Autophagy is an intracellular degradation system conserved among eukaryotes, which utilizes a double-membrane organelle, an autophagosome, to sequester and deliver cytoplasmic materials to lysosomes for degradation [1]. When autophagy is induced by starvation in budding yeast, Atg proteins undergo phase separation to form the preautophagosomal structure (PAS) [2]. Atg9 is the sole transmembrane protein among core Atg proteins and is embedded in a vesicle termed the Atg9 vesicle at the Golgi apparatus, which targets the PAS and functions as the initial membrane source for autophagosome formation [1]. By unknown mechanisms, Atg9 vesicles transform into a cup-shaped isolation membrane (also known as phagophore), which expands and finally seals into an autophagosome.
For expansion, isolation membranes must receive sufficient amounts of phospholipids from endomembranes as a building block. In budding yeast, the expanding edge of the isolation membrane contacts the endoplasmic reticulum (ER), especially the ER exit site (ERES) [3, 4]. Atg9, together with the Atg2–Atg18 complex, localizes at the edge of the isolation membrane and contacts the ERES, where Atg2 functions as a tether between ERES and the isolation membrane [1, 5]. Recently, in vitro analyses have identified the lipid transfer activity of Atg2 between membranes, which proposed an attractive model that Atg2 functions as a conduit and directly provides phospholipids from the ER to the isolation membrane for expansion [6]. However, this model has several serious shortcomings. One of which is the activity of Atg2 to transfer phospholipids only between the cytoplasmic leaflets of the membrane; Atg2 is not a transmembrane protein and cannot transport a phospholipid across the lipid bilayer membrane. As a result, phospholipids transferred from the ER by Atg2 would accumulate at the cytoplasmic leaflet of the isolation membrane, which must be distributed to the luminal leaflet for membrane expansion. Lipid transport between the two leaflets of a membrane is mediated by transmembrane proteins termed lipid translocases, which are further classified into flippases, floppases, and scramblases [7]. Flippases and floppases transport lipids in a unidirectional manner using ATP, whereas scramblases transport lipids bidirectionally without the help of ATP.
Because Atg9 is the sole transmembrane protein among core Atg proteins and localizes at the expanding edge of the isolation membrane, we speculated that Atg9 has some lipid translocating activities. The dithionite assay has been developed for detecting such activities using liposomes incorporating fluorescently labeled phospholipids [8]. Dithionite quenches fluorescently labeled phospholipids located at the outer leaflet of liposomes, reducing the liposome fluorescence by half. In contrast, dithionite diminishes the fluorescence of liposomes incorporating Atg9 almost completely, suggesting that Atg9 transports fluorescently labeled phospholipids from the inner to the outer leaflet of liposomes [9]. This activity is similarly observed for different phospholipid types and independently with ATP, proposing that Atg9 is a lipid scramblase. Phosphatidylinositol 3-phosphate (PI3P) is known to localize at isolation membranes and plays a critical role in autophagosome formation [1]. The distribution of PI3P at the isolation membrane was studied by the quick-freezing and freeze-fracture replica labeling (QF-FRL) method and shown to be distributed at both the cytoplasmic and luminal leaflets of the isolation membrane [10]. We prepared liposomes containing PI3P only at the outer leaflet of liposomes and studied the distribution of PI3P using the QF-FRL method. Notably, the asymmetric distribution of PI3P was destroyed in an Atg9-dependent manner, and PI3P was detected similarly at both leaflets of liposomes [9], suggesting that Atg9 has the activity to transport PI3P from the outer to the inner leaflet of liposomes, further confirming that Atg9 is a lipid scramblase.
To obtain the structural basis of the lipid scramblase activity of Atg9, we then determined the cryo-electron microscopy (cryo-EM) structure of fission yeast Atg9 at 3.0 Å resolution [9]. The membrane-buried region of the Atg9 protomer is composed of four transmembrane helices and two helices buried in the cytoplasmic leaflet of the membrane in a lateral direction, which, together with the cytoplasmic domain, constitute the lateral pore (LP) that is laterally open to the membrane. By sharing two transmembrane helices between protomers, Atg9 forms a trimer, which constructs the vertical pore (VP) at the trimer center that is vertical to the membrane. LP and VP are connected, thereby creating a path between the two leaflets of membranes. Mutational analyses revealed that both LP and VP are essential for autophagosome formation and autophagy activity but not for PAS targeting of Atg9 or Atg2 [9]. Importantly, autophagy-defective mutations at LP and VP also impaired the activity of Atg9 to transport PI3P from the outer to the inner leaflet of liposomes in vitro [9]. These data suggest that the lipid scramblase activity of Atg9 is mediated by LP and VP and that the activity is directly involved in autophagosome formation. These data also propose a model that Atg9 translocates phospholipids at the cytoplasmic leaflet of the isolation membrane, which are delivered from the ER by Atg2, to the luminal leaflet, thereby enabling the expansion of the isolation membrane (Fig. 1).
