Using super-resolution fluorescence microscopy and in silico simulations, Rahmanseresht et al. demonstrate that the N terminus of myosin-binding protein C (MyBP-C) tends to bind to actin filaments in both active and relaxed muscle. Binding to the myosin head also appears possible but only when the myosin head is near the actin filament.
Abstract
Myosin and actin filaments are highly organized within muscle sarcomeres. Myosin-binding protein C (MyBP-C) is a flexible, rod-like protein located within the C-zone of the sarcomere. The C-terminal domain of MyBP-C is tethered to the myosin filament backbone, and the N-terminal domains are postulated to interact with actin and/or the myosin head to modulate filament sliding. To define where the N-terminal domains of MyBP-C are localized in the sarcomere of active and relaxed mouse myocardium, the relative positions of the N terminus of MyBP-C and actin were imaged in fixed muscle samples using super-resolution fluorescence microscopy. The resolution of the imaging was enhanced by particle averaging. The images demonstrate that the position of the N terminus of MyBP-C is biased toward the actin filaments in both active and relaxed muscle preparations. Comparison of the experimental images with images generated in silico, accounting for known binding partner interactions, suggests that the N-terminal domains of MyBP-C may bind to actin and possibly the myosin head but only when the myosin head is in the proximity of an actin filament. These physiologically relevant images help define the molecular mechanism by which the N-terminal domains of MyBP-C may search for, and capture, molecular binding partners to tune cardiac contractility.
Introduction
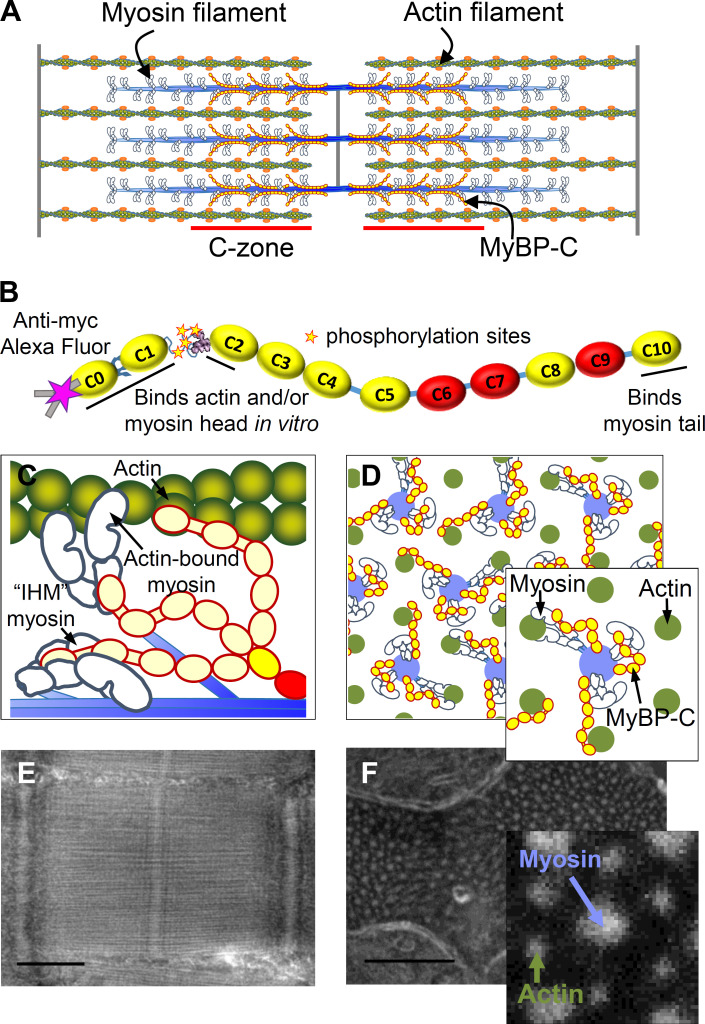
Cardiac contractility is powered by sarcomeres (Fig. 1 A), being the elemental contractile units within muscle cells. Within each sarcomere, myosin molecular motors are organized into thick filaments that bind to and slide actin thin filaments to shorten muscle length. Both sarcomere shortening and relengthening are modulated by the presence of myosin-binding protein C (MyBP-C), a flexible rod-like protein (Fig. 1 B) located within the C-zone of the sarcomere (Moss et al., 2015). The C-terminal C10 domain of MyBP-C is strongly bound to the thick filament backbone (Flashman et al., 2007). The N-terminal C0-C2 domains of MyBP-C bind to the myosin head (Ratti et al., 2011) and/or actin filaments (Mun et al., 2011; Razumova et al., 2006) in vitro. Such binding has been postulated to fine-tune cardiac contractility by modulating actomyosin activation and thin filament sliding velocities (Moss et al., 2015; Oakley et al., 2007; Pfuhl and Gautel, 2012; Previs et al., 2014). Proteolytic removal of the N-terminal (C0C1f) domains of MyBP-C in isolated thick filaments ablated the ability of MyBP-C to modulate actin filament sliding (Previs et al., 2012). The importance of MyBP-C to proper cardiac function is underscored by truncation mutations in the MYBPC3 gene (Alfares et al., 2015) that reduce MyBP-C content in human myocardium (O’Leary et al., 2019), being the leading cause of hypertrophic cardiomyopathy.
Figure 1.
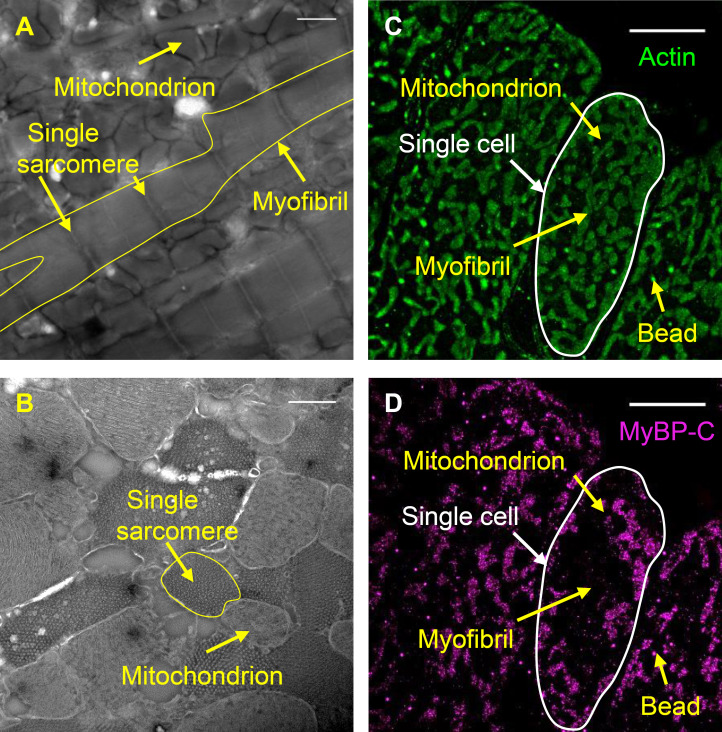
Longitudinal and cross-sectional organization of the cardiac muscle sarcomere. (A) Illustrative representation of myosin and actin, which form interdigitating filaments organized within a sarcomere. MyBP-C is bound within the ∼387-nm-long C-zones of the 1.57-µm bipolar myosin filaments (Zoghbi, et al., 2008). (B) Schematic representation of MyBP-C with 11 Ig-like (yellow) and Fn-like (red) domains. The location of the Myc-tag for Alexa Fluor (magenta star), two internal hinges, four phosphorylation sites, and the actin- and myosin-binding regions are underlined. (C) Illustrative representations of a longitudinal view of part of a single myosin thick filament showing potential interactions of MyBP-C with myosin and actin. (D) Illustrative representations of cross-sectional views of myosin and actin filaments organized within a hexagonal lattice. Note: each myosin filament (blue) is surrounded by six actin filaments (green). (E and F) Negative-stain EM images of longitudinal (E) and transverse (F) cryo-sections of mouse papillary muscle sarcomeres from an active and relaxed preparation, respectively. Note: MyBP-C is not visible in the micrograph. Scale bars, 500 nm (E) and 250 nm (F).
Based on the ability of the N-terminal domains of MyBP-C to bind both the myosin head and actin filaments in vitro (Mun et al., 2011; Ratti et al., 2011; Razumova et al., 2006), multiple models have been proposed to describe the mechanisms by which MyBP-C can modulate muscle contractility under active and relaxed conditions. Specifically, in active muscle, myosin heads are freed from the thick filament surface (Kampourakis et al., 2014) so they can bind to and slide the actin filament. The 43-nm contour length of MyBP-C (Hartzell and Sale, 1985; Previs et al., 2016) combined with in vitro data (Mun et al., 2011; Ratti et al., 2011; Razumova et al., 2006) allows for a model where the N-terminal domains extend away from the thick filament backbone and bind to the myosin head and/or the actin filaments (Fig. 1, C and D). Binding to the actin filament may help displace tropomyosin to activate thin filament sliding and then modulate sliding velocities by acting as an internal load (Kampourakis et al., 2014). In relaxed muscle, the two heads of each myosin molecule can adopt the interacting heads motif (IHM) and fold onto the myosin filament surface (Zoghbi et al., 2008; Al-Khayat et al., 2013) to prevent actomyosin interactions. Data demonstrate that MyBP-C regulates the super-relaxed (SRX) biochemical state of myosin in vitro (McNamara et al., 2016; Nelson et al., 2020), and this state has been attributed to the formation of the IHM. Due to the importance of the N-terminal domains of MyBP-C for regulating actomyosin interactions (Moss et al., 2015; Oakley et al., 2007; Pfuhl and Gautel, 2012; Previs et al., 2014), models have been proposed suggesting the N-terminal domains lie along the thick filament surface (Brunello et al., 2020; Nag et al., 2017; Sarkar et al., 2020; Trivedi et al., 2018) to stabilize the myosin heads in the IHM to account for the SRX state of myosin (Fig. 1, B and C).
The direct visualization of the N-terminal domains of MyBP-C within cardiac muscle is challenging for several reasons. Specifically, the contractile filament environment is crowded, with the distance between the centers of the thick and thin filaments being <30 nm (Matsubara and Millman, 1974; Millman, 1998). MyBP-C is a relatively thin, flexible protein (Hartzell and Sale, 1985; Previs et al., 2016), and the binding partner interactions with MyBP-C are likely transient (Weith et al., 2012). Despite these challenges, determining the localization of the N-terminal domains of MyBP-C is paramount to understanding the mechanisms by which this portion of MyBP-C modulates sarcomere contractility and relaxation.
To begin defining the orientation of the N-terminal domains of MyBP-C in situ, Luther et al. (2011) took advantage of the 3-D crystalline-like organization of myosin and actin filaments (Fig. 1 D) in traverse sections of relaxed frog skeletal muscle. This geometric organization allowed for the generation of an averaged 3-D electron tomogram of the simplest unit of the sarcomere, being a myosin-thick filament surrounded by six actin thin filaments (Fig. 1 D, inset). The tomogram demonstrated faint radial densities that spanned between the myosin and actin filaments within the C-zone of the sarcomere (Fig. 1 A). Luther et al. (2011) concluded this was direct evidence for MyBP-C binding to the actin filaments in relaxed muscle. However, recent models suggesting that the N-terminal domains of MyBP-C lie along the thick filament backbone (Fig. 1, C and D) to stabilize the myosin heads in the IHM (Brunello et al., 2020; Nag et al., 2017; Sarkar et al., 2020; Trivedi et al., 2018) are not supported by this conclusion. If the N-terminal domains of MyBP-C stabilize the IHM, they should appear close to the myosin filament surface in traverse sections of muscle (Fig. 1, C and D). However, it is possible that such density was lost in the average electron tomogram.
To enhance the localization of the N terminus of MyBP-C in situ, transgenic mice were generated that express MyBP-C with a myc-tag on its N terminus, and hearts from these mice were fixed under active and relaxed contractile conditions. Papillary muscle from these hearts was fixed and cryopreserved, and thin transverse sections were cut. Fluorescent probes were attached to actin and the myc-tag on the N terminus of MyBP-C. The fluorophores were visualized using stochastic optical reconstruction microscopy (STORM). The 25-nm localization precision of STORM (Bates et al., 2007; Schermelleh et al., 2010) was improved by taking advantage of the crystalline-like organization of the myosin and actin filaments to apply particle averaging. The approach was advantageous for visualizing the position of the N terminus of MyBP-C because the fluorophore provided a high intensity signal for localization. The experimental STORM images suggest that the position of the N terminus of MyBP-C is disordered but biased toward the actin filaments in situ under both active and relaxed contractile conditions. Quantitative comparison of the STORM images to those generated in silico, with defined binding partner interactions, demonstrated that the N-terminal domains appear capable of binding to actin and/or the myosin head (in the latter case, only when the myosin head is in proximity to the actin filaments). These data suggest that the N-terminal domains of the MyBP-C molecule may not lie along the surface of the myosin filament under active or relaxed contractile conditions. These findings may limit the mechanistic models by which the N-terminal domains of MyBP-C may be operative.
Materials and methods
Mice
All protocols complied with the Guide for the Use and Care of Laboratory Animals published by the National Institutes of Health and were approved by the institutional animal care and use committees at University of Vermont Medical School and Cincinnati Children’s Hospital Medical Center. MyBP-Ct/t mice were backcrossed for more than six generations in the FVB/N background (McConnell et al., 1999). These mice were bred with a mouse that expresses the normal cardiac MyBP-C isoform with a sequence encoding the myc epitope incorporated between the initiator methionine residue and first amino acid of the C0 domain (Yang et al., 1998). The phenotype, ultrastructure, and localization of MyBP-C within the C-zone of cardiac muscle sarcomeres from these mice were demonstrated to be indistinguishable from those of nontransgenic mouse lines (Sadayappan et al., 2005).
Quantification of MyBP-C phosphorylation
An ≈4-mg piece of mouse cardiac muscle was removed from the exterior of a mouse heart before infusion with the fixative. The muscle was cut into multiple pieces. Two pieces were digested in solution as described (O’Leary et al., 2019), and the remaining two pieces were dissolved in SDS-PAGE sample prep buffer (Bio-Rad) and the proteins were separated on a 12% SDS-PAGE gel (Invitrogen). The gel was stained with SimplyBlue Safe Stain and destained with water. The bands corresponding to MyBP-C were excised from the gel, placed in a 1.5-ml microfuge tube, minced with forceps, and then destained with a wash in 50% acetonitrile in distilled water (22°C, 5 min). The gel pieces were then dehydrated in two successive washes of 100% acetonitrile (22°C, 5 min each) and dried completely in a speed vacuum device. A 40-µl aliquot of Milli-Q water was added to each tube along with 5 µl 10X NEBuffer for Protein MetalloPhosphatases (New England Biolabs) and 5 µl of 10 mM MnCl2. A 5-µl aliquot of λ-phosphatase (New England Biolabs) was added to half of the tubes to dephosphorylate the MyBP-C, and 5 µl of Milli-Q water was added to the other tubes. The tubes were heated (30°C, 1 h), and then 10 µl of alkaline-phosphatase (Sigma) was added to the dephosphorylated sample and 10 µl of Milli-Q water was added to the control sample. The tubes were heated (37°C, 1 h). Following incubation, the samples were dried in a speed vacuum device. The samples were reduced by adding 100 µl of 10 mM dithiothreitol (DTT) and heating (50°C, 45 min) and then alkylated with 100 µl of 55 mM iodoacetamide in 50 mM ammonium bicarbonate with incubation in the dark (22°C, 30 min). The excess solution was discarded, and the remaining gel pieces were washed in three consecutive 400-µl rinses of 50% acetonitrile in distilled water while on a shaker (22°C, 15 min each). The gel pieces were dried completely in a speed vacuum device.
