in a disorder for which highly tailored medical therapy is a mainstay.
Mavacamten as a treatment for HCM
Mavacamten is currently in clinical trials for treating HCM (Green et al., 2016). Recent results of the first phase III randomized, double blind, placebo-controlled trial (EXPLORER-HCM, NCT03470545) conducted in hypertrophic obstructive cardiomyopathy patients exhibited encouraging safety and tolerability profiles. Importantly, minor improvements in the New York Heart Association functional class, left ventricular outflow tract obstruction, and health status were observed in this well-designed multicenter trial (Olivotto et al., 2020). The compound was originally identified via a high-throughput screen for molecules that decrease the steady-state, actin-activated myosin ATPase rate. Mavacamten causes a dose-dependent reduction in the ATPase rate, with an EC50 of 300 nM, due to a reduction in the rate of actin-activated phosphate release. By reducing the rate of actin-activated phosphate release, mavacamten slows the transition into the strongly bound state, reducing the duty ratio (Eq. 3). The drug did not affect the calcium dependence of muscle contraction or the calcium transients, an important concern in a disorder that impacts myocardial energetics (Spindler et al., 1998; Crilley et al., 2003).
In contrast to OM, mavacamten promotes the formation of the SRX (Anderson et al., 2018; Rohde et al., 2018), and therefore reduces the number of cross-bridges available to interact with the thin filament. This mavacamten-induced change in the SRX has also been seen in single-molecule studies of ATP turnover (Nelson et al., 2020). By reducing the duty ratio and the number of available myosin heads, mavacamten reduces force production (Eq. 4), and this was seen in treated mice (Green et al., 2016; Mamidi et al., 2018).
Administration of mavacamten to mice with HCM mutations before the development of overt ventricular remodeling prevented cardiac hypertrophy and fibrosis (Green et al., 2016). Of note, while treatment after the development of hypertrophy and fibrosis slowed disease progression, it did not reverse the disease phenotype. Fibrosis and myocyte disarray can serve as substrates for arrhythmias; therefore, this is an important consideration for eventual treatment regimens. HCM is a highly dynamic disorder, and treatments may have particular time windows during which they are most effective. Determining the optimal timing of treatment to ameliorate symptoms and alter the natural history of pathogenic remodeling remains a key goal of further studies.
Prospects for the future
Taken together, the field has made incredible progress in understanding these complex and progressive diseases; however, there is still much work that must be done. Continued progress will require the field to consider the nuances in the molecular mechanisms that drive the disease pathogenesis, as well as the dynamic adaptive and maladaptive pathways that are activated in these progressive and heterogeneous diseases.
The development of novel effective therapeutics will require the field to move beyond the current one-size-fits-all approaches to these complex progressive disorders. While the proliferation of next-generation and whole-genome sequencing technologies have revealed insights into the genetic underpinnings of the diseases, more work is needed to translate these discoveries to the bedside. In particular, it will be important to understand the complex networks of genetic modifiers that give rise to patterns of remodeling with variable progression and incomplete penetrance. Realizing the power of precision medicine for managing and treating cardiomyopathies will require pairing genomics with functional assays and animal models to investigate integrated disease mechanisms. Translational advances will require the field to embrace the nuances of these highly complex clinical disorders.
We propose that it might be possible to improve outcomes in patient subgroups by focusing on shared primary molecular mechanisms among groups of mutations or the earliest stages of compensatory ventricular remodeling (Fatkin et al., 2019; Lavine and Greenberg, 2020). Precision therapeutics will likely be effective for subgroups of patients with mutations producing common molecular insults (Lynn et al., 2018). By deciphering the molecular triggers that drive disease pathogenesis, it should be possible to further refine rigorous biophysical bins that can be coupled to discrete, targetable pathways.
While the field has developed many excellent biophysical tools for examining the molecular underpinnings of cardiomyopathies, new basic science tools will be necessary to realize our proposed precision medicine approach. Our current biophysical tools have provided us with deep insights into the mechanisms of individual mutants, but most lack the throughput necessary for examining thousands of variants across organizational and temporal scales. There is an outstanding need to identify robust biophysical signatures for each of the proposed bins and to develop high-throughput assays with sufficient resolution and signal to screen multiple variants. Moreover, computational techniques will likely contribute to screening of variants; however, this will require advances in computing power and methods for probing longer time scales and accurately measuring macromolecular interactions.
While we have made significant advances over the past few decades, there is still work to be done. Taken together, this is an exciting time for both basic and clinical researchers interested in the mechanisms and care of patients with genetic cardiomyopathies.
Acknowledgments
Néstor Saiz served as editor.
This work was supported by National Institutes of Health grant R01HL141086 (to M.J. Greenberg) and the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital grant PM-LI-2019-829 (to M.J. Greenberg), as well as by National Institutes of Health grants R01HL075619, R01HL107046, and R01HL137375 (to J.C. Tardiff), and by the Steven M. Gootter Foundation (to J.C. Tardiff).
All experiments were conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. J.C. Tardiff is a member of the Scientific Advisory Board for Cytokinetics. The authors declare no competing financial interests.
Author contributions: M.J. Greenberg and J.C. Tardiff wrote the manuscript together.
References