Fig. 1. Model of the Atg2 and Atg9-mediated lipid transfer from the ER to the isolation membrane for expansion.
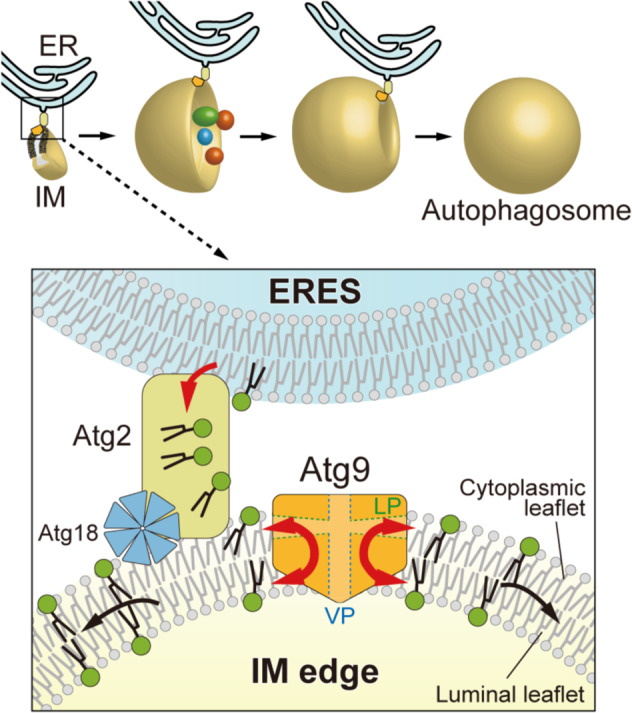
During autophagosome formation, the Atg2–Atg18 complex and Atg9 localize at the expanding edge of the isolation membrane and make contact with the ERES in the ER. Atg2 transfers phospholipids from the ER to the cytoplasmic leaflet of the isolation membrane, which are then distributed to the luminal leaflet by the scramblase activity of Atg9, enabling the expansion of isolation membranes.
Mammals conserve two Atg2 orthologs (ATG2A and ATG2B) and one Atg9 ortholog (ATG9A). ATG2A has been shown to localize at the contact between the ER and the isolation membrane [11] and ATG2A and ATG2B have been shown to possess the lipid transfer activity between membranes in vitro [11–13], both consistent with yeast Atg2. In contrast, ATG9A seems not to be incorporated into the isolation membrane; only transient contacts were observed between them [1]. Nevertheless, ATG9A has also been shown to possess a lipid scramblase activity using the dithionite assay by Dr. Otomo’s group [14] and our PI3P transport assay [9]. Moreover, cryo-EM analysis of ATG9A revealed that ATG9A forms a trimer with LP and VP, quite similar to the structure of yeast Atg9 [14, 15]. Consistent with yeast Atg9, VP in ATG9A is important for lipid scramblase activity [9, 14] and for producing normal-sized autophagosomes in human cells [14]. These observations make us speculate that the collaboration model of Atg2 and Atg9 in isolation membrane expansion (Fig. 1) is also applicable to human ATG2 and ATG9. A more detailed localization analysis of ATG9A in mammalian cells is required for validating this speculation.
There remain many unanswered questions. The mechanism of how Atg9 scrambles phospholipids using LP and VP is still obscure. For membrane expansion, phospholipids must be transferred from the ER to the isolation membrane unidirectionally, the mechanism of which is an essential issue that must be resolved. The degree of contribution of the Atg2 and Atg9-mediated lipid transfer to the completion of autophagosomes is another important question. Several other pathways, such as vesicular transport, have also been proposed to deliver phospholipids to the isolation membrane [1]. The combined usage of cells and in vitro reconstitution systems is essential for future studies to address these critical issues.
