Abstract
Early studies indicated that the androgen receptor (AR) might play important roles in the regulating of the initiation and progression of hepatocellular carcinoma (HCC), but its linkage to the surrounding macrophages and their impacts on the HCC progression remain unclear. Here we found that macrophages in liver cancer might function via altering the microRNA, miR-92a-2-5p, in exosomes to decrease liver cancer cells AR expression, which might then lead to increase the liver cancer cells invasion. Mechanism dissection revealed that miR-92a-2-5p from the exosomes could target the 3′UTR of AR mRNA to suppress AR translation, altering the PHLPP/p-AKT/β-catenin signaling to increase liver cancer cells invasion. Preclinical studies demonstrated that targeting this newly identified signaling with miR-92a-2-5p inhibitors led to suppress liver cancer progression. Together, these findings suggest that macrophages in the liver cancer tumor microenvironment may function via exosomes to regulate liver cancer progression, and targeting this newly identified macrophages/exosomes-miR-92a-2-5p/AR/PHLPP/p-AKT/β-catenin signaling may help in the development of novel treatment strategies to better suppress liver cancer progression.
Subject terms: Cancer microenvironment, Oncogenes
Introduction
Primary liver cancer (PLC) is one of the most important public health issues facing the world. Among which, hepatocellular carcinoma (HCC) accounts for more than 90% of PLC. HCC is the fifth most commonly diagnosed tumor and the second leading cause of cancer death in the world [1]. In the past 30 years, the incidence of HCC has almost tripled in the United States, making it the fastest rising incidence of cancer in both women and men [2]. According to an epidemiological survey of the US population, the incidence of HCC was at least 6 per 100,000 in 2010 [3]. Worldwide incidence of HCC is more than twice as high in males as in females. In 2019, there were 42,030 estimated new liver and intra hepatic bile duct cancer cases and 31,780 estimated deaths in the USA. Among which, the incidence rate of men will be probably three times higher than that of women, and the male patients with HCC usually have a poorer prognosis than female patients [4].
The apparent male prevalence of HCC incidence suggests that sex hormones (androgens and estrogens) may be involved in the regulation of HCC initiation, progression, and metastasis [5]. The androgen receptor (AR) roles in regulating HCC initiation and progression have been examined and the results appear to be complex, as AR can promote HCC initiation and progression at early stage, yet suppress its invasion in the later stages [6–8].
Tumor initiation, progression, and metastasis are significantly influenced by the tumor microenvironment, and infiltration of inflammatory cells, including macrophages, has been proven to be one of the key factors [9, 10]. Macrophages are the most common immune-related cells and can be broadly divided into two subtypes, M1 type (classical) and M2 type (alternative), under different environments [11]. M2 macrophages can secrete multiple types of cytokines and growth factors as well as being involved in extracellular matrix remodeling, angiogenesis, and immunosuppression, which are typical features important for promoting tumor growth and metastasis. Tumor-associated macrophages (TAMs), which exhibit the M2 phenotype, have been proven to be related to the progression and metastasis of many tumors [12].
Exosomes are secreted by a variety of cells, including macrophages, and can migrate from macrophages to tumor cells to promote tumor progression, invasion, and metastasis [13–15]. Studies have shown that exosomes can create an immunosuppressive environment through the signaling between stromal cells and tumor cells to impact tumor progression.
Here, we found that macrophages can promote liver cancer cells invasion through secretion of the exosomes containing the microRNA (miR), miR-92a-2-5p, to liver cancer cells via regulating AR/PHLPP/p-AKT/β-catenin signaling. Thus, we delineated a novel pathway for macrophages’ effects on liver cancer progression, as well as provided mechanistic explanations for seemingly contrasting roles of AR in liver cancer development and progression.
Materials and methods
Cell culture and co-culture
Human liver cancer SK-HEP-1 and Hep G2 cell lines, human HEK 293 T-cell line, monocytic leukemia cell line THP-1, and mouse HCC Hepa 1-6 cell lines were purchased from ATCC. The HA22T cell line (BCRC No. 60168) was a gift from Professor Yuh-Shan Jou (Academia Sinica, Taiwan). SK-HEP-1, HA22T, Hep G2, Hepa 1-6, and HEK 293 T cells were cultured in DMEM. THP-1 cells were cultured in RPMI-1640 media. To induce differentiation into macrophages, THP-1 cells were cultured with 100 ng/ml PMA (Sigma) for 48 h.
We used 0.4-μm-pore-size transwell chambers (Corning Life Science) for the co-culture system. 1 × 105 induced THP-1 cells in 1.5 ml media were added to the upper chambers, and 1 × 105 liver cancer cells in 2.5 ml media were added to the bottom chambers of the six-well plates. For the control group, we only added 1.5 ml of the media to the upper chamber, and both groups were treated with the same conditions. After 48 h of co-culture, the liver cancer cells were collected for further experiments.
Exosomes isolation and identification
THP-1 cells were cultured with 100 ng/ml PMA in normal RPMI-1640 media for 48 h, after which the media was refreshed with RPMI-1640 containing 10% FBS with exosomes removed. After 3 days, the cell culture media was collected and centrifuged at 300 × g for 10 min, 16,500 × g for 20 min, and then filtered through a 0.22 μm filter to remove residual cells and debris. The pellet was collected by ultracentrifugation at 120,000 × g for 70 min (Beckman Coulter) and then resuspended in 50–100 μl PBS. The protein markers, CD9 and CD63, were used to identify exosomes by western blot analysis.
Quantitative real-time PCR
Total RNA was extracted by Trizol reagent (Invitrogen) and reverse transcription was performed using 2 μg of total RNA. The mRNA or miRNA expression levels were conducted and calculated by the Bio-Rad CFX96 system. Expression levels were normalized to GAPDH (for mRNA) or U6 (for miRNA) and relative expression was assessed by ΔΔCt values. All primers were purchased from Integrated DNA Technologies Company. The primers we used for PCR are listed in Supplementary Table 1.
Western blot assay
Cells or exosomes were lysed in RIPA lysis buffer and 30 µg protein was run on SDS/PAGE gels and then transferred to PDVF membranes (Millipore). After blocking for 1 h and rinsed with TBST three times, the membranes were incubated in relevant primary antibodies at 4 °C for overnight. Thereafter, the membranes were rinsed three times and incubated with secondary antibodies, and finally imaged by the ECL system (Thermo Fisher Scientific). The AR (N-20), GAPDH (6c5), CD9 (C-4), CD63 (E-12), β-catenin (E-5), and p-AKT primary antibodies were purchased from Santa Cruz Biotechnology, the PH domain leucine-rich repeat protein phosphatase (PHLPP) primary antibody was purchased from Bethyl Laboratories.
Cell invasion assay
Transwell chambers (Corning Life Science) at 8 μm pore size were used for invasion assay in 24-well plates. First, the upper chamber was filled with 100 μl of diluted Matrigel (1:20 dilution) and incubated for 2 h at 37 °C. Then, 150 μl of serum-free media containing 5 × 104 liver cancer cells was added to the upper chambers, while 750 μl of media containing 10% FBS was added to the lower chambers. The noninvasive cells were then removed and invaded cells were fixed with methanol and stained with 0.1% crystal violet for 30 min after 24 h of incubation and six random fields were counted under a microscope. Each sample was run in triplicate and repeated three times.
