Correction to: Journal of Applied Genetics (2020).
10.1007/s13353-020-00586-0
The original version on this paper contained an error. Figure 5 was published with the same image of figure 4. The correct figure 5 is presented here. The original article has been corrected.
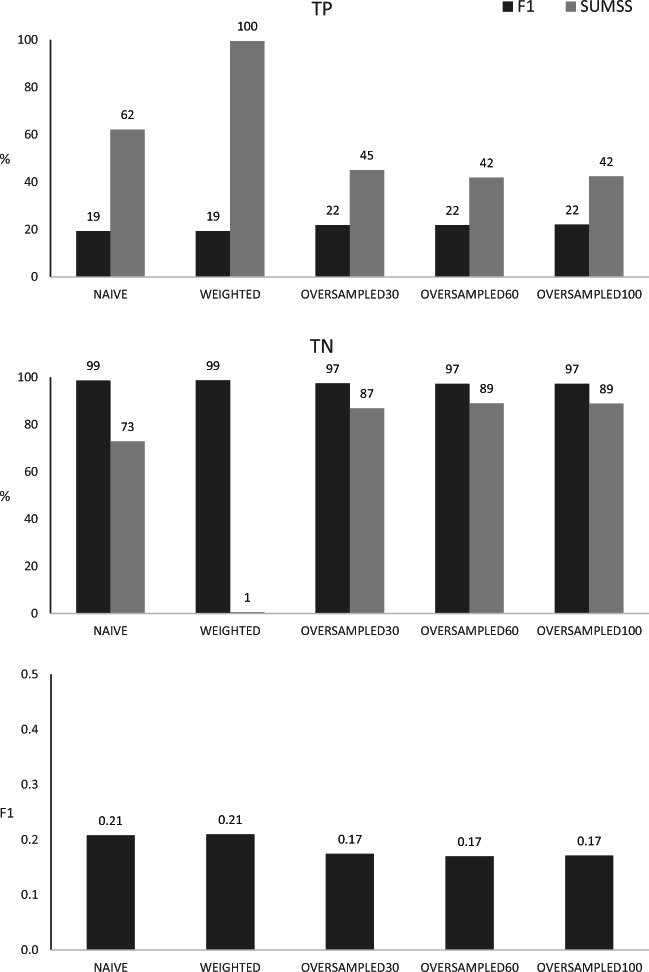
Fig. 5.
Classification of validation data by the different algorithms, based on the probability cutoff thresholds estimated for the F1 or SUMSS metrics. The numbers above columns represent TP—percentages of true positive results, TN—percentages of true negative results, F1—values of the F1 metric
Footnotes
The online version of the original article can be found at 10.1007/s13353-020-00586-0
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.