Abstract
Background
Epidemic arbovirus transmission occurs among humans by mosquito bites and the sylvatic transmission cycles involving non-human primates (NHPs) still exists. However, limited data are available on the extent in NHPs infections and their role. In this study, we have developed and validated a high-throughput serological screening tool to study the circulation of multiple arboviruses that represent a significant threat to human health, in NHPs in Central Africa.
Methodology/Principal findings
Recombinant proteins NS1, envelope domain-3 (DIII) for the dengue (DENV), yellow fever (YFV), usutu (USUV), west nile (WNV) and zika (ZIKV) and envelope 2 for the chikungunya (CHIKV) and o'nyong-nyong (ONNV) were coupled to Luminex beads to detect IgG directed against these viruses. Evaluation of test performance was made using 161 human sera of known arboviral status (66 negative and 95 positive). The sensitivity and specificity of each antigen were determined by statistical methods and ROC curves (except for ONNV and USUV). All NS1 antigens (except NS1-YFV), CHIKV-E2 and WNV-DIII had sensitivities and specificities > 95%. For the other DIII antigens, the sensitivity was low, limiting the interest of their use for seroprevalence studies. Few simultaneous reactions were observed between the CHIKV+ samples and the NS1 antigens to the non-CHIKV arboviruses. On the other hand, the DENV+ samples crossed-reacted with NS1 of all the DENV serotypes (1 to 4), as well as with ZIKV, USUV and to a lesser extent with YFV. A total of 3,518 samples of 29 species of NHPs from Cameroon and the Democratic Republic of Congo (DRC) were tested against NS1 (except YFV), E2 (CHIKV/ONNV) and DIII (WNV) antigens. In monkeys (n = 2,100), the global prevalence varied between 2 and 5% for the ten antigens tested. When we stratified by monkey’s biotope, the arboreal species showed the highest reactivity. In monkeys from Cameroon, the highest IgG prevalence were observed against ONNV-E2 and DENV2-NS1 with 3.95% and 3.40% respectively and in DRC, ONNV-E2 (6.63%) and WNV-NS1 (4.42%). Overall prevalence was low in apes (n = 1,418): ranging from 0% for USUV-NS1 to 2.6% for CHIKV-E2. However, a very large disparity was observed among collection site and ape species, e.g. 18% (9/40) and 8.2% (4/49) of gorillas were reactive with CHIKV-E2 or WNV-NS1, respectively in two different sites in Cameroon.
Conclusions/Significance
We have developed a serological assay based on Luminex technology, with high specificity and sensitivity for simultaneous detection of antibodies to 10 antigens from 6 different arboviruses. This is the first study that evaluated on a large scale the presence of antibodies to arboviruses in NHPs to evaluate their role in sylvatic cycles. The overall low prevalence (<5%) in more than 3,500 NHPs samples from Cameroon and the DRC does not allow us to affirm that NHP are reservoirs, but rather, intermediate hosts of these viruses.
Author summary
In the last decades, chikungunya, zika, yellow fever, usutu and dengue viruses have (re)-emerged in different parts of the world and many of these outbreaks occur in resource-limited countries with limited or under-equipped health facilities and where endemic malaria with very similar clinical symptoms confounds surveillance. Most arboviruses that circulate today likely originated in Africa where sporadic human outbreaks occur. In this work, we developed a serological tool that allows simultaneous detection of IgG antibodies to multiple arbovirus in a biological sample. With this highly sensitive and specific multiplex assay, we screened more than 3,500 samples collected from 29 species of monkeys and apes in Africa. We found a global IgG antibody prevalence of less than 5%. However, this seroprevalence varied by collection site, NPHs species and virus type. Given these findings, we concluded that African non-human primates are most likely not the reservoirs, but rather are intermediate hosts.
Introduction
Ecology and human behavior play a major role in the increasing emergence and re-emergence of infectious diseases and several factors are required for this to occur [1]. These include prevalence of pathogens in the natural host, transmission mode of the pathogens, frequent contact between humans and wildlife, capacity to adapt to a new host and conditions for subsequent epidemic spread into the human population [2,3]. In particular, the increased contact between humans and wildlife lead to increased risk for disease emergence in humans [4,5].
In the last decades, chikungunya (CHIKV), zika (ZIKV), yellow fever (YFV), usutu (USUV) and dengue (DENV) viruses have (re)-emerged in different parts of the world [6–8], many of these outbreaks occur in resource-limited countries with limited or under-equipped health facilities and where endemic malaria with very similar clinical symptoms confounds surveillance [9]. Most arboviruses that circulate today likely originated in Africa where sporadic human outbreaks occur: YFV is known to circulate endemically in Sub-Saharan Africa for centuries [10]; west nile virus (WNV) and ZIKV were first identified in 1937 and 1947, respectively, in Uganda [11]; CHIKV in 1952 in Tanzania [12] USUV in 1959 in South Africa. The precise origins of DENV remain unknown but it is widely prevalent in East, Central and South Africa.
While large outbreaks of arbovirus diseases in human populations are well documented [13–15], many questions remain unanswered on their sylvatic-cycles, in particular, which wildlife species could be involved. In almost any review or research article, the zoonotic cycles for ZIKV, DENV, YFV CHIKV are presented with arboreal mosquitos feeding on non-human primates (NHPs) but this is based on very limited data. There is only very limited evidence of arbovirus infection or exposure in NHPs but also in wildlife in general. Studying the animal reservoir of arboviruses is a challenging question, because (i) there are difficulties inherent in wildlife sampling, (ii) there are limited high throughput screening technologies, (iii) the antigenic proximity among arboviruses hinders specificity and (iv) arboviruses cause acute infections which limits the detection of the virus to a very narrow window of time, rarely exceeding 3 weeks [16]. The detection of antibodies against arboviruses antigens represent an alternative to virus detection but information on seroprevalence in wildlife are very limited. A few studies reported prevalence of antibodies (mostly IgG) in different African non-human primates (NHPs) species (reviewed in Valentine and colleagues in 2019) [17], which varied from 0% to 100%, depending on the species, the targeted arbovirus, the detection method used, the country of sample’s origin and the number of samples tested.
In the present work, we have addressed some of these challenges. We first developed a high throughput serological screening tool based on the Luminex technology. Next, we screened more than 3,500 samples of a wide diversity of NHPs species from Cameroon and the Democratic Republic of Congo (DRC) for presence of antibodies to multiple arboviruses in order to evaluate their potential role in sylvatic cycles. Our data show an overall low seroprevalence of IgG antibodies to arboviruses unevenly distributed according to NHPs species and to sample collection site.
Materials and methods
Human panel samples and ethics statement
We used a panel of 161 samples of known arbovirus serostatus to validate our Luminex based serological test (S1 Table). All the human samples used in this study were anonymized, and there is no way to link back these leftovers to the 161 patients. The panel consisted of 66 arbovirus negative leftover plasma samples from the Virology department of the University Hospital, Montpellier, France and the Institute of Tropical Medicine, Antwerp, Belgium. The patients were referred to these laboratories for various illnesses and were tested for the presence of an arboviral infection by IgM and IgG Immunofluorescence assay or ELISA assays, PCR and sero-neutralization for some of them. All 66 negative control samples used in the present study were negative with all these assays. Positive control samples for CHIKV, DENV, WNV, YFV and ZIKV originated from patients in France, Belgium, Colombia and the DRC. The samples were obtained as follows: during outbreaks (CHIKV and YFV in the DRC, DENV in Colombia and WNV in France); returning travelers in Europe from countries with outbreaks (ZIKV, DENV) or vaccinees (YFV in Colombia). DENV positive sera consisted of six DENV-1, seven DENV-2, four DENV-3, three DENV-4 and three DENV-1,2,3,4 reactive samples. Outbreak and returning travelers’ samples were confirmed positive by PCR and follow-up serum samples, confirmed by commercial serological assays, were used in the present study. For USUV, we had only three samples from experimentally infected mice that were serially bleeded. We were not able to get positive controls for o’nyong nyong virus (ONNV).
Samples from monkeys and apes
We tested samples from bushmeat and pet monkeys from studies that were conducted between 1999 and 2016 on simian retroviruses and the origin of HIV [18–20] in 14 different sites in southern Cameroon and the DRC (Fig 1 and S2A Table). Whole blood was collected from monkey bushmeat, either by intracardiac puncture and subsequent storage at –20°C, or as a dried blood spot (DBS) on Whatman 903 filter paper (GE Healthcare) at the points of hunting injury and spotting, as described previously [21]. Blood was drawn on EDTA tubes from pet monkeys by venipuncture after tranquilization with ketamine [20]. Species were visually identified in the field and confirmed on a subset of samples by 12S sequence analysis, as previously described [22]. Fecal samples were collected between 2005 and 2017 from wild common chimpanzees (Pan troglodytes troglodytes), western lowland gorillas (Gorilla gorilla gorilla), eastern lowland gorillas (Gorilla beringei graueri) and bonobos (Pan paniscus) at 18 different sites in Cameroon and DRC as part of studies on the origin of HIV [23] (Fig 1 and S2B Table). Feces were stored in RNA-later (Sigma-Aldrich, Saint-Quentin Fallavier, France), kept at ambient temperature in the field for a maximum of three weeks, and then stored at –20°C or –80°C in central repository laboratory.