Luciferase assay
The 2000 bp wild-type or mutant 3′UTR region of AR was inserted into psiCHECK2-basic vector (Promega) and transduced into pLKO/miR-92a-2-5p inhibitor and pLKO/oemiR-92a-2-5p stable cells using Lipofectamine 3000 (Invitrogen). Luciferase activities were measured 48 h after transfection by a Dual-Luciferase Assay (Promega). The sequence for AR 3′UTR wild type and mutant plasmids is listed in Supplementary Table 1.
In vivo studies
Male nude mice at 5–7 weeks old were purchased from NCI and we randomly divided the animals into three groups according to the random number table (eight in each group) for xenografting as follows, (Group 1) SK-HEP-1-Luc co-injected with Matrigel, (Group 2) SK-HEP-1-Luc co-injected with THP-1-plko cells, (Group 3) SK-HEP-1-Luc cells co-injected with THP-1-plko-miR-92a-2-5p inhibitor. All cells were injected into left lobe of the liver. Tumor growth and metastasis were monitored weekly via in vivo imaging system (IVIS) analysis. Eight weeks later, we sacrificed the mice, removed tumors and metastases for counting, and further studies for statistical analysis. The animal experiments were approved and conducted under the supervision of the University Committee on Animal Resources of University of Rochester Medical Center.
Immunohistochemical (IHC) staining
Mice liver tumor tissues were embedded in paraffin after 24 h of fixation in 4% paraformaldehyde. The tissues were cut to a thickness of 5 μm and dewaxed in a 65 °C incubator for 30 min and then dewaxed in xylene. After that, the tissues were hydrated in ethanol, then tissues were placed in sodium citrate (pH = 6) for half an hour in a microwave for antigen retrieval, and then 3% H2O2-methanol was used to inactivate endogenous peroxidase. After blocking for 30 min in goat serum, tissues were incubated with primary antibody overnight at 4 °C, then rinsed with PBS, incubated with secondary antibody, and then incubated with the enzyme conjugate horseradish peroxidase-streptavidin and detected by DAB (Zymed). Finally, the tissues were counterstained with hematoxylin and mounted with aqueous mounting gel. After the sealant dried, positive cells in the tissues were observed and calculated under a microscope.
Statistics analysis
Experiments were performed at least three separate times with data points in triplicate. SPSS 19.0 software (SPSS Inc) was used to perform statistical analyses. The experimental results were showed as the mean ± SD. Differences in different groups were analyzed by Student’s t test or one way ANOVA. P values ≤ 0.05 were considered statistically significant.
Results
The invasion capacity of liver cancer cells increases after co-culture with macrophages
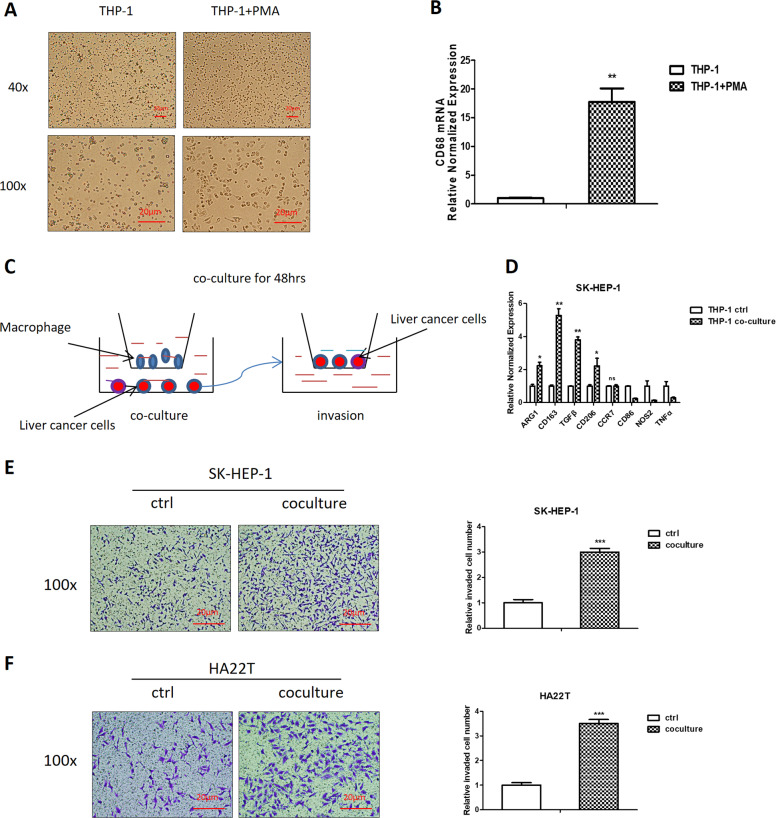
To study the potential impact of infiltrating macrophages on the liver cancer progression, we first applied 100 ng/ml PMA to induce the macrophages in THP-1 cells (Fig. 1a), which are characterized by the expression of recognized macrophage marker CD68 (Fig. 1b). We then applied a co-culture system to culture THP-1 macrophages together with liver cancer cells (Fig. 1c) for 48 h. The results revealed that the co-culture led to increase the M2 markers (CD206, CD163, TGFβ, Arginase-1) in macrophages (Fig. 1d), and the invasion capacity of liver cancer SK-HEP-1 and HA22T cells was also increased (Fig. 1e, f). At the same time, to exclude the role of proliferation in invasion process, we performed MTT assay, and the results showed that the proliferation of liver cancer cells decreased in response to macrophage co-culture, therefore it is unlikely that increased invasion is due to increased proliferation (Supplementary Fig. S3a).
Fig. 1. The invasion capacity of liver cancer cells increases after co-culture with macrophages.
a THP-1 cells were cultured with phorbol-12-myristate-13-acetate (PMA) for 48 h. Representative images of the two groups (THP-1, THP-1+PMA) were shown. b CD68 (macrophages marker) expression was checked by qRT-PCR assay. c Schematic diagram of co-culture and invasion system. d The qRT-PCR was used in macrophages to check M2 markers (Arginase-1, CD163, TGFβ, CD206) and M1 markers (CD86, NOS2, TNFα, CCR7) expression levels after co-culture with SK-HEP-1 and HA22T. e, f Invasion capacity was measured in SK-HEP-1 and HA22T after co-culture compared with controls (ctrl), quantitations are at the right. Data are presented as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
Together, the data from Figs. 1a–f and S3a indicate that infiltrating macrophages can increase liver cancer cells invasion.
