The loss of cardiomyocytes following myocardial infarction (MI) in adult mammals leads to heart failure. We previously reported a regenerative role for the miR-17–92 cluster1 in cardiac repair post-MI by repressing phosphatase and tensin homolog (PTEN)2. Despite favorable effects of PTEN inactivation in heart disease and the suggestion that it is involved in hypertrophy3, 4, whether the loss of PTEN directly promotes cardiomyocyte proliferation to enhance myocardial repair in response to MI remains unknown.
We generated cardiac-specific Pten knockout mice with a tamoxifen-inducible Cre-LoxP system and confirmed no significant differences in cardiac function or morphology between Pten conditional knock-out (cKO) mice (Ptenflox/flox;α-MHC- MerCreMer) and control (Ptenflox/flox) mice (Figure A), which is consistent with previous reports4. While control mice exhibited heart failure 2, 3, 4, 5, and 12 weeks after MI, cardiac function was preserved in Pten cKO mice (Figure B). Histological analyses revealed substantial reductions in infarct sizes in Pten cKO hearts 15 weeks post-MI (Figure C). Cardiomyocyte size was markedly decreased in Pten cKO hearts 12 weeks post-MI compared to the control group (Figure D).
Figure. Loss of PTEN promotes cardiac regeneration and protects the heart from myocardial infarction.
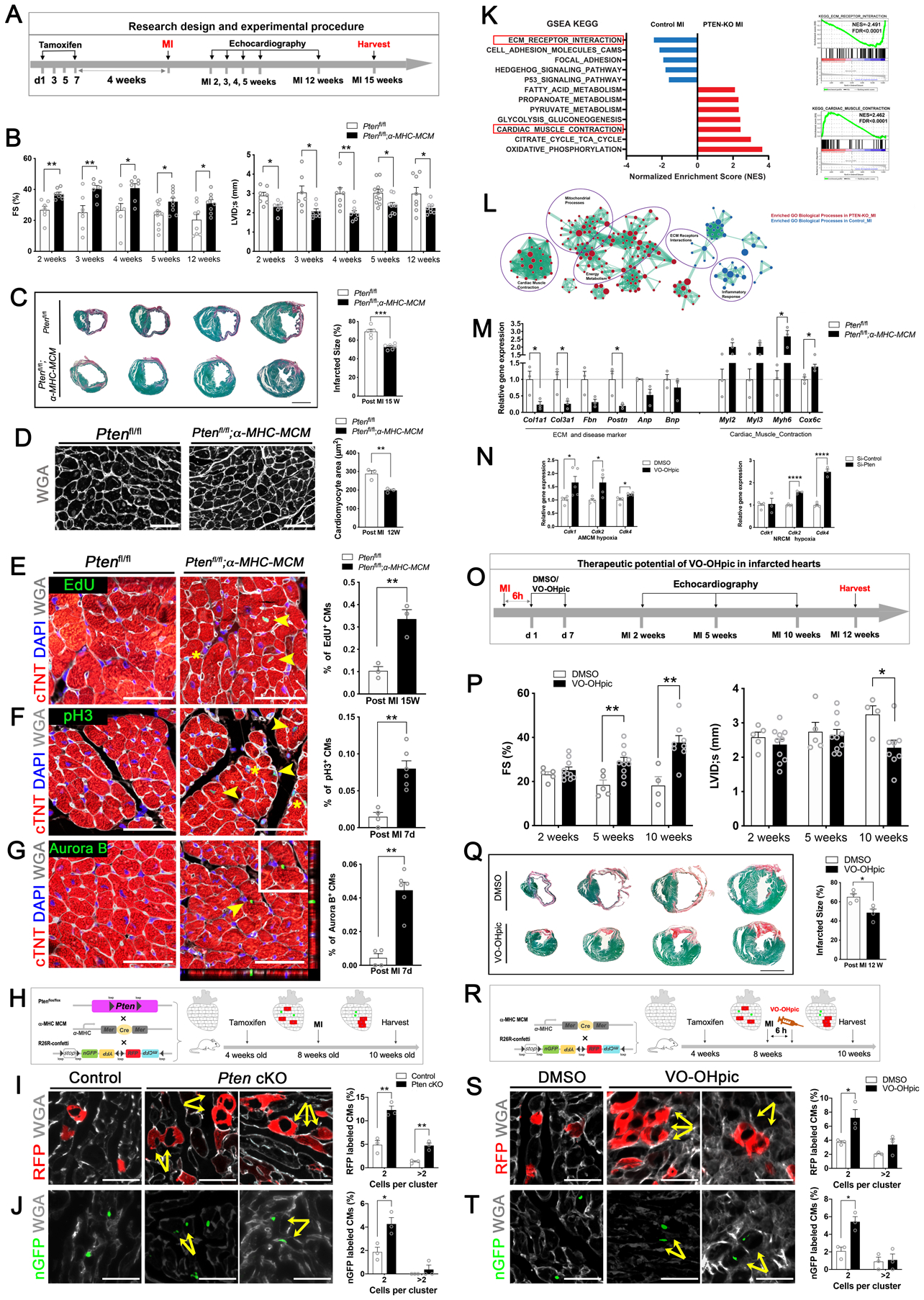
A, A schematic diagram of the research design and experimental procedures. Four-week-old Ptenfl/fl; α-MHC-MerCreMer (Pten cKO) mice, and Ptenfl/fl (control) mice were intraperitoneally injected with tamoxifen (75mg/kg body weight). Four weeks later, these mice were subjected to myocardial infarction (MI) and assessed for cardiac function by echocardiography at the indicated time points prior to harvesting samples for analysis.
B, Echocardiography of the Pten cKO group and control group at 2–5 weeks and 12 weeks post MI (n = 7 to 11 mice per group). Each data point represents an individual mouse. Data is represented as the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p< 0.01). Values of p<0.05 were considered statistically significant. FS: fractional shortening (%); LVID;s: left ventricular internal dimension at end-systole.
C, (Left) Representative images of serial transverse sections from the control and Pten cKO groups at 15 weeks after MI. Sirius red/fast green collagen staining marks the myocardium (green) and scar (red) (scale bar = 2 mm). (Right) Quantification of the scar size (n = 5 hearts for control group, n = 6 hearts for Pten cKO group). Each data point represents an individual mouse with analysis of 4 sections below the ligation for each heart. Infarct size (%) was calculated according to the formula: [length of coronal infarct perimeter (epicardial + endocardial)/ total left ventricle coronal perimeter (epicardial + endocardial)] × 100. Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01, ***p<0.001). Values of p<0.05 were considered statistically significant.