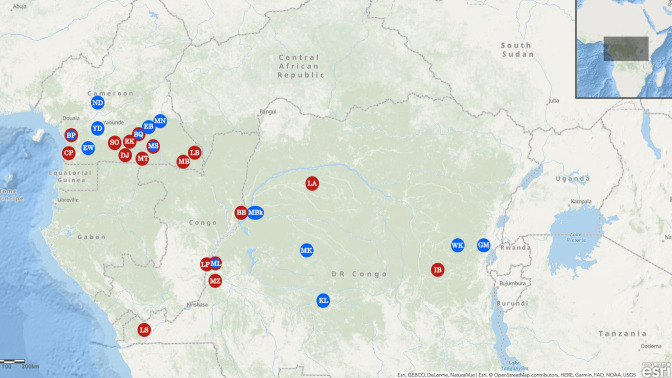
Fig 1. Sample collection sites.
Sites where samples from non-human primates (NHPs) were collected are highlighted with circles on the maps, as follows: blue indicates sites where bushmeat samples from monkeys were collected; red, sites where fecal samples from apes were collected; blue and red, sites where bushmeat samples from monkeys and fecal samples from apes were collected. Abbreviations of sites are as follows: BP, Bipindi; BQ, north of Dja; EB, Eboumetoum; EW, Ebolowa; GM, Goma; KL, Kole; MBk, Mbandaka; MK, Monkoto; ML, Malebo; MN, Mindourou; MS, Messok; ND, Nditam; WK, Walikale; YD, Yaoundé.
Screening for IgG antibodies to arboviruses
Recombinant proteins
We used different commercially available recombinant proteins derived from the envelope or non-structural proteins of CHIKV, ONNV, DENV, ZIKV, USUV, YFV and WNV viruses (S3 Table). The proteins were purchased already purified (all above 95% of purity, except DENV4_NS1 and CHIKV_NSP at >90%) as lyophilized powders, and resuspended in a buffer at concentration as per manufacturer’s instructions, aliquoted and stored frozen at -20°C until use.
Protein coupling to Luminex beads
We used our previously described protocol for coupling primary amines bearing moieties (peptides and proteins) to Luminex beads [21,24]. Briefly, recombinant proteins (1–4μg/1.25 x106 beads) were covalently coupled on carboxyl functionalized fluorescent magnetic beads (Luminex Corp., Austin, TX) with the BioPlex amine coupling kit (Bio-Rad Laboratories, Marnes-la-Coquette, France) according to the manufacturer’s instructions. We blocked unreacted sites with blocking buffer from the amine coupling kit. Protein-coupled microsphere preparations were washed with PBS, and stored in storage buffer (Bio-Rad) at 4°C in the dark until use.
Multiplex screening for IgG antibodies to arbovirus in plasma and fecal dialysates
Before use, recombinant protein-coupled beads were vortexed for 30s and diluted to 2,000 beads/μl of assay buffer (Phosphate Buffered Saline (PBS) containing 0.75 mol/L NaCl, 1% (wt/vol) bovine serum albumin (Sigma Aldrich, Saint-Quentin Fallavier, France), 5% (vol/vol) heat-inactivated fetal bovine serum (Gibco-Invitrogen, Cergy Pontoise, France), and 0.2% (vol/vol) Tween-20 (Sigma-Aldrich). Tests were performed in 96-well flat-bottom chimney plates (Greiner bio one, Frickenhausen, Germany). Fifty microliters of bead mixture were added to each well. Preliminary experiments on different plasma dilutions (1/50-1/1,000) and different incubation times and temperatures showed that the dilution 1/200 and +4°C overnight incubation gave the best signal to noise ratio. Liquid was aspirated with an automatic plate washer (BioTek 405TS Microplate washer) and wells were then incubated with 100 μl of plasma (diluted 1/200 in assay buffer) for 16h at 4°C in the dark on a plate shaker at 300 rpm/min. After 3 washings with 100 μl of assay buffer, 50 μl of biotin-labeled anti-human IgG was added (BD-Pharmingen, Le Pont De Claix, France) at a concentration of 4 μg/ml in each well and incubated for 30 min in the dark while shaking at 300 rpm. Plates were washed 3 times as above, and 50 μl of streptavidin-R-phycoerythrin (Fisher Scientific/Life Technologies, Illkirch, France) at 1 μg/ml were added per well and incubated for ten min with shaking at 300 rpm. As in previous studies from our group and others, anti-human IgG was also used for NHPs [21,25]. Antigen-antibody reactions were then read on BioPlex-200 equipment (Bio-Rad, Marnes-la-Coquette. France). At least 100 events were read for each bead set, and the results were expressed as median fluorescence intensity (MFI) per 100 beads. To detect IgG antibodies to arboviruses in fecal samples, RNA-later-precipitated immunoglobulins were first resolubilized by diluting the fecal/RNA-later mixture (2 mL) with PBS–0.05% Tween 20 (7 mL), followed by incubation for 1 hour at 60°C, centrifugation (3900g for 10 minutes) to clarify the solution, and dialysis against PBS overnight at 4°C under a continuous stirring. The reconstituted extracts were then tested in the Luminex (diluted three volumes of dialysate for one volume of buffer) as previously described [24].
Calculation of cut-off, sensitivity, specificity and accuracy
For the samples of the panel, we used receiver operating characteristics (ROC) curve analysis to determine the cut-off values for each antigen, its sensitivity, specificity and accuracy which corresponds to the area under the curve (AUC). The ROC curve analysis was performed with the Life module of XLSTAT (Addinsoft, Paris, France) implemented in Microsoft Excel. We also determined the sensitivity, specificity and accuracy by calculating the mean MFI of negative controls for each antigen, the standard deviation to the mean (SD) and using as cut-off the mean plus three times the SD because for ONNV and USUV, no or only limited positive control samples were available. We used the Wilson method [26] to calculate online the 95% confidence intervals (CI) around the proportions (http://ww3.ac-poitiers.fr/math/prof/resso/cali/ic_phrek.html).
In the absence of positive controls for NHPs samples, we analyzed the data obtained from plasma and DBS samples with different statistical methods to determine MFI cut-off values for each antigen as reported in our previous studies on ebolavirus in NHPs [27]. We used a change-point analysis with the R package “changepoint” and calculated one single shift in the arithmetic mean with the AMOC (at most one change) method. We also fitted univariate distributions to our data and defined the cut-off based on a 0.05 risk of error [28]. The set of candidate distributions was reduced with a bootstrapped skewness-kurtosis analysis [29]. Maximum likelihood estimation was performed to select the best-fit distribution based on AIC (Akaike information criterion) using the R library “fitdistrplus” [30]. The best-fit distributions were negative binomial and negative exponential distributions and both were considered in data analyses. Data were bootstrapped 10,000 times and averaged for each antigen. Analyses were done with R software version 3.3.6. We then compared the cut-off values identified by the 3 different methods and calculated their mean as a consensus cut-off that we used in this study (S4 Table). We calculated separately cut-off values for samples collected as DBS (samples from the DRC) and those collected as blood in EDTA tubes (samples from Cameroon) because of the wide disparity of blood quantity collected with DBS. We considered a sample antigen reactive if MFI was above the cut-off value. Likewise, for samples collected as feces, we calculate MFI cut-off values separately on the data generate from the dialysates (S5 Table). We considered samples positive for a given antigen if they presented MFI above the cut-off value for this antigen.
Results
Performance of the arbovirus Luminex assay on a reference panel of human samples
Before proceeding to the screening of NHPs samples, we first evaluated the performance of the novel Luminex-based serological assay on the panel of human samples with known status (S1 Table). To determine the assay performances, we first calculated cut-off values by two methods for 15 of the 17 antigens included. For USUV and ONNV, it was not possible to use ROC analysis because of the absence of positive controls and cut-off values were determined as 3XSD of negative samples. Results of cut-off determinations are summarized in Table 1. Regardless of the cut-off method used to determine the sensitivity, specificity and accuracy of each antigen, the specificity is in general high, > 95%, for the majority of the antigens. The sensitivity, on the contrary, depended on the virus and on the recombinant protein used. The sensitivity of NS1 recombinant proteins as determined by the ROC analysis method was 100% for WNV, DENV-2, DENV-3, ZIKV and >95% for DENV-1; 87% for DENV-4 and only 44% for YFV proteins. We also observed a 100% sensitivity for WNV-DIII protein. Sensitivity of CHIKV-E2 envelope protein was also high, >95% by ROC analysis. Of note, DIII recombinant proteins from the envelope, except WNV-DIII, presented weaker sensitivity, compared to NS1 recombinant proteins.
Table 1. Cut-off, Sensitivity, Specificity and Accuracy of antibody detection to the 17 different arbovirus recombinant proteins on the reference panel of 161 human samples.
