Abstract
Mechanistic target of rapamycin complex 1 (mTORC1) regulates CD8+ T-cell differentiation and function. Despite the links between PI3K-AKT and mTORC1 activation in CD8+ T cells, the molecular mechanism underlying mTORC1 activation remains unclear. Here, we show that both the kinase activity and the death domain of DAPK1 are required for maximal mTOR activation and CD8+ T-cell function. We found that TCR-induced activation of calcineurin activates DAPK1, which subsequently interacts with TSC2 via its death domain and phosphorylates TSC2 to mediate mTORC1 activation. Furthermore, both the kinase domain and death domain of DAPK1 are required for CD8+ T-cell antiviral responses in an LCMV infection model. Together, our data reveal a novel mechanism of mTORC1 activation that mediates optimal CD8+ T-cell function and antiviral activity.
Keywords: mTORC1; CD8+ T cells, antiviral function
Subject terms: CD8-positive T cells, Acute inflammation
Introduction
The mTOR signaling pathway senses environmental cues, such as the availability of nutrients, growth factors, and cytokines, to regulate various aspects of T-cell biology, including activation, proliferation, differentiation, effector versus memory generation, and metabolism.1,2 There are two complexes of mTOR that share key components, including mTOR and mLST8.3 mTOR complex 1 (mTORC1) contains the regulatory protein raptor, and mTOR complex 2 contains the alternate member Rictor.3
The role of mTORC1 in T cells has been elucidated by the use of genetic deletion of raptor or the pharmacological inhibitor rapamycin. mTORC1 enhances the differentiation of Th1, Th2, and Th17 cells,4,5 while both positive and negative roles have been reported for Treg and T follicular helper cell differentiation.6–9 Proliferating CD8+ cytotoxic T lymphocytes (CTLs) have asymmetric inheritance of mTORC1 activity, which regulates the expression of the transcription factors T-bet and eomesodermin.10,11 This has been shown to determine the CD8+ T-cell fate, with cells maturing into either short-lived effector or long-lived quiescent memory cells.11 Inhibition of mTOR signaling by rapamycin enhances the generation of CD8+ memory T (Tmem) cells and antiviral activity.12,13 However, rapamycin treatment can also prevent the accumulation of tissue-resident CD8+ Tmem cells in tissues.14 Rheb deletion, a member of mTORC1, prevents CD8+ effector T cell (Teff) development and allows Tmem cell expansion, although these cells are incapable of developing into secondary Teff cells upon antigen challenge.15 mTOR is inhibited by tuberous sclerosis complex (TSC) 1/2 protein heterodimer. Phosphorylation of TSC2 at Ser939 leads to its disassociation with TSC1, which results in the activation of the mTOR complex.16
Despite the important roles of mTOR in T-cell differentiation and function, deletion of mTOR or raptor in T cells has no impact on T-cell development and homeostasis, as peripheral numbers and subsets of CD4+ and CD8+ T cells are largely unaffected.4,17 In T cells, mTORC1 is activated in response to various stimuli, including nutrients, growth factors, antigen receptor stimulation, and cytokines.2 Among these activators, TCR and IL-2 stimulation are two major signal inputs that are fundamental and instructive for determining the T-cell fate, including activation, differentiation, and memory generation.18 In addition to the Jak-STAT, PI3K-AKT, and MAPK pathways, IL-2 also activates the mTOR pathway. Aside from the regulation of CD4+ T-cell differentiation, IL-2 signals promote both effector T-cell generation and differentiation into memory CD8+ T cells.19–21
The conventional view of mTORC1 activation is that stimulation of TCR, as well as IL-2, induces mTORC1 activation via PI3K-PDK1-AKT signaling. This is largely based on data obtained with wortmannin and LY294002, inhibitors of the PI3K signaling pathway.11,12,22 However, wortmannin and LY294002 have well-documented off-target effects, and seminal studies by Cantrell’s group, either using genetic deletion of the p110 subunit of PI3K or specific pharmaceutical inhibitors of PI3K and AKT, showed that activation of PI3K/AKT signaling is not necessary for mTORC1 activation in CTLs.23,24 In contrast, deletion of pdrk1, which encodes PDK1, or the PDK1 inhibitor OSU-03012 abolished mTORC1 activation in CTLs.24 In addition, MALT1 and Carma1, adapter proteins that form a complex with Bcl10 to activate NF-κB, have been shown to regulate the mTORC1 pathway in an AKT-independent manner upon antigen receptor stimulation.25 Although AKT does not directly control mTORC1 activation, its activity controls key T effector gene expression, including chemokines, IFN-γ, and perforin, in CD8+ T cells.23 Thus, despite many discoveries, the detailed molecular mechanism of mTORC1 activation in T cells remains incompletely understood.
Death associated protein kinase 1 (DAPK1) belongs to a calmodulin (CaM)-regulated death associated protein-like serine/threonine kinase superfamily.26 It contains an N-terminal kinase domain, followed by an autoregulatory Ca2+/CaM-binding domain that suppresses the catalytic activity by binding to the catalytic cleft and functions as a pseudosubstrate.27–29 DAPK1 is activated by the binding of Ca2+-activated CaM to the autoregulatory/CaM-binding domain, exposing the catalytic site of the kinase. The interaction between CaM and DAPK1 is enhanced by the dephosphorylation of DAPK1 at Ser308, which serves as a marker for DAPK1 activation.28,29 In addition to the highly homologous kinase domain in the N-terminal shared with other members in the family, DAPK1 possesses a unique death domain in the C-terminal, which mediates important protein–protein interactions with other signaling molecules, including UNC5H2, KLHL20, and TSC2.30,31 Recently, it has been shown that DAPK1 mediates breast cancer cell apoptosis via p53 but activates mTORC1 and promotes tumor cell growth in p53-mutant breast cancer cells.32 This suggests that DAPK1 has a context-dependent role on cell apoptosis or cell growth. Both DAPK1 and another member of this kinase family, DRAK2, have been implicated in the regulation of antigen receptor signaling and T-cell biology.33–35 Thus, we speculated that DAPK1 may play a role in TCR- and IL-2-mediated mTORC1 activation in T cells.
In this study, we demonstrated that DAPK1 promotes TCR- and IL-2-mediated mTORC1 activation in CD8+ T cells. Both the kinase activity and the death domain of DAPK1 are required for full activation of mTORC1. DAPK1 is activated by calcineurin, and activated DAPK1 interacts with TSC2 via its death domain and phosphorylates TSC2, which results in mTORC1 activation. Deletion of either the kinase domain or death domain of DAPK1 in the T-cell compartment led to impaired lymphocytic choriomeningitis virus (LCMV) antiviral activity. These findings identify a novel molecular mechanism of mTORC1 activation in T cells and its functional significance in antiviral infection.
Materials and methods
Mice, viruses, and infections
Dapk1-KDfl/fl and Dapk1-DDfl/fl mice were generated as previously described.36,37 CD4-Cre; Dapk1-KDfl/fl (KD-deficient), CD4-Cre; Dapk1-DDfl/fl mice (DD-deficient) and P14; CD4-Cre; Dapk1-KDfl/fl mice (P14-KD-deficient) were generated in house by crossing Dapk1-KDfl/fl mice or Dapk1-DDfl/fl mice with CD4-Cre or P14 mice. C57BL/6J (CD45.2 and CD45.1) mice were purchased from Huafukang Company (Beijing, China). Mice were infected intraperitoneally (i.p.) with LCMV-Armstrong (2 × 105 PFU) at 6–8 weeks of age, and both sexes were included with randomization. All mice were housed in a specific pathogen-free animal facility, and the mouse studies were approved by the Institutional Animal Care and Use Committee.
Cell culture
CD8+ T cells were separated from the spleens and lymph nodes of 6–8-week-old mice using a mouse CD8+ T Cell Isolation Kit (Miltenyi) according to the manufacturer’s instructions. Cells were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (Gibco), 1× antibiotic with anti-mycotic (Gibco), 1 mM pyruvate (Invitrogen), 1× NEAA (Invitrogen) and 50 μM β-mercaptoethanol (Sigma) and were activated with 0.5 μg/ml anti-CD3 antibody (2C11, BioXell). After 2 days, cells were washed and resuspended in complete media containing 20 ng/ml recombinant human IL-2 (PeproTech) for 6 days to generate CTLs. For western blot analysis without inhibitor, CTLs were starved overnight with serum-free RPMI 1640 and then stimulated with either 2 μg/ml anti-CD3 (2C11, BioXell) and 2 μg/ml anti-CD28 (37.51, BioXell) or 10 ng/ml rhIL-2 for the indicated time period. For the experiments with inhibitors, CTLs were washed with RPMI 1640 medium, resuspended in RPMI 1640 medium containing 10% FBS, and preincubated with the inhibitors (2 μM AKTi-1/2, 2 μM IC87114 or LY294002, 100 nM rapamycin, 2 μM OSU, 2 μM DAPK1i) for 15 min, followed by 20 ng/ml rhIL-2 stimulation for 60 min. All inhibitors were purchased from Sigma.
