Patients infected with P. ovale wallikeri displayed deeper thrombocytopenia and a shorter latency period.
Keywords: Plasmodium ovale curtisi, Plasmodium ovale wallikeri, imported malaria, diagnosis, malaria-endemic countries, parasites, France, malaria
Abstract
We retrospectively analyzed epidemiologic, clinical, and biologic characteristics of 368 Plasmodium ovale wallikeri and 309 P. ovale curtisi infections treated in France during January 2013–December 2018. P. ovale wallikeri infections displayed deeper thrombocytopenia and shorter latency periods. Despite similar clinical manifestations, P. ovale wallikeri–infected patients were more frequently treated with artemisinin-based combination therapy. Although the difference was not statistically significant, P. ovale wallikeri–infected patients were 5 times more frequently hospitalized in intensive care or intermediate care and had a higher proportion of severe thrombocytopenia than P. ovale curtisi–infected patients. Rapid diagnostic tests that detect aldolase were more efficient than those detecting Plasmodium lactate dehydrogenase. Sequence analysis of the potra gene from 90 P. ovale isolates reveals an insufficient polymorphism for relapse typing.
Malaria is a vectorborne disease caused by Plasmodium, a parasite transmitted by Anopheles mosquitoes. In 2018, malaria was responsible for ≈228 million cases and 405,000 deaths worldwide (1). Plasmodium ovale is endemic in Africa and represents the main agent of relapsing malaria (2). In mainland France, P. ovale was responsible for ≈6% of imported malaria cases in 2018 (3). Since the 2017 Frances updates for Plasmodium infection management recommendations, first-line treatment of P. ovale infections is based on chloroquine- or artemisinin-based combination therapy (ACT), instead of atovaquone/proguanil (4).
Because of low parasite density and poor efficiency of rapid diagnostic test (RDT) detection (5), P. ovale infections are difficult to diagnose. Consequently, infections caused by P. ovale remain poorly studied, and little is known about the global burden of the disease worldwide or its geographic distribution.
Since 2010, P. ovale has been divided into 2 species, Plasmodium ovale wallikeri and P. ovale curtisi, on the basis of gene polymorphisms (6–8). P. ovale wallikeri appears to cause malaria infections with a shorter latency period (9,10) and with deeper thrombocytopenia than P. ovale curtisi (11,12). Both P. ovale wallikeri (13) and P. ovale curtisi (14) can be responsible for a clinical relapse event, defined as renewed asexual parasitemia originating from liver dormancies (2). Relapse characterization relies on microscopic diagnosis and medical history. No consensus molecular method for P. ovale spp. relapse typing is reported. However, P. ovale tryptophan–rich antigen (potra) gene sequencing has previously been used for genotyping purpose (13,14).
At the microscopic level, the only observable difference between the species is a lack of Schüffner granulations in P. ovale wallikeri infected erythrocytes (15). However, this feature is rare and difficult to see, which makes P. ovale species distinction almost impossible even for an experienced microscopist. Molecular biology is a promising tool and is both sensitive and specific for the differentiation of P. ovale wallikeri from P. ovale curtisi. The first nested PCR that discriminates P. ovale wallikeri and P. ovale curtisi was developed in 2007 (16), and the first quantitative PCR (qPCR) was developed in 2013 (17).
In this study, we conducted a large retrospective multicenter analysis of imported P. ovale cases. Epidemiologic, clinical, and biologic characteristics of 309 P. ovale curtisi– and 368 P. ovale wallikeri–infected patients treated in France during January 2013–December 2018 were analyzed. The effectiveness of Rapid Diagnostic Test (RDT) and the polymorphism of potra gene were also investigated.
Methods
Sample Selection
France’s National Malaria Reference Center (FNMRC) is in charge of epidemiologic surveillance of imported malaria in France. Whole blood samples of patients with Plasmodium infections were received from hospital correspondents in France. FNMRC correspondents also reported demographic, epidemiologic, clinical, and biologic data through a reporting website. We retrospectively selected all the reported and PCR-confirmed P. ovale infections that occurred during January 2013–December 2018.
DNA Extraction
DNA was extracted from 200 μL of whole blood samples by using Magnapure automaton (Roche Diagnostics, https://diagnostics.roche.com) and eluted in 100 μL of elution buffer, according to the manufacturer’s instructions. DNA was stored at –20°C until further analysis.
Diagnosis of P. ovale Infection
The diagnosis of Plasmodium ovale infection was made by the hospital correspondent and confirmed by FNMRC with a thin blood smear reading, a thick blood smear reading, or both. Thick blood smears were considered positive if >1 trophozoïtes was visualized after examination of 1,000 leukocytes. Thin blood smears were used to confirm Plasmodium species identification. Parasite density was calculated by using the formula parasite density (parasites per μL) = patient leukocyte count (per µL) × (no. parasites counted)/(no. leucocytes counted), according to World Health Organization (WHO) recommendations (18). Parasitemia was calculated by counting the percentage of infected red blood cells on thin blood smears according to WHO recommendations (18). All P. ovale infections were confirmed with nested PCR (19,20) during 2013–2014, with qPCR–Taqman (Launch Diagnostics, https://www.launchdiagnostics.com) during 2015–2017, and with Bio-Evolution (https://www.bio-evolution.net/index.php) in 2018.
P. ovale curtisi and P. ovale wallikeri differentiation
qPCR–high-resolution melting (HRM) targeting the 18S rRNA gene was performed to differentiate P. ovale wallikeri from P. ovale curtisi by using Plasmo1_F and Plasmo2_R primers. The method development and validation was described previously (21). In brief, qPCR-HRM results were compared with nested PCR results from Calderaro et al. (16), and they displayed similar species determination. In all studied samples, P. ovale wallikeri and P. ovale curtisi melting plots displayed 2 specific melting temperatures (Tm) as Tm1 and Tm2, and the ΔTm between the 2 Tm was calculated.
For uncertain results (i.e., only 1 Tm on melting plot analysis [21]), nested PCR was performed by using rPLU1 and rPLU5 primers in the first PCR reaction and rOVA1/rOVA2 for P. ovale curtisi amplification or rOVA1v/rOVA2v for P. ovale wallikeri amplification in second PCR reaction (16). PCR products were visualized on 1% agarose gel stained with GelRed (https://biotium.com). We used P. ovale wallikeri and P. ovale curtisi isolates as positive controls and water as a negative control for each qPCR-HRM run.
RDT Efficiency in P. ovale wallikeri and P. ovale curtisi Detection
We evaluated the efficiency of 4 different RDTs detecting pan-Plasmodium proteins (aldolase or Plasmodium lactate dehydrogenase [pLDH]) for the detection of P. ovale wallikeri and P. ovale curtisi. Vikia Malaria Ag Pf/Pan (bioMérieux, https://www.biomerieux.com) (22) and Binax Now Pf/Pan (Abbott, https://www.abbott.com) (23) were used for aldolase detection (aldolase-RDT). Palutop+4 Pan/Pv/Pf (Biosynex, https://www.biosynex.com) (24) and Core Malaria Pan/Pv/Pf (Core Diagnostics, https://www.corediagnostics.net) were used for pLDH protein detection (pLDH-RDT). Results were interpreted according to the manufacturer’s instructions.