| Antigens | Cut-off calculation method | Cut-off value (MFIa) | N tested/ N negative | Specificity (%) | 95% CIb | N tested/ N negative | Sensitivity (%) | 95% CI | Accuracy (%) | 95% CI |
|---|---|---|---|---|---|---|---|---|---|---|
| CHIKV_E2 | Mean+3SD | 324 | 66/65 | 98.48 | 91.0–99.0 | 27/24 | 88.89 | 71.0–96.0 | 96 | 92.0–97.0 |
| ROC | 229 | 66/63 | 95.45 | 87.0–98.0 | 27/26 | 96.3 | 40.0–75.0 | 96 | 92.0–97.0 | |
| CHIKV_NSP | Mean+3SD | 395 | 66/65 | 98.48 | 0.91–99.0 | 27/12 | 44.44 | 27.0–62.0 | 83 | 75.0–84.0 |
| ROC | 246 | 66/65 | 98.48 | 0.91–99.0 | 27/20 | 74.07 | 55.0–86.0 | 91 | 86.0–93.0 | |
| ONNV_E2 | Mean+3SD | 362 | 66/64 | 96.97 | 0.91–99.0 | NA/NA | NA | NA | NA | NA |
| ZIKV_DIII | Mean+3SD | 485 | 66/65 | 98.48 | 0.91–99.0 | 16/0 | 0 | 0.0–19.0 | 79 | 70.0–80.0 |
| ROC | 145 | 66/59 | 89.39 | 79.0–94.0 | 16/02 | 12.5 | 03.0–36.0 | 74 | 65.0–75.0 | |
| ZIKV_NS1 | Mean+3SD | 74 | 66/65 | 98.48 | 91.0–99.0 | 16/16 | 100 | 80.0–100 | 0.99 | 96.0–100 |
| ROC | 345 | 66/66 | 100 | 94.0–100 | 16/16 | 100 | 80.0–100 | 100 | 100–100 | |
| YFV_NS1 | Mean+3SD | 421 | 66/65 | 98.48 | 91.0–99.0 | 18/5 | 27.78 | 12.0–50.0 | 83 | 75.0–85.0 |
| ROC | 176 | 66/62 | 93.94 | 85.0–97.0 | 18/8 | 44.44 | 24.0–66.0 | 83 | 75.0–85.0 | |
| DENV1_DIII | Mean+3SD | 134 | 66/66 | 100 | 94.0–100 | 23/17 | 73.91 | 53.0–87.0 | 93 | 88.0–95.0 |
| ROC | 132 | 66/66 | 100 | 94.0–100 | 23/18 | 78.26 | 58.0–91.0 | 94 | 90.0–96.0 | |
| DENV2_DIII | Mean+3SD | 842 | 66/65 | 98.48 | 91.0–99.0 | 23/4 | 17.39 | 06.0–37.0 | 78 | 69.0–78.0 |
| ROC | 631 | 66/63 | 95.45 | 87.0–98.0 | 23/8 | 34.78 | 18.0–55.0 | 80 | 71.0–81.0 | |
| DENV3_DIII | Mean+3SD | 197 | 66/63 | 95.45 | 87.0–98.0 | 23/17 | 73.91 | 53.0–87.0 | 90 | 84.0–91.0 |
| ROC | 79 | 66/57 | 86.36 | 76.0.-92.0 | 23/21 | 91.3 | 76.0–97.0 | 88 | 81.0–89.0 | |
| DENV4_DIII | Mean+3SD | 544 | 66/64 | 96.97 | 91.0–99.0 | 23/6 | 26.09 | 12.0–46.0 | 79 | 70.0–80.0 |
| ROC | 112 | 66/53 | 80.3 | 69.0–88.0 | 23/18 | 78.26 | 58.0–91.0 | 80 | 71.0–81.0 | |
| DENV1_NS1 | Mean+3SD | 673 | 66/65 | 98.48 | 91.0–99.0 | 23/21 | 91.3 | 73.0–97.0 | 97 | 93.0–98.0 |
| ROC | 550 | 66/64 | 96.97 | 91.0–99.0 | 23/22 | 95.65 | 79.0–99.0 | 97 | 93.0–98.0 | |
| DENV2_NS1 | Mean+3SD | 812 | 66/64 | 96.97 | 91.0–99.0 | 23/23 | 100 | 85.0–100 | 98 | 95.0–99.0 |
| ROC | 996 | 66/66 | 100 | 94.0–100 | 23/23 | 100 | 85.0–100 | 100 | 100–100 | |
| DENV3_NS1 | Mean+3SD | 54 | 66/64 | 96.97 | 91.0–99.0 | 23/23 | 100 | 85.0–100 | 98 | 95.0–99.0 |
| ROC | 200 | 66/66 | 100 | 94.0–100 | 23/23 | 100 | 85.0–100 | 100 | 100–100 | |
| DENV4_NS1 | Mean+3SD | 471 | 66/65 | 98.48 | 91.0–99.0 | 23/19 | 82.61 | 62.0–93.0 | 94 | 90.0–96.0 |
| ROC | 462 | 66/65 | 98.48 | 91.0–99.0 | 23/20 | 86.96 | 67.0–95.0 | 96 | 91.0–97.0 | |
| USUV_NS1 | Mean+3SD | 89 | 66/65 | 98.48 | 91.0–99.0 | NA/NA | NA | NA | NA | NA |
| WNV_NS1 | Mean+3SD | 100 | 66/64 | 96.97 | 91.0–99.0 | 11/11 | 100 | 74.0–100 | 97 | 94.0–99.0 |
| ROC | 1297 | 66/66 | 100 | 94.0–100 | 11/11 | 100 | 74.0–100 | 100 | 100–100 | |
| WNV_DIII | Mean+3SD | 309 | 66/65 | 98.48 | 91.0–99.0 | 11/11 | 100 | 74.0–100 | 99 | 96.0–100 |
| ROC | 817 | 66/66 | 100 | 94.0–100 | 11/11 | 100 | 74.0–100 | 100 | 100–100 |
(CHIKV: Chikungunya virus); (ZIKV: Zika virus); (DENV: Dengue virus); (USUV: Usutu virus); (WNV: West Nile virus); (YFV: Yellow Fever virus); (ONNV: O’Nyong Nyong virus).
a MFI: median fluorescence intensity
b CI: confidence interval.
One hallmark of sero-detection of antibodies to arboviruses is cross-reaction due to antigenic homology between the different viruses. For the evaluation of the level of cross-reaction on our panel of 95 positive samples, we used cutoff values obtained with the ROC analysis method (except for ONNV and USUV). S6 Table shows the proportion of CHIKV, ZIKV, DENV, WNV and YFV positive samples that also react present with heterologous antigens. By doing so, 14/27 (52%) CHIKV+ positive samples cross-reacted with closely related ONNV-E2, and also with others flavivirus, varying from 2/27 (7.4%) to 16/27 (60%). On the other hand, flavivirus positive samples presented no or weak cross-reactions with antigens from the alphaviruses, CHIKV and ONNV, and variable rates of cross-reactions were observed among the different flaviviruses. Unsurprisingly, DENV+ positive samples highly cross-reacted with all DENV-NS1 proteins, regardless the dengue virus serotype not allowing to differentiate among dengue serotypes. Cross reactivity was also seen between the DIII proteins of the dengue serotypes; for example, of the six DENV-1+ samples, five reacted with DENV-1, DENV-3 and DENV-4 DIII and only DENV-2 DIII antigen presented no reaction with the six DENV-1 positive samples. DENV+ samples also reacted at high proportion with NS1 antigens from ZIKV (14/23, 60%), YFV (19/23, 82.6%) and WNV (9/23, 39%). All WNV+ samples cross-reacted with NS1 antigens from USUV and at different levels (0–36%) with other arboviral antigens. Finally, YFV+ samples cross-reacted with other NS1 antigens at levels equal or superior to that of cognate YFV NS1 antigen. To summarize, cross-reactions in our control panel with the different arbovirus antigens is variable and ranged between 0% and 60%.
Overall, accuracy of the different antigens evaluated was excellent (>95%) for all NS1, except YFV-NS1. CHIKV-E2 and WNV-DIII recombinant proteins also presented high accuracy. We thus decided to use only antigens with high sensitivity and specificity (i.e. CHIKV-E2, ONNV, ZIKV-NS1, DENV1-4-NS1, WNV-NS1 and DIII, USUV-NS1) for the screening of wildlife samples.
Seroprevalence of IgG antibodies to arboviruses in multiple monkey species from Cameroon and the DRC
Overall, 2,100 samples collected from 26 different monkey species were screened (S2A Table and S1 Data). Total IgG antibodies to the ten selected recombinant proteins varied between 2% to 5% (Table 2). We analyzed more in detail the seroprevalence of IgG antibodies to the different arbovirus antigens, stratifying by monkey species (Table 2). Seven of 26 species screened were IgG negative towards all the antigens tested, i.e. red-capped mangabey (n = 7), Hamlyn’s monkey (n = 6), L’Hoest’s monkey (n = 38), Preuss’s monkey (n = 1), drill (n = 1), mandrills (n = 24) and olive baboon (n = 16). It should be noted that for four of these seven species, less than ten samples were tested. For the remaining 19 monkey species presenting IgG reaction against at least one antigen, seroprevalence varied greatly by monkey species and virus antigen. Hence, the blue monkey presented the highest proportion of positive samples, 31% (16/51), against CHIKV and were only marginally positive (1/51) against ZIKV and DENV-3 NS1 recombinant proteins. The next monkey species presenting high proportions of reactive samples are black colobus with 28% (2/7) reactive against CHIKV-E2 and DENV-NS1 recombinant proteins, angolan colobus and northern talapoin with 8% (2/25) and 11% (2/18) reactive samples against DENV-NS1 and WNV-DIII antigens, respectively. Red-tailed and mona monkeys presented 12% (23/181) and 11% (1/9) reactive samples against ONNV_E2 antigen. The remaining 13 monkey species presented less than 8% of reactive samples against any of the tested antigens.