Immunoblot analysis
CTL cells (1 × 107) were lysed in RIPA lysis buffer containing PMSF, proteinase inhibitor cocktail, and phosphatase inhibitor cocktail (Roche, 1 mM of each) for 30 min on ice and then centrifuged at 12,000 × g for 10 min at 4 °C. Cell lysates were separated by 10% SDS/PAGE and transferred onto PVDF membranes (Millipore). The blots were probed with the following primary antibodies overnight at 4 °C: p-p70S6KThr389 (108D2, CST), p-S6Ser235/Ser236 (D57.2.2E, CST), p-DAPK1Ser308 (DKPS308, Sigma), p-AKTThr308 (244F9, CST), p-AKTSer473 (D9E, CST), p-PLC-γTyr783 (CST), p-STAT5Tyr694 (C71E5, CST), p-NFAT/NFAT (25A10.D6. D2, Abcam), p-p65Ser536 (93H1, CST), p-Erk1/2Thr202/Tyr204 (D13.14.4E, CST), p-p38Thr180/Tyr182 (D3F9, CST), p-TSC2Ser939 (CST), p70S6K (49D7, CST), S6 (54D2, CST), DAPK1 (DAPK17, Santa), DAPK1 (Sigma), AKT (40D4, CST), PLC-γ (D9H10, CST), STAT5 (D2O6Y, CST), p65 (D14E12, CST), Erk1/2 (137F5, CST), p38 (D13E1, CST), TSC1 (D43E2, CST), TSC2 (D93F12, CST), and β-actin (13E5, CST). Afterward, the immune complexes on the membrane were incubated with a horseradish peroxidase-conjugated secondary antibody (CST) for 1 h, followed by detection using a chemiluminescence assay kit (Tiangen, Beijing).
Retroviral shRNA transduction
The shRNA sequences for PI3K, PDK1, DAPK1, calcineurin, and scrambled controls are shown in Supplementary Table 1. Complementary oligos were annealed and ligated into shRNA vector (pSIREN-RetroQ-ZsGreen) from Clontech. Positive colonies were verified by BamHI/EcoRI digestion and sequencing. The knockdown efficiency for each shRNA was measured in B16 melanoma cells by immunoblotting. Plate-E cells (Clontech) were cultured and transfected with 40 μg of plasmid using Lipofectamine 2000 for 16 h. Fresh medium was replaced and cultured for another 24 h. Retroviral supernatants were collected by centrifugation. CD8+ T cells were purified from the spleens of WT mice using a CD8+ T Cell Isolation kit (Miltenyi) and activated with plate-bound anti-CD3/CD28 (5 μg/ml of each) for 24 h and then transduced with control retrovirus or shRNA retrovirus. After 24 h, cells were restimulated with soluble anti-CD3/CD28 (2 μg/ml of each) for 60 min, and phosphorylation of S6 was determined by phosflow.
Immunofluorescence and confocal microscopy
WT CTLs were starved overnight and then stimulated with or without anti-CD3/CD28 (2 μg/ml of each) for 30 min. CTLs were adhered to polylysine-coated glass slides (Sigma) for 30 min at 37 °C. Cells were fixed in 4% paraformaldehyde and permeabilized in 0.5% Triton X-100 buffer. Cells were stained with anti-DAPK1 (DAPK17, Santa), anti-TSC2 (D93F12, CST), and anti-calcineurin (CST) antibodies and revealed by FITC-conjugated goat anti-rabbit or Cy3-conjugated donkey anti-mouse immunoglobulins (CST). DAPI was used for nuclear staining. Slides were then mounted with antifade mounting medium (Beyotime). Images were obtained with a confocal microscope (Olympus) under a 60× oil objective.
Transfection and co-immunoprecipitation
HEK293T cells were cotransfected with pRK5 DAPK1-Flag and pcDNA3 TSC2-HA plasmids for 16 h or different mutated forms of pRK5 DAPK1 together with pcDNA3.1 calcineurin-HA for 16 h. Approximately 36 h after transfection, cells were harvested and lysed in NP40 lysis buffer containing 1 mM PMSF (Roche), 1 mM protein inhibitor cocktail (Roche), and 1 mM phosphatase inhibitor cocktail (Roche) for 30 min and then centrifuged at 12,000 rpm for 10 min at 4 °C. Approximately 1 mg of cell protein lysates was precleared with protein A/G-PLUS-Agarose beads (Santa Cruz) and incubated with 2 μg of anti-Flag (Sigma) or anti-HA (Abcam) overnight. The resulting complex was pulled down with protein A/G-PLUS-Agarose beads. The pellets were washed in NP40 lysis buffer before resuspension and denatured with 2× loading buffer. The proteins were then analyzed by 10% SDS/PAGE gel electrophoresis and immunoblotted with appropriate antibodies.
Flow cytometry
Dead cells were identified using Live/Dead Fixable Dead Cell stain (Invitrogen). Surface staining was performed in PBS containing 0.5% BSA. The following antibodies were used: anti-CD4 (RM4-5), anti-CD8 (53-6.7), anti-CD44 (IM7), anti-CD45.1 (A20), anti-CD45.2 (104), anti-CD62L (MEL-14), anti-CD127 (SB/199), anti-KLRG1 (2F1/KLRG1), anti-CCR7 (4B12), anti-CXCR3 (CXCR3-173), and anti-TCRVα2 (B20.1). Following surface staining, intracellular staining was performed with a Cytofix/Cytoperm Fixation/Permeabilization Kit (BD) according to the manufacturer’s recommendations. For intracellular cytokine production analysis, splenocytes were first stimulated by GP33 (amino acid sequence: KAVYNFATM) peptides (0.2 μg/ml, absin) and Golgiplug (BD) for 5 h at 37 °C. The following antibodies were used: anti-IFN-γ (XMG1.2), anti-TNF-α (MP6-XT22), anti-IL-2 (JES6-5H4), and anti-granzyme B (GB11). Staining of transcription factors was performed with the Foxp3/Transcription Factor Staining Buffer Set (eBioscience) according to the manufacturer’s recommendations. The following antibodies were used: anti-Foxp3 (FJK-16s), anti-Eomes (Dan11mag), anti-TCF-1 (C63D9), anti-Bcl-2 (3F11), anti-T-bet (4B10), and anti-Ki-67 (SolA15). When required, cells were washed and then incubated for 30 min with secondary antibody (Alexa Fluor 488/647-conjugated goat antibody to rabbit immunoglobulin G (Abcam)). For phosphorylation-specific flow cytometry, cells were fixed and permeabilized with Phosflow Lyse/Fix (BD) and Phosflow Perm Wash Buffer (BD) according to the manufacturer’s recommendations and were subsequently stained for 30 min at room temperature with phosphorylation-specific antibodies. The following antibodies were used: anti-p-S6 (D57.2.2E) and anti-p-4E-BP1 (236B4). When required, cells were washed and then incubated for 30 min with secondary antibody (Alexa Fluor 488/647-conjugated goat antibody to rabbit immunoglobulin G). All fluorochrome-conjugated antibodies were purchased from BD PharMingen, eBioscience, Biolegend, or CST. Data acquisition and analysis were performed on a FACS LSR II or FACS Verse (BD) and with FlowJo software (Tree Star).
Quantitative RT-PCR
WT and KD-deficient mice were infected with LCMV. At 8 dpi, splenic CD44+CD8+ T cells were purified with negative selection. Spleen cells were incubated with biotin-conjugated antibody cocktail for CD19, B220, CD11b, CD11c, CD4, Ter119, and CD62L for 30 min and then incubated with magnetic beads coated with anti-biotin Abs (Miltenyi) for 30 min, followed by negative selection. Total RNA was extracted by Trizol (Life Technologies).
To determine the tissue virus load, tissues were ground, and viral RNAs were extracted using a TIANamp Virus DNA/RNA Kit (TIANGEN). A reverse-transcriptase reaction was performed with a ReverTra Ace qPCR RT Kit (TOYOBO). The resulting cDNA was analyzed for the expression of various genes with SYBR Green Realtime PCR Master Mix (TOYOBO) on a CFX96 Touch Real-Time System (Bio-Rad).
Gene expression was normalized to that of the control gene Gapdh. The following primer sets were used, as given in Supplementary Table 1.
Adoptive transfer
P14+CD8+ T cells were purified from P14; CD45.2; CD4-Cre; Dapk1-KDfl/fl mice (P14-KD-deficient) and P14; CD45.2 WT mice with biotin-conjugated CD8 antibodies and Streptavidin MicroBeads (Miltenyi Biotec). A total of 5 × 105 P14+CD8+ T cells were injected intravenously (i.v.) into C57/B6J CD45.1 or CD4-Cre; Dapk1-KDfl/fl mice (KD-deficient) recipient mice, and 12 h later, they were infected i.p. with LCMV-Armstrong (2 × 105 PFU). After 8 days, mice were killed for experiments.
Statistical analysis
Statistical analysis was performed with Prism Software (GraphPad 7.0) by paired or unpaired t test or one-way ANOVA. A p value of < 0.05 was considered significant.
