Abstract
Introduction
Since December 2019, severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) has caused the coronavirus disease 2019 (COVID-19) pandemic in China and worldwide. New drugs for the treatment of COVID-19 are in urgent need. Considering the long development time for new drugs, the identification of promising inhibitors from FDA-approved drugs is an imperative and valuable strategy. Recent studies have shown that the S1 and S2 subunits of the spike protein of SARS-CoV-2 utilize human angiotensin-converting enzyme 2 (hACE2) as the receptor to infect human cells.
Methods
We combined molecular docking and surface plasmon resonance (SPR) to identify potential inhibitors for ACE2 from available commercial medicines. We also designed coronavirus pseudoparticles that contain the spike protein assembled onto green fluorescent protein or luciferase reporter gene-carrying vesicular stomatitis virus core particles.
Results
We found that thymoquinone, a phytochemical compound obtained from the plant Nigella sativa, is a potential drug candidate. SPR analysis confirmed the binding of thymoquinone to ACE2. We found that thymoquinone can inhibit SARS-CoV-2, SARS-CoV, and NL63 pseudoparticles infecting HEK293-ACE2 cells, with half-maximal inhibitory concentrations of 4.999, 7.598, and 6.019 μM, respectively. The SARS-CoV-2 pseudoparticle inhibition had half-maximal cytotoxic concentration of 35.100 μM and selection index = 7.020.
Conclusion
Thymoquinone is a potential broad-spectrum inhibitor for the treatment of coronavirus infections.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-021-00400-2.
Keywords: Broad-spectrum inhibitor, Coronavirus, NL63-CoV, SARS-CoV, SARS-CoV-2, Thymoquinone
Key Summary Points
| Combined computer-based virtual screening and experimental assay for ACE2 inhibitors. |
| Verify that thymoquinone is a potential broad-spectrum inhibitor of coronavirus infection, which has been verified by the inhibition of pseudotyped SARS-CoV-2, SARS-CoV, and NL63-CoV viruses. |
| Thymoquinone may prevent viruses entering cells by binding ACE2 and interfering S1–ACE2 interaction. |
Digital Features
This article is published with digital features, including a summary slide, to facilitate understanding of the article. To view digital features for this article go to https://doi.org/10.6084/m9.figshare.13547726.
Introduction
The recent coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) infection, has spread to more than 200 countries. There are more than 30 million people infected, with a death toll greater than 1,026,284, and these numbers are increasing [1]. Given the unfortunate fact that COVID-19 cannot be effectively treated, there is apparently an urgent need of drugs against SARS-CoV-2. Hindered by the long development time for new drugs, the identification of promising inhibitors from approved drugs is an imperative and valuable drug development strategy for COVID-19 [2, 3].
SARS-CoV-2 belongs to the clade betacoronaviruses and has about 82% of its RNA genome identical to that of SARS-CoV, which broke out in 2003 [4]. The approximately 30,000 nucleotide genome of SARS-CoV-2 encodes two overlapping polyproteins, pp1a and pp1ab, which are hydrolyzed into functional polypeptides for viral replication and transcription [5]. For both SARS-CoV-2 and SARS-CoV, the S1 and S2 subunits of the spike glycoprotein (S-protein) utilize the human angiotensin-converting enzyme 2 (hACE2) as the receptor to infect human cells, with the receptor-binding domain (RBD) in the S1 subunit initiating binding with ACE2 and the S2 subunit in particular mediating the membrane fusion [6–8]. It should be noted that the RBD-ACE2 binding affinity of SARS-CoV-2 is 10–20 times higher than that of SARS-CoV, which may be attributed to the higher infectivity and transmissibility of SARS-CoV-2 [9].
Seven human coronaviruses (HCoVs) have been identified thus far: human coronavirus OC43 (OC43, discovered in the 1960s), human coronavirus 229E (229E, discovered in the 1960s), human coronavirus NL63 (NL63, discovered in 2004), human coronavirus HKU1 (HKU1, discovered in 2005), SARS-CoV (discovered in 2002), Middle East respiratory syndrome-related coronavirus (MERS, discovered in 2012), and SARS-CoV-2. Among them, SARS-CoV-2, SARS-CoV, and NL63-CoV use ACE2 as the receptor for the virus entry into the host cell. Hence, drugs targeting ACE2 potentially have broad-spectrum antiviral activities against SARS-CoV-2, SARS, and NL63 coronaviruses.