The proteins were digested into tryptic peptides with the addition of 100 µl of 0.02 µg/µl trypsin (Promega). The samples were cooled (4°C, 30 min) to aid in the trypsin absorption and then incubated (37°C, 18 h). The gel pieces were dried in a speed vacuum device. A 100-µl aliquot of 7% formic acid in 50 mM ammonium bicarbonate was added to deactivate the trypsin (22°C, 15 min). The excess liquid was removed from the tube and placed into a fresh labeled microcentrifuge tube. This solution contained the initial extract of MyBP-C peptides. Additional peptides were extracted from the gel pieces with three sequential dehydrating washes in 50% acetonitrile in distilled water (22°C, 5 min each). After each wash, the excess liquid was removed from the tube and added to the labeled microcentrifuge tube. The resultant peptides were dried down and reconstituted for a final time in 40 µl of 0.1% trifluoroacetic acid. The tubes were then centrifuged at 14,000 rpm for 5 min to pellet the surfactant. The top 35 µl of solution was transferred into a mass spectrometry (MS) analysis vial.
For both the in-solution and in-gel preparations, the peptides were separated by liquid chromatography (LC), and their abundances were measured by MS. A 20-µl aliquot of each sample was injected onto an Acquity UPLC HSS T3 column (100 Å, 1.8 µm, 1 × 150 mm; Waters Corporation) attached to an UltiMate 3000 ultra-high–pressure LC system (Dionex). The ultra-high–pressure LC effluent was directly infused into a Q Exactive Hybrid Quadrupole-Orbitrap mass spectrometer through an electrospray ionization source (Thermo Fisher Scientific). Data were collected in data-dependent MS/MS mode with the top five most abundant ions being selected for fragmentation.
The resultant RAW files containing the mass spectra were processed using the SEQUEST algorithm in the Proteome Discoverer 2.2 software package (Thermo Fisher Scientific). The MS/MS spectra were matched to theoretical spectra generated using the human proteome database (downloaded from UniProt). Variable mass changes accounting for the loss of methionine from the N terminus of each protein with the addition of acetylation (−89.16 D), the carbamidomethylation of cysteine (+57.02 D), oxidation of methionine and proline (+15.99 D and +32.00 D, respectively), and phosphorylation of serine, threonine, and tyrosine (+79.98 D) were added to the search parameters. The area of the LC-MS peak for each peptide detected in the MS/MS spectra was determined using Proteome Discoverer 2.2. The Minora Feature Detector was enabled to allow for quantification of LC peak areas for peaks lacking MS/MS spectra in a RAW file but having the same exact mass, charge state, elution time, and isotope pattern as those detected in other RAW files.
The relative abundance of α-myosin heavy chain to MyBP-C was determined from the abundance of the top three ionizing peptides from each protein in the in-solution preparations. The degree of phosphorylation of MyBP-C at serines 273, 282, 302, and 307 in the non–phosphatase-treated gel samples was determined from the phosphatase-treated gel samples (which contained no phosphorylated MyBP-C peptides) using a mass-balance approach as previously described for wild-type FVB mice (Previs et al., 2012).
Single ATP turnover measurements
The SRX state of myosin was measured using permeabilized multicellular ventricular preparations as previously described (McNamara et al., 2016; McNamara et al., 2019). Briefly, a small piece of left ventricular myocardium was permeabilized on ice for 6 h using skinning buffer (100 mM NaCl, 8 mM MgCl2, 5 mM EGTA, 5 mM K2HPO4, 5 mM KH2PO4, 3 mM NaN3, 5 mM ATP, 1 mM DTT, 20 mM 2,3-Butanedione monoxime (BDM), and 0.1% [vol/vol] Triton X-100, pH 7) with solution changes every 2 h. Next, the tissue was glycerinated overnight (∼17 h) on ice in glycerinating buffer (120 mM K-acetate, 5 mM Mg-acetate, 5 mM EGTA, 2.5 mM K2HPO4, 2.5 mM KH2PO4, 50 mM MOPS, 5 mM ATP, 20 mM BDM, 2 mM DTT, and 50% [vol/vol] glycerol, pH 6.8). The following morning, the glycerinating buffer was refreshed and the samples were stored at −20°C for up to 5 d before use.
On the day of the experiment, thin strips of skinned ventricular muscle were dissected in ice-cold glycerinating buffer and loaded into flow chambers constructed using two layers of double-sided tape sandwiched between a coverslip and glass slide. Flow cells were kept on ice until imaged. Prior to imaging, cells were extensively washed with rigor buffer (120 mM K-acetate, 5 mM Mg-acetate, 5 mM EGTA, 2.5 mM K2HPO4, 2.5 mM KH2PO4, 50 mM MOPS, and 2 mM fresh DTT, pH 6.8) to remove ATP, BDM, and glycerol. Rigor buffer containing 250 µM 2′/3′-O-(N-methyl-anthraniloyl)-adenosine-5′-triphosphate (mant-ATP; Sapphire Biosciences; NU-202S) was flowed into the flow chamber. Fibers were located using bright-field illumination with a Plan Apo 20× objective on a Nikon 90i upright microscope. Fluorescence images (excitation 360 nm, emission 460 nm) were acquired every 5 s using a Photometrics CoolSNAP HQ2 camera. After ∼60 s of imaging, the flow cell was perfused with rigor buffer that contained 4 mM ATP, and imaging continued for at least 6 min.
Fluorescence intensity was measured over the course of the acquired images by subtracting background fluorescence from the fiber fluorescence using multiple regions of interest (ROIs) for each. The fluorescence intensity for each time point was normalized to the intensity measured immediately before ATP perfusion. Each trace was exported to GraphPad Prism, and the following double exponential equation was used to fit the data using nonlinear regression analysis: I = 1 − P1 [1 − e(−t/T1)] − P2 [1 − e(−t/T2)]. In this equation, I denotes the fluorescence intensity at a given time, t. The first exponential describes the initial rapid decay in fluorescence intensity proposed to result from nucleotide exchange by myosin in the disordered relaxed state and diffusion of unbound mant-ATP out of the fiber. The second exponential is proposed to result from the slow nucleotide turnover by myosin in the SRX state. P represents the proportion of fluorescence intensity attributed to its given exponent, while T is the lifetime of nucleotide turnover.
Intact muscle preparation for EM and STORM imaging
Mice were anesthetized with isoflurane and euthanized by cervical dislocation. The chest cavity was opened, and a needle was inserted through the apex into the left ventricle. For the active muscle preparations, fixative solution (3% paraformaldehyde and 0.1% glutaraldehyde in PBS) was infused via a syringe pump at 0.8 ml/min for 1 h. Beating ceased within the first 10 min of infusion. The heart was then infused with PBS for 30 min to wash out excess fixative solution. For the relaxed muscle preparations, the heart was first infused with 10 µM blebbistatin (Thermo Fisher Scientific) in PBS for 30 min. During this infusion, the syringe was wrapped in aluminum foil to block ambient light and prevent degradation of the myosin inhibitor. The heart was then infused for 1 h with a solution containing 10 µM blebbistatin in 3% paraformaldehyde and 0.1% glutaraldehyde in PBS. This was followed by a 30-min infusion of PBS to remove the fixative solution.
The whole hearts were placed in PBS and shipped on ice overnight from the University of Vermont to the University of Massachusetts Medical School for dissection and cryo-sectioning. The papillary muscle was dissected from the heart, fixed with 3% paraformaldehyde and 0.1% glutaraldehyde in PBS buffer (pH 7.4), infiltrated with 2.3 M sucrose solution as a cryo-protectant to prevent ice crystal formation, and flash frozen by immersion in liquid nitrogen (Tokuyasu, 1973). Both 100- and 400-nm thick longitudinal and transverse cryo-sections were cut on a cryo-ultramicrotome (Leica EM UC7 with FC7 cryo-chamber attachment) at −120°C and placed on 35-mm glass-bottom culture dishes (MatTek Corporation; MatTek 10 mm Microwell) at room temperature for STORM imaging.
Negative stain transmission EM
Longitudinal and transverse cryo-sections of the muscle (100 nm thick) were placed on EM grids at room temperature for negative staining. The sections were stained with 0.5% ammonium molybdate or 1% uranyl acetate. Images were obtained on a Tecnai G2 spirit transmission EM (FEI) at 120 kV using a Gatan Erlangshen charge-coupled device (CCD) camera (Gatan Inc.).
Measurement of filament distances in EM images
The distance between two neighboring actin filaments along the vertices of actin hexagons was measured from negative-stained EM images of transverse muscle cryo-sections. The distance measurements were performed in ImageJ (Schneider et al., 2012) by drawing straight lines between the centers of adjacent actin filaments using the line tool. The length of the line was recorded in nanometers. Multiple measurements were made, and the average (±SD) was reported as the interactin filament distance. The same method was used to measure the interfilament distance between the centers of actin and myosin filaments in transverse muscle sections. The length of sarcomeres in longitudinal sections was measured by drawing a line connecting the center of two consecutive Z-bands. The length of the line was recorded in micrometers. Multiple measurements were made, and the average (±SD) was reported as the length of the sarcomere.
Immunostaining for STORM imaging
Cross sections of the muscle (400 nm thick) on the glass coverslip of a MatTek dish with PBS buffer were shipped to University of Vermont overnight at 0°C. To fluorescently label actin filaments and the N terminus of MyBP-C, the free aldehyde groups of the fixatives (3% paraformaldehyde and 0.1% glutaraldehyde in PBS) were blocked by incubating the section first with 0.1% sodium borohydride (Sigma) and then 0.1% glycine (Sigma) in PBS (5 and 10 min each, respectively). After the sections were washed with PBS several times, they were incubated for 1 h with 1% BSA (Sigma) in PBS to prevent nonspecific binding. Next, the sections were incubated with 1 µM mouse monoclonal anti-Myc-Alexa Fluor 647 (or 488) antibody (Cell Signaling) for 1 h and then fixed with 3% paraformaldehyde (Sigma) and 0.1% glutaraldehyde (Sigma) in PBS. The fixed sections were incubated with sodium borohydride (0.1%) and glycine (0.1%) in PBS for 5 min. The sections were washed with PBS, and the actin filaments were labeled with 100 nM Alexa Fluor 488 (or 647) phalloidin (Invitrogen) for 1 h. The Alexa Fluor dye was preconjugated to the phalloidin. Sections were washed with PBS several times and fixed with 3% paraformaldehyde and 0.1% glutaraldehyde. Sections were treated a final time with sodium borohydride (0.1%) and glycine (0.1%) in PBS (5 min) and then washed with PBS. Multicolor fluorescent beads (Invitrogen; 100 nm, Tetraspeck) were added to sections and incubated for 5 min. The fluorescent beads served as fiducial markers for drift correction and image registration between image stacks. After the sections were washed with PBS multiple times, 1 ml of the STORM buffer containing 0.7 mg/ml glucose oxidase (Sigma), 40 µg/ml catalase (Sigma), 20 mM Cysteamine (Sigma), and 50 mM 2-Mercaptoethanolc (Sigma) in PBS, pH 7.4, was added to the MatTek dish for STORM imaging.
STORM imaging
Two-color STORM movies were acquired using a Nikon N-STORM system consisting of a Nikon Ti-E TIRF inverted microscope with a 100× 1.49 NA oil immersion lens. The N terminus of MyBP-C was labeled with anti-Myc-antibody Alexa Fluor 647 (or 488), and actin filaments were labeled with Alexa Fluor 488 (or 647) phalloidin. In all experiments, the Alexa Fluor 647 was excited first with the 647-nm laser light to initiate stochastic blinking, and 80,000 images were collected at 50 Hz using an Andor iXON3 DU897 electron multiplying CCD camera. Next, the Alexa Fluor 488 was excited with the 488-nm laser light to initiate stochastic blinking, and 80,000 images were collected at 50 Hz using the electron multiplying CCD camera.
Rationale for selection of fluorophores
Two-color STORM is challenging because the blinking rates of both fluorophores and photon emission need to be optimized to achieve high localization precision (Tam et al., 2014). Alexa Fluor 647 demonstrates optimal blinking, meaning it spends a long time in the dark state, and produces high photon counts under a range of buffer conditions (Dempsey et al., 2011). Thus, Alexa Fluor 647 was selected for the analyses. Alexa Fluor 568, 555, and 488 were empirically tested using multiple combinations of 2-aminoethanethiol and β-mercaptoethanol in the imaging buffer. Samples were tested with and without paraformaldehyde and glutaraldehyde fixation and sodium borohydride and glycine treatment. The blinking rate of neither Alexa Fluor 568 nor 555 could be optimized in samples colabeled with Alexa Fluor 647 under any of the conditions tested. Alexa Fluor 488 and 647 were the only pair to demonstrate optimal blinking rates and photon counts for the analyses. This combination has been used by others (Zessin et al., 2013). The photon count of Alexa Fluor 488 (∼1,200) was approximately half that of Alexa Fluor 647 (∼2,500), but this was sufficient to achieve <13 nm of localization uncertainty.
STORM localization, drift correction, localization averaging, and image reconstruction
Low-resolution image reconstructions of whole fields of view of muscle cells were generated using Nikon N-STORM software to locate sarcomeres. High-resolution image reconstructions of single sarcomeres were generated using a custom, semiautomated analysis routine. In this custom routine, single molecule localizations were identified by fitting a 2-D Gaussian function to fluorescent spots in the individual STORM images using the open source package RapidSTORM (Wolter et al., 2012). The localization file created by RapidSTORM contains the x and y positions of the centroid of each fluorescence spot and their intensity and width in each frame.
A threshold of 500 photons was used to reject spots that were too dim. Fluorescence spots with point spread functions (PSFs) >400 nm were rejected because they could be from multiple molecules (Schermelleh et al., 2010). Buffer conditions were empirically adjusted during initial experiments (see above), and the excitation laser power was optimized during each experiment to ensure the majority of fluorophores were in the dark state and there was good separation between blinking molecules in each acquisition frame. These optimizations were performed to limit the possibility of overlap between the PSFs of two adjacent molecules in any given frame. Such overlap is a limitation of STORM in densely labeled samples (van de Linde et al., 2010).
Correction of instrumental drift in the x and y positions of the fluorescence spots was applied using a custom software package provided by the Heilemann group (Goethe-Universität, Frankfurt, Germany). This package uses the coordinates of the multicolor fluorescent beads that are found in a large number of frames and affine matrices to correct for the drift. Image registration was performed using coordinates of beads that were present in both color channels and the LocAlization Microscopy Analyzer package (Malkusch and Heilemann, 2016).