D, (Left) Immunofluorescence staining of Wheat Germ Agglutinin (WGA) in cardiomyocytes of the papillary muscles in transverse sections from post-MI 12-week mouse hearts from control and Pten cKO mice. WGA marks cell membranes (white) for detecting cardiomyocyte size (scale bar = 50 μm). (Right) Quantification of cardiomyocyte size (n = 3 hearts for each group, each data point represents an individual mouse, enumerating 250–300 cells per heart). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
E, (Left) Immunofluorescence staining of EdU incorporation in the MI border region in transverse sections from post-MI 15-week mouse hearts from control and Pten cKO mice. EdU labels proliferating cells (green); cardiac troponin T (cTNT) marks cardiomyocytes (red); WGA marks cell membranes (white), and DAPI labels nuclei (blue). Arrowheads point to EdU-positive signal in cardiomyocytes, asterisks mark EdU-positive signal in non-cardiomyocytes (scale bars = 50 μm). (Right) Quantification of the percentage of EdU-positive cardiomyocytes (n = 3 hearts for each group, enumerating 2☓103-3☓103 myocytes per section, 2 sections from the border zone per heart, and each data point represents an individual mouse). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
F, (Left) Immunofluorescence staining of phospho-Histone H3 (pH3) in the border zone of 7-day post-MI mouse hearts from control and Pten cKO mice (PH3 [green]; cTNT [red]; WGA [white], and DAPI [blue]). Arrowheads point to pH3-positive signal in cardiomyocytes, asterisks mark pH3-positive signal in non-cardiomyocytes (scale bars = 50 μm). (Right) Quantification of pH3-positive cardiomyocytes (n = 4 hearts for control group, n = 6 hearts for Pten cKO group, enumerating 2☓103-3☓103 myocytes per section, 3–4 sections from the border zone in each heart). Each data point represents an individual mouse. Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
G, (Left) Immunofluorescence staining of Aurora B kinase in the border zone using transverse sections from 7-day post-MI mouse hearts from control and Pten cKO mice. Aurora B (green); cTNT (red); WGA (white), and DAPI (blue). Arrowhead points to Aurora B kinase-positive signal in cardiomyocytes, which is enlarged in the boxed area. Representative Z-stack 3-D confocal microscopy showing a cytokinetic cardiomyocyte in a Pten cKO heart section (scale bars = 50 μm). (Right) Quantification of Aurora B kinase-positive cardiomyocytes (n = 4 hearts for control group, n = 6 hearts for Pten cKO group, enumerating 2☓103-3☓103 myocytes per section, 3–4 sections from the border zone of each heart). Each data point represents an individual mouse. Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