Results
DAPK1 activity, but not PI3K and AKT activity, is required for mTORC1 activation in cytolytic T cells
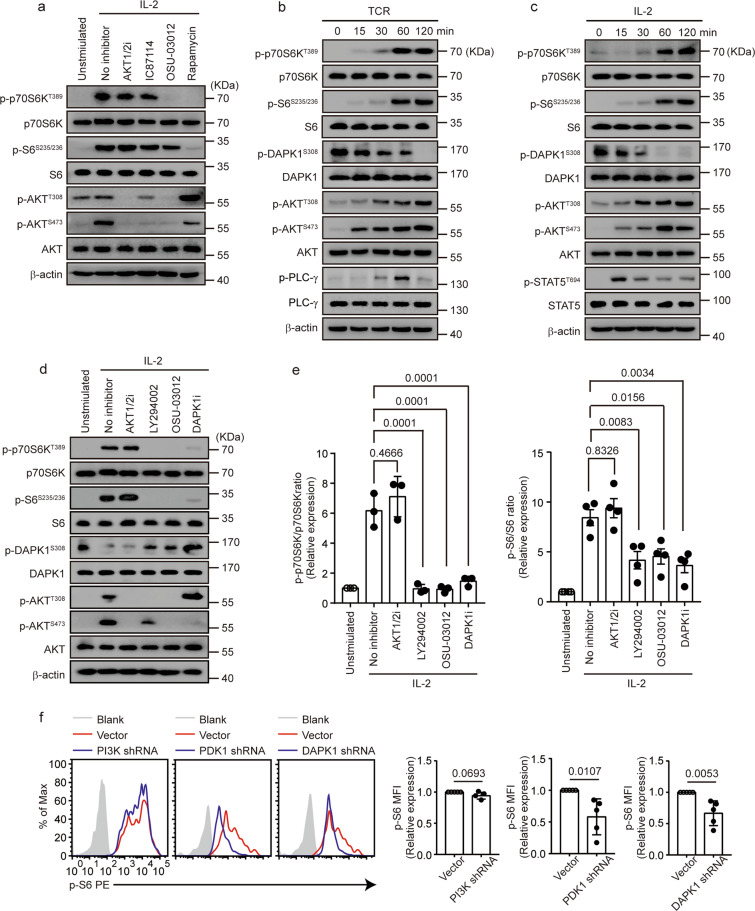
To clarify the roles of PI3K and AKT in mTORC1 activation, we measured the phosphorylation of ribosomal protein S6 kinase β1 (known as p70S6K) and ribosomal protein S6, two targets downstream of mTORC1, in the presence or absence of multiple pharmaceutical inhibitors in CTLs upon IL-2 stimulation. Inhibitors of AKT (AKT1/2i), PI3K (IC87114), and PDK1 (OSU-03012) suppressed phosphorylation of AKT at the threonine 308 and serine 473 sites (Fig. 1a). Consistent with previous reports,23,24 inhibitors of AKT and PI3K did not significantly inhibit mTORC1 activation, whereas the addition of the PDK1 inhibitor OSU-03012 and rapamycin noticeably suppressed both p70S6K and S6 phosphorylation (Fig. 1a). In line with a previous study,38 rapamycin suppressed IL-2-induced AKT phosphorylation at serine 473 but enhanced phosphorylation of AKT at 308 (Fig. 1a), probably due to reduced expression of PTEN. DAPK1 activation requires the dephosphorylation of Ser308 within the autoinhibitory domain.28,29 To probe the potential role of DAPK1 in mTORC1 activation in CTLs, we assessed DAPK1 activation upon TCR and IL-2 stimulation in CTLs by measuring the phosphorylation status of DAPK1 at Ser308. TCR and IL-2 stimulation induced either PLC-γ phosphorylation or STAT5 phosphorylation, respectively. In contrast, both induced p70S6K and S6 phosphorylation and AKT phosphorylation at both the Thr308 and Ser473 sites (Fig. 1b, c). In addition, both TCR and IL-2 induced rapid dephosphorylation of DAPK1 within 15 min, which was prolonged upon further stimulation (Fig. 1b, c), suggesting that both stimuli activate DAPK1. Next, we used a DAPK1 inhibitor to evaluate whether the kinase activity of DAPK1 is required for IL-2-mediated mTORC1 activation. LY294002, a commonly used less specific PI3K inhibitor, significantly inhibited both p70S6K and S6 phosphorylation (Fig. 1d, e). Similarly, OSU-03012 and DAPK1i suppressed p70S6K and S6 phosphorylation in a dose-dependent manner (Fig. 1d, e and Supplementary Fig. 1a, b). In contrast to AKTi-1/2, LY294002 and OSU-03012 partially reversed the dephosphorylation of DAPK1 at Ser308 (Fig. 1d), indicating that these two inhibitors also inhibited DAPK1 activation. The presence of DAPK1i suppressed AKT phosphorylation at Ser473 but enhanced the phosphorylation of AKT at Thr308 (Fig. 1d and Supplementary Fig. 1b), similar to rapamycin (Fig. 1a). To further substantiate this, we transduced retrovirus-encoding shRNAs against the PI3K subunit p100γ, PDK1, and DAPK1 into activated CTLs and measured S6 phosphorylation upon IL-2 stimulation. We found that suppression of PDK1 and DAPK1, but not the PI3K subunit p110γ, significantly inhibited S6 phosphorylation (Fig. 1f). Together, these data suggest that DAPK1 activity, but not that of PI3K and AKT, contributes to TCR- and IL-2-induced mTORC1 activation.
Fig. 1.
TCR- and IL-2-induced DAPK1 activity is required for mTORC1 activation in CTLs. a–e WT CTLs were starved overnight with serum-free RPMI 1640. a WT CTLs were preincubated with or without inhibitors (2 μM AKTi-1/2, 2 μM IC87114, 2 μM OSU-03012, 100 nM rapamycin) for 15 min, followed by or without 10 ng/ml rhIL-2 stimulation for 60 min. Phosphorylated p70S6K, S6, AKT-T308 and -S473, and total proteins were determined by immunoblot analysis. WT CTLs were stimulated with either anti-CD3/CD28 (2 μg/ml of each) (b) or rhIL-2 (20 ng/ml) (c) for the indicated times. Phosphorylation of PLC-γ, STAT5, p70S6K, S6, AKT-T308 and -S473, and DAPK1-S308 and loading controls were determined by immunoblot analysis. d, e WT CTLs were preincubated with or without the inhibitors (2 μM AKTi-1/2, 2 μM LY294002, 2 μM OSU-03012, 2 μM DAPK1i) for 15 min, followed by or without 10 ng/ml rhIL-2 stimulation for 60 min. Phosphorylated p70S6K, S6, AKT-T308 and -Ser473, and DAPK1-S308 and total proteins were determined by immunoblot analysis. Representative blots are shown in d, and the band intensities were quantified and analyzed for statistical significance in e. f Naive CD44lowCD62Lhigh CD8+ T cells were purified from the spleens of WT mice and then stimulated with anti-CD3/CD28 (2 μg/ml of each) in complete RPMI 1640 media for 24 h. Activated CD8+ T cells were transduced with control retrovirus, or retrovirus-encoding PI3K shRNA, PDK1 shRNA, or DAPK1 shRNA, cultured for 24 h and restimulated with anti-CD3/CD28 (2 μg/ml) for 60 min. Phosphorylation of S6 in the transduced CD8+ T cells was measured by flow cytometry. The data shown are representative of at least three independent experiments. Significance was determined by the unpaired t test (e, f). Error bars are the mean ± SD
Both the kinase domain and death domain of DAPK1 are required for mTORC1 activation
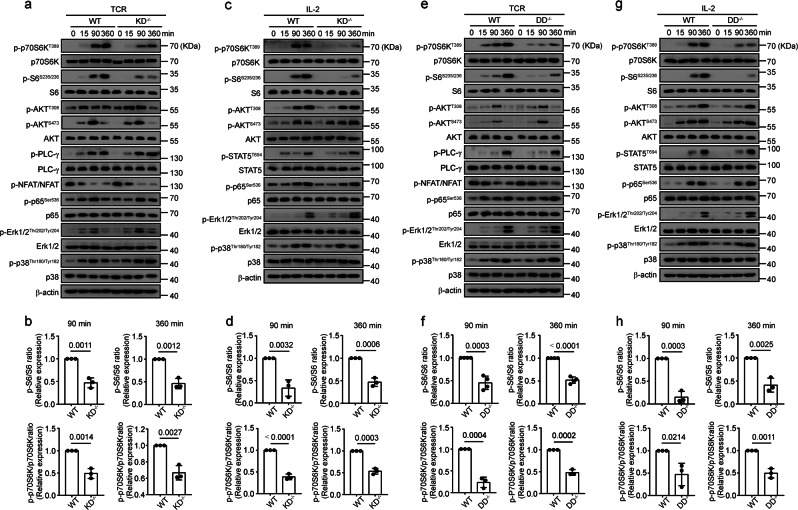
To address the limitations of inhibitors and further substantiate the potential function of DAPK1 in mTORC1 activation, we generated T-cell-specific Dapk1 kinase domain-deficient mice (KD−/−) and Dapk1 death domain-deficient mice (DD−/−) by crossing CD4-Cre transgenic mice with DAPK1 kinase domain floxed mice (KDfl/fl mice) or DAPK1 death domain floxed mice (DDfl/fl mice). These mice showed no obvious abnormalities in development and inflammation at steady state (data not shown); both DAPK1-KD-deficient mice and DD-deficient mice contained normal percentages and distributions of peripheral and thymic CD4+ and CD8+ T cells compared with those of WT mice (data not shown).