Only fluorescence spots that appeared in consecutive frames (two or more frames) and had a displacement of <1 pixel (160 nm) were considered a single fluorophore. To improve the localization precision of a single fluorophore, only the average of the centroid position of spots appearing in two or more consecutive frames was used as the final localization. This average was weighted by the number of detected photons for each spot in the consecutive frames, with the final intensity for each averaged spot being the sum of intensities (detected photons) in each frame (Bates et al., 2007). Finally, only averaged fluorescence spots with an intensity of >1,000 photons and an SD of <25 nm were kept using a custom-written Python code. These selection criteria ultimately served as additional filters to help ensure that each fluorescence spot belonged to a single fluorophore (Alexa Fluor 647 or 488) being on one actin filament or one MyBP-C molecule.
Image reconstructions were generated using a custom-written Python code to render each localization with a normalized 2-D Gaussian distribution. The average x and y position of each fluorophore was used as the center of each Gaussian, and the width of each Gaussian was calculated from the theoretical localization accuracy, which is dependent on the number of photons detected in each spot (Betzig et al., 2006). This results in the spots with the highest localization accuracy in the reconstructed images being brighter and sharper. The average localization accuracy in the reconstructed images was 12 nm for Alexa Fluor 647 and 13 nm for Alexa Fluor 488.
STORM imaging of DNA origami nanorulers
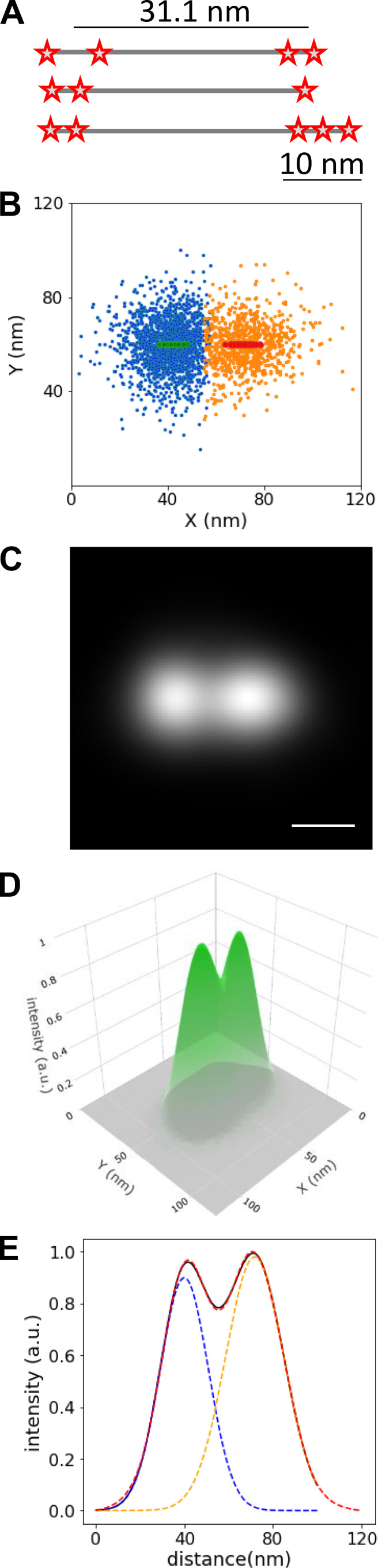
DNA origami nanorulers (GATTAquant DNA Nanotechnologies, GmbH) were used to test the resolution of our microscope and image analysis methods. The nanorulers are fluorescently labeled DNA origami scaffolds with two fluorescent marks, containing three to four Alexa Fluor 647 molecules spaced 31.1 nm between their centers (see Fig. S4 A). The nanorulers were attached to a coverslip using BSA–biotin-neutravidin chemistry (protocol provided by the manufacturer). Multicolor fluorescent beads were added as fiducial markers for drift correction. STORM buffer was added, and movies were acquired using a laser excitation of 647 nm. A total of 10,000 images were collected at 50 Hz. STORM localization and drift correction were performed using the custom routine (see above).
Figure S4.
Geometry and reconstructed images of DNA origami scaffolds used to evaluate the resolution of the STORM analyses. (A) Illustrative representation of Alexa Fluor 647–labeled DNA origami scaffolds (i.e., nanorulers) having 3–4 Alexa Fluor 647 molecules spaced at 6-nm intervals, with their centers being separated by 31.1 nm. (B) Orange and blue points (n = 2,671) are the average centroid position of stochastic signals that persisted for two or more consecutive frames when imaging the nanorulers. Red and green symbols are the centroid positions determined for each half of 68 different nanorulers by STORM analysis. (C) The reconstructed image for the orange and blue points shown in B, rendered with Gaussian distributions representing each point’s localization uncertainty. This image demonstrates the average positions of the Alexa Fluor 647 molecules on each side of the 68 nanorulers. Scale bar, 25 nm. (D) 3-D plot of the average reconstructed image shown in C, with the z axis being intensity, representing localization uncertainty. (E) The intensity profile along the x axis of the average reconstructed image (black), fitted with sum (red dashed line) of two single Gaussian (blue and orange dashed lines) functions. The distance between the centers of the dual Gaussians was 32.0 ± 1 nm. a.u., arbitrary unit.
To identify DNA origami nanorulers in the image, a custom Python code was written. The super-resolution image reconstruction (25 nm/pixel) for nanorulers was generated using a 2-D histogram of the localization points. To identify the bright features in the image, a binary image was created by thresholding. The Python image processing toolbox (Skimage, morphology and measure modules) was used to find the position of bright features in the image. From the detected bright features, those with an area <16 pixel2 were rejected. For the remaining features, the number of localizations within a 4 × 4-pixel box around the centroid of each feature was counted; features with <30 points or >5,000 points (possibly beads) were rejected, and the rest were kept as ROIs that possibly include a nanoruler. For each ROI, localizations that persisted for two or more frames and were closer than 1 pixel were considered to belong to the same fluorophore and were averaged. The average localization precision measured from the uncertainty on these averages was 7 nm. Meanshift clustering algorithm (Cluster module in the Python sklearn library) was applied to the average localizations within each ROI to identify clusters of localizations. ROIs that included two clusters with a centroid distance between 25 and 35 nm (close to the expected value of 31.1 nm) were kept as nanorulers. All detected nanorulers were aligned by defining the long axis of the ruler as the new x axis and transforming the localizations to the new x and y coordinate system. Next, the aligned nanorulers were overlaid (see Fig. S4 B). To reconstruct the average image from all nanorulers, all localizations were rendered with a 2-D Gaussian distribution (see Fig. S4, C and D). A fixed uncertainty (σ) of 7 nm was used for the Gaussian functions. To measure the distance between the two sides of the nanoruler, the intensity profile along the x axis of the nanoruler was fitted with a mixture of two Gaussian functions, and the distance between the mean values for the peaks was measured as the length of the nanoruler (see Fig. S4 E).
Defining the best templates for template-matching with the actin images
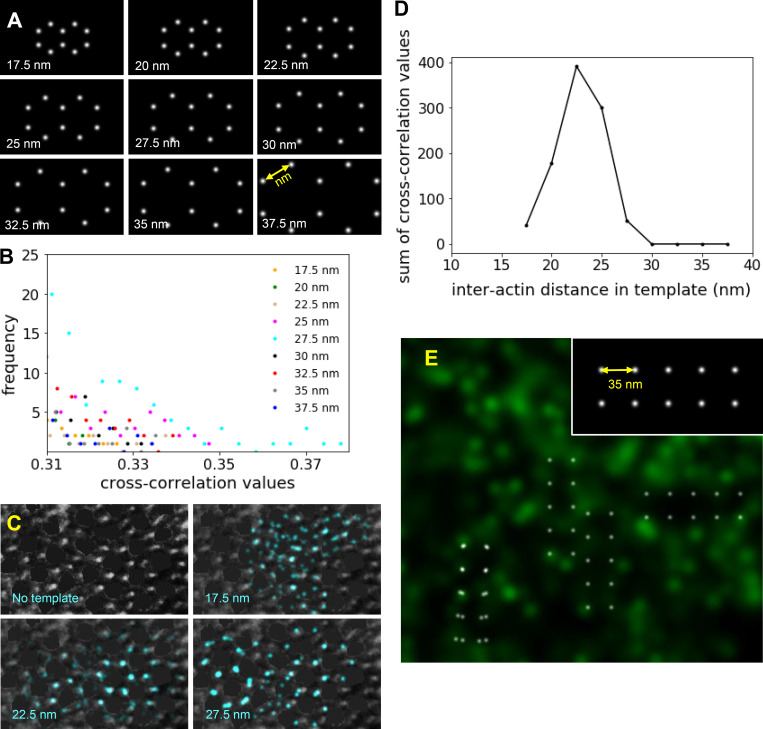
The optimal template size for template-matching was identified by cross-correlating templates, composed of two identical conjoined hexagons (two-actin hexagons) of variable sizes, with the reconstructed images of single myofibrils. The interactin filament spacing along the vertices of the hexagon varied from 17.5 to 37.5 nm (see Fig. S3 A) to encompass the interactin filament spacing measured in EM images. Each template was rastered along STORM images on a pixel-by-pixel basis and rotated at 5° intervals (from 0° to 180°) in each location. Therefore, for each template, 36 rotational orientations were rastered across the image and cross-correlated on a pixel-by-pixel basis. Cross-correlation values between the templates of each size and reconstructed images of each myofibril were calculated, and their frequency distribution was generated (see Fig. S3 C). The highest cross-correlation values represent the best matches; thus, only matches with correlation values higher than a specific threshold were kept. This threshold was defined based on the maximum cross-correlation value and uncertainty (σ) of the distribution where threshold = maximum cross correlation − σ. A custom Python code was implemented for these analyses. The cross-correlation values higher than this threshold were summed for each template size for 45 different myofibrils, and their frequency distribution was plotted (see Fig. 2 F).
Figure S3.
Identification of myosin and actin lattice and quantification of interactin spacing in actin STORM and EM images. (A) Templates containing two identical adjoining hexagons with different interactin spacing (17.5–37.5 nm) used in template matching by cross-correlation analysis. (B) The tail of the distribution of cross-correlation values indicative of cross correlations with the highest values. Such values were summed from analysis of 45 sarcomeres and plotted to determine the best fits (Fig. 2 F). (C) The highest cross-correlation values from the cross correlation of a negatively stained EM image of transverse papillary muscle with 17.5-, 22.5-, and 27.5-nm hexagonal templates. The intensities from the thick filaments were removed from the image using the brush tool in ImageJ (Schneider et al., 2012) to mimic the STORM images. The template matches with 22.5-nm interactin distances (blue points) overlapped with one another, highlighting the region of the sarcomere with particularly good preservation of the myosin and actin filament lattice. (D) The sum of greatest cross-correlation values resulting from cross correlation of the two-hexagonal templates of various sizes (range, 17.5–37.5 nm) with the EM images demonstrated a maximum for the template with 22.5-nm interactin spacing. (E) The highest cross-correlation values between an actin STORM image and a four-square template with a side length of 35 nm (inset) did not overlap with one another, as shown in Fig. 2 F for the two-hexagonal template.
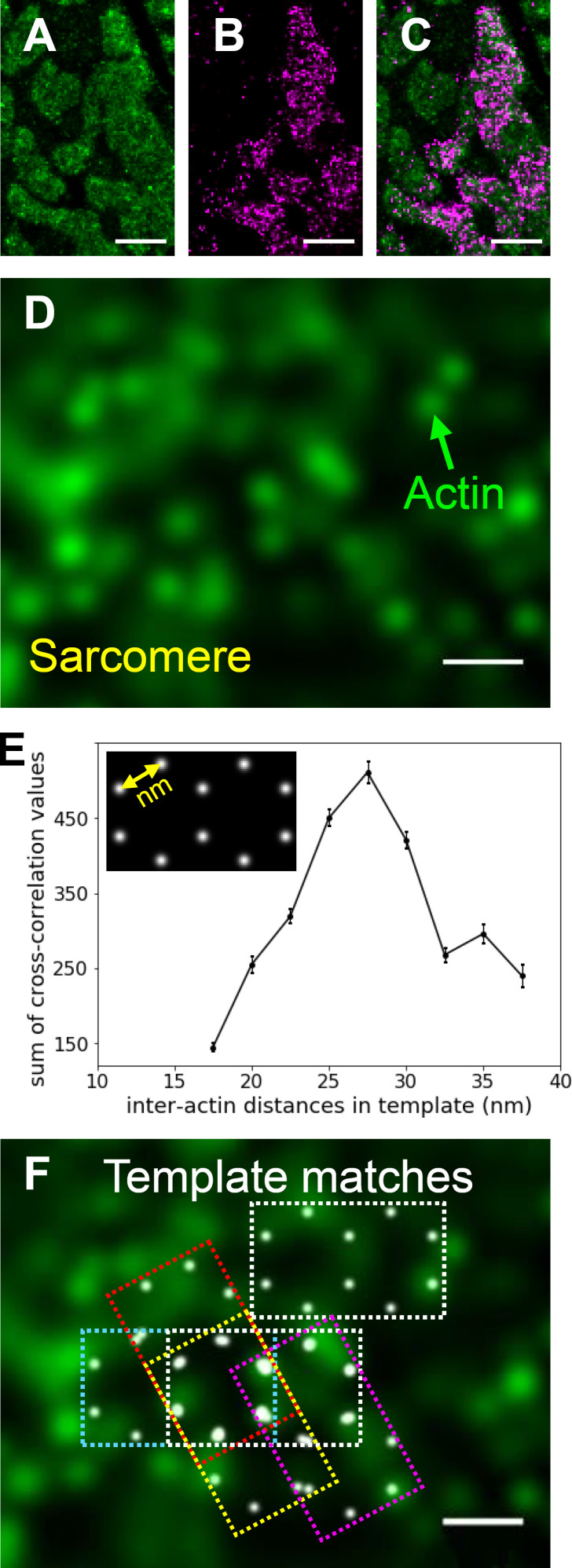
Figure 2.
Visualization of actin and MyBP-C within individual sarcomeres by STORM and detection of the actin hexagonal lattice. (A and B) Reconstructed images (see Materials and methods) of Alexa Fluor 488-phalloidin (actin filaments; A) and Alexa Fluor 647 anti-myc (MyBP-C’s C0 domain; B) within 400-nm transverse cryo-sections of mouse papillary muscle imaged by STORM. Images show part of one cell. Fluorescently labeled polygonal structures within the cell are myofibrils, and dark spaces are mostly mitochondria (Fig. S1). Scale bar, 1 µm. (C) Merged reconstructed images in A and B. Scale bar, 1 µm. (D) Individual actin filaments (green) within a single sarcomere. Scale bar, 50 nm. (E) The sum of greatest cross-correlation values resulting from cross correlation of fluorescence images with templates containing two identical conjoined hexagons (inset) of various sizes (range, 17.5–37.5 nm, n = 45 myofibrils). (F) Only templates with 25–30-nm interactin distances (white points in dashed rectangles) overlapped with one another within each sarcomere, suggesting that these were regions of particularly good preservation of the actin filament lattice. Scale bar, 50 nm.
As a positive control, an identical protocol was performed on the negatively stained EM images of the traverse sections of the muscle (see Fig. S3 C). The thick filaments were manually omitted from the images using the brush tool in ImageJ (Tokuyasu, 1973). As a negative control, a template composed of four conjoined squares (four-square template) was cross-correlated with the sarcomere images using the same routine discussed above. The side length for each square was 35 nm (see Fig. S3 E).