Table 2. Seroprevalence of IgG antibodies to Chikungunya (CHIKV), O’nyong nyong (ONNV), Zika (ZIKV), Dengue (DENV), Usutu (USUV) and West Nile (WNV) viruses stratified by monkey species.
| Species | Total | CHIKV E2 | ONNV E2* | ZIKV NS1 | DENV1 NS1 | DENV2 NS1 | DENV3 NS1 | DENV4 NS1 | At least 1 DENV NS1 | USUV_NS1* | WNV NS1* | WNV DIII* | Range (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allan swamp monkey | 41 | - a | - | 1/41 (2.4)b | - | - | - | - | - | 1/41 (2.4) | 2/41 (4.8) | 3/41 (7.3) | (0.0–7.3) |
| Agile mangabey | 128 | 3/128 (2.3) | 4/61 (6.5) | 2/128 (1.5) | 2/128 (1.5) | - | 1/128 (0.8) | 2/128 (1.5) | 3/128(2.3) | 1/112 (0.8) | 1/112 (0.8) | 2/112 (1.7) | (0.0–6.5) |
| Red capped mangabey | 7 | - | - | - | - | - | - | - | - | - | - | - | (0.0–0.0) |
| Angolan colobus | 25 | - | - | 1/25 (4.0) | 2/25 (8.0) | 1/25 (4.0) | 2/25 (8.0) | 2/25 (8.0) | 2/25 (8.0) | 2/21 (9.5) | 2/21(9.5) | - | (0.0–9.5) |
| Mantled guereza | 34 | 1/34 (2.9) | 1/27 (3.7) | 1/34 (2.9) | 1/34 (2.9) | 1/34 (2.9) | 1/34 (2.9) | 1/34 (2.9) | 1/34 (2.9) | 1/34 (2.9) | 1/34 (2.9) | 1/34 (2.9) | (0.0–3.7) |
| Black colobus | 7 | 2/7 (28.0) | nt c | 1/7(14) | 1/7 (14) | 2/7 (28.0) | 1/7 (14) | 1/7 (14) | 2/7 (28.0) | - | - | - | (0.0–28.0) |
| Tsuapa red colobus | 86 | 2/86 (2.3) | 3/85 (3.5) | 1/86 (1.1) | 1/86 (1.1) | 1/86 (1.1) | - | 1/86 (1.1) | 3/86 (3.4) | 3/85 (3.4) | 1/85 (1.1) | 5/85 (5.8) | (0.0–5.8) |
| Red tailed monkey | 234 | 8/234 (3.4) | 23/181 (12) | 11/234 (4.7) | 6/234 (2.6) | 6/234 (2.6) | 5/234 (2.1) | 7/234 (3.0) | 12/234 (5.1) | 8/181 (4.5) | 8/181 (4.5) | 13/181 (7.1) | (2.6–12.0) |
| Mustached monkey | 504 | 21/504 (4.1) | 7/148 (4.7) | 12/504 (2.3) | 7/504 (1.3) | 15/504 (2.9) | 6/504 (1.2) | 9/504 (1.7) | 20/504 (3.9) | 7/369 (1.8) | 5/369 (1.3) | 10/369 (2.7) | (1.2–4.7) |
| Hamlyn’s monkey | 6 | - | nt | - | - | - | - | - | - | nt | nt | nt | (0.0–0.0) |
| l’Hoest’s monkey | 38 | - | nt | - | - | - | - | - | - | nt | nt | nt | (0.0–0.0) |
| Blue monkey | 51 | 16/51 (31.0) | nt | 1/51 (1.9) | - | - | 1/51 (1.9) | - | 1/51 (1.9) | nt | nt | nt | (0.0–31.0) |
| Mona monkey | 9 | - | 1/9 (11) | - | - | - | - | - | - | - | - | - | (0.0–11.0) |
| De Brazza monkey | 59 | - | 2/43 (4.6) | 2/59 (3.3) | - | - | - | 1/59 (1.7) | 1/59 (1.7) | 2/53 (3.7) | 3/53 (5.6) | 4/53 (7.5) | (0.0–7.5) |
| Greater spot-nosed | 385 | 6/385 (1.5) | 9/210 (4.2) | 17/385 (4.4) | 7/385 (1.8) | 18/385 (4.7) | 9/385 (2.3) | 10/385 (2.6) | 29/385 (7.5) | 12/315 (3.8) | 6/315 (1.9) | 7/315 (2.2) | (1.5–7.5) |
| Crested mona monkey | 182 | 2/182 (1.0) | 3/77 (3.8) | 6/182 (3.3) | 7/182 (3.8) | 8/182 (4.4) | 5/182 (2.7) | 5/182 (2.7) | 9/182 (4.9) | 3/137 (2.1) | 4/137 (2.9) | 2/137 (1.4) | (1.0–4.9) |
| Preuss monkey | 1 | - | - | - | - | - | - | - | - | - | - | - | (0.0–0.0) |
| Wolf’s monkey | 71 | 1/71 (1.4) | 2/55 (3.6) | 1/71 (1.4) | 2/71 (2.8) | 2/71 (2.8) | 3/71 (4.2) | 1/71 (1.4) | 5/71 (7.0) | 1/55 (1.8) | 1/55 (1.8) | - | (0.0–7.0) |
| Tantalus monkey | 14 | - | - | - | - | - | - | - | - | - | - | 1/14 (7.1) | (0.0–7.1) |
| Patas monkey | 16 | - | - | - | - | - | - | - | - | - | - | 1/16 (6.2) | (0.0–6.2) |
| Grey cheecked mangabey | 110 | 5/110 (4.5) | - | 7/110 (6.3) | 5/110 (4.5) | 5/110 (4.5) | 5/110 (4.5) | 8/110 (7.2) | 9/110 (8.1) | 2/74 (2.7) | 1/74 (1.3) | 3/74 (4.0) | (0.9–8.1) |
| Black mangabey | 33 | - | 1/29 (3.4) | - | - | - | - | 1/33 (3.0) | 1/33 (3.0) | - | - | - | (0.0–3.0) |
| Drill | 1 | - | - | - | - | - | - | - | - | - | - | - | (0.0–0.0) |
| Mandrill | 24 | - | - | - | - | - | - | - | - | - | - | - | (0.0–0.0) |
| Northern talapoin | 18 | - | - | 1/18 (5.5) | 2/18 (11.0) | 1/18 (5.5) | 1/18 (5.5) | 1/18 (5.5) | 2/18 (11) | 1/18 (5.5) | - | 2/18 (11.1) | (0.0–11.0) |
| Olive baboon | 16 | - | - | - | - | - | - | - | - | - | - | - | (0.0–0.0) |
| Total | 2100 | 67/2100 (3.2) | 56/1109 (5.0) | 65/2100 (3.0) | 43/2100 (2.0) | 60/2100 (2.8) | 40/2100 (1.9) | 50/2100 (2.3) | 100/2100 (4.6) | 44/1613 (2.7) | 37/1613 (2.3) | 54/1613 (3.3) | (1.9–4.9) |
a -: no positive samples detected
b n/N tested (percentages)
c: nt, not tested.
*The total of samples tested on ONNV, Usutu and WNV antigens are different to those tested on the other antigens for certain species.
Thereafter, IgG antibodies were stratified by collection sites and prevalences showed variation between 1.46% (WNV_NS1 in Cameroon) and 6.63% (ONNV_E2 in the DRC) (Table 3). The proportions of ZIKV and DENV positive samples were in general higher in samples collected in Cameroon than in DRC, while IgG antibodies against CHIKV, ONNV, USUV and WNV antigens were higher in samples collected in DRC than in Cameroon (Table 3). For DENV-2 and WNV, the difference was statistically significant (p = 0.0321 for DENV-2, 0.0007 and 0.0039 for WNV-NS1 and WNV-DIII, respectively). The proportion of positive samples is also unevenly distributed among the sampling sites within countries. A clear difference was observed between samples collected from pets and those obtained from feral monkeys. Hence 8/175 (4.5%) samples from pets, all from infant or juvenile monkeys living in urban areas, reacted with at least one of the antigens, while 5.2–46.6% of samples reacted with at least one antigen in samples collected from feral monkeys (>90% adults). IgG antibodies to CHIKV-E2 protein were detected at high proportion (35%) in WK, eastern DRC and in lesser extent, in GM also in eastern DRC and in EB, EW and BP in Cameroon.
Table 3. Seroprevalence of IgG antibodies to Chikungunya (CHIKV), O’nyong nyong (ONNV), Zika (ZIKV), Dengue (DENV), Usutu (USUV) and West Nile (WNV) viruses in monkeys stratified by collection site shown in Fig 1.