Deletion of the kinase domain of DAPK1 resulted in significantly reduced phosphorylation of p70S6K and S6 at 90 and 360 min after stimulation, but the activation of AKT-T308, AKT-S473, NFAT, p65, Erk, and p38 was not affected upon either TCR or IL-2 stimulation (Fig. 2a–d). This was associated with reduced CD8+ T-cell activation, proliferation, and enhanced apoptosis in vitro (data not shown). TCR-stimulated PLC-γ activation and IL-2-induced STAT5 phosphorylation were not affected by the deletion of the DAPK1 kinase domain (Fig. 2a–d). Next, we measured the effect of TCR and IL-2 stimulation in the absence of the DAPK1 death domain. Similarly, the DD-deficient mouse CTLs demonstrated a significant reduction in the phosphorylation of p70S6K and S6 but no effects on other signaling pathways upon either TCR or IL-2 stimulation compared with wild-type animals (Fig. 2e–h). We obtained similar results with flow cytometry on TCR-induced S6 phosphorylation at 15 and 30 min in both KD-deficient and DD-deficient CD8+ T cells (data not shown).
Fig. 2.
Both the kinase domain and death domain of DAPK1 are required for mTORC1 activation. WT and KD-deficient CTLs were starved overnight with serum-free RPMI 1640 overnight and then stimulated with anti-CD3/CD28 (2 μg/ml of each) (a, b) or rhIL-2 (20 ng/ml) (c, d) for the indicated times. Expression of phosphorylated forms and total proteins of p70S6K, S6, AKT-T308 and -S473, NFAT, p65, Erk1/2, and p38 were determined by immunoblot analysis. PLC-γ and STAT5 activation were measured in a and c, respectively. Representative graphs are shown in a and c, and the band intensities were quantified and analyzed for statistical significance in b and d. To detect NFAT phosphorylation, cytosolic fractions were isolated using a cytosolic/nuclear fractionation kit, and NFAT phosphorylation was determined by immunoblot. WT and DD-deficient CTLs were starved with serum-free RPMI 1640 media and then stimulated with anti-CD3/CD28 (2 μg/ml of each) (e, f) or rhIL-2 (20 ng/ml) (g, h) for the indicated times. Immunoblot analysis was performed as in a–d
Together, these data demonstrate that both the kinase domain and the death domain of DAPK1 contribute to TCR- and IL-2-induced mTORC1 activation.
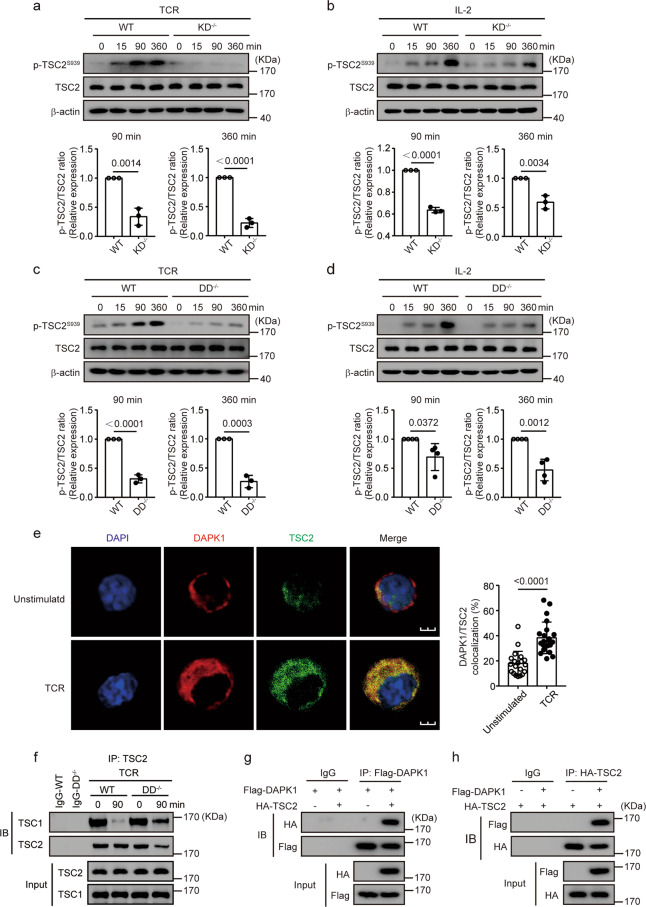
DAPK1 interacts with TSC2 via its death domain and mediates TSC2-S939 phosphorylation
Phosphorylation of TSC2 at serine 939 promotes its disassociation with TSC1, which activates mTOR. To investigate whether DAPK1 is involved in TSC2 phosphorylation, we first compared the phosphorylation of TSC2-S939 after TCR stimulation in KD-deficient CTLs with wild-type CTLs. While both TCR and IL-2 induced TSC2-S939 phosphorylation in WT CTLs, TSC2-S939 phosphorylation was significantly reduced in KD-deficient cells (Fig. 3a, b). In addition, we found that DD-deficient CTLs had significantly reduced phosphorylation of TSC2 at Ser939 upon either TCR or IL-2 stimulation compared with that of wild-type CTLs (Fig. 3c, d). Next, we used immunofluorescence staining to determine whether DAPK1 is colocalized with TSC2 upon TCR stimulation. TCR stimulation resulted in significantly enhanced colocalization of DAPK1 with TSC2 (Fig. 3e). Furthermore, stimulation of TCR for 90 min led to a reduction in the amount of TSC1 that could be precipitated with antibodies against TSC2 in wild-type CTLs (Fig. 3f, lanes 3 and 4). This was significantly impaired in DAPK1-DD CTLs (Fig. 3f, lanes 5 and 6). To further substantiate this, we overexpressed Flag-tagged DAPK1 and HA-tagged TSC2 in HEK293T cells. Immunoprecipitation of Flag-DAPK1 with anti-Flag antibodies could pull down HA-tagged TSC2; conversely, immunoprecipitation of HA-TSC2 with anti-HA antibodies could pull down Flag-tagged DAPK1 (Fig. 3g, h), indicating the association of DAPK1 with TSC2.
Fig. 3.
DAPK1 interacts with TSC2 in CTLs via its death domain. WT and DAPK1-KD-deficient CTLs were starved overnight with serum-free RPMI 1640 and then stimulated with 2 μg/ml anti-CD3/CD28 (a) or 20 ng/ml rhIL-2 (b) for the indicated times. WT and DAPK1-DD-deficient CTLs were starved overnight with serum-free RPMI 1640 and then stimulated with 2 μg/ml anti-CD3/CD28 (c) or 20 ng/ml rhIL-2 (d) for the indicated times. Phosphorylation of TSC2-Ser939, TSC2, and β-actin was determined by immunoblot analysis (a–d). The band intensities were quantified and analyzed for statistical significance. e WT CTLs were starved overnight with serum-free RPMI 1640 and then stimulated with or without plate-bound anti-CD3/CD28 (2 μg/ml of each) for 60 min. Representative immunofluorescence images of DAPK1 and TSC2 (n = 3 independent experiments with similar results). The percentages of colocalization areas of DAPK1 and TSC2 were quantified and analyzed for statistical significance (20 cells per group). f WT and DAPK1-DD-deficient CTLs were starved overnight with serum-free RPMI 1640. After that, cells were unstimulated or stimulated with anti-CD3/CD28 (2 μg/ml of each) for 90 min. Cell lysates were precipitated with anti-TSC2 antibodies, and immune complexes were analyzed with antibodies against TSC1 and TSC2. g, h HEK293T cells were cotransfected with plasmids of Flag-DAPK1 and HA-TSC2. Cells were cultured for another 36 h after transfection. Cell lysates were harvested and precipitated with anti-Flag (g) or anti-HA (h) antibodies. The immune complexes were separated by 10% SDS-PAGE, followed by immunoblot analysis with anti-Flag or anti-HA antibodies. The data shown are representative of three independent experiments. Statistical significance was measured by the unpaired Student’s t test (a–e). Error bars represent the mean ± SD
Together, these data suggest that DAPK1 and its death domain are required for optimal IL-2- and TCR-driven TSC2-S939 phosphorylation.