We aimed to find potential drug candidates by combining computer-based virtual screening and experimental assay. We have identified more than 20 commercial medicines that may form hydrogen bonds with the key residues within the binding pocket of ACE2 protein, including thymoquinone, a phytochemical compound obtained from the plant Nigella sativa. Thymoquinone holds promising pharmacological properties against several diseases. It exhibits outstanding anti-oxidant, anti-inflammatory, anticancer, and other important biological activities [10]. Thymoquinone effectively transforms cancer progression signaling pathways. It not only improves anticancer activity of chemotherapeutic drugs but also attenuates their side effects [11]. In brief, thymoquinone provides an exciting opportunity for the treatment of COVID-19 through blocking virus entry into the host cell.
Methods
Materials
Thymoquinone and eugenol were purchased from Aladdin (Beijing, China); glycyrrhizin and hesperidin were purchased from Targetmol (Shanghai, China); hispidulin, artemisinin, and emodin were purchased from Solarbio (Beijing, China); cirsimaritin was purchased from Esite (Chengdu, China); and hesperetin, curcuma, and MLN-4760 were purchased from Med Chem Express (Monmouth Junction, NJ, USA). All compounds were dissolved in dimethyl sulfoxide and stored as 10 mM stock solutions at 4 °C.
The Trans1-T1 strain, F-φ80(lacZ)△M15△lacX74hsdR(rk,m)△recA1398end AltonA (TransGen Biotech, Beijing, China), was used to clone and propagate plasmid DNA. Miniprep and Maxiprep kits (Axygen, USA) were used to harvest and purify plasmid DNA. ACE2 protein was purchased from novoprotein (Shanghai, China).
HEK 293T (human embryonic kidney; ATCC CRL-11268) was maintained in high-glucose Dulbecco’s modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and 1% antibiotic/antimycotic.
The vesicular stomatitis virus (VSV) exogenously expressing EGFP (VSV-dG-GFP) and Firefly Luciferase (VSV-dG-fLuc) are both recombinant vesicular stomatitis virus variants with G protein deletion, expressing green fluorescent protein (GFP) and firefly luciferase (fLuc), respectively. The VSV-dG-GFP and VSV-dG-fLuc pseudoviruses were generated through a plasmid-based reverse genetics system. The pBluescript-N (pBS-N), pBS-P, pBS-L, and pBS-G are helper vectors for the first-round virus rescue. The plasmids pVSV-ΔG-GFP and pVSV-dG-fLuc were purchased from Kerafast (Boston, MA, USA).
The Prediction of Binding Sites
The ACE2 (pdb: 6vw1) PDB file was downloaded from the protein data bank (http://www.rcsb.org/). All the heterogeneous atoms were removed and 6vw1 chain A was selected for the subsequent molecular docking. The ACE2 docking grid was maximized for thymoquinone. The PDB file (6vw1 chain A) was converted to the PDBQT format before virtual screening. The grid (ligand docking search space) was located as described above. Then, Autodock Vina 1.1.2 [6] was used for the subsequent molecular docking. Protein–ligand interactions were visualized using PyMoL v.1.7.4.5. The amino acid residues of S-protein close to the hit ligand (≤ 1 Å) were highlighted as potential residues involved in the protein–ligand interaction.
Surface Plasmon Resonance (SPR) Analysis
The affinity of thymoquinone and MLN-4760 with ACE2 was examined by the Biacore T200 System (GE Healthcare, Uppsala, Sweden) at 25 °C. ACE2 was immobilized on a CM5 chip with a coupling method to about 14,564.5 response units (RU). Serial dilutions of the compounds were prepared (100, 50, 25, 12.500, 6.250, 3.125, 1.563, 0.782, and 0 μM) and sequentially injected at a rate of 30 mL/min, over 180 s for buffer injection and 300 s for dissociation. A blank injection was used to check the carryover effects. The signal was adjusted for nonspecific binding of the samples to the dextran matrix by subtracting the signal in the reference channel from the signal in the active channel.
