Abstract
Non-cyanobacterial diazotrophs (NCDs) have recently emerged as potentially important contributors to marine nitrogen fixation. One of the most widely distributed NCDs is Gamma-A, yet information about its autecology is still scarce and solely relies on the PCR-based detection of its nitrogenase (nifH) gene in seawater, since previous metagenomic surveys targeting free-living planktonic size fractions (<3 μm) have not detected it. Here, we explore the diversity, biogeography, size-distribution, and nitrogenase gene expression of Gamma-A across four larger planktonic size-fractions (0.8–5, 5–20, 20–180, and 180–2000 μm) using metagenomes and metatranscriptomes from the Tara Oceans. We detected a single variant of a complete Gamma-A nifH gene along with other nitrogenase-related genes (nifKDT) within a metatranscriptomic-based contig of the Marine Atlas of Tara Ocean Unigenes. Gamma-A was detected in tropical and subtropical oceanic regions across all the size-fractions. However, the highest gene and transcript abundances were found in the 0.8–5 and 5–20 μm size-fractions at the surface, whereas abundances at the deep chlorophyll maximum were lower and similar across all size-fractions. The ubiquitous presence of active Gamma-A in large planktonic size-fractions suggests a filamentous or particle-attached lifestyle and places its potential to fix nitrogen in larger planktonic compartments.
Subject terms: Microbial biooceanography, Microbial ecology, Biodiversity
Introduction
The production of bioavailable nitrogen (N) supplied via biological dinitrogen (N2) fixation, i.e., the reduction of N2 gas to ammonium, supports primary productivity in wide areas of the oligotrophic oceans [1]. Besides N2-fixing (or diazotrophic) cyanobacteria, recent studies have also suggested that non-cyanobacterial diazotrophs (NCDs) might play key roles in marine N2 fixation [2]. Within the diversity of the nitrogenase-encoding nifH gene, a marker gene frequently used to assess the diversity of diazotrophs, a group of Gammaproteobacteria known as Gamma-A (also γ-24774A11 or UMB) [3, 4] has been suggested to be one of the most important NCDs [5, 6]. However, all current knowledge of Gamma-A is solely based on amplification of a small fragment of its nifH gene, which can be detected by degenerate PCR-primers [7] or quantified using specific primers and probes in quantitative PCR surveys [4, 8, 9]. Applying these approaches, Gamma-A has been found in the warm, oligotrophic, and fully oxygenated surface waters of tropical and subtropical latitudes [3–6, 8, 10–12], leading to the hypothesis that this diazotroph might rely on either a light-driven machinery or on photosynthetic products from other organisms [13]. However, the lack of genomic information has prevented further insights about the metabolism and the ecology of Gamma-A.
One of the many unresolved questions about Gamma-A concerns its cell-size distribution. The size-class where N2 fixation occurs is important because it determines, for example, the efficiency of sinking and sequestration of fixed N into deep waters [14]. Despite the widespread distribution of Gamma-A, none of the recent global metagenomic studies that explore marine diazotrophic diversity within the free-living size-fraction (<3 μm) have detected Gamma-A-related nifH sequences [15, 16]. Indeed, among previous PCR-based studies that quantified the abundance/expression of the Gamma-A nifH gene (Table S1), the only two studies that explicitly distinguished between the <3 and >3 μm size-fractions showed that Gamma-A was exclusively present in the >3 μm size-fractions [17, 18]. Furthermore, Gamma-A-related nifH sequences have recently been associated with sinking particles ranging in size from 50 to 200 μm [14] and the gut content of copepods [19]. Altogether this suggests that Gamma-A may either be large in size or attached to particles or other organisms, but a thorough global analysis of the distribution of Gamma-A across size fractions has never been conducted.
Here, we aimed to gain further knowledge of Gamma-A by searching for Gamma-A nifH-related sequences within the Marine Atlas of Tara Ocean Unigenes (MATOU) database [20] and by exploring their presence and activity (via expression of nif transcripts) in metagenomes and metatranscriptomes across four planktonic size-fractions (0.8–5, 5–20, 20–180, and 180–2000 µm) from the sunlit ocean sampled during the global Tara Oceans expedition [20]. Methods used for the identification and quantification of Gamma-A genes in the Tara Oceans dataset are described in the Supplementary information.