- Adhikari, A.S., Trivedi D.V., Sarkar S.S., Song D., Kooiker K.B., Bernstein D., Spudich J.A., and Ruppel K.M.. 2019. β-Cardiac myosin hypertrophic cardiomyopathy mutations release sequestered heads and increase enzymatic activity. Nat. Commun. 10:2685. 10.1038/s41467-019-10555-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad, F., Banerjee S.K., Lage M.L., Huang X.N., Smith S.H., Saba S., Rager J., Conner D.A., Janczewski A.M., Tobita K., et al. 2008. The role of cardiac troponin T quantity and function in cardiac development and dilated cardiomyopathy. PLoS One. 3:e2642. 10.1371/journal.pone.0002642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Qi D., Wriggers W., Pinto A., Zhu J., Bilbao A., Gillilan R.E., Hu S., and Padrón R.. 2016. Conserved Intramolecular Interactions Maintain Myosin Interacting-Heads Motifs Explaining Tarantula Muscle Super-Relaxed State Structural Basis. J. Mol. Biol. 428:1142–1164. 10.1016/j.jmb.2016.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo, L., Ware J.S., Pinto A., Gillilan R.E., Seidman J.G., Seidman C.E., and Padrón R.. 2017. Effects of myosin variants on interacting-heads motif explain distinct hypertrophic and dilated cardiomyopathy phenotypes. eLife. 6:e24634. 10.7554/eLife.24634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert, N.R., Brosseau C., Federico A., Krenz M., Robbins J., and Warshaw D.M.. 2002. Molecular mechanics of mouse cardiac myosin isoforms. Am. J. Physiol. Heart Circ. Physiol. 283:H1446–H1454. 10.1152/ajpheart.00274.2002 [DOI] [PubMed] [Google Scholar]
- Andersen, P.S., Havndrup O., Bundgaard H., Larsen L.A., Vuust J., Pedersen A.K., Kjeldsen K., and Christiansen M.. 2004. Genetic and phenotypic characterization of mutations in myosin-binding protein C (MYBPC3) in 81 families with familial hypertrophic cardiomyopathy: total or partial haploinsufficiency. Eur. J. Hum. Genet. 12:673–677. 10.1038/sj.ejhg.5201190 [DOI] [PubMed] [Google Scholar]
- Anderson, R.L., Trivedi D.V., Sarkar S.S., Henze M., Ma W., Gong H., Rogers C.S., Gorham J.M., Wong F.L., Morck M.M., et al. 2018. Deciphering the super relaxed state of human β-cardiac myosin and the mode of action of mavacamten from myosin molecules to muscle fibers. Proc. Natl. Acad. Sci. USA. 115:E8143–E8152. 10.1073/pnas.1809540115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, J.E., Brosseau C., Joel P.B., and Warshaw D.M.. 2002. The biochemical kinetics underlying actin movement generated by one and many skeletal muscle myosin molecules. Biophys. J. 82:2134–2147. 10.1016/S0006-3495(02)75560-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bárány, M. 1967. ATPase activity of myosin correlated with speed of muscle shortening. J. Gen. Physiol. 50:197–218. 10.1085/jgp.50.6.197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick, S.K., Clippinger S.R., Greenberg L., and Greenberg M.J.. 2019. Computational Tool to Study Perturbations in Muscle Regulation and Its Application to Heart Disease. Biophys. J. 116:2246–2252. 10.1016/j.bpj.2019.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart, N.K., Thomas L., Korczyk D., Atherton J.J., Stewart G.J., and Fatkin D.. 2020. Amyloid Cardiomyopathy. Heart Lung Circ. 29:575–583. 10.1016/j.hlc.2019.11.019 [DOI] [PubMed] [Google Scholar]
- Bell, G.I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. 10.1126/science.347575 [DOI] [PubMed] [Google Scholar]
- Blair, E., Redwood C., de Jesus Oliveira M., Moolman-Smook J.C., Brink P., Corfield V.A., Ostman-Smith I., and Watkins H.. 2002. Mutations of the light meromyosin domain of the beta-myosin heavy chain rod in hypertrophic cardiomyopathy. Circ. Res. 90:263–269. 10.1161/hh0302.104532 [DOI] [PubMed] [Google Scholar]
- Boothe, S.D., Myers J.D., Pok S., Sun J., Xi Y., Nieto R.M., Cheng J., and Jacot J.G.. 2016. The Effect of Substrate Stiffness on Cardiomyocyte Action Potentials. Cell Biochem. Biophys. 74:527–535. 10.1007/s12013-016-0758-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brizendine, R.K., Alcala D.B., Carter M.S., Haldeman B.D., Facemyer K.C., Baker J.E., and Cremo C.R.. 2015. Velocities of unloaded muscle filaments are not limited by drag forces imposed by myosin cross-bridges. Proc. Natl. Acad. Sci. USA. 112:11235–11240. 10.1073/pnas.1510241112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporizzo, M.A., Chen C.Y., and Prosser B.L.. 2019. Cardiac microtubules in health and heart disease. Exp. Biol. Med. (Maywood). 244:1255–1272. 10.1177/1535370219868960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporizzo, M.A., Chen C.Y., Bedi K., Margulies K.B., and Prosser B.L.. 2020. Microtubules Increase Diastolic Stiffness in Failing Human Cardiomyocytes and Myocardium. Circulation. 141:902–915. 10.1161/CIRCULATIONAHA.119.043930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra, M., Rundell V.L., Tardiff J.C., Leinwand L.A., De Tombe P.P., and Solaro R.J.. 2001. Ca(2+) activation of myofilaments from transgenic mouse hearts expressing R92Q mutant cardiac troponin T. Am. J. Physiol. Heart Circ. Physiol. 280:H705–H713. 10.1152/ajpheart.2001.280.2.H705 [DOI] [PubMed] [Google Scholar]
- Chen, C.Y., Caporizzo M.A., Bedi K., Vite A., Bogush A.I., Robison P., Heffler J.G., Salomon A.K., Kelly N.A., Babu A., et al. 2018. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat. Med. 24:1225–1233. 10.1038/s41591-018-0046-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou, K.K., Rocks J.W., Chen C.Y., Cho S., Merkus K.E., Rajaratnam A., Robison P., Tewari M., Vogel K., Majkut S.F., et al. 2016. Mechanical signaling coordinates the embryonic heartbeat. Proc. Natl. Acad. Sci. USA. 113:8939–8944. 10.1073/pnas.1520428113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, K.W., Lee J., and Kim Y.. 2016. Genetic Variations Leading to Familial Dilated Cardiomyopathy. Mol. Cells. 39:722–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho, S., Vashisth M., Abbas A., Majkut S., Vogel K., Xia Y., Ivanovska I.L., Irianto J., Tewari M., Zhu K., et al. 2019. Mechanosensing by the Lamina Protects against Nuclear Rupture, DNA Damage, and Cell-Cycle Arrest. Dev. Cell. 49:920–935.e5. 10.1016/j.devcel.2019.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, A., Kutys M.L., Zhang K., Polacheck W.J., Sheng C.C., Luu R.J., Eyckmans J., Hinson J.T., Seidman J.G., Seidman C.E., and Chen C.S.. 2018. Force Generation via β-Cardiac Myosin, Titin, and α-Actinin Drives Cardiac Sarcomere Assembly from Cell-Matrix Adhesions. Dev. Cell. 44:87–96.e5. 10.1016/j.devcel.2017.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clippinger, S.R., Cloonan P.E., Greenberg L., Ernst M., Stump W.T., and Greenberg M.J.. 2019. Disrupted mechanobiology links the molecular and cellular phenotypes in familial dilated cardiomyopathy. Proc. Natl. Acad. Sci. USA. 116:17831–17840. 10.1073/pnas.1910962116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clippinger, S.R., Cloonan P.E., Wang W., Greenberg L., Stump W.T., Angsutararux P., Nerbonne J.M., and Greenberg M.J.. 2020. Mechanical dysfunction induced by a hypertrophic cardiomyopathy mutation is the primary driver of cellular adaptation. bioRxiv. (Preprint posted May 5, 2020). 10.1101/2020.05.04.067181 [DOI] [Google Scholar]
- Crilley, J.G., Boehm E.A., Blair E., Rajagopalan B., Blamire A.M., Styles P., McKenna W.J., Ostman-Smith I., Clarke K., and Watkins H.. 2003. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J. Am. Coll. Cardiol. 41:1776–1782. 10.1016/S0735-1097(02)03009-7 [DOI] [PubMed] [Google Scholar]
- Cuda, G., Fananapazir L., Epstein N.D., and Sellers J.R.. 1997. The in vitro motility activity of beta-cardiac myosin depends on the nature of the beta-myosin heavy chain gene mutation in hypertrophic cardiomyopathy. J. Muscle Res. Cell Motil. 18:275–283. 10.1023/A:1018613907574 [DOI] [PubMed] [Google Scholar]
