Abstract
The various malformations of the aerodigestive tract collectively known as esophageal atresia/tracheoesophageal fistula (EA/TEF) constitute a rare group of birth defects of largely unknown etiology. Previous studies have identified a small number of rare genetic variants causing syndromes associated with EA/TEF. We performed a pilot exome sequencing study of 45 unrelated simplex trios (probands and parents) with EA/TEF. Thirteen had isolated and 32 had nonisolated EA/TEF; none had a family history of EA/TEF. We identified de novo variants in protein-coding regions, including 19 missense variants predicted to be deleterious (D-mis) and 3 likely gene-disrupting (LGD) variants. Consistent with previous studies of structural birth defects, there is a trend of increased burden of de novo D-mis in cases (1.57-fold increase over the background mutation rate), and the burden is greater in constrained genes (2.55-fold, p = 0.003). There is a frameshift de novo variant in EFTUD2, a known EA/TEF risk gene involved in mRNA splicing. Strikingly, 15 out of 19 de novo D-mis variants are located in genes that are putative target genes of EFTUD2 or SOX2 (another known EA/TEF gene), much greater than expected by chance (3.34-fold, p value = 7.20e−5). We estimated that 33% of patients can be attributed to de novo deleterious variants in known and novel genes. We identified APC2, AMER3, PCDH1, GTF3C1, POLR2B, RAB3GAP2, and ITSN1 as plausible candidate genes in the etiology of EA/TEF. We conclude that further genomic analysis to identify de novo variants will likely identify previously undescribed genetic causes of EA/TEF.
Subject terms: Diseases, Genetics
Introduction
Esophageal atresia/tracheoesophageal fistula (EA/TEF) is a rare, complex congenital aerodigestive anomaly with an estimated incidence of 1 in 2500 to 1 in 4000 live births [1, 2]. Almost half of infants born with this congenital anomaly have associated congenital malformations of other organ systems, most commonly cardiovascular, digestive [1], urogenital, and musculoskeletal [3]. These defects have been observed together as the vertebral defects, anal atresia, cardiac defects, tracheoesophageal fistula, renal anomalies, and limb abnormalities (VACTERL) association [4]. While there have been rare reports of variants in FOXF1 and ZIC3 in VACTERL-association patients [5], the molecular etiology for the majority of VACTERL cases remains unknown. Chromosome anomalies including aneuploidies and microdeletions are observed in 6–10% of nonisolated EA/TEF [3, 5] patients. These anomalies include trisomy 13, 18, and 21, monosomy X [6], and several copy number variants (CNVs). Several monogenic causes of syndromes that include EA/TEF have also been elucidated and include variants in MYCN, SOX2, CHD7, and MID1. Monogenetic causes account for only about 5% of EA/TEF cases, and are mostly de novo (with the exception of variants in recessive Fanconi anemia-related genes) [5–7].
SOX2 has been reported as an important gene for esophagus and anterior stomach development [8]. SOX2 is involved in Wnt signaling by binding β-catenin, a central mediator of the Wnt pathway [9]. Deletion of the Wnt signaling downstream mediator β-catenin leads to lung agenesis, and the foregut fails to separate [10]. EFTUD2 is associated with esophageal atresia and other developmental disorders such as mandibulofacial dysostosis with microcephaly with the heterozygous loss of function variants [11–13]. EFTUD2 is required for pre-mRNA splicing as component of the spliceosome [14, 15].
There have been few studies investigating the genetic causes of nonisolated EA/TEF, and it is still widely considered to have a multifactorial etiology. Small scale twin studies, however, have shown a higher concordance rate between monozygotic twins (67%) compared to dizygotic twins (42%), suggesting a genetic contribution [16, 17]. Animal studies have identified genes in several developmental pathways associated with tracheoesophageal anomalies, among them sonic hedgehog pathway genes. Murine models with homozygous deficiencies of SHH and GLI2 exhibit foregut anomalies including EA, TEF, and tracheoesophageal stenosis and hypoplasia [18]. Other developmental genes involved with foregut development in animal studies include transcription factors Foxf1, vitamin A effectors (Rarα, Rarβ) homeobox-containing transcription factors and their regulators (Nkx2.1 [19], Hoxc4, Pcsk5), and developmental transcriptional regulators (Tbx4, Sox2) [3, 20].
EA/TEF is identified prenatally in about 50% of cases. When the diagnosis is suspected (usually by sonographic findings of polyhydramnios and a small stomach), prognostic clinical information about associated birth defects is commonly sought. Definitive prognostic information is usually limited unless a chromosomal anomaly is identified. In an effort to identify novel genetic variants associated with EA/TEF, we studied 45 individuals with EA/TEF and their biological parents, none of whom had a family history of EA/TEF. We sought to identify novel genetic causes of EA/TEF using exome sequencing (WES). Our goal is to understand the genomic architecture of EA/TEF, and to better characterize the syndromes and conditions associated with EA/TEF. We designed this pilot study to assess whether genomic characterization of EA/TEF would provide more accurate prognostic information and help tailor therapy based on predicted phenotype. We plan to combine these data with that of other congenital malformations to provide a more comprehensive understanding of human development.