Mechanism dissection of how infiltrating macrophages can increase the liver cancer cells invasion: via altering the AR expression
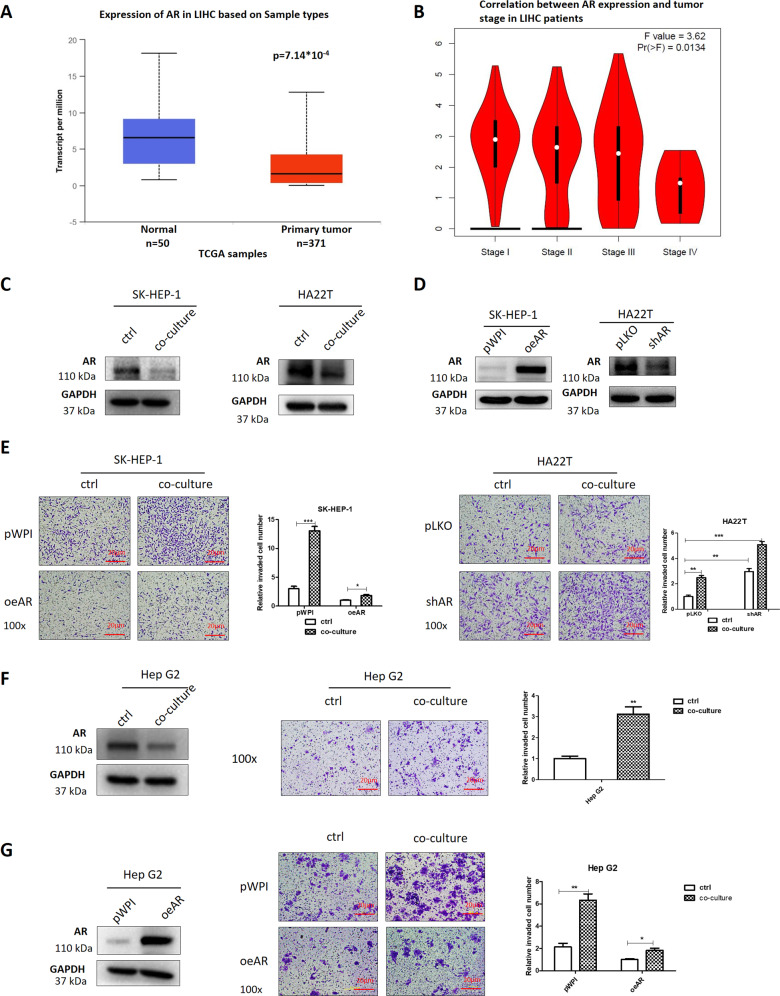
To dissect the mechanism of how infiltrating macrophages can increase liver cancer cells invasion, we focused on the AR signaling, as recent studies indicated that AR might play a key role to alter the liver cancer progression. We first analyzed the clinical significance of AR in HCC samples through UALCAN (http://ualcan.path.uab.edu/) and GEPIA (http://gepia.cancer-pku.cn/) websites and clinical data from TCGA database samples showed a decrease in AR expression in the primary tumors (Fig. 2a), with a decreased AR expression as tumor staging increased (Fig. 2b). We also collected tumors from ten male HCC patients that went under surgical resection and were diagnosed Stage II after surgery, the IHC assay showed that AR expression was lower in tumor tissues than in para-tumor tissues (Supplementary Fig. S3b).
Fig. 2. Mechanism dissection of how macrophages increase liver cancer cells invasion: via altering the AR expression.
a Expression of AR in liver hepatocellular carcinoma (LIHC) based on different sample types (normal and primary tumor) in TCGA database. b Correlation between AR expression and tumor stage in LIHC patients from the database. c AR protein expression was checked by western blots in SK-HEP-1 and HA22T cells after co-culture compared with controls (ctrl). d AR expression was detected by western blots after overexpressing AR in SK-HEP-1 cells and knocking down AR in HA22T cells. e Chamber-transwell invasion assays were performed using SK-HEP-1 cells (left) transfected with AR-cDNA (oeAR) and pWPI control vector and HA22T cells (right) transfected with AR-shRNA (shAR) and pLKO control vector after co-culture with macrophages for 48 h compared with ctrls. f AR protein expression (left) and invasion capacity (middle) were measured in Hep G2 cells after co-culture with macrophages for 48 h, quantitation at right. g AR protein expression (left) and invasion capacity (middle) were measured after overexpressing AR or pWPI in Hep G2 cells in co-culture system compared with ctrls. For (e–g) quantitations are at the right. Data are presented as mean ± SD. *p < 0.05; **p < 0.01; ***p < 0.001.
We then examined the co-culture’s influence on the AR expression, and found that AR protein expression in the SK-HEP-1 and HA22T cells decreased after co-culture with THP-1 macrophages (Fig. 2c).
To examine the potential impact of altered AR on the liver cancer cells invasion, we found that increasing AR via adding AR-cDNA decreased the infiltrating macrophages-increased liver cancer cells invasion (Fig. 2d, e, left). In contrast, decreasing AR via adding AR-shRNA led to increase more significantly the liver cancer cells invasion (Fig. 2d, e, right). Similar results were also obtained when we replaced SK-HEP-1 cells with the HCC Hep G2 cells (Fig. 2f, g). Moreover, this enhanced liver cancer cell invasion can also be found in another co-culture system using mouse macrophages (Raw 264.7) and mouse HCC cells (Hepa 1–6) (Supplementary Fig. S1).
Together, the data from Figs. 2a–g, S1, and S3b indicate that infiltrating macrophages may increase the liver cancer cells invasion via suppressing liver cancer cells AR expression.
Mechanism dissection of how infiltrating macrophages can decrease the AR expression in the liver cancer cells: via altering the miRNAs
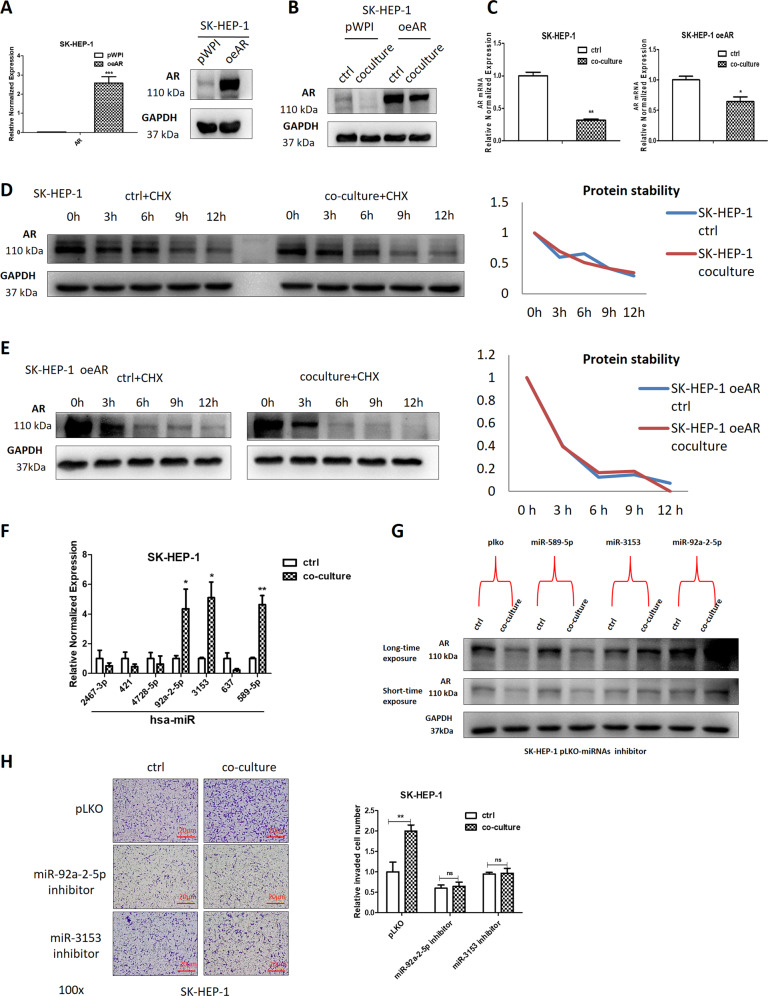
As shown in Fig. 2c, AR overexpression in liver cancer cells could partly reverse co-culture’s function, therefore we examined AR expression in liver cancer oeAR cells, the results showed that co-culture still resulted in a decrease of AR expression in liver cancer oeAR cells (Fig. 3a, b). The AR mRNA level was also decreased in SK-HEP-1 and SK-HEP-1 oeAR cells after co-culture (Fig. 3c). To exclude the possibility that a decreased AR was due to a decreased protein stability, we examined AR protein level in response to cycloheximide to block de novo protein synthesis in liver cancer cells, and results showed that there is no difference between control (not co-cultured) and co-culture groups, both in SK-HEP-1 and SK-HEP-1 oeAR cells (Fig. 3d, e). These results suggest that macrophages may regulate liver cancer cells AR mRNA level either through transcriptional or posttranscriptional regulation.