- Dai, Y., Amenov A., Ignatyeva N., Koschinski A., Xu H., Soong P.L., Tiburcy M., Linke W.A., Zaccolo M., Hasenfuss G., et al. 2020. Troponin destabilization impairs sarcomere-cytoskeleton interactions in iPSC-derived cardiomyocytes from dilated cardiomyopathy patients. Sci. Rep. 10:209. 10.1038/s41598-019-56597-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J., and Molkentin J.D.. 2014. Myofibroblasts: trust your heart and let fate decide. J. Mol. Cell. Cardiol. 70:9–18. 10.1016/j.yjmcc.2013.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis, J., Davis L.C., Correll R.N., Makarewich C.A., Schwanekamp J.A., Moussavi-Harami F., Wang D., York A.J., Wu H., Houser S.R., et al. 2016. A Tension-Based Model Distinguishes Hypertrophic versus Dilated Cardiomyopathy. Cell. 165:1147–1159. 10.1016/j.cell.2016.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, S.M. 2013. The ubiquitin proteasome system in human cardiomyopathies and heart failure. Am. J. Physiol. Heart Circ. Physiol. 304:H1283–H1293. 10.1152/ajpheart.00249.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, S.M. 2019. Nonobstructive Hypertrophic Cardiomyopathy-The High-Hanging Fruit. JAMA Cardiol. 4:235–236. 10.1001/jamacardio.2018.4953 [DOI] [PubMed] [Google Scholar]
- De La Cruz, E.M., and Ostap E.M.. 2004. Relating biochemistry and function in the myosin superfamily. Curr. Opin. Cell Biol. 16:61–67. 10.1016/j.ceb.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Deacon, J.C., Bloemink M.J., Rezavandi H., Geeves M.A., and Leinwand L.A.. 2012. Identification of functional differences between recombinant human α and β cardiac myosin motors. Cell. Mol. Life Sci. 69:2261–2277. 10.1007/s00018-012-0927-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debold, E.P., Schmitt J.P., Patlak J.B., Beck S.E., Moore J.R., Seidman J.G., Seidman C., and Warshaw D.M.. 2007. Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse alpha-cardiac myosin in the laser trap assay. Am. J. Physiol. Heart Circ. Physiol. 293:H284–H291. 10.1152/ajpheart.00128.2007 [DOI] [PubMed] [Google Scholar]
- Dominguez, R., Freyzon Y., Trybus K.M., and Cohen C.. 1998. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: visualization of the pre-power stroke state. Cell. 94:559–571. 10.1016/S0092-8674(00)81598-6 [DOI] [PubMed] [Google Scholar]
- DuFort, C.C., Paszek M.J., and Weaver V.M.. 2011. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12:308–319. 10.1038/nrm3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezekian, J.E., Clippinger S.R., Garcia J.M., Yang Q., Denfield S., Jeewa A., Dreyer W.J., Zou W., Fan Y., Allen H.D., et al. 2020. Variant R94C in TNNT2-Encoded Troponin T Predisposes to Pediatric Restrictive Cardiomyopathy and Sudden Death Through Impaired Thin Filament Relaxation Resulting in Myocardial Diastolic Dysfunction. J. Am. Heart Assoc. 9:e015111. 10.1161/JAHA.119.015111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatkin, D., Huttner I.G., Kovacic J.C., Seidman J.G., and Seidman C.E.. 2019. Precision Medicine in the Management of Dilated Cardiomyopathy: JACC State-of-the-Art Review. J. Am. Coll. Cardiol. 74:2921–2938. 10.1016/j.jacc.2019.10.011 [DOI] [PubMed] [Google Scholar]
- Fenn, W.O. 1923. A quantitative comparison between the energy liberated and the work performed by the isolated sartorius muscle of the frog. J. Physiol. 58:175–203. 10.1113/jphysiol.1923.sp002115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrante, M.I., Kiff R.M., Goulding D.A., and Stemple D.L.. 2011. Troponin T is essential for sarcomere assembly in zebrafish skeletal muscle. J. Cell Sci. 124:565–577. 10.1242/jcs.071274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford, S.J., Mamidi R., Jimenez J., Tardiff J.C., and Chandra M.. 2012. Effects of R92 mutations in mouse cardiac troponin T are influenced by changes in myosin heavy chain isoform. J. Mol. Cell. Cardiol. 53:542–551. 10.1016/j.yjmcc.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forissier, J.F., Carrier L., Farza H., Bonne G., Bercovici J., Richard P., Hainque B., Townsend P.J., Yacoub M.H., Fauré S., et al. 1996. Codon 102 of the cardiac troponin T gene is a putative hot spot for mutations in familial hypertrophic cardiomyopathy. Circulation. 94:3069–3073. 10.1161/01.CIR.94.12.3069 [DOI] [PubMed] [Google Scholar]
- Frank, S., and Braunwald E.. 1968. Idiopathic hypertrophic subaortic stenosis. Clinical analysis of 126 patients with emphasis on the natural history. Circulation. 37:759–788. 10.1161/01.CIR.37.5.759 [DOI] [PubMed] [Google Scholar]
- Gangadharan, B., Sunitha M.S., Mukherjee S., Chowdhury R.R., Haque F., Sekar N., Sowdhamini R., Spudich J.A., and Mercer J.A.. 2017. Molecular mechanisms and structural features of cardiomyopathy-causing troponin T mutants in the tropomyosin overlap region. Proc. Natl. Acad. Sci. USA. 114:11115–11120. 10.1073/pnas.1710354114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisterfer-Lowrance, A.A., Kass S., Tanigawa G., Vosberg H.P., McKenna W., Seidman C.E., and Seidman J.G.. 1990. A molecular basis for familial hypertrophic cardiomyopathy: a beta cardiac myosin heavy chain gene missense mutation. Cell. 62:999–1006. 10.1016/0092-8674(90)90274-I [DOI] [PubMed] [Google Scholar]
- Geisterfer-Lowrance, A.A., Christe M., Conner D.A., Ingwall J.S., Schoen F.J., Seidman C.E., and Seidman J.G.. 1996. A mouse model of familial hypertrophic cardiomyopathy. Science. 272:731–734. 10.1126/science.272.5262.731 [DOI] [PubMed] [Google Scholar]
- Gersh, B.J., Maron B.J., Bonow R.O., Dearani J.A., Fifer M.A., Link M.S., Naidu S.S., Nishimura R.A., Ommen S.R., Rakowski H., et al. American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines . 2011. 2011 ACCF/AHA Guideline for the Diagnosis and Treatment of Hypertrophic Cardiomyopathy: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Developed in collaboration with the American Association for Thoracic Surgery, American Society of Echocardiography, American Society of Nuclear Cardiology, Heart Failure Society of America, Heart Rhythm Society, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J. Am. Coll. Cardiol. 58:e212–e260. 10.1016/j.jacc.2011.06.011 [DOI] [PubMed] [Google Scholar]