Actin image analysis by template matching
Based on the initial cross-correlation analyses using two identical conjoined hexagons with interactin spacing ranging from 17.5 to 37.5 nm (previous section), only the best-fitting templates were used (those with 25-, 27.5-, and 30-nm interactin spacing) to identify areas of the STORM images with good preservation of the actin hexagonal lattice in the remainder of the analyses. An identical routine to that described above was performed for all myofibrils within the actin fluorescence STORM images.
Particle selection and averaging
From each sarcomere, two-actin hexagons were picked from the area of the sarcomere where multiple hexagons overlapped, being indicative of an area with good preservation of the myosin and actin lattice. This approach ensured that the hexagon selected did not represent random signal, as could be the case by application of a single hexagonal template and lack of observation of lattice spacing. Next, the two-hexagon selection was rotated around its center to place its long axis along the x axis (angle = 0°). This put all the hexagons from different sarcomeres in the same orientation. One hexagon (particle) with at least three fluorescent spots was selected by cropping a 90 × 90-pixel box around one of the actin hexagons (n = 120) using a custom Python code. The center of the 90 × 90 box was at the center of the hexagon, being the center of the myosin thick filament. An average reconstructed actin image was generated by summing the cropped actin images using the ImageJ Z-projection tool (Schneider et al., 2012).
ROIs identical to the actin ROIs were cropped from the images reconstructed from the MyBP-C color channel. The MyBP-C ROIs were rotated with the same angle as the actin ROIs, and an identical 90 × 90-pixel box was cropped from the MyBP-C image. An average reconstructed MyBP-C image was generated by summing the cropped MyBP-C images using the ImageJ Z-projection tool (Schneider et al., 2012). The 3-D plots for the average actin and MyBP-C images were generated using the Python graphing library Plotly.
Measurement of interactin filament distances from the average actin reconstruction image and estimation of accuracy
The interactin distance for actins along the vertices of the hexagon in the average reconstructed image for actin was calculated from an average intensity profile. This was generated by averaging six line scans between every two neighboring actin filaments. The average intensity profile was fitted with the sum of two Gaussians using a least-square method (Python lmfit library), as demonstrated in Fig. S4 E. The distance between the center of two adjacent actins was calculated from the distance between the center of the Gaussian peaks, and the uncertainty of this distance represents the accuracy of the measurement. The uncertainty of the distance was calculated from the SD in the location of each Gaussian peak
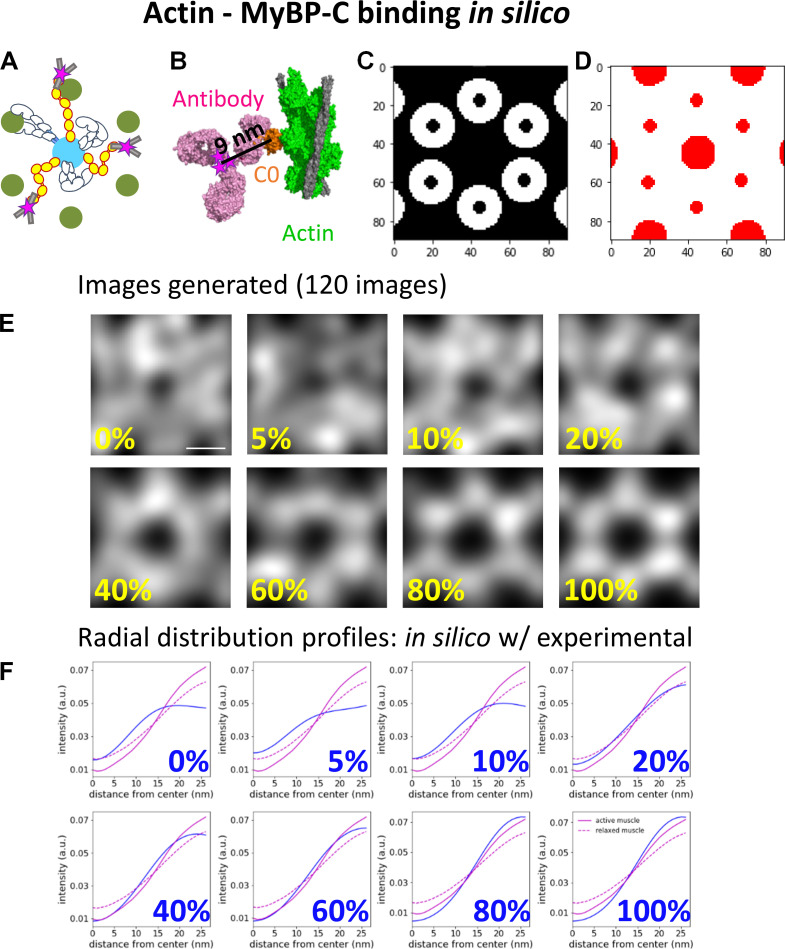
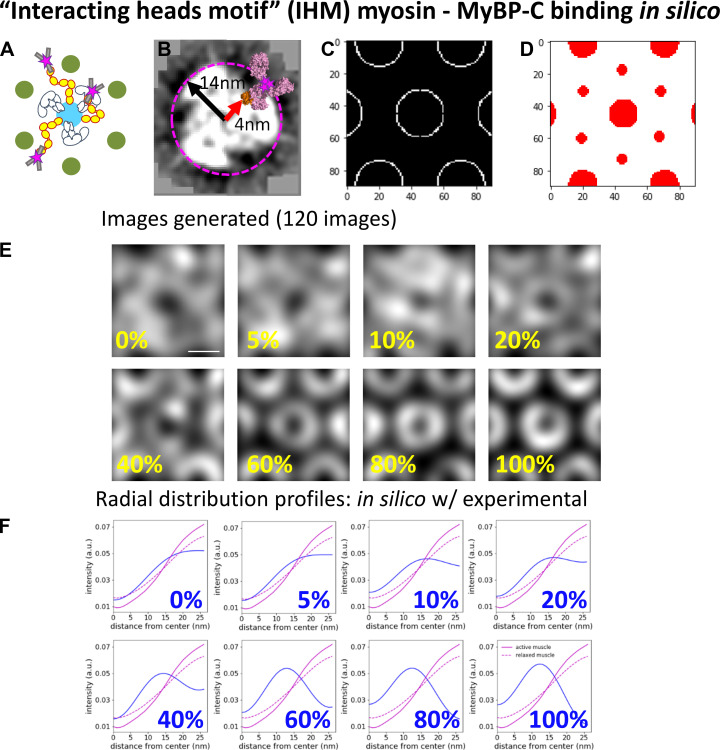
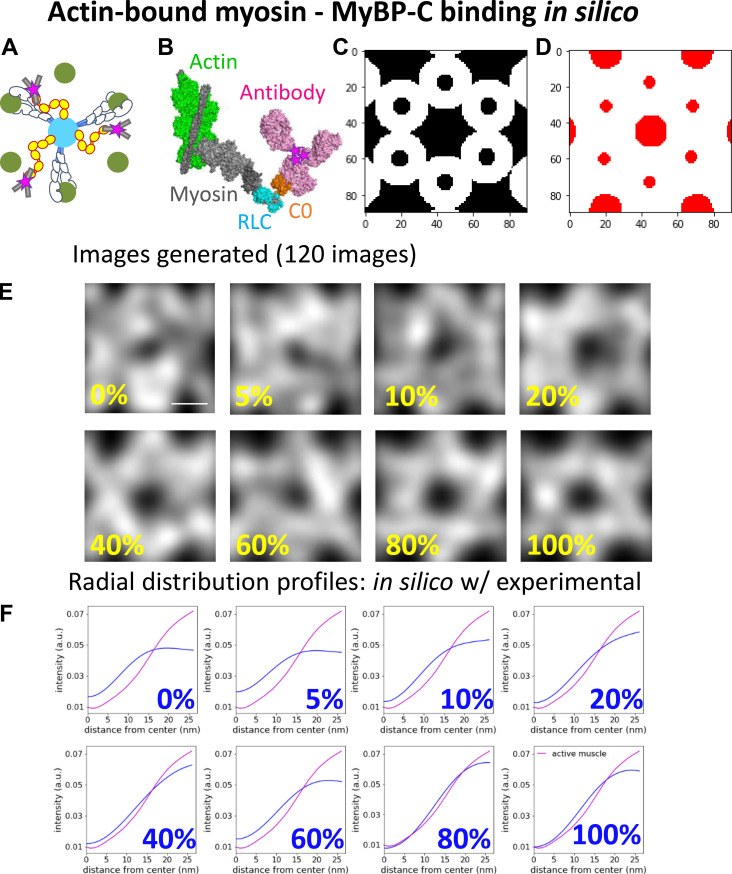
Generation of in silico fluorescence images
In silico fluorescence images were generated to define the localization of MyBP-C N termini within the actin hexagon when the C0 domain of MyBP-C was bound to actin or the myosin head under varied contractile states. Three binding scenarios were considered for the position of the N-terminal C0 domain of MyBP-C relative to its potential binding partners. These included the C0 domain being bound to (1) actin regardless of the contractile state of the muscle (see Fig. S7), (2) the regulatory light chain (RLC) region of the myosin head, when the myosin head adopts the IHM and lays along the myosin filament surface (see Fig. S8), and (3) the RLC of myosin, when the myosin heads are extended away from the myosin filament and bound to the actin filament (see Fig. S9). For each of the binding partner scenarios, the C0 domain was either bound to its specified partner or fixed randomly in locations in the space between the myosin and actin filament surfaces.
Figure S7.
Comparison of the radial intensity profile of MyBP-C fluorescence from the center of the actin hexagon in active and relaxed sarcomeres to in silico models with MyBP-C bound to actin. (A) Illustrative representations of MyBP-C bound to actin. (B) The high-resolution cryo-EM structure of MyBP-C’s C0 domain (orange) bound to a cardiac thin filament (green; PDB accession no. 6CXI). The binding domain of an antibody (magenta; PDB accession no. 1IGT) was placed in close proximity to the C0 domain, and the fluorophores (represented as magenta stars) were added to the cysteines presumed to be accessible for direct conjugation of the Alexa Fluor dyes. Six fluorophores were expected to be bound to a single antibody. (C) White areas are target zones for random placement of the fluorophores for when MyBP-C is bound to the actin, as shown in A, for in silico models. (D) Red areas are regions of the image in which fluorophores cannot be randomly placed due to steric exclusion from the myosin and actin filaments. (E) Average in silico reconstructed images generated with a fraction of the fluorophores bound to actin and the remaining fluorophores randomly distributed throughout the image except for the red areas shown in C. Each image is an average of 120 individual simulations. Scale bar, 25 nm. (F) The radial intensity profiles for the position of MyBP-C relative to the center of the average actin hexagon in the active (solid magenta) and relaxed (dotted magenta) sarcomeres and in silico images shown in E (blue); cf. Fig. 4 C. a.u., arbitrary unit.
Figure S8.
Comparison of radial intensity profile of MyBP-C fluorescence from the center of the actin hexagon in active and relaxed sarcomeres to in silico models with MyBP-C bound to myosin, when myosin adopts the IHM and is stabilized on the surface of the myosin thick filament. (A) Illustrative representations of MyBP-C bound to myosin heads, which adopt the IHM. (B) Projection of a cross section from the 3-D EM reconstruction of cardiac muscle thick filament (Zoghbi, et al., 2008; EMD-1465). MyBP-C’s C0 domain (orange; from PDB accession no. 6CXI) with an antibody (magenta; PDB accession no. 1IGT) was placed in close proximity to the outer surface of the RLC of myosin thick filament (4). The red arrow indicates the 4-nm distance between the center of the thick filament and the surface of the C0 domain. The black arrow indicates the 14-nm distance between the center of the thick filament and centroid position of fluorophore with the antibody attached to the C0 domain. The pink dashed circle indicates the potential localization of the fluorophore around each thick filament due to this binding geometry. (C) White areas are target zones for random placement of the fluorophores for when MyBP-C is bound to myosin, which adopts the IHM, as shown in A, for in silico models. (D) Red areas are regions of the image in which fluorophores cannot be randomly placed due to steric exclusion from the myosin and actin filaments. (E) Average in silico reconstructed images generated with a fraction of the fluorophores bound to myosin when the myosin heads are stabilized along the surface of the myosin thick filament and the remaining fluorophores are randomly distributed throughout the image, except for the red areas shown in C. Each image is an average of 120 individual simulations. Scale bar, 25 nm. (F) The radial distribution profiles for the position of MyBP-C relative to the center of the average actin hexagon in the active and relaxed sarcomeres and in silico images shown in E; color scheme as in Fig. S5 F. a.u., arbitrary unit.
Figure S9.
Comparison of radial intensity profile of MyBP-C fluorescence from the center of the actin hexagon in active sarcomeres to in silico models with MyBP-C bound to myosin, when myosin is bound to actin. (A) Illustrative representations of MyBP-C bound to myosin, which is attached to actin. (B) Theoretical model generated using the high-resolution cryo-EM structure of myosin (grey and blue) bound to an actin thin filament (green and grey) in rigor (PDB accession no. 5H53). MyBP-C’s C0 domain (orange; from PDB accession no. 6CXI) with an antibody (magenta; PDB accession no. 1IGT) was placed in close proximity to the RLC (blue) attached to myosin heavy chain (grey) based on in vitro structural evidence (Ratti et al., 2011). The pink stars indicate the centroid position of the fluorophores attached to the antibody. (C) White areas are target zones for random placement of the fluorophores for when MyBP-C is bound to myosin with myosin bound to actin, as shown in A, for in silico models. (D) Red areas are regions of the image in which fluorophores cannot be randomly placed due to steric exclusion from the myosin and actin filaments. (E) Average in silico reconstructed images generated with a fraction of the fluorophores bound to myosin when myosin is bound to actin and the remaining fluorophores are randomly distributed throughout the image, except for red areas shown in C. Each image is an average of 120 individual simulations. Scale bar, 25 nm. (F) The radial intensity profiles for the position of MyBP-C relative to the center of the average actin hexagon in the active sarcomere and in silico images shown in E; color scheme as in Fig. S6 F. a.u., arbitrary unit.
The in silico images were generated using an array of myosin thick and actin thin filaments (Fig. 1 D), with spacing and geometry to mimic the experimental STORM images. Each in silico image was 90 × 90 pixels. Each image contained a myosin thick filament at its center, surrounded by six actin filaments organized in a hexagonal array. Each actin filament was neighbored by two partial thick filaments, as is the case in the myosin and actin lattice (Fig. 1 D).
The 3-D geometries between the C0 domains and the binding partners were based on high-resolution structural data from EM studies (Fujii and Namba, 2017; Harris et al., 1997; Ratti et al., 2011; Risi et al., 2018; Zoghbi et al., 2008). Each in silico–generated image contained five MyBP-C molecules per 90 × 90-pixel area to mimic the number of MyBP-C molecules observed in the experimental acquisitions. The five MyBP-C molecules were either considered bound to the specified binding partner, as described below and in the legends of Fig. S7, Fig. S8, and Fig. S9, or randomly located in the spaced between myosin and actin filaments. In scenarios where the localization was random, the centroid of each MyBP-C spot was excluded from being within the space occupied by the myosin (18 nm) and actin (7 nm) filaments. For each condition, 120 in silico images were simulated and summed to generate the average image as described for the experimental data.