Fig. 3. Mechanism dissection of how macrophages decrease AR expression in liver cancer cells: via altering the miRNAs.
a The qRT-PCR and western blot were used to detect the efficiency of overexpressing AR (oeAR) in SK-HEP-1 cells. b Western bolt was used to detect the AR expression in SK-HEP-1 oeAR cells in non co-culture control (crtl) and co-culture groups. c The qRT-PCR was used to show AR mRNA expression in SK-HEP-1 and SK-HEP-1 oeAR cells both in ctrl and co-culture groups. d Western blots were used to determine AR protein stability in SK-HEP-1 cells in ctrl and co-culture groups. e Western blots were used to check AR protein stability in SK-HEP-1 oeAR cells in ctrl and co-culture groups. f The qRT-PCR was used to show the seven miRNAs expression in SK-HEP-1 cells in ctrl and co-culture groups. g AR expression was checked by western blot after transducing pLKO-miR-589-5p/pLKO-miR-3153/pLKO-miR-92a-2-5p inhibitor into SK-HEP-1 cells in ctrl and co-culture groups. h Invasion capacity of SK-HEP-1 cells was measured after transducing pLKO-miR-3153/pLKO-miR-92a-2-5p inhibitor in ctrl and co-culture groups, quantitation at right. Data are presented as mean ± SD. ns not significant. *p < 0.05; **p < 0.01.
As the exogenous AR expression was controlled by a promoter that is different than the endogenous chromosomal AR locus, it is more likely that macrophages may impact AR expression in liver cancer cells through posttranscriptional regulation. Particular miRNAs that could target both endogenous and vector-expressed AR mRNA could be used to mediate this process. The bioinformatic analysis led to candidate miRNAs that are capable of doing so.
Further testing of their expression in response to co-culture showed that miR-92a-2-5p, miR-3153, miR-589-5p increased after co-culture in liver cancer cells (Fig. 3f). Inhibition of these miRNAs by vector-based miRNA inhibitors indicated that miR-92a-2-5p and miR-3153 could reverse co-culture’s impact on AR (Fig. 3g), as well as the invasion capacity of liver cancer cells after co-culture (Fig. 3h).
Together, the data from Fig. 3a–h indicate that macrophages may function via increasing miR-92a-2-5p and miR-3153 expression to decrease AR expression of liver cancer cells.
Mechanism dissection of how macrophages increase miRNAs expression in liver cancer: via transportation by exosomes from macrophages to liver cancer cells
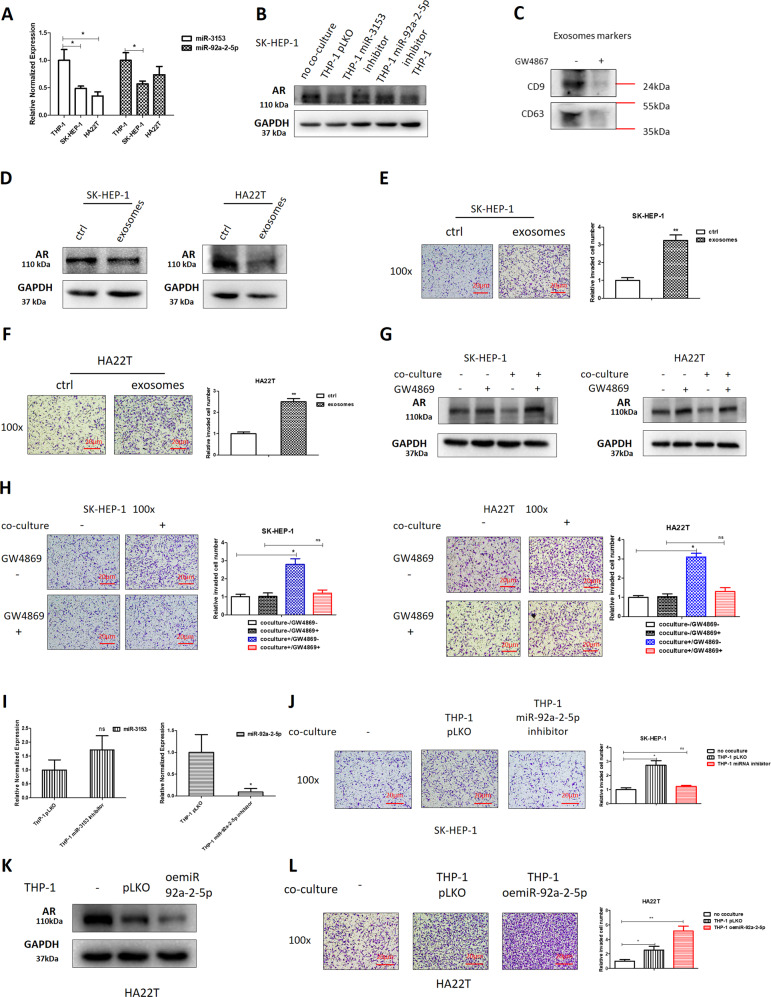
To dissect the mechanism how macrophages increase miRNAs expression in liver cancer, we focused on exosomes, as recent studies indicated that miRNAs can be transported by exosomes from macrophages to neighboring tumor cells [16]. Indeed, the miR-92a-2-5p and miR-3153 expression in THP-1 macrophages were higher than in the liver cancer cells (Fig. 4a). When we inhibited the two miRNAs separately in macrophages and co-cultured these macrophages with liver cancer cells, results showed that it could reverse the co-culture impact on liver cancer cells (Fig. 4b).
Fig. 4. Mechanism dissection of how macrophages increase miRNAs level in liver cancer: via exosomes secretion from macrophages to liver cancer cells.
a The qRT-PCR was used to show the miR-3153 and miR-92a-2-5p expression in SK-HEP-1, HA22T, and THP-1 cells. b Western bolt was used to determine AR expression after transducing miR-3153/miR-92a-2-5p inhibitor into THP-1 cells in no co-culture (ctrl) and co-culture groups. c Exosomes markers (CD9 and CD63) were detected by western blot, and treatment with GW4869 (exosomes inhibitor) was used as a control group to check the extraction efficiency of exosomes. d Western blots were used to determine AR expression in liver cancer cells in ctrl group and exosomes group. Invasion capacity of SK-HEP-1 (e) and HA22T (f) cells was measured in ctrl groups and exosomes groups. g Western blots were used to determine AR expression after treating with exosomes inhibitor (GW4869) with/without co-culture. h Invasion capacity of liver cancer cells after treating with exosome inhibitor (GW4869) with/without co-culture. i The qRT-PCR was used to show the miR-92a-2-5p and miR-3153 levels in exosomes after transducing related miRNA inhibitors in macrophages. j Invasion capacity of liver cancer cells was measured after transducing miR-92a-2-5p inhibitor into macrophages with/without co-culture. k AR expression was checked by western blot after overexpressing miR-92a-2-5p into macrophages with/without co-culture. l Invasion capacity of liver cancer cells was measured after overexpressing miR-92a-2-5p in macrophages with/without co-culture. For (e, f, h, j, l) quantitations are at the right. Data are presented as mean ± SD. ns not significant. *p < 0.05; **p < 0.01.