Data Collection
Each hospital correspondent sent an EDTA blood sample of a patient infected with P. ovale to FNMRC. This process was completed by using the online patient form containing multiple data, including demographic data (place of birth, ethnicity, age, and sex), epidemiologic data (trip purpose, visited country, duration of travel, and use of prophylaxis or bed nets), biologic data (parasite count, RDT results, leukocytes, hemoglobin and platelet counts, with severe thrombopenia defined as <50 G/L [25], and date of diagnosis), and clinical data (date of symptom onset, fever, headache, asthenia, and arthralgia or myalgia, as well as free symptomatology description for other symptoms, antimalarial treatment used, hospital or ambulatory regimen, and duration of hospitalization). Severe malaria biologic and clinical signs, adapted from the severe P. falciparum WHO recommendations (4,26), and relapsing P. ovale infection, defined as new P. ovale infection after a first completed and effective antimalarial treatment (27), were reported.
The latency period was calculated for each infection by subtracting the date of return from travel to the onset of the symptoms as defined by Rojo-Marcos et al. (11,12). The period of high malaria transmission in West Africa was defined as August–November on the basis of Nabarro et al. definition (10). The delay between symptom onset and diagnosis was also determined. We looked for false or incomplete microscopic diagnosis (Plasmodium spp.) to estimate the potential effect on P. ovale microscopic diagnosis of the described lack of Schüffner granulations in P. ovale wallikeri–infected erythrocytes (15).
No specific consent was required from patients because the parasitologic data were collected from the FNMRC database and analyzed in accordance with the common public health mission of all National Reference Centers in France, in coordination with the Santé Publique France organization for malaria surveillance and care. The study of the biologic samples obtained from routine medical care was considered as noninterventional research accordingly to article L1221–1.1 of the public health code in France and only requires the nonopposition of the patient during sampling (per article L1211–2 of the public health code). All data collected were anonymized before analysis.
potra Sequencing and Analysis
We amplified potra fragments as previously described (28). Bidirectional sequencing reaction was performed for the secondary potra fragment. Gene sequences were analyzed with Sequencher 5.0 (Genecodes, http://www.genecodes.com). Isolates from GenBank under accession nos. HM594183 (28), MG588152, and MG588154 (29) were used as P. ovale curtisi reference sequences; HM594180 (28) and MG588148–150 (29) were used as P. ovale wallikeri reference sequences.
Statistical Analysis
P. ovale wallikeri and P. ovale curtisi infections were compared in terms of demographic, epidemiologic, clinical, and biologic characteristics. The Kolmogorov-Smirnov test with the Lilliefors correction was used to verify the normality of variables distributions, and the Levene test was used to verify the homogeneity of the variances. If both criteria were validated, a Student t-test was used; otherwise, a Mann-Whitney U-test was performed to compare medians. Proportions were compared by using the χ2 or Fisher exact test according to sample size (>5 or <5). R software was used to perform statistical tests (30).
Results
P. ovale Sample Selection
During January 2013–December 2018, 15,028 Plasmodium spp. infection cases were reported to NMRC, including 765 P. ovale infections. Seventeen cases were excluded from the analysis because blood sample were unavailable. After exclusion of co-infections and inclusion of 59 P. ovale initially misdiagnosed (confirmed by PCR), 677 P. ovale cases from 63 different hospitals in France were finally included (Figure 1). By using qPCR-HRM for species differentiation, we identified 368 P. ovale wallikeri and 309 P. ovale curtisi infections. The 2 species segregated perfectly in qPCR-HRM; P. ovale wallikeri had a ΔTm of 1.62–2.69, and P. ovale curtisi had a ΔTm of 2.84–4.22.
Figure 1.
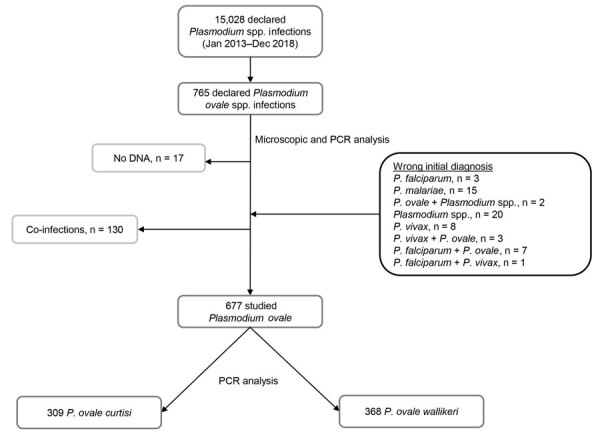
Flow-chart of the retrospective study analyzing characteristics of Plasmodium ovale wallikeri and P. ovale curtisi infections treated in France during January 2013–December 2018. All reported P. ovale infection cases were confirmed with microscopy and PCR analysis, and co-infections were excluded. A total of 59 P. ovale isolates initially misdiagnosed by the hospital correspondent were added. A total of 677 P. ovale infection cases were included in the study.
Patients’ Demographic and Epidemiologic Characteristics
P. ovale wallikeri and P. ovale curtisi showed similar repartition by month, except for October, which showed an increase in P. ovale wallikeri infections and a decrease in P. ovale curtisi cases (Figure 2, panel A). Among P. ovale cases, the proportion of P. ovale wallikeri infections increased from 44% to 59% during January 2013–December 2018 (Figure 2, panel B).
Figure 2.
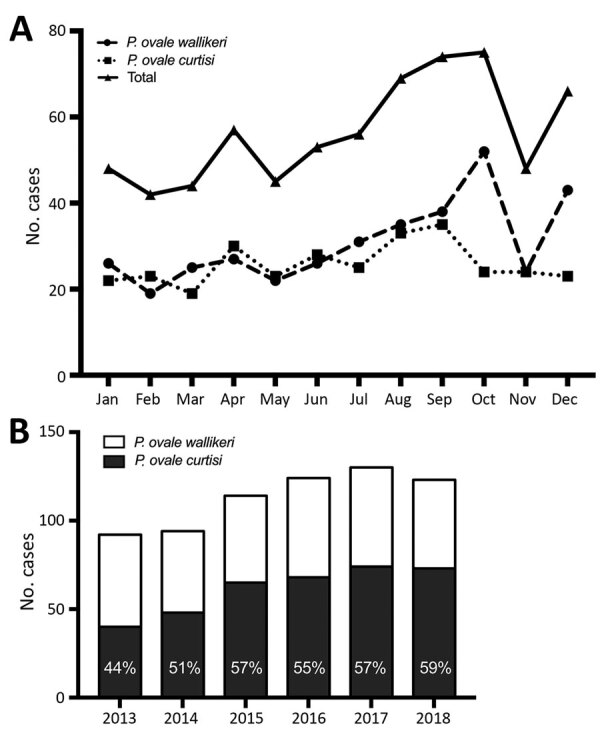
Number of Plasmodium ovale infection cases included in a study analyzing characteristics of P. ovale wallikeri and P. ovale curtisi infections treated in France during January 2013–December 2018, by month of inclusion (A) and year of inclusion (B).
P. ovale wallikeri– and P. ovale curtisi–infected patients did not display any differences in demographic and epidemiologic characteristics (Table 1). Countries of contamination were not statistically different between imported P. ovale curtisi and P. ovale wallikeri cases (p = 0.52) (Figure 3; Appendix Table 1).