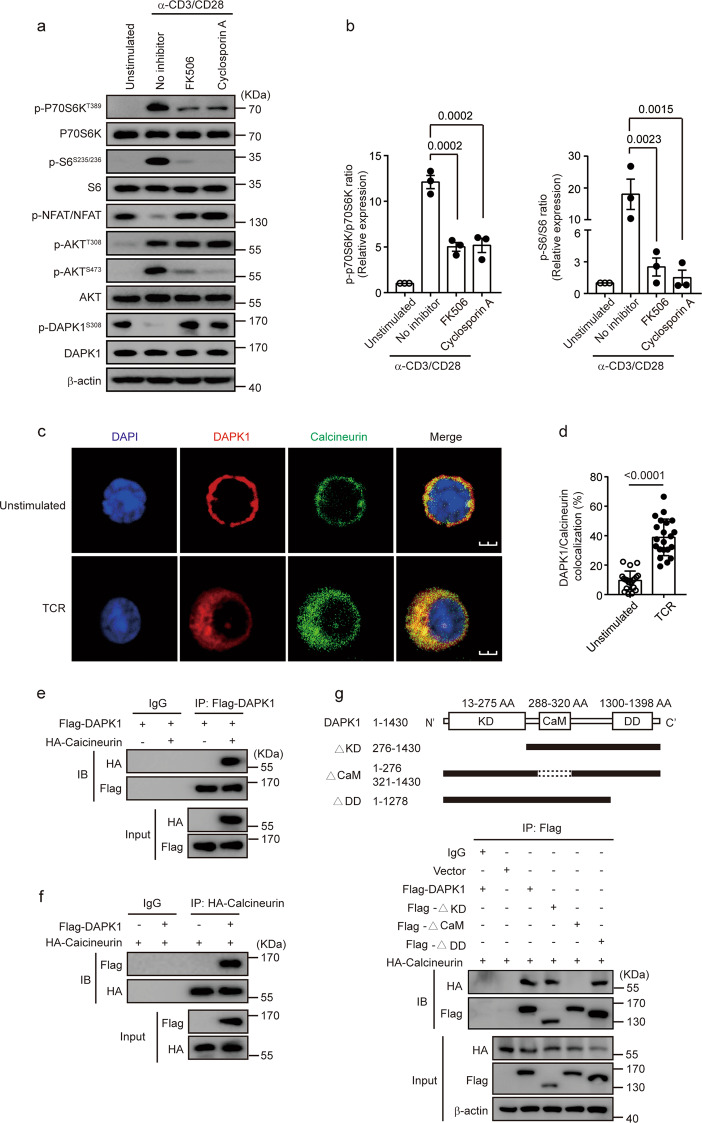
DAPK1 is activated by calcineurin upon TCR stimulation
Next, we investigated whether calcineurin, a phosphatase downstream of CaM, is required for DAPK1 activation upon TCR stimulation. TCR stimulation induced dephosphorylation of both NFAT and DAPK1 at Ser308 (Fig. 4a). Inhibition of calcineurin by either FK506 or cyclosporin A (CSA) suppressed the phosphorylation of p70S6K and S6, while TCR-induced dephosphorylation of both NFAT and DAPK1 was reversed by either FK506 or CSA (Fig. 4a, b). The inhibition of CSA suppressed NFAT dephosphorylation and S6 and p70S6K phosphorylation in a dose-dependent manner (Supplementary Fig. 2a). Knockdown of calcineurin expression with retroviral shRNA against calcineurin resulted in similar inhibition of TCR-induced S6 phosphorylation (Supplementary Fig. 2b). Next, we investigated whether DAPK1 was associated with calcineurin upon TCR stimulation by immunofluorescence staining. TCR stimulation of CTLs led to significant colocalization of DAPK1 with calcineurin (Fig. 4c, d). Next, we overexpressed Flag-tagged DAPK1 and HA-calcineurin in HEK293T cells and tested whether DAPK1 was associated with calcineurin. Reciprocal co-immunoprecipitation assays revealed that DAPK1 was associated with calcineurin (Fig. 4e, f). To further map the binding domain of DAPK1 with calcineurin, we generated full-length Flag-tagged DAPK1 or different truncated forms lacking the kinase domain, the CaM-binding domain, or the death domain (Fig. 4g). We cotransfected the HA-calcineurin plasmid with either full-length Flag-tagged DAPK1 or the truncated forms. The full length of DAPK1, as well as the kinase domain and death domain truncated forms of DAPK1, but not the CaM truncated form, were able to precipitate calcineurin (Fig. 4h).
Fig. 4.
Calcineurin interacts with DAPK1 and mediates DAPK1 activation in CTLs. a, b WT CTLs were starved overnight with serum-free RPMI 1640. Then, CTLs were preincubated with the inhibitors (2 μM FK506 and 500 nM cyclosporin A) for 15 min and stimulated without or with anti-CD3/CD28 (2 μg/ml of each) for 60 min. Phosphorylation of p70S6K, S6, NFAT, AKT-T308 and -S473, and DAPK1-S308 and total proteins were determined by immunoblot. Representative graphs are shown in a, and the western blot results were quantified and analyzed for statistical significance in b. c, d WT CTLs were starved overnight with serum-free RPMI 1640 and then stimulated with or without anti-CD3/CD28 (2 μg/ml of each) for 60 min. Representative immunofluorescence images of DAPK1 and calcineurin (n = 3 independent experiments with similar results). The percentages of colocalization areas of DAPK1 and calcineurin were quantified and analyzed for statistical significance (20 cells per group). e, f HEK293T cells were cotransfected with Flag-DAPK1 and HA-calcineurin. Cells were cultured for another 24 h after transfection. Cell lysates were harvested and precipitated with anti-Flag (e) or anti-HA (f) antibodies. The immune complexes were separated by 10% SDS-PAGE, followed by immunoblot analysis with anti-Flag or anti-HA antibodies. g Schematic representatives of full-length DAPK1 and mutated DAPK1 forms. HEK293T cells were cotransfected with either full-length DAPK1 or mutated DAPK1, together with HA-calcineurin. Cells were cultured for another 36 h after transfection. Cell lysates were harvested and precipitated with anti-Flag antibodies. The immune complexes were separated by 10% SDS-PAGE, followed by immunoblot analysis with anti-Flag or anti-HA antibodies. The data shown are representative of three independent experiments. Statistical significance was measured by the unpaired Student’s t test (b, d). Error bars represent the mean ± SD
Together, these data suggest that TCR stimulation leads to the binding of DAPK1 with calcineurin via its CaM-binding domain but that the death domain is dispensable for the interaction of DAPK1 with calcineurin.
Both the kinase domain and death domain of DAPK1 are required for the antiviral CD8+ T-cell response
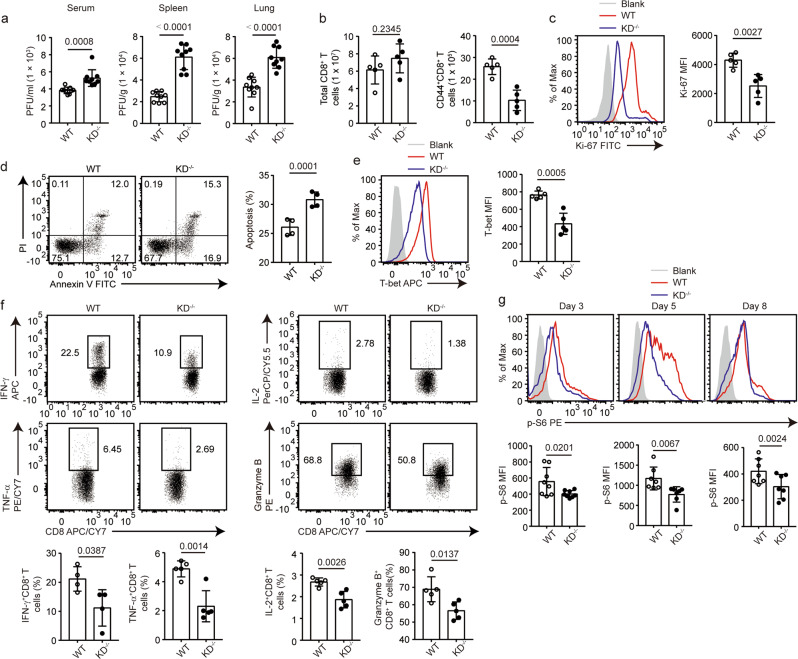
To examine the role of DAPK1 in the antiviral CD8+ T-cell response, we infected WT mice and DAPK1-KD−/− mice with the Armstrong strain of LCMV. The KD-deficient mice had enhanced viral titers in serum, spleen, and lung (Fig. 5a), suggesting that the kinase activity of DAPK1 is required for antiviral activity. Although the total amounts of CD8+ T cells were comparable in the spleens of WT and KD-deficient mice, the numbers of activated CD44+CD8+ T cells were reduced in the KD-deficient mice compared with those in WT control mice (Fig. 5b). This was accompanied by reduced Ki-67 staining in the CD44+CD8+ compartments of the DAPK1-KD-deficient mice compared with staining in the control mice (Fig. 5c), suggesting that DAPK1 activity is required for CD8+ T-cell proliferation upon virus infection. Furthermore, deletion of the kinase domain of DAPK1 led to enhanced CD8+ T-cell apoptosis (Fig. 5d). This was associated with a significant reduction in Bcl-2 mRNA and protein expression in KD-deficient CD8+ T cells (Supplementary Fig. 3a).
Fig. 5.
DAPK1 regulates proliferation, apoptosis, and cytotoxicity of CTLs during acute viral infection. WT and DAPK1-KD-deficient mice were infected with 2 × 105 PFU LCMV-Armstrong virus i.p. a At 5 dpi, sera, spleens, and lungs were collected from infected WT and KD-deficient mice (n = 9). Viral RNAs were extracted, and tissue viral loads were determined by quantitative RT-PCR. b Numbers of splenic total CD8+ T cells and CD44+CD8+ T cells in WT and KD-deficient mice at 8 dpi with LCMV. The expression of Ki-67 (c), percentage of apoptosis (d), expression of T-bet (e), and the frequencies of IFN-γ, TNF-α, IL-2, and granzyme B (f)-producing cells in splenic CD44+CD8+ T cells at 8 dpi with LCMV were determined by flow cytometry. Phosphorylation of S6 (g) in splenic CD44+CD8+ T cells was determined by phosflow staining at 3, 5, and 8 dpi with LCMV. The data shown are representative of at least three independent experiments (b–g, n = 5). Statistical significance was measured by the unpaired Student’s t test. Error bars are the mean ± SD
Next, we compared the expression of CD8+ effector molecules between WT and DAPK1-KD-deficient mice. Compared with CD44+CD8+ T cells isolated from WT mice, CD44+CD8+ T cells from DAPK1-KD-deficient mice expressed less T-bet by flow cytometry as measured by mean fluorescence intensity (Fig. 5e). Deletion of the DAPK1 kinase domain significantly reduced the percentage of IFN-γ, TNF-α, IL-2, and granzyme B-expressing cells by flow cytometry (Fig. 5f), and granzyme b, granzyme k, Ifng, Tbx21, and Klrg1 mRNA expression was reduced in KD-deficient CD8+ T cells compared with expression in wild-type controls (Supplementary Fig. 3b). Furthermore, the CD44+CD8+ T cells isolated from the KD-deficient mice had a significant reduction in mTORC1 activation as measured by the phosphorylation of protein S6 on days 3, 5, and 8 post infection (Fig. 5g). In line with this, at 8 dpi (day post infection), the KD-deficient CD44+CD8+ T cells had reduced expression of genes related to glycolysis compared with WT counterparts (Supplementary Fig. 3c).