Methods
Subject recruitment
Patients with isolated and nonisolated EA/TEF were recruited from two medical centers—Columbia University Medical Center (CUMC) in New York, USA and Cairo University General Hospital in Cairo, Egypt. Subjects eligible for the study included individuals diagnosed with known forms of EA/TEF and no family history of EA/TEF, based upon medical record review. All participants provided informed consent. The study was approved by the Columbia University institutional review board. Blood and/or saliva samples were obtained from the probands and both biological parents. A three-generation family history was taken at the time of enrollment and clinical data were extracted from the medical records and by patient and parental interview.
Exome sequencing
Exome sequencing was performed at Novogene Genome Sequencing Company (Chula Vista, CA). A total of 1.0 μg genomic DNA was used as input material. Sequencing libraries were generated using Agilent SureSelect Human All ExonV6 kit (Agilent Technologies, CA, USA) following manufacturer’s recommendations. Briefly, fragmentation was carried out by hydrodynamic shearing system (Covaris, MA, USA) to generate 180–280 bp fragments. Remaining overhangs were converted into blunt ends via exonuclease/polymerase activities, and enzymes were removed. After adenylation of 3′ ends of DNA fragments, adapter oligonucleotides were ligated. DNA fragments with ligated adapter molecules on both ends were selectively enriched in a PCR reaction. Captured libraries were enriched in a PCR reaction to add index tags to prepare for hybridization. Products were purified using AMPure XP system (Beckman Coulter, Beverly, USA) and quantified using the Agilent high sensitivity DNA assay on the Agilent Bioanalyzer 2100 system. The qualified libraries were sequenced on an Illumina HiSeq sequencer after pooling according to effective concentration and expected data volume. Read length were paired-end 150 bp.
Bioinformatics analysis and calling of de novo variants
We used GATK-recommended best practices for calling single nucleotide variants (SNVs) and short insertions and deletions (indels) from exome sequencing data. Specifically, we used BWA-mem [21] to align reads to human reference genome (GRCh37), Picard Tools to mark PCR duplicates, and GATK [22] haplotypeCaller for calling variants jointly from all sequenced samples, and GATK variant quality score recalibration (VQSR) to recalibrate variant quality. We applied multiple heuristic filtering rules to remove potential technical artifacts as previously described [23, 24]. Specifically, we only retained variants that met all the following criteria: GQ ⩾ 30, FS ⩽ 25, QD ⩾ 2 (SNV), QD ⩾ 1 (INDEL), ReadPosRankSum ⩾ −3 (INDEL), read depth on alt allele ⩾ 5, alt allele depth to total depth ⩾ 0.1, VQSRSNP ⩽ 99.80, VQSRINDEL ⩽ 99.70 and mappability (based on 200 insert length) = 1.
To call de novo variants, we applied a previously published procedure [23, 24] and used IGV [25] to visualize candidate de novo variants and remove potential artifacts. All nonsynonymous de novo variants were sanger confirmed. In addition, we used PLINK to infer population structure and kinship. We used xHMM [26] to infer large CNVs to ruled out patients who potentially get EA/TEF due to chromosomal anomalies.
Annotation and in silico prediction
We used ANNOVAR [27] to annotate variants and aggregate population frequency (Exome Aggregation Consortium (ExAC)) and Genome Aggregation Database [28], protein-coding consequence, and multiple in silico predictions on genetic variants, including CADD [29] and REVEL [30].
Putative targets of EFTUD2 or SOX2
We obtained putative targets of EFTUD2 based on RNA binding protein (RBP) binding sites profiled by eCLIP in a HepG2 cell line from ENCODE [31] and processed using a recently published pipeline [32]. We selected the genes for which the peak count is equal to or greater than 2. We obtained target genes of transcription factor SOX2 based on ChIP data from glandular mouse stomach [33] curated by ChEA [34].
Statistical analysis
For de novo variants, we determined the overall burden of four variant types including synonymous, likely gene disrupting (LGD, i.e., stop gain, frameshift, and splice site), missense and deleterious missense (D-mis, defined by REVEL ≥ 0.5 or CADD Phred score ≥ 25) in all genes and constrained genes (defined by ExAC [28] pLI ≥ 0.5). We used a less stringent pLI threshold for defining constrained genes, because it captures more known haploinsufficient genes [35]. We obtained estimated background mutation rate in previous publications calibrated for exome sequencing data [36]. The expected number of variants in different gene sets were calculated by summing up the background mutation rate of the specific variant class in the gene-set multiplied by twice the number of cases. We then test the burden of de novo variants in a gene set by a Poisson test with the baseline expectation as the mean under the null model. To estimate the proportion of cases that can be attributed to de novo deleterious variants, the difference between the observed number and expected number of de novo deleterious variants is divided by the number of cases [37].