H, Lineage-tracing strategy and experimental design for assessing adult cardiomyocyte proliferation in the Pten genetic-deletion in vivo model. Ptenfl/fl; α-MHC-MerCreMer (Pten cKO) and α-MHC-MerCreMer (control) mice were bred with R26R-Confetti Cre-reporter mice. When treated with a low dose of tamoxifen, the expression of α-MHC led to Cre–loxP recombination and random labeling of cardiomyocytes with a single color.
I, Quantification of the clusters of two or more RFP+ cardiomyocyte clones in control and Pten cKO mouse hearts (scale bars = 50 μm, n = 3 hearts per group). Quantification showing the ratio of the cells in clusters of two or more RFP+ cardiomyocytes/total RFP+ cardiomyocytes in Pten cKO and control groups (each data point represents an individual mouse, counting 3 sections per heart, enumerating 300 ± 100 colored cells per section). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
J, Quantification of the clusters of two or more nuclear GFP+ (nGFP) cardiomyocytes in control and Pten cKO mouse hearts (scale bars = 50 μm, n = 3 hearts per group). Quantification showing the ratio of cells in clusters of two or more nGFP+ cardiomyocytes/total nGFP+ cardiomyocytes in Pten cKO and control groups (each data point represents an individual mouse, counting 3 sections per heart, enumerating 300 ± 100 colored cells per section). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
K, (Left) RNA-sequencing analysis of gene expression and enrichment differences between Pten cKO and control mouse hearts 7-days after MI. (Right) Representative enrichment plots for perturbed ECM Receptor Interaction and Cardiac Muscle Contraction (KEGG) (n = 4 hearts per group).
L, Enrichment network map for perturbed Gene Ontology (GO) biological processes gene sets in Pten-cKO-MI versus control-MI samples. Key clusters of the GO network were annotated in purple circles. The color of the circle denotes the direction of enrichment and the size of the circle denotes the size of the respective gene set.
M, Quantitative RT-PCR (qRT-PCR) to verify gene expression from ECM- and contraction- associated as well as cardiac disease-associated genes (n = 3 heart per group, each data point represents an individual mouse). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05). Values of p<0.05 were considered statistically significant.
N, (Left) Eight-week-old C57 mice were injected with the PTEN inhibitor VO-OHpic (10 μg/kg body weight) or DMSO (control). Cardiomyocytes were isolated and cultured in hypoxia (5% O2) and quantitative RT-PCR (qRT-PCR) was used to detect the expression of cell cycle genes (n = 4 hearts in DMSO group, n = 5 hearts in VO-OHpic group, each data point represents an individual mouse). (Right) qRT-PCR detection of the expression of cell cycle genes in neonatal rat cardiomyocytes cultured in hypoxia (0.1% O2) (n = 4 wells in siRNA-control group, n = 3 wells in siRNA-Pten group). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Values of p<0.05 were considered statistically significant.