To estimate the distance between the C0 domain of MyBP-C and the position of the Alexa Fluor on the anti-myc antibody, the binding domain of a monoclonal antibody (Harris et al., 1997; PDB accession no. 1IGT) was placed near the N terminus of the C0 domain from a high-resolution cryo-EM structure (Risi et al., 2018; PDB accession no. 6CXI). Six fluorophores were expected to bind to the surface-accessible lysine residues, which are randomly distributed on the antibody. Thus, the average position of the Alexa Fluor molecules was ≈7 nm from the binding site of the antibody to the surface of the C0 domain and ≈9 nm from the center of the C0 domain (see Fig. S7 B). The rotational uncertainty in the position of the antibody on the C0 domain was accounted for by adding an additional 7 nm of uncertainty to the localization.
For the actin-binding model (see Fig. S7 A), the expected distances between the fluorophore and the center of the actin filament were determined using the high-resolution cryo-EM structure (Risi et al., 2018; PDB accession no. 6CXI) of the C0 domain bound to an actin filament. Based on this structure, the average position of the fluorophores was estimated to be 11.5 nm from the center of the actin filament (see Fig. S7 A) when accounting for the rotational uncertainty in the position of the antibody on the C0 domain. Thus, the rendered Gaussian spots were placed within 11.5 nm of the actin filament to generate actin–MyBP-C binding in silico images.
For the relaxed myosin-binding model (see Fig. S8 A), the distance between the center of the myosin thick filament backbone and RLC region of the myosin heads was measured from the 3-D reconstructed EM images of blebbistatin-treated cardiac myosin thick filaments (Zoghbi et al., 2008). The distance from the center of the thick filament to the RLC of the myosin heads was ≈4 nm (see Fig. S8 A). Therefore, the average position of the fluorophores on the antibody was ≈14 nm from the center of the myosin backbone. Thus, the Gaussian spots were randomly distributed within a ring, ≈14 nm from the center of the thick filament.
For the myosin-binding model, when the myosin head is bound to the actin filament (see Fig. S9 A), the position of the RLC region of myosin relative to actin was determined from the high-resolution cryo-EM structure of myosin bound to a skeletal thin filament (Fujii and Namba, 2017; PDB accession no. 5h53). The C0 domain was placed near the RLC region in this structure, and the antibody was given free rotation around the C0 domain but restricted to being oriented toward the actin filament. Therefore, the average position of the antibody was estimated to be within an ≈15-nm distance from the center of actin (see Fig. S9 A). Thus, the Gaussian spots were placed within ≈15 nm of the center of the actin filament.
Radial intensity profile and goodness-of-fit calculation
To quantify the spatial distribution of the Alexa Fluor–labeled antibody attached to the C0 domain from the center of the myosin thick filament within the reconstructed images, the average radial intensity profile of MyBP-C in the experimental and in silico images was calculated. The mean radial intensity profile was generated using the ImageJ Polar Transformer plugin (Dempsey et al., 2011). The average MyBP-C image was transferred to polar coordinates, and the 360 radial line scans beginning at the center of the hexagon were summed to produce an average intensity profile (see Fig. 4 C).
Figure 4.
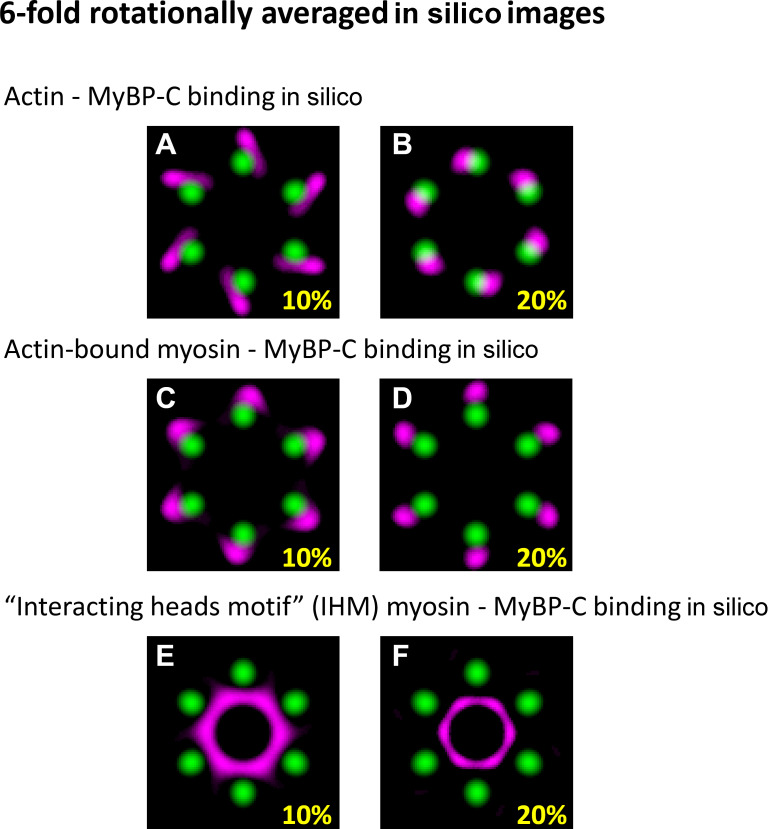
Comparison of the localization N terminus of MyBP-C within the actin lattice in active and relaxed muscle sarcomeres with in silico models with known binding geometries. (A) Illustrative representations of MyBP-C’s three binding partner interactions considered using in silico models. (B) In silico image generated with 20% of MyBP-C N termini bound to actin filaments and the remaining molecules randomly distributed in the space between the thick and thin filament surfaces. Dashed line and yellow arrow demonstrate the generation of the radial intensity profile plotted in C. (C) Radial intensity profiles of the fluorescence intensity of MyBP-C’s N terminus between the center of the actin hexagon and vertices of the actin filaments in the in silico image shown in B (dashed line) and the experimental data for the active (red line) and relaxed (black line) muscle preparations. (D) RMSDs between the radial intensity profiles, as shown in C, of the active (red) and relaxed (black) muscle images and the in silico images generated with a fraction of MyBP-C N termini bound to a binding partner (Fig. S6, Fig. S7, and Fig. S8). Error bars represent the SEM of the RMSDs from six in silico images generated for each condition. (E) Merged images for sixfold rotational average of all actin (green) and MyBP-C (magenta) images. The brightness and contrast of the image were adjusted to present the brightest spots in the images. Scale bars, 25 nm. a.u., arbitrary unit.
The goodness of fit between the experimental and in silico MyBP-C radial intensity profiles was determined using the RMSD. A smaller RMSD represents better agreement between the data and the model.
Sixfold rotational averaging
To visualize the MyBP-C molecules bound to the actin filaments in the experimental and in silico generated images, the signal-to-noise ratio in images was improved by sixfold rotational averaging. The 60° rotation was chosen because of the sixfold symmetry of actin filament arrangement within the hexagon. The brightness and contrast were adjusted in each rotationally averaged image to highlight the location of the maximal intensity.
Data availability
Data described in this paper are presented in the supplementary materials.
Online supplemental material
Fig. S1 contains the single ATP turnover measurements from multicellular ventricular preparations from wild-type and transgenic mice expressing myc-tag labeled MyBP-C. Fig. S2 shows the longitudinal and cross-sectional views of mouse cardiac papillary muscle. Fig. S3 identifies the myosin and actin lattice and quantification of interactin spacing in actin STORM and EM images. Fig. S4 contains the geometry and reconstructed images of DNA origami scaffolds used to evaluate the resolution of the STORM analyses. Fig. S5 compares high-resolution STORM images reconstructed with Alexa Fluor 647 versus Alexa Fluor 488 for actin and MyBP-C. Fig. S6 shows the relative positions of actin and MyBP-C in sarcomeres from relaxed and active muscle preparations determined by combining STORM with particle averaging. Fig. S7 compares the radial intensity profile of MyBP-C fluorescence from the center of the actin hexagon in active and relaxed sarcomeres to in silico models with MyBP-C bound to actin. Fig. S8 compares the radial intensity profile of MyBP-C fluorescence from the center of the actin hexagon in active and relaxed sarcomeres to in silico models with MyBP-C bound to myosin, when myosin adopts the IHM and is stabilized on the surface of the myosin thick filament. Fig. S9 compares the radial intensity profile of MyBP-C fluorescence from the center of the actin hexagon in active sarcomeres to in silico models with MyBP-C bound to myosin, when myosin is bound to actin. Fig. S10 compares the sixfold rotationally averaged in silico images. Table S1 shows the degree of cMyBP-C phosphorylation quantified by LC-MS.
Figure S1.
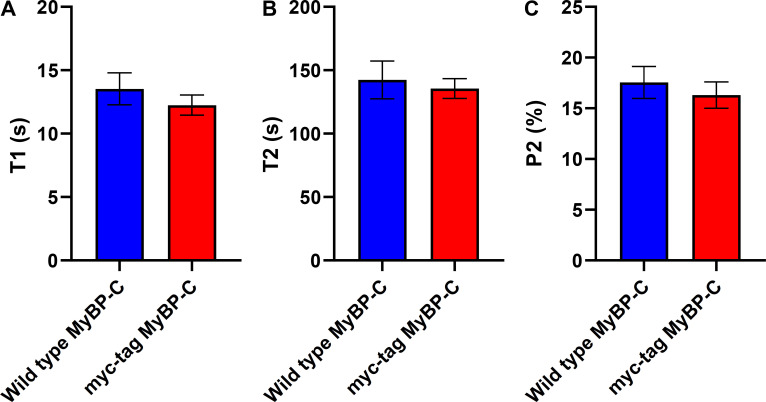
Single ATP turnover measurements from multicellular ventricular preparations from wild-type and transgenic mice expressing myc-tag–labeled MyBP-C. (A) The lifetime of nucleotide turnover associated with fast phase of fluorescence decay. (B) The lifetime of nucleotide turnover associated with the slow (SRX) phase of fluorescence decay. (C) The amplitude of the slow phase of fluorescence decay, indicating the fraction of myosin molecules that adopted the SRX state. n = 4 wild-type mice (14 technical replicates) and n = 2 myc-tag–labeled MyBP-C mice (12 technical replicates). Error bars indicate ±SEM (P > 0.4).
Figure S2.
Longitudinal and cross-sectional views of mouse cardiac papillary muscle. (A and B) Negative-stain EM images of 100-nm-thick cryo-sections, longitudinal (A) and transverse (B), of mouse papillary muscle from active and relaxed muscle preparations, respectively, demonstrating the actin and myosin filament lattice within the myofibrils. Scale bars, 1 µm (A) and 500 nm (B). (C and D) Reconstructed images of Alexa Fluor 488-phalloidin (actin) filaments (C) and Alexa Fluor 647 anti-myc (MyBP-C’s C0 domain; D) within 400-nm-thick transverse cryo-sections of mouse papillary muscle imaged by STORM. Scale bars, 5 µm.
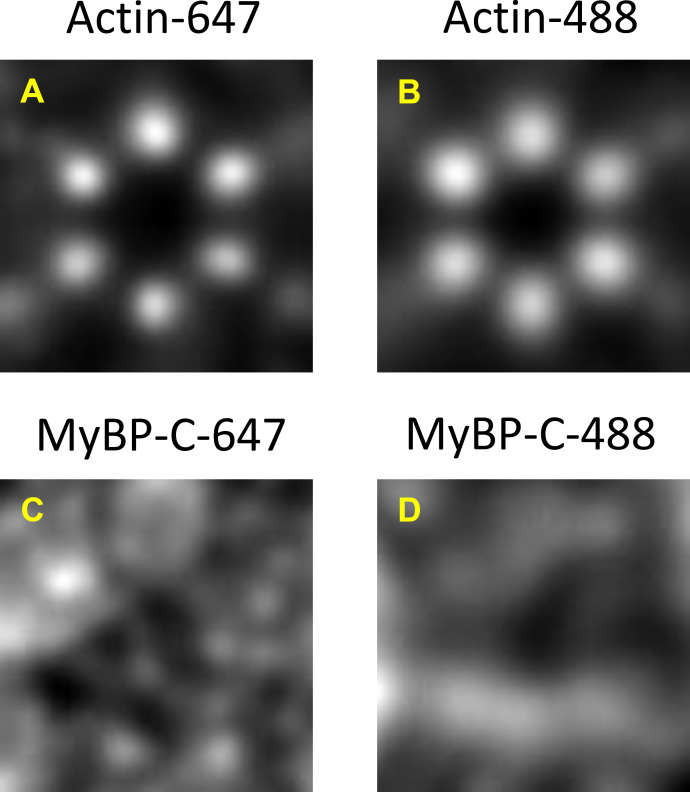
Figure S5.
Comparison of high-resolution STORM images reconstructed with Alexa Fluor 647 versus Alexa Fluor 488 for actin and MyBP-C. (A–D) Actin (A and B) and MyBP-C (C and D) are shown. A broad, toroid signal was observed for MyBP-C using both color fluorophores.
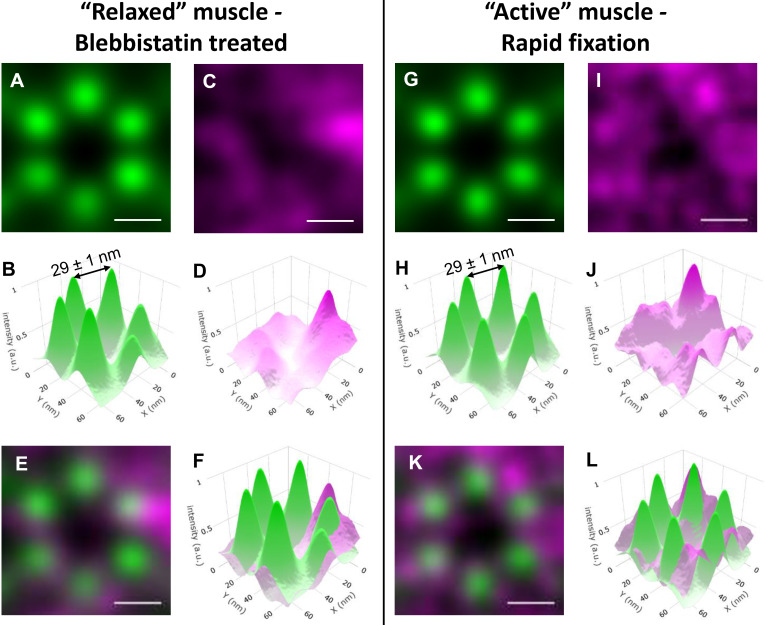
Figure S6.
Relative positions of actin and MyBP-C in sarcomeres from relaxed and active muscle preparations determined by combining STORM with particle averaging. (A and B) Average reconstructed actin image (A) and 3-D plot (B) generated from 134 actin hexagons originating from different relaxed muscle sarcomeres in the hearts treated with blebbistatin before fixation. (C and D) Average reconstructed MyBP-C image (C) and 3-D plot (D) from areas of the MyBP-C images corresponding to those shown for actin in A. The z axis is intensity, representing localization uncertainty. (E) Merged images shown in A and C. Actin (green) and MyBP-C (magenta). (F) Merged 3-D images shown in C and D. Actin (green) and MyBP-C (magenta). (G–L) Similar data to that shown in A–F but from active muscle sarcomeres. 120 individual actin reconstructions were used to generate the average images. The interactin distances and uncertainties were similar between the active and relaxed sarcomeres. a.u., arbitrary unit.
Figure S10.