Results
Exome sequencing data
A total of 45 individuals with EA/TEF were enrolled into the study. Probands were between the ages of 1.5 years and 55.7 years with an average of 10.2 years old (Table 1). Thirteen probands had isolated EA/TEF and 32 probands had neurodevelopmental delay and/or at least one additional congenital defect and were classified as nonisolated. Fourteen of the probands had congenital heart defects, 8 had neurodevelopmental delay, 4 had gastrointestinal defects, 12 had genitourinary defects (nonrenal), 8 had skeletal defects, 2 had craniofacial defects and 2 had other defects. The majority of probands were of European ancestry (60%), and the remaining were of African-American (15%), Egyptian (15%), and Asian (10%) ancestry. None of the 45 probands reported a family history of EA/TEF.
Table 1.
Patient characteristics of 45 patients with esophageal atresia.
| N = 45 | |
|---|---|
| Mean age (range) | 10.2 years (1.5–55.7 years) |
| Sex | |
| Male | 25 (56%) |
| Female | 20 (44%) |
| Type of EA | |
| Type A | 3 (7%) |
| Type C | 11 (24%) |
| Type D | 1 (2%) |
| Type H (TEF only) | 3 (7%) |
| Unknown | 27 (60%) |
| Failure to thrive | 8 (18%) |
| Associated anomalies | 13 (65%) |
| Nonisolated cases | 32 (71%) |
| Developmental delay | 8 (18%) |
| Other congenital defects | 28 (64%) |
Overall burden of de novo variants
We identified 57 de novo variants in 45 probands (Supplementary Table 1). We compared overall burden of de novo variants in 45 cases to expectations from a background mutation model [36]. We classified protein-coding variants into four groups: synonymous, missense, D-mis, and LGD. Overall the frequency of synonymous variants in cases is close to expectation from background mutation rate (p value = 0.68, enrichment rate = 1.1×). There is a trend of enrichment of missense variants (p = 0.12, enrichment rate = 1.3×) and D-mis variants (p = 0.06, enrichment rate = 1.6×) in cases compared to expectation (Table 2).
Table 2.
Overall burden of de novo heterozygous variants.
| Gene sets | Variant class | Obs_Num | Obs_Rate | Exp_Num | Exp_Rate | Enrichment | p value |
|---|---|---|---|---|---|---|---|
| All genes | Synonymous | 15 | 0.333 | 13.7 | 0.304 | 1.1 | 0.68 |
| Missense | 39 | 0.867 | 30.2 | 0.671 | 1.29 | 0.12 | |
| D-mis | 19 | 0.422 | 12.1 | 0.269 | 1.57 | 0.06 | |
| LGD | 3 | 0.066 | 4.04 | 0.089 | 0.743 | 0.81 | |
| Constrained genes | Synonymous | 8 | 0.178 | 4.98 | 0.111 | 1.61 | 0.17 |
| Missense | 16 | 0.356 | 11.06 | 0.246 | 1.45 | 0.13 | |
| D-mis | 12 | 0.267 | 4.71 | 0.105 | 2.55 | 0.003 | |
| SOX2 or EFTUD2 targets | Synonymous | 8 | 0.178 | 4.84 | 0.108 | 1.65 | 0.16 |
| Missense | 19 | 0.422 | 10.76 | 0.24 | 1.77 | 2.2e−16 | |
| D-mis | 15 | 0.333 | 4.49 | 0.099 | 3.34 | 6.6e−05 |
LGD likely gene disrupting, including frameshift, stop gain, and variants at canonical splice site, D-mis predicted deleterious missense variants.
Exp_Rate and Obs_Rate are respectively the expected and observed fraction of genes with a specific type of de novo mutation. Exp_Num and Obs_Num are the expected and observed number of genes with a specific type of de novo mutation, respectively. Constrained genes are defined by ExAC_pLI > 0.5.
Consistent with previous studies of other types of birth defects [24, 38, 39], the enrichment of D-mis variants is more pronounced (p value = 0.003, enrichment rate = 2.6×) in constrained genes that are intolerant of loss of function variants (ExAC pLI ≥ 0.5) (Table 2).