| CHIKV_E2 | ONNV_E2# | ZIKV_NS1 | DENV1_NS1 | DENV2_NS1 | DENV3_NS1 | DENV4_NS1 | > 1 DENV NS1 | USUV_NS1* | WNV_NS1* | WNV_DIII* | Totala (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cameroon (n = 1470) | ||||||||||||
| Pets (n = 175) | -b | 2 (1.1%) c | - | 2 (1.1) | - | 1 (0.5) | 1 (0.5) | 2 (1.1) | - | 1 (0.5) | 5 (2.8) | 8/175 (4.5) |
| BP (n = 19) | 1 (5.2) | 1 (5.2%) | 1 (5.2) | 1 (5.2) | 1 (5.2) | - | 1 (5.2) | 1 (5.2) | 1 (5.2) | - | - | 3/19 (15.7) |
| ND (n = 65) | - | 1 (1.5%) | 1 (1.5) | 1 (1.5) | 1 (1.5) | 1 (1.5) | 2 (3.0) | 1 (5.2) | 1 (1.5) | 1 (1.5) | 1 (1.5) | 5/65 (7.69) |
| YD (n = 96) | - | - | 2 (2.0) | 1 (1.0) | 1 (1.0) | - | 3 (3.1) | 2 (2.0) | 1 (1.0) | 1 (1.0) | - | 5/96 (5.20) |
| BQ (n = 51) | 1 (1.9) | 4 (7.8%) | 2 (3.9) | 3 (5.9) | 5 (9.8) | 3 (5.9) | 5 (9.8) | 4 (4.2) | 2 (3.9) | 4 (7.8) | - | 13/51 (25.4) |
| EW (n = 228) | 12 (5.2) | nt$ | 10 (4.4) | 5 (2.1) | 14 (6.1) | 6 (2.6) | 5 (2.1) | 8 (3.5) | 4 (2.9)* | 3 (1.72)* | 10 (5.7)* | 44/228(19.2) |
| EB (n = 236) | 11 (4.66) | nt | 7 (3.0) | 3 (1.2) | 5 (2.1) | 3 (1.2) | 4 (1.7) | 4 (4.5) | 2 (1.6)* | 1 (0.6)* | 3 (1.8)* | 25/236(10.5) |
| MS (n = 94) | 3 (3.2) | 6 (6.4%) | 7 (7.4) | 4 (4.2) | 6 (6.4) | 4 (4.2) | 4 (4.2) | 4 (4.5) | 6 (6.4) | 5 (5.4) | 1 (1.0) | 17/94 (18.0) |
| MN (n = 506) | 12 (2.3) | 12 (7.6%)# | 17 (3.3) | 12 (2.3) | 17 (3.3) | 10 (1.9) | 12 (2.3) | 14 (2.8) | 9 (2.7)* | 1 (0.3)* | 9 (2.7)* | 68/506(13.4) |
| Total | 40 (2.72) | 26 (3.95) | 47 (3.19) | 32 (2.17) | 50 (3.40) | 28 (1.90) | 37 (2.51) | 40 (2.72) | 26 (2.23) | 17 (1.46) | 29 (2.49) | |
| DRC (n = 630) | ||||||||||||
| MBk(n = 125) | 2 (1.6) | 6 (4.8%) | 3 (2.4) | - | 2 (1.6) | 1 (0.8) | - | - | 5 (4.0) | 8 (6.4) | 6 (4.8) | 20/125 (16.0) |
| ML (n = 45) | - | 12 (2.7%) | 5 (11) | 2 (4.4) | 1 (2.2) | 2 (4.4) | 1 (2.2) | 2 (4.4) | 3 (6.6) | 4 (8.8) | 10 (22) | 21/45 (46.6) |
| MK (n = 29) | - | 1 (3.4%) | 1 (3.4) | 1 (3.4) | 2 (6.8) | 1 (3.4) | 4 (13) | 1 (3.4) | 2 (6.8) | 2 (6.8) | 2 (6.8) | 8/29 (27.5) |
| KL (n = 280) | 5 (1.8) | 11 (4.3%)# | 7 (2.5) | 7 (2.5) | 5 (1.8) | 6 (2.1) | 8 (2.8) | 6 (2.1) | 8 (3.1)* | 6 (2.3)* | 7 (2.7)* | 32/280 (11.4) |
| WK (n = 39) | 14 (35) | nt | 2 (5.1) | 1 (2.5) | - | 1 (2.5) | - | - | nt | nt | nt | 15/39 (38.4) |
| GM (n = 112) | 6 (5.3) | nt | - | - | - | 1 (0.9) | - | - | nt | nt | nt | 7/112 (6.2) |
| Total | 27 (4.28) | 30 (6.63) | 18 (2.85) | 11 (1.74) | 10 (1.58) | 12 (1.89) | 13 (2.06) | 9 (1.42) | 18 (3.96) | 20 (4.42) | 25 (5.53) | |
a Total number of samples reactive with one or more antigens.
b -: no positive samples were identified.
c number of positives (percentages).
* Total numbers tested are different: Cameroon: n = 1161(EW = 174; EB = 158; MN = 329); DRC. n = 452 (KL = 253).
# Total numbers tested are different: Cameroon: n = 657 (MN = 157); DRC: n = 452 (KL = 253).
$: nt = not tested.
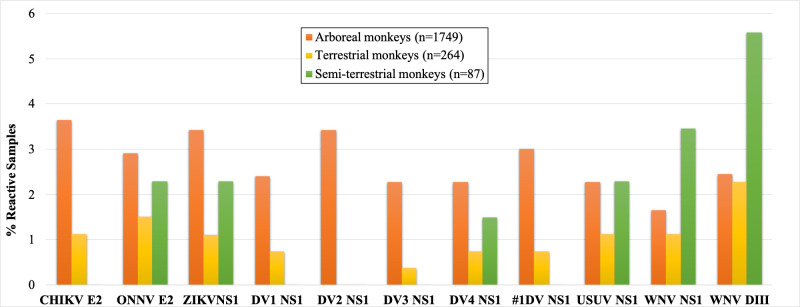
We then stratified the proportion of positive samples by NHP biotopes and split NHPs into three groups: arboreal (n = 1,749), terrestrial (n = 264) and semi-terrestrial (n = 87) species (Fig 2 and S7 Table). Following this grouping, in the arboreal group positive samples were detected against all the ten antigens tested. Another remarkable observation is the quasi-absence of DENV positive samples in semi-terrestrial monkeys, with the exception of one sample (1/87) with antibodies against DENV-4 NS1 antigen. Finally, in this group of semi-terrestrial NHPs, despite the relative low number of samples tested (n = 87), high proportions of reactive samples were observed against WNV and USUV virus antigens.
Fig 2. Proportions of positive samples to several arboviruses and stratified by monkey biotope/habitat.
The figure shows the proportion (vertical axis) of samples positive to the different antigens reported in the horizontal axis, in the different monkey species biotopes as detailed in supplementary S7 Table.
We also checked for simultaneous reactions to multiple viruses (Table 4). For example, in Cameroon, 10% of CHIKV-E2 positive samples (n = 40), were also reactive with ONNV-E2, ZIKV-NS1 and DENV-2 NS1 antigens; 40% of USUV NS1 reactive samples (n = 22) reacted also with WNV NS1 antigen. In DRC, 18% of CHIKV-E2 reactive samples (n = 27) were reactive with ONNV-E2 and 11% with ZIKV-NS1 antigens. In both countries, DENV-NS1 reactive samples reacted with other NS1 antigens in proportions ranging between 27 and 63%. Among the USUV-NS1 reactive samples (n = 18), 72% reacted with WNV-NS1 antigen.
Table 4. Percentages of monkey samples reacting simultaneously with different arboviral recombinant proteins.
| Cameroon | Number of reactive samples | ONNV_E2 | ZIKV_NS1 | DENV1_NS1 | DENV2_NS1 | DENV3_NS1 | DENV4_NS1 | USUV_NS1 | WNV_NS1 | WNV_DIII |
|---|---|---|---|---|---|---|---|---|---|---|
| CHIKV_E2 | 40 | 10.0 | 10.0 | 2.5 | 10.0 | 2.5 | 2.5 | 2.5 | 2.5 | 2.5 |
| ONNV_E2 | 26 | 7.7 | 3.8 | 3.8 | 3.8 | 3.8 | 7.7 | 7.7 | 7.7 | |
| ZIKV_NS1 | 47 | 27.7 | 36.2 | 31.9 | 27.7 | 29.8 | 12.8 | 2.1 | ||
| DENV1_NS1 | 26 | 69.2 | 50.0 | 80.8 | 23.1 | 19.2 | 3.8 | |||
| DENV2_NS1 | 39 | 41.0 | 41.0 | 17.9 | 10.3 | 2.6 | ||||
| DENV3_NS1 | 22 | 50.0 | 22.7 | 22.7 | 0.0 | |||||
| DENV4_NS1 | 37 | 21.6 | 16.2 | 2.7 | ||||||
| USUV_NS1 | 22 | 40.9 | 0.0 | |||||||
| WNV_NS1 | 10 | 0.0 | ||||||||
| WNV_DIII | 15 | |||||||||
| DRC | Number of reactive samples | ONNV_E2 | ZIKV_NS1 | DENV1_NS1 | DENV2_NS1 | DENV3_NS1 | DENV4_NS1 | USUV_NS1 | WNV_NS1 | WNV_DIII |
| CHIKV_E2 | 27 | 18.5 | 11.1 | 3.7 | 3.7 | 3.7 | 3.7 | 3.7 | 3.7 | 0.0 |
| ONNV_E2 | 30 | 13.3 | 6.7 | 10.0 | 3.3 | 10.0 | 13.3 | 16.7 | 23.3 | |
| ZIKV_NS1 | 18 | 22.2 | 16.7 | 27.8 | 27.8 | 33.3 | 33.3 | 11.1 | ||
| DENV1_NS1 | 11 | 54.5 | 63.6 | 63.6 | 36.4 | 27.3 | 9.1 | |||
| DENV2_NS1 | 10 | 50.0 | 50.0 | 40.0 | 40.0 | 20.0 | ||||
| DENV3_NS1 | 12 | 50.0 | 41.7 | 41.7 | 8.3 | |||||
| DENV4_NS1 | 13 | 30.8 | 30.8 | 7.7 | ||||||
| USUV_NS1 | 18 | 72.2 | 16.7 | |||||||
| WNV_NS1 | 20 | 10.0 | ||||||||
| WNV_DIII | 25 |
Low seroprevalence of IgG antibodies to arboviruses antigens in apes from Cameroon and the DRC
African apes, including gorillas, chimpanzees and bonobos are endangered species and are on the red list of IUCN. Thus, only non-invasive sampling methods are allowed as alternative methods to study pathogens in these animals. Here we tested dialysates of 1,418 fecal samples from bonobos, chimpanzees and gorillas on the same antigens used for samples from monkeys (S1 Data). Overall, the proportions of IgG positive samples against any given arbovirus antigen was low (<5%) (Table 5). Gorillas, with 4.35% (35/803) reactive samples against CHIKV-E2 antigen, presented the highest proportion of IgG positive samples. There was no DENV positive sample in the 263 chimpanzees’ fecal dialysates tested. Many samples collected in the remaining 12 collection sites presented IgG antibodies against multiples arboviral antigens. For example, samples collected from gorillas in SO (Cameroon) reacted against all the ten antigens. As for monkeys, simultaneous reactions to multiple virus antigens were also observed with samples from apes. For example, 59%, 19% and 24% of CHIKV-E2 positive samples reacted also with ONNV-E2, ZIKV-NS1 and WNV-DIII antigens, respectively (Table 6). Likewise, DENV-1 NS1 reactive samples reacted with 33%-100% of other DENV, WNV and USUV NS1 antigens.