- Green, E.M., Wakimoto H., Anderson R.L., Evanchik M.J., Gorham J.M., Harrison B.C., Henze M., Kawas R., Oslob J.D., Rodriguez H.M., et al. 2016. A small-molecule inhibitor of sarcomere contractility suppresses hypertrophic cardiomyopathy in mice. Science. 351:617–621. 10.1126/science.aad3456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.J., Kazmierczak K., Szczesna-Cordary D., and Moore J.R.. 2010. Cardiomyopathy-linked myosin regulatory light chain mutations disrupt myosin strain-dependent biochemistry. Proc. Natl. Acad. Sci. USA. 107:17403–17408. 10.1073/pnas.1009619107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.J., Shuman H., and Ostap E.M.. 2014. Inherent force-dependent properties of β-cardiac myosin contribute to the force-velocity relationship of cardiac muscle. Biophys. J. 107:L41–L44. 10.1016/j.bpj.2014.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, M.J., Arpağ G., Tüzel E., and Ostap E.M.. 2016. A Perspective on the Role of Myosins as Mechanosensors. Biophys. J. 110:2568–2576. 10.1016/j.bpj.2016.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinto, P.J., Manning E.P., Schwartz S.D., and Tardiff J.C.. 2007. Computational Characterization of Mutations in Cardiac Troponin T Known to Cause Familial Hypertrophic Cardiomyopathy. J. Theor. Comput. Chem. 6:413–419. 10.1142/S0219633607003271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther, L.K., Rohde J.A., Tang W., Walton S.D., Unrath W.C., Trivedi D.V., Muretta J.M., Thomas D.D., and Yengo C.M.. 2019. Converter domain mutations in myosin alter structural kinetics and motor function. J. Biol. Chem. 294:1554–1567. 10.1074/jbc.RA118.006128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S.P., Bartley C.R., Hacker T.A., McDonald K.S., Douglas P.S., Greaser M.L., Powers P.A., and Moss R.L.. 2002. Hypertrophic cardiomyopathy in cardiac myosin binding protein-C knockout mice. Circ. Res. 90:594–601. 10.1161/01.RES.0000012222.70819.64 [DOI] [PubMed] [Google Scholar]
- He, H., Javadpour M.M., Latif F., Tardiff J.C., and Ingwall J.S.. 2007. R-92L and R-92W mutations in cardiac troponin T lead to distinct energetic phenotypes in intact mouse hearts. Biophys. J. 93:1834–1844. 10.1529/biophysj.107.107557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, A.S., and Day S.M.. 2016. Hypertrophic cardiomyopathy: single gene disease or complex trait? Eur. Heart J. 37:1823–1825. 10.1093/eurheartj/ehv562 [DOI] [PubMed] [Google Scholar]
- Helms, A.S., Alvarado F.J., Yob J., Tang V.T., Pagani F., Russell M.W., Valdivia H.H., and Day S.M.. 2016. Genotype-Dependent and -Independent Calcium Signaling Dysregulation in Human Hypertrophic Cardiomyopathy. Circulation. 134:1738–1748. 10.1161/CIRCULATIONAHA.115.020086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, A.S., Tang V.T., O’Leary T.S., Friedline S., Wauchope M., Arora A., Wasserman A.H., Smith E.D., Lee L.M., Wen X.W., et al. 2020a. Effects of MYBPC3 loss-of-function mutations preceding hypertrophic cardiomyopathy. JCI Insight. 5:e133782. 10.1172/jci.insight.133782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms, A.S., Thompson A.D., Glazier A.A., Hafeez N., Kabani S., Rodriguez J., Yob J.M., Woolcock H., Mazzarotto F., Lakdawala N.K., et al. 2020b. Spatial and Functional Distribution of MYBPC3 Pathogenic Variants and Clinical Outcomes in Patients With Hypertrophic Cardiomyopathy. Circ. Genom. Precis. Med. 13:396–405. 10.1161/CIRCGEN.120.002929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, A.V. 1938. The heat of shortening and the dynamic constants of muscle. Proceedings of the Royal Society Series B-Biological Sciences. 126:136–195. [DOI] [PubMed] [Google Scholar]
- Hinson, J.T., Chopra A., Nafissi N., Polacheck W.J., Benson C.C., Swist S., Gorham J., Yang L., Schafer S., Sheng C.C., et al. 2015. HEART DISEASE. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 349:982–986. 10.1126/science.aaa5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, C.Y., Day S.M., Ashley E.A., Michels M., Pereira A.C., Jacoby D., Cirino A.L., Fox J.C., Lakdawala N.K., Ware J.S., et al. 2018. Genotype and Lifetime Burden of Disease in Hypertrophic Cardiomyopathy: Insights from the Sarcomeric Human Cardiomyopathy Registry (SHaRe). Circulation. 138:1387–1398. 10.1161/CIRCULATIONAHA.117.033200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooijman, P., Stewart M.A., and Cooke R.. 2011. A new state of cardiac myosin with very slow ATP turnover: a potential cardioprotective mechanism in the heart. Biophys. J. 100:1969–1976. 10.1016/j.bpj.2011.02.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, X., Margadant F.M., Yao M., and Sheetz M.P.. 2017. Molecular stretching modulates mechanosensing pathways. Protein Sci. 26:1337–1351. 10.1002/pro.3188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley, H.E. 1990. Sliding filaments and molecular motile systems. J. Biol. Chem. 265:8347–8350. 10.1016/S0021-9258(19)38888-X [DOI] [PubMed] [Google Scholar]
- Iskratsch, T., Wolfenson H., and Sheetz M.P.. 2014. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15:825–833. 10.1038/nrm3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janota, C.S., Calero-Cuenca F.J., and Gomes E.R.. 2020. The role of the cell nucleus in mechanotransduction. Curr. Opin. Cell Biol. 63:204–211. 10.1016/j.ceb.2020.03.001 [DOI] [PubMed] [Google Scholar]
- Javadpour, M.M., Tardiff J.C., Pinz I., and Ingwall J.S.. 2003. Decreased energetics in murine hearts bearing the R92Q mutation in cardiac troponin T. J. Clin. Invest. 112:768–775. 10.1172/JCI15967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampourakis, T., Zhang X., Sun Y.B., and Irving M.. 2018. Omecamtiv mercabil and blebbistatin modulate cardiac contractility by perturbing the regulatory state of the myosin filament. J. Physiol. 596:31–46. 10.1113/JP275050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor, P.F., Abraham J.R., Dipchand A.I., Benson L.N., and Redington A.N.. 2010. The impact of changing medical therapy on transplantation-free survival in pediatric dilated cardiomyopathy. J. Am. Coll. Cardiol. 55:1377–1384. 10.1016/j.jacc.2009.11.059 [DOI] [PubMed] [Google Scholar]
- Kerr, T.P., Sewry C.A., Robb S.A., and Roberts R.G.. 2001. Long mutant dystrophins and variable phenotypes: evasion of nonsense-mediated decay? Hum. Genet. 109:402–407. 10.1007/s004390100598 [DOI] [PubMed] [Google Scholar]
- Kerr, J.P., Robison P., Shi G., Bogush A.I., Kempema A.M., Hexum J.K., Becerra N., Harki D.A., Martin S.S., Raiteri R., et al. 2015. Detyrosinated microtubules modulate mechanotransduction in heart and skeletal muscle. Nat. Commun. 6:8526. 10.1038/ncomms9526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieu, T.T., Awinda P.O., and Tanner B.C.W.. 2019. Omecamtiv Mecarbil Slows Myosin Kinetics in Skinned Rat Myocardium at Physiological Temperature. Biophys. J. 116:2149–2160. 10.1016/j.bpj.2019.04.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankford, E.B., Epstein N.D., Fananapazir L., and Sweeney H.L.. 1995. Abnormal contractile properties of muscle fibers expressing beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. J. Clin. Invest. 95:1409–1414. 10.1172/JCI117795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavine, K.J., and Greenberg M.J.. 2020. Beyond genomics-technological advances improving the molecular characterization and precision treatment of heart failure. Heart Fail. Rev. 10.1007/s10741-020-10021-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Termglinchan V., Diecke S., Itzhaki I., Lam C.K., Garg P., Lau E., Greenhaw M., Seeger T., Wu H., et al. 2019. Activation of PDGF pathway links LMNA mutation to dilated cardiomyopathy. Nature. 572:335–340. 10.1038/s41586-019-1406-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, W., Craig R., and Vibert P.. 1994. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 368:65–67. 10.1038/368065a0 [DOI] [PubMed] [Google Scholar]