Most of genes with deleterious de novo variants are putative targets of EFTUD2 or SOX2
One patient has a de novo frameshift deletion (c.2314del, p.(Gln772ArgfsTer21)) in EFTUD2 (elongation factor Tu GTP binding domain containing 2). The phenotype of the patient includes EA/TEF, bilateral clubfoot, hydrocele, atrial septal defect, and pyepylectasislectasis, which overlaps with features of Guion-Almeida type of mandibulofacial dysostosis caused by heterozygous EFTUD2 variants [13]. De novo variants in EFTUD2 are known to be associated with EA [11, 12]. EFTUD2 encodes a component of the spliceosome complex that regulates mRNA splicing, a master regulator that potentially regulates the expression of thousands of genes. We hypothesized that genes regulated by EFTUD2 and other master regulators relevant to EA/TEF (such as SOX2 [8]) are more likely to be EA/TEF risk genes and therefore enriched with de novo variants. To test this, we obtained putative targets of EFTUD2 based on eCLIP data in a HepG2 cell line from ENCODE [31] and targets of SOX2 based on ChIP-seq data in mouse stomach [33]. There are 1629 and 4463 targets of SOX2 and EFTUD2, respectively; and the union of the targets is 5454. Among 19 genes with D-mis de novo variants, 15 are targets of SOX2 or EFTUD2, much larger than expected by background (enrichment rate = 3.34×, p value = 6.6e−05). Overall, the burden indicates that 33% of EA/TEF patients are attributable to deleterious de novo variants in genes that are SOX2 or EFUD2 targets.
Table 3 summarizes the associated clinical features and variants in candidate genes prioritized by intolerance to loss of function variants and biological pathways implicated in developmental disorders. Seven genes, ADD1, APC2, GLS, SMAD6, RAB3GAP2, PTPN14, and EFTUD2 are OMIM genes and are associated with Mendelian diseases (Table 3). ITSN1 was recently discovered as a risk gene for autism spectrum disorder [40]. The ITSN1 variant carrier was only 18 months at the time of enrollment, which is too young to make the diagnosis of autism.
Table 3.
De novo heterozygous variants in candidate genes.
| Study ID | Chr | Pos | Gene (OMIM#) | pLI | Coding sequence change (HGVSc) | Coding sequence change (HGVS) | Type | REVEL | CADD | EA/TEF | Additional anomalies | OMIM condition (Inheritance) (OMIM#) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 230 | 4 | 2877643 | ADD1 (102680) | 0.61 | NM_001119.4:c.1A>G | NP_001110.2:p.(Met1?) | D-mis | 0.53 | 25.1 | EA + TEF | Extra wedge-shaped vertebrae, extra ribs, horseshoe kidney, bilateral radial hypoplasia with associated thumb and wrist anomaly | |
| 48 | 2 | 131521881 | AMER3 (NA) | 0.00 | NM_001105193.1:c.2236C>T | NP_001098663.1:p.(Arg746Ter) | LGD | N/A | 35 | EA + TEF | Atrial septal defect, bilateral clubfoot, hydrocele, renal pyelectasis | |
| 15 | 14 | 24035494 | AP1G2 (603534) | 0.00 | NM_001282475.1:c.77G>A | NP_001269404.1:p.(Arg26His) | D-mis | 0.39 | 25.9 | EA + TEF | Atrial septal defect, patent ductus arteriosus, short stature, small kidneys, hiatal hernia | |
| 101 | 19 | 1456100 | APC2 (612034) | 0.99 | NM_005883.2:c.665T>C | NP_005874.1:p.(Ile222Thr) | D-mis | 0.65 | 26.4 | EA + TEF | Pierre Robin sequence, solitary kidney, cleft palate | Sotos syndrome 3 (AR) (617169); Cortical dysplasia, complex, with other brain malformations 10 (AR) (618677) |
| 4 | 1 | 109795559 | CELSR2 (604265) | 0.99 | NM_001408.2:c.2858A>G | NP_001399.1:p.(Asn953Ser) | D-mis | 0.57 | 24.1 | EA + TEF | Duodenal atresia, Wolf-Parkinson White syndrome | |
| 48 | 17 | 42930931 | EFTUD2 (603892) | 0.99 | NM_001142605.1:c.2314del | NP_001136077.1:p.(Gln772ArgfsTer21) | LGD | N/A | N/A | EA + TEF | Atrial septal defect, bilateral clubfoot, hydrocele, renal pyelectasis | Mandibulofacial dysostosis, Guion-Almeida type (AD) (610536) |