To directly implicate exosomes from macrophages in regulating liver cancer cell invasion, we cultured the macrophage with addition of DMSO to promote the M2 polarization, and we collected the media from these M2-type macrophages (Fig. 4c), added them to liver cancer cells, and found that macrophages exosomes led to a reduction of AR expression in liver cancer cells (Fig. 4d) and an increase of cell invasion (Fig. 4e, f). Furthermore, if exosomes secretion in macrophages was inhibited by an inhibitor (GW4869) in the co-culture system, macrophages lost the ability to impact both SK-HEP-1 and HA22T cells (Fig. 4g, h).
As it was shown in Fig. 4b, inhibition of the two miRNAs separately in macrophages can reverse co-culture’s function, so we checked the miRNA expression in exosomes after transducing pLKO-miR-92a-2-5p inhibitor or pLKO-miR-3153 inhibitor, the results showed that the miR-92a-2-5p in exosomes was lower in the THP-1 miR-92a-2-5p inhibitor group than in the THP-1 pLKO control group, but the miR-3153 level was higher than that in THP-1 pLKO control groups (Fig. 4i), suggesting that exosomes can transport miR-92a-2-5p from macrophages to liver cancer cells, but not miR-3153. When we added pLKO-miR-92a-2-5p inhibitor into macrophages, the invasion capacity of liver cancer cells also can be reversed (Fig. 4j). Furthermore, when we overexpressed miR-92a-2-5p in macrophages and co-cultured with liver cancer cells, the results showed decreased AR expression and increased invasion capacity of HA22T cells (Fig. 4k, l).
Together, results from Fig. 4a–l suggest that macrophages decrease AR expression of liver cancer cells to increase invasion capacity likely via transporting miR-92a-2-5p from macrophages to liver cancer cells through exosomes.
Mechanism study of how miR-92a-2-5p alters the AR expression under the co-culture condition: via directly targeting the 3′UTR of AR mRNA
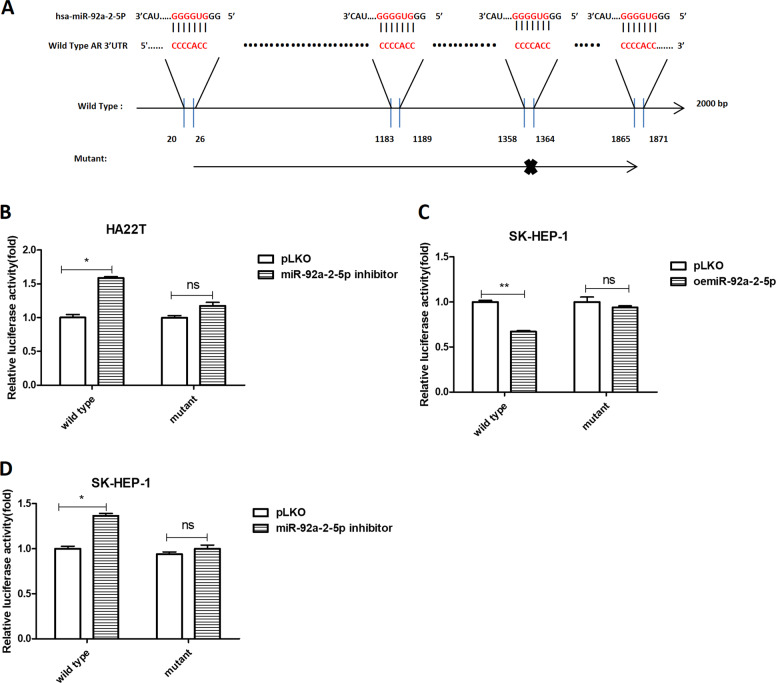
To dissect the mechanism of how macrophages/exosomes/miR-92a-2-5p can decrease the AR expression, we searched the website (http://www.targetscan.org/vert_72/) to identify some potential targeting sites of miR-92a-2-5p that are located on the 3′UTR of AR mRNA. We then constructed a wild-type 3′UTR or a mutant with deletion of target sites of miR-92a-2-5p into psiCHECK2 vector (Fig. 5a). Results from luciferase report assay showed that when transfected with the wild-type AR 3′UTR, the luciferase activity was increased in HA22T cells after inhibiting miR-92a-2-5p and decreased in SK-HEP-1 cells after addition of miR-92a-2-5p. But when transfected with the mutant AR 3′UTR, the luciferase activity had no significant differences (Fig. 5b, c). We also tested knocking down miR-92a-2-5p in the SK-HEP-1 cells, and the results were consistent with those in HA22T cells (Fig. 5b, d), suggesting that miR-92a-2-5p can directly target the 3′UTR of AR mRNA to suppress its protein expression.
Fig. 5. Mechanism dissection of how miR-92a-2-5p alters the AR expression under the co-culture condition: via direct targeting of the 3′UTR of AR mRNA.
a Sequence of 2000 bp wild-type and mutant AR 3′UTR. Luciferase reporter activity was measured after transfection of wild-type or mutant AR 3′UTR in HA22T cells transduced with pLKO or miR-92a-2-5p inhibitor (b) and SK-HEP-1 cells transduced with pLKO or oemiR-92a-2-5p (c). d Luciferase reporter activity was measured after transfection of wild-type or mutant AR 3′UTR in SK-HEP-1 cells transduced with pLKO or miR-92a-2-5p inhibitor. Data are presented as mean ± SD. ns not significant. *p < 0.05; **p < 0.01.