Table 5. Seroprevalence of IgG antibodies to Chikungunya (CHIKV), Zika (ZIKV), Dengue (DENV), Usutu (USUV), O’Nyong nyong (ONNV) and West Nile (WNV) viruses stratified by ape species and collection site as shown in Fig 1.
| Country | Sampling site | CHIKV | ONNV | ZIKV | DV1 | DV2 | DV3 | DV4 | USUV | WNV | WNV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| E2 | E2 | NS1 | NS1 | NS1 | NS1 | NS1 | NS1 | NS1 | DIII | ||
| Bonobo | |||||||||||
| DRCa | LP (n = 14) | - b | - | - | - | 1 (7.14) c | - | - | - | 2 (14.2) | - |
| DRC | ML (n = 18) | - | - | 1 (5.55) | - | - | - | - | - | - | - |
| DRC | MZ (n = 183) | 1 (0.54) | - | - | - | - | - | - | - | - | - |
| DRC | LA (n = 137) | - | - | - | - | 3 (2.1) | - | 2 (1.4) | - | 12 (8.75) | - |
| Subtotal | n = 352 | 1 (0.28) | - | 1 (0.28) | - | 4 (1.13) | - | 2 (0.56) | - | 14 (3.97) | - |
| Chimpanzee | |||||||||||
| CMR | EK (n = 35) | - | 2 (5.71) | 1 (2.85) | - | - | - | - | - | - | - |
| CMR | BQ (n = 29) | - | 2 (6.89) | 2 (6.9) | - | - | - | - | - | 2 (6.89) | - |
| CMR | CP (n = 5) | - | - | - | - | - | - | - | - | - | - |
| CMR | MB (n = 113) | 1 (0.88) | - | - | - | - | - | - | - | 1 (0.88) | - |
| DRC | BB (n = 10) | - | - | - | - | - | - | - | - | 2 (20.0) | - |
| DRC | LS (n = 71) | - | - | - | - | - | - | - | - | - | - |
| Subtotal | n = 263 | 1 (0.38) | 4 (1.52) | 3 (1.14) | - | - | - | - | - | 5 (1.90) | - |
| Gorilla | |||||||||||
| DRC | IB (n = 47) | - | - | - | - | - | - | - | - | 2 (4.25) | - |
| CMR | EK (n = 65) | - | - | - | - | - | - | - | - | - | - |
| CMR | BQ (n = 127) | 9 (7.08) | 7 (5.51) | 10 (7.8) | - | - | - | 1 (0.78) | - | - | 3 (2.36) |
| CMR | SO (n = 49) | 3 (6.12) | 2 (4.08) | 2 (4.08) | 2 (4.08) | 2 (4.08) | 2 (4.08) | 2 (4.08) | 1 (2.04) | 4 (8.16) | 2 (4.08) |
| CMR | BP (n = 85) | - | - | - | - | - | - | - | - | - | - |
| CMR | CP (n = 195) | 6 (3.07) | 3 (1.53) | 2 (1.02) | 1 (0.51) | 1 (0.51) | - | - | - | - | 1 (0.51) |
| CMR | MS (n = 48) | - | 1 (2.08) | 2 (4.16) | - | - | - | - | - | - | - |
| CMR | DJ (n = 104) | 6 (5.76) | 2 (1.92) | - | - | - | - | - | - | - | - |
| CMR | MT (n = 40) | 9 (18.3) | 7 (17.5) | 2 (5.00) | - | - | - | - | - | - | 3 (7.5) |
| CMR | LB (n = 20) | - | - | - | - | - | - | - | - | - | - |
| CMR | MB (n = 23) | 2 (8.69) | 1 (4.34) | 1 (4.34) | - | - | - | - | - | - | - |
| Subtotal | n = 803 | 35 (4.35) | 23 (2.82) | 19 (2.36) | 3 (0.37) | 3 (0.37) | 2 (0.25) | 3 (0.37) | 1 (0.12) | 6 (0.74) | 9 (1.12) |
| Total | n = 1418 | 37 (2.60) | 27 (1.90) | 23 (1.62) | 3 (0.21) | 7 (0.49) | 2 (0.14) | 5 (0.35) | 1 (0.07) | 25 (1.76) | 9 (0.63) |
a CMR, Cameroon; DRC, Democratic Republic of Congo
b no positive samples were identified
c number of positives (percentages)
Table 6. Percentages of ape samples reacting simultaneously with different arboviral recombinant proteins.
| Number of reactive samples | ONNV_E2 | ZIKV_NS1 | DENV1_NS1 | DENV2_NS1 | DENV3_NS1 | DNV4_NS1 | USUV_NS1 | WNV_NS1 | WNV_DIII | |
|---|---|---|---|---|---|---|---|---|---|---|
| CHIKV_E2 | 37 | 59.5 | 18.9 | 2.7 | 2.7 | 2.7 | 2.7 | 2.7 | 8.1 | 24.3 |
| ONNV_E2 | 27 | 23.1 | 3.8 | 3.8 | 3.8 | 3.8 | 3.8 | 11.5 | 34.6 | |
| ZIKV_NS1 | 23 | 13.0 | 13.0 | 8.7 | 8.7 | 4.3 | 8.7 | 17.4 | ||
| DENV1_NS1 | 3 | 100 | 66.7 | 66.7 | 33.3 | 66.7 | 33.3 | |||
| DENV2_NS1 | 7 | 28.6 | 57.1 | 14.3 | 85.7 | 14.3 | ||||
| DENV3_NS1 | 2 | 100 | 50.0 | 100 | 50.0 | |||||
| DENV4_NS1 | 5 | 20.0 | 80.0 | 20.0 | ||||||
| USUV_NS1 | 1 | 100 | 4.5 | |||||||
| WNV_NS1 | 25 | 8.0 | ||||||||
| WNV_DIII | 9 |
Discussion
In this study, we used a novel high throughput multiplex immunoassay that included ten antigens derived from six different arboviruses, including the four Dengue serotypes, to evaluate prevalence of IgG antibodies to these viruses in a large number of samples from NHPs species from Cameroon and the DRC. Overall, we showed that IgG antibodies to arboviral antigens are low and unevenly distributed in a wide diversity of the different NHPs species and in the different sites of the two countries where the samples were collected.
Validation of a serological screening tool
First, we developed a screening tool to detect IgG antibodies to CHIKV, ONNV, DENV, ZIKV, USUV, YFV and WNV viruses using 17 purified recombinant viral proteins. After performance evaluation, we kept only ten of these antigens for the screening of wildlife samples that presented the highest sensitivities and/or with high specificities (Table 3). In our assay, we found that the commonly used NS1 (DENV1-4, ZIKV, WNV) and E2 (CHIKV) antigens together with WNV-DIII recombinant proteins are the most sensitive antigens to detect IgG antibodies in our panel, with sensitivity and specificity ≥95% (Table 1). DIII antigens from ZIKV and DENV1-4 viruses and NSP for CHIKV were excluded because of low sensitivities. This could be explained by several parameters, including antibody kinetics, specificity restricted to one or two epitopes or recombinant protein structure impairment due to in vitro production issues. YFV-NS1 was not further used because of its low sensitivity, however it cannot be excluded that positive controls from vaccinated people (15/18) are not the most appropriate controls for assay development.
Sensitivity and cross-reactions are the recurrent concerns in the detection of antibodies to arbovirus infection. ELISAs, Immunofluorescence tests (IFT), multiplex microspheres assays (MIA) or Rapid Diagnostic Tests (RDT) have been used for the detection of IgG or IgM to various arbovirus [16,31]. Reported sensitivity and specificity varied greatly (39–100%) by the assay, the antibody isotype detected, the geographical environment, the time since clinical symptom onset of the infection [16]. For example, in a study performed in French Guiana in 2019 on a panel of 199 samples, including 90 ZIKV positives [32], the authors evaluated two commercially available ELISA assays and observed sensitivities of 71% and 79% and specificities of 70% and 62% for IgG detection. In our current work, the level of cross-reaction varied between 0% and 60%, depending on the antigen (Table 3) and correspond to what was observed in previous studies [33,34].