| 48 | 2 | 191827642 | GLS (138280) | 0.99 | NM_014905.4:c.1940C>T | NP_055720.3:p.(Thr647Ile) | D-mis | 0.169 | 27.1 | EA + TEF | Atrial septal defect, bilateral clubfoot, hydrocele, renal pyelectasis | Epileptic encephalopathy, early infantile, 71 (AR) (618328); Infantile cataract, skin abnormalities, glutamate excess, and impaired intellectual development (AD) (618339); Global developmental delay, progressive ataxia, and elevated glutamine (AR) (618412) |
| 17 | 16 | 27473692 | GTF3C1 (603246) | 0.99 | NM_001286242.1:c.5965C>A | NP_001273171.1:p.(Pro1989Thr) | D-mis | 0.60 | 26.4 | EA + TEF | Multiple congenital hemangiomas | |
| 2–8 | 21 | 35254586 | ITSN1 (602442) | 0.99 | NM_001331010.1:c.4366C>T | NP_001317939.1:p.(Arg1456Cys) | D-mis | 0.22 | 33 | EA | Tetralogy of Fallot | |
| 267 | 1 | 899892 | KLHL17 (NA) | 0.00 | NM_198317.2:c.1682C>A | NP_938073.1:p.(Ala561Glu) | D-mis | 0.81 | 31 | EA + TEF | Heart defect, kyphosis, tracheomalacia, right leg hemihypertrophy | |
| 95 | 5 | 141248684 | PCDH1 (603626) | 0.87 | NM_001278613.1:c.401A>G | NP_001265542.1:p.(Glu134Gly) | D-mis | 0.66 | 26.7 | EA + TEF | None | |
| 125 | 12 | 18439865 | PIK3C2G (609001) | 0.00 | NM_001288772.1:c.761 + 2T>C | LGD | .N/A | 22.5 | EA + TEF | Coarctation of aorta, total anomalous pulmonary venous return, congenital stricture in distal esophagus, hypospadias | ||
| 275 | 4 | 57883376 | POLR2B (180661) | 0.99 | NM_001303268.1:c.1898C>T | NP_001290197.1:p.(Ala633Val) | D-mis | 0.92 | 34 | EA + TEF | Left multicystic dysplastic kidney, aortic plexus | |
| 248 | 1 | 214557279 | PTPN14 (603155) | 0.99 | NM_005401.4:c.1919G>A | NP_005392.2:p.(Arg640His) | D-mis | 0.119 | 25 | EA + TEF | Dilated cardiomyopathy (not congenital-diagnosed in 30 s) | Choanal atresia and lymphedema (AR) (613611) |
| 2–6 | 1 | 220364518 | RAB3GAP2 (609275) | 0.99 | NM_012414.3:c.1379G>A | NP_036546.2:p.(Arg460Gln) | D-mis | 0.32 | 35 | EA + TEF | None | Martsolf syndrome (AR)(212720), Warburg micro syndrome 2 (AR) (614225) |
| 275 | 15 | 67073475 | SMAD6 (602931) | 0.00 | NM_005585.4:c.1093G>A | NP_005576.3:p.(Gly365Ser) | D-mis | 0.85 | 32 | EA + TEF | Left multicystic dyplastic kidney, aortic plexus | Aortic valve disease 2 (AD)(614823) |
| 15 | 7 | 97854186 | TECPR1 (614781) | 0.00 | NM_015395.2:c.2617G>A | NP_056210.1:p.(Asp873Asn) | D-mis | 0.74 | 35 | EA + TEF | Atrial septal defect, patent ductus arteriosus, short stature, small kidneys, hiatal hernia |
LGD likely gene disrupting, including frameshift, stop gain and variants at canonical splice site. D-mis predicted deleterious missense variants, EA/TEF esophageal atresia/tracheoesophageal fistula, AR autosomal recessive, AD autosomal dominant.
Eleven of these genes (CELSR2, PCDH1, APC2, GLS, GTF3C1, ITSN1, MAP4K3, ADD1, POLR2B, PTPN14, RAB3GAP2) are constrained genes with a D-mis variant. Three genes AP1G2, KLHL17, SMAD6, and TECPR1 are nonconstrained genes with D-mis variants. EFTUD2, PIK3C2G, and AMER3 have LGD variants and EFTUD2 is a known candidate gene for EA.
Discussion
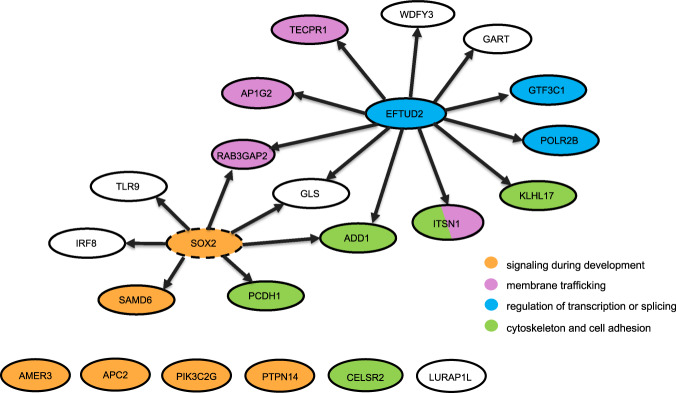
In this pilot study, we report exome sequencing results on 45 proband-parent trios with isolated or nonisolated EA/TEF with no family history of EA/TEF. We identified 22 LGD or D-mis de novo variants. Consistent with previous studies of structural birth defects or developmental disorders, genes that are constrained are enriched with deleterious variants, likely due to a historical reduction of reproductive fitness by such predicted deleterious variants. The majority of the genes with deleterious de novo variants are putative targets of SOX2 or EFTUD2, two master regulators that are known to cause EA/TEF through haploinsufficiency and may provide a biological mechanism for the etiology of some EA/TEF. Figure 1 shows genes with LGD or D-mis de novo variants and their relationships with EFTUD2 and SOX2. We did not identify any de novo variants in SOX2 gene in our small cohort. Given the overall high enrichment rate of 3.34, we expect that more than half of target genes of SOX2 or EFTUD2 with de novo predicted pathogenic variants are candidate EA/TEF risk genes [37, 41].