VSV Based Pseudovirus Production
VSV pseudotypes coated with the S-protein of SARS-CoV-2/SARS-CoV/NL63-CoV were generated based on a previously described protocol with slight modification [12]. In brief, HEK-293T or Vero-E6 cells were transfected with a plasmid overexpressing the codon-optimized (namely, the deletion of the C-terminal 18 amino acids) S-protein, which was able to significantly improve the pseudotypes packaging efficiency [13, 14]. Twenty-four hours later, the cells were transduced at MOI = 10 IU with the VSV-G glycoprotein-deficient VSV-dG-GFP or VSV-dG-fLuc. After 1 h incubation at 37 °C, the cells were washed by culture medium once and then replenished with fresh culture medium with anti-VSV rat serum to eliminate the infectivity of the residual input viruses. The S-protein pseudotyped VSV viruses were harvested 24 h after transduction by the clarification at 12,000 rpm for 2 min and then stored at − 80 °C. The virus titer (infectious unit) was determined by a plaque assay on BHK21-hACE2 cells with serial-diluted inocula.
Determination of the Half Maximal Inhibitory Concentration (IC50)
HEK293T-ACE2 cells were seeded in 96-well plates. Twenty-four hours later, the cells were challenged with SARS-CoV-2/SARS-CoV/NL63-CoV pseudotyped VSV (MOI = 0.100 IU) expressing GFP or luciferase, which were diluted using DMEM with 10% FBS in the presence of varying concentrations (100 μM, 33 μM, 11 μM, 3.700 μM, 1.230 μM, 410 nM, 137 nM) of the tested compounds. Next, 16–20 h after the infection, the cells infected with the GFP-expressing pseudoviruses were imaged with a fluorescence microscope; the cells infected with pseudoviruses expressing firefly luciferase were lyzed by 1 × passive lysis buffer (Promega) at room temperate for 15 min, and the luciferase activity was evaluated by One-Glo luciferase assay kit (Promega) through a GloMax® 20/20 Luminometer. The cytotoxicity of the tested chemicals was evaluated by bright field images before fluorescence imaging or cell lysis. The IC50 value was determined by nonlinear regression analysis.
Determination of the Half Maximal Cytotoxic Concentration (CC50)
The CC50 of the compounds was determined by the CellTiter-Glo 2 (Promega) cell viability assay kit. In brief, 293T-hACE2 cells were seeded in 96-well plates with three-fold serial-diluted compounds (from 100 to 0.137 μM). Twenty-four hours later, the cell viability was detected by CellTiter-Glo 2 reagent following the manufacturer’s instructions, and the luminescence was determined with a Spectra MaxiD3 multi-well Luminometer 458 (Molecular Devices, USA). The CC50 value was determined by nonlinear regression analysis.
Determination of the Toxicity of Thymoquinone to the BHK21-ACE2 Cells
To prove that the effects of thymoquinone upon the pseudoviruses were not due to thymoquinone toxifying the cells, we performed lactate dehydrogenase (LDH) tests by measuring the LDH released from the cells following thymoquinone action. The LDH-release assay kit was purchased from Beyotime (Shanghai, China). The optimal number of BHK21-ACE2 cells/well were plated in 100 µL of medium in triplicate wells in a 96-well plate. Various concentrations (10 μM, 3.3 μM, 1.1 μM, 0.37 μM, 0.12 μM, 0 μM) of thymoquinone were added to one set of triplicate wells. The LDH released into the medium was transferred to a new plate and mixed with the reaction mixture. After 30 min of room-temperature incubation, the absorbance at 490 nm and 600 nm was measured using a plate-reading spectrophotometer to determine the LDH activity.
Compliance with Ethics Guidelines
This study did not involve any human material, data, or participants, so no institutional review board (IRB) approval was required.
Results
Predicted Binding Site of Thymoquinone With ACE2
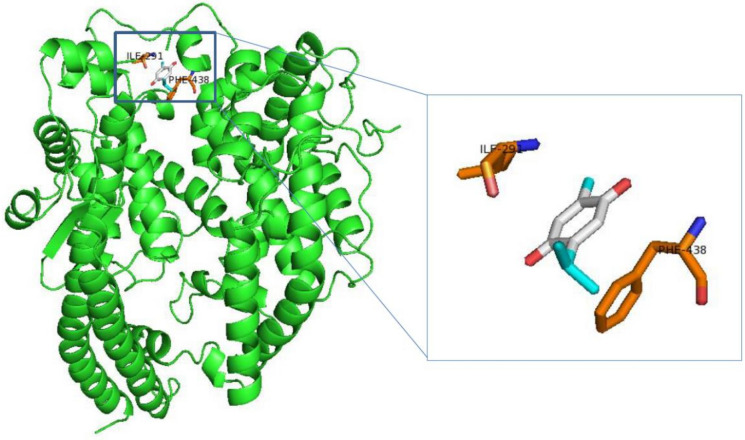
The chain A of 6vw1 (ACE2) was extracted to perform molecular docking with thymoquinone. Autodock Vina revealed that thymoquinone binds to residues ILE291 and PHE438 of ACE2 (Fig. 1).