We obtained similar results using DD-deficient mice (Supplementary Fig. 4a). The percentages and numbers of viral antigen-specific gp33+CD44+CD8+ T cells recovered from the spleens of DD-deficient mice were significantly reduced compared with cells isolated from WT mice (Supplementary Fig. 4b). CD44+CD8+ T cells from DD-deficient mice expressed less granzyme B (Supplementary Fig. 4c), while the phosphorylation of S6 was reduced in the activated DD-deficient CD44+CD8+ T cells upon virus infection (Supplementary Fig. 4d) compared with those of the WT cells.
Together, these data demonstrate that both the kinase domain and the death domain of DAPK1 in T cells are required for mounting an efficient antiviral response during an acute viral infection.
A cell-intrinsic requirement of DAPK1 activity in the antiviral CD8+ T-cell response
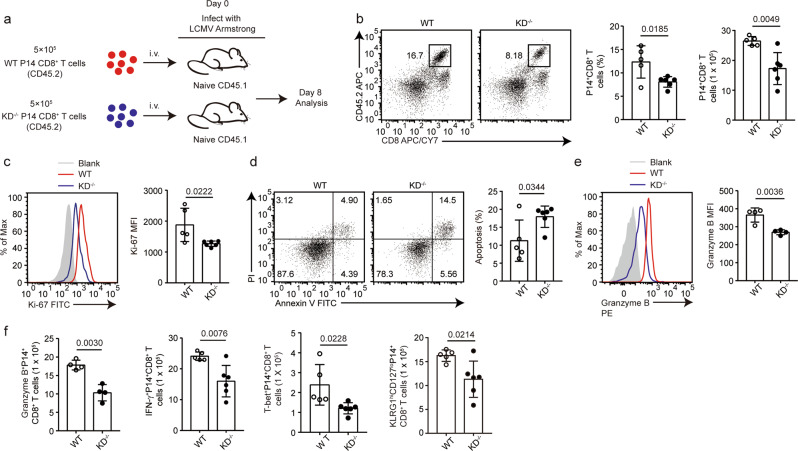
To determine if the requirement of DAPK1 activity for antiviral CD8+ T-cell response is intrinsic to T cells, we injected WT P14 CD8+ T cells or P14 CD4-Cre; KDfl/fl CD8+ T cells into congenic WT mice and infected the mice with the LCMV-Armstrong strain (Fig. 6a). At 8 dpi, the percentages and numbers of antigen-specific KD-deficient P14+CD44+CD8+ T cells were significantly reduced compared with those of WT control P14+CD44+CD8+ T cells (Fig. 6b). Deletion of the kinase domain of DAPK1 led to reduced proliferation and enhanced apoptosis of P14+CD8+ T cells (Fig. 6c, d). Furthermore, KD-deficient P14+CD8+ T cells recovered from the WT recipient mice expressed less granzyme B compared with that of the wild-type counterpart cells, as measured by percentage and mean fluorescence intensity (Fig. 6e). In addition, the total numbers of KD-deficient GZB+P14+ CD8+, IFN-γ+P14+CD8+, T-bet+P14+CD8+, and KLRG1highCD127lowP14+CD8+ T cells recovered from the WT recipient mice were significantly less than those of the WT counterpart cells (Fig. 6f).
Fig. 6.
DAPK1 promotes antigen-specific CD8+ T-cell responses during acute viral infection. a Antigen-specific P14+CD8+ T cells were purified with magnetic beads from P14; CD45.2 (P14-WT) and P14; CD45.2; CD4-Cre; Dapk1-KDfl/fl mice (P14-KD-deficient) mice. On day 0, 5 × 105 WT P14+CD8+ T cells or KD-deficient P14+CD8+ T cells were injected i.v. into C57/B6J CD45.1 recipient mice. After 12 h, the recipient mice were infected i.p. with LCMV-Armstrong (2 × 105 PFU) virus for 8 days. b Frequency and number of splenic P14+CD8+ T cells in recipient mice. The expression of Ki-67 (c), percentage of apoptosis (d), and expression of granzyme B (e) in WT P14+CD8+ T cells or KD−/− P14+CD8+ T cells recovered from recipient mice were determined by flow cytometry. f The numbers of granzyme B+, IFN-γ+, T-bet+, and terminal effective (KLRG1highCD127low) WT P14+CD8+ T cells or KD−/− P14+CD8+ T cells were quantified. The data shown are representative of at least two independent experiments (n ≧ 5 per group). Statistical significance was measured by the unpaired Student’s t test. Error bars are the mean ± SD
Next, we injected WT P14 CD8+ T cells into congenic WT or CD4-Cre+; DAPK1-KDfl/fl recipient mice, followed by LCMV infection (Supplementary Fig. 5a). The percentages of P14+CD44+CD8+ T cells at 8 dpi recovered from either WT or KD-deficient mice were similar (Supplementary Fig. 5b). These cells showed similar proliferative activity and apoptosis (Supplementary Fig. 5c, d). The expression levels of granzyme B were comparable between cells recovered from WT and KD-deficient recipient mice (Supplementary Fig. 5e).
Taken together, these data demonstrate that the requirement of DAPK1 activity for the CD8+ antiviral T-cell response is T-cell intrinsic.
DAPK1 activity in T cells promotes CD8+ Tmem-cell formation after virus infection
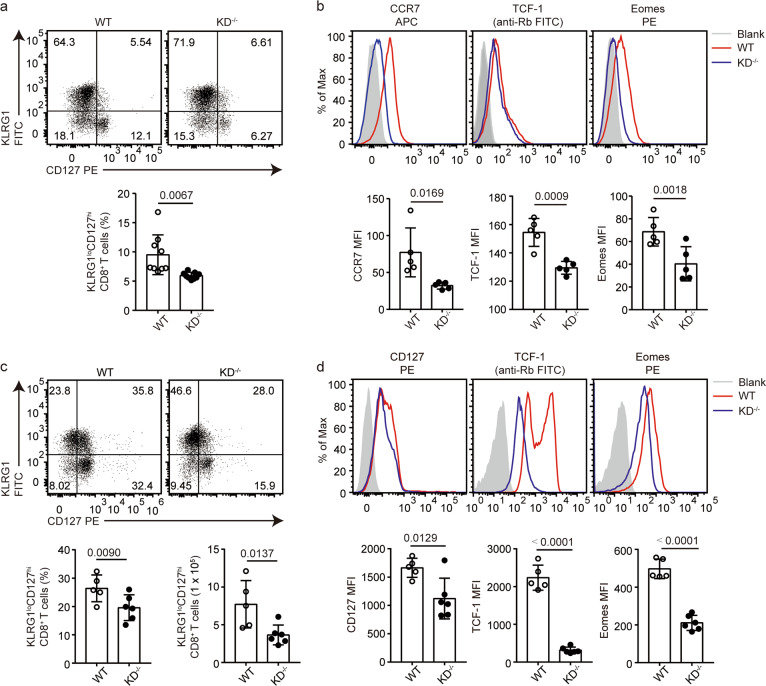
Next, we asked whether DAPK1 activity in T cells plays a role in CD8+ T-cell memory formation. To test this hypothesis, we infected WT and KD-deficient mice with the LCMV-Armstrong strain and compared the percentages of CD127highKLRG1low memory precursor cells (MPSCs) in the population of CD44+CD8+ T cells at 8 dpi. The KD-deficient mice had significantly reduced CD127highKLRG1low MPSCs compared with levels in WT mice (Fig. 7a). Activated KD-deficient CD44+CD8+ T cells expressed less of the memory-related markers CCR7, TCF-1, and Eomes (Fig. 7b).
Fig. 7.