Fig. 1. Genes with LGD or D-mis de novo variants and their relationship with EFTUD2 and SOX2.
Each gene is represented by a circle. Arrows indicate putative TF-target or RBP-target relationships. We did not observe de novo variants in SOX2 (dashed circle) in our cohort. Genes are colored by biological pathways. Only the pathways with at least three genes with LGD or D-mis variants are shown.
Three genes, ADD1, GLS, and RAB3GAP2, are putative targets of both EFTUD2 and SOX2 [31, 33]. Notably, ITSN1, AP1G2, TECPR1, and RAB3GAP2 are involved in membrane trafficking pathway or autophagy [42–45]. KLHL17, ADD1, CELSR2, PCDH1, and ITSN1 are involved in cytoskeleton or cell adhesion [42, 46, 47]. AMER3 and APC2 are both key regulators in Wnt signaling, a process known to be implicated in EA/TEF and other birth defects [48]. A few other genes, SMAD6, PTPN14, and PIK3C2G, are involved in signaling pathways that are critical during development [46, 49, 50].
Our current analysis is limited by the source of ChIP-seq of SOX2 from stomach [33] and eCLIP of EFTUD2 from a liver cancer cell line [31]. The availability of data from relevant tissues, e.g., ChIP-seq of SOX2 and eCLIP-seq of EFTUD2 in developing foregut, will enable more precise analysis of de novo and rare variants. In addition, gene expression data, especially single cell sequencing data, of developing esophagus and trachea, will also allow us to refine the analysis and improve the ability to identify the most relevant EA/TEF genes.
Finally, it will be important to increase the sample size of future genomic studies to more precisely estimate the contribution of de novo variants to EA/TEF, and to identify novel risk genes with high confidence and relate the genetic factors to clinical outcomes.
Supplementary information
Acknowledgements
We would like to acknowledge the patients and their families who participated in the study and are grateful for their tremendous contribution. We thank Steve Wyles and Sue Paul from EA for adults, as well as the TOFS UK, OARA, Bridging the Gap of EA/TEF and the Canadian EA network organizations for publicizing the study and assisting with recruitment. We are also appreciative for the technical assistance provided by Patricia Lanzano, Jiangyuan Hu, Liyong Deng, Nikita Chintalapudi, and Charles LeDuc from Columbia University and the study team at Cairo University General Hospital. We thank Na Zhu for help with the calculation of background mutation rate. Funding support provided by P01HD093363 (JW, YS, and WKC) and R01GM120609 (YS).
Data availability
All likely pathogenic variants are in ClinVar submission number SUB7053346. Accession numbers of submitted variants can be found in Supplemental Table 1.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jiayao Wang, Priyanka R. Ahimaz, Somaye Hashemifar
Contributor Information
Yufeng Shen, Email: ys2411@cumc.columbia.edu.
Wendy K. Chung, Email: wkc15@cumc.columbia.edu
Supplementary information
The online version of this article (10.1038/s41431-020-0680-2) contains supplementary material, which is available to authorized users.
References
- 1.Pinheiro PFM, e Silva ACS, Pereira RM. Current knowledge on esophageal atresia. World J Gastroenterol. 2012;18:3662. doi: 10.3748/wjg.v18.i28.3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krishnan U, Mousa H, Dall’Oglio L, Homaira N, Rosen R, Faure C, et al. ESPGHAN-NASPGHAN guidelines for the evaluation and treatment of gastrointestinal and nutritional complications in children with esophageal atresia-tracheoesophageal fistula. J Pediatr Gastroenterol Nutr. 2016;63:550–70. doi: 10.1097/MPG.0000000000001401. [DOI] [PubMed] [Google Scholar]
- 3.Stoll C, Alembik Y, Dott B, Roth M-P. Associated malformations in patients with esophageal atresia. Eur J Med Genet. 2009;52:287–90. doi: 10.1016/j.ejmg.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Shaw-Smith C. Genetic factors in esophageal atresia, tracheo-esophageal fistula and the VACTERL association: roles for FOXF1 and the 16q24. 1 FOX transcription factor gene cluster, and review of the literature. Eur J Med Genet. 2010;53:6–13. doi: 10.1016/j.ejmg.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geneviève D, de Pontual L, Amiel J, Lyonnet S. Genetic factors in isolated and syndromic esophageal atresia. J Pediatr Gastroenterol Nutr. 2011;52:S6–8. doi: 10.1097/MPG.0b013e318213316a. [DOI] [PubMed] [Google Scholar]