O, Schematic diagram of the therapeutic experimental design. 8-week-old C57BL6J mice were subjected to surgically-induced MI and then intraperitoneally injected with DMSO (control) or PTEN inhibitor VO-Ohpic (10 μg/kg body weight) 6 hours after surgery. Cardiac function was assessed by echocardiography at the indicated times before harvesting heart samples for analysis.
P, Echocardiography of mice treat with DMSO or VO-Ohpic at 2, 5, and 10 weeks post-MI (n = 4 to 10 mice per group, each data point represents an individual mouse). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant. FS: fractional shortening (%); LVID;s: left ventricular internal dimension at end-systole.
Q, (Left) Representative images of serial transverse sections from the control and VO-OHpic group at 12 weeks after MI. Sirius red/fast green collagen staining of myocardium (green) and scar (red) (scale bar = 2mm). (Right) Quantification of the scar size (n = 4 mice for each group, each data point represents an individual mouse, summarizing analysis of 5 sections for each sample). Infarct size was calculated according to the formula: [length of coronal infarct perimeter (epicardial + endocardial)/ total left ventricle coronal perimeter (epicardial + endocardial)] × 100. Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05). Values of p<0.05 were considered statistically significant.
R, The Lineage-tracing strategy and experimental design for assessing adult cardiomyocyte proliferation in the PTEN-pharmacological inhibition in vivo model. α-MHC MerCreMer;R26R-Confetti mice were used to assess cardiomyocyte proliferation following tamoxifen activation of Cre-loxP recombination in the DMSO or VO-OHpic group post-MI.
S, Quantification of the clusters of two or more RFP+ cardiomyocytes in DMSO and VO-OHpic treated hearts (scale bars = 50 μm, n = 3 mice per group). Quantification showing the ratio of the cells in clusters of two or more RFP+ cardiomyocytes / total RFP+ cardiomyocytes in DMSO and VO-OHpic group (each data point represents an individual mouse, counting 3 sections per heart, 300 ± 100 colored cells per section). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
T, Quantification of the clusters of two or more nuclear GFP+ (nGFP) cardiomyocytes in DMSO and VO-OHpic treated hearts (scale bars = 50 μm, n = 3 mice per group). Quantification showing the ratio of the cells in clusters of two or more nGFP+ cardiomyocytes / total nGFP+ cardiomyocytes in DMSO and VO-OHpic groups (each data point represents an individual mouse, counting 3 sections per heart, 300 ± 100 colored cells per section). Data represents the mean ± SEM. Statistical analysis was performed using an unpaired, two-tailed Student’s t-test between two independent groups (*p<0.05, **p<0.01). Values of p<0.05 were considered statistically significant.
We detected a substantial increase in EdU incorporation in adult cardiomyocytes from Pten cKO hearts (Figure E). This observation was supported by increased staining of the mitosis marker phospho-Histone H3 (pH3) (Figure F), as well as that of the cytokinesis marker Aurora B kinase, in adult cardiomyocytes of Pten cKO mice following MI (Figure G).
To provide independent evidence that loss of Pten stimulated cardiomyocyte proliferation after MI, we employed a lineage tracing strategy using a R26R-Confetti Cre-reporter system with a loxP-flanked multicolor fluorescent protein5. We bred Ptenfl/fl; α-MHC-MerCreMer (Pten cKO) mice and α-MHC-MerCreMer (control) mice with R26R-Confetti Cre-reporter mice, and used low-dose tamoxifen to induce Cre recombination, to ensure Pten-deleted cardiomyocyte labeling with a single color. Four weeks later, we induced MI and assessed the amount of adjacent daughter cardiomyocytes having the same color (Figure H). Quantification revealed that the clusters of two or more RFP+ (Figure I) and nuclear GFP+ (Figure J) cardiomyocytes were more frequently observed in Pten cKO hearts compared to control hearts. Together, these results demonstrate the loss of Pten induces adult cardiomyocyte proliferation following MI.