Results and discussion
We recruited only one Gamma-A nifH-containing contig (contig ID “MATOU-v1_23614344”) from MATOU that shared 99.7% nucleotide identity with the Gamma-A nifH gene (AY896371.1). The MATOU-v1_23614344 contig (hereafter Gamma-A-MATOU) had a length of 4737 nucleotides and we predicted the complete nifH, nifK, nifD and nifT gene sequences (Table 1). The low percent identity that the Gamma-A-MATOU had compared with current sequenced genomes highlights the gap of representative genomes within this Gammaproteobacterial diazotrophic cluster.
Table 1.
Gene annotation of the Gamma-A-MATOU contig.
| Nucleotide position | Best BLASTn hit in NCBI | |||||
|---|---|---|---|---|---|---|
| MATOU sequence ID | # ORF | Gene Annotation | Start | End | Hit description [ACCN] | % identity |
| MATOU-v1_23614344 | 1 | nifH | 83 | 976 | Uncultured bacterium clone 9260A2D08 dinitrogenase reductase (nifH) gene, partial cds [KC013050.1] | 99.72 |
| MATOU-v1_23614344 | 2 | nifDa | 1115 | 2615 | Uncultured prokaryote clone EGM_33 nitrogenase molybdenum-iron protein (nifD) gene, partial cds [KY441249.1] | 78.39 |
| MATOU-v1_23614344 | 3 | nifK | 2768 | 4339 | Gynuella sunshinyii YC6258, complete genome [CP007142.1] | 73.02 |
| MATOU-v1_23614344 | 4 | nifT | 4427 | 4639 | Gamma proteobacterium BW-2 chromosome, complete genome [CP032507.1] | 68.93 |
aIt contains a possible insertion of 22 nucleotides that breaks the reading frame possibly due to assembly or sequencing errors.
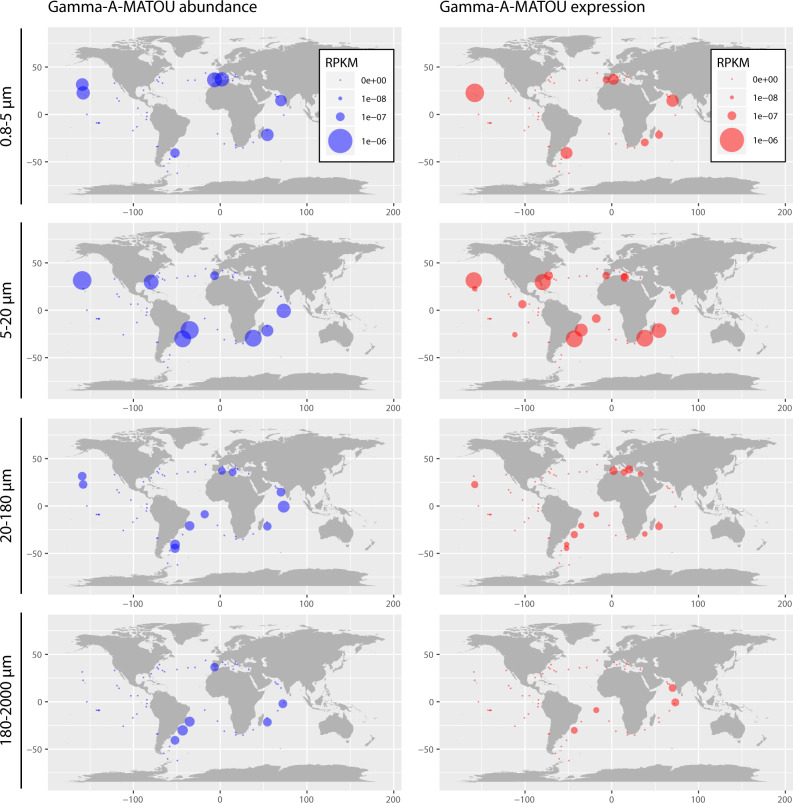
We analyzed the abundance and the expression of the Gamma-A-MATOU across size-fractions in surface (Fig. 1) and deep chlorophyll maximum (DCM) (Fig. S1) waters of the global ocean. Gamma-A-MATOU was detected in 49 (out of 355) metagenomes and 65 (out of 354) metatranscriptomes, corresponding to 24 stations located in the Mediterranean Sea, Indian Ocean, North, and South Atlantic Oceans and North and South Pacific Oceans (Figs. 1 and S1). Within the 5–20 µm size-fraction, the number of metatranscriptomes with presence of Gamma-A-MATOU (n = 27) exceeded the number of metagenomes (n = 15). Whereas the relative abundance of genes and transcripts of the Gamma-A-MATOU was remarkably constant across the four size-fractions at the DCM, it was much more variable between size-fractions in the surface layer, being higher in the 0.8–5 and 5–20 µm size-fractions than in the two largest size-fractions (Fig. 2).