- 6.Felix JF, Tibboel D, de Klein A. Chromosomal anomalies in the aetiology of oesophageal atresia and tracheo-oesophageal fistula. Eur J Med Genet. 2007;50:163–75. doi: 10.1016/j.ejmg.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 7.Murphy AJ, Li Y, Pietsch JB, Chiang C, Lovvorn HN. Mutational analysis of NOG in esophageal atresia and tracheoesophageal fistula patients. Pediatr Surg Int. 2012;28:335–40. doi: 10.1007/s00383-011-3022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Que J, Okubo T, Goldenring JR, Nam K-T, Kurotani R, Morrisey EE, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–31. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kormish JD, Sinner D, Zorn AM. Interactions between SOX factors and Wnt/β-catenin signaling in development and disease. Dev Dyn. 2010;239:56–68. doi: 10.1002/dvdy.22046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morrisey EE, Hogan BL. Preparing for the first breath: genetic and cellular mechanisms in lung development. Dev Cell. 2010;18:8–23. doi: 10.1016/j.devcel.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gordon CT, Petit F, Oufadem M, Decaestecker C, Jourdain AS, Andrieux J, et al. EFTUD2 haploinsufficiency leads to syndromic oesophageal atresia. J Med Genet. 2012;49:737–46. doi: 10.1136/jmedgenet-2012-101173. [DOI] [PubMed] [Google Scholar]
- 12.Voigt C, Mégarbané A, Neveling K, Czeschik JC, Albrecht B, Callewaert B, et al. Oto-facial syndrome and esophageal atresia, intellectual disability and zygomatic anomalies-expanding the phenotypes associated with EFTUD2 mutations. Orphanet J Rare Dis. 2013;8:110. doi: 10.1186/1750-1172-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lines MA, Huang L, Schwartzentruber J, Douglas SL, Lynch DC, Beaulieu C, et al. Haploinsufficiency of a spliceosomal GTPase encoded by EFTUD2 causes mandibulofacial dysostosis with microcephaly. Am J Hum Genet. 2012;90:369–77. doi: 10.1016/j.ajhg.2011.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Yan C, Hang J, Finci LI, Lei J, Shi Y. An atomic structure of the human spliceosome. Cell. 2017;169:918–29e.14. doi: 10.1016/j.cell.2017.04.033. [DOI] [PubMed] [Google Scholar]
- 15.Bertram K, Agafonov DE, Dybkov O, Haselbach D, Leelaram MN, Will CL, et al. Cryo-EM structure of a pre-catalytic human spliceosome primed for activation. Cell. 2017;170:701–13.e11. doi: 10.1016/j.cell.2017.07.011. [DOI] [PubMed] [Google Scholar]
- 16.Schulz AC, Bartels E, Stressig R, Ritgen J, Schmiedeke E, Mattheisen M, et al. Nine new twin pairs with esophageal atresia: a review of the literature and performance of a twin study of the disorder. Birth Defects Res Part A: Clin Mol Teratol. 2012;94:182–6. doi: 10.1002/bdra.22879. [DOI] [PubMed] [Google Scholar]
- 17.Maroszyńska I, Fortecka-Piestrzeniewicz K, Niedźwiecka M, Żarkowska-Szaniawska A. Isolated esophageal atresia in both premature twins. Pediatr Pol. 2015;90:91–3. doi: 10.1016/j.pepo.2014.10.003. [DOI] [Google Scholar]
- 18.Shaw-Smith C. Oesophageal atresia, tracheo-oesophageal fistula, and the VACTERL association: review of genetics and epidemiology. J Med Genet. 2006;43:545–54. doi: 10.1136/jmg.2005.038158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Jiang M, Kim E, Lin S, Liu K, Que J, et al. Development and stem cells of the esophagus. Semin Cell Dev Biol. 2017;66:25–35. doi: 10.1016/j.semcdb.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Salem AH, Kothari M, Oquaish M, Khogeer S, Desouky MS. Morbidity and mortality in esophageal atresia and tracheoesophageal fistula: a 20-year review. Ann Pediatr Surg. 2013;9:93–8. doi: 10.1097/01.XPS.0000430524.83127.5d. [DOI] [Google Scholar]
- 21.Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:13033997. 2013. https://arxiv.org/abs/1303.3997.
- 22.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–6. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qi H, Yu L, Zhou X, Wynn J, Zhao H, Guo Y, et al. De novo variants in congenital diaphragmatic hernia identify MYRF as a new syndrome and reveal genetic overlaps with other developmental disorders. PLoS Genet. 2018;14:e1007822. doi: 10.1371/journal.pgen.1007822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–92. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fromer M, Moran JL, Chambert K, Banks E, Bergen SE, Ruderfer DM, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kircher M, Witten DM, Jain P, O’roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, et al. REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet. 2016;99:877–85. doi: 10.1016/j.ajhg.2016.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van Nostrand EL, Freese P, Pratt GA, Wang X, Wei X, Blue SM, et al. A large-scale binding and functional map of human RNA binding proteins. bioRxiv. 2018. 10.1101/179648.