Table 1. Demographic and epidemiologic characteristics of patients infected with Plasmodium ovale wallikeri and P. ovale curtisi, France, January 2013–December 2018* .
| Characteristic | P. ovale curtisi, n = 309 | P. ovale wallikeri, n = 368 | p value |
|---|---|---|---|
| Age, y, median (IQR) |
31 (21–47) |
34 (21–47) |
0.973 |
| Sex, % | 0.716 | ||
| M | 63.4 | 61.4 | |
| F |
36.6 |
38.6 |
|
| Ethnicity | 0.502 | ||
| Black | 200 (74.3) | 239 (75.7) | |
| White | 64 (23.8) | 68 (21.5) | |
| Asian | 2 (0.7) | 1 (0.3) | |
| Other |
3 (1.2) |
8 (2.5) |
|
| If African, place of birth | 0.420 | ||
| Africa | 144 (83.2) | 164 (80) | |
| Nonendemic country |
29 (16.8) |
41 (20) |
|
| Type of patient | 0.192 | ||
| Immigrant† | 23 (11.6) | 21 (8.6) | |
| Traveler‡ | 137 (68.8) | 187 (77.3) | |
| Visiting friends or relatives | 109 (79.6) | 152 (81.3) | |
| Tourism | 6 (4.4) | 8 (4.3) | |
| Work | 22 (16) | 27 (14.4) | |
| Resident | 19 (9.5) | 20 (8.3) | |
| Expatriate | 6 (38.6) | 10 (50) | |
| Humanitarian | 13 (61.4) | 10 (50) | |
| Military |
20 (10.1) |
14 (5.8) |
|
| Duration of travel, d, median (IQR) |
58 (29–91) |
50 (24–91) |
0.106 |
| Chemoprophylaxis | 0.882 | ||
| Yes | 97 (40) | 123 (39.3) | |
| Complete | 35 (44.9) | 42 (43.8) | |
| Incomplete | 43 (55.1) | 54 (56.2) | |
| Prematurely stopped | 26 (60.5) | 36 (66.7) | |
| Occasionally taking | 17 (39.5) | 18 (33.3) | |
| No data | 19 (NA) | 27 (NA) | |
| No |
146 (60) |
190 (60.7) |
|
| Using bed nets | 0.119 | ||
| Yes | 48 (26.7) | 41 (20.2) | |
| No | 130 (73.3) | 162 (79.8) |
*Values are no. (%) patients except as indicated. IQR, interquartile range; NA, not available. †A person who was born and lived in Africa. ‡A person who lived in a non–Plasmodium-endemic country.
Figure 3.
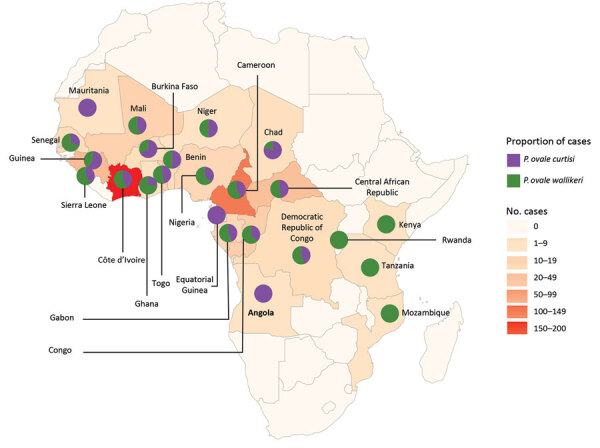
Geographic repartition of the origin countries of imported Plasmodium ovale wallikeri and P. ovale curtisi infection cases into France, January 2013–December 2018. Pie charts showed the repartition of cases between both species in each country.
For well-followed chemoprophylaxis (n = 77), the main treatments used were doxycycline (48%), atovaquone/proguanil (25%), and mefloquine (18%). No statistically significant differences were observed in the percentage of infection between those treatments.
P. ovale Diagnosis
Parasite densities for P. ovale curtisi and P. ovale wallikeri infections were similar (median 4,500 parasites/µL, [interquartile range (IQR) 1,094–10,197 parasites/µL] for P. ovale curtisi vs. median 3,970 parasites/µL [IQR 598–9,240 parasites/µL] for P. ovale wallikeri). We noted 8.5% of species misidentification for P. ovale curtisi and 9% for P. ovale wallikeri (Figure 1).
Aldolase and pLDH-RDT Efficiency
We compared the diagnostic performance of aldolase-RDTs and pLDH-RDTs for P. ovale diagnosis. Aldolase-RDTs detection were more efficient in P. ovale spp. detection than pLDH-RDTs (p<0.001); no differences between the 2 species were observed. P. ovale wallikeri was more frequently detected with pLDH-RDT than P. ovale curtisi (p<0.001) (Table 2). The positivity of aldolase and pLDH-RDTs were strongly associated with parasite density. Percentage of positive RDT results increased with parasite density for both pLDH-RDT and aldolase-RDT (Table 2). A positive aldolase-RDT result was associated with a parasite density significantly higher than with a negative aldolase-RDT result for both species (median 6,612 parasites/µL [IQR 2,410–14,175 parasites/µL] for P. ovale wallikeri vs. median 1,287 parasites/μL [IQR 450–4,500 parasites/μL] for P. ovale curtisi; p<0.001) (Figure 4). Similarly, the parasite density of positive pLDH-RDT P. ovale wallikeri samples were significantly higher than those of negative pLDH-RDT (median 11,000 parasites/µL [IQR 3,960–52,910 parasites/µL] vs. median 3,227 parasites/µL [IQR 551–7,118] parasites/µL; p<0.001). Vikia (bioMérieux) aldolase-RDT had a greater accuracy for detecting P. ovale infections compared than did Binax Now (Abbott) (59.3% vs. 40.9%; p<0.001) and a better sensitivity (median 4,230 parasites/µL [IQR 1,205–9,450 parasites/µL] for positive Vikia vs. median 8350 parasites/µL [IQR 4,032–16,166 parasites/µL] for positive Binax Now; p<0.001).
Table 2. Comparison of aldolase and pLDH-RDT efficiency in Plasmodium ovale wallikeri and P. ovale curtisi infection diagnosis, France, January 2013–December 2018*.
| RDT result | Parasite density, parasites/μL | LDH |
Aldolase |
|||||
|---|---|---|---|---|---|---|---|---|
| P. ovale | P. ovale wallikeri | P. ovale curtisi | P. ovale | P. ovale wallikeri | P. ovale curtisi | |||
| Positive | 55 (10.6) | 45 (16) | 10 (4.2) | 211 (47.8) | 120 (50) | 91 (45.3) | ||
| <1,000 | 5 (3.9) | 3 (3.9) | 2 (3.9) | 25 (19.5) | 16 (20) | 9 (17.6) | ||
| 1,000–5,000 | 15 (9.4) | 14 (15) | 1 (1.5) | 65 (40.6) | 42 (54.5) | 23 (33.8) | ||
| 5,000–10,000 | 6 (7.8) | 5 (12) | 1 (2.8) | 44 (57.1) | 24 (66.7) | 20 (57.1) | ||
| 10,000–50,000 | 16 (16.2) | 11 (20) | 5 (11.4) | 67 (67.7) | 29 (78.4) | 38 (86.4) | ||
|
|
>50,000 |
13 (86.7) |
12 (86) |
1 (100) |
|
10 (100) |
9 (100) |
1 (100) |
| Negative |
|
465 (89.4) |
237 (84) |
228 (95.8) |
|
230 (52.2) |
120 (50) |
110 (54.7) |
| p value | <0.001 |
0.322 |
||||||
*Values are no. (%) patients except as indicated. LDH, lactate dehydrogenase; pLDH, plasmodium lactate dehydrogenase; RDT, rapid diagnostic test. †Proportions of positive and negative LDH or aldolase-RDT were compared for P. ovale wallikeri and P. ovale curtisi by using a χ2 test.