To gain a greater understanding of the effect of Pten deficiency on heart protection and regeneration after MI, we performed unbiased genome-wide transcriptional profiling. 1,734 genes and 197 genes were down- and up-regulated in the Pten cKO group, respectively, compared with the control group (q<0.05; fold change >0.5 log2 scale). Gene Set Enrichment Analysis (GSEA) using the KEGG pathway database revealed that downregulated genes were enriched for functional terms associated with the extracellular matrix (ECM) and cell adhesion, whereas upregulated genes were associated with cardiac muscle contraction and metabolism (Figure K). In addition, the enrichment network for perturbed Gene Ontology (GO) biological process gene-sets showed the key clusters annotated with ECM and inflammatory responses in the control group and with cardiac muscle contraction, energy metabolism, and mitochondrial processes in the Pten cKO group after MI (Figure L). Subsequent quantitative RT- PCR verified this differential expression (Figure M). We found increased expression of cell cycle genes in isolated adult and neonatal cardiomyocytes with PTEN depletion, upon hypoxia treatment (Figure N). Collectively, these findings demonstrate that loss of PTEN protects the heart after MI injury by attenuating the inflammatory response and fibrotic remodeling in addition to enhancing contraction.
To determine if PTEN could be a potential therapeutic target, we treated mice with the PTEN inhibitor VO-OHpic following MI. Eight-week-old C57BL/6J mice were intraperitoneally injected with a low dose of VO-OHpic or DMSO six hours after inducing MI (Figure O). PTEN inactivation by VO-OHpic significantly improved cardiac function (Figure P) and reduced scar formation (Figure Q). Overall, our data show that PTEN inhibition could be an effective therapeutic approach for treating infarcted hearts.
Finally, using the R26R-Confetti Cre-reporter lineage tracing system (Figure R), we demonstrated that PTEN inhibition by VO-OHpic stimulated cardiomyocyte proliferation in MI injured adult mouse hearts (Figure S, T). Together, our studies have uncovered an important and previously unrecognized role for PTEN inhibition in cardiac regeneration and repair, which may represent a therapeutic approach to protect the heart from ischemic injury and heart failure.
All animals in this study conformed to the Public Health Service Guide for Care and Use of Laboratory Animals and was approved by the Institutional Animal Care and Use Committee (IACUC) of Zhejiang University.
Acknowledgments
We thank the Core Facilities of Zhejiang University School of Medicine. We thank the Laboratory Animal Center of Zhejiang University.
Sources of Funding:
This work was supported by the National Key R&D Program of China (2017YFA0103700), the National Natural Science Foundation of China (Nos. 81470382, 81670257, 81970227 to J. Chen) and Zhejiang Provincial NSF project (LZ20H020001 to J. Chen). Work from the Wang laboratory was supported by NIH grant HL125925.
Footnotes
Disclosures:
None.
Data sharing: The data that support the findings of this study, and experimental methods and materials, are available from the corresponding authors upon reasonable request by email.
References
- 1.Chen J, Huang ZP, Seok HY, Ding J, Kataoka M, Zhang Z, Hu X, Wang G, Lin Z, Wang S, Pu WT, Liao R and Wang DZ. mir-17–92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res. 2013;112:1557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao F, Kataoka M, Liu N, Liang T, Huang ZP, Gu F, Ding J, Liu J, Zhang F, Ma Q, Wang Y, Zhang M, Hu X, Kyselovic J, Hu X, Pu WT, Wang J, Chen J and Wang DZ. Therapeutic role of miR-19a/19b in cardiac regeneration and protection from myocardial infarction. Nat Commun. 2019;10:1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oudit GY, Kassiri Z, Zhou J, Liu QC, Liu PP, Backx PH, Dawood F, Crackower MA, Scholey JW and Penninger JM. Loss of PTEN attenuates the development of pathological hypertrophy and heart failure in response to biomechanical stress. Cardiovasc Res. 2008;78:505–14. [DOI] [PubMed] [Google Scholar]
- 4.Ruan H, Li J, Ren S, Gao J, Li G, Kim R, Wu H and Wang Y. Inducible and cardiac specific PTEN inactivation protects ischemia/reperfusion injury. J Mol Cell Cardiol. 2009;46:193–200. [DOI] [PubMed] [Google Scholar]
- 5.Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon- Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD and Clevers H. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–44. [DOI] [PubMed] [Google Scholar]