DAPK1 activity in T cells promotes CD8+ Tmem-cell formation after virus infection. WT and DAPK1-KD-deficient mice were infected with 2 × 105 PFU LCMV-Armstrong virus i.p. a At 8 dpi, the expression of KLRG1 and CD127 was determined by flow cytometry in WT and DAPK1-KD-deficient splenic CD44+CD8+ T cells, and the histograms depict the percentage of KLRG1lowCD127high cells. b At 8 dpi, expression of CCR7, TCF-1, and Eomes in WT and DAPK1-KD-deficient splenic CD44+CD8+ T cells was analyzed by flow cytometry. c At 30 dpi, the expression of KLRG1 and CD127 was determined in WT and DAPK1-KD-deficient splenic CD44+CD8+ T cells, and the percentage and number of KLRG1lowCD127high cells are depicted. d At 30 dpi, WT and DAPK1-KD-deficient splenic CD44+CD8+ T cells were analyzed for their expression of CD127, TCF-1, and Eomes. The data shown are representative of at least three experiments (n ≧ 5 per group). Statistical significance was measured by the unpaired Student’s t test. Error bars are the mean ± SD
We compared CD127highKLRG1low memory cells in WT or KD-deficient mice at 30 dpi with the LCMV-Armstrong strain. Similarly, the KD-deficient mice had significantly reduced CD127highKLRG1low cells compared with levels in WT mice (Fig. 7c). In addition, the mean fluorescence intensities of CCR7, TCF-1, and Eomes were reduced in the KD-deficient CD44+CD8+ T cells compared with those in WT CD44+CD8+ T cells (Fig. 7d). Together, these data demonstrate that DAPK1 activity in T cells promotes CD8+ T-cell memory formation.
Discussion
The mTOR signaling pathway plays multifaceted regulatory roles in T-cell biology, including T-cell activation, differentiation, effector function, and memory formation. In this study, we found that DAPK1 is activated by calcineurin in addition to Ca2+/CaM upon TCR stimulation and identified that DAPK1 rather than AKT mediates TCR- and IL-2-induced mTORC1 activation via interaction with TSC2 through its death domain and phosphorylation of TSC2 via its kinase activity. Furthermore, we found that both the kinase domain and the death domain of DAPK1 were required to promote optimal CD8+ T-cell antiviral activity.
Although it is well appreciated that DAPK1 activation is subjected to multiple regulatory mechanisms, including CaM binding and Ser308 dephosphorylation, in the autoregulatory domain by mutagenesis studies, little is known about how DAPK1 is activated in T cells. Our data indicate that the phosphatase calcineurin interacts with and activates DAPK1. These data suggest that calcineurin not only dephosphorylates and activates NFAT, an important transcription factor for T-cell activation, but also dephosphorylates and activates DAPK1 to drive the mTOR pathway. Calcineurin lies downstream of CaM, itself a key activator of DAPK1, and both are dependent on TCR-dependent release of Ca2+ stores. In contrast, it remains unclear how IL-2 activates DAPK1, as this is likely to occur in both a CaM- and calcineurin-independent manner.39 It is tempting to speculate that IL-2 activates DAPK1 via other unknown phosphatases, and identification of which warrants further study.
The association of DAPK1 and mTOR has been previously documented in vitro and in nonimmune cells.30,32 TSC2 has been identified to interact with the death domain of DAPK1 in vitro, and recombinant DAPK1 phosphorylates and inactivates TSC2,30 a role that has also been attributed to AKT.40 Furthermore, Dapk1−/− MEFs have reduced mTORC1 activation upon EGF stimulation.30 In p53-mutated human breast cancer cells, DAPK1 phosphorylates TSC2 to induce mTORC1 activation to mediate cancer cell growth, while in p53 intact breast cancer cells, DAPK1 activates p53 to induce apoptosis. Using a pharmacological inhibitor of DAPK1 and genetic approaches, our data showed that both DAPK1 kinase activity and the death domain are required for TCR- and IL-2-induced mTORC1 activation. Thus, the mode of DAPK1-mediated mTORC1 activation may represent a general mechanism in both nonimmune and immune cells. The assumption that AKT drives mTORC1 via phosphorylation of TSC2 may have led to an underappreciation of the importance of DAPK1.
Carma1 and Bcl10, two proteins required for NF-κB activation in immune cells, have been suggested to mediate TCR-induced mTORC1 activation in a PI3K-independent manner.25 DAPK1 has also been implicated in NF-κB activation in T cells.33 Thus, it is intriguing to speculate that the CARM complex may interact with DAPK1 to regulate not only NF-κB activation but also mTORC1 activation.
Although S6 can be phosphorylated by the PI3K, mTOR and MAPK pathways and p70S6K and S6 may play differential roles in T-cell biology,41,42 we found similar regulatory patterns of P70S6K and S6 by DAPK1. Notably, there is residual P70S6K and S6 activation in either KD-deficient or DD-deficient T cells, suggesting that other kinases may have compensatory roles in the phosphorylation of TSC2 and mTORC1 activation. Another possibility is that although PI3K/AKT is not necessary for mTORC1 activation in T cells, this signaling axis may nevertheless contribute to mTORC1 activation and related cellular events. Deletion of DAPK1 has been reported to enhance T-cell activation, accumulation of HIF-1α, and enhanced Th17 differentiation.33,34 However, in addition to mTORC1 activation, we did not find any effects of the loss of DAPK1 kinase activity and death domain on TCR- and IL-2-induced proximal signaling pathways or CD4+ T-cell lineage differentiation, including Th17 cells (data not shown).
Recently, it has been reported that pegylated IFN-α induces DAPK1 and mTOR expression, the latter activating downstream PAK-1 to mediate anti-HCV responses in primary human hepatocytes and HCVcc.43 Our work does not explain how DAPK1 mediates the expression rather than the activation of mTOR. Knockdown of mTOR expression impaired IFN-α- and DAPK1-mediated antiviral activity in hepatocytes.43 Consistent with this, both the kinase domain and death domain of DAPK1 in T cells are required for CD8+ T-cell antiviral responses in the LCMV model in our hands. In contrast to the inhibition of mTOR by rapamycin, the lack of DAPK1 activity resulted in impaired T effector expansion and memory formation. A fine-tuned regulation of mTOR activity is required for the proper function of T cells, and an emerging concept in mTOR signaling is that the magnitude, duration, and oscillation of mTORC1 activation may determine the outcome of T-cell responses.22,44 Further detailed studies on how DAPK1 and other serine kinases affect the intensity and kinetics of mTORC1 in different immune models are warranted.
In summary, our studies identified that DAPK1 is a key factor driving AKT-independent activation of mTORC1 downstream of TCR stimulation in CTLs and is required for optimal antiviral immunity in these cells.
Supplementary information
Acknowledgements
This work was supported by grants from the National Scientific Foundation of China to X.-P.Y. (81671539, 31470851, and 31870892) and Z.H.T. (81873870), and the Integrated Innovative Team for Major Human Diseases Program of Tongji Medical College, HUST (2019kfyXKJC066) to X.-P.Y.
Author contributions
Z.W., P.L., and X.-P.Y. conceived and designed the study and wrote the manuscript with critical input from G.W., Y.L., Z.L., and A.L.; Z.W., P.L., R.H., H.C.L., N.L., Y.X., G.B., Q.D., M.X., J.W., L.P., and Z.-H.T. performed the experiments and analyzed the data; Z.L. and A.L. helped analyze the data and assisted with the experimental design; X.C., H.B.L., and Y.L. provided essential reagents and assisted with experimental design and data analysis. X.-P.Y. wrote the paper and supervised the project.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Zhengping Wei, Pingfei Li
Supplementary information
The online version of this article (10.1038/s41423-019-0293-2) contains supplementary material.