Mechanism dissection of how macrophages/exosomes/miR-92a-2-5p/AR axis alters the invasion capacity of liver cancer cells: via altering the PHLPP/p-AKT/β-catenin signaling
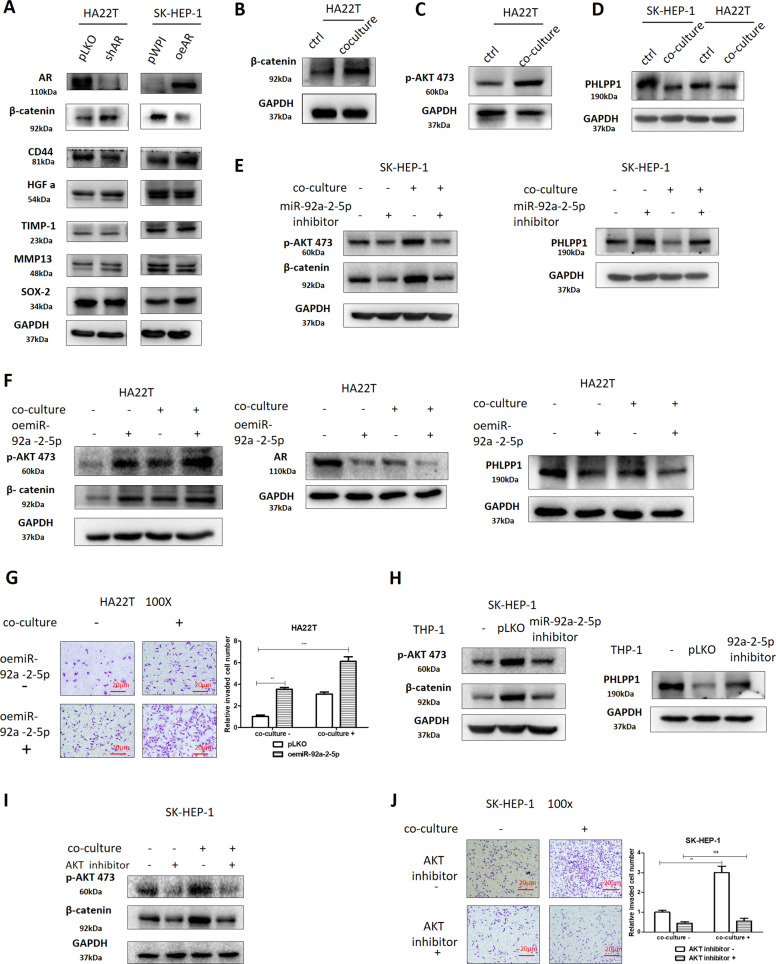
To examine the detailed mechanisms underlying AR’s role in regulating liver cancer cells invasion, we tested the expression of proteins that have been connected with cancer cells migration and invasion in both culture alone or in co-culture. The exploratory experiments led to the focus on β-catenin as it is mostly consistent with the change in AR expression induced by co-culture with macrophages (Fig. 6a, b). At the same time, we constructed the β-catenin wild type and mutant plasmid, then we conducted luciferase assay to check β-catenin activation, the result was consistent with western blot assay (Supplementary Fig. S3c). We also checked the clinical significance of β-catenin in HCC in the UALCAN (http://ualcan.path.uab.edu/) website, which showed an increase in β-catenin expression in primary tumors (Supplementary Fig. S2a), and a higher β-catenin expression meant a lower survival rate (Supplementary Fig. S2b). A survey of literature indicated that AR could transcriptionally regulate PHLPP that can inactivate AKT through dephosphorylation to increase invasion capacity of prostate cancer [17], while p-AKT/β-catenin signaling was related to tumor progression [18]. Indeed, similar results were found in liver cancer cells when we examined the p-AKT and PHLPP levels after co-culture, with increased p-AKT expression in HA22T cells (Fig. 6c), and a decrease of PHLPP level in both cell lines after co-culture (Fig. 6d).
Fig. 6. Mechanism dissection of how the macrophage/miR-92a-2-5p/AR axis alters the invasion capacity of liver cancer cells: through the PHLPP/p-AKT/β-catenin pathway.
a Western blots were used to determine AR downstream genes expression in pLKO/shAR group and pWPI/oeAR group. b Western blot was used to check β-catenin expression in HA22T cells after co-culture. c Western blot was used to check p-AKT expression in HA22T cells with/without co-culture. d Western blot was performed to check PHLPP expression in control (ctrl) and co-culture groups. e Western blot was used to determine β-catenin, p-AKT, and PHLPP expression after transducing miR-92a-2-5p inhibitor into SK-HEP-1 with/without co-culture. f Western blots were used to check β-catenin, p-AKT, AR, and PHLPP expression after overexpressing miR-92a-2-5p in HA22T cells with/without co-culture. g Invasion capacity of HCC cells after overexpressing miR-92a-2-5p in HA22T cells with/without co-culture. h Western blots were used to check β-catenin, p-AKT, and PHLPP expression in SK-HEP-1 cells after transducing pLKO-miR-92a-2-5p inhibitor into macrophages with/without co-culture. i Western blots were used to determine β-catenin and p-AKT expression in SK-HEP-1 cells after treating with/without AKT inhibitor with/without co-culture. j Invasion capacity of liver cancer cells after treating with/without adding AKT inhibitor in SK-HEP-1 cells with/without co-culture. For (g, j) quantitations are at the right. Data are presented as mean ± SD. ns not significant. *p < 0.05; **p < 0.01.
To directly implicate the role of miR-92a-5p in these regulations, we also checked the impact of knocking down/overexpressing miR-92a-2-5p in liver cancer cells in the co-culture system. The results revealed that knocking down the miRNA could reverse macrophages’ influence on liver cancer cells and overexpressing the miRNA could strengthen macrophages’ impact (see Figs. 3h, 6e–g, and S2c). Furthermore, changing the miR-92a-5p expression in macrophages resulted in similar changes in liver cancer cells in invasion as well as protein expression (see Figs. 4j–l, 6h, and S2d). Consistent with the pivotal role of AKT in this process, when we treated the co-culture system with the AKT inhibitor, MK-2206, the impact of macrophages on liver cancer cells was blocked (Figs. 6i, j and S2e, f). Similarly, treatment with an inhibitor of β-catenin, ICG-001, in the co-culture system resulted in blocking the effect of co-culture on liver cancer cells (Supplementary Fig. S2g, h).
Together, results from Figs. 6a–j, S2a–h, and S3c suggest that the macrophages/exosomes/miR-92a-2-5p/AR axis can regulate liver cancer cell invasion via altering the AR/PHLPP/p-AKT/β-catenin signaling.
Preclinical studies targeting this newly identified signaling with miR-92a-2-5p inhibitor with suppression of liver cancer progression
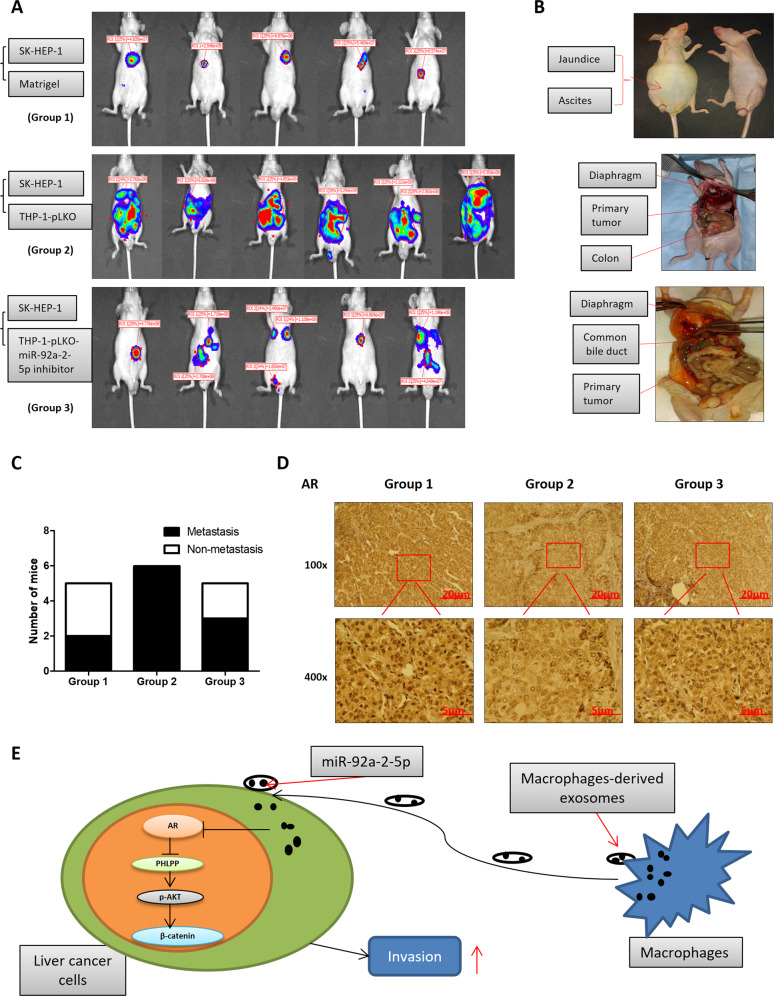
To test whether the mechanisms that we uncovered in vitro similarly play a significant role in vivo, we examined whether the miR-92a-2-5p inhibitor can suppress liver cancer progression in a xenograft animal model. It was found that inhibition of miR-92a-2-5p by vector-based miRNA inhibitor in macrophages could reverse co-culture’s impact on AR (see Fig. 4b), as well as the invasion capacity of liver cancer cells after co-culture (see Fig. 4j). We first generated SK-HEP-1 cells with luciferase reporter gene to check tumor progression in mice using the noninvasive IVIS and we also transduced THP-1 cells with pLKO vector or pLKO-miR-92a-2-5p inhibitor and these SK-HEP-1-Luc cells were co-injected with/without the prepared THP-1 cells. Then 24 mice were divided into three groups (eight in each group) for xenografting as follows, (Group 1) SK-HEP-1-Luc cells diluted 1:1 with Matrigel, (Group 2) SK-HEP-1-Luc co-injected with THP-1-pLKO cells, and (Group 3) SK-HEP-1-Luc co-injected with THP-1-pLKO-miR-92a-2-5p inhibitor. All cells were injected into the left lobes of liver capsules.