- 32.Feng H, Bao S, Rahman MA, Weyn-Vanhentenryck SM, Khan A, Wong J, et al. Modeling RNA-binding protein specificity in vivo by precisely registering protein-RNA crosslink sites. Mol Cell. 2019;74:1189–1204.e6. doi: 10.1016/j.molcel.2019.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarkar A, Huebner AJ, Sulahian R, Anselmo A, Xu X, Flattery K, et al. Sox2 suppresses gastric tumorigenesis in mice. Cell Rep. 2016;16:1929–41. doi: 10.1016/j.celrep.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 34.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–44. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han X, Chen S, Flynn E, Wu S, Wintner D, Shen Y. Distinct epigenomic patterns are associated with haploinsufficiency and predict risk genes of developmental disorders. Nat Commun. 2018;9:2138. doi: 10.1038/s41467-018-04552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walsh R, Mazzarotto F, Whiffin N, Buchan R, Midwinter W, Wilk A, et al. Quantitative approaches to variant classification increase the yield and precision of genetic testing in Mendelian diseases: the case of hypertrophic cardiomyopathy. Genome Med. 2019;11:5. doi: 10.1186/s13073-019-0616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jin SC, Homsy J, Zaidi S, Lu Q, Morton S, DePalma SR, et al. Contribution of rare inherited and de novo variants in 2871 congenital heart disease probands. Nat Genet. 2017;49:1593–601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deciphering Developmental Disorders S. Prevalence and architecture of de novo mutations in developmental disorders. Nature. 2017;542:433–8. doi: 10.1038/nature21062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Feliciano P, Zhou X, Astrovskaya I, Turner T, Wang T, Brueggeman L, et al. Exome sequencing of 457 autism families recruited online provides evidence for novel ASD genes. bioRxiv. 2019: 516625. https://www.biorxiv.org/content/10.1101/516625v1. [DOI] [PMC free article] [PubMed]
- 41.He X, Sanders SJ, Liu L, De Rubeis S, Lim ET, Sutcliffe JS, et al. Integrated model of de novo and inherited genetic variants yields greater power to identify risk genes. PLoS Genet. 2013;9:e1003671. doi: 10.1371/journal.pgen.1003671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hussain NK, Jenna S, Glogauer M, Quinn CC, Wasiak S, Guipponi M, et al. Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3:927–32. doi: 10.1038/ncb1001-927. [DOI] [PubMed] [Google Scholar]
- 43.Takatsu H, Sakurai M, Shin HW, Murakami K, Nakayama K. Identification and characterization of novel clathrin adaptor-related proteins. J Biol Chem. 1998;273:24693–700. doi: 10.1074/jbc.273.38.24693. [DOI] [PubMed] [Google Scholar]
- 44.Ogawa M, Yoshikawa Y, Kobayashi T, Mimuro H, Fukumatsu M, Kiga K, et al. A Tecpr1-dependent selective autophagy pathway targets bacterial pathogens. Cell Host Microbe. 2011;9:376–89. doi: 10.1016/j.chom.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 45.Spang N, Feldmann A, Huesmann H, Bekbulat F, Schmitt V, Hiebel C, et al. RAB3GAP1 and RAB3GAP2 modulate basal and rapamycin-induced autophagy. Autophagy. 2014;10:2297–309. doi: 10.4161/15548627.2014.994359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–62. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mische SM, Mooseker MS, Morrow JS. Erythrocyte adducin: a calmodulin-regulated actin-bundling protein that stimulates spectrin-actin binding. J Cell Biol. 1987;105:2837–45. doi: 10.1083/jcb.105.6.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brauburger K, Akyildiz S, Ruppert JG, Graeb M, Bernkopf DB, Hadjihannas MV, et al. Adenomatous polyposis coli (APC) membrane recruitment 3, a member of the APC membrane recruitment family of APC-binding proteins, is a positive regulator of Wnt-beta-catenin signalling. FEBS J. 2014;281:787–801. doi: 10.1111/febs.12624. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Zhang J, Bauer A, Zhang L, Selinger DW, Lu CX, et al. Fine-tuning BMP7 signalling in adipogenesis by UBE2O/E2-230K-mediated monoubiquitination of SMAD6. EMBO J. 2013;32:996–1007. doi: 10.1038/emboj.2013.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Au AC, Hernandez PA, Lieber E, Nadroo AM, Shen YM, Kelley KA, et al. Protein tyrosine phosphatase PTPN14 is a regulator of lymphatic function and choanal development in humans. Am J Hum Genet. 2010;87:436–44. doi: 10.1016/j.ajhg.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All likely pathogenic variants are in ClinVar submission number SUB7053346. Accession numbers of submitted variants can be found in Supplemental Table 1.