Fig. 1.
Thymoquinone (cyan stick) binds to ACE2 protein (green) at residues ILE291 (orange stick) and PHE438 (orange stick)
SPR Technology–Based Binder Identification
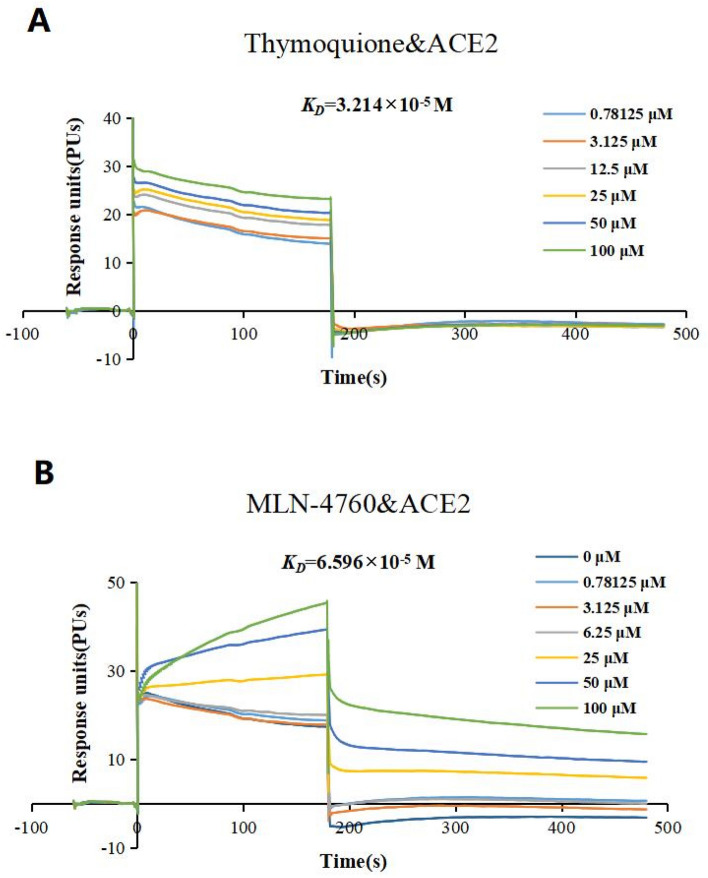
To confirm the interaction between thymoquinone and ACE2, we tested whether thymoquinone has a high affinity with the target protein using the SPR assay. The ACE2 protein was immobilized on a CM5 chip with thymoquinone flowing across the surface. We found that the affinity of thymoquinone–ACE2 binding depends strongly on the dosage, with KD (equilibrium dissociation constant) being 32.140 μM (Fig. 2a). Since MLN-4760 is a reported ACE2 inhibitor [15], we used MLN-4760 as a positive control. SPR showed that MLN-4760 binds to ACE2 with KD = 65.960 μM (Fig. 2b). These results, summarized in Table 1, suggested that thymoquinone can, through interfering with the S1–ACE2 interaction, block the virus entry into the host cell.
Fig. 2.
Sensograms of ACE2 interacting with thymoquinone (a) and MLN-4760 (b)
Table 1.
Affinity of compounds to ACE2 at 25 °C
| Compound | KD (M) | Rmax (RU) | Chi2 (RU2) | Chi |
|---|---|---|---|---|
| Thymoquinone | 3.214 × 10−5 | 11.580 | 0.328 | 0.573 |
| MLN-4760 | 6.596 × 10−5 | 48.410 | 2.760 | 1.660 |
Determination of the IC50 and CC50 of Thymoquinone for SARS-CoV-2 Pseudovirus
To alleviate safety concerns and immediately initiate work in our BSL2 laboratory, we designed SARS-CoV-2 pseudoparticles (CoV-2pp) that contain the SARS-CoV-2 S-protein assembled onto luciferase reporter gene-carrying VSV core particles, based on the genome sequence of SARS-CoV-2 released on January 12, 2020 [16, 17]. These pseudotyped viral particles faithfully reflect key aspects of native SARS-CoV-2 entry into host cells via binding to its ACE2 receptor [18]; they can thus be used to test potential entry inhibitors of SARS-CoV-2.