Figure 4.
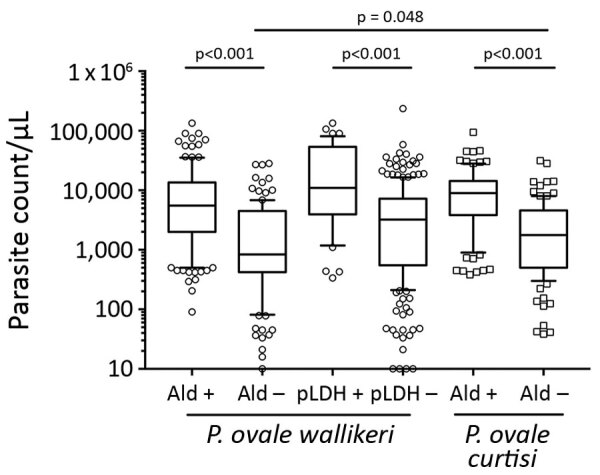
Comparison of parasite count according to RDT results in study analyzing characteristics of Plasmodium ovale wallikeri and P. ovale curtisi infections treated in France during January 2013–December 2018. Ald, aldolase RDTs; pLDH, plasmodium lactate dehydrogenase RDTs; Poc, P. ovale curtisi; Pow, P. ovale wallikeri; RDT, rapid diagnostic test.
Biologic and Clinical Characteristics
Patients infected with P. ovale wallikeri displayed deeper thrombocytopenia than those with P. ovale curtisi (Table 3), but reported symptomatology and disease severity did not differ. P. ovale wallikeri infections had shorter latency periods and a higher proportion of latency periods <50 days (p<0.001) (Table 3). Compared with patients who did not take prophylactic treatment, patients who reported well-managed prophylactic treatment had longer latency periods (median 90 days [IQR 47–177 days] vs. median 30 days [IQR 8–125 days]; p<0.001). Uncompleted prophylactic treatment did not extend latency period (median 33 days [IQR 17–112 days] vs. median 30 days [IQR 8–125 days]; p = 0.34). Military patients had longer latency periods than other patients (median 109 days [IQR 57–159 days] vs. median 40 days [IQR 12–142 days]; p = 0.0018), as did Caucasian versus African patients (median 84 days [IQR 28–140 days] vs. median 42 days [IQR 12–147 days]; p = 0.005 days). In the African population, no differences were found between African-born patients and others (mean 53 days [IQR 12–170 days] vs. mean 35 days [IQR 11–117 days]). The latency period was shorter in symptomatic patients returning from West Africa during the malaria season than in low-transmission or no-transmission seasons (median 27 days [IQR 10–67 days] vs. medina 90 days [IQR 17–158 days]; p<0.001) (Appendix Figure). P. ovale wallikeri infections and P. ovale curtisi infections were each responsible for 16 reported clinical relapses.
Table 3. Biologic and clinical characteristics of Plasmodium ovale wallikeri and P. ovale curtisi infections, France, January 2013–December 2018*.
| Characteristic | P. ovale curtisi, n = 309 | P. ovale wallikeri, n = 368 | p value |
|---|---|---|---|
| Parasite density, parasites/µL, median (IQR) | 4,500 (1,094–10,197) | 3,970 (598–9,240) | 0.112 |
| Leucocyte count, G/L, median (IQR) | 5.6 (4.4–7.1) | 5.2 (4.1–6.5) | 0.0501 |
| Hemoglobin, g/L, median (IQR) |
127 (113–140) |
126 (114–139) |
0.855 |
| Platelet count, G/L, median (IQR) | 111 (84–145) | 94 (70–130) | <0.001 |
| <75 | 56 (19.4) | 104 (31) | |
| 75–150 | 168 (58.1) | 174 (51.9) | 0.003 |
| >150 |
65 (22.5) |
57 (17.1) |
|
| Severe thrombocytopenia | 13 (4.5) | 25 (7.5) | 0.123 |
| Diagnostic delay, d, median (IQR) |
5 (3–7) |
4 (2–7) |
0.583 |
| Delay between return from endemic country and onset of symptoms, d, median (IQR) | 72 (18–208) | 34 (10–95) | <0.001 |
| <50 days |
87 (42.4) |
150 (59.5) |
<0.001 |
| Symptoms | |||
| Fever | 262 (95.6) | 316 (97.8) | 0.125 |
| Arthralgia or myalgia | 120 (54.8) | 138 (57.7) | 0.525 |
| Asthenia | 108 (58) | 133 (61.3) | 0.506 |
| Headache | 151 (68.6) | 201 (75.3) | 0.103 |
| Anorexia | 5 | 4 | |
| Diarrhea | 13 | 18 | |
| Abdominal pain | 28 | 29 | |
| Nausea | 16 | 20 | |
| Vomiting | 24 | 13 | |
| Cough |
6 |
12 |
|
| Clinical categorization | 0.927 | ||
| Uncomplicated malaria | 293 (97.7) | 335 (97.4) | |
| Severe malaria | 3 (1) | 5 (1.5) | |
| Asymptomatic |
4 (1.3) |
4 (1.1) |
|
| Admission to hospital | 158 (55.4) | 196 (60.3) | 0.243 |
| Duration of hospitalization, d, median (IQR) | 2 (1–3) | 3 (1–4) | 0.0732 |
| Intensive- or intermediate-care hospitalization | 1 (2.2) | 7 (11.3) | 0.134 |
| Conventional hospitalization |
46 (97.8) |
55 (88.7) |
|
| Treatment | 0.00359 | ||
| Chloroquine | 147 (54.8) | 152 (47.8) | |
| Artemisinin therapy | 46 (17.1) | 93 (29.2) | |
| Artemeter/lumefantrine | 11 (25.5) | 39 (41.9) | |
| Artesunate | 2 (4.3) | 5 (5.4) | |
| Arteminol/piperaquine | 33 (70.2) | 49 (52.7) | |
| Atovaquone/proguanil | 64 (23.9) | 64 (20.1) | |
| Mefloquine | 3 (1.2) | 0 (0) | |
| Quinine | 8 (3) | 9 (2.9) |
*Values are no. (%) patients except as indicated. IQR, interquartile range.