References
- 1.Chi H. Regulation and function of mTOR signalling in T cell fate decisions. Nat. Rev. Immunol. 2012;12:325–338. doi: 10.1038/nri3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Powell JD, Pollizzi KN, Heikamp EB, Horton MR. Regulation of immune responses by mTOR. Annu. Rev. Immunol. 2012;30:39–68. doi: 10.1146/annurev-immunol-020711-075024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saxton RA, Sabatini DM. mTOR Signaling in growth, metabolism, and disease. Cell. 2017;169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 4.Delgoffe GM, Kole TP, Zheng Y, Zarek PE, Matthews KL, Xiao B, et al. The mTOR kinase differentially regulates effector and regulatory T cell lineage commitment. Immunity. 2009;30:832–844. doi: 10.1016/j.immuni.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delgoffe GM, Pollizzi KN, Waickman AT, Heikamp E, Meyers DJ, Horton MR, et al. The kinase mTOR regulates the differentiation of helper T cells through the selective activation of signaling by mTORC1 and mTORC2. Nat. Immunol. 2011;12:295–303. doi: 10.1038/ni.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu L, Huang Q, Wang H, Hao Y, Bai Q, Hu J, et al. The kinase mTORC1 promotes the generation and suppressive function of follicular regulatory T cells. Immunity. 2017;47:538–551. doi: 10.1016/j.immuni.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 7.Zeng H, Yang K, Cloer C, Neale G, Vogel P, Chi H. mTORC1 couples immune signals and metabolic programming to establish T(reg)-cell function. Nature. 2013;499:485–490. doi: 10.1038/nature12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeng H, Cohen S, Guy C, Shrestha S, Neale G, Brown SA, et al. mTORC1 and mTORC2 kinase signaling and glucose metabolism drive follicular helper T cell differentiation. Immunity. 2016;45:540–554. doi: 10.1016/j.immuni.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ray JP, Staron MM, Shyer JA, Ho PC, Marshall HD, Gray SM, et al. The interleukin-2-mTORc1 kinase axis defines the signaling, differentiation, and metabolism of T helper 1 and follicular B helper T cells. Immunity. 2015;43:690–702. doi: 10.1016/j.immuni.2015.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rao RR, Li Q, Odunsi K, Shrikant PA. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and eomesodermin. Immunity. 2010;32:67–78. doi: 10.1016/j.immuni.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pollizzi KN, Sun IH, Patel CH, Lo YC, Oh MH, Waickman AT, et al. Asymmetric inheritance of mTORC1 kinase activity during division dictates CD8(+) T cell differentiation. Nat. Immunol. 2016;17:704–711. doi: 10.1038/ni.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araki K, Turner AP, Shaffer VO, Gangappa S, Keller SA, Bachmann MF, et al. mTOR regulates memory CD8 T-cell differentiation. Nature. 2009;460:108–112. doi: 10.1038/nature08155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Rao RR, Araki K, Pollizzi K, Odunsi K, Powell JD, et al. A central role for mTOR kinase in homeostatic proliferation induced CD8+ T cell memory and tumor immunity. Immunity. 2011;34:541–553. doi: 10.1016/j.immuni.2011.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sowell RT, Rogozinska M, Nelson CE, Vezys V, Marzo AL. Cutting edge: generation of effector cells that localize to mucosal tissues and form resident memory CD8 T cells is controlled by mTOR. J. Immunol. 2014;193:2067–2071. doi: 10.4049/jimmunol.1400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollizzi KN, Patel CH, Sun IH, Oh MH, Waickman AT, Wen J, et al. mTORC1 and mTORC2 selectively regulate CD8(+) T cell differentiation. J. Clin. Investig. 2015;125:2090–2108. doi: 10.1172/JCI77746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang J, Kim J, Alexander A, Cai S, Tripathi DN, Dere R, et al. A tuberous sclerosis complex signalling node at the peroxisome regulates mTORC1 and autophagy in response to ROS. Nat. Cell Biol. 2013;15:1186–1196. doi: 10.1038/ncb2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang K, Shrestha S, Zeng H, Karmaus PW, Neale G, Vogel P, et al. T cell exit from quiescence and differentiation into Th2 cells depend on Raptor-mTORC1-mediated metabolic reprogramming. Immunity. 2013;39:1043–1056. doi: 10.1016/j.immuni.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaech SM, Cui W. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 2012;12:749–761. doi: 10.1038/nri3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boyman O, Sprent J. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 2012;12:180–190. doi: 10.1038/nri3156. [DOI] [PubMed] [Google Scholar]
- 20.Liao W, Lin JX, Leonard WJ. Interleukin-2 at the crossroads of effector responses, tolerance, and immunotherapy. Immunity. 2013;38:13–25. doi: 10.1016/j.immuni.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones RG, Pearce EJ. MenTORing immunity: mTOR signaling in the development and function of tissue-resident immune cells. Immunity. 2017;46:730–742. doi: 10.1016/j.immuni.2017.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pollizzi KN, Powell JD. Regulation of T cells by mTOR: the known knowns and the known unknowns. Trends Immunol. 2015;36:13–20. doi: 10.1016/j.it.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Macintyre AN, Finlay D, Preston G, Sinclair LV, Waugh CM, Tamas P, et al. Protein kinase B controls transcriptional programs that direct cytotoxic T cell fate but is dispensable for T cell metabolism. Immunity. 2011;34:224–236. doi: 10.1016/j.immuni.2011.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finlay DK, Rosenzweig E, Sinclair LV, Feijoo-Carnero C, Hukelmann JL, Rolf J, et al. PDK1 regulation of mTOR and hypoxia-inducible factor 1 integrate metabolism and migration of CD8+ T cells. J. Exp. Med. 2012;209:2441–2453. doi: 10.1084/jem.20112607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hamilton KS, Phong B, Corey C, Cheng J, Gorentla B, Zhong X, et al. T cell receptor-dependent activation of mTOR signaling in T cells is mediated by Carma1 and MALT1, but not Bcl10. Sci. Signal. 2014;7:ra55. doi: 10.1126/scisignal.2005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deiss LP, Feinstein E, Berissi H, Cohen O, Kimchi A. Identification of a novel serine/threonine kinase and a novel 15-kD protein as potential mediators of the gamma interferon-induced cell death. Genes Dev. 1995;9:15–30. doi: 10.1101/gad.9.1.15. [DOI] [PubMed] [Google Scholar]
- 27.Singh P, Ravanan P, Talwar P. Death associated protein kinase 1 (DAPK1): a regulator of apoptosis and autophagy. Front. Mol. Neurosci. 2016;9:46. doi: 10.3389/fnmol.2016.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shohat G, Spivak-Kroizman T, Cohen O, Bialik S, Shani G, Berrisi H, et al. The pro-apoptotic function of death-associated protein kinase is controlled by a unique inhibitory autophosphorylation-based mechanism. J. Biol. Chem. 2001;276:47460–47467. doi: 10.1074/jbc.M105133200. [DOI] [PubMed] [Google Scholar]
- 29.Shohat G, Shani G, Eisenstein M, Kimchi A. The DAP-kinase family of proteins: study of a novel group of calcium-regulated death-promoting kinases. Biochim. Biophys. Acta. 2002;1600:45–50. doi: 10.1016/s1570-9639(02)00443-0. [DOI] [PubMed] [Google Scholar]
- 30.Stevens C, Lin Y, Harrison B, Burch L, Ridgway RA, Sansom O, et al. Peptide combinatorial libraries identify TSC2 as a death-associated protein kinase (DAPK) death domain-binding protein and reveal a stimulatory role for DAPK in mTORC1 signaling. J. Biol. Chem. 2009;284:334–344. doi: 10.1074/jbc.M805165200. [DOI] [PubMed] [Google Scholar]
- 31.Shiloh R, Bialik S, Kimchi A. The DAPK family: a structure-function analysis. Apoptosis. 2014;19:286–297. doi: 10.1007/s10495-013-0924-5. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J, Zhao D, Poage GM, Mazumdar A, Zhang Y, Hill JL, et al. Death-associated protein kinase 1 promotes growth of p53-mutant cancers. J. Clin. Investig. 2015;125:2707–2720. doi: 10.1172/JCI70805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chuang YT, Fang LW, Lin-Feng MH, Chen RH, Lai MZ. The tumor suppressor death-associated protein kinase targets to TCR-stimulated NF-kappa B activation. J. Immunol. 2008;180:3238–3249. doi: 10.4049/jimmunol.180.5.3238. [DOI] [PubMed] [Google Scholar]
- 34.Chou TF, Chuang YT, Hsieh WC, Chang PY, Liu HY, Mo ST, et al. Tumour suppressor death-associated protein kinase targets cytoplasmic HIF-1alpha for Th17 suppression. Nat. Commun. 2016;7:11904. doi: 10.1038/ncomms11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGargill MA, Wen BG, Walsh CM, Hedrick SM. A deficiency in Drak2 results in a T cell hypersensitivity and an unexpected resistance to autoimmunity. Immunity. 2004;21:781–791. doi: 10.1016/j.immuni.2004.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pei L, Wang S, Jin H, Bi L, Wei N, Yan H, et al. A novel mechanism of spine damages in stroke via DAPK1 and Tau. Cereb. Cortex. 2015;25:4559–4571. doi: 10.1093/cercor/bhv096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu S, Zhu H, Tang N, Chen W, Li X, Li H, et al. Selective degeneration of entorhinal-CA1 synapses in Alzheimer’s disease via activation of DAPK1. J. Neurosci. 2016;36:10843–10852. doi: 10.1523/JNEUROSCI.2258-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hukelmann JL, Anderson KE, Sinclair LV, Grzes KM, Murillo AB, Hawkins PT, et al. The cytotoxic T cell proteome and its shaping by the kinase mTOR. Nat. Immunol. 2016;17:104–112. doi: 10.1038/ni.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ross SH, Rollings C, Anderson KE, Hawkins PT, Stephens LR, Cantrell DA. Phosphoproteomic analyses of interleukin 2 signaling reveal integrated JAK kinase-dependent and -independent networks in CD8(+) T cells. Immunity. 2016;45:685–700. doi: 10.1016/j.immuni.2016.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat. Cell Biol. 2002;4:658–665. doi: 10.1038/ncb840. [DOI] [PubMed] [Google Scholar]
- 41.Salmond RJ, Emery J, Okkenhaug K, Zamoyska R. MAPK, phosphatidylinositol 3-kinase, and mammalian target of rapamycin pathways converge at the level of ribosomal protein S6 phosphorylation to control metabolic signaling in CD8 T cells. J. Immunol. 2009;183:7388–7397. doi: 10.4049/jimmunol.0902294. [DOI] [PubMed] [Google Scholar]
- 42.Salmond RJ, Brownlie RJ, Meyuhas O, Zamoyska R. Mechanistic target of rapamycin complex 1/S6 kinase 1 signals influence T cell activation independently of ribosomal protein S6 phosphorylation. J. Immunol. 2015;195:4615–4622. doi: 10.4049/jimmunol.1501473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu WL, Yang HC, Hsu CS, Wang CC, Wang TS, Kao JH, et al. Pegylated IFN-alpha suppresses hepatitis C virus by promoting the DAPK-mTOR pathway. Proc. Natl Acad. Sci. USA. 2016;113:14799–14804. doi: 10.1073/pnas.1618517114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng H, Chi H. mTOR signaling in the differentiation and function of regulatory and effector T cells. Curr. Opin. Immunol. 2017;46:103–111. doi: 10.1016/j.coi.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.