We then used the HEK293T-ACE2 cell model in a 96-well format to test the effects of multiple drugs that had been in silico predicted binding to ACE2, including glycyrrhizin, eugenol, hesperetin, hesperidin, hispidulin, artemisinin, curcuma, cirsimaritin, MLN-4760, emodin, and thymoquinone. These drugs were chosen based on their capability of binding to ACE2 as predicted by molecular docking.
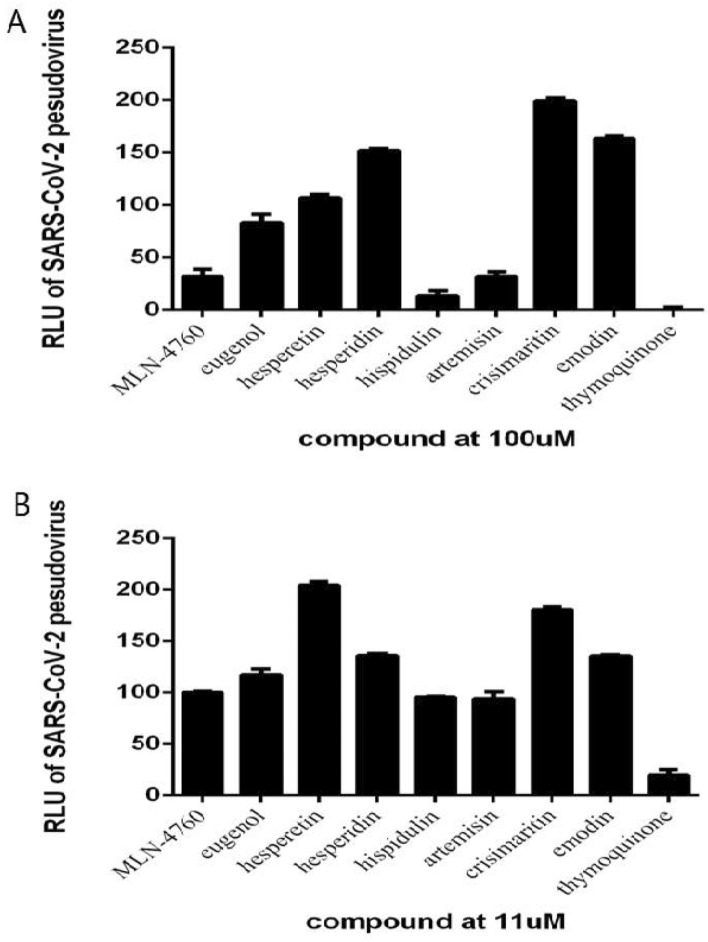
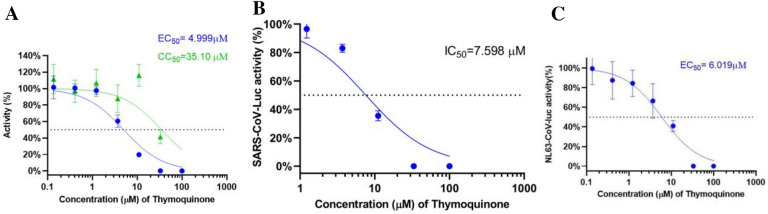
Although MLN-4760, hispidulin, artemisinin, and thymoquinone can all inhibit SARS-CoV-2 pseudovirus at 100 μM, which is consistent with their affinities with ACE2 (Supplemental Material 1), only thymoquinone can inhibit SARS-CoV-2 pseudovirus at 11 μM (Fig. 3a, b). By adding different concentrations of thymoquinone ranging from 100 μM to 137 nM, we found that the inhibition of thymoquinone on the entry of SARS-CoV-2 pseudovirus into the HEK293T-ACE2 cells is strongly dose-dependent. When the concentration exceeds 11 μM, thymoquinone can inhibit more than 80% of virus entry (Fig. 3B). The indices are as follows: half-maximal inhibitory concentration (IC50) = 4.999 μM (Fig. 4a); half-maximal cytotoxic concentration (CC50) = 35.100 μM (Fig. 4a); selectivity index = 7.020; toxicity was observed at 11 μM; cells died at 33 μM.
Fig. 3.