- Lehman, S.J., Tal-Grinspan L., Lynn M.L., Strom J., Benitez G.E., Anderson M.E., and Tardiff J.C.. 2019. Chronic Calmodulin-Kinase II Activation Drives Disease Progression in Mutation-Specific Hypertrophic Cardiomyopathy. Circulation. 139:1517–1529. 10.1161/CIRCULATIONAHA.118.034549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linari, M., Brunello E., Reconditi M., Fusi L., Caremani M., Narayanan T., Piazzesi G., Lombardi V., and Irving M.. 2015. Force generation by skeletal muscle is controlled by mechanosensing in myosin filaments. Nature. 528:276–279. 10.1038/nature15727 [DOI] [PubMed] [Google Scholar]
- Liu, B., Tikunova S.B., Kline K.P., Siddiqui J.K., and Davis J.P.. 2012. Disease-related cardiac troponins alter thin filament Ca2+ association and dissociation rates. PLoS One. 7:e38259. 10.1371/journal.pone.0038259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y., White H.D., Belknap B., Winkelmann D.A., and Forgacs E.. 2015. Omecamtiv Mecarbil modulates the kinetic and motile properties of porcine β-cardiac myosin. Biochemistry. 54:1963–1975. 10.1021/bi5015166 [DOI] [PubMed] [Google Scholar]
- Liu, C., Kawana M., Song D., Ruppel K.M., and Spudich J.A.. 2018. Controlling load-dependent kinetics of β-cardiac myosin at the single-molecule level. Nat. Struct. Mol. Biol. 25:505–514. 10.1038/s41594-018-0069-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenzini, M., Norrish G., Field E., Ochoa J.P., Cicerchia M., Akhtar M.M., Syrris P., Lopes L.R., Kaski J.P., and Elliott P.M.. 2020. Penetrance of Hypertrophic Cardiomyopathy in Sarcomere Protein Mutation Carriers. J. Am. Coll. Cardiol. 76:550–559. 10.1016/j.jacc.2020.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey, S. 2002. Functional consequences of mutations in the myosin heavy chain at sites implicated in familial hypertrophic cardiomyopathy. Trends Cardiovasc. Med. 12:348–354. 10.1016/S1050-1738(02)00181-0 [DOI] [PubMed] [Google Scholar]
- Lowey, S., Lesko L.M., Rovner A.S., Hodges A.R., White S.L., Low R.B., Rincon M., Gulick J., and Robbins J.. 2008. Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an alpha- or beta-myosin heavy chain backbone. J. Biol. Chem. 283:20579–20589. 10.1074/jbc.M800554200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey, S., Bretton V., Gulick J., Robbins J., and Trybus K.M.. 2013. Transgenic mouse α- and β-cardiac myosins containing the R403Q mutation show isoform-dependent transient kinetic differences. J. Biol. Chem. 288:14780–14787. 10.1074/jbc.M113.450668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey, S., Bretton V., Joel P.B., Trybus K.M., Gulick J., Robbins J., Kalganov A., Cornachione A.S., and Rassier D.E.. 2018. Hypertrophic cardiomyopathy R403Q mutation in rabbit β-myosin reduces contractile function at the molecular and myofibrillar levels. Proc. Natl. Acad. Sci. USA. 115:11238–11243. 10.1073/pnas.1802967115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lymn, R.W., and Taylor E.W.. 1971. Mechanism of adenosine triphosphate hydrolysis by actomyosin. Biochemistry. 10:4617–4624. 10.1021/bi00801a004 [DOI] [PubMed] [Google Scholar]
- Lynn, M.L., Lehman S.J., and Tardiff J.C.. 2018. Biophysical Derangements in Genetic Cardiomyopathies. Heart Fail. Clin. 14:147–159. 10.1016/j.hfc.2017.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maass, A.H., Ikeda K., Oberdorf-Maass S., Maier S.K., and Leinwand L.A.. 2004. Hypertrophy, fibrosis, and sudden cardiac death in response to pathological stimuli in mice with mutations in cardiac troponin T. Circulation. 110:2102–2109. 10.1161/01.CIR.0000144460.84795.E3 [DOI] [PubMed] [Google Scholar]
- Madan, A., Viswanathan M.C., Woulfe K.C., Schmidt W., Sidor A., Liu T., Nguyen T.H., Trinh B., Wilson C., Madathil S., et al. 2020. TNNT2 mutations in the tropomyosin binding region of TNT1 disrupt its role in contractile inhibition and stimulate cardiac dysfunction. Proc. Natl. Acad. Sci. USA. 117:18822–18831. 10.1073/pnas.2001692117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majkut, S., Dingal P.C., and Discher D.E.. 2014. Stress sensitivity and mechanotransduction during heart development. Curr. Biol. 24:R495–R501. 10.1016/j.cub.2014.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, F.I., Hartman J.J., Elias K.A., Morgan B.P., Rodriguez H., Brejc K., Anderson R.L., Sueoka S.H., Lee K.H., Finer J.T., et al. 2011. Cardiac myosin activation: a potential therapeutic approach for systolic heart failure. Science. 331:1439–1443. 10.1126/science.1200113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamidi, R., Li J., Doh C.Y., Verma S., and Stelzer J.E.. 2018. Impact of the Myosin Modulator Mavacamten on Force Generation and Cross-Bridge Behavior in a Murine Model of Hypercontractility. J. Am. Heart Assoc. 7:e009627. 10.1161/JAHA.118.009627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning, E.P., Guinto P.J., and Tardiff J.C.. 2012. Correlation of molecular and functional effects of mutations in cardiac troponin T linked to familial hypertrophic cardiomyopathy: an integrative in silico/in vitro approach. J. Biol. Chem. 287:14515–14523. 10.1074/jbc.M111.257436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marian, A.J., Zhao G., Seta Y., Roberts R., and Yu Q.T.. 1997. Expression of a mutant (Arg92Gln) human cardiac troponin T, known to cause hypertrophic cardiomyopathy, impairs adult cardiac myocyte contractility. Circ. Res. 81:76–85. 10.1161/01.RES.81.1.76 [DOI] [PubMed] [Google Scholar]
- Mazzarotto, F., Olivotto I., Boschi B., Girolami F., Poggesi C., Barton P.J.R., and Walsh R.. 2020a. Contemporary Insights Into the Genetics of Hypertrophic Cardiomyopathy: Toward a New Era in Clinical Testing? J. Am. Heart Assoc. 9:e015473. 10.1161/JAHA.119.015473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzarotto, F., Tayal U., Buchan R.J., Midwinter W., Wilk A., Whiffin N., Govind R., Mazaika E., de Marvao A., Dawes T.J.W., et al. 2020b. Reevaluating the Genetic Contribution of Monogenic Dilated Cardiomyopathy. Circulation. 141:387–398. 10.1161/CIRCULATIONAHA.119.037661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell, M., Tal Grinspan L., Williams M.R., Lynn M.L., Schwartz B.A., Fass O.Z., Schwartz S.D., and Tardiff J.C.. 2017. Clinically Divergent Mutation Effects on the Structure and Function of the Human Cardiac Tropomyosin Overlap. Biochemistry. 56:3403–3413. 10.1021/acs.biochem.7b00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKillop, D.F., and Geeves M.A.. 1993. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys. J. 65:693–701. 10.1016/S0006-3495(93)81110-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally, E.M., and Mestroni L.. 2017. Dilated Cardiomyopathy: Genetic Determinants and Mechanisms. Circ. Res. 121:731–748. 10.1161/CIRCRESAHA.116.309396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Singh R.R., and Sadayappan S.. 2019. Cardiac myosin binding protein-C phosphorylation regulates the super-relaxed state of myosin. Proc. Natl. Acad. Sci. USA. 116:11731–11736. 10.1073/pnas.1821660116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memo, M., Leung M.C., Ward D.G., dos Remedios C., Morimoto S., Zhang L., Ravenscroft G., McNamara E., Nowak K.J., Marston S.B., and Messer A.E.. 2013. Familial dilated cardiomyopathy mutations uncouple troponin I phosphorylation from changes in myofibrillar Ca2+ sensitivity. Cardiovasc. Res. 99:65–73. 10.1093/cvr/cvt071 [DOI] [PubMed] [Google Scholar]