After 8 weeks, 16 of the 24 mice generated tumors, 5 in Group 1, 6 in Group 2, and 5 in Group 3, with tumor formation rates at 62.5%, 75%, and 62.5%, respectively. The results of IVIS scans showed that the tumor growth of liver cancer cells with macrophages (THP-1 pLKO) group (Group 2) was significantly faster than that of the control group (Group 1), and the addition of pLKO-miR-92a-2-5p inhibitor to macrophages (Group 3) could partially inhibit tumor growth when compared with the THP-1 pLKO group (Fig. 7a). We sacrificed the mice after 8 weeks and counted the metastasis of each mouse (Fig. 7b). The results showed that the SK-HEP-1-Luc plus THP-1-pLKO co-culture group had more mice with metastasis than the other two groups (Fig. 7c). IHC data also confirmed that when macrophages and liver cancer cells were co-injected into mouse livers, AR expression in tissues was reduced, which was consistent with our in vitro data (Fig. 7d).
Fig. 7. Preclinical studies targeting this newly identified signaling with miR-92a-2-5p inhibitor with suppression of liver cancer progression.
a Tumor size and metastasis in mice were measured by IVIS imaging. b Typical symptoms (ascites and jaundice in upper panel), primary tumor and metastasis (middle and lower panels) in mice. c Histogram of number of mice with metastasis and non-metastasis comparing the three groups. d IHC staining of AR in three groups. The IHC data showed that AR expression was decreased in THP-1 pLKO group (Group 2) compared with the control group (Group 1), and transducing miR-92a-2-5p inhibitor into THP-1 cells (Group 3) could partly reverse AR’s expression. e Schematic model of macrophages-derived exosomes (MDE) mediated transfer of miR-92a-2-5p to increase liver cancer cell invasion via the AR/PHLPP/p-AKT/β-catenin signaling axis.
At the same time, we analyzed miR-92a expression in HCC clinical samples from TCGA database, the result showed that miR-92a expression was increased in tumor tissues when compared with normal tissues (Supplementary Fig. S3d, left). What is more, the data from ENCORI Pan-Cancer Analysis Platform (http://starbase.sysu.edu.cn/panCancer.php) also showed that HCC patients with low miR-92a-2-5p expression had longer survival time than patients with high miR-92a-2-5p expression (Supplementary Fig. S3d, right), which also confirmed the importance of miR-92a-2-5p function in vivo.
In summary, the data in Figs. 7 and S3d demonstrated that miR-92a-2-5p from macrophages played an important role in promoting liver cancer progression via altering AR signaling, and the inhibition of miR-92a-2-5p may suppress the function of macrophages to liver cancer progression.
Discussion
The apparent male prevalence of HCC occurrence suggests that HCC may be regulated by sex hormones, both androgen and estrogen, during its initiation, progression, and metastasis [5]. Studies have shown AR can engage cycle-related kinase to promote HBV-associated hepatocarcinogenesis through HBx-AR signaling. More recently, it was shown that AR signaling can regulate HCC metabolism to promote tumor initiation [19]. On the other hand, it is possible that AR at a molecular level may play different roles during the progression of HCC and AR has been shown to suppress HCC invasion and metastasis [8]. Our study is another example where AR can play a suppressive role for liver cancer invasion and likely tumor metastasis.
More and more evidence demonstrates that the tumor microenvironment may play important roles in the regulation of tumor progression, metastasis, and drug resistance [20, 21]. In addition to the influence of the individual’s genetic background [22, 23], the immune status between different genders is largely regulated by sex hormones such as estrogen, androgen, and progesterone. Sex hormones interact with immune cells such as lymphocytes, monocytes, and dendritic cells through the sex hormone receptors expressed on immune cells, regulating the release of cytokines and chemokines, thereby affecting the proliferation, differentiation and maturation of immune cells [24]. The clinical associations between M2 macrophages and poor prognosis of tumors have been reported in many tumors, including M2 macrophages promoting tumor growth and invasiveness in liver cancer [25]. Despite the reported clinical significances of TAMs, the intrinsic signaling pathways between macrophages and liver cancer cells and how they influence liver cancer progression remains poorly understood. In our study, we found that macrophages can decrease AR expression to increase invasion ability of liver cancer.
Exosomes are 30–150 nm vesicles and formed in the process of endocytosis [26]. By transferring genetic contents through intercellular communication, exosomes play an important role in the progression of many tumors.
The content of exosomes has been shown to include peptides and nucleic acids, such as ncRNAs including miRNAs that can regulate gene expression by binding to the 3′UTR of specific target mRNAs to inhibit translation or to induce degradation. The expression of miRNAs is strictly controlled and expression dysregulation of miRNAs is usually associated with cancers. The miR-92a-2-5p belongs to the miR-106a–363 cluster that can be potentially used as biomarkers of breast cancer and small cell lung cancer [27]. In this study, we demonstrated that macrophages transmitted miR-92a-2-5p to liver cancer cells through exosomes to increase the invasion capacity of liver cancer. Macrophages-derived exosomes (MDE) mediated transfer of miR-92a-2-5p to increase liver cancer cells invasion, while transfection of antagomiR-92a-2-5p to liver cancer cells can partially block the effect of MDE on invasion capacity, suggesting that MDE might contain additional molecules that can regulate liver cancer behaviors in addition to miR-92a-5p, the identity of which remains to be examined.
The PHLPP is a member of the Serine/Threonine protein phosphatase family and consists of two isoforms, PHLPP1 and PHLPP2. PHLPP functions as a negative regulator of PI3K/Akt and RAF/RAS/ERK signals, making it a protective factor in the process of tumor metastasis and resistance [28, 29]. In addition, PHLPP expression is significantly associated with differentiation and local tumor T-stage of lung adenocarcinoma, while low expression of PHLPP is associated with poor prognosis of resectable lung adenocarcinoma [30]. In prostate cancer, inhibition of AR would reduce the expression of FKBP5 and then decrease PHLPP levels to increase phosphorylation of AKT [17, 31]. In our study, we examined the PHLPP pathway in our liver cancer co-culture system, and results showed that the AR decreased by co-culture system can also go through the PHLPP/p-AKT pathway to influence the invasion capacity of liver cancer cells. Consistent with this, using an AKT inhibitor in the co-culture system could block AR’s capacity to regulate liver cancer invasion. Consistent with the role of AKT in regulating Wnt/β-catenin pathway in other tissues [32, 33], in our co-culture system, we also found that this signaling pathway was responsible for the change of β-catenin expression level as well as HCC cells invasion. This conclusion, as shown in Fig. 7e, was strengthened by the outcome in response to the β-catenin inhibitor that could block the biochemical and cell behavior induced by macrophages in the liver cancer tumor microenvironment.