Fig. 1. Distribution of Gamma-A-MATOU in the surface ocean.
Abundance (metagenome-based; left panel) and expression (metatranscriptome-based; right panel) of Gamma-A-MATOU across size fractions are shown. The area of the bubble is proportional to the abundance of metagenomic reads (blue) or transcripts (red) of Gamma-A-MATOU for each sample. Abundances of metagenomic and metatranscriptomic reads are expressed as RPKM (Reads Per Kilobase covered per Million of mapped reads).
Fig. 2. Abundance and expression of the Gamma-A-MATOU nitrogenase gene cluster across size fractions and depths.
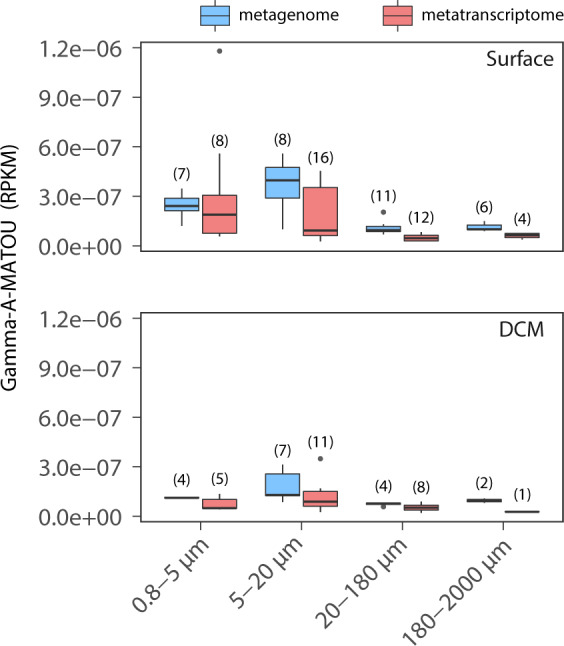
Boxplots representing the abundance (blue) and expression (red) of Gamma-A-MATOU. The number of samples used for the calculation of each boxplot is indicated between parentheses. Abundances of metagenomic and metatranscriptomic reads are expressed as RPKM (Reads Per Kilobase covered per Million of mapped reads).
This global distribution of active Gamma-A in tropical and subtropical photic waters is consistent with previous primer-based studies [5, 6] but further shows its ubiquitous occurrence across planktonic size-fractions spanning at least three orders of magnitude in size (Figs. 1 and S1). The presence of diazotrophs in large planktonic size-fractions, which might include sinking particles and the guts of copepods [14, 19], has been linked to a more efficient sinking and sequestration of fixed N in deep waters [21]. Hence, our results suggest that Gamma-A may be one of the most important NCD in marine nitrogen fixation. Its presence in the 0.8–5 µm size-fraction, together with its previously reported absence in size-fractions <3 µm [15–18], suggests that most of the signal from the 0.8–5 µm size-fraction was coming from particles larger than 3 µm. Furthermore, the relative abundance of genes and transcripts of the Gamma-A-MATOU were generally higher in the 0.8–5 and 5–20 µm size-fractions, indicating that Gamma-A might be fixing N2 preferentially in these fractions, although transcripts were detected even in the largest size-fraction (180–2000 µm) but with lower abundances (Fig. 2). Our analysis further suggests that Gamma-A might possess the ability to attach to different marine phytoplankton species spanning sizes from 3 to 2000 µm, or to be associated with organic particles of various sizes. Sinking particles have been suggested to be potential niches for heterotrophic N2 fixation [2, 14, 22] due to microaerobic environments generated within these particles by microbial remineralization of organic matter [23, 24] and may provide suitable conditions for oxygen-sensitive nitrogenases. Gamma-A might also form aggregates of cells like those observed in other N2-fixing gammaproteobacterial species such as Pseudomonas stutzeri, which might be a mechanism to control O2 diffusion and facilitate N2 fixation in oxic environments [25]. Alternatively, Gamma-A might be a filamentous N2-fixing microorganism such as the diazotrophic cyanobacterium Trichodesmium [26], as Trichodesmium nifH gene sequences were also recruited across different size-fractions from this same dataset, thus pointing to the presence of filaments of different sizes (data not shown).