- Messer, A.E., Bayliss C.R., El-Mezgueldi M., Redwood C.S., Ward D.G., Leung M.C., Papadaki M., Dos Remedios C., and Marston S.B.. 2016. Mutations in troponin T associated with Hypertrophic Cardiomyopathy increase Ca(2+)-sensitivity and suppress the modulation of Ca(2+)-sensitivity by troponin I phosphorylation. Arch. Biochem. Biophys. 601:113–120. 10.1016/j.abb.2016.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, D.E., Tardiff J.C., and Chandra M.. 2001. Cardiac troponin T mutations: correlation between the type of mutation and the nature of myofilament dysfunction in transgenic mice. J. Physiol. 536:583–592. 10.1111/j.1469-7793.2001.0583c.xd [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, B.P., Muci A., Lu P.P., Qian X., Tochimoto T., Smith W.W., Garard M., Kraynack E., Collibee S., Suehiro I., et al. 2010. Discovery of omecamtiv mecarbil the first, selective, small molecule activator of cardiac Myosin. ACS Med. Chem. Lett. 1:472–477. 10.1021/ml100138q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto, S., Yanaga F., Minakami R., and Ohtsuki I.. 1998. Ca2+-sensitizing effects of the mutations at Ile-79 and Arg-92 of troponin T in hypertrophic cardiomyopathy. Am. J. Physiol. 275:C200–C207. 10.1152/ajpcell.1998.275.1.C200 [DOI] [PubMed] [Google Scholar]
- Mun, J.Y., Previs M.J., Yu H.Y., Gulick J., Tobacman L.S., Beck Previs S., Robbins J., Warshaw D.M., and Craig R.. 2014. Myosin-binding protein C displaces tropomyosin to activate cardiac thin filaments and governs their speed by an independent mechanism. Proc. Natl. Acad. Sci. USA. 111:2170–2175. 10.1073/pnas.1316001111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muretta, J.M., Rohde J.A., Johnsrud D.O., Cornea S., and Thomas D.D.. 2015. Direct real-time detection of the structural and biochemical events in the myosin power stroke. Proc. Natl. Acad. Sci. USA. 112:14272–14277. 10.1073/pnas.1514859112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, S., Sommese R.F., Ujfalusi Z., Combs A., Langer S., Sutton S., Leinwand L.A., Geeves M.A., Ruppel K.M., and Spudich J.A.. 2015. Contractility parameters of human β-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Sci. Adv. 1:e1500511. 10.1126/sciadv.1500511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, S., Trivedi D.V., Sarkar S.S., Adhikari A.S., Sunitha M.S., Sutton S., Ruppel K.M., and Spudich J.A.. 2017. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24:525–533. 10.1038/nsmb.3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, L., Kovács Á., Bódi B., Pásztor E.T., Fülöp G.A., Tóth A., Édes I., and Papp Z.. 2015. The novel cardiac myosin activator omecamtiv mecarbil increases the calcium sensitivity of force production in isolated cardiomyocytes and skeletal muscle fibres of the rat. Br. J. Pharmacol. 172:4506–4518. 10.1111/bph.13235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura, M., and Sadoshima J.. 2018. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 15:387–407. 10.1038/s41569-018-0007-y [DOI] [PubMed] [Google Scholar]
- Nakano, S.J., Walker J.S., Walker L.A., Li X., Du Y., Miyamoto S.D., Sucharov C.C., Garcia A.M., Mitchell M.B., Ambardekar A.V., and Stauffer B.L.. 2019. Increased myocyte calcium sensitivity in end-stage pediatric dilated cardiomyopathy. Am. J. Physiol. Heart Circ. Physiol. 317:H1221–H1230. 10.1152/ajpheart.00409.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, S.R., Li A., Beck-Previs S., Kennedy G.G., and Warshaw D.M.. 2020. Imaging ATP Consumption in Resting Skeletal Muscle: One Molecule at a Time. Biophys. J. 119:1050–1055. 10.1016/j.bpj.2020.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer, S., Kolm P., Ho C.Y., Kwong R.Y., Desai M.Y., Dolman S.F., Appelbaum E., Desvigne-Nickens P., DiMarco J.P., Friedrich M.G., et al. HCMR Investigators . 2019. Distinct Subgroups in Hypertrophic Cardiomyopathy in the NHLBI HCM Registry. J. Am. Coll. Cardiol. 74:2333–2345. 10.1016/j.jacc.2019.08.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishii, K., Morimoto S., Minakami R., Miyano Y., Hashizume K., Ohta M., Zhan D.Y., Lu Q.W., and Shibata Y.. 2008. Targeted disruption of the cardiac troponin T gene causes sarcomere disassembly and defects in heartbeat within the early mouse embryo. Dev. Biol. 322:65–73. 10.1016/j.ydbio.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Nyitrai, M., and Geeves M.A.. 2004. Adenosine diphosphate and strain sensitivity in myosin motors. Philos. Trans. R. Soc. Lond. B Biol. Sci. 359:1867–1877. 10.1098/rstb.2004.1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivotto, I., Oreziak A., Barriales-Villa R., Abraham T.P., Masri A., Garcia-Pavia P., Saberi S., Lakdawala N.K., Wheeler M.T., Owens A., et al. EXPLORER-HCM study investigators . 2020. Mavacamten for treatment of symptomatic obstructive hypertrophic cardiomyopathy (EXPLORER-HCM): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 396:759–769. 10.1016/S0140-6736(20)31792-X [DOI] [PubMed] [Google Scholar]
- Palmiter, K.A., Tyska M.J., Haeberle J.R., Alpert N.R., Fananapazir L., and Warshaw D.M.. 2000. R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. J. Muscle Res. Cell Motil. 21:609–620. 10.1023/A:1005678905119 [DOI] [PubMed] [Google Scholar]
- Papadaki, M., Holewinski R.J., Previs S.B., Martin T.G., Stachowski M.J., Li A., Blair C.A., Moravec C.S., Van Eyk J.E., Campbell K.S., et al. 2018. Diabetes with heart failure increases methylglyoxal modifications in the sarcomere, which inhibit function. JCI Insight. 3:e121264. 10.1172/jci.insight.121264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavadai, E., Rynkiewicz M.J., Ghosh A., and Lehman W.. 2020. Docking Troponin T onto the Tropomyosin Overlapping Domain of Thin Filaments. Biophys. J. 118:325–336. 10.1016/j.bpj.2019.11.3393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planelles-Herrero, V.J., Hartman J.J., Robert-Paganin J., Malik F.I., and Houdusse A.. 2017. Mechanistic and structural basis for activation of cardiac myosin force production by omecamtiv mecarbil. Nat. Commun. 8:190. 10.1038/s41467-017-00176-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter, J.R., Meller A., Zimmerman M.I., Greenberg M.J., and Bowman G.R.. 2020. Conformational distributions of isolated myosin motor domains encode their mechanochemical properties. eLife. 9:e55132. 10.7554/eLife.55132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preller, M., and Manstein D.J.. 2012. 4.8 Myosin Motors: Structural Aspects and Functionality. In Comprehensive Biophysics. Egelman E.H., editor. Elsevier, Amsterdam. 118–150. 10.1016/B978-0-12-374920-8.00410-0 [DOI] [Google Scholar]