Acknowledgements
This work was supported in part by JSPS KAKENHI Grant Number 18H03989, 19H05707 (to NNN), 15K21608, 18K06097 (to KM), JST CREST Grant Number JPMJCR13M7 (to NNN), grants from the Takeda Science Foundation (to NNN), and from the Naito Foundation (to NNN).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nakatogawa H. Mechanisms governing autophagosome biogenesis. Nat Rev Mol Cell Biol. 2020;21:439–58. doi: 10.1038/s41580-020-0241-0. [DOI] [PubMed] [Google Scholar]
- 2.Fujioka Y, Alam JM, Noshiro D, Mouri K, Ando T, Okada Y, et al. Phase separation organizes the site of autophagosome formation. Nature. 2020;578:301–5. doi: 10.1038/s41586-020-1977-6. [DOI] [PubMed] [Google Scholar]
- 3.Graef M, Friedman JR, Graham C, Babu M, Nunnari J. ER exit sites are physical and functional core autophagosome biogenesis components. Mol Biol Cell. 2013;24:2918–31. doi: 10.1091/mbc.e13-07-0381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. J Cell Sci. 2013;126:2534–44. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 5.Gomez-Sanchez R, Rose J, Guimaraes R, Mari M, Papinski D, Rieter E, et al. Atg9 establishes Atg2-dependent contact sites between the endoplasmic reticulum and phagophores. J Cell Biol. 2018;217:2743–63. doi: 10.1083/jcb.201710116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Osawa T, Kotani T, Kawaoka T, Hirata E, Suzuki K, Nakatogawa H, et al. Atg2 mediates direct lipid transfer between membranes for autophagosome formation. Nat Struct Mol Biol. 2019;26:281–8. doi: 10.1038/s41594-019-0203-4. [DOI] [PubMed] [Google Scholar]
- 7.Sharom FJ. Flipping and flopping–lipids on the move. IUBMB Life. 2011;63:736–46. doi: 10.1002/iub.515. [DOI] [PubMed] [Google Scholar]
- 8.Ploier B, Menon AK. A fluorescence-based assay of phospholipid scramblase activity. J Vis Exp. 2016;115:54635. doi: 10.3791/54635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matoba K, Kotani T, Tsutsumi A, Tsuji T, Mori T, Noshiro D, et al. Atg9 is a lipid scramblase that mediates autophagosomal membrane expansion. Nat Struct Mol Biol. 2020. 10.1038/s41594-020-00518-w. [DOI] [PubMed]
- 10.Cheng J, Fujita A, Yamamoto H, Tatematsu T, Kakuta S, Obara K, et al. Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat Commun. 2014;5:3207. doi: 10.1038/ncomms4207. [DOI] [PubMed] [Google Scholar]
- 11.Valverde DP, Yu S, Boggavarapu V, Kumar N, Lees JA, Walz T, et al. ATG2 transports lipids to promote autophagosome biogenesis. J Cell Biol. 2019;218:1787–98. doi: 10.1083/jcb.201811139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osawa T, Ishii Y, Noda NN. Human ATG2B possesses a lipid transfer activity which is accelerated by negatively charged lipids and WIPI4. Genes Cells. 2020;25:65–70. doi: 10.1111/gtc.12733. [DOI] [PubMed] [Google Scholar]
- 13.Maeda S, Otomo C, Otomo T. The autophagic membrane tether ATG2A transfers lipids between membranes. Elife. 2019;8:e45777. doi: 10.7554/eLife.45777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maeda S, Yamamoto H, Kinch LN, Garza CM, Takahashi S, Otomo C, et al. Structure, lipid scrambling activity and role in autophagosome formation of ATG9A. Nat Struct Mol Biol. 2020. 10.1038/s41594-020-00520-2. [DOI] [PMC free article] [PubMed]
- 15.Guardia CM, Tan XF, Lian T, Rana MS, Zhou W, Christenson ET, et al. Structure of human ATG9A, the only transmembrane protein of the core autophagy machinery. Cell Rep. 2020;31:107837. doi: 10.1016/j.celrep.2020.107837. [DOI] [PMC free article] [PubMed] [Google Scholar]