Patient Care
A similar proportion of patients were hospitalized in the P. ovale curtisi and P. ovale wallikeri groups. Eight malaria case-patients with WHO-defined severe criteria (26) were reported during the period analysis (Table 3). P. ovale wallikeri–infected patients were 5 times more likely to be hospitalized in intensive or intermediate care than P. ovale curtisi–infected patients (Table 3). A higher percentage of P. ovale wallikeri infections were treated with ACT (29.2% vs. 17.1%; p<0.001), but no association was found between ACT treatment and parasite density, between ACT treatment and platelet count, or between ACT treatment and positive and negative RDTs. Patients treated with ACT did have shorter latency periods than other patients (median 33 days [IQR 11–111 days] vs. 54 days [IQR 15–170 days]; p = 0.025) and patients with latency periods <50 days were more often treated with ACT than others (28.6% vs. 20.3%; p = 0.048). This high proportion of ACT prescription was highest in patients with latency periods <50 days and platelet counts <60 G/L (52.3% vs. 22.7%; p = 0.002).
New recommendations from the Infectious Diseases Society in France (La Société de Pathologie Infectieuse de Langue Française) edited in 2017 (4) had a clear effect on P. ovale infection treatment (Figure 5), including replacement of atovaquone/proguanil by artemisinin-based combination therapy. However, little change in rates of chloroquine prescription occurred (52.5% before the revisions and 47.2% after).
Figure 5.
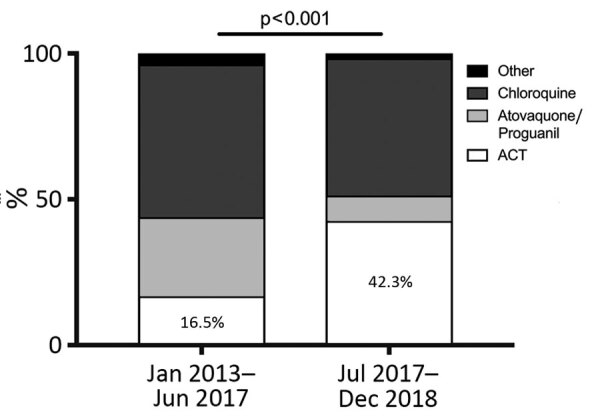
Effects of the new ACT treatment recommendations for Plasmodium spp. infections from La Société de Pathologie Infectieuse de Langue Française, revised in June 2017. ACT, chloroquine- or artemisinin-based combination therapy.
For the period analyzed, no statistically significant relationship was found between the number of included P. ovale infection cases per hospital and the percentage of patients receiving ACT treatment. We also analyzed the relation between the total number of included Plasmodium infection cases per hospital and the percentage of intensive care or intermediate care hospitalizations and did not find any statistically significant relation (data not shown).
potra Sequencing and Analysis
In total, 49 potra genes were sequenced from P. ovale wallikeri and 41 potra genes were sequenced from P. ovale curtisi. Three different genotypes (299, 317, and 335 bp) were identified in P. ovale curtisi and 4 different genotypes (245, 263, 263′, and 281 bp) in P. ovale wallikeri (Table 4). The major genotypes were (MANPIN)1(AITPIN)2 for P. ovale wallikeri and (TINPIN)3(TITPIS)1 for P. ovale curtisi. No association was found between country of contamination and potra genotype.
Table 4. Analysis of the potra fragment polymorphisms sequenced for Plasmodium ovale wallikeri and P. ovale curtisi, France, January 2013–December 2018.
| Species | Size, bp | Dominant amino acid repeat | No. (%) samples | GenBank accession no. of reference sequence |
|---|---|---|---|---|
| P. ovale wallikeri | 245 | (MANPIN)1(AITPIN)2 | 43 (88) | HMG594180 |
| 263 | (MANPIN)1(AITPIN)3 | 2 (4) | MG588149 | |
| 263 | (MANPIN)2(AITPIN)2 | 1 (2) | MG588148 | |
|
|
281 |
(MANPIN)2(AITPIN)3 |
3 (6) |
MG588150 |
| P. ovale curtisi | 299 | (TINPIN)3(TITPIS)1 | 26 (63) | MG588152 |
| 317 | (TINPIN)3(TITPIS)2 | 13 (32) | HM594183 | |
| 335 | (TINPIN)4(TITPIS)2 | 2 (5) | MG588154 |
Discussion
Our findings show that patients infected with P. ovale wallikeri displayed deeper thrombocytopenia than those infected with P. ovale curtisi (p<0.001) and had a shorter latency period (p<0.001). Those features of P. ovale wallikeri infection are currently debated in the literature, with some studies describing deeper thrombocytopenia (11,12) and shorter latency periods (9) and other finding refuting any differences between the 2 species (31).
We reported 1.2% of patients with diagnosed P. ovale infection having severe criteria of malaria (26), a similar percentage to the data reported by the malaria surveillance in the United States (32) or by Kotepui et al. (33). Seven P. ovale wallikeri– and 1 P. ovale curtisi–infected patients were hospitalized in intensive or intermediate care. Six of those patients did not have WHO-defined severe malaria criteria (26). Hospitalization in intensive or intermediate care for non–WHO-defined severe malaria was previously described in uncomplicated malaria patients with P. falciparum (34) or P. vivax (35) infections. We examined the hospitalization information of 5,227 uncomplicated malaria patients (all infected with Plasmodium species) for the study period in the NMRC database. Among these patients, 180 (3.6%) were hospitalized in intensive or intermediate care with a median length of hospital stay shorter to that observed with severe malaria patients (median 2 days [IQR 1–3 days] vs. median 3 days [IQR 2–4 days]; p<0.001).
In June 2017, La Société de Pathologie Infectieuse de Langue Française updated malaria management recommendations (4) and proposed the use of ACT as first-line treatment for all Plasmodium spp. infections and placed atovaquone/proguanil as a second-line treatment. Our data confirmed that physicians followed the new guidelines with a clear change between ACT and atovaquone/proguanil prescription frequency (Figure 5). P. ovale wallikeri infections were treated more often with ACT. To explain this phenomenon, we compared the antimalarial treatment used according to the platelet counts, parasite density, pLDH-RDTs results, and latency period duration. No association was observed between the type of antimalarial treatment and platelet counts, parasite density, or pLDH-RDTs results, but we highlighted a relationship between ACT treatment and shorter latency period (p = 0.048). The combination of low platelet count and short latency delay in Plasmodium infections are suggestive of P. falciparum infection (36). In the context of emergency care before species confirmation, those features might have influenced the prescription of ACT. Because they were seen more frequently in P. ovale wallikeri infections, we assumed that this tendency could partially explain that most of the ACT treatment administered occurred in the P. ovale wallikeri group.
About 44% of patients that took a prophylactic treatment reported taking their medication regularly, as prescribed. The latency period was longer in those patients (p<0.001). Because prophylactic treatments are not effective against liver-dormant forms of P. ovale (2) and did not protect patients from relapsing malaria, those results are not surprising. This phenomenon is well-illustrated in military patients, a population with a higher rate of chemoprophylaxis treatment (85%) and greater compliance with the drug regimen (62%) who had longer latency periods than other patients (p<0.001).