- Pruitt, B.L., Dunn A.R., Weis W.I., and Nelson W.J.. 2014. Mechano-transduction: from molecules to tissues. PLoS Biol. 12:e1001996. 10.1371/journal.pbio.1001996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment, I., Holden H.M., Whittaker M., Yohn C.B., Lorenz M., Holmes K.C., and Milligan R.A.. 1993a. Structure of the actin-myosin complex and its implications for muscle contraction. Science. 261:58–65. 10.1126/science.8316858 [DOI] [PubMed] [Google Scholar]
- Rayment, I., Rypniewski W.R., Schmidt-Bäse K., Smith R., Tomchick D.R., Benning M.M., Winkelmann D.A., Wesenberg G., and Holden H.M.. 1993b. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 261:50–58. 10.1126/science.8316857 [DOI] [PubMed] [Google Scholar]
- Rice, R., Guinto P., Dowell-Martino C., He H., Hoyer K., Krenz M., Robbins J., Ingwall J.S., and Tardiff J.C.. 2010. Cardiac myosin heavy chain isoform exchange alters the phenotype of cTnT-related cardiomyopathies in mouse hearts. J. Mol. Cell. Cardiol. 48:979–988. 10.1016/j.yjmcc.2009.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, P., Griffiths P.J., Watkins H., and Redwood C.S.. 2007. Dilated and hypertrophic cardiomyopathy mutations in troponin and alpha-tropomyosin have opposing effects on the calcium affinity of cardiac thin filaments. Circ. Res. 101:1266–1273. 10.1161/CIRCRESAHA.107.156380 [DOI] [PubMed] [Google Scholar]
- Robinson, P., Liu X., Sparrow A., Patel S., Zhang Y.H., Casadei B., Watkins H., and Redwood C.. 2018. Hypertrophic cardiomyopathy mutations increase myofilament Ca2+ buffering, alter intracellular Ca2+ handling, and stimulate Ca2+-dependent signaling. J. Biol. Chem. 293:10487–10499. 10.1074/jbc.RA118.002081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robison, P., Caporizzo M.A., Ahmadzadeh H., Bogush A.I., Chen C.Y., Margulies K.B., Shenoy V.B., and Prosser B.L.. 2016. Detyrosinated microtubules buckle and bear load in contracting cardiomyocytes. Science. 352:aaf0659. 10.1126/science.aaf0659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, J.A., Thomas D.D., and Muretta J.M.. 2017. Heart failure drug changes the mechanoenzymology of the cardiac myosin powerstroke. Proc. Natl. Acad. Sci. USA. 114:E1796–E1804. 10.1073/pnas.1611698114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohde, J.A., Roopnarine O., Thomas D.D., and Muretta J.M.. 2018. Mavacamten stabilizes an autoinhibited state of two-headed cardiac myosin. Proc. Natl. Acad. Sci. USA. 115:E7486–E7494. 10.1073/pnas.1720342115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust, E.M., Albayya F.P., and Metzger J.M.. 1999. Identification of a contractile deficit in adult cardiac myocytes expressing hypertrophic cardiomyopathy-associated mutant troponin T proteins. J. Clin. Invest. 103:1459–1467. 10.1172/JCI6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S.S., Trivedi D.V., Morck M.M., Adhikari A.S., Pasha S.N., Ruppel K.M., and Spudich J.A.. 2020. The hypertrophic cardiomyopathy mutations R403Q and R663H increase the number of myosin heads available to interact with actin. Sci. Adv. 6:eaax0069. 10.1126/sciadv.aax0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucerman, J.J., Tan P.M., Buchholz K.S., McCulloch A.D., and Omens J.H.. 2019. Mechanical regulation of gene expression in cardiac myocytes and fibroblasts. Nat. Rev. Cardiol. 16:361–378. 10.1038/s41569-019-0155-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semsarian, C., Ingles J., Maron M.S., and Maron B.J.. 2015. New perspectives on the prevalence of hypertrophic cardiomyopathy. J. Am. Coll. Cardiol. 65:1249–1254. 10.1016/j.jacc.2015.01.019 [DOI] [PubMed] [Google Scholar]
- Shaffer, J.F., Kensler R.W., and Harris S.P.. 2009. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J. Biol. Chem. 284:12318–12327. 10.1074/jbc.M808850200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemankowski, R.F., Wiseman M.O., and White H.D.. 1985. ADP dissociation from actomyosin subfragment 1 is sufficiently slow to limit the unloaded shortening velocity in vertebrate muscle. Proc. Natl. Acad. Sci. USA. 82:658–662. 10.1073/pnas.82.3.658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, L., Cho S., and Discher D.E.. 2017. Mechanosensing of matrix by stem cells: From matrix heterogeneity, contractility, and the nucleus in pore-migration to cardiogenesis and muscle stem cells in vivo. Semin. Cell Dev. Biol. 71:84–98. 10.1016/j.semcdb.2017.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommese, R.F., Sung J., Nag S., Sutton S., Deacon J.C., Choe E., Leinwand L.A., Ruppel K., and Spudich J.A.. 2013. Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human β-cardiac myosin motor function. Proc. Natl. Acad. Sci. USA. 110:12607–12612. 10.1073/pnas.1309493110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spindler, M., Saupe K.W., Christe M.E., Sweeney H.L., Seidman C.E., Seidman J.G., and Ingwall J.S.. 1998. Diastolic dysfunction and altered energetics in the alphaMHC403/+ mouse model of familial hypertrophic cardiomyopathy. J. Clin. Invest. 101:1775–1783. 10.1172/JCI1940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirito, P., Maron B.J., Chiarella F., Bellotti P., Tramarin R., Pozzoli M., and Vecchio C.. 1985. Diastolic abnormalities in patients with hypertrophic cardiomyopathy: relation to magnitude of left ventricular hypertrophy. Circulation. 72:310–316. 10.1161/01.CIR.72.2.310 [DOI] [PubMed] [Google Scholar]
- Spudich, J.A. 2014. Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophys. J. 106:1236–1249. 10.1016/j.bpj.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J.A. 2015. The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochem. Soc. Trans. 43:64–72. 10.1042/BST20140324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam, R., and Winkelmann D.A.. 2004. Chaperone-mediated folding and assembly of myosin in striated muscle. J. Cell Sci. 117:641–652. 10.1242/jcs.00899 [DOI] [PubMed] [Google Scholar]
- Stewart, M.A., Franks-Skiba K., Chen S., and Cooke R.. 2010. Myosin ATP turnover rate is a mechanism involved in thermogenesis in resting skeletal muscle fibers. Proc. Natl. Acad. Sci. USA. 107:430–435. 10.1073/pnas.0909468107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, J., Nag S., Mortensen K.I., Vestergaard C.L., Sutton S., Ruppel K., Flyvbjerg H., and Spudich J.A.. 2015. Harmonic force spectroscopy measures load-dependent kinetics of individual human β-cardiac myosin molecules. Nat. Commun. 6:7931. 10.1038/ncomms8931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney, H.L., Feng H.S., Yang Z., and Watkins H.. 1998. Functional analyses of troponin T mutations that cause hypertrophic cardiomyopathy: insights into disease pathogenesis and troponin function. Proc. Natl. Acad. Sci. USA. 95:14406–14410. 10.1073/pnas.95.24.14406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swenson, A.M., Tang W., Blair C.A., Fetrow C.M., Unrath W.C., Previs M.J., Campbell K.S., and Yengo C.M.. 2017. Omecamtiv Mecarbil Enhances the Duty Ratio of Human β-Cardiac Myosin Resulting in Increased Calcium Sensitivity and Slowed Force Development in Cardiac Muscle. J. Biol. Chem. 292:3768–3778. 10.1074/jbc.M116.748780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesna, D., Zhang R., Zhao J., Jones M., Guzman G., and Potter J.D.. 2000. Altered regulation of cardiac muscle contraction by troponin T mutations that cause familial hypertrophic cardiomyopathy. J. Biol. Chem. 275:624–630. 10.1074/jbc.275.1.624 [DOI] [PubMed] [Google Scholar]