Variable prevalence of IgG antibodies to arboviral antigens in monkeys from Cameroon and DRC
The overall IgG prevalence is <5% for all the six arboviruses tested in both Cameroon and the DRC. Combining all the DENV serotype NS1 antigens as one target increased IgG prevalence for some monkey species (Table 2). This strategy could thus be a valuable alternative of screening, especially if ELISA, not MIA, is to be used. This sero-prevalence to arboviruses in general is lower than in other published papers. For example, a work by Diallo and colleagues [35] reported 58.8% (10/17) prevalence of DENV in African Green Monkeys (AGMs) in Senegal using an ELISA assay for screening. In Zambia 34.4% (33/96) of samples from NHPs were positive for ZIKV using Plaque Reduction Neutralization Test (PRNT) [36]. Another study investigated the enzootic cycle of CHIKV in monkeys (C. sabaeus, E. patas, and P. papio) collected over 3 years in Senegal. They used PRNT and found 72% (479/667) seropositivity for CHIKV, and 40% of 42 randomly selected samples were also positive for ONNV [37]. Buechler et al investigated the prevalence of ZIKV exposure in wild baboons and AGMs from South Africa by ELISA assay and found that 4.9% (2/41) and 16% (4/25) had antibodies against ZIKV, respectively [38]. Another work, performed on samples collected in diverse African mammals including mandrills from Gabon, Kading [39] and colleagues used PRNT to evaluate the presence of antibodies to flaviviruses (DENV-2, WNV, YFV) and alphaviruses (CHIKV, ONNV). In the set of 25 mandrills tested, the authors found that 80% and 76% presented neutralizing antibodies to flaviviruses and alphaviruses, respectively. Proportion of neutralizing antibodies as determined PRNT, at the level of different virus was also high, ranging from 16% for ONNV to 48% for YFV. However, sample dilutions to reach 80% reduction of plaque formation are often low (1/10 to 1/100) for most of the antigens tested. In our current work, we used stringent cut-off calculation methods to determine positivity criteria and the observed prevalence could thus underestimate the actual situation in wildlife. The difference in the proportion of positive samples in mandrills from our study and the one reported by Kading and colleagues [39] in Gabon, could also be due to the location or the detection assay used (Luminex versus PRNT). While we only detect IgG antibodies, PRNT rely on the reaction of all immunoglobulin isotypes, including IgM. In other monkey species, we observed high prevalence of IgG antibodies to some antigens. For example, 31% (16/51) (Table 2) samples from blue monkeys, presented antibodies to CHIKV-E2 recombinant protein, 7.3% (3/41) samples from Allan swamp monkey presented IgG to WNV-DIII proteins. Variability by monkey species could be due to the ecology of the monkey species. For example, the blue monkey is an arboreal species that dwells high in the canopy of contiguous and fragmented lowland and montane tropical moist forests, riverine and gallery forests and could thus be exposed to different mosquito vectors than baboons. At higher level of classification (by biotope, S7 Table and Fig 2), lower prevalence was observed in terrestrial and semi-terrestrial NHPs species, probably reflecting the ecology of vectors of studied viruses. However, this can only be ascertained in conjunction with future entomological investigations. The variability we observed in the proportion of positive samples by monkey species could also be due to geography, i.e, collection site, and different ecological environment ranging from forest savannah to tropical rain forest. Age can also play a role, as can be the case for pets whose low prevalence could be rather explained by their younger age. Indeed, monkeys kept as pets are generally juveniles whose mothers have been killed by hunters in the wild. In addition, these pets were mainly living in urban areas. On the other hand, the large majority (> 90%) of bushmeat samples are from adult animals. While one would be expecting significant differences between USUV and WNV viruses and the other arboviruses because birds instead of NHP, are thought to be the reservoirs of USUV and WNV viruses, no differences were observed in the present work.
Low prevalence of IgG antibodies to arboviruses in apes from Cameroon and the DRC
In apes, the overall IgG prevalence to the different arbovirus antigens we tested in the present work was generally lower than what was observed for monkeys. A primary possibility to explain this difference is the biological source of the sample. Hence, samples from monkeys were plasma or dried blood spots, while ape samples were feces dialysates and IgG concentration in this latter medium is lower than in plasma. Nevertheless, we showed in our earlier works on HIV/SIV and Ebola that IgG antibodies to these viruses can be detected in fecal dialysates of apes for SIV [19,23] and of survivors to Ebola virus disease, although at lower sensitivities [25]. The lower prevalence in apes can also be due to their terrestrial and semi-terrestrial behavior as observed in monkeys. In apes, we observed two distinct patterns of IgG reactivity towards arboviral antigens. In one side, in bonobos and chimpanzees, antibodies were mainly directed against WNV-NS1 antigen, and in gorillas against CHIKV-E2 antigen (Table 5). Significant difference for CHIKV IgG prevalence was also observed between chimpanzees and gorillas’ samples collected on the same site (MB site in Cameroon, 0.88% vs 8.69%, p = 0.039). This dichotomy could be explained by differential behaviors between the Pan genus and gorillas. Indeed, while gorillas spend their nights in nests on the ground, common chimpanzees and bonobos have their nests in the trees. They are thus probably exposed to different mosquito vectors. Like for monkeys, seroprevalence of IgG positive samples is also unevenly distributed in sampling sites and can thus also explain differences in species. For example, most of WNV-NS1 positive bonobos samples were from LA site. This variability by sampling site warrants further investigation, especially at the entomological level.
Limitations of the present study
A first limitation of our study is the absence of control panel with a set of well documented samples for each of the viruses we studied here, e.g. USUV and ONNV. This could have improved the evaluation of the sensitivity of our assay. A second limitation of this novel assay is the absence of sylvatic virus antigens. Including these antigens in our Multiplex assay could have increased the chances to detect antibodies against viruses circulating in wildlife. However, DENV serotypes for which sylvatic counterparts have been identified (DENV-1,-2, and 4), amino acids identity in NS1 ranged from 91% to 97% and for the envelope, between 93% and 98% (S8 Table). Thus, based on these sequence identities, it is likely that sylvatic infections have been detected with our assay. Finally, for some species (e.g: drills and Preuss’s monkey) the sample size was very limited, making it impossible to estimate prevalence of arboviruses in these monkey species.
In summary, this is the first study that evaluated on a large scale the presence of antibodies to arboviruses in NHPs to evaluate their role in sylvatic cycles. We used in this work archived samples from a diverse set of NHPs from Cameroon and the DRC to investigate the circulation of different arboviruses in these wild animals. Archived samples have helped to address multiple questions relevant to the field of infectious diseases [37]. Because of the overall low IgG prevalence observed in our study, we cannot conclude that monkeys are an important wildlife reservoir for arboviruses to maintain the sylvatic viral cycle, especially given the relative low numbers of NHPs living in close proximity to humans as compared to birds, bats or rodents which are also suspected to play a role in the sylvatic life cycle for some of this viruses. Rather, NHPs might be intermediate hosts of these pathogens. Additional research is still needed in the improvement of detection tools and to elucidate the sylvatic reservoirs of arboviruses and their potential impact on human health.
Supporting information
(DOCX)
A: Number of monkey samples collected from different sites in Cameroon and the Democratic Republic of Congo (DRC). Location of sites are shown in Fig 1 with the same abbreviations. B: Number of bonobo, chimpanzee and gorilla fecal samples collected from different sites in Cameroon and the Democratic Republic of Congo (DRC). Location of sites are shown in Fig 1 with the same abbreviations.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(PDF)
(XLSX)
Acknowledgments
We thank the staff and SIV team from Projet PRESICA (Innocent Ndong Bass, Aimé Mebenga, Joseph Moudindo, and Thomas Atemkem) and Donald Mbohli from Projet Grand Singes for the collection of samples and logistical support in Cameroon. In addition, we thank the field staff from DRC (Mubonga Mukulumanya, Lunguya-Metila Octavie, Guy Midingi and Mbenzo-Abokome Valentin); Dr Mazongo, Dr Abanda, Dr Jonnhy, and all the local staff in Equateur and Nord-Kivu provinces for their collaboration and participation in this study; the staff of the World Wildlife Fund for Nature/DRC; the Institut National de Recherches Biomédicales (Kinshasa, DRC); the Bonobo Conservation Initiative; and Vie Sauvage, Didier Mazongo, Octavie Lunguya, Muriel Aloni, and Valentin Mbenzo-Abokome for field work in DRC. We are thankful to Dr. Marjan Van Esbroeck and Johan Michiels from the Belgian National Reference for Arboviruses for providing patient sera.
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
RR received the PhD grant from INSERM and MUSE. MP and EMN received funding from US National Institutes of Health (grant number RO1 AI 50529) and Agence Nationale de Recherches sur le SIDA (grant numbers ANRS 12125, 12182, and 12325). AA acknowledges ArboSud grant from Montpellier University of Excellence (MUSE). KKA was supported by Research Fund Flanders (FWO, grant number G054820N). The funders had no role in study design, data collection and analysis, d ecision to publish, or preparation of the manuscript.