Most of the P. ovale cases we analyzed were originally diagnosed by microscopic analysis. Species misidentification occurred for 8.8% of the samples, and the main misidentification was between P. malariae and P. ovale. In endemic settings, microscopic analysis or PCR diagnosis are not always available in remote setting. Simple and affordable point-of-care compatible diagnostic tools are required. Although RDTs are widely spread nowadays in malaria-endemic countries, their efficiency for P. ovale diagnosis is not sufficiently studied compared with that for P. falciparum of P. vivax diagnosis. To supplement this deficiency, we analyzed the ability of aldolase and pLDH-RDTs to detect P. ovale wallikeri and P. ovale curtisi infection (Table 2). Aldolase-RDTs detection was definitively more accurate for P. ovale diagnosis than pLDH-RDTs (p<0.001). pLDH-RDTs used in this study (Palutop+4 [Biosynex] and Core Malaria [Core Diagnostics, https://www.corediagnostics.net]) were more efficient in diagnosing P. ovale wallikeri than P. ovale curtisi infection, but their performance remained extremely low (≈16% of infections diagnosed). This discrepancy might be explained by lactate dehydrogenase protein polymorphisms in P. ovale (37) affecting affinity of RDT-antibodies for P. ovale lactate dehydrogenase (38). Tang et al. (39) compared the efficiency of several pLDH-RDTs and confirmed variable diagnostic performance for P. ovale. In contrast, aldolase-RDTs had similar efficiency in detection of both species (50% for P. ovale wallikeri and 41.2% for P. ovale curtisi) that increased with parasite density (Table 2; Figure 4). Vikia demonstrated better performances than BinaxNow in P. ovale spp. detection (p<0.001).
The ability of P. ovale to establish liver-dormant forms (hypnozoïtes) induces relapse episodes of fever and parasitemia (2,40). Relapsing malaria was observed in only 3.5% of the included patients, a lower prevalence than previously reported (14). This difference is probably linked to the recommendations in France that advises systematic primaquine treatment of all P. ovale–infected patients, even for the first episode (except for major contraindication such as G6PD deficiency, pregnancy, and breastfeeding) (4). Currently, diagnosis of P. ovale infection relapse is mainly based on clinical data. potra gene sequencing has been used to distinguish reinfection from relapse by genotyping the initial and corresponding relapse sample (13,14). We evaluated the polymorphism of potra genes in 80 samples and, as previously described, identified a limited number of polymorphisms (Table 4) (28). Our results, combined with those of Zhou et al. (29), demonstrate that the potra gene is not a satisfying genetic marker of relapse. New genetic markers, such as microsatellite typing, need to be developed for P. ovale genotyping, as was previously done for P. falciparum (41,42) and P. vivax (43,44).
A limitation of our study is that, because of uncompleted online patient form filling (Appendix Table 2), we might lack statistical power to highlight differences in some rare infections features, such as hospitalization in intensive or intermediate care. In addition, our study is retrospective and might suffer from missing data about infection characteristics. Furthermore, we collected P. ovale isolates from Africa only.
In conclusion, our large retrospective study on P. ovale wallikeri and P. ovale curtisi infections confirmed that patients infected with P. ovale wallikeri display deeper thrombocytopenia and shorter latency periods. In addition, we found that physicians in France used more ACT to treat P. ovale wallikeri than P. ovale curtisi infections. This difference might be linked to the lower platelet level and shorter latency period seen with P. ovale wallikeri infections. In addition, we described a higher in intensive or intermediate care admission in P. ovale wallikeri–infected patients. Because of missing data and lack of power, this observation was not statistically significant and needs to be confirmed by a large, prospective study.
Additional information about Plasmodium ovale wallikeri and P. ovale curtisi infections and diagnostic approaches to imported malaria, France, 2013–2018.
Acknowledgments
We thank the hospital correspondents who sent us P. ovale samples and filled in the online patient forms. We also thank Claire Kamaliddin and Alexandre Lampros for their assistance in translation and review of the manuscript.
Additional members of the French National Reference Center for Imported Malaria Study Group who contributed data: Chantal Garabedian (Aix-en-Provence), Alain Domergue (Aix-en-Provence), Sylvain Clauser (Boulogne), Patrice Agnamey (Amiens), Céline Damiani (Amiens), Ludovic de Gentile (Angers), Marc Pihet (Angers), Anne Marfaing-Koka (Clamart), Anthony Marteau (Avicenne Hospital, Bobigny), Izri Arezki (Avicenne Hospital, Bobigny), Cecile Ficko (Bégin Hospital, Vincennes), Sébastien Larréché (Bégin Hospital, Vincennes), Adela Enache Angoulvant (Kremlin-Bicêtre), Nadia Guennouni (Kremlin-Bicêtre), Thierry Pistone (Bordeaux), Valérie Fuster-Dumas (Bordeaux), Denis Malvy (Bordeaux), Dorothée Quinio (Brest), Gilles Nevez (Brest), Didier Raffenot (Chambéry), Olivier Rogeaux (Chambéry), Céline Nourrisson (Clermont-Ferrand), Naima Dahane (Cochin Hospital, Paris), Angèle Li (Creil), Bernadette Cuisenier (Dijon), Louise Basmacyan (Dijon), Annie Motard-Picheloup (Fréjus St-Raphaël), Cécile Garnaud (Grenoble), Céline Dard (Grenoble), Dominique Maubon (Grenoble), Eric Dannaoui (Hôpital Européen Georges Pompidou Hospital, Paris), Françoise Botterel (Créteil), Dieudonné Bemba (Bondy), Isabelle Poilane (Bondy), Ana Mendes-Moreira (La Rochelle), Gauthier Pean-de-Ponfilly (Lariboisière Hospital, Paris), Bruno Megarbane (Lariboisière Hospital, Paris), Céline Mesnil (Lariboisière Hospital, Paris), Muriel Silva (Le Havre), Céline Malassigne (Le Havre), Pascal Penn (Le Mans), Anne-Sophie Deleplancque (Lille), Boualem Sendid (Lille), Marie-Laure Darde (Limoges), Marie-Fleur Durieux (Limoges), Martine Bloc’h (Colombes), Luce Landraud (Colombes), Stéphane Picot (Lyon), Pauline Tirard-Collet (Lyon), Léo Vidoni (Lyon), Marie-Laure Bigel (Mantes-la-Jolie), Hélène Savini (HIA Laveran, Marseille), Coralie l’Ollivier (Marseille), Nicole Desbois-Nogard Nicole (Martinique), Hélène Broutier (Meaux), Patrick Bastien (Montpellier), Alain Gravet (Mulhouse), Caroline Lohmann (Mulhouse), Anne Debourgogne (Nancy), Fakhri Jeddi (Nantes), Rose-Anne Lavergne (Nantes), Marie-Elisabeth Bougnoux (Necker Hospital, Paris), Emilie Sitterle (Necker Hospital, Paris), Christelle Pomares Estran (Nice), Pascal Delaunay (Nice), Milène Sasso (Nîmes), Victor Mercier (Nîmes), Laurence Lachaud (Nîmes) Bernadette Buret (Niort), Didier Poisson (Orléans), Jérôme Guinard (Orléans), Aurélie Guigon (Orléans), Oussama Mouri (Pitié-Salpétrière Hospital, Paris), Estelle Perraud-Cateau (Poitiers), Gwénaël le Moal (Poitiers), Antoine Huguenin (Reims), Sorya Belaz (Rennes), Anne Delaval (Aulnay-sous-bois), Cécile Leprince (Aulnay-sous-bois), Jean-Yves Siriez (Robert Debré Hospital, Paris), Lauren Pull (Robert Debré Hospital, Paris), Odile Fenneteau (Robert Debré Hospital, Paris), Loïc Favennec (Rouen), Gilles Gargala (Rouen), Ghania Belkadi (Saint-Antoine Hospital, Paris), Cécile Tournus (Saint-Denis), Samia Hamane (Saint-Louis Hospital, Paris), Stéphane Bretagne (Saint-Louis Hospital, Paris), Julie Brunet (Strasbourg), Ahmed Abou-Bacar (Strasbourg), Guillaume Menard (Toulon), Pamela Chauvin (Toulouse), Faïza Ajana (Tourcoing), Pierre Patoz (Tourcoing), Nathalie Desuremain (Trousseau Hospital, Paris), Pierre Mornand (Trousseau Hospital, Paris), Farida Moreau-Benaoudia (Troyes), Maxime Thouvenin (Troyes), Gisèle Dewulf (Valenciennes), Edith Mazars (Valenciennes), Odile Eloy (Le Chesnay), Sylvie Maurellet Evrard (Villeneuve St-Georges), Alice Raffetin (Villeneuve St-Georges), Pauline Caraux-Paz (Villeneuve St-Georges).