The new information of Gamma-A nitrogenase-related genes provided here might help in the design of new molecular probes that combined with visualization techniques such as geneFISH [27] and cell-sorting techniques [28] will yield insight into the genome and the lifestyle of this cosmopolitan diazotroph. Future efforts focused on reconstructing prokaryotic genomes of larger planktonic size-fractions might help to reveal the metabolic potential of some of the currently elusive uncultured diazotrophic microorganisms such as Gamma-A.
Supplementary information
Acknowledgements
We specially thank Ana M. Cabello, Rosie Gradoville, Katie Harding, Marine Landa, Kendra Turk-Kubo and Clara Ruiz-Gonzalez for their helpful comments and suggestions on the manuscript. JPZ was supported by Simons Collaboration on Ocean Processes and Ecology (SCOPE, grant 329108). FMC-C was supported by a Marie Curie Individual Global Fellowship—Horizon 2020 European Framework Programme (UCYN2PLAST, grant 749380).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Francisco M. Cornejo-Castillo, Email: frcornej@ucsc.edu
Jonathan P. Zehr, Email: zehrj@ucsc.edu
Supplementary information
The online version of this article (10.1038/s41396-020-00765-1) contains supplementary material, which is available to authorized users.
References
- 1.Karl D, Michaels A, Bergman B, Capone D, Carpenter E, Letelier R, et al. Dinitrogen fixation in the world’s oceans. Biogeochemistry. 2002;57/58:47–98. doi: 10.1023/A:1015798105851. [DOI] [Google Scholar]
- 2.Bombar D, Paerl RW, Riemann L. Marine non-cyanobacterial diazotrophs: moving beyond molecular detection. Trends Microbiol. 2016;24:916–27.. doi: 10.1016/j.tim.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Bird C, Martinez JM, O’Donnell AG, Wyman M. Spatial distribution and transcriptional activity of an uncultured clade of planktonic diazotrophic γ-proteobacteria in the Arabian Sea. Appl Environ Microbiol. 2005;71:2079–85.. doi: 10.1128/AEM.71.4.2079-2085.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moisander PH, Beinart RA, Voss M, Zehr JP. Diversity and abundance of diazotrophic microorganisms in the South China Sea during intermonsoon. ISME J. 2008;2:954–67. doi: 10.1038/ismej.2008.51. [DOI] [PubMed] [Google Scholar]
- 5.Moisander PH, Serros T, Paerl RW, Beinart RA, Zehr JP. Gammaproteobacterial diazotrophs and nifH gene expression in surface waters of the South Pacific Ocean. The. ISME J. 2014;8:1962–73. doi: 10.1038/ismej.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Langlois R, Großkopf T, Mills M, Takeda S, LaRoche J. Widespread distribution and expression of Gamma A (UMB), an uncultured, diazotrophic, γ-proteobacterial nifH phylotype. PLoS ONE. 2015;10:e0128912. doi: 10.1371/journal.pone.0128912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehr JP, Turner PJ. Nitrogen fixation: nitrogenase genes and gene expression. Methods Microbiol. 2001;30:271–86.. doi: 10.1016/S0580-9517(01)30049-1. [DOI] [Google Scholar]
- 8.Church MJ, Short CM, Jenkins BD, Karl DM, Zehr JP. Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl Environ Microbiol. 2005;71:5362–70.. doi: 10.1128/AEM.71.9.5362-5370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Langlois RJ, Hümmer D, LaRoche J. Abundances and distributions of the dominant nifH phylotypes in the Northern Atlantic Ocean. Appl Environ Microbiol. 2008;74:1922–31.. doi: 10.1128/AEM.01720-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bonnet S, Rodier M, Turk-Kubo KA, Germineaud C, Menkes C, Ganachaud A, et al. Contrasted geographical distribution of N2 fixation rates and nifH phylotypes in the Coral and Solomon Seas (southwestern Pacific) during austral winter conditions. Glob Biogeochem Cycles. 2015;29:1874–92. doi: 10.1002/2015GB005117. [DOI] [Google Scholar]
- 11.Gradoville MR, Bombar D, Crump BC, Letelier RM, Zehr JP, White AE. Diversity and activity of nitrogen-fixing communities across ocean basins. Limnol Oceanogr. 2017;62:1895–909.. doi: 10.1002/lno.10542. [DOI] [Google Scholar]
- 12.Chen TY, Chen YL, Sheu DS, Chen HY, Lin YH, Shiozaki T. Community and abundance of heterotrophic diazotrophs in the northern South China Sea: revealing the potential importance of a new alphaproteobacterium in N2 fixation. Deep Sea Res Part I. 2019;143:104–14.. doi: 10.1016/j.dsr.2018.11.006. [DOI] [Google Scholar]
- 13.Moisander PH, Benavides M, Bonnet S, Berman-Frank I, White AE, Riemann L. Chasing after non-cyanobacterial nitrogen fixation in marine pelagic environments. Front Microbiol. 2017;8:1736. doi: 10.3389/fmicb.2017.01736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnelid H, Turk-Kubo K, Ploug H, Ossolinski JE, Collins JR, Van Mooy BA, et al. Diverse diazotrophs are present on sinking particles in the North Pacific Subtropical Gyre. ISME J. 2019;13:170–82. doi: 10.1038/s41396-018-0259-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornejo-Castillo FM. Diversity, ecology and evolution of marine diazotrophic microorganisms. Barcelona: Universitat Politècnica de Catalunya; 2017.