References
- 1.Guégan J-F, Ayouba A, Cappelle J, de Thoisy B. Forests and emerging infectious diseases: unleashing the beast within. Environmental Research Letters. 2020;15(8):083007 10.1088/1748-9326/ab8dd7 [DOI] [Google Scholar]
- 2.Sutherst RW. Global change and human vulnerability to vector-borne diseases. Clin Microbiol Rev. 2004;17(1):136–73. Epub 2004/01/17. 10.1128/cmr.17.1.136-173.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fouque F, Reeder JC. Impact of past and on-going changes on climate and weather on vector-borne diseases transmission: a look at the evidence. Infect Dis Poverty. 2019;8(1):51 Epub 2019/06/15. 10.1186/s40249-019-0565-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gottdenker NL, Streicker DG, Faust CL, Carroll CR. Anthropogenic land use change and infectious diseases: a review of the evidence. EcoHealth. 2014;11(4):619–32. Epub 2014/05/24. 10.1007/s10393-014-0941-z . [DOI] [PubMed] [Google Scholar]
- 5.Keesing F, Belden LK, Daszak P, Dobson A, Harvell CD, Holt RD, et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature. 2010;468(7324):647–52. Epub 2010/12/03. 10.1038/nature09575 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang YS, Higgs S, Vanlandingham DL. Emergence and re-emergence of mosquito-borne arboviruses. Current opinion in virology. 2019;34:104–9. Epub 2019/02/12. 10.1016/j.coviro.2019.01.001 . [DOI] [PubMed] [Google Scholar]
- 7.Weaver SC, Charlier C, Vasilakis N, Lecuit M. Zika, Chikungunya, and Other Emerging Vector-Borne Viral Diseases. Annual review of medicine. 2018;69:395–408. Epub 2017/08/29. 10.1146/annurev-med-050715-105122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young PR. Arboviruses: A Family on the Move. Advances in experimental medicine and biology. 2018;1062:1–10. Epub 2018/05/31. 10.1007/978-981-10-8727-1_1 . [DOI] [PubMed] [Google Scholar]
- 9.Prasad N, Murdoch DR, Reyburn H, Crump JA. Etiology of Severe Febrile Illness in Low- and Middle-Income Countries: A Systematic Review. PloS one. 2015;10(6):e0127962 Epub 2015/07/01. 10.1371/journal.pone.0127962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shope RE. The discovery of arbovirus diseases. Ann N Y Acad Sci. 1994;740:138–45. Epub 1994/12/15. 10.1111/j.1749-6632.1994.tb19864.x . [DOI] [PubMed] [Google Scholar]
- 11.Dick GW. Epidemiological notes on some viruses isolated in Uganda; Yellow fever, Rift Valley fever, Bwamba fever, West Nile, Mengo, Semliki forest, Bunyamwera, Ntaya, Uganda S and Zika viruses. Trans R Soc Trop Med Hyg. 1953;47(1):13–48. Epub 1953/01/01. 10.1016/0035-9203(53)90021-2 . [DOI] [PubMed] [Google Scholar]
- 12.Mason PJ, Haddow AJ. An epidemic of virus disease in Southern Province, Tanganyika Territory, in 1952–53; an additional note on Chikungunya virus isolations and serum antibodies. Trans R Soc Trop Med Hyg. 1957;51(3):238–40. Epub 1957/05/01. 10.1016/0035-9203(57)90022-6 . [DOI] [PubMed] [Google Scholar]
- 13.Casey RM, Harris JB, Ahuka-Mundeke S, Dixon MG, Kizito GM, Nsele PM, et al. Immunogenicity of Fractional-Dose Vaccine during a Yellow Fever Outbreak—Final Report. The New England journal of medicine. 2019;381(5):444–54. Epub 2018/02/15. 10.1056/NEJMoa1710430 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ingelbeen B, Weregemere NA, Noel H, Tshapenda GP, Mossoko M, Nsio J, et al. Urban yellow fever outbreak-Democratic Republic of the Congo, 2016: Towards more rapid case detection. PLoS neglected tropical diseases. 2018;12(12):e0007029 Epub 2018/12/12. 10.1371/journal.pntd.0007029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otshudiema JO, Ndakala NG, Mawanda EK, Tshapenda GP, Kimfuta JM, Nsibu LN, et al. Yellow Fever Outbreak—Kongo Central Province, Democratic Republic of the Congo, August 2016. MMWR Morbidity and mortality weekly report. 2017;66(12):335–8. Epub 2017/03/31. 10.15585/mmwr.mm6612a5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kerkhof K, Falconi-Agapito F, Van Esbroeck M, Talledo M, Arien KK. Reliable Serological Diagnostic Tests for Arboviruses: Feasible or Utopia? Trends in microbiology. 2019. Epub 2019/12/23. 10.1016/j.tim.2019.11.005 . [DOI] [PubMed] [Google Scholar]
- 17.Valentine MJ, Murdock CC, Kelly PJ. Sylvatic cycles of arboviruses in non-human primates. Parasites & vectors. 2019;12(1):463 Epub 2019/10/04. 10.1186/s13071-019-3732-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aghokeng AF, Liu W, Bibollet-Ruche F, Loul S, Mpoudi-Ngole E, Laurent C, et al. Widely varying SIV prevalence rates in naturally infected primate species from Cameroon. Virology. 2006;345(1):174–89. Epub 2005/11/01. 10.1016/j.virol.2005.09.046 . [DOI] [PubMed] [Google Scholar]
- 19.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, et al. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science (New York, NY). 2006;313(5786):523–6. Epub 2006/05/27. 10.1126/science.1126531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peeters M, Courgnaud V, Abela B, Auzel P, Pourrut X, Bibollet-Ruche F, et al. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg Infect Dis. 2002;8(5):451–7. Epub 2002/05/09. 10.3201/eid0805.010522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahuka-Mundeke S, Ayouba A, Mbala-Kingebeni P, Liegeois F, Esteban A, Lunguya-Metila O, et al. Novel multiplexed HIV/simian immunodeficiency virus antibody detection assay. Emerg Infect Dis. 2011;17(12):2277–86. Epub 2011/12/17. 10.3201/eid1712.110783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neel C, Etienne L, Li Y, Takehisa J, Rudicell RS, Bass IN, et al. Molecular epidemiology of simian immunodeficiency virus infection in wild-living gorillas. Journal of virology. 2010;84(3):1464–76. Epub 2009/11/13. 10.1128/JVI.02129-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Arc M, Ayouba A, Esteban A, Learn GH, Boue V, Liegeois F, et al. Origin of the HIV-1 group O epidemic in western lowland gorillas. Proc Natl Acad Sci U S A. 2015;112(11):E1343–52. 10.1073/pnas.1502022112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayouba A, Touré A, Butel C, Keita AK, Binetruy F, Sow MS, et al. Development of a Sensitive and Specific Serological Assay Based on Luminex Technology for Detection of Antibodies to Zaire Ebola Virus. J Clin Microbiol. 2017;55(1):165–76. Epub 2016/11/01. 10.1128/JCM.01979-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ayouba A, Ahuka-Mundeke S, Butel C, Mbala Kingebeni P, Loul S, Tagg N, et al. Extensive Serological Survey of Multiple African Nonhuman Primate Species Reveals Low Prevalence of Immunoglobulin G Antibodies to 4 Ebola Virus Species. The Journal of infectious diseases. 2019;220(10):1599–608. Epub 2019/01/19. 10.1093/infdis/jiz006 . [DOI] [PubMed] [Google Scholar]
- 26.Wilson EB. Probable inference, the law of succession, and statiscal inference Journal of the American Statistical Association. 1927;22:209–12. [Google Scholar]
- 27.De Nys HM, Kingebeni PM, Keita AK, Butel C, Thaurignac G, Villabona-Arenas CJ, et al. Survey of Ebola Viruses in Frugivorous and Insectivorous Bats in Guinea, Cameroon, and the Democratic Republic of the Congo, 2015–2017. Emerg Infect Dis. 2018;24(12):2228–40. Epub 2018/10/12. 10.3201/eid2412.180740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laing ED, Mendenhall IH, Linster M, Low DHW, Chen Y, Yan L, et al. Serologic Evidence of Fruit Bat Exposure to Filoviruses, Singapore, 2011–2016. Emerg Infect Dis. 2018;24(1):114–7. Epub 2017/12/21. 10.3201/eid2401.170401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bogen KT, Cullen AC, Frey HC, Price PS. Probabilistic exposure analysis for chemical risk characterization. Toxicological sciences: an official journal of the Society of Toxicology. 2009;109(1):4–17. Epub 2009/02/19. 10.1093/toxsci/kfp036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Delignette-Muller ML, Dutang C. fitdistrplus: An R Package for Fitting Distributions. 2015. 2015;64(4):34 Epub 2015-03-20. 10.18637/jss.v064.i04 [DOI] [Google Scholar]
- 31.Basile AJ, Horiuchi K, Panella AJ, Laven J, Kosoy O, Lanciotti RS, et al. Multiplex microsphere immunoassays for the detection of IgM and IgG to arboviral diseases. PloS one. 2013;8(9):e75670 Epub 2013/10/03. 10.1371/journal.pone.0075670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matheus S, Talla C, Labeau B, de Laval F, Briolant S, Berthelot L, et al. Performance of 2 Commercial Serologic Tests for Diagnosing Zika Virus Infection. Emerg Infect Dis. 2019;25(6):1153–60. Epub 2019/05/21. 10.3201/eid2506.180361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mora-Cárdenas E, Aloise C, Faoro V, Knap Gašper N, Korva M, Caracciolo I, et al. Comparative specificity and sensitivity of NS1-based serological assays for the detection of flavivirus immune response. PLoS neglected tropical diseases. 2020;14(1):e0008039 Epub 2020/01/30. 10.1371/journal.pntd.0008039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong SJ, Boyle RH, Demarest VL, Woodmansee AN, Kramer LD, Li H, et al. Immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections and from flavivirus vaccination. J Clin Microbiol. 2003;41(9):4217–23. Epub 2003/09/06. 10.1128/jcm.41.9.4217-4223.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diallo M, Ba Y, Sall AA, Diop OM, Ndione JA, Mondo M, et al. Amplification of the sylvatic cycle of dengue virus type 2, Senegal, 1999–2000: entomologic findings and epidemiologic considerations. Emerg Infect Dis. 2003;9(3):362–7. Epub 2003/03/20. 10.3201/eid0903.020219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wastika CE, Sasaki M, Yoshii K, Anindita PD, Hang’ombe BM, Mweene AS, et al. Serological evidence of Zika virus infection in non-human primates in Zambia. Archives of Virology. 2019;164(8):2165–70. 10.1007/s00705-019-04302-0 [DOI] [PubMed] [Google Scholar]
- 37.Althouse BM, Guerbois M, Cummings DAT, Diop OM, Faye O, Faye A, et al. Role of monkeys in the sylvatic cycle of chikungunya virus in Senegal. Nat Commun. 2018;9(1):1046 Epub 2018/03/15. 10.1038/s41467-018-03332-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buechler CR, Bailey AL, Weiler AM, Barry GL, Breitbach ME, Stewart LM, et al. Seroprevalence of Zika Virus in Wild African Green Monkeys and Baboons. mSphere. 2017;2(2). Epub 2017/03/16. 10.1128/mSphere.00392-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kading RC, Borland EM, Cranfield M, Powers AM. Prevalence of antibodies to alphaviruses and flaviviruses in free-ranging game animals and nonhuman primates in the greater Congo basin. Journal of wildlife diseases. 2013;49(3):587–99. Epub 2013/06/20. 10.7589/2012-08-212 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
A: Number of monkey samples collected from different sites in Cameroon and the Democratic Republic of Congo (DRC). Location of sites are shown in Fig 1 with the same abbreviations. B: Number of bonobo, chimpanzee and gorilla fecal samples collected from different sites in Cameroon and the Democratic Republic of Congo (DRC). Location of sites are shown in Fig 1 with the same abbreviations.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
(PDF)
(XLSX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.