Biography
Dr. Joste is resident in pathology and laboratory medicine and is a medical microbiologist. At the time of this study, he worked at the French National Malaria Reference Center at Bichat Hospital, Paris, France. His research interests include the diagnosis and epidemiology of Plasmodium ovale spp. infection.
Footnotes
Suggested citation for this article: Joste V, Bailly J, Hubert V, Pauc C, Gendrot M, Guillochon E, et al. Plasmodium ovale wallikeri and P. ovale curtisi infections and diagnostic approaches to imported malaria, France, 2013–2018. Emerg Infect Dis. 2021 Feb [date cited]. https://doi.org/10.3201/eid2702.202143
Additional members of the French National Reference Center for Imported Malaria Study Group who contributed data are listed at the end of this article.
References
- 1.World Health Organization. World malaria report 2019. [cited 2020 Jan 7]. https://www.who.int/publications-detail/world-malaria-report-2019
- 2.Markus MB. Do hypnozoites cause relapse in malaria? Trends Parasitol. 2015;31:239–45. 10.1016/j.pt.2015.02.003 [DOI] [PubMed] [Google Scholar]
- 3.Centre National de Référence du Paludisme. Rapport d’activité annuel 2018. [cited 2020 Jan 7]. https://anofel.net/wp-content/uploads/2019/07/ra_cnr_2018_CNPaludisme.pdf
- 4.Bouchaud O, Bruneel F, Caumes E, Houzé S, Imbert P, Pradines B, et al. Management and prevention of imported malaria. 2018 update of the 2007 French clinical guidelines. Med Mal Infect. 2020;50:161–93. 10.1016/j.medmal.2019.10.009 [DOI] [PubMed] [Google Scholar]
- 5.Yerlikaya S, Campillo A, Gonzalez IJ. A systematic review: performance of rapid diagnostic tests for the detection of Plasmodium knowlesi, Plasmodium malariae, and Plasmodium ovale monoinfections in human blood. J Infect Dis. 2018;218:265–76. 10.1093/infdis/jiy150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sutherland CJ, Tanomsing N, Nolder D, Oguike M, Jennison C, Pukrittayakamee S, et al. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J Infect Dis. 2010;201:1544–50. 10.1086/652240 [DOI] [PubMed] [Google Scholar]
- 7.Tachibana M, Tsuboi T, Kaneko O, Khuntirat B, Torii M. Two types of Plasmodium ovale defined by SSU rRNA have distinct sequences for ookinete surface proteins. Mol Biochem Parasitol. 2002;122:223–6. 10.1016/S0166-6851(02)00101-9 [DOI] [PubMed] [Google Scholar]
- 8.Win TT, Jalloh A, Tantular IS, Tsuboi T, Ferreira MU, Kimura M, et al. Molecular analysis of Plasmodium ovale variants. Emerg Infect Dis. 2004;10:1235–40. 10.3201/eid1007.030411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nolder D, Oguike MC, Maxwell-Scott H, Niyazi HA, Smith V, Chiodini PL, et al. An observational study of malaria in British travellers: Plasmodium ovale wallikeri and Plasmodium ovale curtisi differ significantly in the duration of latency. BMJ Open. 2013;3:e002711. 10.1136/bmjopen-2013-002711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nabarro LEB, Nolder D, Broderick C, Nadjm B, Smith V, Blaze M, et al. Geographical and temporal trends and seasonal relapse in Plasmodium ovale spp. and Plasmodium malariae infections imported to the UK between 1987 and 2015. BMC Med. 2018;16:218. 10.1186/s12916-018-1204-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rojo-Marcos G, Rubio-Muñoz JM, Ramírez-Olivencia G, García-Bujalance S, Elcuaz-Romano R, Díaz-Menéndez M, et al. Comparison of imported Plasmodium ovale curtisi and P. ovale wallikeri infections among patients in Spain, 2005-2011. Emerg Infect Dis. 2014;20:409–16. 10.3201/eid2003.130745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rojo-Marcos G, Rubio-Muñoz JM, Angheben A, Jaureguiberry S, García-Bujalance S, Tomasoni LR, et al. ; TropNet Plasmodium ovale investigator group. Prospective comparative multi-centre study on imported Plasmodium ovale wallikeri and Plasmodium ovale curtisi infections. Malar J. 2018;17:399. 10.1186/s12936-018-2544-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veletzky L, Groger M, Lagler H, Walochnik J, Auer H, Fuehrer H-P, et al. Molecular evidence for relapse of an imported Plasmodium ovale wallikeri infection. Malar J. 2018;17:78. 10.1186/s12936-018-2226-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groger M, Veletzky L, Lalremruata A, Cattaneo C, Mischlinger J, Manego Zoleko R, et al. Prospective Clinical and Molecular Evaluation of Potential Plasmodium ovale curtisi and wallikeri Relapses in a High-transmission Setting. Clin Infect Dis. 2019;69:2119–26. 10.1093/cid/ciz131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phuong MS, Lau R, Ralevski F, Boggild AK. Parasitological correlates of Plasmodium ovale curtisi and Plasmodium ovale wallikeri infection. Malar J. 2016;15:550. 10.1186/s12936-016-1601-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Calderaro A, Piccolo G, Perandin F, Gorrini C, Peruzzi S, Zuelli C, et al. Genetic polymorphisms influence Plasmodium ovale PCR detection accuracy. J Clin Microbiol. 2007;45:1624–7. 10.1128/JCM.02316-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Calderaro A, Piccolo G, Gorrini C, Rossi S, Montecchini S, Dell’Anna ML, et al. Accurate identification of the six human Plasmodium spp. causing imported malaria, including Plasmodium ovale wallikeri and Plasmodium knowlesi. Malar J. 2013;12:321. 10.1186/1475-2875-12-321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. Basic malaria microscopy. Part I: learner’s guide. Second edition [cited 2020 Jun 15]. https://www.who.int/malaria/publications/atoz/9241547820
- 19.Snounou G, Singh B. Nested PCR analysis of Plasmodium parasites. Methods Mol Med. 2002;72:189–203. [DOI] [PubMed] [Google Scholar]