Comparison of the sixfold rotationally averaged in silico images. (A and B) Rotational averages for in silico images representing 10% and 20% of MyBP-C molecules bound to actin. (C and D) The myosin heads when myosin is attached to actin. (E and F) Myosin heads adopting the IHM. The unbound MyBP-C molecules were randomly distributed in the free space between the myosin thick and actin thin filament surfaces under all conditions. Brightness and contrast in all images were adjusted to highlight the regions with highest probability of occupancy for MyBP-C.
Results
Expression and function of N-terminal myc-tagged cardiac MyBP-C in mice
A critical aspect for accurately localizing the N terminus of MyBP-C in situ was to limit the distance between the N terminus and the fluorescent reporter dyes. This distance was limited to 7 nm by creating a transgenic mouse that expressed cardiac MyBP-C with an N-terminal myc-tag that allowed for the attachment of a fluorescently labeled primary antibody to the protein in fixed muscle samples. The gross cardiac morphology and physiology of these transgenic mice were previously demonstrated to be identical to that of wild-type mice (Sadayappan et al., 2005). Neither the abundance of expressed myc-tagged MyBP-C (one MyBP-C for 11.8 myosin molecules) nor its 64% ± 26% average phosphorylation level (Table S1) differed from that of wild-type mice, as determined by quantitative MS (Previs et al., 2012).
To evaluate whether the myc-tag disrupted the ability of myosin to adopt the SRX biochemical state, single ATP turnover measurements were made using relaxed, permeabilized multicellular ventricular preparations, as previously described (McNamara et al., 2016; McNamara et al., 2019). Neither the lifetime of the slow, SRX nucleotide turnover (142 ± 28 versus 135 ± 28 [SEM]) nor the fraction of myosin heads that adopted the SRX state (17.5 ± 3.0% versus 16.3 ± 3.2% [SEM]) differed (P > 0.4) between the preparations from the hearts of wild-type (n = 4) or myc-tag MyBP-C (n = 2) mice (Fig. S1). These data demonstrate collectively that the presence of the myc-tag does not affect sarcomere protein stoichiometry or the ability of myosin to adopt the SRX biochemical state within the muscle.
EM of sarcomere structure
Papillary muscles were initially fixed in situ via left ventricular pressurization as the heart was still beating. This was done in an effort to capture some of the myosin molecules while bound to the actin filaments, to mimic active contractile conditions. An additional preparation included the pretreatment of the heart with blebbistatin, a myosin inhibitor that stabilizes the myosin heads along the myosin filament surface in the IHM (Al-Khayat et al., 2013; Kampourakis et al., 2014; Wilson et al., 2014; Zoghbi et al., 2008), to mimic fully relaxed contractile conditions. The unique 3-D ultrastructure of cardiac muscle sarcomeres was evident in EM images of 100-nm thick, negatively stained, longitudinal and transverse cryo-sections of the papillary muscles from both preparative conditions (Fig. 1, E and F; and Fig. S2, A and B). In longitudinal views (Fig. 1 E and Fig. S2 A), 2.1 ± 0.2-µm (SD; n = 33) and 2.1 ± 0.1-µm (SD; n = 24) sarcomeres were observed within myofibrils in both the active and relaxed muscle preparations. These lengths demonstrate that in situ fixation during ventricular pressurization results in sarcomere lengths similar to those observed during cardiac filling in live mouse hearts (Kobirumaki-Shimozawa et al., 2016).
In cross-sectional views of the relaxed muscle preparations (Fig. 1 F and Fig. S2 B), the known hexagonal organization of six actin filaments surrounding a myosin filament was clearly apparent (Fig. 1 F, inset) in small, limited regions of each sarcomere (Fig. S2 B). The difference in thickness of the thick and thin filaments in the relaxed muscle preparation is consistent with the density of the myosin heads being folded on the myosin filament surface (Huxley, 1968). The lack of full preservation of the entire sarcomere lattice within preparations likely resulted from the collapse of the filaments during the cutting of the cryo-sections and/or drying of the negative stain. The center-to-center distance between actin filaments along the vertices of the hexagon was 22.4 ± 3.9 nm (SD; n = 49), being similar to the 21.5 ± 1.9-nm (SD; n = 48) center-to-center distance between the actin filaments and the myosin thick filament at the center of each actin hexagon. Due to the limited thickness of single MyBP-C molecules (≈4 nm) and their inherent flexibility (Colson et al., 2016; Hartzell and Sale, 1985; Previs et al., 2016), MyBP-C was not visible in the cross-sectional views of the negatively stained EM images (Fig. 1 F and Fig. S2 B).
STORM imaging of actin and MyBP-C in cardiac muscle cross sections
Transverse cryo-sections (400 nm thick) of the fixed papillary muscles were cut for fluorescence STORM imaging. Actin and the N terminus of MyBP-C were labeled with phalloidin (actin) and an anti-myc primary antibody (MyBP-C), respectively. Each molecule was preconjugated to separate color Alexa Fluor dyes. Multicolor 100-nm beads were added to the sections for correction of instrumental drift and two-color channel alignment. Multicolor STORM was used to determine the localization of fluorophores in the traverse heart sections (see Materials and methods). Low-resolution reconstructed images of actin fluorescence (see Materials and methods) demonstrated that the signal was restricted to small areas (Fig. S2 C), being of similar size and shape as the sarcomeres observed in the traverse EM images (Fig. S2 B). Dark spaces were also present in the reconstructed images of actin fluorescence (Fig. S2 C), which were of similar size and shape as those of the mitochondria in the EM images (Fig. S2 B).
Low-resolution reconstructed images of MyBP-C fluorescence demonstrated the signal was also restricted to within the sarcomeres (Fig. S2 D). However, when merged with the actin images, it appeared that MyBP-C was not in every sarcomere sampled (Fig. 2, A–C). This was because only some of the ∼2.1-µm-long sarcomeres within the 400-nm thick sections were anticipated to have their C-zone (∼387 nm in length) sampled, where MyBP-C is localized (Fig. 1 E).
High-resolution reconstructed images of actin fluorescence (see Materials and methods) contained bright puncta (Fig. 2 D), which were consistent with single actin filaments being viewed in cross section (Fig. 1 D). However, the myosin and actin filament lattice observed in the small, limited regions of the EM images (Fig. 1 F and Fig. S2 B), were not readily detected by eye in the STORM images. This was most likely due to the minimal number of reconstructed actin filaments in each image, resulting from the stringent selection criteria required to achieve 12-nm localization precision (see Materials and methods) and the collapse of myosin and actin filaments from cutting, as observed in the EM images (Fig. 1 F and Fig. S2 B).
Identification of the myosin and actin lattice by template matching
To determine whether regions of the STORM images had good preservation of the myosin and actin lattice, the images were interrogated with theoretical templates. Images of individual sarcomeres were scanned using templates composed of two identical, conjoined hexagons (Fig. S3 A), and cross-correlation values for each location were determined (Fig. S3 B). The interactin filament spacing along the vertices of the hexagon in the templates was varied (17.5–37.5 nm; Fig. S3 A) to encompass the interactin filament spacing initially measured in EM images (Fig. 1 F and Fig. S2 B). Each template was rastered along the image on a pixel-by-pixel basis and rotated at 5° intervals (from 0° to 180°) in each location. Templates with 25–30-nm interactin filament spacing (Fig. S3 A) demonstrated maxima in cross-correlation values with the underlying fluorescence image (Fig. 2 E). The template matches with the highest cross-correlation values were unique in that they typically overlapped with one another within a sarcomere (Fig. 2 F). This overlap suggested that small regions of each sarcomere contained good preservation of the myosin and actin filament lattice, being similar to that observed within the EM images of the traverse muscle sections (Fig. 1 F and Fig. S2 B).
As a positive control, the template-matching strategy was applied to the transverse EM images in which the signal from the thick filaments was removed to best mimic the STORM images of actin (Fig. S3 C). The two-hexagon template with 22.5-nm interactin filament spacing demonstrated a maximum in cross-correlation value with the underlying EM image (Fig. S3 D). This was similar to the 22.4 ± 3.9-nm (SD; n = 49) interactin filament spacing initially measured from the EM images using ImageJ (see Materials and methods). Like the best matches observed in the actin fluorescence images (Fig. 2 F), the templates with 22.5-nm spacing overlapped with each other in EMs to identify the well-preserved region of the myosin and actin lattice (Fig. S3 C).
As a negative control, a rectangular template was also applied to the STORM images (Fig. S3 E). The template matches with the highest cross-correlation values to the fluorescence image were randomly distributed throughout the sarcomeres. These matches did not overlap to identify larger areas of lattice spacing (Fig. S3 E).
Visualization of an actin hexagon by particle averaging
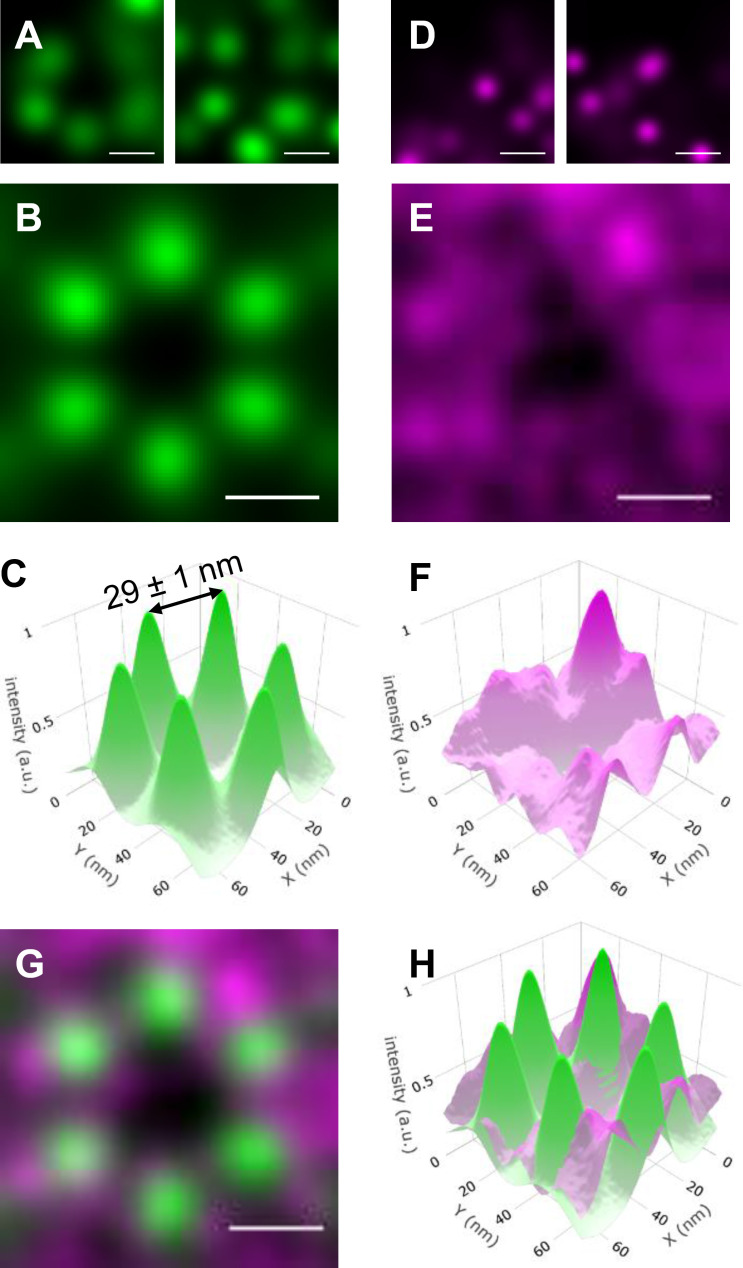
Next, a single ROI, identified by the template matching, was cropped from each of the actin fluorescence images. Each ROI (n = 120) contained 5 ± 1 (SD) distinct fluorescence spots of actin (Fig. 3 A), which presumably contained a myosin filament at their center (Fig. 1 F). The lack of detection of all six actin filaments was likely due to the STORM selection criteria to ensure low localization uncertainty. The ROIs (n = 120) were aligned based on the orientation of the hexagonal template used for its identification. The ROIs were summed to produce an averaged reconstructed image (Fig. 3, B and C). This high-resolution image was composed of a hexagonal array of actin filaments, with 29 ± 1-nm interactin spacing along the vertices of the hexagon (Fig. 3 C). The distribution of fluorescence intensity at each vertex (Fig. 3 C; full width at half-maximum = 16.5 ± 1 nm) reflects the uncertainty in the position of six individual spots within each ROI, coupled with the physical distance between the Alexa Fluor on the phalloidin molecules bound to each actin filament.
Figure 3.
Relative positions of actin and MyBP-C in sarcomeres from active muscle preparations determined by combining STORM with particle averaging. (A) Examples of partial hexagons cropped from active muscle sarcomeres in the actin STORM images. (B) Average reconstructed actin fluorescence image generated from 120 actin ROIs originating from different sarcomeres. (C) 3-D plot of the average actin image shown in B, with the z axis (intensity) representing localization certainty. The distance between two neighboring actin filaments was 29 ± 1 nm. (D) ROIs of the MyBP-C reconstructed images cropped from areas identical to those shown for actin in A. (E) Average reconstructed MyBP-C image, as in B for the actin. (F) 3-D plot of the average MyBP-C image shown in E. (G) Merged images shown in B and E. Actin (green) and MyBP-C (magenta). (H) Merged 3-D images shown in C and F. Actin (green) and MyBP-C (magenta). Scale bars, 25 nm. a.u., arbitrary unit.
The resolution of the closely spaced fluorophore on neighboring actin filaments is at the boundary of what is currently possible using conventional STORM imaging (Bates et al., 2007; Schermelleh et al., 2010). To obviate the possibility that the result was an artifact of particle averaging, the STORM and particle averaging approaches were used to image Alexa Fluor clusters, separated by 31.1 nm, on DNA origami scaffolds. The ability to resolve the average position (32 ± 1 nm) of closely spaced Alexa Fluors on the DNA origami scaffolds (Fig. S4, A–E) was similar to that observed for the actin filaments in the muscle preparations.
Localization of the N terminus of MyBP-C within the actin hexagon of active muscle
To identify whether the N-terminal domains of MyBP-C are preferentially colocalized with actin or lie close to the thick filament backbone, the position of the N terminus of MyBP-C within the hexagonal array of actin filaments was determined. ROIs (n = 120) identical to those in the actin images were cropped from the corresponding high-resolution reconstructed fluorescence images of MyBP-C N termini. The individual ROIs (n = 120) from the MyBP-C fluorescence images contained 5 ± 3 (SD) discrete fluorescence spots (Fig. 3 D). The spots were of similar quality to those from the actin filaments (Fig. 3 A). The MyBP-C ROIs were then rotated based on the orientation of the actin ROIs that they were associated with, using the approach developed for actin. Because the contour length of MyBP-C is 43 nm (Hartzell and Sale, 1985; Previs et al., 2016), the C terminus of the MyBP-C molecules in each image could be attached to the myosin filament at the center of the actin hexagon or to either of the two myosin filaments that neighbored each actin filament distal to the hexagon (Fig. 1 D). While the individual MyBP-C ROIs contained bright, discrete spots (Fig. 3 D), indicative of single molecules, the summed MyBP-C image demonstrated a broad, diffuse, toroid-like appearance (Fig. 3, E and F). This appearance was suggestive of a disordered localization of MyBP-C N termini within the myosin and actin lattice.