- 16.Delmont TO, Quince C, Shaiber A, Esen ÖC, Lee ST, Rappé MS, et al. Nitrogen-fixing populations of planctomycetes and proteobacteria are abundant in surface ocean metagenomes. Nat Microbiol. 2018;3:804–13. doi: 10.1038/s41564-018-0176-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benavides M, Moisander PH, Daley MC, Bode A, Aristegui J. Longitudinal variability of diazotroph abundances in the subtropical North Atlantic Ocean. J Plankton Res. 2016;38:662–72.. doi: 10.1093/plankt/fbv121. [DOI] [Google Scholar]
- 18.Gradoville MR, Farnelid H, White AE, Turk-Kubo KA, Stewart B, Ribalet F, et al. Latitudinal constraints on the abundance and activity of the cyanobacterium UCYN-A and other marine diazotrophs in the North Pacific. Limnol Oceanogr. 2020. 10.1002/lno.11423.
- 19.Scavotto RE, Dziallas C, Bentzon-Tilia M, Riemann L, Moisander PH. Nitrogen-fixing bacteria associated with copepods in coastal waters of the North Atlantic Ocean. Environ Microbiol. 2015;17:3754–65.. doi: 10.1111/1462-2920.12777. [DOI] [PubMed] [Google Scholar]
- 20.Carradec Q, Pelletier E, Da Silva C, Alberti A, Seeleuthner Y, Blanc-Mathieu R, et al. A global ocean atlas of eukaryotic genes. Nat Commun. 2018;9:1–3. doi: 10.1038/s41467-017-02342-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karl DM, Church MJ, Dore JE, Letelier RM, Mahaffey C. Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc Natl Acad Sci. 2012;109:1842–9. doi: 10.1073/pnas.1120312109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pedersen JN, Bombar D, Paerl RW, Riemann L. Diazotrophs and N2-fixation associated with particles in coastal estuarine waters. Front Microbiol. 2018;9:2759. doi: 10.3389/fmicb.2018.02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploug H, Kühl M, Buchholz-Cleven B, Jørgensen BB. Anoxic aggregates-an ephemeral phenomenon in the pelagic environment? Aquat Microb Ecol. 1997;13:285–94.. doi: 10.3354/ame013285. [DOI] [Google Scholar]
- 24.Ploug H. Small-scale oxygen fluxes and remineralization in sinking aggregates. Limnol Oceanogr. 2001;46:1624–31.. doi: 10.4319/lo.2001.46.7.1624. [DOI] [Google Scholar]
- 25.Bentzon-Tilia M, Severin I, Hansen LH, Riemann L. Genomics and ecophysiology of heterotrophic nitrogen-fixing bacteria isolated from estuarine surface water. MBio. 2015;6:e00929–15. doi: 10.1128/mBio.00929-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergman B, Sandh G, Lin S, Larsson J, Carpenter EJ. Trichodesmium—a widespread marine cyanobacterium with unusual nitrogen fixation properties. FEMS Microbiol Rev. 2013;37:286–302. doi: 10.1111/j.1574-6976.2012.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moraru C, Lam P, Fuchs BM, Kuypers MM, Amann R. GeneFISH—an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol. 2010;12:3057–73.. doi: 10.1111/j.1462-2920.2010.02281.x. [DOI] [PubMed] [Google Scholar]
- 28.Batani G, Bayer K, Böge J, Hentschel U, Thomas T. Fluorescence in situ hybridization (FISH) and cell sorting of living bacteria. Sci Rep. 2019;9:1–3. doi: 10.1038/s41598-019-55049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.