Correction to: Plant Molecular Biology 10.1007/s11103-020-01041-8
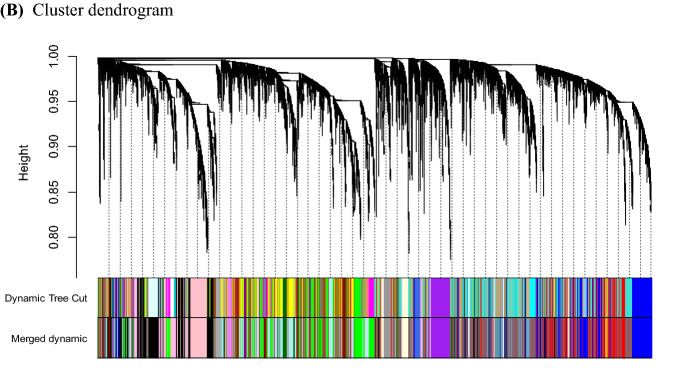
In the above mentioned publication, part of Fig. 6B was distorted (extra diagonal lines appeared). The original article has been corrected and the proper version of Fig. 6B is also published here.
Fig. 6.
WGCNA. Clustered module eigengenes identified by WGCNA and cutoff for merged modules (a). Hierarchical cluster gene tree showing co-expression modules (b). The major tree branches form 38 merged modules that are labeled with different colors. Heat map where each cell color shows the correlation of a trait to each WGCNA module eigengene (c). GO enrichment analysis of genes in green module (d) and genes in module pink (e) according biological processes visualized with REVIGO. Expression profile of all genes of the module green correlating to sucrose loss (f) and module pink correlating to invert sugar (g). The median of variance stabilizing transformed expression values for all samples belonging to one storability group of each gene were determined and the z-score computed. Plots show mean z-score values of all genes and error bars show the standard deviation
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.