When the summed MyBP-C image was merged with the actin image, the toroid of MyBP-C fluorescence overlapped with the six discrete spots of actin fluorescence, and the intensity of the MyBP-C signal was minimal near the center of the actin hexagonal array where the myosin filament was located (Fig. 3, G and H). The lack of six discrete spots of MyBP-C fluorescence in the summed image demonstrated that <100% of the MyBP-C N termini were bound to the actin filaments. The lack of a single signal of MyBP-C fluorescence near the center of the actin hexagon demonstrated that 100% of the MyBP-C N termini were not localized toward the backbone of the myosin filament.
Two-color STORM is challenging because the blinking rates of the fluorophore and photon emission need to be optimized to achieve high localization precision in each color channel (Tam et al., 2014). Optimal blinking is especially critical for STORM of densely labeled samples because there is potential for overlap between the PSFs of neighboring molecules (van de Linde et al., 2010). Several combinations of fluorophores and STORM buffers were initially tested to achieve optimal imaging conditions (see Materials and methods). Regardless of this optimization, STORM images were collected with both phalloidin–Alexa Fluor 647/anti-myc–Alexa Fluor 488 and phalloidin–Alexa Fluor 488/anti-myc–Alexa Fluor 647 labeling strategies to be certain that the results were not biased by the selection of the fluorophore. The summed images (Fig. 3, B, C, E, and F) were created using a mixture of images generated with both labeling strategies. To test whether the diffuse nature of the MyBP-C signal resulted from differences in localization precision between Alexa Fluor 647 and 488, summed actin and MyBP-C images were generated using only data from a single fluorophore (Fig. S5). The summed actin images demonstrated six discrete spots organized in a hexagon (Fig. S5, A and B), while the summed MyBP-C images demonstrated a broad, diffuse, toroid-like appearance (Fig. S5, C and D) regardless of the labeling strategy. These data demonstrated that the diffuse signal for MyBP-C was not due to an artifact of two-color STORM imaging.
Localization of the N terminus of MyBP-C in relaxed myocardium
To determine if the lack of MyBP-C fluorescence near the myosin filament was due to the extension of the myosin heads toward the actin filaments in active muscle, the entire series of imaging was repeated for a relaxed heart, which was pretreated with blebbistatin before fixation. The thickness of the myosin filaments in traverse EM images (Fig. 1 F) suggested the myosin heads were stabilized along the thick filament surface in the relaxed muscle preparation (Huxley, 1968). If the N terminus of MyBP-C interacted with myosin heads adopting the IHM, then the MyBP-C fluorescence would be shifted toward the center of the actin hexagon in this muscle preparation. Despite the potential difference in position of the myosin heads between these preparations, only subtle differences were visually apparent in the toroid-like appearance of MyBP-C and the overlap between the MyBP-C fluorescence and that of the actin hexagonal array (Fig. S6). The lack of MyBP-C fluorescence toward the myosin filament in either preparation suggested that the N-terminal domains of MyBP-C were extended toward the actin filaments in both physiological states examined.
Quantitative interpretation of STORM images using in silico modeling
To quantify how the fluorescence intensity profile of the N terminus of MyBP-C relates to the position of actin and the myosin head in the active and relaxed muscle preparations, in silico fluorescence-based structural models were developed with theoretical binding partner interactions predicted from in vitro studies (see Materials and methods). These models consisted of an array of myosin thick and actin thin filaments (Fig. 1 D) with the geometries determined from the STORM images. Three scenarios were considered with the position of the N-terminal C0 domain of MyBP-C bound to its potential binding partners under varying contractile states (Fig. 4 A, Fig. S7, Fig. S8, and Fig. S9). These included the C0 domain being (1) bound to the actin filaments (Fig. S7); (2) bound to the head region of myosin only when the myosin heads adopt the IHM and thus are positioned along the myosin thick filament surface (Fig. S8); and (3) bound to the myosin head region only when the myosin heads are extended away from the thick filament and are bound to actin (Fig. S9). For each of the in silico images of MyBP-C fluorescence, the C0 domain was either bound to its specified partner or fixed randomly in the space between the myosin and actin filaments.
To compare the distribution of MyBP-C fluorescence within the actin hexagonal array between the experimental and in silico images, radial intensity profiles were generated from each image (Fig. 4, B and C; Fig. S7 F; Fig. S8 F; and Fig. S9 F). In the experimental muscle preparations, the MyBP-C radial intensity profile was lowest near the center of the actin hexagon and increased monotonically to a maximum near the actin vertices (Fig. 4 C, Fig. S7 F, Fig. S8 F, and Fig. S9 F). The goodness of fit between the radial intensity profiles of the experimental and in silico images was empirically determined as the lowest RMSD (Fig. 4 D). The experimental images for the active muscle preparations were best fitted by models in which 60–80% of the MyBP-C N termini were bound to actin and/or the myosin head, but only if the myosin head was bound to the actin filament (Fig. 4 D, Fig. S7 F, and Fig. S9 F). The poorest fits were with MyBP-C N termini bound to myosin heads adopting the IHM along the thick filament surface (Fig. 4 D and Fig. S8 F).
Was the bias in MyBP-C fluorescence toward the actin filaments (Fig. 3, G and H) due to the absence of myosin heads in the IHM (i.e., near the myosin filament backbone) in the active muscle preparations? In the blebbistatin-treated preparation, the diameter of the myosin filaments was large (Fig. 1 F), and thus the myosin heads were presumably stabilized in the IHM on the thick filament surface (Al-Khayat et al., 2013; Kampourakis et al., 2014; Wilson et al., 2014; Zoghbi et al., 2008). Even under these extreme conditions of muscle relaxation, the radial intensity profiles of MyBP-C fluorescence were most similar to in silico images, with 20% of the MyBP-C N termini bound to the actin filaments (Fig. 4 D and Fig. S7 F) and the remaining 80% of the N termini randomly located within the space between the actin and myosin filament surfaces. The fits were poorest when even a small (<10%) fraction of the MyBP-C N termini were bound to the myosin filament surface (Fig. 4 D and Fig. S8 F).
Refinement of the summed STORM images with rotational averaging
To visualize MyBP-C N termini that may be bound to an actin filament and/or myosin head in the experimental data, sixfold rotationally averaged images were generated using all of the actin and MyBP-C ROIs (n = 254; Fig. 4 E). Six discrete spots of actin fluorescence were observed, with a maximum in MyBP-C fluorescence 11 nm from the center of each actin filament (Fig. 4 E). These images were similar to sixfold rotational averages of the in silico images with 10–20% of the MyBP-C molecules bound to actin (Fig. S10, A and B) or the myosin head, when myosin is attached to the actin filament (Fig. S10, C and D). In both the experimental (Fig. 4 E) and in silico images (Fig. S10, A–D), the maxima in fluorescence were localized toward the exterior of the actin filaments. This apparent localization was because one of the N termini could be coming from the myosin filament at the center of the actin hexagon, and two N termini could be coming from the two myosin filaments that neighbor the actin filament distal to the center of the hexagon (Fig. 1 D). In the in silico images, a ring in fluorescence intensity was observed toward the center of the actin hexagon when even 10% of the N termini were bound to myosin heads that adopted the IHM (Fig. S10, E and F). No such peak in fluorescence was observed in the experimental image (Fig. 4 E). The lack of such a peak demonstrated that the N terminus of MyBP-C may not lie along the surface of the myosin filament in these muscle preparations.
Discussion
The relative positions of the N terminus of MyBP-C and actin were visualized in active and relaxed mouse papillary muscle using STORM. The resolution of the reconstructed images was enhanced by taking advantage of the native organization of myosin and actin filaments in traverse sections of muscle (Fig. 1, D and F) to apply particle averaging. The 29-nm distance between neighboring actin filaments (Fig. 3 C) was similar to the 26-nm distance measured in x-ray studies of intact cat (Matsubara and Millman, 1974) and rat (Ait-Mou et al., 2016) cardiac muscles. The interactin distance measured by STORM was 22% greater than that measured in the EM images (Fig. 1 F and Fig. S1 B); this is probably due to the lack of sample shrinkage in the absence of negative stain (Millman, 1998). This powerful combination of approaches allowed for the direct visualization of a hexagonal array of actin filaments using light microscopy. While such an approach was previously used to visualize nuclear pores distributed throughout detergent-skinned cells (Szymborska et al., 2013), this is the first demonstration of this technique in a densely packed cytoplasmic environment. This was made possible by the crystalline-like organization of the actin and myosin filaments, which allowed for particle averaging.
With the actin filaments resolved, the relative position of the N terminus of MyBP-C molecules was determined from the fluorescence of a second color fluorophore. The localization of the bright fluorophore enhanced the detection of N termini despite their minimal density in muscle sections and high degree of intrinsic flexibility (Colson et al., 2016; Hartzell and Sale, 1985; Previs et al., 2016), which likely limits visualization by EM imaging. The experimental data for the average position of MyBP-C N termini in the traverse sections of muscle demonstrated diffuse localization of the molecules. Discrete puncta in the average image would only be seen if the molecules were bound to the actin filaments with high probability (Fig. S7 E). The experimental data were best described by structural models in which the majority of the N termini were fixed in random locations in the space between the myosin and actin filament surfaces. Such random localization is consistent with the elongated MyBP-C molecules (Hartzell and Sale, 1985; Previs et al., 2016) undergoing Brownian motion to explore the free space between the myosin and actin filament surfaces before fixation.
In-depth quantitative comparison of the diffuse MyBP-C signal in the experimental data with in silico images demonstrated a bias in the position of MyBP-C N termini toward the actin filaments, independent of the contractile state. The orientation of the N terminus of MyBP-C away from the myosin filament backbone is likely facilitated by the C-terminal domain being bound to the thick filament backbone (Flashman et al., 2007) and the presence of two flexible, internal hinges (Fig. 1 B) between the more rigid elongated segments of the molecule (Colson et al., 2016; Previs et al., 2016). Moreover, the in silico images also demonstrated that the bias in the position toward the actin filaments was enhanced by a fraction of the MyBP-C N termini being bound to the actin filaments and/or the myosin head when the myosin head is bound to the actin filament (Fig. S7 and Fig. S9).
The binding of MyBP-C N termini to actin in the active muscle is consistent with models that suggest the N-terminal domains bind to the thin filament to help initiate muscle contraction (Pfuhl and Gautel, 2012; Previs et al., 2014). Binding of the N termini to actin while the filament is sliding may also impose a resistive load on the contraction. This is because the C terminus attaches the other end of the molecule to the myosin filament, creating a mechanical tether between the myosin and actin filaments. Similarly, binding of the N termini to the myosin head may impose a mechanical load as the myosin head goes through the power stroke to slow filament sliding. Due to the uncertainty in the position of the myosin heads in this preparation, further distinction between these two potential models is not possible.
In contrast, even under conditions where the myosin heads appeared to be stabilized along the myosin filament surface (Fig. 1 F), the MyBP-C N termini do not appear to be enriched near the center of the actin hexagon (Fig. 4 E), as demonstrated with as little as 10% of the N termini bound to the myosin filament surface in silico (Fig. S10). Thus, if MyBP-C regulates the SRX state of myosin by stabilizing the IHM (Brunello et al., 2020; Nag et al., 2017; Sarkar et al., 2020; Trivedi et al., 2018), either a very small fraction (i.e., <10%) of the N termini (Fig. 1 B) participate in such structural interactions in muscle or the internal domains of MyBP-C are responsible for this stabilization. Based on the potential geometry of MyBP-C within the sarcomere, stabilization of the IHM may result from interactions with the C4–C6 domains of MyBP-C.The affinity of MyBP-C for the actin filament in relaxed cardiac muscle (i.e., in the absence of calcium) is dependent on the extent of MyBP-C phosphorylation, with higher affinities associated with dephosphorylated MyBP-C (Belknap et al., 2014; Ponnam et al., 2019; Shaffer et al., 2009). Given that MyBP-C appears to have relatively high levels of phosphorylation under normal conditions (Table S1), MyBP-C dephosphorylation, which occurs with heart failure and genetic cardiomyopathies, may increase the affinity of MyBP-C N termini for the thin filament and thus pathologically sensitize the thin filament to calcium, causing diastolic dysfunction (Oakley et al., 2007; Rosas et al., 2015). Once cardiac muscle is activated, calcium levels are high, and calcium appears to ablate the ability of phosphorylation to tune MyBP-C function (Previs et al., 2016). Under fully active conditions, the myosin heads would be in close proximity to the actin filament (Brunello et al., 2020; Kampourakis et al., 2014), presenting the MyBP-C N termini with an additional binding partner (i.e., the myosin head). Due to the low (micromolar) in vitro affinity constants for MyBP-C to bind both actin (Belknap et al., 2014) and myosin (Ponnam et al., 2019), the N-terminal domains of MyBP-C may be in a dynamic binding equilibrium between these two binding partners. In conclusion, the data generated from this powerful single-molecule fluorescence approach may contribute to a mechanistic understanding of the role of MyBP-C in modulating myofilament contractility through its specific binding partner interactions under varied physiological states.
Supplementary Material
shows the degree of cMyBP-C phosphorylation quantified by LC-MS.
Acknowledgments
Henk L. Granzier served as editor.
We thank D. Taatjes, N. Bishop, and N. Bouffard from the University of Vermont Microscopy Imaging Center and G. Kennedy from the Instrumentation and Modeling Facility for imaging expertise. We thank M. Heilemann and S. Malkusch for providing the packages PyMalk and Localization Microscopy Analyzer necessary for instrument drift correction and alignment of color channels. We are grateful to the Core Electron Microscope Facility at UMass Medical School and its staff for use of their resources.
We acknowledge funding from the National Institutes of Health grant 1S10ODO21580-01 for the ultramicrotome. This paper was funded by National Institutes of Health funds, which supported S. Rahmanseresht, T.S. O’Leary, and M.J. Previs (HL124041); K.H. Lee, J. Robbins, D.M. Warshaw, and R. Craig (HL059408, AR067279, HL126909, HL139883, and HL150953); R. Craig (AR072036); and S. Sadayappan (AR078001, HL130356, HL139680, AR067279, HL105826, and HL143490). J.W. McNamara was supported by an American Heart Association postdoctoral fellowship (17POST33630095). This work is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Dr. Sadayappan provides consulting and collaborative research studies to the Leducq Foundation (CURE-PLAN), Red Saree Inc., Greater Cincinnati Tamil Sangam, AstraZeneca, MyoKardia, Merck, and Amgen, but such work is unrelated to the content of this manuscript. The remaining authors declare no competing financial interests.
Author contributions: M.J. Previs, D.M. Warshaw, and R. Craig designed the research; S. Rahmanseresht, K.H. Lee, T.S. O’Leary, and J.W. McNamara preformed experiments; S. Rahmanseresht, K.H. Lee, T.S. O’Leary, J.W. McNamara, S. Sadayappan, D.M. Warshaw, R. Craig, and M.J. Previs analyzed data; S. Sadayappan and J. Robbins contributed essential reagents; S. Rahmanseresht, K.H. Lee, T.S. O’Leary, J.W. McNamara, D.M. Warshaw, R. Craig, and M.J. Previs wrote the manuscript.