The inhibition ratio of the indicated compounds against SARS-CoV-2 pseudovirus at 100 μM (a) and 11 μM (b)
Fig. 4.
Determination of the IC50 of thymoquinone for SARS-CoV-2 (a), SARS-CoV (b), and NL63-CoV (c) pseudoviruses. The CC50 of thymoquinone for SARS-CoV-2 was also determined (a)
For glycyrrhizin and curcuma, we found that they were not effective in inhibiting SARS-CoV-2 pseudovirus-expressing VSV-dG-GFP (Supplementary Material 2).
Determination of the IC50 of Thymoquinone for SARS-CoV and NL63 Pseudoviruses
SARS-CoV and NL63-CoV also use ACE2 as the receptor for entry into the host cell [19, 20]. Therefore, the extensive researches on SARS-CoV-2 may be applicable to SARS-CoV and NL63-CoV. We designed SARS-CoV and NL63-CoV pseudoparticles (CoV-2pp) that contain their S-protein assembled onto luciferase reporter gene-carrying VSV core particles. We found that thymoquinone has IC50 = 7.598 μM for SARS-CoV pseudovirus (Fig. 4b) and IC50 = 6.019 μM for NL63-CoV pseudovirus (Fig. 4c). Taken together, thymoquinone potentially has broad-spectrum antiviral activity against SARS-CoV-2, SARS, and NL63 coronaviruses.
Inhibition of the Pseudoviruses Was Not Due to Thymoquinone Toxifying the BHK21-ACE2 Cells
To prove that the effects of thymoquinone upon the pseudoviruses were direct, namely, not by toxifying the cells, we added various concentrations (10 μM, 3.3 μM, 1.1 μM, 0.37 μM, 0.12 μM, 0 μM) of thymoquinone to the wells of BHK21-ACE2 cells. We subsequently measured the activity of LDH released from the cells. Apparently, thymoquinone had relatively low toxicity to the BHK21-ACE2 cells (Fig. 5). Even with a concentration as large as 10 μM, the cell mortality rate is only 13.23%. This result is consistent with the 35.1 μM CC50 value tested by the CellTiter-Glo cell viability assay.
Fig. 5.
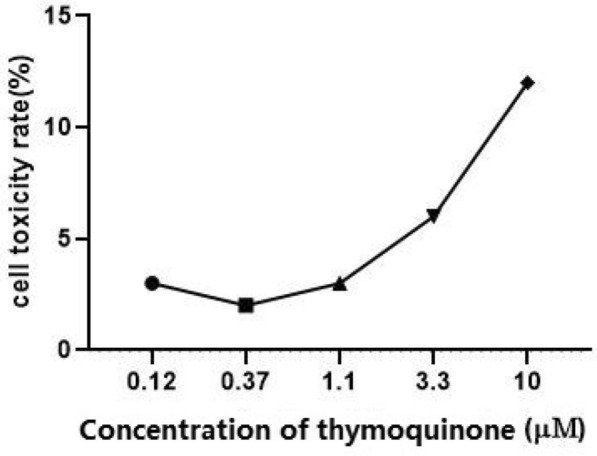
Determination of the toxicity of thymoquinone to the BHK21-ACE2 cells
Discussion
With the global spread of new coronavirus diseases, it is increasingly difficult to control the epidemics; the development of effective drugs targeting the diseases is urgently needed. However, the development of new drugs is a very complicated, expensive, and long process. A novel drug usually costs USD2.6 billion and takes on average 12 years. Drug repositioning greatly shortens the time and cost of R&D; it is thus the most effective method to fight such a sudden disease. This strategy may lead to the alleviation of the current epidemic. In the present study, we adopted a computational approach to screen available commercial medicines which may function as inhibitors for ACE2. By further experimental screening of the in silico predicted ACE2 inhibitors, we identified that thymoquinone, extracted from the annual flowering plant Nigella sativa (family Ranunculaceae), is efficacious in the inhibition of SARS-CoV-2, SARS-CoV, and NL63-CoV pseudoviruses. The black seeds and their oil of Nigella sativa have been previously reported for a range of medicinal applications, including for rheumatoid arthritis, asthma, inflammatory diseases, diabetes, and digestive diseases [21–25].