In conclusion, macrophages transmit miR-92a-2-5p to liver cancer cells by shuttling exosomes to increase the invasion capacity of liver cancer, and a potential therapy to target this newly identified signal may help in the improvement of treatments to better suppress the liver cancer progression.
Supplementary information
Acknowledgements
This work was supported by NIH grant (CA155477) and George Whipple Professorship and National Natural Science Foundation of China (81903004). We thank Karen Wolf for editing the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by R. De Maria
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Guodong Liu, Xiwu Ouyang
Contributor Information
Yixiong Li, Email: liyixiong2011@163.com.
Chawnshang Chang, Email: chang@urmc.rochester.edu.
Supplementary information
The online version of this article (10.1038/s41418-020-0575-3) contains supplementary material, which is available to authorized users.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go? Hepatology. 2014;60:1767–75. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Jemal A. Cancer Statistics. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 5.Li Z, Tuteja G, Schug J, Kaestner KH. Foxa1 and Foxa2 are essential for sexual dimorphism in liver. Cancer Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ma W, Lai H, Yeh S, Cai X, Chang C. Androgen receptor roles in hepatocellular carcinoma, fatty liver, cirrhosis and hepatitis. Endocr Relat Cancer. 2014;21:R165–82. doi: 10.1530/ERC-13-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma WL, Hsu CL, Wu MH, Wu CT, Wu CC, Lai JJ, et al. Androgen receptor is a new potential therapeutic target for the treatment of hepatocellular carcinoma. Gastroenterology. 2008;135:947–55. doi: 10.1053/j.gastro.2008.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma W, Hsu C, Yeh C, Wu M, Huang C, Jeng L, et al. Hepatic androgen receptor suppresses hepatocellular carcinoma metastasis through modulation of cell migration and anoikis. Hepatology. 2012;56:176–85. doi: 10.1002/hep.25644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ackerman D, Simon MC. Hypoxia, lipids, and cancer: surviving the harsh tumor microenvironment. Trends Cell Biol. 2014;24:472–8. doi: 10.1016/j.tcb.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–12. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 11.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32:593–604. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 12.Talmadge JE, Donkor M, Scholar E. Inflammatory cell infiltration of tumors: Jekyll or Hyde. Cancer Metastasis Rev. 2007;26:373–400. doi: 10.1007/s10555-007-9072-0. [DOI] [PubMed] [Google Scholar]
- 13.Melo SA, Luecke LB, Kahlert C, Fernandez AF, Gammon ST, Kaye J, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523:177–82. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Allenson K, Castillo J, San Lucas FA, Scelo G, Kim DU, Bernard V, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol. 2017;28:741–7. doi: 10.1093/annonc/mdx004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, et al. Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer. 2015;136:2616–27. doi: 10.1002/ijc.29324. [DOI] [PubMed] [Google Scholar]
- 16.Binenbaum Y, Fridman E, Yaari Z, Milman N, Schroeder A, Ben David G, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. 2018;78:5287–99. doi: 10.1158/0008-5472.CAN-18-0124. [DOI] [PubMed] [Google Scholar]
- 17.Mulholland DJ, Tran LM, Li Y, Cai H, Morim A, Wang S, et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell. 2011;19:792–804. doi: 10.1016/j.ccr.2011.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ma J, Guo X, Zhang J, Wu D, Hu X, Li J, et al. PTEN gene induces cell invasion and migration via regulating AKT/GSK-3β/β-catenin signaling pathway in human gastric cancer. Dig Dis Sci. 2017;62:3415–25. doi: 10.1007/s10620-017-4764-y. [DOI] [PubMed] [Google Scholar]
- 19.Sun H, Yang W, Tian Y, Zeng X, Zhou J, Mok MTS, et al. An inflammatory-CCRK circuitry drives mTORC1-dependent metabolic and immunosuppressive reprogramming in obesity-associated hepatocellular carcinoma. Nat Commun. 2018;9:5214. doi: 10.1038/s41467-018-07402-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Binenbaum Y, Na Ara S, Gil Z. Gemcitabine resistance in pancreatic ductal adenocarcinoma. Drug Resist Updat. 2015;23:55–68. doi: 10.1016/j.drup.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 21.McMillin DW, Negri JM, Mitsiades CS. The role of tumour–stromal interactions in modifying drug response: challenges and opportunities. Nat Rev Drug Discov. 2013;12:217–28. doi: 10.1038/nrd3870. [DOI] [PubMed] [Google Scholar]
- 22.Patin E, Hasan M, Bergstedt J, Rouilly V, Libri V, Urrutia A, et al. Natural variation in the parameters of innate immune cells is preferentially driven by genetic factors. Nat Immunol. 2018;19:302–14. doi: 10.1038/s41590-018-0049-7. [DOI] [PubMed] [Google Scholar]
- 23.Aguirre-Gamboa R, Joosten I, Urbano PCM, van der Molen RG, van Rijssen E, van Cranenbroek B, et al. Differential effects of environmental and genetic factors on T and B Cell immune traits. Cell Rep. 2016;17:2474–87. doi: 10.1016/j.celrep.2016.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bhatia A, Sekhon HK, Kaur G. Sex Hormones and Immune Dimorphism. Sci World J. 2014;2014:1–8. doi: 10.1155/2014/159150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, et al. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol. 2015;62:607–16. doi: 10.1016/j.jhep.2014.10.029. [DOI] [PubMed] [Google Scholar]
- 26.Wahlgren J, Karlson TDL, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li M, Zhou Y, Xia T, Zhou X, Huang Z, Zhang H, et al. Circulating microRNAs from the miR-106a–363 cluster on chromosome X as novel diagnostic biomarkers for breast cancer. Breast Cancer Res Treat. 2018;170:257–70. doi: 10.1007/s10549-018-4757-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao T, Furnari F, Newton AC. PHLPP: a phosphatase that directly dephosphorylates Akt, promotes apoptosis, and suppresses tumor growth. Mol Cell. 2005;18:13–24. doi: 10.1016/j.molcel.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Stevens PD, Liu J, Yang H, Wang W, Wang C, et al. PHLPP is a negative regulator of RAF1, which reduces colorectal cancer cell motility and prevents tumor progression in mice. Gastroenterology. 2014;146:1301–12. doi: 10.1053/j.gastro.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Y, Lv D, Wang W, Ye M, Chen X, Yang H. High PHLPP1 expression levels predicts longer time of acquired resistance to EGFR tyrosine kinase inhibitors in patients with lung adenocarcinoma. Oncotarget. 2017;8:59000–7. doi: 10.18632/oncotarget.19777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19:575–86. doi: 10.1016/j.ccr.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–99.. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 33.Steinhart Z, Angers S. Wnt signaling in development and tissue homeostasis. Development. 2018;145:v146589. doi: 10.1242/dev.146589. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.