References
- Ait-Mou, Y., Hsu K., Farman G.P., Kumar M., Greaser M.L., Irving T.C., and de Tombe P.P.. 2016. Titin strain contributes to the Frank-Starling law of the heart by structural rearrangements of both thin- and thick-filament proteins. Proc. Natl. Acad. Sci. USA. 113:2306–2311. 10.1073/pnas.1516732113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khayat, H.A., Kensler R.W., Squire J.M., Marston S.B., and Morris E.P.. 2013. Atomic model of the human cardiac muscle myosin filament. Proc. Natl. Acad. Sci. USA. 110:318–323. 10.1073/pnas.1212708110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfares, A.A., Kelly M.A., McDermott G., Funke B.H., Lebo M.S., Baxter S.B., Shen J., McLaughlin H.M., Clark E.H., Babb L.J., et al. 2015. Results of clinical genetic testing of 2,912 probands with hypertrophic cardiomyopathy: expanded panels offer limited additional sensitivity. Genet. Med. 17:880–888. 10.1038/gim.2014.205 [DOI] [PubMed] [Google Scholar]
- Bates, M., Huang B., Dempsey G.T., and Zhuang X.. 2007. Multicolor super-resolution imaging with photo-switchable fluorescent probes. Science. 317:1749–1753. 10.1126/science.1146598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap, B., Harris S.P., and White H.D.. 2014. Modulation of thin filament activation of myosin ATP hydrolysis by N-terminal domains of cardiac myosin binding protein-C. Biochemistry. 53:6717–6724. 10.1021/bi500787f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig, E., Patterson G.H., Sougrat R., Lindwasser O.W., Olenych S., Bonifacino J.S., Davidson M.W., Lippincott-Schwartz J., and Hess H.F.. 2006. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 313:1642–1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Brunello, E., Fusi L., Ghisleni A., Park-Holohan S.J., Ovejero J.G., Narayanan T., and Irving M.. 2020. Myosin filament-based regulation of the dynamics of contraction in heart muscle. Proc. Natl. Acad. Sci. USA. 117:8177–8186. 10.1073/pnas.1920632117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson, B.A., Thompson A.R., Espinoza-Fonseca L.M., and Thomas D.D.. 2016. Site-directed spectroscopy of cardiac myosin-binding protein C reveals effects of phosphorylation on protein structural dynamics. Proc. Natl. Acad. Sci. USA. 113:3233–3238. 10.1073/pnas.1521281113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempsey, G.T., Vaughan J.C., Chen K.H., Bates M., and Zhuang X.. 2011. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat. Methods. 8:1027–1036. 10.1038/nmeth.1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flashman, E., Watkins H., and Redwood C.. 2007. Localization of the binding site of the C-terminal domain of cardiac myosin-binding protein-C on the myosin rod. Biochem. J. 401:97–102. 10.1042/BJ20060500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii, T., and Namba K.. 2017. Structure of actomyosin rigour complex at 5.2 Å resolution and insights into the ATPase cycle mechanism. Nat. Commun. 8:13969. 10.1038/ncomms13969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, L.J., Larson S.B., Hasel K.W., and McPherson A.. 1997. Refined structure of an intact IgG2a monoclonal antibody. Biochemistry. 36:1581–1597. 10.1021/bi962514+ [DOI] [PubMed] [Google Scholar]
- Hartzell, H.C., and Sale W.S.. 1985. Structure of C protein purified from cardiac muscle. J. Cell Biol. 100:208–215. 10.1083/jcb.100.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huxley, H.E. 1968. Structural difference between resting and rigor muscle; evidence from intensity changes in the lowangle equatorial x-ray diagram. J. Mol. Biol. 37:507–520. 10.1016/0022-2836(68)90118-6 [DOI] [PubMed] [Google Scholar]
- Kampourakis, T., Yan Z., Gautel M., Sun Y.B., and Irving M.. 2014. Myosin binding protein-C activates thin filaments and inhibits thick filaments in heart muscle cells. Proc. Natl. Acad. Sci. USA. 111:18763–18768. 10.1073/pnas.1413922112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobirumaki-Shimozawa, F., Oyama K., Shimozawa T., Mizuno A., Ohki T., Terui T., Minamisawa S., Ishiwata S., and Fukuda N.. 2016. Nano-imaging of the beating mouse heart in vivo: Importance of sarcomere dynamics, as opposed to sarcomere length per se, in the regulation of cardiac function. J. Gen. Physiol. 147:53–62. 10.1085/jgp.201511484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther, P.K., Winkler H., Taylor K., Zoghbi M.E., Craig R., Padrón R., Squire J.M., and Liu J.. 2011. Direct visualization of myosin-binding protein C bridging myosin and actin filaments in intact muscle. Proc. Natl. Acad. Sci. USA. 108:11423–11428. 10.1073/pnas.1103216108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkusch, S., and Heilemann M.. 2016. Extracting quantitative information from single-molecule super-resolution imaging data with LAMA - LocAlization Microscopy Analyzer. Sci. Rep. 6:34486. 10.1038/srep34486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubara, I., and Millman B.M.. 1974. X-ray diffraction patterns from mammalian heart muscle. J. Mol. Biol. 82:527–536. 10.1016/0022-2836(74)90246-0 [DOI] [PubMed] [Google Scholar]
- McConnell, B.K., Jones K.A., Fatkin D., Arroyo L.H., Lee R.T., Aristizabal O., Turnbull D.H., Georgakopoulos D., Kass D., Bond M., et al. 1999. Dilated cardiomyopathy in homozygous myosin-binding protein-C mutant mice. J. Clin. Invest. 104:1235–1244. 10.1172/JCI7377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Li A., Smith N.J., Lal S., Graham R.M., Kooiker K.B., van Dijk S.J., Remedios C.G.D., Harris S.P., and Cooke R.. 2016. Ablation of cardiac myosin binding protein-C disrupts the super-relaxed state of myosin in murine cardiomyocytes. J. Mol. Cell. Cardiol. 94:65–71. 10.1016/j.yjmcc.2016.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara, J.W., Singh R.R., and Sadayappan S.. 2019. Cardiac myosin binding protein-C phosphorylation regulates the super-relaxed state of myosin. Proc. Natl. Acad. Sci. USA. 116:11731–11736. 10.1073/pnas.1821660116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millman, B.M. 1998. The filament lattice of striated muscle. Physiol. Rev. 78:359–391. 10.1152/physrev.1998.78.2.359 [DOI] [PubMed] [Google Scholar]
- Moss, R.L., Fitzsimons D.P., and Ralphe J.C.. 2015. Cardiac MyBP-C regulates the rate and force of contraction in mammalian myocardium. Circ. Res. 116:183–192. 10.1161/CIRCRESAHA.116.300561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun, J.Y., Gulick J., Robbins J., Woodhead J., Lehman W., and Craig R.. 2011. Electron microscopy and 3D reconstruction of F-actin decorated with cardiac myosin-binding protein C (cMyBP-C). J. Mol. Biol. 410:214–225. 10.1016/j.jmb.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag, S., Trivedi D.V., Sarkar S.S., Adhikari A.S., Sunitha M.S., Sutton S., Ruppel K.M., and Spudich J.A.. 2017. The myosin mesa and the basis of hypercontractility caused by hypertrophic cardiomyopathy mutations. Nat. Struct. Mol. Biol. 24:525–533. 10.1038/nsmb.3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, S.R., Li A., Beck-Previs S., Kennedy G.G., and Warshaw D.M.. 2020. Imaging ATP consumption in resting skeletal muscle: one molecule at a time. Biophys. J. 119:1050–1055. 10.1016/j.bpj.2020.07.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary, T.S., Snyder J., Sadayappan S., Day S.M., and Previs M.J.. 2019. MYBPC3 truncation mutations enhance actomyosin contractile mechanics in human hypertrophic cardiomyopathy. J. Mol. Cell. Cardiol. 127:165–173. 10.1016/j.yjmcc.2018.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, C.E., Chamoun J., Brown L.J., and Hambly B.D.. 2007. Myosin binding protein-C: enigmatic regulator of cardiac contraction. Int. J. Biochem. Cell Biol. 39:2161–2166. 10.1016/j.biocel.2006.12.008 [DOI] [PubMed] [Google Scholar]
- Pfuhl, M., and Gautel M.. 2012. Structure, interactions and function of the N-terminus of cardiac myosin binding protein C (MyBP-C): who does what, with what, and to whom? J. Muscle Res. Cell Motil. 33:83–94. 10.1007/s10974-012-9291-z [DOI] [PubMed] [Google Scholar]
- Ponnam, S., Sevrieva I., Sun Y.B., Irving M., and Kampourakis T.. 2019. Site-specific phosphorylation of myosin binding protein-C coordinates thin and thick filament activation in cardiac muscle. Proc. Natl. Acad. Sci. USA. 116:15485–15494. 10.1073/pnas.1903033116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previs, M.J., Beck Previs S., Gulick J., Robbins J., and Warshaw D.M.. 2012. Molecular mechanics of cardiac myosin-binding protein C in native thick filaments. Science. 337:1215–1218. 10.1126/science.1223602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previs, M.J., Michalek A.J., and Warshaw D.M.. 2014. Molecular modulation of actomyosin function by cardiac myosin-binding protein C. Pflugers Arch. 466:439–444. 10.1007/s00424-013-1433-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Previs, M.J., Mun J.Y., Michalek A.J., Previs S.B., Gulick J., Robbins J., Warshaw D.M., and Craig R.. 2016. Phosphorylation and calcium antagonistically tune myosin-binding protein C’s structure and function. Proc. Natl. Acad. Sci. USA. 113:3239–3244. 10.1073/pnas.1522236113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratti, J., Rostkova E., Gautel M., and Pfuhl M.. 2011. Structure and interactions of myosin-binding protein C domain C0: cardiac-specific regulation of myosin at its neck? J. Biol. Chem. 286:12650–12658. 10.1074/jbc.M110.156646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razumova, M.V., Shaffer J.F., Tu A.Y., Flint G.V., Regnier M., and Harris S.P.. 2006. Effects of the N-terminal domains of myosin binding protein-C in an in vitro motility assay: Evidence for long-lived cross-bridges. J. Biol. Chem. 281:35846–35854. 10.1074/jbc.M606949200 [DOI] [PubMed] [Google Scholar]
- Risi, C., Belknap B., Forgacs-Lonart E., Harris S.P., Schröder G.F., White H.D., and Galkin V.E.. 2018. N-terminal domains of cardiac myosin binding protein C cooperatively activate the thin filament. Structure. 26:1604–1611.e4. 10.1016/j.str.2018.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosas, P.C., Liu Y., Abdalla M.I., Thomas C.M., Kidwell D.T., Dusio G.F., Mukhopadhyay D., Kumar R., Baker K.M., Mitchell B.M., et al. 2015. Phosphorylation of cardiac myosin-binding protein-C is a critical mediator of diastolic function. Circ. Heat Fail. 8:582–594. 10.1161/CIRCHEARTFAILURE.114.001550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadayappan, S., Gulick J., Osinska H., Martin L.A., Hahn H.S., Dorn G.W. II, Klevitsky R., Seidman C.E., Seidman J.G., and Robbins J.. 2005. Cardiac myosin-binding protein-C phosphorylation and cardiac function. Circ. Res. 97:1156–1163. 10.1161/01.RES.0000190605.79013.4d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar, S.S., Trivedi D.V., Morck M.M., Adhikari A.S., Pasha S.N., Ruppel K.M., and Spudich J.A.. 2020. The hypertrophic cardiomyopathy mutations R403Q and R663H increase the number of myosin heads available to interact with actin. Sci. Adv. 6:eaax0069. 10.1126/sciadv.aax0069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schermelleh, L., Heintzmann R., and Leonhardt H.. 2010. A guide to super-resolution fluorescence microscopy. J. Cell Biol. 190:165–175. 10.1083/jcb.201002018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider, C.A., Rasband W.S., and Eliceiri K.W.. 2012. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9:671–675. 10.1038/nmeth.2089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer, J.F., Kensler R.W., and Harris S.P.. 2009. The myosin-binding protein C motif binds to F-actin in a phosphorylation-sensitive manner. J. Biol. Chem. 284:12318–12327. 10.1074/jbc.M808850200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymborska, A., de Marco A., Daigle N., Cordes V.C., Briggs J.A.G., and Ellenberg J.. 2013. Nuclear pore scaffold structure analyzed by super-resolution microscopy and particle averaging. Science. 341:655–658. 10.1126/science.1240672 [DOI] [PubMed] [Google Scholar]
- Tam, J., Cordier G.A., Borbely J.S., Sandoval Álvarez A., and Lakadamyali M.. 2014. Cross-talk-free multi-color STORM imaging using a single fluorophore. PLoS One. 9:e101772. 10.1371/journal.pone.0101772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu, K.T. 1973. A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 57:551–565. 10.1083/jcb.57.2.551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi, D.V., Adhikari A.S., Sarkar S.S., Ruppel K.M., and Spudich J.A.. 2018. Hypertrophic cardiomyopathy and the myosin mesa: viewing an old disease in a new light. Biophys. Rev. 10:27–48. 10.1007/s12551-017-0274-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Linde, S., Wolter S., Heilemann M., and Sauer M.. 2010. The effect of photoswitching kinetics and labeling densities on super-resolution fluorescence imaging. J. Biotechnol. 149:260–266. 10.1016/j.jbiotec.2010.02.010 [DOI] [PubMed] [Google Scholar]
- Weith, A., Sadayappan S., Gulick J., Previs M.J., Vanburen P., Robbins J., and Warshaw D.M.. 2012. Unique single molecule binding of cardiac myosin binding protein-C to actin and phosphorylation-dependent inhibition of actomyosin motility requires 17 amino acids of the motif domain. J. Mol. Cell. Cardiol. 52:219–227. 10.1016/j.yjmcc.2011.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, C., Naber N., Pate E., and Cooke R.. 2014. The myosin inhibitor blebbistatin stabilizes the super-relaxed state in skeletal muscle. Biophys. J. 107:1637–1646. 10.1016/j.bpj.2014.07.075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolter, S., Löschberger A., Holm T., Aufmkolk S., Dabauvalle M.C., van de Linde S., and Sauer M.. 2012. rapidSTORM: accurate, fast open-source software for localization microscopy. Nat. Methods. 9:1040–1041. 10.1038/nmeth.2224 [DOI] [PubMed] [Google Scholar]
- Yang, Q., Sanbe A., Osinska H., Hewett T.E., Klevitsky R., and Robbins J.. 1998. A mouse model of myosin binding protein C human familial hypertrophic cardiomyopathy. J. Clin. Invest. 102:1292–1300. 10.1172/JCI3880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zessin, P.J.M., Krüger C.L., Malkusch S., Endesfelder U., and Heilemann M.. 2013. A hydrophilic gel matrix for single-molecule super-resolution microscopy. Opt. Nanoscopy. 2:4. 10.1186/2192-2853-2-4 [DOI] [Google Scholar]
- Zoghbi, M.E., Woodhead J.L., Moss R.L., and Craig R.. 2008. Three-dimensional structure of vertebrate cardiac muscle myosin filaments. Proc. Natl. Acad. Sci. USA. 105:2386–2390. 10.1073/pnas.0708912105 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows the degree of cMyBP-C phosphorylation quantified by LC-MS.
Data Availability Statement
Data described in this paper are presented in the supplementary materials.