It is well known that thymoquinone belongs to the so-called pan-assay interference compounds (PAINS) [26], which bind promiscuously with many proteins because it has a high propensity to be redox active as well as being reactive to nucleophiles present in the side chains of proteins such as cysteine and lysine. To prove that thymoquinone is a genuine drug candidate, a direct binding assay is needed, and the SPR technology was particularly recommended [27].
In this study, we used SPR to confirm the affinity of thymoquinone with ACE2 and found its KD to be 32.140 μM, which is consistent with the results obtained from molecular docking. These results imply that thymoquinone might inhibit SARS-CoV-2 infection by blocking the binding of the SARS-CoV-2 S-protein to the cellular receptor ACE2. Because the relevant experimental researches on SARS-CoV-2 need to be conducted in a BSL-3 laboratory, we used SARS-CoV-2 pseudovirus to test thymoquinone’s effects at the cellular level. Thymoquinone inhibits the SARS-CoV-2 pseudovirus in a dose-dependent manner with ideal IC50 and CC50 values. This study therefore supports the recent clinical trials (ClinicalTrials.gov Identifier: NCT04553705) treating COVID-19 patients with Nigella sativa. The safe dosage of thymoquinone must be rigorously determined to reduce its toxicity.
A previous study [28] showed that selective ACE2 inhibitor MLN-4760 neither blocks nor increases the binding of SARS-CoV-2 spike RBD to human ACE2, and probably has no effect on the viral entry. We confirmed this result: MLN-4760 did not inhibit SARS-CoV-2 pseudovirus at 11 μM. Since ACE2 is also the receptor of SARS-CoV and NL63-CoV, we also tested whether or not thymoquinone is effective against SARS-CoV and NL63-CoV. We found that thymoquinone does inhibit the two pseudoviruses.
A limitation of this study, as already stated, is that more rigorous testing will be needed in future to establish the drug candidacy of thymoquinone, partly due to the fact that thymoquinone, as a famous PAINS, binds promiscuously with many proteins, because it has a high propensity to be redox active as well as being reactive to nucleophiles present in the side chains of proteins such as cysteine and lysine. Another limitation was that, without a BSL-3 laboratory, we could not use living viruses to perform experiments. We will pursue experiments on living coronaviruses in the future.
Conclusion
Thymoquinone is potentially a broad-spectrum inhibitor for the treatment of coronavirus infections by blocking the binding of the viral S-protein to the cellular receptor ACE2, thus blocking viral entry into the host cell. The inhibition is strongly dose-dependent with ideal IC50 and CC50 values. Further testing of thymoquinone is needed to establish its drug candidacy because it is a well-known PAINS.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Funding
This work was partly supported by National Science and Technology Major project (2017ZX09303008), National Natural Science Foundation of China (81973251, 61773196, 32070681), Hebei NSF (H2019206445), Guangdong Provincial Special Projects on COVID-19 (2020KZDZX1182), Guangdong Provincial Key Laboratory of Computational Science and Material Design (2019B030301001), Shenzhen Research Funds (JCYJ20170817104740861), Shenzhen Peacock Plan (KQTD2016053117035204), and by Center for Computational Science and Engineering of Southern University of Science and Technology, in addition to the National Science and Technology Major Project (2017ZX09303008), and Shenzhen Bay laboratory start up fund (21230071). The journal's Rapid Service Fee was funded by the authors.
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Disclosures
Huan Xu, Bing Liu, Zhen Xiao, Meiling Zhou, Lin Ge, Fan Jia, Yanling Liu, Hongshan Jin, Xiuliang Zhu, Jian Gao, Javed Akhtar, Bai Xiang, Ke Tan, Guanyu Wang have nothing to disclose.
Compliance with Ethics Guidelines
This study didn't involve any human material, data or participants and therefore meant no IRB approval was required.
Data Availability
The datasets generated during and analyzed during the current study are available from the corresponding authors on reasonable request.
Footnotes
Huan Xu, Bing Liu and Zhen Xiao contributed equally to this article.
Contributor Information
Bai Xiang, Email: baixiang@hebmu.edu.cn.
Ke Tan, Email: tanke@hebtu.edu.cn.
Guanyu Wang, Email: wanggy@sustech.edu.cn.