- Takagi, Y., Homsher E.E., Goldman Y.E., and Shuman H.. 2006. Force generation in single conventional actomyosin complexes under high dynamic load. Biophys. J. 90:1295–1307. 10.1529/biophysj.105.068429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda, S., Yamashita A., Maeda K., and Maéda Y.. 2003. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 424:35–41. 10.1038/nature01780 [DOI] [PubMed] [Google Scholar]
- Tardiff, J.C., Hewett T.E., Palmer B.M., Olsson C., Factor S.M., Moore R.L., Robbins J., and Leinwand L.A.. 1999. Cardiac troponin T mutations result in allele-specific phenotypes in a mouse model for hypertrophic cardiomyopathy. J. Clin. Invest. 104:469–481. 10.1172/JCI6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teare, D. 1958. Asymmetrical hypertrophy of the heart in young adults. Br. Heart J. 20:1–8. 10.1136/hrt.20.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teerlink, J.R., Felker G.M., McMurray J.J., Solomon S.D., Adams K.F. Jr., Cleland J.G., Ezekowitz J.A., Goudev A., Macdonald P., Metra M., et al. COSMIC-HF Investigators . 2016. Chronic Oral Study of Myosin Activation to Increase Contractility in Heart Failure (COSMIC-HF): a phase 2, pharmacokinetic, randomised, placebo-controlled trial. Lancet. 388:2895–2903. 10.1016/S0140-6736(16)32049-9 [DOI] [PubMed] [Google Scholar]
- Teerlink, J.R., Diaz R., Felker G.M., McMurray J.J.V., Metra M., Solomon S.D., Adams K.F., Anand I., Arias-Mendoza A., Biering-Sørensen T., et al. GALACTIC-HF Investigators . 2020. Cardiac Myosin Activation with Omecamtiv Mecarbil in Systolic Heart Failure. N. Engl. J. Med. 10.1056/NEJMoa2025797 [DOI] [PubMed] [Google Scholar]
- Tobacman, L.S., Nihli M., Butters C., Heller M., Hatch V., Craig R., Lehman W., and Homsher E.. 2002. The troponin tail domain promotes a conformational state of the thin filament that suppresses myosin activity. J. Biol. Chem. 277:27636–27642. 10.1074/jbc.M201768200 [DOI] [PubMed] [Google Scholar]
- Toepfer, C.N., Wakimoto H., Garfinkel A.C., McDonough B., Liao D., Jiang J., Tai A.C., Gorham J.M., Lunde I.G., Lun M., et al. 2019. Hypertrophic cardiomyopathy mutations in MYBPC3 dysregulate myosin. Sci. Transl. Med. 11:eaat1199. 10.1126/scitranslmed.aat1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, D.V., Adhikari A.S., Sarkar S.S., Ruppel K.M., and Spudich J.A.. 2018. Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light. Biophys. Rev. 10:27–48. 10.1007/s12551-017-0274-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucholski, T., Cai W., Gregorich Z.R., Bayne E.F., Mitchell S.D., McIlwain S.J., de Lange W.J., Wrobbel M., Karp H., Hite Z., et al. 2020. Distinct hypertrophic cardiomyopathy genotypes result in convergent sarcomeric proteoform profiles revealed by top-down proteomics. Proc. Natl. Acad. Sci. USA. 117:24691–24700. 10.1073/pnas.2006764117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska, M.J., Hayes E., Giewat M., Seidman C.E., Seidman J.G., and Warshaw D.M.. 2000. Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ. Res. 86:737–744. 10.1161/01.RES.86.7.737 [DOI] [PubMed] [Google Scholar]
- van Steensel, B., and Belmont A.S.. 2017. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell. 169:780–791. 10.1016/j.cell.2017.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren, P., Harris D.E., Alpert N.R., and Warshaw D.M.. 1995. Cardiac V1 and V3 myosins differ in their hydrolytic and mechanical activities in vitro. Circ. Res. 77:439–444. 10.1161/01.RES.77.2.439 [DOI] [PubMed] [Google Scholar]
- Vera, C.D., Johnson C.A., Walklate J., Adhikari A., Svicevic M., Mijailovich S.M., Combs A.C., Langer S.J., Ruppel K.M., Spudich J.A., et al. 2019. Myosin motor domains carrying mutations implicated in early or late onset hypertrophic cardiomyopathy have similar properties. J. Biol. Chem. 294:17451–17462. 10.1074/jbc.RA119.010563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, M.R., Lehman S.J., Tardiff J.C., and Schwartz S.D.. 2016. Atomic resolution probe for allostery in the regulatory thin filament. Proc. Natl. Acad. Sci. USA. 113:3257–3262. 10.1073/pnas.1519541113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann, D.A., Forgacs E., Miller M.T., and Stock A.M.. 2015. Structural basis for drug-induced allosteric changes to human β-cardiac myosin motor activity. Nat. Commun. 6:7974. 10.1038/ncomms8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead, J.L., Zhao F.Q., Craig R., Egelman E.H., Alamo L., and Padrón R.. 2005. Atomic model of a myosin filament in the relaxed state. Nature. 436:1195–1199. 10.1038/nature03920 [DOI] [PubMed] [Google Scholar]
- Woody, M.S., Greenberg M.J., Barua B., Winkelmann D.A., Goldman Y.E., and Ostap E.M.. 2018. Positive cardiac inotrope omecamtiv mecarbil activates muscle despite suppressing the myosin working stroke. Nat. Commun. 9:3838. 10.1038/s41467-018-06193-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody, M.S., Winkelmann D.A., Capitanio M., Ostap E.M., and Goldman Y.E.. 2019. Single molecule mechanics resolves the earliest events in force generation by cardiac myosin. eLife. 8:e49266. 10.7554/eLife.49266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, Y., Namba K., and Fujii T.. 2020. Cardiac muscle thin filament structures reveal calcium regulatory mechanism. Nat. Commun. 11:153. 10.1038/s41467-019-14008-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaga, F., Morimoto S., and Ohtsuki I.. 1999. Ca2+ sensitization and potentiation of the maximum level of myofibrillar ATPase activity caused by mutations of troponin T found in familial hypertrophic cardiomyopathy. J. Biol. Chem. 274:8806–8812. 10.1074/jbc.274.13.8806 [DOI] [PubMed] [Google Scholar]
- Yancy, C.W., Jessup M., Bozkurt B., Butler J., Casey D.E. Jr., Colvin M.M., Drazner M.H., Filippatos G.S., Fonarow G.C., Givertz M.M., et al. 2017. 2017 ACC/AHA/HFSA Focused Update of the 2013 ACCF/AHA Guideline for the Management of Heart Failure: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of America. Circulation. 136:e137–e161. 10.1161/CIR.0000000000000509 [DOI] [PubMed] [Google Scholar]
- Yang, Z., Stull J.T., Levine R.J., and Sweeney H.L.. 1998. Changes in interfilament spacing mimic the effects of myosin regulatory light chain phosphorylation in rabbit psoas fibers. J. Struct. Biol. 122:139–148. 10.1006/jsbi.1998.3979 [DOI] [PubMed] [Google Scholar]
- Yin, Z., Ren J., and Guo W.. 2015. Sarcomeric protein isoform transitions in cardiac muscle: a journey to heart failure. Biochim. Biophys. Acta. 1852:47–52. 10.1016/j.bbadis.2014.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]