- 20.Fuehrer H-P, Stadler M-T, Buczolich K, Bloeschl I, Noedl H. Two techniques for simultaneous identification of Plasmodium ovale curtisi and Plasmodium ovale wallikeri by use of the small-subunit rRNA gene. J Clin Microbiol. 2012;50:4100–2. 10.1128/JCM.02180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joste V, Kamaliddin C, Kendjo E, Hubert V, Argy N, Houzé S. Distinction of Plasmodium ovale wallikeri and Plasmodium ovale curtisi using quantitative Polymerase Chain Reaction with High Resolution Melting revelation. Sci Rep. 2018;8:300. 10.1038/s41598-017-18026-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou M, Kim S, Khim N, Chy S, Sum S, Dourng D, et al. Performance of “VIKIA Malaria Ag Pf/Pan” (IMACCESS®), a new malaria rapid diagnostic test for detection of symptomatic malaria infections. Malar J. 2012;11:295. 10.1186/1475-2875-11-295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nkrumah B, Acquah SE, Ibrahim L, May J, Brattig N, Tannich E, et al. Comparative evaluation of two rapid field tests for malaria diagnosis: Partec Rapid Malaria Test® and Binax Now® Malaria Rapid Diagnostic Test. BMC Infect Dis. 2011;11:143. 10.1186/1471-2334-11-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dijk DPJ, Gillet P, Vlieghe E, Cnops L, van Esbroeck M, Jacobs J. Evaluation of the Palutop+4 malaria rapid diagnostic test in a non-endemic setting. Malar J. 2009;8:293. 10.1186/1475-2875-8-293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lampah DA, Yeo TW, Malloy M, Kenangalem E, Douglas NM, Ronaldo D, et al. Severe malarial thrombocytopenia: a risk factor for mortality in Papua, Indonesia. J Infect Dis. 2015;211:623–34. 10.1093/infdis/jiu487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severe malaria. Trop Med Int Health. 2014;19(Suppl 1):7–131. 10.1111/tmi.12313_2 [DOI] [PubMed] [Google Scholar]
- 27.White NJ, Imwong M. Relapse. Adv Parasitol. 2012;80:113–50. 10.1016/B978-0-12-397900-1.00002-5 [DOI] [PubMed] [Google Scholar]
- 28.Oguike MC, Betson M, Burke M, Nolder D, Stothard JR, Kleinschmidt I, et al. Plasmodium ovale curtisi and Plasmodium ovale wallikeri circulate simultaneously in African communities. Int J Parasitol. 2011;41:677–83. 10.1016/j.ijpara.2011.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou R, Liu Y, Li S, Zhao Y, Huang F, Yang C, et al. Polymorphisms analysis of the Plasmodium ovale tryptophan-rich antigen gene (potra) from imported malaria cases in Henan Province. Malar J. 2018;17:127. 10.1186/s12936-018-2261-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wickham H, Henry L. tidyr: easily tidy data with “spread()” and “gather()” functions. R package version 0.8.1 [cited yyyy mmm dd]. https://CRAN.R-project.org/package = tidyr
- 31.Frickmann H, Wegner C, Ruben S, Loderstädt U, Tannich E. A comparison of two PCR protocols for the differentiation of Plasmodium ovale species and implications for clinical management in travellers returning to Germany: a 10-year cross-sectional study. Malar J. 2019;18:272. 10.1186/s12936-019-2901-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mace KE, Arguin PM, Lucchi NW, Tan KR. Malaria Surveillance - United States, 2016. MMWR Surveill Summ. 2019;68:1–35. 10.15585/mmwr.ss6805a1 [DOI] [PubMed] [Google Scholar]
- 33.Kotepui M, Kotepui KU, Milanez GD, Masangkay FR. Severity and mortality of severe Plasmodium ovale infection: A systematic review and meta-analysis. PLoS One. 2020;15:e0235014. 10.1371/journal.pone.0235014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwake L, Streit JP, Edler L, Encke J, Stremmel W, Junghanss T. Early treatment of imported falciparum malaria in the intermediate and intensive care unit setting: an 8-year single-center retrospective study. Crit Care. 2008;12:R22. 10.1186/cc6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lança EFC, Magalhães BML, Vitor-Silva S, Siqueira AM, Benzecry SG, Alexandre MAA, et al. Risk factors and characterization of Plasmodium vivax-associated admissions to pediatric intensive care units in the Brazilian Amazon. PLoS One. 2012;7:e35406. 10.1371/journal.pone.0035406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson P, Jenney AW, Tachado M, Yung A, Manitta J, Taylor K, et al. Imported malaria treated in Melbourne, Australia: epidemiology and clinical features in 246 patients. J Travel Med. 2001;8:76–81. 10.2310/7060.2001.24309 [DOI] [PubMed] [Google Scholar]
- 37.Talman AM, Duval L, Legrand E, Hubert V, Yen S, Bell D, et al. Evaluation of the intra- and inter-specific genetic variability of Plasmodium lactate dehydrogenase. Malar J. 2007;6:140. 10.1186/1475-2875-6-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bauffe F, Desplans J, Fraisier C, Parzy D. Real-time PCR assay for discrimination of Plasmodium ovale curtisi and Plasmodium ovale wallikeri in the Ivory Coast and in the Comoros Islands. Malar J. 2012;11:307. 10.1186/1475-2875-11-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang J, Tang F, Zhu H, Lu F, Xu S, Cao Y, et al. Assessment of false negative rates of lactate dehydrogenase-based malaria rapid diagnostic tests for Plasmodium ovale detection. PLoS Negl Trop Dis. 2019;13:e0007254. 10.1371/journal.pntd.0007254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson LJ, Wampfler R, Betuela I, Karl S, White MT, Li Wai Suen CSN, et al. Strategies for understanding and reducing the Plasmodium vivax and Plasmodium ovale hypnozoite reservoir in Papua New Guinean children: a randomised placebo-controlled trial and mathematical model. PLoS Med. 2015;12:e1001891. 10.1371/journal.pmed.1001891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leclerc MC, Durand P, de Meeûs T, Robert V, Renaud F. Genetic diversity and population structure of Plasmodium falciparum isolates from Dakar, Senegal, investigated from microsatellite and antigen determinant loci. Microbes Infect. 2002;4:685–92. 10.1016/S1286-4579(02)01587-3 [DOI] [PubMed] [Google Scholar]
- 42.Su X, Wellems TE. Toward a high-resolution Plasmodium falciparum linkage map: polymorphic markers from hundreds of simple sequence repeats. Genomics. 1996;33:430–44. 10.1006/geno.1996.0218 [DOI] [PubMed] [Google Scholar]
- 43.Gunawardena S, Karunaweera ND, Ferreira MU, Phone-Kyaw M, Pollack RJ, Alifrangis M, et al. Geographic structure of Plasmodium vivax: microsatellite analysis of parasite populations from Sri Lanka, Myanmar, and Ethiopia. Am J Trop Med Hyg. 2010;82:235–42. 10.4269/ajtmh.2010.09-0588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Orjuela-Sánchez P, Brandi MC, Ferreira MU. Microsatellite analysis of malaria parasites. Methods Mol Biol. 2013;1006:247–58. 10.1007/978-1-62703-389-3_17 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional information about Plasmodium ovale wallikeri and P. ovale curtisi infections and diagnostic approaches to imported malaria, France, 2013–2018.