References
- 1.Gabutti G, d’Anchera E, Sandri F, Savio M, Stefanati A. Coronavirus: update related to the current outbreak of COVID-19. Infect Dis Therapy. 2020;2020:1–13. doi: 10.1007/s40121-020-00295-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wei T, et al. In Silico screening of potential spike glycoprotein inhibitors of SARS-CoV-2 with drug repurposing strategy. Chin J Integr Med. 2020;26:663–669. doi: 10.1007/s11655-020-3427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altay O, et al. Current status of COVID-19 therapies and drug repositioning applications. Iscience. 2020;23:101303. doi: 10.1016/j.isci.2020.101303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goyal B, Goyal D. Targeting the dimerization of the main protease of coronaviruses: a potential broad-spectrum therapeutic strategy. ACS Comb Sci. 2020;22:297–305. doi: 10.1021/acscombsci.0c00058. [DOI] [PubMed] [Google Scholar]
- 5.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosch BJ, Van der Zee R, De Haan CA, Rottier PJ. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gheblawi M, et al. Response by Gheblawi et al to letter regarding article, “angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2”. Circ Res. 2020;127:e46–e47. doi: 10.1161/CIRCRESAHA.120.317332. [DOI] [PubMed] [Google Scholar]
- 8.Jaimes JA, Millet JK, Whittaker GR. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. Iscience. 2020;23:101212. doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darakhshan S, Bidmeshki-Pour A, Hosseinzadeh-Colagar A, Sisakhtnezhad S. Thymoquinone and its therapeutic potentials. Pharmacol Res. 2015;95–96:138–158. doi: 10.1016/j.phrs.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Thymoquinone inhibits the metastasis of renal cell cancer cells by inducing autophagy via AMPK/mTOR signaling pathway. Cancer Sci. 2018;109:3865–3873. doi: 10.1111/cas.13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitt MA. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J Virol Methods. 2010;169:365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukushi S, et al. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J Gen Virol. 2005;86:2269–2274. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- 14.Schwegmann-Weßels C, et al. Comparison of vesicular stomatitis virus pseudotyped with the S proteins from a porcine and a human coronavirus. J Gen Virol. 2009;90:1724–1729. doi: 10.1099/vir.0.009704-0. [DOI] [PubMed] [Google Scholar]
- 15.Towler P, et al. ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem. 2004;279:17996–18007. doi: 10.1074/jbc.M311191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu F, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu N, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou X, et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nature Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li W, et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith MK, et al. Human angiotensin-converting enzyme 2 (ACE2) is a receptor for human respiratory coronavirus NL63. Adv Exp Med Biol. 2006;581:285–288. doi: 10.1007/978-0-387-33012-9_48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahmad A, et al. A review on therapeutic potential of Nigella sativa: a miracle herb. Asian Pac J Trop Biomed. 2013;3:337–352. doi: 10.1016/s2221-1691(13)60075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Butt MS, Sultan MT. Nigella sativa: reduces the risk of various maladies. Crit Rev Food Sci Nutr. 2010;50:654–665. doi: 10.1080/10408390902768797. [DOI] [PubMed] [Google Scholar]
- 23.Ijaz H, et al. Review: nigella sativa (prophetic medicine): a review. Pak J Pharm Sci. 2017;30:229–234. [PubMed] [Google Scholar]
- 24.Kooti W, Hasanzadeh-Noohi Z, Sharafi-Ahvazi N, Asadi-Samani M, Ashtary-Larky D. Phytochemistry, pharmacology, and therapeutic uses of black seed (Nigella sativa) Chin J Nat Med. 2016;14:732–745. doi: 10.1016/s1875-5364(16)30088-7. [DOI] [PubMed] [Google Scholar]
- 25.Padhye S, Banerjee S, Ahmad A, Mohammad R, Sarkar FH. From here to eternity—the secret of Pharaohs: therapeutic potential of black cumin seeds and beyond. Cancer therapy. 2008;6:495–510. [PMC free article] [PubMed] [Google Scholar]
- 26.Baell JB. Screening-based translation of public research encounters painful problems. ACS Med Chem Lett. 2015;6:229–234. doi: 10.1021/acsmedchemlett.5b00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Baell J, Walters MA. Chemistry: chemical con artists foil drug discovery. Nature News. 2014;513:481. doi: 10.1038/513481a. [DOI] [PubMed] [Google Scholar]
- 28.Nami B, Ghanaiyan A, Ghanaeian K, Nami N. The effect of ACE2 inhibitor MLN-4760 on the interaction of SARS-CoV-2 spike protein with human ACE2: a molecular dynamics study. 2020. https://chemrxiv.org.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and analyzed during the current study are available from the corresponding authors on reasonable request.